Abstract
A bioactive platform for the quantitative observation of cell migration is presented by 1) presenting migration factors in a well-defined manner on 2-D substrates, and 2) enabling continuous cell tracking. Well-defined substrate presentation is achieved by correctly orienting immobilized proteins (chemokines and cell adhesion molecules), such that the active site is accessible to cell surface receptors. A thiol-terminated self-assembled monolayer on a silica slide was used as a base substrate for subsequent chemistry. The thiol-terminated surface was converted to an immobilized metal ion surface using a maleimido-nitrilotriacetic acid (NTA) cross-linker that bound Histidine-tagged recombinant proteins on the surface with uniform distribution and specific orientation. This platform was used to study the influence of surface-immobilized chemokine SDF-1α and cell adhesion molecule ICAM-1 on murine splenic B lymphocyte migration. While soluble SDF-1α induced trans-migration in a Boyden Chamber type chemotaxis assay, immobilized SDF-1α alone did not elicit significant surface-migration on our test-platform surface. Surface-immobilized cell adhesion protein, ICAM-1, in conjunction with activation enabled migration of this cell type on our surface. Controlled exposure to UV light was used to produce stable linear gradients of His-tagged recombinant SDF-1α co-immobilized with ICAM-1 following our surface chemistry approach. XPS and antibody staining showed defined gradients of outwardly oriented SDF-1α active sites. This test platform can be especially valuable for investigators interested in studying the influence of surface-immobilized factors on cell behavior and may also be used as a cell migration enabling platform for testing the effects of various diffusible agents.
Keywords: Haptokinesis, chemotaxis, chemokine, cell adhesion molecule, surface immobilization, surface gradients, B lymphocyte, SDF-1α, CXCL12a, ICAM-1
INTRODUCTION
Cell migration is fundamental to a wide variety of phenomena such as homing of lymphocytes to lymphoid organs [1], movement of leukocytes towards sites of infection [2], tissue morphogenesis [3], movement of metastatic cells towards sources of growth factors [4], orchestration of neuronal wiring during brain development [5], angiogenesis [6], and wound healing [7]. The incessant “random” migration of B lymphocytes within lymphoid organs, evolved to maximize the probability that rare antigen-specific B cells encounter their cognate antigen, [8],[9] is tightly regulated via micro-anatomic localization, differentiation states, receptor engagement, and coordinated interactions of adhesion molecules and chemokines, making them migratory only under specialized conditions [10,11]. It is not surprising then that since the late 1800s investigators have consistently noted the difficulty of B lymphocytes to migrate on 2-D surfaces in vitro that lack some of these key elements [12–15].
Lymphocyte migration studies have typically used [16] the Boyden Chamber type transmigration chemotaxis assays [17] that have several limitations [18]. First, they lack the ability to dissect the roles of autocrine and paracrine signaling. Second, they do not allow discernment of cell migration parameters such as cell displacement, track length, translocation speed, directional persistence time, chemotactic/haptotactic index, and turning behavior because this assay-type monitors a population of cells after exposure to a chemoattractant in a steep gradient across a very thin porous mesh, a process which is not directly viewable and thus allows data acquisition only at the end points of experiments. Third, this type of assay is prone to the influence of interfering artifacts and is less conservative at distinguishing between chemotaxis and chemokinesis because the pore size and thickness of the trans-migration mesh/membrane are of the same order of magnitude as the characteristic dimension of the migrating cell body. Finally, they do not allow the study of the effects of surface-immobilized factors such as chemokines and cell adhesion molecules (either solo or concurrently with other immobilized and diffusible factors) on cell migration.
The Zigmond [19] and Dunn [20] chambers and other approaches including the ibidi® cell migration slide/chambers have been developed and used [21–24] to directly visualize the cell migration process via time-lapse imaging on a 2-D substrate, enabling researchers to avoid some of the limitations of the Boyden Chamber type assays. Much like the Boyden Chamber though, the Zigmond and Dunn Chamber methods utilize a quasi-static diffusive gradient (that is sensitive to fluid flow fluctuations), and are not optimized for the presentation of surface-immobilized factors that influence cell migration. Perhaps cell migration researchers have yet to embrace techniques developed for immobilizing and orienting protein, as has been done in other fields, particularly biosensors, proteomics, protein adsorption, and cell adhesion [25,26]. Table 1 summarizes some of the techniques developed to immobilize proteins on surface.
Table 1.
Different approaches to immobilize proteins on surfaces.
| Immobilization | Mechanism | Orientation | Stability | Ref. |
|---|---|---|---|---|
| Adsorption | Hydrophobic, hydrogen-bonding, van der Waals, and/or electrostatic interactions of proteins with surface | Random | Poor | [27] |
| Assembly | Hydrophobic interactions used to create hybrid lipid bilayer that present protein on surface | Random | Poor | [28] |
| Covalent Binding | Lysine-residues reaction with N-hydroxysuccinimide (NHS) ester groups on surface | Random | Excellent | [29] |
| Lysine-residues reaction with aldehyde groups on surface | Random | Excellent | [30] | |
| Cysteine-residues reaction with maleimide groups on surface | Random | Excellent | [31] | |
| Cysteine-residues reaction with disulfide-derivatized surface | Random | Excellent | [32] | |
| Affinity | Interaction of Biotin-tag on protein with streptavidin on surface | Oriented | Excellent | [33] |
| Interaction of His-tag on protein with metal-ion chelated surface | Oriented | Good | [34] | |
| Interaction of Leucine zipper-tag on protein with complementary zipper on surface | Oriented | Good | [35] | |
| Interaction of Glutathione S-Transferase (GST)-tag on protein with glutathione on surface | Oriented | Good | [36] | |
| Interaction of Fc-tag on protein with Protein A or Protein G on surface | Oriented | Good | [37] |
Because proteins have complex structures and activities, an immobilization approach that preserves a protein’s native state and orients it for optimal target interactions over an extended time period would be ideal. Past attempts on protein immobilization have mainly used nonspecific adsorption [27,28], or covalent bond formation between available functional groups (e.g., amines, thiols) on protein molecules and complementary coupling groups (e.g., aldehyde, maleimides) on the surface [29–32]. A major concern with both these approaches is the random orientation of proteins on the surfaces [25,26]. This precludes the active sites of a substantial population of immobilized protein molecules from being accessible to targets such as cell surface receptors [38,39]. In addition, the possibility of protein denaturation exists upon strong interaction between randomly immobilized protein and the surface [26,39]. Although, the covalently attached protein is more permanently bound as compared to the physisorbed protein, the latter also risks desorption [25,40]. Therefore, surface-techniques to control the orientation and stability of immobilized protein molecules on solid surfaces would be very useful [25,26,41,42]. To solve this problem, affinity based interaction such as streptavidin-biotin [33], leucine zipper tags [35], Glutathione-Glutathione S-Transferase (GST) tag [36], Nickel-His-tag [34], Protein A-Fc tag [37] have been applied in the past.
Developing a much needed robust in vitro test-platform for studying the migration of “fickle” cell types such as B lymphocytes applying some of these techniques from surface science would be useful. In this study a cell migration testing platform was developed that (1) utilized surfaces of defined chemistry to stably present an adhesive protein [8,43] in conjunction with a chemokine [10,44] (proteins associated with B lymphocyte migration) in an orientation that promotes interactions with the cognate cell receptor, (2) enabled direct and continuous visualization of cell migratory behavior, (3) showed amenability of this surface immobilization approach to patterning via UV to form surface gradients of proteins of interest in a tunable fashion. As proof-of-principle this platform cell culture substrate was used to probe the effect of surface-immobilized chemokine SDF-1α and cell adhesion molecule ICAM-1 on the migration of B lymphocytes.
MATERIALS AND METHODS
Fabrication and Characterization of Histidine-Tagged SDF-1α
Expression Plasmid Vector and Bacterial Transformation
A pET32 plasmid encoding C-terminal 6X His-tagged SDF-1α was kindly provided by Dr. Ghalib Alkhatib from Texas Tech University Health Science Center. 50 μL of BL21(DE3)pLysS cells (Strategene, Santa Clara, CA) was transformed with 50–100ng of plasmid and grown on antibiotic selective agar plates (100 μg/mL carbenicillin, BioPioneer Inc., San Diego, CA).
Recombinant Protein Expression, Purification, and Refolding
A bacteria culture originating from a single colony on the agar plate was grown in LB broth with 100 μg/mL carbenicillin at 37°C until an OD600 of 0.6 was achieved. Culture was then induced with 1mM isopropyl β-D-thiogalactoside for 5h at 30°C and protein expression was confirmed via SDS-PAGE.
The bacterial pellet was harvested via centrifugation and resuspended in lysis buffer to break open the bacterial cell wall and membrane. The suspension was rotated (30 min, RT), sonicated (50% duty and 60 pulses/cycle for 3 cycles), rotated again (30 min, RT), and centrifuged (15,000g for 20 min at 4°C). The pellet was resuspended in 6M Guanidine in base phosphate buffer to solubilize the inclusion bodies containing the recombinant His-tagged SDF-1α protein, rotated, sonicated, and rotated again. The lysate was centrifuged and protein supernatant was mixed overnight at 4°C with a column of primed Ni-NTA agarose (Qiagen, Germantown, MD).
The next day, the protein-agarose mixture was washed with decreasing guanidine concentration (6M, 4M, 2M, 0M) pH 7.8 in base phosphate buffer and 20 mM imidazole, respectively. The protein was then eluted with 0.5M Imidazole in base phosphate buffer (pH 7.2) and diluted with Round 1 refolding buffer (55mM Tris Base, 10mM NaCl, 0.4mM KCl, 2mM MgCl2, 2mM CaCl2, 550mM Arginine, 1mM DTT, pH 8.2). Protein sample was then subjected to 3 rounds of dialysis in refolding buffer at 4°C with progressively decreasing concentration of Arginine (550 mM to 55 mM). Protein sample was finally concentrated using Amicon Ultra-15 column (10 kDa MWCO, EMD Millipore, Billerica, MA) and concentration was determined via A280 and Bradford Assay.
Trans-well Migration Assay to Assess Chemotactic Activity of Recombinant Protein
Chemotactic activity of the purified recombinant His-tagged protein was assessed using the Neuroprobe 96-Trans-Well Migration System (Neuroprobe, Gaithersburg, MD) and compared to commercial, non-tagged SDF-1α (R&D Systems, Minneapolis, MN) as a positive control, or a negative control lacking the presence of chemokine. 3.3×106 cells/ml suspended in pre-warmed RPMI-1640 containing 20 mM HEPES and 1% BSA were placed on the upper chambers. The lower chamber contained in the same media SDF-1α at 100 ng/mL (recombinant or commercial) or no SDF-1α. The amount of cells transmigrated to the lower chamber over 3 hours at 37°C was quantified for the different conditions. To further confirm that cell migration was attributed solely to the recombinant SDF-1α, the recombinant SDF-1α was also incubated with a 14:1 molar excess of anti-SDF antibody (R&D Systems, MAB310) for 30 min at RT prior to introduction in a separate trans-well migration experiment.
Fabrication of Bioactive Cell Migration Substrates with Uniformly Immobilized Proteins
Formation of Silane-Based Monolayers
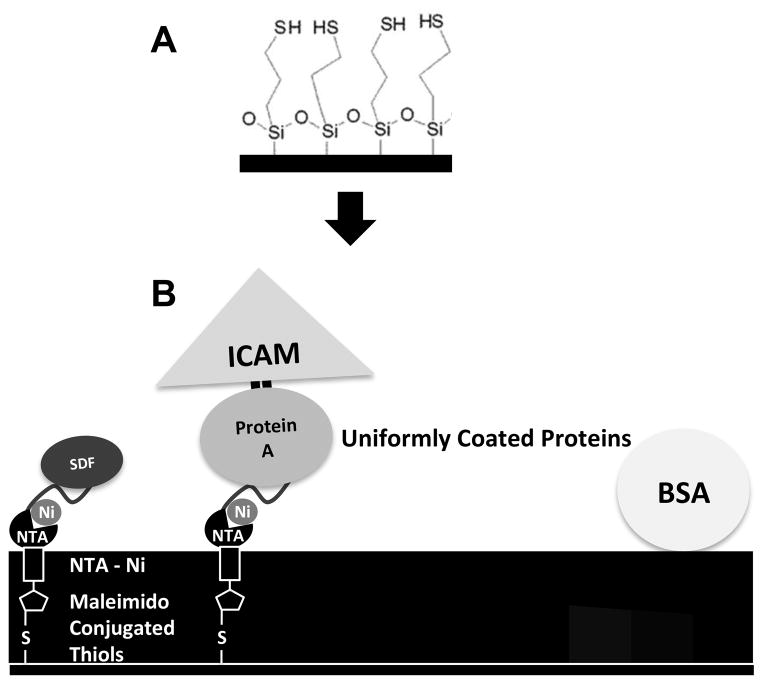
Silica slides (Ted Pella, Inc., Redding, CA), piranha etched followed by oxygen plasma treatment, were treated with a 0.1% solution of 3-mercaptopropyltrimethoxy-silane (MTS) (UCT, Inc., Bristol, PA) in dry toluene (VWR, Radnor, PA) [45,46]. The formed self-assembled monolayer, on the silica slides, presenting a uniform surface of reactive thiols (Figure 1A) was rinsed sequentially with toluene, acetone, ethanol, and DI water, and dried with nitrogen gas.
Figure 1. Formation of uniformly coated protein surfaces.
(A) Silica microscope slides were treated with MTS to form a uniform, thiol-terminated self-assembled monolayer on the surface. (B) A nitrilotriacetic acid (NTA)-containing cross-linker with a reactive maleimido group on one end (Maleimido-C3-NTA) enabled attachment of cross-linker to surface thiols. The NTA chelated Nickel. The Ni -- His-tag affinity enabled immobilization of both His-tagged SDF-1α (SDF-His) and His-tagged Protein A (PA-His), which were contacted on the surface at the same time for competitive binding. Following rinsing, cell adhesion molecule ICAM-1/Fc (ICAM-Fc) fusion chimera was introduced, which bound via affinity of the Fc region to Protein A. Once the proteins of interest were immobilized the surfaces were blocked with BSA. This process allowed the bioactive presentation of the active site (near N-terminus) of SDF-1α by attachment via C-terminus His-tag to Ni, and ICAM-1 by attachment via the Fc region to Protein A; and the formation of uniform surfaces of randomly distributed proteins.
IMAC Chemistry to Specifically Immobilize Proteins in Correct Orientation
To properly orient proteins on the surface, we chose the immobilized metal ion affinity chromatography (IMAC) chemistry [26,34,47–50]. In 50 mM Tris-HCl buffer pH 7.4 (VWR, Radnor, PA), the maleimide group on the end of a maleimido-C3-NTA (Dojindo Molecular Technologies Inc., Rockville, MD) molecule was covalently bound to an available surface thiol. On the opposite end of the molecule, the chelator, nitrilotriacetic acid (NTA), was subsequently loaded with a nickel ion (NiCl2 in Tris-HCl buffer, pH 7.4). Histidine residues on the C-terminus of the protein SDF-1α eventually filled the coordination sites of the chelator-metal complex binding the protein to the surface in an orientation that preserved its function by keeping the active site near the N-terminus accessible to cell surface receptors. Subsequently, blocking was done using 1% BSA.
Based on the concentration range of chemokines in biological fluids (1–5 μg/ml) [51], His-tagged SDF-1α (SDF-His) was initially contacted over a range of concentrations on the surface (0–5 μg/ml) and tested for eliciting migratory behavior of murine B lymphocytes. Murine B lymphocytes did not show much migration on these surfaces. Next, ICAM-1, an important component of the interstitial cell migration surfaces in the secondary lymphoid organs, was immobilized on the surface. This was done by first contacting the surface with well-mixed solutions of different concentrations of the His-tagged SDF-1α and His-tagged Protein A (PA-His, 10 μg/ml) (Abcam, Cambridge, MA) for 2–4 hours at room temperature followed by rinsing gently with PBS (Ca2+ and Mg2+ free) for 5 mins on a rocker. This produced a surface with competitively bound SDF-1α and Protein A that was next contacted with a 5 μg/ml solution of ICAM-1/Fc Chimera (ICAM-Fc, R&D Systems, Minneapolis, MN) for 2–4 hours at room temperature (See Figure 1B). The ICAM-1/Fc Chimera fusion protein has the extracellular LFA-1 binding segment of amino acids 28–485 fused to an Fc antibody-segment that binds to substrate-immobilized Protein A. Any loosely bound protein was gently rinsed away with PBS for 5 mins on a rocker, followed by blocking any remaining non-specific sites with 1% BSA in PBS. All solutions (His-tagged SDF-1α, His-tagged Protein A, ICAM-1/Fc Chimera, and BSA) were made fresh in PBS (Ca2+ and Mg2+ free).
Antibody Binding to Confirm Surface Immobilization and Orientation of Proteins
Surfaces with uniformly immobilized His-tagged SDF-1α and ICAM-1/Fc were visualized via fluorescence intensity measurement at 532 nm and 635 nm (GenePixPro4200A Microarray Scanner, Molecular Devices, LLC, Sunnyvale, CA) upon binding with fluorescently tagged anti-SDF-1α and anti-ICAM-1 monoclonal antibodies (R&D systems, Minneapolis, MN), respectively. To verify specific binding of His-tagged SDF-1α, after fluorescence scanning, the slide was incubated with 1M Imidazole (Sigma, St. Louis, MO) for 10 mins on a rocker, followed by a deionized water rinse. Imidazole competes with the His-tag of protein for binding with surface-immobilized nickel, displacing the protein [34]. A second fluorescence intensity measurement of the slide (at the same laser power and gain settings as before) was conducted to investigate reduction in surface fluorescence via displacement of His-tagged protein-antibody complex by Imidazole.
Cell Migration Studies
Cell Harvest, Culture, and Activation
Spleens were harvested from Bcl2 transgenic mice (6–12 weeks of age), following procedures approved by the Institutional Animal Care and Use Committee of Duke University. Cells were isolated mechanically from the spleens by grinding between frosted slides. Residual tissue clumps were removed by filtering through a 40 μm nylon filter. Next, red blood cells within cell fraction were lysed (RBC Lysing Buffer, St. Louis, MO). Finally, B lymphocytes were isolated using Dynabeads® Mouse CD43 Untouched™ B Cells kit (Life Technologies, Grand Island, NY) in isolation buffer (Ca2+ and Mg2+ free PBS supplemented with 0.1% BSA and 2 mM EDTA, Sigma-Aldrich, St. Louis, MO). We confirmed > 95% cell viability and purity on the day of harvest. Cells were subsequently cultured in suspension in RPMI media with final concentration of following supplements: 20% FCS, 40 nM beta-mercaptoethanol, 1% Pen-Strep, 0.8 mM Sodium Pyruvate, 1% non-essential amino acids, and 10mM HEPES buffer – B cell media. Under such culturing conditions, murine B lymphocytes did not show much migratory behavior. Therefore, migration studies were next conducted using harvested B lymphocytes with activation over 48 hours. For activation, 20 μg/ml Anti-IgM (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA) and 20 μg/ml Anti-CD40 (BD Biosciences, San Jose, CA) antibodies were used per 2 million B cells in B cell media [52,53]. All ingredients were from Invitrogen, except where specified.
Cell Migration Experiments
All cell migration experiments were conducted at 37°C. Activated B lymphocytes were washed in DPBS containing Ca++ and Mg++, re-suspended uniformly in 1 ml of cell migration media (1% BSA in RPMI-1640 (both from Sigma-Aldrich, St. Louis, MO) with 25 mM HEPES), and plated on a cell migration surface at a density of 50,000 cells/cm2. This plating density provided about 30–40 cells in a field of view, which resulted in few cell-cell interactions, yet a good number of migrating cells to observe in a single recording. Since cell migration was observed outside the incubator at 37°C, a CO2 independent buffer was used in the media. Imaging was started after a 10 min equilibration period for cells to settle on migration surface; phase contrast images were taken in a time series every 10 seconds for a period of 15 minutes using a Nikon Eclipse TE2000-U microscope for each cell migration surface.
Data Quantification and Analysis
IMARIS software (Bitplane AG, Zurich, Switzerland) was used to track and quantify the cell migration data. Only those cells that were present in the field of view and plane of focus throughout the microscopic recording period were tracked. Cells that showed no signs of active movement and cells or smaller round entities that moved in almost a straight line along a perceived direction of convection (as was noted by movement of any non-cellular particulates) were not tracked. Two parameters were quantified per cell that was tracked; track length and displacement. Track length is the magnitude of the actual distance travelled by the cell based on its meandering path, whereas the scalar component of displacement provides the straight distance between the start and end point of a cell track, and the vector component provides information about the net direction in which a cell travelled. (This is illustrated in Supplementary Material, Figure S1.)
Between 3 to 14 independent surfaces (replicates) were tested for cell migration for each of the treatments. All tracked cells from the multiple replicate surfaces tested for each treatment were pooled together, with each migrating cell considered as an N of 1. An average of 67.6 ± 14.3 cells were tracked per condition tested. Numerical data, presented as mean ± standard error of the mean, were analyzed (as indicated in the respective figure captions) using Student’s t-test, one-way analysis of variance (ANOVA), or ANOVA on Ranks, followed by a post hoc Tukey’s or Dunn’s test (p < 0.05 considered statistically significant) using SigmaPlot 11.0 software (Systat Software Inc., San Jose, CA).
Fabrication of Immobilized Protein Gradients
UV Burning to Form Gradients
To demonstrate further the amenability of our surface chemistry approach to correctly orient proteins on surfaces for surface patterning, we created linear gradients of oriented proteins. Chemical gradients were formed on the silanized silica surfaces using a previously developed controlled oxidation process [54–57]. The surface thiols were selectively oxidized by moving a mask over the surface in a controlled manner during exposure to UV light (290 nm) resulting in a gradient of reactive surface thiols and a counter gradient of nonreactive sulfonates on the surface. Gradient steepness was specified by the velocity at which UV mask retracted over a distance of 5 mm; lower velocities allowed for more UV exposure across the ‘burnt’ half of the slide, thus increasing the thiol gradient steepness. Subsequently, via the same chemistry as described earlier for the creation of uniformly immobilized protein surfaces, the thiol gradient was translated via a nickel gradient to gradients of proteins. (This process is illustrated in Supplementary Material, Figure S2.)
XPS Characterization of Gradient Surfaces
X-ray photoelectron spectroscopy (XPS, Kratos Axis Ultra Spectrometer hν= 1486.6 eV, Manchester, U.K.) was used to characterize the UV burned chemical surface gradients on the silica slides. A series of XPS atomic intensity signals at discrete spots along the gradient were measured. For each X-ray spot (100 μm diameter) on a sample, general survey scans (analyzer pass energy of 160 eV) followed by high resolution scans (analyzer pass energy of 20 eV) were obtained. XPS also characterized a 5 mm thiol gradient (1 mm/min mask withdrawal velocity) that was reacted with the maleimido-C3-NTA and loaded with nickel. Scaled areas under the thiol and nickel peaks were calculated at 6 points across the gradient as a quantitative measure of the relative surface amounts of these species.
Antibody Binding to Confirm Surface Binding of Proteins on Gradients
Gradients of His-tagged SDF-1α were visualized upon specific fluorescent monoclonal antibody binding via fluorescence intensity measurement at 532 nm as described previously. Likewise, specific binding of His-tagged SDF-1α on the gradient was verified via the previously described competitive binding procedure using Imidazole. For comparison of gradient surfaces formed from the same concentration of SDF-1α, fluorescence intensity for each gradient surface was normalized. This was done by dividing each graph by its own maximum intensity that was measured as the average intensity at the plateau (not the peaks of noisy spikes in the signal).
RESULTS
Characterization of Recombinant SDF-1α Fabrication
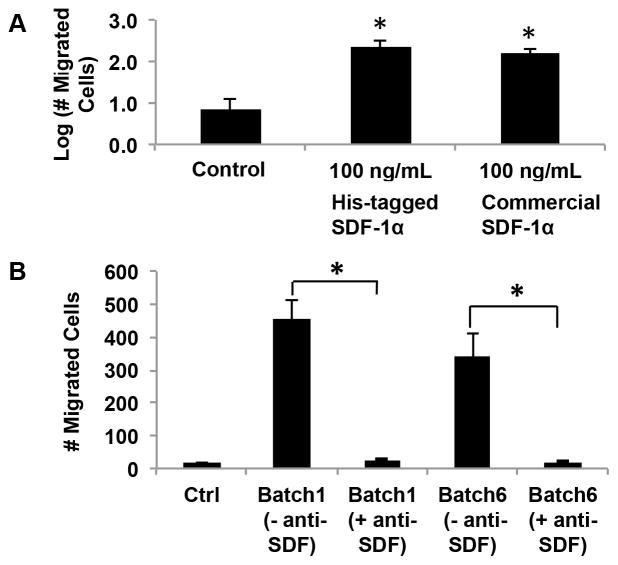
The bioactivity of the in-house produced recombinant SDF-1α was tested via a standard trans-well migration assay. The cells that trans-migrated into the lower chambers of the Trans-Well Migration System were quantified via fluorescence microscopy for the different conditions tested. Figure 2A shows statistically significant increase in trans-migration of SDF-1α exposed cells over the control (no chemokine) for recombinant His-tagged SDF-1α, which was comparable to commercial SDF, verifying the bioactivity of the recombinantly produced protein.
Figure 2. In-house produced recombinant His-tagged SDF-1α is bioactive.
(A) The bioactivity of His-tagged SDF-1α was verified via a trans-well migration assay, in which commercially available SDF-1α, His-tagged SDF-1α, or no chemokine control conditions were maintained in the lower chamber, whereas migrating cells were placed in the upper chamber separated by a thin porous membrane. The number of cells that crossed the membrane was quantified. A statistically significant increase in trans-migration of cells over the control case was observed for both in-house produced His-tagged SDF-1α and commercially purchased SDF-1α, and the latter two were not statistically distinguishable (One-way ANOVA, *P<0.05). (B) The specificity of His-tagged SDF-1α was confirmed, via an antibody neutralization assay, in which migration was tested with SDF-1α in the lower chamber that was equilibrated with or without anti-SDF antibody. Excess antibody neutralization (Ab:SDF-1α of 14:1) of two recombinant batches showed complete abolishment of chemotactic behavior to levels comparable to the control case (no chemokine).
Antibody treatment of recombinantly produced SDF-1α showed complete abolishment of chemotactic behavior to levels comparable to the control (no chemokine) (Figure 2B). This confirmed the bioactivity and specificity of our in-house produced protein. Another observation is that there were differing batch-to-batch activities of the His-tagged SDF-1α, although these differences were not statistically significant, and in all batches the SDF-1α showed complete abolishment of chemotactic behavior upon antibody incubation.
Fabrication of Uniformly Immobilized Protein Surfaces for Cell Migration Studies
The bioactive immobilization of proteins His-tagged SDF-1α (SDF-His) and ICAM-1/Fc (ICAM-Fc) was confirmed via significant binding of fluorescently tagged anti-SDF-1α and ICAM-1 antibodies to the nickel coated chemistry surfaces with these proteins attached in comparison to BSA coated control chemistry surfaces that did not show appreciable antibody binding. Figure 3 (Columns 1 and 2) shows antibody binding to His-tagged SDF-1α and ICAM-1/Fc in a concentration dependent manner. For 0 μg/ml His-tagged SDF-1α exposure, there was insignificant SDF-1α fluorescent antibody binding (Figure 3, Column 1, Row 1), which increased appreciably upon exposure of these surfaces to 5 μg/ml His-tagged SDF-1α (Figure 3, Column 2, Row 1). Likewise, exposure of these chemistry surfaces to the same concentration of ICAM-1/Fc (5 μg/ml), with or without His-tagged SDF-1α, resulted in similar ICAM-1 fluorescent antibody binding, showing reproducibility (Figure 3, Columns 1 and 2, Row 2). Finally, Figure 3 (Column 3) demonstrates that the binding of His-tagged SDF-1α and ICAM-1/Fc via Protein A to these nickel coated chemistry surfaces is mostly specific via significant abolishment of fluorescence upon Imidazole incubation of protein bound substrates (Imidazole competes with the His-tag of proteins for affinity to the surface-immobilized nickel). This also implies that the surface-immobilized protein is correctly oriented keeping the active site accessible.
Figure 3. Characterization of uniformly coated protein surfaces by fluorescent antibody binding.
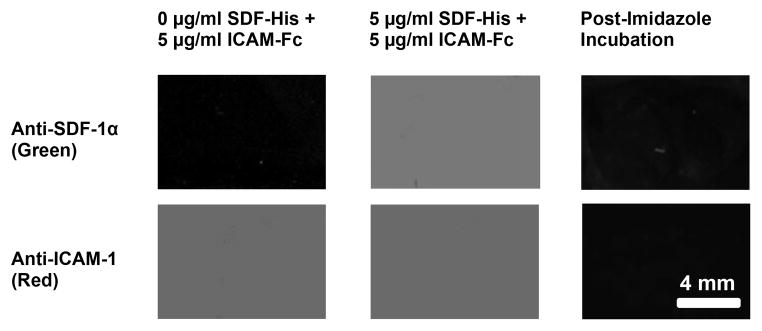
Fluorescent antibody binding to the surface was specific; insignificant monoclonal anti-SDF-1 antibody binding (green) was seen for a surface contacted with 0 μg/ml SDF-His (Column 1, Row 1), whereas significant antibody binding (green) was observed on surfaces contacted with 5 μg/ml SDF-His (Column 2, Row 1). Likewise, fluorescent antibody binding to the surface was reproducible; significant and comparable monoclonal anti-ICAM-1 antibody binding (red) was seen on different substrates contacted with the same 5 μg/ml ICAM-Fc concentration (Column 1, Row 2; and Column 2, Row 2). Finally, Significant abolishment of fluorescence (Column 3) upon Imidazole incubation of fluorescent antibody bound protein immobilized substrates (5 μg/ml ICAM-Fc + 5 μg/ml SDF-His contacted) demonstrated that the binding of His-tagged SDF-1α to the nickel coated surfaces was mostly specific, implying correct orientation of the protein with active site accessible.
Cell Activation and Migration
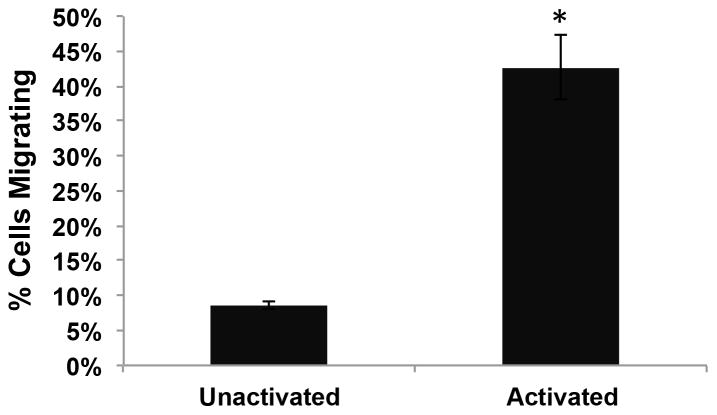
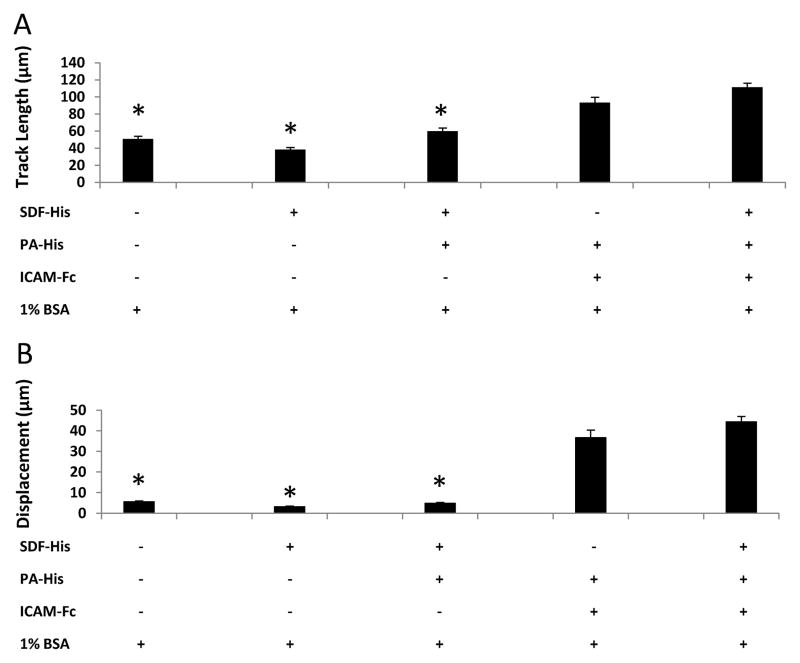
Unactivated murine splenic B lymphocytes failed to migrate on surfaces presenting His-tagged SDF-1α (SDF-His) and ICAM-1/Fc (ICAM-Fc). However, activation of these cells by a combination treatment of 20 μg/ml anti-IgM and 20 μg/ml anti-CD40 antibodies over 48 hours resulted in robust migratory behavior in over 40% of the cells (Figure 4). Comparison of the activated B cell migration on different uniformly coated surfaces revealed differences in track length (TL) and displacement (D). Surfaces presenting both His-tagged SDF-1α and ICAM-1/Fc induced extensive migration (TL = 111.5 ± 4.6 μm, D = 44.3 ± 2.6 μm), compared to surfaces presenting SDF-1α only (TL = 38.5 ± 2.1 μm, D = 3.2 ± 0.3 μm) or BSA-coated control surfaces (TL = 50.8 ± 3.2 μm, D = 5.5 ± 0.4 μm) (Figure 5). Comparison of the TL and the D values for the latter two surfaces indicates that a TL value can be generated even if the cell is not significantly displaced several cell body lengths from the start point, but is simply “dancing on the spot.” Since short-term hovering of the cell centroid about a location can give a “high” track length value, the metric of displacement was included. However, the displacement metric can also be misleading (by itself) in the case of cells that may meander significant distance, but may end up in a spot very close to where they started. (Supplementary Material, Figure S3 illustrates the cell migratory behavior comparison between SDF-His vs. PA-His + ICAM-Fc contacted surfaces, respectively.)
Figure 4. Activation of cells is necessary for migration.
The number of cells in 48–72 hour-old cultures that included 20μg/ml Anti-IgM and 20μg/ml Anti-CD40 in culture media exhibiting activated profiles and migratory behavior was significantly higher than in control cultures without these activating agents (Student’s t-test, *P = 0.002).
Figure 5. ICAM-1 is necessary for activated murine B lymphocyte surface migration.
A one-way ANOVA on Ranks failed to show significant differences in cell migration on (10 μg/ml PA-His + 5 μg/ml ICAM-Fc + BSA) and (10 μg/ml PA-His + 5 μg/ml ICAM-Fc + 5 μg/ml SDF-His + BSA) contacted conditions for (A) track length and (B) displacement. On the other hand, displacement on BSA-contacted control surfaces and surfaces without ICAM-Fc was barely more than a cell body length and was significantly lower than the conditions containing ICAM-Fc (One way ANOVA of Ranks, * P ≤ 0.001). The same statistical differences were seen for track length, although the values for track length for these three non-ICAM-Fc conditions are greater than the characteristic cell body length; this is because the track length accrues even as a cell “dances” on the spot without much net displacement.
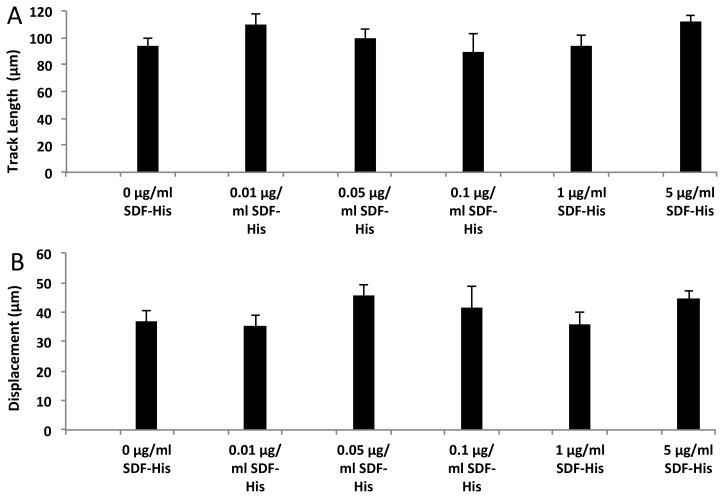
No significant differences in the two parameters were noted between the surfaces presenting both His-tagged SDF-1α and ICAM-1/Fc (TL = 111.5 ± 4.6 μm, D = 44.3 ± 2.6 μm) and the surfaces presenting ICAM-1/Fc only (TL = 93.5 ± 6.2 μm, D = 36.7 ± 3.6 μm). Furthermore, varying the concentration of His-tagged SDF-1α for a relatively constant concentration of ICAM-1/Fc did not significantly affect cell migration (Figure 6). Therefore, SDF-1α that influenced trans-migration when present in a diffusive form in solution did not appear to influence cell migration when surface-immobilized. The average migration speed of the activated murine B lymphocytes on our His-tagged SDF-1α and ICAM-1/Fc immobilized surfaces was 6.6 ± 0.5 μm/min.
Figure 6. Immobilized SDF-1α does not significantly influence activated murine B lymphocyte surface migration beyond ICAM-1.
Surfaces for each condition compared above were exposed to 10 μg/ml PA-His competitively with the specified concentrations of SDF-His followed by exposure to 5 μg/ml ICAM-Fc and blocking with 1% BSA. A one-way ANOVA of Ranks failed to show significant differences in (A) track length (* P = 0.098) and (B) displacement (* P = 0.296) between any of the concentrations of SDF-His for surfaces co-immobilized with ICAM-Fc via PA-His.
Fabrication of Immobilized Protein Gradients
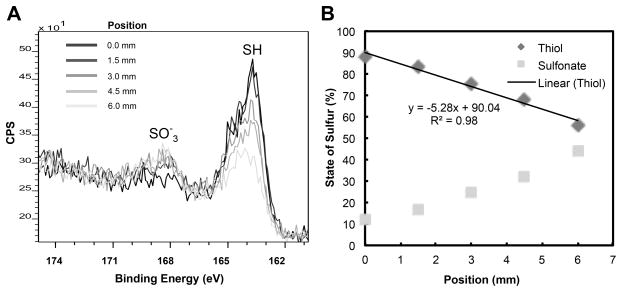
The different stages in the creation of protein gradients on silica slides using the controlled UV exposure set-up were characterized using XPS. XPS analysis of an MTS treated slide without UV exposure showed that the sulfur 2p region spectra had only one peak at 163 eV, indicative of the binding energy of thiols; whereas after 5 minutes of UV exposure, this thiol peak was reduced and a sulfonate peak appeared yielding a roughly 50-50 split of thiols and sulfonates on the surface. (Supplementary Material, Figure S4.A presents corresponding XPS plot.) XPS measuring the sulfur state at multiple locations along a 5 mm gradient formed via graded UV exposure showed a linear gradient of thiols (percent thiol per mm, Figure 7A and B). Finally, XPS analysis of a 5 mm linear thiol gradient that was reacted with maleimido-C3-NTA and loaded with nickel verified the creation of a gradient of nickel ions on top of the gradient of thiols; the nickel gradient follows the thiol gradient very closely as noted by the similar slopes between the two (−116.76 for the nickel gradient vs −110.77 for the thiol gradient in terms of total counts per mm in Supplementary Material Figure S4.B).
Figure 7. Characterization of the chemical surface gradient by XPS.
(A) Graded exposure of the thiol surface to UV light by withdrawing a mask over 5 minutes across 6 mm resulted in opposing gradients of thiols and sulfonates. This is demonstrated by the progressively reducing SH peak and increasing SO3− peak (in the sulfur 2p region spectra, CPS = counts per second) as position on surface increases from 0 mm (end position of mask) to 6 mm (start position of mask). (B) The scaled area under the peaks was used to quantify the % of thiol and sulfonates with respect to the distance on surface that was exposed to UV.
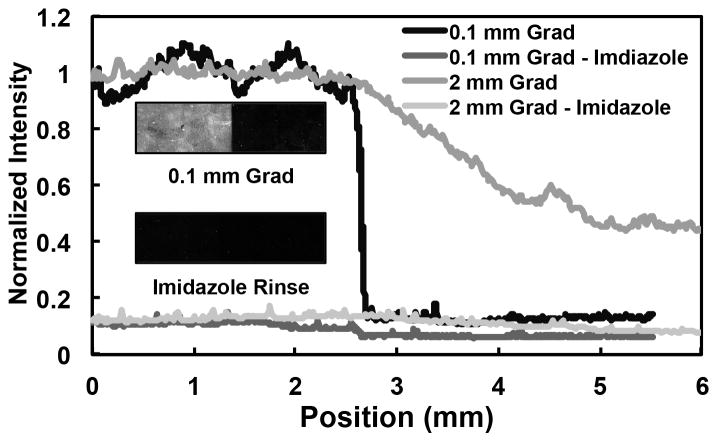
Fluorescent antibody binding to surface bound SDF-1α helped visualize SDF-1α gradients. Figure 8 shows two different gradient slopes formed by altering the removal speed of the UV mask over different distances. As seen in the inset image, fluorescent antibody binding was negligible on the sulfonate side of the gradient (dark region on right), which lacked the presentation of Nickel for binding of His-tagged protein. Bhatia et al. have also shown that the sulfonate group works well to inhibit non-specific adsorption [58]. On the other hand, antibody binding was robust on the thiol side of the gradient (bright region on left) that allowed specific binding of His-tagged protein to the immobilized Nickel. Furthermore, almost all of this fluorescence was abolished after incubation with imidazole that displaces the His-tag from the nickel (image inset, Figure 8). This validated the specific binding of our C-terminally His-tagged SDF-1α to the gradient surface. Since the affinity of specific His-tag-based binding is much higher than non-specific adsorption, it can be concluded that the relative amount of nonspecific adsorption is negligible after following rigorous rinsing steps and blocking in the fabrication process. The specifically bound His-tagged protein has its active site at the N-terminus free; therefore, it is oriented for accessibility.
Figure 8. Verification of specifically immobilized SDF-1α protein gradient.
SDF-1α gradients (via binding of His-tagged SDF-1α to the Ni gradients) with varying slopes were formed by altering the removal speed of the UV mask and then stained with a fluorescent antibody against SDF-1α. The measured fluorescence intensities of (1) a steep gradient across 0.1 mm and (2) a shallow gradient across 2 mm were abolished with an imidazole rinse (which competes with the His-tag for affinity with Ni), demonstrating that SDF-1α binding to gradient is specific via the His-tag. This is visually demonstrated via the inset showing a scanned fluorescent image of the steep gradient before and after imidazole rinse.
DISCUSSION
This work presents a test-platform culture substrate to track and study cell migration in real-time via the integration of several enabling technologies: (1) His-tag placement on protein in desired location via recombinant protein fabrication, (2) defined surface functionalization via use of self-assembled monolayers, (3) correct and stable multiple-protein orientation via IMAC based protein immobilization, (4) production of tunable and continuous protein gradients on surfaces (patterning) via graded UV exposure, and (5) continuous visualization and analysis of cell migratory behavior via a 2-D substrate-based assay and tracking.
The active site of SDF-1α, the chemokine used in this study, is at the N-terminal residues [59–61]. To ensure that the protein’s N-terminus was available for interaction with the cognate lymphocyte receptor we sought to attach the protein to the surface of our test-platform via the C-terminus. However, tethering the C terminus of a protein is very difficult to achieve with control, ease, and reliability using most available crosslinking techniques. Histidine residues attached to the C-terminus could bind to surface-immobilized metal ions via IMAC chemistry, thereby correctly orienting the protein. However, most commercially available His-tagged proteins, including SDF-1α have a His-tag at the N-terminus. Therefore, we produced C terminally His-tagged SDF-1α in house. We demonstrated that our recombinant His-tagged SDF-1α was specific and bioactive. Whereas the effect of batch-to-batch variability can be attributed to slight differences in production run conditions, to remove this variability, all subsequent experiments only used His-tagged SDF-1α from Batch 1.
Silica slides were used as a model substrate in the development of the 2-D cell migration test-platform. Silane-based self-assembled monolayers formed on these slides created uniform surfaces presenting reactive thiol groups following previously published methods [45,46]. The reactive thiols enabled covalent binding to the maleimide group of linker maleimide-C3-NTA molecules, the chelator nitrilotriacetic acid (NTA) groups of which were loaded with nickel ions, enabling the specific binding of His-tagged proteins of interest as described previously [26,34,47–50]. Such NTA-Ni immobilization surfaces are known to provide a biocompatible environment for preserving bound protein functionality [25]. The different surface fabrication steps are reproducible and have also been separately characterized in the cited literature. Cha et al., have demonstrated that whereas oriented protein selectively immobilized on a surface via the 6X His-tag faithfully reflected the activities of solution phase proteins, those with random orientation on the surface did not [62].
Conditions to enable in vitro migration of murine B lymphocytes needed to be established. We found that B lymphocytes (harvested from 6–12 week old mice) required activation via application of anti-IgM and anti-CD40 to show any significant surface migration. Resting B cells have been shown previously to respond to either T-independent IgM-mediated or T-dependent CD40-mediated activation via increased viability and migration [16,53]. The antibody binding are thought to mimic the binding of the respective cognate ligands to the cell surface receptors IgM and CD40, respectively, thereby initiating downstream activation pathways. B lymphocytes require induced activation to show migratory behavior in vitro potentially because they are taken out of their in vivo environment, where they naturally receive “activation” signals.
Initially, it was found that surfaces with His-tag immobilized SDF-1α were not conducive to migration of these activated cells. Spatio-temporal cell attachments/detachments regulate cell motility. Therefore, the adhesiveness of the substratum is a critical parameter to be considered while designing substrates to study cell migration [63]. This issue was addressed by incorporating Intercellular Adhesion Molecule-1 (ICAM-1) into our surface scheme and was co-immobilized on the surface using His-tagged Protein A to preserve its orientation. ICAM-1 was selected as the co-immobilant because it is ubiquitously present on the cell membranes that form the interstitial migration surfaces in the secondary lymphoid organs and influences lymphocyte trans-endothelial migration by providing adhesion cues [8,52,64–68]. Upon incorporation of ICAM-1 the activated B lymphocytes showed robust migratory behavior (Figure 4). The average migration speed of the B lymphocytes on our SDF-1α and ICAM-1 immobilized surfaces was 6.6 ± 0.5 μm/min, which is in close agreement with the ~ 6 μm/min ‘random walk’ speed at which these cells move within secondary lymphoid organ follicles [8,9,69], suggesting that our cell migration model system may elicit in vivo relevant migratory behavior of B lymphocytes.
This observation fits well with work by Dang and Rock that showed the engagement of the surface Ig receptor with anti-IgM antibodies (as in our cell activation protocol) stimulated murine B lymphocytes to markedly increase their expression of the cell adhesion molecules ICAM-1 and Lymphocyte function-associated antigen 1 (LFA-1) [52]. We postulate that the increased concentration of LFA-1 and it’s binding to the surface-immobilized ICAM-1 (ligand to LFA-1) was responsible for initiating cell adhesion and motility on our cell migration test-platform substrates. The ICAM-1/LFA-1 interactions are not stable enough to arrest cell movement, but are low-affinity adhesions that are transient enough (rapid making and breaking of adhesions) to give the cells the traction necessary to move given a large enough area of contact (Kd of ~ 130 nM) [70]. Using planar lipid bilayers containing ICAM-1 and CXCL13, Carrasco et al. have also seen this dependence of B-cell migration on ICAM-1 [8,43,71].
Although our recombinant SDF-1α influenced trans-well migration of cells when present in a diffusive form in solution, when immobilized on our nickel coated chemistry surfaces it failed (over a wide range of concentrations) to influence migration of activated splenic murine B lymphocytes beyond the influence of ICAM-1 alone (Figures 5 and 6). The fact that cells did not migrate in the absence of surface-immobilized ICAM-1, irrespective of the presence or absence of SDF-1α indicates that ICAM-1 and not SDF-1α is necessary for activated B lymphocyte migration in our cell migration system. It is possible that the SDF-1α when co-immobilized along with the bulkier combination of ICAM-1 via Protein-A is inaccessible to the cells because of steric hindrance; however, this possibility is less likely because we also observed accessible positive fluorescent antibody binding of the proteins when co-immobilized on the surface. Assuming that cells were able to interact with surface-immobilized SDF-1α, the question arises why they still not respond to SDF-1α presence. There can be several possibilities; For example, upon immobilization using our chemistry approach the bioactive SDF-1α may become ineffective as it may need to be internalized, which may be impeded by the immobilization scheme, or cells may need an additional missing co-factor along with SDF-1α. We do not know the exact reason why immobilized SDF-1α did not affect cell migration in our system. Our results bring into question the effectiveness of surface-immobilized SDF-1α in controlling activated splenic murine B lymphocyte migration.
Compared to trans-well migration studies, there are few studies on cell migration and related processes over surfaces, still fewer on protein-functionalized surfaces, and hardly any on functionalized surfaces with oriented and patterned protein. Table 2 summarizes some of these studies. Cell migration surfaces have been investigated using the microfluidic delivery of ligands to adsorb onto the substrate to form a gradient [21,22] and by covalent ligand immobilization using other gradient-forming techniques [29,72] that are discussed as follows. Microfluidics presents a controlled and reproducible approach to gradient formation. However, immobilization based on physisorption [21,22,73–76] provides less stable surfaces as proteins may denature and/or desorb over a period of time. Furthermore, it is impossible to control for packing density, conformation, and orientation of the physiosorbed protein [25,26,41]. Moreover, if the different component streams in a microfluidics mixer do not get enough contact time, then the resulting gradients have a more discrete step profile [21]. Likewise, physisorption based gradients often are constrained by the requirement of a mixture of two non-interacting proteins (e.g. protein of interest and BSA) to be introduced at the same time to avoid the possibility of a single protein saturating the surface even at low concentration [21]. In contrast, functionalizable alkanethiol gradients formed by cross diffusion [29] and a surface electrochemistry approach [72] to covalently attach proteins provide tunable means of forming continuous and stable protein gradients, although the surface immobilized protein may not be correctly oriented or active. More recently, Hjorto et al. used a microcontact printing approach to create gradients of active chemokine in the correct orientation [77]; however, their gradients were discontinuous, and chemokine presentation via an antibody lead to depletion of surface-immobilized ligand as cells internalized the gradient. Recent in vitro studies of B cell migration on 2-D surfaces by Carrasco et al. used planar lipid bilayers containing ICAM-1 and chemokines for the study of cell migration [8,43,71]. However, these surfaces have fabrication, stability, and protein orientation challenges, in addition to not being easily amenable to oriented, continuous, and stable protein gradient formation. Therefore, a new approach to stably orient multiple proteins on surfaces that is also amenable to surface patterning (e.g. gradients) would be a valuable enabling technology.
Table 2.
Different surfaces patterning approaches to study cellular behavior.
| Patterning | Method | Protein Orientation | Stability | Ref. |
|---|---|---|---|---|
| Uniform Distribution | Planar lipid bilayer containing GPI-linked adhesive proteins, biotinylated antigens, and a chemokine coat | Oriented/Random | Poor | [73–75] |
| Discrete Gradient | Micro-contact printing created chemokine gradient via antibody-based immobilization | Oriented | Poor | [77] |
| Discrete Gradient | Micro-fluidic network delivered protein gradient onto a silicone stamp transferred to a substrate via micro-contact printing | Random | Poor | [22] |
| Continuous Gradient | Micro-fluidic network created adhesive protein gradient adsorbed directly from a solution onto a substrate | Random | Poor | [21] |
| Continuous Gradient | Electrochemistry created surface gradients of covalently attached adhesive proteins and growth factors | Random | Excellent | [72] |
| Continuous Gradient | Cross diffusion created surface gradients of covalently-attached adhesive proteins | Random | Excellent | [29] |
| Continuous Gradient | Plasma corona treatment created wettability gradients converted to surface protein gradient via adsorption | Random | Poor | [76] |
Therefore, we extended the applicability of our surface chemistry approach to design specific cell migration surfaces in vitro to create surface gradients of immobilized and oriented proteins of interest. Oxidation of the surface thiols via controlled exposure to UV was used to first form a reactive surface thiols gradient using methods developed by Hlady et al. [55–57]. Subsequently, this gradient was translated via the maleimide-NTA-Ni chemistry to gradients of proteins of interest. Whereas XPS analysis confirmed every stage of the gradient formation process (Figure 7; and Supplementary Material, Figure S4), antibody-based immunofluorescence confirmed the presence of specifically bound protein gradients on the substrates (Figure 8).
In our system, by altering the removal speed of the UV mask we can tune the slope of the resulting protein gradients, which can be applied to the study of haptotaxis, the directed migration of cells up or down immobilized protein gradients. Furthermore, the His-tag to nickel interaction can be reversibly broken via imidazole, making these gradient as well as uniformly coated surfaces amenable to regeneration in a spatio-temporal manner, a property that can be exploited for different applications. In general, our protocol for immobilizing potentially any His- or Fc-tagged proteins (through His-tagged Protein A) and bio-functional protein patterning (via UV exposure) could be useful for a variety of different applications. Particularly, any silica-substrate based devices such as the Zigmond and Dunn chambers or silica-substrate analogs of the ibidi® cell migration slides, which presently do not present surface active factors (for example, BCA-1and VCAM-1 amongst others) in the correct orientation with active sites accessible in a stable manner, can be modified and patterned using our methodology for cell migration study. This methodology can be adapted to better model biointerfaces to suit the specific migration requirements of cell types such as endothelial cells, fibroblasts, neurons, neutrophils, T cells, and particularly those that are challenging such as B cells.
CONCLUSION
As proof-of-principle, the utility of a cell migration test platform presenting proteins of interest (such as SDF-1α and ICAM-1) in an orientated and stable form was demonstrated by studying the migration of primary splenic murine B lymphocytes, a difficult cell type with few documented 2-D surfaces migration studies in vitro. Using this platform, we showed that murine B lymphocytes needed activation and immobilized ICAM-1 to exhibit any surface migratory behavior. Although SDF-1α solutions influenced trans-migration, surfaces with uniform coatings of immobilized SDF-1α over a range of concentration failed to influence migration of this activated cell type. The findings indicate a potentially limited influence of immobilized SDF-1α on activated cell migration in our system. These interesting results merit further investigation. In principle, this modular, tunable, and controlled experimental system amenable to gradient fabrication can be applied to essentially any combination of protein or peptide surface immobilization, orientation, and patterning for a variety of biochemical, biotechnological, and biomedical applications beyond studying the migration behavior of any cell type.
Supplementary Material
Highlights.
A bioactive platform for detailed observation of 2-D cell migration is presented.
Migration factors are oriented on surface via His- and Fc- tags and IMAC chemistry.
ICAM-1 surface immobilization along with activation enabled B cell migration.
No influence of surface immobilized SDF-1α on activated B cell migration was seen.
Controlled exposure to UV enabled patterning via linear protein gradient fabrication.
Acknowledgments
The authors thank Ghalib Alkhatib from Texas Tech University Health Sciences Center for the His-tagged SDF-1α plasmid; Charlie Gersbach, Lauren Barth, and Pablo Perez-Pinera from the Department of Biomedical Engineering at Duke University for initial His-tagged SDF-1α purification and training; Vladimir Hlady and Lindsey Corum from the Department of Bioengineering at the University of Utah for training on surface gradient fabrication; and Michael Nichols for critical feedback and editorial assistance. This work was funded by NIH NIAID HHSN#272201000053C, NIH R01-DK-054932, and the Donald Alstadt Funds for SMIF Undergraduate User Program at Duke University.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Singer S, Kupfer A. Annual Review of Cell Biology. 1986;2:337. doi: 10.1146/annurev.cb.02.110186.002005. [DOI] [PubMed] [Google Scholar]
- 2.Thelen M. Nature Immunology. 2001;2:129. doi: 10.1038/84224. [DOI] [PubMed] [Google Scholar]
- 3.Locascio A, Nieto MA. Current Opinion in Genetics & Development. 2001;11:464. doi: 10.1016/s0959-437x(00)00218-5. [DOI] [PubMed] [Google Scholar]
- 4.Condeelis J, Singer RH, Segall JE. Annu Rev Cell Dev Biol. 2005;21:695. doi: 10.1146/annurev.cellbio.21.122303.120306. [DOI] [PubMed] [Google Scholar]
- 5.Chao DL, Ma L, Shen K. Nature Reviews Neuroscience. 2009;10:262. doi: 10.1038/nrn2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sahni A, Francis CW. Blood. 2000;96:3772. [PubMed] [Google Scholar]
- 7.Stroncek JD, Reichert WM. Indwelling Neural Implants: Strategies for Contending With the In Vivo Environment. 2008 [PubMed] [Google Scholar]
- 8.de Guinoa JS, Barrio L, Mellado M, Carrasco YR. Blood. 2011;118:1560. doi: 10.1182/blood-2011-01-332106. [DOI] [PubMed] [Google Scholar]
- 9.Pereira JP, Kelly LM, Cyster JG. International Immunology. 2010;22:413. doi: 10.1093/intimm/dxq047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bleul CC, Schultze JL, Springer TA. The Journal of Experimental Medicine. 1998;187:753. doi: 10.1084/jem.187.5.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Butcher EC, Picker LJ. Science. 1996;272:60. doi: 10.1126/science.272.5258.60. [DOI] [PubMed] [Google Scholar]
- 12.Lewis WH, Webster LT. The Journal of Experimental Medicine. 1921;33:261. doi: 10.1084/jem.33.2.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lewis WH. Bull Johns Hopkins Hosp. 1933;53:147. [Google Scholar]
- 14.Haston WS, Shields JM, Wilkinson PC. The Journal of Cell Biology. 1982;92:747. doi: 10.1083/jcb.92.3.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harris H. British Journal of Experimental Pathology. 1953;34:599. [PMC free article] [PubMed] [Google Scholar]
- 16.Brandes M, Legler DF, Spoerri B, Schaerli P, Moser B. International Immunology. 2000;12:1285. doi: 10.1093/intimm/12.9.1285. [DOI] [PubMed] [Google Scholar]
- 17.Boyden S. The Journal of Experimental Medicine. 1962;115:453. doi: 10.1084/jem.115.3.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mosadegh B, Saadi W, Wang SJ, Jeon NL. Biotechnology and Bioengineering. 2008;100:1205. doi: 10.1002/bit.21851. [DOI] [PubMed] [Google Scholar]
- 19.Zigmond SH, Hirsch JG. The Journal of Experimental Medicine. 1973;137:387. doi: 10.1084/jem.137.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zicha D, Dunn GA, Brown AF. Journal of Cell Science. 1991;99:769. doi: 10.1242/jcs.99.4.769. [DOI] [PubMed] [Google Scholar]
- 21.Dertinger SK, Jiang X, Li Z, Murthy VN, Whitesides GM. Proceedings of the National Academy of Sciences. 2002;99:12542. doi: 10.1073/pnas.192457199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lang S, von Philipsborn AC, Bernard A, Bonhoeffer F, Bastmeyer M. Analytical and Bioanalytical Chemistry. 2008;390:809. doi: 10.1007/s00216-007-1363-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanley PJ, Xu Y, Kronlage M, Grobe K, Schön P, Song J, Sorokin L, Schwab A, Bähler M. Proceedings of the National Academy of Sciences. 2010;107:12145. doi: 10.1073/pnas.0911986107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suraneni P, Rubinstein B, Unruh JR, Durnin M, Hanein D, Li R. The Journal of Cell Biology. 2012;197:239. doi: 10.1083/jcb.201112113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frasconi M, Mazzei F, Ferri T. Analytical and Bioanalytical Chemistry. 2010;398:1545. doi: 10.1007/s00216-010-3708-6. [DOI] [PubMed] [Google Scholar]
- 26.Nakanishi K, Sakiyama T, Kumada Y, Imamura K, Imanaka H. Current Proteomics. 2008;5:161. [Google Scholar]
- 27.DiMilla PA, Stone JA, Quinn JA, Albelda SM, Lauffenburger DA. The Journal of Cell Biology. 1993;122:729. doi: 10.1083/jcb.122.3.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Su L, Kelly JB, Hawkridge FM, Rhoten MC, Baskin SI. Journal of Electroanalytical Chemistry. 2005;581:241. [Google Scholar]
- 29.Smith JT, Tomfohr JK, Wells MC, Beebe TP, Kepler TB, Reichert WM. Langmuir. 2004;20:8279. doi: 10.1021/la0489763. [DOI] [PubMed] [Google Scholar]
- 30.Kyprianou D, Guerreiro AR, Nirschl M, Chianella I, Subrahmanyam S, Turner AP, Piletsky S. Biosensors and Bioelectronics. 2010;25:1049. doi: 10.1016/j.bios.2009.09.030. [DOI] [PubMed] [Google Scholar]
- 31.Houseman BT, Gawalt ES, Mrksich M. Langmuir. 2003;19:1522. [Google Scholar]
- 32.Smith EA, Thomas WD, Kiessling LL, Corn RM. Journal of the American Chemical Society. 2003;125:6140. doi: 10.1021/ja034165u. [DOI] [PubMed] [Google Scholar]
- 33.Xu F, Zhen G, Yu F, Kuennemann E, Textor M, Knoll W. Journal of the American Chemical Society. 2005;127:13084. doi: 10.1021/ja050818q. [DOI] [PubMed] [Google Scholar]
- 34.Sigal GB, Bamdad C, Barberis A, Strominger J, Whitesides GM. Analytical Chemistry. 1996;68:490. doi: 10.1021/ac9504023. [DOI] [PubMed] [Google Scholar]
- 35.Zhang K, Diehl MR, Tirrell DA. Journal of the American Chemical Society. 2005;127:10136. doi: 10.1021/ja051457h. [DOI] [PubMed] [Google Scholar]
- 36.Murray AM, Kelly CD, Nussey SS, Johnstone AP. J Immunol Methods. 1998;218:133. doi: 10.1016/s0022-1759(98)00114-8. [DOI] [PubMed] [Google Scholar]
- 37.Johnson CP, Jensen IE, Prakasam A, Vijayendran R, Leckband D. Bioconjugate Chemistry. 2003;14:974. doi: 10.1021/bc034063t. [DOI] [PubMed] [Google Scholar]
- 38.Garca AJ, Vega MaD, Boettiger D. Molecular Biology of the Cell. 1999;10:785. doi: 10.1091/mbc.10.3.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Michael KE, Vernekar VN, Keselowsky BG, Meredith JC, Latour RA, García AJ. Langmuir. 2003;19:8033. [Google Scholar]
- 40.Vernekar VN, Latour RA., Jr Materials Research Innovations. 2005;9:53. [Google Scholar]
- 41.Mrksich M. Chem Soc Rev. 2000;29:267. [Google Scholar]
- 42.Blawas A, Reichert W. Biomaterials. 1998;19:595. doi: 10.1016/s0142-9612(97)00218-4. [DOI] [PubMed] [Google Scholar]
- 43.Barrio L, de Guinoa JS, Carrasco YR. The Journal of Immunology. 2013;191:3867. doi: 10.4049/jimmunol.1301623. [DOI] [PubMed] [Google Scholar]
- 44.Nagasawa T, Hirota S, Tachibana K, Takakura N, Nishikawa S-i, Kitamura Y, Yoshida N, Kikutani H, Kishimoto T. Nature. 1996;382:635. doi: 10.1038/382635a0. [DOI] [PubMed] [Google Scholar]
- 45.Hu M, Noda S, Okubo T, Yamaguchi Y, Komiyama H. Applied Surface Science. 2001;181:307. [Google Scholar]
- 46.Ulman A. Chemical Reviews. 1996;96:1533. doi: 10.1021/cr9502357. [DOI] [PubMed] [Google Scholar]
- 47.Kim KH, Kim JD, Kim YJ, Kang SH, Jung SY, Jung H. Small. 2008;4:1089. doi: 10.1002/smll.200700519. [DOI] [PubMed] [Google Scholar]
- 48.Laura EV, Elisa H, María Fernanda S, Vitor LM, Roberto MT, Carla EG. Proteins at Interfaces III State of the Art. American Chemical Society; 2012. p. 37. [Google Scholar]
- 49.Shen J, Ahmed T, Vogt A, Wang J, Severin J, Smith R, Dorwin S, Johnson R, Harlan J, Holzman T. Analytical Biochemistry. 2005;345:258. doi: 10.1016/j.ab.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 50.Porath J, Carlsson J, Olsson I, Belfrage G. Nature. 1975;258:598. doi: 10.1038/258598a0. [DOI] [PubMed] [Google Scholar]
- 51.Poznansky MC, Olszak IT, Foxall R, Evans RH, Luster AD, Scadden DT. Nature Medicine. 2000;6:543. doi: 10.1038/75022. [DOI] [PubMed] [Google Scholar]
- 52.Dang LH, Rock KL. The Journal of Immunology. 1991;146:3273. [PubMed] [Google Scholar]
- 53.Wortis HH, Teutsch M, Higer M, Zheng J, Parker DC. Proceedings of the National Academy of Sciences. 1995;92:3348. doi: 10.1073/pnas.92.8.3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu J, Hlady V. Colloids and Surfaces B: Biointerfaces. 1996;8:25. doi: 10.1016/S0927-7765(96)01298-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Corum LE, Hlady V. Biomaterials. 2010;31:3148. doi: 10.1016/j.biomaterials.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ding YX, Streitmatter S, Wright BE, Hlady V. Langmuir. 2010;26:12140. doi: 10.1021/la101674b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ding YX, Hlady V. Croatica Chemica Acta Arhiv za kemiju. 2011;84:193. doi: 10.5562/cca1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bhatia SK, Shriver-Lake LC, Prior KJ, Georger JH, Calvert JM, Bredehorst R, Ligler FS. Analytical Biochemistry. 1989;178:408. doi: 10.1016/0003-2697(89)90662-3. [DOI] [PubMed] [Google Scholar]
- 59.Crump MP, Gong JH, Loetscher P, Rajarathnam K, Amara A, Arenzana-Seisdedos F, Virelizier JL, Baggiolini M, Sykes BD, Clark-Lewis I. The EMBO Journal. 1997;16:6996. doi: 10.1093/emboj/16.23.6996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huang X, Shen J, Cui M, Shen L, Luo X, Ling K, Pei G, Jiang H, Chen K. Biophysical Journal. 2003;84:171. doi: 10.1016/S0006-3495(03)74840-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Luo J, Luo Z, Zhou N, Hall JW, Huang Z. Biochemical and Biophysical Research Communications. 1999;264:42. doi: 10.1006/bbrc.1999.1476. [DOI] [PubMed] [Google Scholar]
- 62.Cha T, Guo A, Zhu XY. Proteomics. 2005;5:416. doi: 10.1002/pmic.200400948. [DOI] [PubMed] [Google Scholar]
- 63.Palecek SP, Loftus JC, Ginsberg MH, Lauffenburger DA, Horwitz AF. Nature. 1997;385:537. doi: 10.1038/385537a0. [DOI] [PubMed] [Google Scholar]
- 64.Peled A, Grabovsky V, Habler L, Sandbank J, Arenzana-Seisdedos F, Petit I, Ben-Hur H, Lapidot T, Alon R. Journal of Clinical Investigation. 1999;104:1199. doi: 10.1172/JCI7615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Peled A, Kollet O, Ponomaryov T, Petit I, Franitza S, Grabovsky V, Slav MM, Nagler A, Lider O, Alon R. Blood. 2000;95:3289. [PubMed] [Google Scholar]
- 66.Ding Z, Issekutz TB, Downey GP, Waddell TK. Blood. 2003;101:4245. doi: 10.1182/blood-2002-06-1782. [DOI] [PubMed] [Google Scholar]
- 67.Gretz JE, Anderson AO, Shaw S. Immunological Reviews. 1997;156:11. doi: 10.1111/j.1600-065x.1997.tb00955.x. [DOI] [PubMed] [Google Scholar]
- 68.Woolf E, Grigorova I, Sagiv A, Grabovsky V, Feigelson SW, Shulman Z, Hartmann T, Sixt M, Cyster JG, Alon R. Nature Immunology. 2007;8:1076. doi: 10.1038/ni1499. [DOI] [PubMed] [Google Scholar]
- 69.Miller MJ, Wei SH, Parker I, Cahalan MD. Science. 2002;296:1869. doi: 10.1126/science.1070051. [DOI] [PubMed] [Google Scholar]
- 70.Hogg N, Laschinger M, Giles K, McDowall A. Journal of Cell Science. 2003;116:4695. doi: 10.1242/jcs.00876. [DOI] [PubMed] [Google Scholar]
- 71.de Guinoa JS, Barrio L, Carrasco YR. The Journal of Immunology. 2013;191:2742. doi: 10.4049/jimmunol.1300684. [DOI] [PubMed] [Google Scholar]
- 72.Liu L, Ratner BD, Sage EH, Jiang S. Langmuir. 2007;23:11168. doi: 10.1021/la701435x. [DOI] [PubMed] [Google Scholar]
- 73.Groves JT, Dustin ML. Journal of Immunological Methods. 2003;278:19. doi: 10.1016/s0022-1759(03)00193-5. [DOI] [PubMed] [Google Scholar]
- 74.Kam L, Boxer SG. Journal of Biomedical Materials Research. 2001;55:487. doi: 10.1002/1097-4636(20010615)55:4<487::aid-jbm1041>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 75.Holden MA, Jung SY, Yang T, Castellana ET, Cremer PS. Journal of the American Chemical Society. 2004;126:6512. doi: 10.1021/ja048504a. [DOI] [PubMed] [Google Scholar]
- 76.Lee JH, Lee HB. Journal of Biomedical Materials Research. 1998;41:304. doi: 10.1002/(sici)1097-4636(199808)41:2<304::aid-jbm16>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 77.Hjortø GM, Hansen M, Larsen NB, Kledal TN. Biomaterials. 2009;30:5305. doi: 10.1016/j.biomaterials.2009.06.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.