Abstract
We compared transcriptomes of terminally differentiated mouse 3T3-L1 and human adipocytes to identify cell-specific differences. Gene expression and high content analysis (HCA) data identified the androgen receptor (AR) as both expressed and functional, exclusively during early human adipocyte differentiation. The AR agonist dihydrotestosterone (DHT) inhibited human adipocyte maturation by downregulation of adipocyte marker genes, but not in 3T3-L1. Interestingly, AR induction corresponded with dexamethasone activation of the glucocorticoid receptor (GR); however, when exposed to the differentiation cocktail required for adipocyte maturation, AR adopted an antagonist conformation and was transcriptionally repressed. To further explore effectors within the cocktail, we applied a novel, image-based support vector machine (SVM) classification scheme to show adipocyte differentiation components inhibit AR action. The results demonstrate human adipocyte differentiation, via GR activation, upregulates AR but also inhibits AR transcriptional activity.
INTRODUCTION
White adipose tissue is a central metabolic organ that regulates energy balance by storing and mobilizing lipids. In lean individuals, adipocytes maintain a dynamic equilibrium between triglyceride storage and lipolysis. On the other hand, obese subjects have enlarged adipocytes resulting from high caloric intake and increased triglyceride storage in larger lipid droplets. As the obesity epidemic continues to spread, it is likely a variety of therapeutic intervention strategies will be evaluated, including the targeting of metabolic pathways directly involved in fat synthesis and storage (Guilherme et al., 2008). Thus, the identification of genes associated with human adipocyte differentiation is key to understanding fat deposition and the pathogenesis of obesity.
There is now significant evidence suggesting androgens are important regulators of energy balance, fat deposition, and body composition in males and females (Blouin et al., 2009b), while also influencing other endocrine targets including bone and skeletal muscle (Zitzmann, 2009). It is well established that men experience an increase in body mass index (BMI) as a consequence of hypogonadism and aging, conditions associated with a decreased level of circulating testosterone (Gould et al., 2007). In women, the association between obesity and androgens is more enigmatic and poorly characterized. Although female AR deficiency has not been well studied, women with complete androgen insensitivity syndrome have increased fat mass (Dati et al., 2009). On the other hand, hyperandrogenemia has been known to provoke insulin resistance, independent of obesity (Coviello et al., 2006) through systemic oxidative stress, including disruption of β–cell dysfunction (Liu et al., 2010). However, levels of AR do not predict fat distribution or negatively correlate with BMI in men and women (Wake et al., 2007) suggesting that AR activity per se might be differentially regulated in obese versus lean states.
Androgens influence gene transcription through activation of AR, a member of the nuclear receptor (NR) superfamily of transcription factors (Chang et al., 1988; Lubahn et al., 1988). Upon ligand binding, conformational change, and homodimerization, AR can regulate gene transcription by binding to specific DNA motifs (Schoenmakers et al., 2000) which comprise consensus hormone response elements in AR target genes (HREs). Consensus HREs are also recognized by GR allowing extensive crosstalk between receptors (Lieberman et al., 1993; Nordeen et al., 1990; Roche et al., 1992) and shared target genes, including the immunophilin FKBP5 (Magee et al., 2006). Indeed, recent genome-wide analyses have shown AR and GR binding sites in non-adipocyte cells are enriched in pathways associated with lipid and fatty acid metabolism (Bolton et al., 2007; Massie et al., 2011; Reddy et al., 2009). In 3t3-L1 cells (Yu et al., 2010), primary GR target genes are involved in fatty acid transport (FABP4 (Hotamisligil et al., 1996)), energy storage (CIDEC (Nishino et al., 2008)), and those which are adipocyte-specific (PPARγ2 (Tontonoz et al., 1994)). Genome-wide analysis of AR binding in adipocytes has yet to be performed.
Overall, cell-based studies in human preadipocytes (Blouin et al., 2009a; Blouin et al., 2010; Gupta et al., 2008) and 3T3-L1 (Singh et al., 2006) have shown androgens suppress lipid accumulation during late stage, terminal endpoints. Here, we have analyzed the transcriptomes of terminally differentiated mouse 3T3-L1 and human adipocytes to identify species-specific genes and pathways involved in the adipogenic process. When we analyzed mRNAs changing during adipogenesis in vitro, we identified an increase in AR mRNA specifically in human adipocytes, while being undetectable in differentiating 3T3-L1 cells. Because the regulation of AR and a dominant functional role remains largely undefined, we complemented our transcriptomic discovery with image-based and chemical biology approaches to identify and classify dominant mechanisms of AR regulation in adipocytes.
RESULTS
Identification of genes unique to human adipocyte differentiation
To discover novel genes associated with human adipocyte differentiation in vitro, we used Illumina whole genome arrays to measure gene expression in subcutaneous primary human adipocytes treated for up to 14 days with rosiglitazone, IBMX, dexamethasone, and insulin (MIX). ANOVA analysis of gene expression revealed 2674 differentially regulated array features with many of the standard genes associated with adipocyte differentiation upregulated (Supplemental Figure 1). Next, we compared gene expression data between terminally differentiated mouse 3T3-L1 adipocytes (Schupp et al., 2009) and human adipocytes differentiated for 14 days. As expected, overlapping probe sets (817; Figure 1A) were comprised of transcription factors critical for adipocyte differentiation, including PPARγ (Rosen et al., 1999) and CEBPα (Rosen et al., 2002), and genes classically associated with cholesterol/fat/lipid metabolism programs, including ADIPOQ, FABP4, ABCA1, CD36, and FASN among others (Supplemental Excel File 1). However, we also detected 3496 and 1496 genes uniquely expressed in 3T3-L1 (Supplemental Excel File 1) and human (Supplemental Excel File 1) adipocytes, respectively.
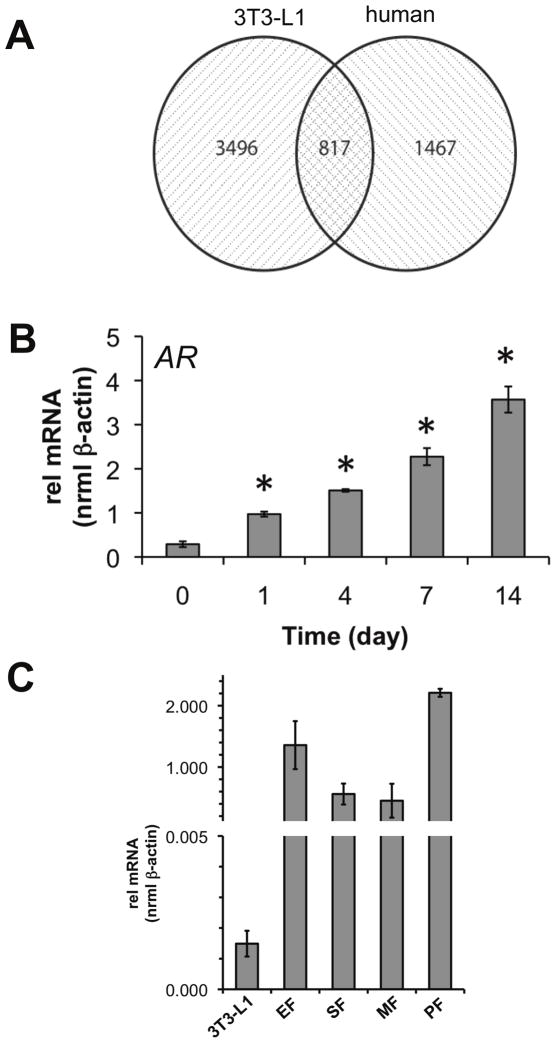
Figure 1.
Microarray analysis identifies unique profiles associated with mouse (3T3-L1) and human adipocyte differentiation models. (A) Comparison of the human expression data to a compatible dataset generated in mouse 3T3-L1 found 3496 and 1467 genes exclusive to mouse and human microarrays, respectively, with an 817 gene overlap. (B) Of the 1467 genes regulated in the human class, human AR was detected and validated by qPCR (*p<0.05 vs day 0). (C) Expression of AR in mouse tissues and cell lines. Total mRNA was isolated and AR transcript levels were compared between epididymal fat (EF), subcutaneous fat (SF), mesenteric fat, peritoneal fat depots (n=3 mice), and terminally differentiated 3T3-L1 (9 d).
Of the genes detected which were changing in the human arrays, AR mRNA was up-regulated 3.5-fold between day 0 and day 14. We validated the induction of AR mRNA using quantitative real-time PCR (qPCR) and measured a 3.35-fold induction as early as day 1, and a 12.3-fold increase over the 14 d differentiation period (Figure 1B). For comparison, we measured AR expression in primary mouse adipose tissue. We found AR mRNA was expressed in epididymal fat (EF), subcutaneous fat (SF), mesenteric fat (MF), and peritoneal fat (PF) at 910-, 376-, 302-, and 1484-times the level, respectively, of AR expression in mature 3T3-L1 adipocytes (Figure 1C). Our initial findings indicated AR was not highly expressed in 3T3-L1 adipocytes.
DHT inhibits human adipocyte differentiation
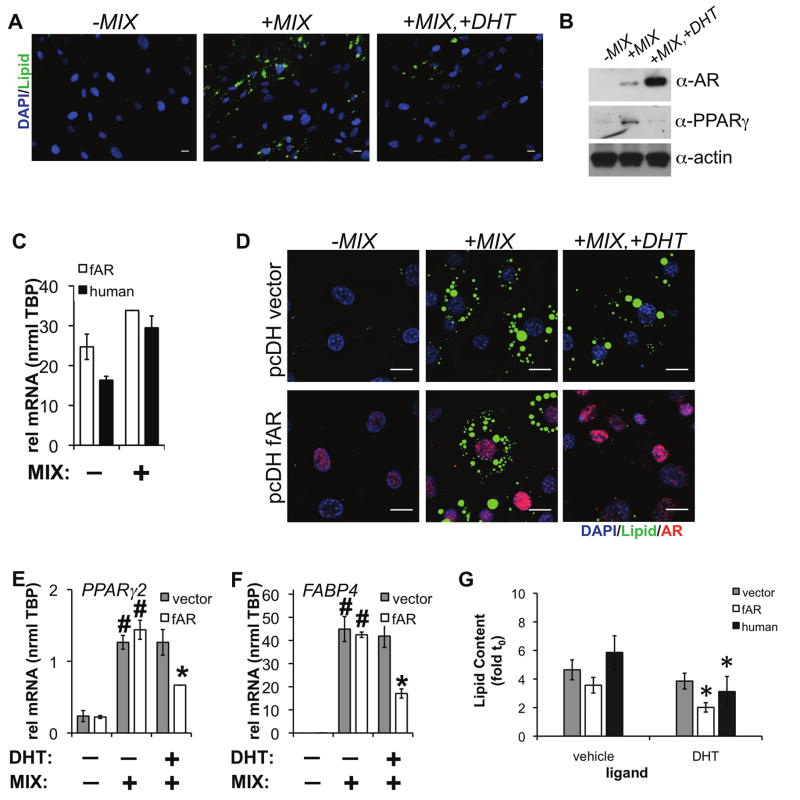
Since AR mRNA was detected as a significantly-expressed and induced gene in human adipocytes, we hypothesized this differentiation program would be sensitive to androgen (dihydrotestosterone, DHT) treatment. To test this hypothesis, we applied a set of previously customized image analysis tools that automatically identify cells and nuclei to extract fluorescence-based measurements (e.g., high content analysis: HCA) of NR and coregulator proteins, and, as a biomarker for differentiation, levels of intracellular lipids (Hartig et al., 2011). As shown in Figure 2A, MIX-induced adipocyte differentiation was reduced by supplementation with 10 nM DHT at 96 h as indicated by reduced lipid accumulation. By Western blotting, MIX supplemented with 10 nM DHT showed both decreased PPARγ expression and increased AR levels (Figure 2B). To further validate the role of AR in adipocyte differentiation, we then compared the effects of DHT on the differentiation of 3T3-L1 adipocytes when human AR was expressed by lentivirus. 3T3-L1 cells were infected with either empty vector (pcDH) or virus encoding human FLAG-AR (pcDH fAR). In response to vehicle (DMSO) or MIX, AR expression in 3T3-L1 was approximately equal to levels found in human adipocytes (Figure 2C). After 48 h, cells were induced to differentiate with standard 3T3-L1 adipogenic cocktail (dexamethasone, IBMX, insulin) and EtOH (vehicle), or 10 nM DHT for 96 h. DHT (10 nM) suppressed lipid accumulation (−44% vs vehicle) only when AR was expressed by lentivirus and not in the vector control (Figure 2D). The adipocyte-specfic genes PPARγ2 (−54% vs vehicle, Figure 2E) and FABP4 (−60% vs vehicle, Figure 2F) were also significantly down-regulated by DHT only in 3T3-L1 cells expressing human AR. Further, we measured a 47% decrease in lipid accumulation in human adipocytes, which compared well with the effect in 3T3-L1 when AR was expressed by lentivirus (Figure 2G). Our results support previous observations demonstrating inhibition of human adipocyte differentiation by AR agonists (Blouin et al., 2009a; Blouin et al., 2010; Gupta et al., 2008). Taken together, we speculated the discrepancy in DHT-mediated inhibition of early adipocyte differentiation between the two cell types is attributable to differences in their AR expression levels. Human adipocytes express sufficient AR at a level where DHT can inhibit early adipogenesis, while extremely low levels of AR in 3T3-L1 cells prohibit any effect of DHT on lipid accumulation.
Figure 2.
(A) Human preadipocytes were induced to differentiate for 96 h with rosiglitazone, insulin, dexamethasone, and IBMX (MIX) in the presence or absence of 10 nM DHT. Cells were then stained with DAPI (DNA), CellMask Blue (CMBl, general protein dye), and LipidTOX (Lipid), followed by HTM. Scale bar is 50 μm. (B) Lysates were prepared from adipocytes differentiated for 4 d in the presence or absence of 10 nM DHT. Protein levels of AR and PPARγ were detected by western blot. (C) Human AR-FLAG (fAR) or vector control lentiviral particles were prepared and introduced into 3T3-L1 preadipocytes for 48 h before chemical induction of differentiation. Human AR expression was detected in both human adipocytes and 3T3L1 expressing fAR before and after 96 h of differentiation. (D) After cell differentiation in the presence or absence of 10 nM DHT for 96 h, cells were processed for immunofluorescence to AR, stained for lipid and DAPI/CMBl, and imaged. Scale bar, 20 μm. (E) PPARγ2 and (F) FABP4 mRNA levels were measured from 3T3L1 cells expressing vector (pcDH) or fAR differentiated in the presence or absence of 10 nM DHT for 96 h. (n=2 independent experiments +/− s.e.m., #p<0.05 compared to no differentiation or *p<0.05 compared to differentiation without DHT). (G) Lipid accumulation was quantified by HCA after lentiviral expression of human AR (n=average of 4 wells+/− s.e.m.) or androgen treatment in human preadipocytes (n=5 independent experiments+/− s.e.m., *p<0.05).
Human adipocyte differentiation is suppressed by AR activation
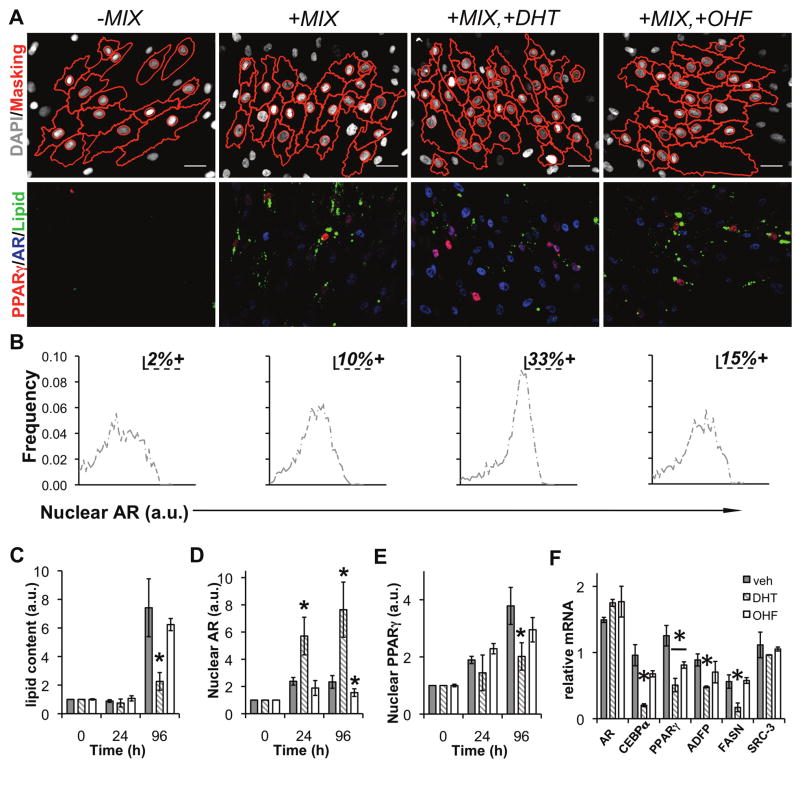
We next examined the effect of 10 nM DHT and 10 μM O-hydroxyflutamide (OHF), an AR antagonist, on expression of adipocyte-specific mRNAs and nuclear levels of PPARγ and AR in human subcutaneous preadipocytes induced to differentiate by MIX. Amongst positive cells, AR and PPARγ exhibited a chiefly nuclear signal (>75%) by immunofluorescence (Figure 3A), which was independent of DHT and OHF. Using AR levels at 96 h (90th percentile) as a threshold to define AR-positive cells, we established that DHT increased the number of AR positive cells by 3.3-fold compared to MIX (Figure 3B). Of note, the range of expression for positive cells was less than one-log between minimum and maximum AR levels. As determined by Pearson’s r calculation (N>3700 cells for all treatments), cells exhibiting higher levels of AR as a result of androgen treatment did not correspond with decreased nuclear PPARγ (PrMIX/DHT=0.105, N=4765 cells). Indeed, no treatment induced a significant correlation between nuclear AR and nuclear PPARγ (Pr-MIX=0.06; PrMIX=0.09; PrMIX/OHF=0.05) suggesting an upstream regulatory event might contribute to the regulation of both receptors. Following 96 h of treatment, differentiation of human preadipocytes in the presence of DHT was associated with reduced lipid accumulation (−32%, Figure 3C), dramatically increased levels of AR protein (10-fold over vehicle, Figure 3D), and reduced levels of nuclear PPARγ (−20%, Figure 3E). AR protein stabilization by androgen appears to be the main effect, as AR mRNA levels were only modestly changed during differentiation in the presence of DHT (+15% vs. no DHT, Figure 3F). Conversely, levels of adipocyte-associated (C/EBPα: −79%, PPARγ: −60%) and lipid storage (ADFP: −46%, FASN: −70%) genes were all reduced under the same conditions (Figure 3F). As a control, the mRNA level of the transcriptional coactivator SRC-3 was not significantly affected by differentiation in the presence of androgen (DHT) or anti-androgen (OHF). We suggest AR activation by DHT leads to an accumulation (stabilization) of nuclear AR (Furutani et al., 2002) which reduces expression of PPARγ mRNA and subsequently slows adipocyte differentiation. Collectively, these results extended the findings of Figure 2 and position the activity of AR as a critical determinant of lipid storage in adipocytes.
Figure 3.
AR activation suppresses human adipogenesis. (A) Images from human adipocytes differentiated for 96 h with the indicated ligand treatments, immunolabeled with antibodies to PPARγ (gray scale), AR (red), and stained with LipidTOX (Lipid, green), and DAPI. On the left-most panels, masks (red) generated by image analysis algorithms were overlaid onto the DAPI (gray scale) images to indicate cell and nucleus borders. (B) Percent AR-positive cells were defined by measuring the 90th percentile expression level for +MIX and applying that threshold to single cell distributions of AR for the other 3 treatments. The effect of androgens on the rate of lipid accumulation, nuclear PPARγ, and nuclear AR was determined by differentiation of human preadipocytes for the indicated time points in the presence of DHT, vehicle (EtOH), or OHF. HCA was used to quantify the induction of (C) lipid accumulation, (D) AR, and (E) PPARγ. In parallel, relative mRNA was determined for (F) AR, C/EBPα, PPARγ, ADFP, FASN, and SRC-3.
GR promotes AR expression during human adipocyte differentiation
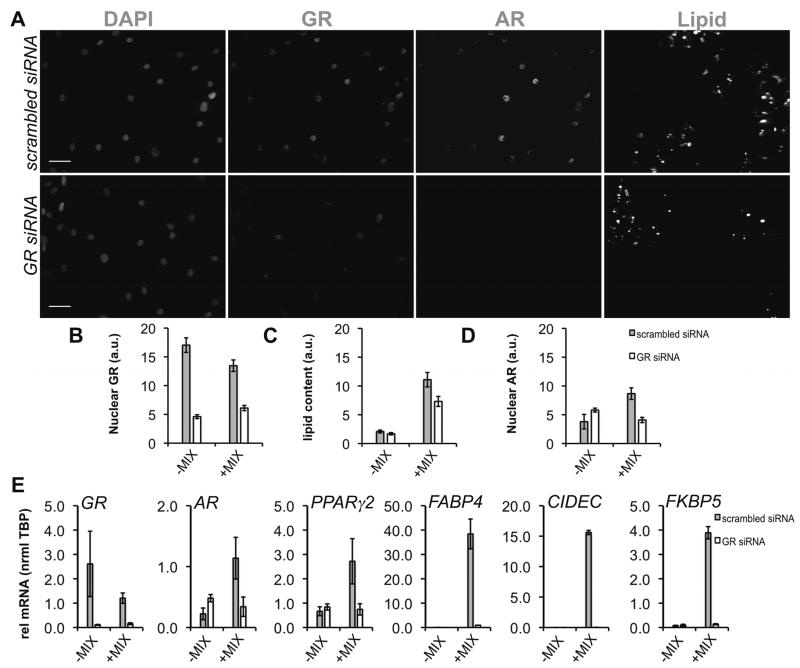
As a pioneer factor in preadipocytes, the glucocorticoid receptor (GR) is required to initiate cell differentiation and activate transcription of pro-adipocyte genes, including PPARγ2 (Steger et al., 2010). Because recent ChIP-Seq experiments in A549 cells found dexamethasone-induced GR binding regions <10 kB upstream of the AR promoter (Reddy et al., 2009) and AR was significantly upregulated early (~24 h after differentiation (Figure 2A)), we reasoned GR might regulate AR expression. To test this hypothesis, GR was downregulated by siRNA in preadipocytes. Subsequently, cells were treated with MIX or vehicle (EtOH) for 96 h. Using immunofluorescence (Figure 4A) and HCA, we detected GR knockdown (−54% vs scR siRNA, Figure 4B) which corresponded to both decreased lipid accumulation (−35% vs scR siRNA, Figure 4C) and reduced nuclear AR (−52% compared to scR siRNA). By qPCR (Figure 4E), GR siRNA inhibited AR mRNA induction with significant downregulation of PPARγ2, adipogenic targets of PPARγ and GR (FABP4, CIDEC), and a canonical GR/AR target (FKBP5). The marked inhibition of AR induction after GR siRNA suggested GR is required for early upregulation of AR in adipocytes, in addition to the established roles of GR as a primary regulator of early adipogenesis (Pantoja et al., 2008; Steger et al., 2010).
Figure 4.
siRNA-mediated depletion of GR inhibits AR upregulation in differentiating human adipocytes. (A) Preadipocytes were transfected with scrambled (scrambled siRNA) or siRNA to GR (GR siRNA) for 48 h followed by induction of differentiation (+MIX) and HTM. Scale bar, 20 μm. (B) HCA detection of nuclear GR, lipid accumulation, and nuclear AR after siRNA transfection (n≥4 experiments) by immunofluorescence and lipid labeling. (C) qPCR was used to validate HCA measurements of GR knockdown and downstream effects on AR, lipid accumulation, and target gene expression: GR, AR, PPARγ2, FABP4, CIDEC, FKBP5. RNA was isolated from 2 independent experiments. Asterisks indicate measured variables statistically different from the non-targeting siRNA control at the 95% confidence level (*p<0.05). Error bars indicate s.e.m.
Dexamethasone is required for AR upregulation in human adipocytes
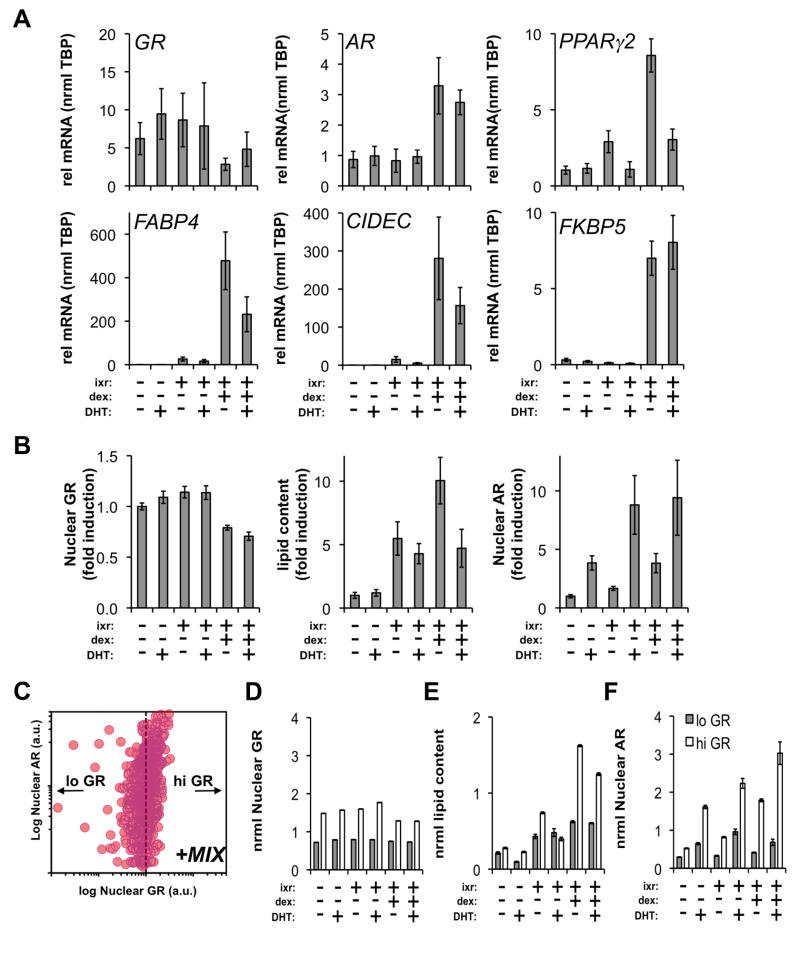
To further understand the role of GR and dexamethasone in driving AR upregulation, we examined the uncoupling of dexamethasone (dex) from the three other components (ixr: insulin, IBMX, rosiglitazone) present in MIX. At the mRNA level (Figure 5A), dex supplementation was required for AR induction (4.5-fold vs EtOH) and maximal upregulation of PPARγ2 (6.4-fold vs EtOH). Target genes of PPARγ2 and GR, FABP4, CIDEC, and FKBP5, respectively, were each coordinately and significantly upregulated when dex was added to ixr. As observed in Figure 3F, the addition of 10 nM DHT to ixr+dex treatments decreased the expression of PPARγ2 (−58%) and genes associated lipid transport (FABP4: −80%) and storage (CIDEC: −60%). Upregulation of FKBP5 by dex supplementation was unaffected by 10 nM DHT indicating a dominant effect of dex on target genes shared by both AR and GR.
Figure 5.
AR expression is correlated with dexamethasone-driven adipocyte differentiation. (A) Preadipocytes were differentiated (ixr: insulin/IBMX/rosiglitazone) for 96 h in the presence or absence of dexamethasone (dex) +/− 10 nM DHT. qPCR was used to measure mRNA expression for GR, AR, PPARγ2, FABP4, CIDEC, FKBP5. RNA was isolated from 2 independent experiments. (B) In parallel experiments, effects of dex and DHT on AR induction and lipid accumulation were evaluated by HCA. Subsequent to the treatments, GR and AR were detected by immunofluorescence in combination with lipid labeling. (C) Cell-to-cell correlation between AR and GR was monitored. One representative experiment is shown for cells differentiated with standard cocktail (MIX) for 96 h. Individual cell measurements of nuclear GR and AR were normalized to the median intensity of standard differentiation cocktail (ixr+d; MIX) at 96 h. GR normalization of all treatments set a threshold (dotted line) to subdivide cells into GRlo and GRhi subpopulations. (D) The median single cell intensities of GR, (E) lipid content and (F) AR were determined for the GRlo and GRhi populations at 96 h (*p<0.05, n≥400 cells/treatment). Error bars indicate SEM.
In parallel experiments, we confirmed the qPCR findings (Figure 5A) and dissected the regulation of AR by dex in human adipocytes using HCA (Figure 5B). Addition of dex to differentiation components (ixr) caused a decrease in GR protein levels (ixr+dex: −21% vs EtOH) consistent with a ligand- and proteasome-dependent downregulation mechanism (Wallace and Cidlowski, 2001). While the mRNA data indicated dex was required to upregulate AR mRNA (Figure 5A), DHT addition to any treatment was sufficient to increase AR protein levels with a maximum observed induction of 8.9-fold (ixr+dex+DHT) providing further evidence of androgen-mediated AR stabilization in adipocytes. Collectively, these data show lipid accumulation requires dex, and it is inhibited by supplementation of differentiation cocktail with DHT (ixr+dex+DHT).
Figure 4 and Figure 5A–B supported the role of both dex and GR action in driving AR induction in human adipocytes. Although there was no clear correlation between AR and PPARγ (Figure 3A), we detected a positive correlation between AR and GR (PrMIX=0.57) as shown in Figure 5C for preadipocytes differentiated (ixr+d; MIX) for 96 h. At the single cell level, we divided the population into low and high GR-expressing cells based on the median level of GR detected after treatment with complete differentiation cocktail (Figure 5D; ixr+dex). Although median AR levels varied as a function of DHT treatment, temporal analysis of the two subpopulations showed cells with higher GR were associated with both higher lipid (Figure 5E) and AR levels (Figure 5F). These results suggest GR (Figure 4) and dex (Figure 5) are needed to regulate AR at the single cell and transcriptional levels in adipocytes. Even though AR expression is increased by dex and GR, we reasoned AR might adopt a repressive conformation that abrogates its ability to prevent adipocyte differentiation.
Adipocyte differentiation reagents modulate AR conformation
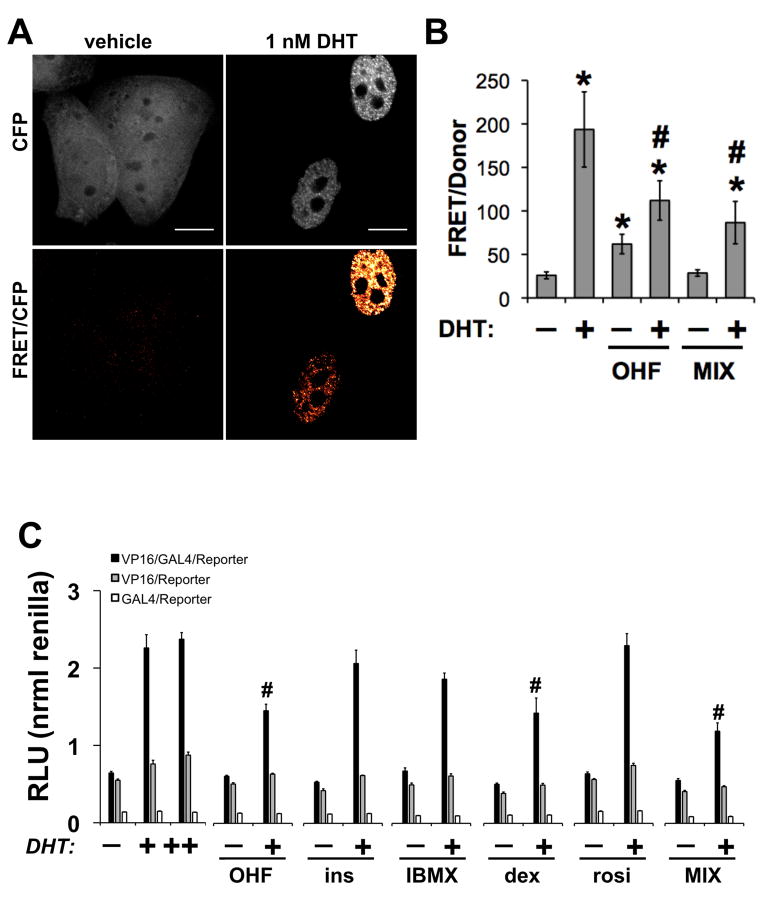
The N-terminal activation function 1 (AF-1) and the C-terminal ligand-dependent (AF-2) domains of AR are largely inactive in the absence of ligand. In the presence of agonists, AR adopts a conformational change allowing communication between AF-1 and AF-2 and subsequent transcriptional activation (Kemppainen et al., 1999; Langley et al., 1995; Schaufele et al., 2005). To test our hypothesis regarding inhibition of AR activity, we used a fluorescence resonance energy transfer (FRET)-based assay (Jones et al., 2009; Klokk et al., 2007; Schaufele et al., 2005) to determine the effect of differentiation cocktail on the intramolecular interaction between CFP-AF-1 and YFP-AF-2 domains in full-length AR. In this assay, agonist binding induces a conformational change bringing the CFP and YFP into close proximity, enabling energy transfer, which precedes nuclear translocation and transcriptional activity. In these experiments, we transiently expressed a CFP-AR-YFP fusion protein in HeLa and exposed the cells to 10 nM DHT, EtOH (vehicle), 10 uM OHF, or MIX for 20 h (Figure 6A). To detect effects on androgen-induced conformation, OHF/DHT and MIX/DHT treatments were applied. Figure 6B shows DHT markedly induced nuclear translocation, altered subnuclear localization, and increased energy transfer (7.5-fold) indicative of intramolecular events bringing the AF-1 and AF-2 domains into close proximity (Figure 6B). Compared to vehicle, OHF stimulated a lesser increase in FRET (2.4-fold), while MIX showed no significant change. However, treatment of cells with OHF or MIX in the presence of 1 nM DHT blocked the androgen-induced conformational change by −43% and −56%, respectively. These experiments suggested one or more components of adipocyte differentiation cocktail antagonize AR activity by blocking ligand-induced N/C terminal interactions.
Figure 6.
Human adipocyte differentiation alters the conformation of AR. (A) HeLa cells transiently transfected with CFP-AR-YFP were treated with vehicle, 10 uM OHF, or differentiation media (MIX) in the presence or absence of 1 nM DHT for 20 h. Scale bar, 10 μm. (B) FRET imaging was used to determine AR conformation (n≥10 cells/condition). (C) An N-terminal region containing AR 1-660 was fused to the VP16 TAD while a C-terminal region containing AR 624-919 (wild-type) was fused to the to the Gal4-DBD domain. HeLa cells were co-transfected with AR fragments and reporter constructs as described in Materials and Methods. After 12 h of transfection, cells were trypsinized and equally seeded into 96-well plates. Cells were allowed to re-attach and subsequently treated with dexamethasone (500 nM), IBMX (250 uM), insulin (200 nM), rosiglitazone (3 uM), MIX, AR ligands, alone or in the presence of 0.1 nM DHT. After 24 h of treatment, luciferase activity was detected. Data represents the units of firefly luciferase corrected for the units of renilla luciferase detected in the same plate. Shown is one representative experiment from 4 replications. p<0.05 was considered significant for treatments compared to vehicle (#) or DHT (*) treatments.
In a complementary approach, we used a mammalian two-hybrid assay to determine whether AR N/C-terminal domain interactions might be altered by treatment with each component of the differentiation cocktail. As the agonists bind the ligand-binding domain of AR, the AR (1-660)-VP16 and AR (624-919)-Gal4 fusion proteins become close enough to activate luciferase activity driven by the Gal4-VP16 interaction (Langley et al., 1995). Expression of individual VP16 or Gal4 plasmids were used as negative controls. When we treated cells with individual or all components combined (MIX), the basal AF-1/AF-2 interaction was not affected (Figure 6C). In complementary experiments, we also treated cells with individual or all components (MIX) combined in the presence of 0.1 nM DHT. Similar to OHF, dexamethasone and MIX inhibited DHT-induced N/C terminal interaction (Figure 6C). Thus, the next step was to determine the extent to which AR transcriptional activity was inhibited by adipocyte differentiation signals.
Inhibition of AR transcriptional activity by chemical inducers of adipocyte differentiation
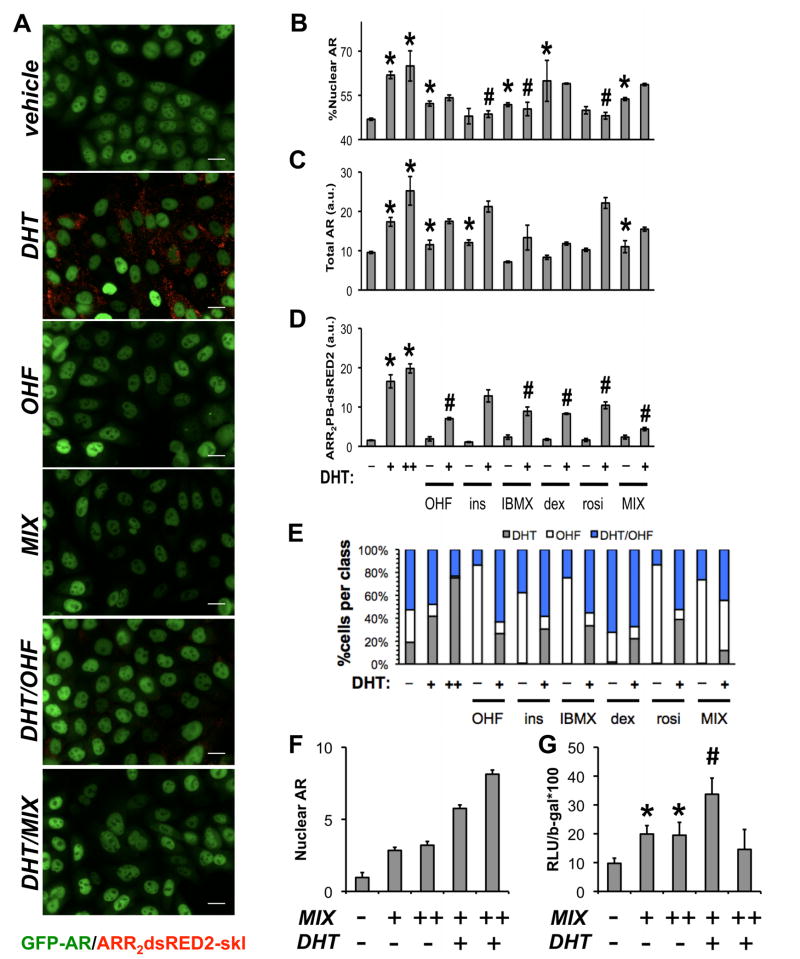
As MIX components were shown to alter the conformation of AR, we next assessed whether AR translocation and transcriptional activity were similarly modulated. To do this, we created a transcriptional biosensor cell line based on a parental GFP-AR HeLa model described previously, which faithfully expresses AR target genes in response to agonist treatments (Szafran et al., 2008). We stably incorporated an ARR2PB-dsRED2-skl reporter construct, which is responsive to AR agonists and inhibited by antagonists (in competition experiments), including MIX (Figure 7A). After image processing, HCA was used to simultaneously quantify GFP-AR subcellular localization (% Nuclear AR; Figure 7B), total AR expression (Figure 7C), and ARR2PB-dsRED2-skl reporter activity (Figure 7D) for compound concentrations used in differentiation cocktail. As expected (Szafran et al., 2008), DHT elicited significant, dose-dependent increases in all 3 features. Dexamethasone, IBMX, and MIX induced AR translocation (Figure 7B), while insulin and MIX upregulated total AR levels (Figure 6C). Although some of the components together (MIX) or individually affected total AR and AR translocation, none increased AR transcriptional activity (Figure 7D). When treatments were performed in the presence of 1 nM DHT (+), several effects were observed. First, insulin, IBMX, and rosiglitazone decreased androgen-induced AR translocation compared to 1 nM DHT (Figure 7B). Second, IBMX, dexamethasone, and rosiglitazone inhibited DHT-induced reporter activity (Figure 7C).
Figure 7.
Adipocyte differentiation reagents repress AR transcriptional activity. (A) GFP-AR:ARR2PB-dsRED2-skl double stable HeLa cells were used to evaluate effects of differentiation reagents (~48 h) on GFP AR nuclear translocation, total AR levels, and androgen-dependent reporter activity encoded by the expression of peroxisome-targeted dsRED2. After treatment, experiments were processed for automated microscopy, imaged, and segmented into single cell regions for feature extraction. Response data is depicted for (B) nuclear translocation, (C) total AR and (D) ARR2PB-dsRED-skl activity for the indicated treatments. Experiments were performed in the presence and absence of 1 nM DHT. One representative experiment from 3 replications is shown; p<0.05 was considered significant for treatments compared to vehicle (#) or DHT (*) treatments. (E) SVM allowed more sensitive detection of AR repression by adipocyte differentiation components. A SVM classifier was trained to recognize OHF, DHT, or DHT/OHF patterns and applied to cell-level responses (from the DAPI, GFP-AR, and ARR2PB-dsRED2-skl channels). Cells were automatically classified into DHT, OHF, or DHT/OHF treatment groups based on selected, robust subcellular measurements used by the SVM classifier. For each subplot, the abscissa represents compound treatments, while the ordinate denotes the percentage of cells assigned to each population. To validate effects on AR-mediated transcription in primary cells, human preadipocytes were differentiated (MIX) for 7 d followed by transfection of cells with pARR2PB-luciferease and β-galactosidase (β-gal). Subsequently, cells were treated with 10 nM DHT alone or MIX +/− 10 nM DHT for 48 h in serum-containing media. Effects of these treatments on (F) AR levels were evaluated by HCA (n≥100 cells/condition). (G) ARR2PB luciferase activity was measured and normalized to β-gal (n=3 +/− SEM). Scale bar, 20 μm.
Although our three basic measurements reporting important mechanisms of AR activity were informative (Figure 7B–D), they do not encompass the wealth of phenotypic data available in the images, which can characterize the biological phenotypes caused by the different compound treatments. Extending our recently developed HCA approaches designed to classify estrogen receptor responses (Ashcroft et al., 2011), we identified single cell-level features that classified MIX components based on their similarity to control treatments. We generated a control dataset consisting of cells treated with 10 nM DHT, 10 uM OHF, or 1 nM DHT+10 uM OHF. After image processing, we extracted intensity- and morphology-based features (276 measurements per cell) from the GFP-AR and ARR2PB-dsRED2-skl channels. These measurements include mechanistic features (e.g. GFP-AR levels and localization), cytological features (e.g. GFP-AR nuclear variance (Szafran et al., 2008)) and statistical features (e.g. cytoplasmic ARR2PB-dsRED2-skl kurtosis). We processed the features and applied stepwise discriminant analysis (Jennrich, 1977) which selected 32 features especially useful in distinguishing between control treatments (Supplemental Table 1). Finally, we trained a classifier on the control data using the SDA-selected features which was used to classify the cells from the compound treatments. This classification framework was validated on the control data to assess classifier performance (Supplemental Table 1). From classification, we were able to identify the percentage of cells exhibiting AR agonism or antagonism given a particular treatment (Figure 7E). Interestingly, by incorporating effects on GFP-AR protein levels, localization, subnuclear pattern, and reporter levels, this approach had high enough sensitivity to detect effects of all MIX components in the absence of DHT, which was impossible by CFP-AR-YFP (Figure 6B) or mammalian two-hybrid assays (Figure 6C).
Figure 7E suggested each MIX compound redistributed cell populations to predominantly OHF or DHT/OHF phenotypes. Also, during DHT treatment, individual addition of IBMX, insulin, or rosiglitazone decreased the number of cells classified as agonist (DHT) and increased the number of cells classified as anti-androgen (DHT/OHF). Furthermore, MIX reflected the individual components, and was classified as a predominantly antagonist treatment (e.g., OHF alone or DHT/OHF) with a complete loss of the DHT class.
To complement our findings in the GFP-AR:ARR2PB-dsRED2 biosensor cell line and verify the inhibition of AR transcriptional activity by adipocyte differentiation reagents, we expressed the AR-specific ARR2PB-luciferase reporter plasmid (Schoenmakers et al., 2000) in terminally differentiated human adipocytes. We found nuclear AR was increased by 2.84-fold in cells differentiated for 7 d (MIX: +, DHT: −), while not significantly increased compared to 96 h (Figure 3D). Treatment with MIX for two additional days (MIX: ++, DHT: −) did not significantly increase nuclear AR. An additional 2 d of treatments with androgen alone (MIX: +, DHT: +), or androgen and MIX (DHT:+, MIX:++), resulted increased nuclear AR 5.8-fold and 8.1-fold compared to vehicle treatment (MIX: −, DHT: −), respectively (Figure 7F). Concomitantly, when ARR2PB-luciferase was expressed in terminally differentiated adipocytes (7 d), androgen-induced transcriptional activity was decreased when cells were treated with both DHT and MIX for an additional 2 d (Figure 7G). These combined experimental approaches show, in human preadipocytes, AR is upregulated by dexamethasone activation of GR, while also transcriptionally inhibited to putatively promote features of adipocyte maturation.
DISCUSSION
The identification of modulators of human adipocyte gene expression is central to understanding the mechanisms of obesity. Our comparative analysis of the transcriptomes from differentiated human adipocytes and terminally differentiated mouse 3T3-L1 adipocytes (Schupp et al., 2009) suggested early, significant AR upregulation and function is a specific feature of the human preadipocyte differentiation model (Figure 1). Recently, AR was reported to be expressed at low (Fu et al., 2005) or undetectable (Lahnalampi et al., 2010) levels in 3T3-L1 cells. In our experiments, DHT only inhibited mouse 3T3-L1 differentiation when AR was exogenously-expressed by lentivirus, whereas lipid accumulation was clearly reduced in human adipocytes 4 d post-induction (Figure 2). Inhibition of human adipocyte differentiation by DHT was further marked by parallel increases in AR protein levels and repression of adipocyte-specific markers (Figure 3) at the protein (PPARγ) and gene (FASN, PPARγ, ADFP, C/EBPα) levels. Indeed, consistent with our effects of DHT on lipid accumulation, DHT has also been found to reduce expression of ACC1 and DGAT2, key enzymes in triacylglycerol synthesis (Gupta et al., 2008),
Upon ligand binding and conformational change, AR, progesterone receptor (PR), mineralocorticoid receptor (MR), and GR can regulate gene transcription by binding a consensus GGT/AACAnnnTGTTCT hormone response element (HRE) (Lieberman et al., 1993; Lombes et al., 1993; Nordeen et al., 1990; Roche et al., 1992). Due to the shared consensus binding motifs for AR, GR, MR, and PR, these receptors can affect the transcriptional activity and regulation of other Type 1 members. For example, both PR and AR have been shown to inhibit GR transcriptional activity at HREs (Archer et al., 1994; Chen et al., 1997; Yen et al., 1997). Therefore, the common DNA-binding site AR/MR/PR/GR share provides a capacity to mediate cellular responses through intra-receptor crosstalk. Specifically, recent work has shown dexamethasone induces GR binding upstream of AR (Reddy et al., 2009). We therefore proposed dexamethasone mobilizes GR to launch adipocyte differentiation, positioning GR as a central regulator of early differentiation and AR expression. Our work shows GR (Figure 4), regulated by dexamethasone (Figure 5), is required to induce AR expression and regulate the adipocyte phenotype. Novel to the understanding of the GR/AR inter-related gene network, we also established nuclear GR levels are tightly correlated with lipid accumulation (Figure 5D) and nuclear AR (Figure 5E) at the single cell level during differentiation.
We sought to identify further elements of the AR regulation circuit by analyzing effects of adipocyte differentiation conditions on AR conformation and transcriptional activity. First, adipocyte differentiation reagents reduce androgen-dependent AR N/C terminal interactions (Figure 6A–B) inducing quantitatively similar results as a classic anti-androgen (OHF). Second, we used a novel AR fluorescent biosensor cell line and HCA to identify how individual components of adipocyte differentiation cocktail antagonize AR activity and affect transcriptional competence (Figure 7A–E). Coupled with a machine-learning algorithm to classify subtle phenotypes into DHT, OHF, and DHT/OHF, we were able to able to establish rosiglitazone and dexamethasone as potent inhibitors of AR activity. Androgen-regulated AR transcriptional activity in human adipocytes was decreased by differentiation cocktail (Figure 7G) which validated our findings in GFP-AR:ARR2PB-dsRED2 biosensor cells. Previous studies have shown dexamethasone and glucocorticoids can inhibit androgen action and downregulate AR activity by competing for androgen response elements (Burnstein et al., 1995; Davies and Rushmere, 1990) without high affinity binding to AR (Wilson and French, 1976). Rosiglitazone, on the other hand, suppresses AR-regulated genes, including the ARR2PB composite reporter, by reduction of agonist-induced receptor binding to DNA (Moss et al., 2010; Yang et al., 2006).
Clinical and epidemiological data implies the existence of a negative cycle between circulating testosterone and obesity, although AR mRNA is unaltered with increasing BMI, independent of sex (Wake et al., 2007). Indeed, further supporting inhibition of AR transcriptional activity during fat maturation, each MIX component has been shown to induce adipogenesis in vivo (De Vos et al., 1996; Fujikura et al., 2005; Madsen et al., 2008; Masuzaki et al., 2001; Masuzaki et al., 2003). Further, testosterone production is suppressed by visceral obesity (Stanworth and Jones, 2009) and patients with visceral obesity and the metabolic syndrome have higher incidence of hypogonadism (Gould et al., 2007).
Androgens have been thought to block adipocyte differentiation (Blouin et al., 2009a; Blouin et al., 2010; Gupta et al., 2008; Singh et al., 2006; Singh et al., 2003) by upregulation of Wnt target genes (Singh et al., 2006). In our transcriptome analysis, we detected 51 Wnt target genes exhibiting at least 2-fold change (Supplemental Excel File 1), including targets upregulated by testosterone (CD44, FST, LEF1). 92% of the Wnt target genes were downregulated between preadipocyte and terminal differentiation, which may correspond to decreased AR activity. PPARγ (Liu and Farmer, 2004; Waki et al., 2007) and GR (Bujalska et al., 2006) activation repress Wnt target genes. Thus, subtle alterations in the activity of pro-adipogenic transcription factors may sustain repression of Wnt signaling via inhibition of AR.
Recent studies (Veilleux et al., 2012) support our findings by showing androgens and glucocorticoids exhibit extensive crosstalk to direct lipid storage in a sex and depot-specific manner by increasing androgen metabolism. Although excess glucocorticoids and androgens can profoundly affect adipocyte function and promote altered metabolism, additional studies are required to better understand how selective manipulation of the AR/GR axis controls fat deposition and improves overall lipid profiles. Finally, the experiments described in this paper propose a feed-forward loop whereby glucocorticoids activate GR to promote AR expression, yet inhibit AR activity. Because peripheral fat tissue contains the enzymatic machinery to synthesize GR agonists, it is possible these steroids produced in loco are responsible for AR antagonism and activation of lipid storage genes that contributes to maintenance of proper energy balance (Lee et al., 2008).
SIGNIFICANCE
The characterization of genes associated with human adipocytes is fundamental to understanding the pathogenesis of obesity. Given this need, we identified specific genes associated with the primary, early human adipocyte differentiation program which are not sufficiently expressed in 3T3-L1. In support of recent epigenomic analyses of mouse 3T3-L1 and human adipocyte systems (Mikkelsen et al., 2010; Soccio et al., 2011), the 3T3-L1 and human adipocyte transcriptomes are also dissimilar. In this study, we used high content analysis and quantitative imaging to show in vitro human adipocytes express AR mRNA and protein, regulated by both GR and dexamethasone action, and GR and AR levels are directly correlated. Of specific importance, we used novel image analysis tools to establish antagonism of AR by adipocyte differentiation components, and identified dexamethasone as the dominant inhibitor of AR transcriptional activity. AR mRNA is not negatively correlated with BMI (Wake et al., 2007), suggesting obesity does not downregulate AR, but negatively modulates its activity. Our data suggests GR and corticosteroids can both positively regulate AR expression while simultaneously decreasing AR activity and alter androgen effects on energy storage. Since androgens favorably direct muscle differentiation (Singh et al., 2003), regulate muscle mass (Chambon et al., 2010), and increase lean body mass in aging males (Tenover, 1992) and females (Rariy et al., 2011), the use of selective GR or AR modulators in combination may play a role in alleviating the consequences of obesity.
MATERIALS AND METHODS
Primary Cell Culture and Differentiation
Cryopreserved, subcutaneous primary human preadipocytes from normal female donors with an average body mass index of 27.51 were provided by Zen-Bio Inc (Research Triangle Park, NC). Cells were received at passage 2 and experiments performed before passage 10. Experiments were performed using pooled human preadipocytes from 5 individual female donors (Lot SL0033). Both human preadipocytes and 3T3-L1 cells were maintained at 5% CO2/37°C in DMEM/F12 (Mediatech, Manassas, VA) with 10% fetal bovine serum (FBS; Gemini Bio-Products, West Sacramento, CA), 100 U/ml penicillin, 100 ug/ml streptomycin (growth media). Medium was replaced during routine maintenance every 2 days.
Confluent cells were differentiated using growth media supplemented with 100 nM human insulin, 0.250 mM 3-isobutyl-1-methylxanthine (IBMX), 500 nM dexamethasone, and 3 uM rosiglitazone (BRL49653, Cayman Chemical Company, Ann Arbor, MI). IBMX, insulin, and dexamethasone were purchased from Sigma Chemical Company, St. Louis, MO.
DNA microarrays
Microarray analysis was performed using the Illumina Sentrix Beadchip Array (Human HT-12) containing ~48,000 probe sequences that spans the human transcriptome. Adipocytes were differentiated for the indicated periods, washed and total RNA was prepared using RNeasy (Qiagen, CA). RNA was purified on a Qiagen Mini spin column. Two hundred nanograms of total RNA were amplified and purified using Illumina TotalPrep RNA Amplification Kit (Ambion, Cat# IL1791) following the manufacturer’s instructions. The first strand cDNA was synthesized by incubating RNA with T7 oligo(dT) primer and reverse transcriptase mix at 42 °C for 2 hours. RNase H and DNA polymerase master mix were immediately added into the reaction mix following reverse transcription and were incubated for 2 hours at 16 °C to synthesize second strand cDNA. In vitro transcription was performed and biotinylated cRNA was synthesized by a 16-hour amplification with dNTP mix containing biotin-dUTP and T7 RNA polymerase. Amplified cRNA was subsequently purified and the concentration was measured by NanoDrop ND-1000 Spectrophotometer (NanoDrop Technologies, DE). An aliquot of 1.5 micrograms of amplified products were loaded onto Illumina Human HT-12 Beadchips and hybridized at 58°C for 17 hours, washed and incubated with straptavidin-Cy3 to detect biotin-labeled cRNA on the arrays. Arrays were dried and scanned using a BeadArray Reader (Illumina, CA). Each microarray was performed, minimally, with three independent RNA isolates. We performed microarrays at 7 time points: day 0 (n=5), day 1 (n=4), day 2 (n=4), day 3 (n=4), day 4 (n=4), day 7 (n=3), day 14 (n=4).
Microarray analysis
Background subtracted and quantile normalized gene expression values were calculated using Illumina BeadStudio Software. Differential gene expression was calculated by fitting a linear model to a group means parameterization coupled to a secondary fit of all possible pair-wise comparisons equating to an ANOVA analysis using R2.10 and the Limma 2.19 analysis package. Controlling the false discovery rate was used to correct for multiple testing. Genes presented in the heatmap are transcripts with a minimum 4-fold change at any given contrast (FDR q<0.0001). Samples were clustered using complete agglomerization and Euclidean distance. Rows were scaled to have a mean of zero and standard deviation of one. 3T3-L1 gene expression data at was obtained from GSE14004 (Schupp et al., 2009).
RNA extraction and qPCR analysis
Total RNA was extracted from cells using the RNeasy kit (Qiagen, Germantown, MD), following the manufacturer’s instructions. To measure relative mRNA expression, qPCR was performed using the Taqman RT-PCR one-step master mix in conjunction with an ABI 7500 real-time PCR system (Applied Biosystems, Foster City, CA). Each sample was tested in duplicate in two independent experiments. β-actin and TBP were used as invariant controls. TaqMan Gene Expression Assays were used for the following human genes: CEBPα, Assay ID Hs00269972_s1; PPARγ2, Assay ID Hs01115513_m1; GR, Assay ID Hs00353740_m1; CIDEC, Assay ID Hs01032998_m1; FABP4, Assay ID Hs01086177_m1; FKBP5, Assay ID Hs01561006_m1; AR, Assay ID Hs00171172_m1. The following primer and probe sets were used to detect human (h) hFASN, hSRC-3, and hADFP. hFASN: cggagtgaatctgggttgat(F), caggcacacacgatggac(R), Roche Universal Probe Library probe #11 (probe); hSRC-3: ggacctggtaagaaggtgtattcag (F), tgcctcttagcataggacacaga (R), tccatgcgcagcatgaaggaga (probe); hADFP: gtgactggcagtgtggagaag (F), tccgactccccaagactgt (R), ccaagtctgtggtcagtggcagca (probe). Murine FABP4 and PPARγ2 were detected with the following primer and probe sets: mPPARg2: gaaagacaacggacaaatcacc (F), gggggtgatatgtttgaacttg (R), Roche Universal Probe Library probe #7 (probe); mFABP4: aagagaaaacgagatggtgacaa (F), cttgtggaagtcacgccttt (L), Roche Universal Probe Library probe #31 (probe).
siRNA Transfection
Before cell plating, optical quality 96-well plates (Greiner SensoPlate Plus, Monroe, NC) were coated with 50 ul of FBS (Gemini Bio-Products) overnight at 37C. Confluent preadipocytes were transfected with GR siRNA (Qiagen, Germantown, MD) or mismatch control at a final concentration of 20 nM using Dharmafect transfection reagent (Dharmacon, Lafayette, CO). After transfection, cells were incubated for 48 h at 5% CO2/37°C before induction of differentiation for 96 h.
Antibodies
The following antibodies were purchased from commercial sources and used for immunofluorescence and western blotting: rabbit polyclonal AR (N-20, Santa Cruz Biotechnology, Santa Cruz, CA), rabbit monoclonal PPARγ (Cell Signaling, Danvers, MA), GR (Genetex, Irvine, CA), and mouse monoclonal PPARγ (clone E-8, Santa Cruz Biotechnology, Santa Cruz, CA). Mouse monoclonal antibody to AR (AR-441 (Nazareth et al., 1999)) was kindly provided by Dean Edwards and Nancy Weigel (Baylor College of Medicine, Houston, TX).
Western Blotting
Cells were collected by scraping, and lysed in RIPA buffer supplemented with the appropriate proteinase and phosphatase inhibitors. Protein concentration was standardized by BCA assay. Western blot analysis was performed with whole cell lysates run on 4–12% Bis-Tris NuPage® (Millipore, Bedford, MA) gels and transferred onto Immobilon-P Transfer Membranes (Millipore). Membranes were blocked 1 h with 5% milk (in TBS with 0.1% Tween-20). Primary antibodies were incubated overnight at 4C, followed by secondary antibodies for 1 hour at room temperature. Immunoreactive bands were visualized by SuperSignal West Femto chemiluminescence reagents (Pierce, Rockford, IL). All membranes were then subjected to stripping buffer (Pierce) for 30 minutes at RT, reblocked, and reprobed for β-actin (mouse monoclonal, Sigma Chemical Co.) as a loading control.
Immunofluorescence
For fluorescence detection of antibodies and neutral lipid content in multi-well plates, the following protocol was carried out on the BioMek NX (Beckman Coulter, Fullerton, CA). Well plate systems used were: 96-well and 384-well (Greiner Sensoplate Plus, Monroe, NC). Aspirations and plate washes were performed with an ELx405 (BioTek, Winooski, VT). Following differentiation, media was aspirated and 4% formaldehyde (sold as ultrapure paraformaldehye, Electron Microscopy Sciences, Hatfield, PA) in PBS was immediately added for 30 minutes at room temperature. Plates were then quenched with 100 mM ammonium chloride. After quenching, plates were washed three times with TBS. Fixed adipocytes were permeabilized with 0.1% Triton X-100 in TBS for 10 minutes and washed three times with TBS. Non-specific antibody binding was blocked by pre-incubating for 30 min in 2% BSA in TBS/0.01% saponin (which was also used as an antibody diluent) at room temperature. Antibodies were then diluted at a 1:200 concentration in antibody diluent and incubated overnight at 4°C. Subsequently, plates were washed with TBS and incubated with secondary antibodies for 1 h at room temperature. AlexaFluor 647-conjugated anti-mouse and AlexaFluor 568-conjugated anti-rabbit secondary antibodies (Molecular Probes, Invitrogen) were used. Cells were then washed 3 times and incubated with CellMask Blue (1 μg/ml, Invitrogen, a general protein dye), LipidTOX green (1:1000, Invitrogen, a non-polar lipid-binding dye), and DAPI (10 μg/ml, a DNA-specific dye) in PBS for 45 minutes at room temperature. Dyes were then aspirated, PBS/0.01% azide added, and plates imaged immediately.
Imaging and Microscopy
For high-speed image acquisition for subsequent analysis, cells were imaged using the Cell Lab IC-100 Image Cytometer (IC-100; Beckman Coulter) equipped with a Nikon S Fluor 20X/0.75NA objective. The imaging camera (Hamamatsu; Bridgewater, NJ) was set to capture 8 bit images at 2×2 binning (672 × 512 pixels, 0.684×0.684 um2/pixel) with 5 images captured per field (DAPI, CMBl, LipidTOX, A568 and A647 secondary antibodies). In general, 12–16 images were captured per well for image analysis. Experiments performed in stable HeLa cell lines and 3T3-L1 were imaged with a 40X/0.90NA objective.
Image processing
Images were analyzed using custom algorithms developed with the Pipeline Pilot (v7.5) software platform (Accelrys, San Diego, CA) in a similar workflow as previously described (Hartig et al., 2011; Szafran et al., 2008). After background subtraction, nuclear and cell masks are generated using a combination of non-linear least squares and watershed-from-markers image manipulations of the DAPI images. Cell populations were filtered to discard events with cell aggregates, mitotic cells, apoptotic cells, cellular debris, or poor segmentation. Applied gates were based upon nuclear area, nuclear circularity, and cell size/nucleus ratio. In general, these filters removed 10% of the population of segmented cells. All events with whole cell masks bordering the edge of the image were additionally eliminated from analysis. Post-analysis measurements were exported to spreadsheet software (Microsoft Excel) for further analysis.
Production of lentiviral particles
Human AR cDNA was cloned into the lentiviral expression vector pCDH–CMV-MCS-EF1-Puro (System Biosciences, Mountain View, CA) by Xba I/Nhe I digestion. Pseudolentiviruses were produced in 293TN cells by co-transfecting lentiviral expression constructs and the pPACK packaging plasmid mix (System Biosciences). Pseudoviral particles were harvested 48 h post-transfection and were concentrated using PEG-it virus precipitation solution kit (System Biosciences).
FRET imaging
CFP/YFP FRET experiments were performed using a CFP-AR-YFP construct and reagents kindly provided by Fred Schaufele (UCSF) and Fahri Saatcioglu (University of Oslo). CFP-AR-YFP was transiently expressed in HeLa grown on standard 12 mm glass coverslips. Constructs were transfected using Lipofectamine 2000 (Invitrogen). Media was removed and replaced with fresh DMEM/F12 with 5% FBS 24 h post-transfection. Cells were then treated overnight with compounds (20 h) prepared in growth media (DMEM/F12, 5% FBS). Treatment was followed with these steps: fixation 4% PFA (30 min), quench 100 mM NH4Cl (10 min), and mount with SlowFade Gold (Invitrogen). After fixation, cells were washed with PBS++ 3 times, while all other wash steps were performed with PIPES/HEPES/EGTA/MgCl2 (PEM) buffer, prepared at a final pH of 6.8.
FRET imaging was performed as described previously with the DeltaVision Core Image Restoration Microscope (Applied Precision, Issaquah, WA). Z-stacks were imaged at 0.2 um separation and a frame size of 1024×1024 pixels at 1×1 binning with an Olympus IX71 microscope using a 60X, 1.42 NA Plan Achromat objective (Olympus, Center Valley, PA), and a Photometrics CoolSnap HQ2 CCD camera. Filter sets were as follows, with a dichroic to split CFP and YFP: excitation 430 nm/emission 470 nm (CFP), excitation 500 nm/emission 535 nm (YFP), and excitation 430 nm/emission 535 nm (FRET). After deconvolution (Softworx, Applied Precision), FRET calculations were performed using the Applied Precision FRET user interface. FRET measurements on individual nuclei were acquired on maximum intensity projections of the derived FRET image. Spectral bleed through was corrected for by acquiring specimens containing only CFP-AR and YFP-AR. Standard values for α and β coefficients were 0.6 (CFP) and 0.12 (YFP) acquired from single donor/acceptor plasmid expression experiments. Additional and supporting analysis was performed using PixFRET (Feige et al., 2005).
AR NH2-COOH domain interaction assay
An interaction between COOH terminal AF2 (activation function 2) and NH2 terminal sequence F23XXLF27 was measured using the CheckMate™/Flexi® Vector Mammalian Two-Hybrid System (Promega, Madison, WI). A segment containing AR 1-660 was fused to the VP16 TAD domain of plasmid pFN10A, while segments AR 624-919 were fused to the to the Gal4-DBD domain of plasmid pFN11A. One day before transfection, 1.5×106 Hela cells were seeded into a 60 mm dish. Cells were co-transfected with 1.3 ug of pFN11A-AR624-919, 1.3 ug of pFN10A-1-660, 1.3 ug of pGL4.31 (containing five GAL4 binding sites upstream of a minimal TATA box, which is upstream of a firefly luciferase gene that acts as a reporter for interactions between proteins), and 1 ng of phRL-TK carrying the Renilla luciferase gene. After 12 hours of transfection, cells were trypsinized and equally seeded into a 96-well plate. Multi-replicate detection of NH2-COOH interaction was evaluated in ligand competition experiments where adipogenic compounds and OHF were titrated against 0.1 nM DHT. After 24 h of treatment, luciferase activity was assayed with the Promega Dual Glo assay kit, using a luminometer (PerkinElmer). Data represent the units of firefly luciferase corrected for the units of renilla luciferase detected in the same plate. As controls to account for basal activity of pFN11A-AR624-919 and pFN10A-AR1-660, parallel experiments were performed with each plasmid expressed individually.
Luciferase expression experiments
We used replication deficient adenoviruses coupled with poly-lysine (Allgood et al., 1997) to express pARR2PB-luciferase in human preadipocytes. Reporter gene activity was detected using the Promega Luciferase Assay Kit. Relative luminescence units were normalized to β-galactosidase activity using a standard 2-Nitrophenyl β-d-galactopyranoside (Sigma) colorimetric assay.
GFP-AR:ARR2PB-dsRED2-skl biosensor experiments
We developed a stable cell line for cell-based HCA of AR translocation and reporter activity under treatment with adipogenic compounds. The parental GFP-AR HeLa cell line (Szafran et al., 2008) was infected with a lentivirus encoding a pARR2PB-dsRED2skl reporter construct, based on the AR-responsive composite probasin promoter. This reporter encodes a dsRED2 protein (Clontech) fused at the C-terminus with a peroxisome targeting sequence (SKL, serine, lysine, leucine) that serves to localize and concentrate the fluorescence signal. Antibiotic resistant cultures were selected with both puromycin (1.5 ug/ml) and hygromycin (200 ug/ml) for two weeks. Subsequently, clones were single cell sorted by FACS where clones were identified based on GFP-AR and ARR2PB-dsRED2-skl responsiveness (e.g. low GFP/high dsRED2). Single cell clones in phenol-red free DMEM (Mediatech) with 5% FBS, 100 U/ml penicillin, 100 ug/ml streptomycin, 1.5 ug/ml puromycin, and 200 ug/ml hygromycin. One single cell clone (GFP-AR:ARR2PB-dsRED2skl) that exhibited maximal DHT-responsive reporter activity was used for detecting androgenic effects of rosiglitazone, dexamethasone, insulin, IBMX, and OHF.
Before experiments, 384 well plates (Greiner Sensoplate Plus) were coated with FBS overnight. Cells were plated (4000 cell/well), after excess FBS was removed from each well, using the TiterTek Multi-Drop Plus (TiterTek, Huntsville, AL). GFP-AR:ARR2PB-dsRED2skl cells were seeded in antibiotic-free, phenol red-free DMEM containing 5% charcoal-stripped, dialyzed (SD) FBS and incubated 48 h. Cells were then exposed cells to single doses of each compound in the presence or absence of 1 nM DHT. Vehicle (EtOH), 10 nM DHT, 10 uM OHF, and 1 nM DHT/10 uM OHF treatments functioned as controls, each in individual columns. Following compound addition, cells were incubated 48 h to allow for simultaneous detection of AR translocation and reporter expression. Plates were processed for imaging as previously described (Szafran et al., 2008).
Support vector machine classification
For each experiment, a validation dataset was created consisting of 16-wells for the following control treatments: vehicle (ethanol), agonist (10 nM DHT), a negative ligand control (10 uM OHF), and antagonist (10 uM OFH/1 nM DHT). After the image analysis and feature extraction steps described above, single-cell-level measurements were used to train a classifier on the control dataset to distinguish between the control treatments. Before training, 1000 single-cell samples were randomly sampled (without replacement) from each control treatment to ensure classes were equally sized in the training process. Next, linearly dependent features were removed. All other features were scaled to a range of [−1,1]. Feature selection was performed on the training data using stepwise discriminant analysis (SDA), which defines a list of the most discriminative feature for classification (Jennrich, 1977). Finally, we used the SDA-selected features to train a support vector machine (SVM) classifier with a radial-basis-function kernel (Cortes and Vapnik, 1995). 10-fold cross-validation on the training data was used with a grid search to optimize the parameters C (slack penalty) and g (kernel parameter).
For validation, we randomly sampled 10 different sets upon which we trained a classifier, and for each corresponding sub-training-set we tested its classifier on the remaining control samples. Resulting classification labels were used to calculate precision and recall scores for each classifier, and these accuracy metrics were averaged across the panel of classifiers to produce a performance measure for the classification approach. The supervised learning approach was implemented in Python 2.6 and utilizes LIBSVM 2.9 (http://www.csie.ntu.edu.tw/~cjlin/libsvm/).
Statistical Analyses
Data presented were acquired from a minimum of 2 (q-RT PCR) or 3 (HCA) independent experiments performed on multiple days, unless otherwise indicated. ANOVA was first used to compare the effects of time or ligand treatment. If significant differences were identified, then data was compared with Tukey’s HSD post hoc tests. All tests were carried out at the 95% confidence interval using JMP-IN 7 (SAS, Cary, NC).
Supplementary Material
Acknowledgments
This work was funded by NIH 5R01DK055622, the Hankamer Foundation, DOD Prostate Cancer Research Program (DAMD W81XWH-10-1-0390), with pilot grant and equipment support from the John S. Dunn Gulf Coast Consortium for Chemical Genomics (MA Mancini). Additional funding was provided by NIH 1F32DK85979 (SM Hartig), 5T32HD007165 (BW O’Malley), 5K01DK081446 (B He), the Caroline Weiss Law Foundation (SE McGuire), 5K12DK083014 (DJ Lamb), and 3U19DK062434 (NURSA). Imaging resources were supported by SCCPR U54 HD-007495 (FJ DeMayo), P30 DK-56338 (MK Estes), P30 CA-125123 (CK Osborne), and the Dan L. Duncan Cancer Center of Baylor College of Medicine. The authors thank Huiying Sun, M.G. Mancini, L. Vergara and J. Broughman for technical resource support. The authors also thank Nancy Weigel and Fabio Stossi for critical review of the manuscript.
Footnotes
ACCESSION NUMBERS
The Gene Expression Omnibus (GEO) accession number for the microarray data reported in this paper is GSE39342.
References
- Allgood VE, Zhang Y, O’Malley BW, Weigel NL. Analysis of chicken progesterone receptor function and phosphorylation using an adenovirus-mediated procedure for high-efficiency DNA transfer. Biochemistry. 1997;36:224–232. doi: 10.1021/bi961125c. [DOI] [PubMed] [Google Scholar]
- Archer TK, Lee HL, Cordingley MG, Mymryk JS, Fragoso G, Berard DS, Hager GL. Differential steroid hormone induction of transcription from the mouse mammary tumor virus promoter. Mol Endocrinol. 1994;8:568–576. doi: 10.1210/mend.8.5.8058066. [DOI] [PubMed] [Google Scholar]
- Ashcroft FJ, Newberg JY, Jones ED, Mikic I, Mancini MA. High content imaging-based assay to classify estrogen receptor-alpha ligands based on defined mechanistic outcomes. Gene. 2011;477:42–52. doi: 10.1016/j.gene.2011.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blouin K, Nadeau M, Mailloux J, Daris M, Lebel S, Van L-T, Tchernof A. Pathways of adipose tissue androgen metabolism in women: depot differences and modulation by adipogenesis. Pathways of adipose tissue androgen metabolism in women: depot differences and modulation by adipogenesis. 2009a;296:E244–E255. doi: 10.1152/ajpendo.00039.2008. [DOI] [PubMed] [Google Scholar]
- Blouin K, Nadeau M, Perreault M, Veilleux A, Drolet R, Marceau P, Mailloux J, Luu-The V, Tchernof A. Effects of androgens on adipocyte differentiation and adipose explant metabolism in men and women. Clin Endocrinol. 2010;72:176–188. doi: 10.1111/j.1365-2265.2009.03645.x. [DOI] [PubMed] [Google Scholar]
- Blouin K, Veilleux A, Luu-The V, Tchernof A. Androgen metabolism in adipose tissue: recent advances. Mol Cell Endocrinol. 2009b;301:97–103. doi: 10.1016/j.mce.2008.10.035. [DOI] [PubMed] [Google Scholar]
- Bolton EC, So AY, Chaivorapol C, Haqq CM, Li H, Yamamoto KR. Cell- and gene-specific regulation of primary target genes by the androgen receptor. Cell- and gene-specific regulation of primary target genes by the androgen receptor. 2007;21:2005–2017. doi: 10.1101/gad.1564207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bujalska IJ, Quinkler M, Tomlinson JW, Montague CT, Smith DM, Stewart PM. Expression profiling of 11beta-hydroxysteroid dehydrogenase type-1 and glucocorticoid-target genes in subcutaneous and omental human preadipocytes. J Mol Endocrinol. 2006;37:327–340. doi: 10.1677/jme.1.02048. [DOI] [PubMed] [Google Scholar]
- Burnstein KL, Maiorino CA, Dai JL, Cameron DJ. Androgen and glucocorticoid regulation of androgen receptor cDNA expression. Androgen and glucocorticoid regulation of androgen receptor cDNA expression. 1995;115:177–186. doi: 10.1016/0303-7207(95)03688-1. [DOI] [PubMed] [Google Scholar]
- Chambon C, Duteil D, Vignaud A, Ferry A, Messaddeq N, Malivindi R, Kato S, Chambon P, Metzger D. Myocytic androgen receptor controls the strength but not the mass of limb muscles. Myocytic androgen receptor controls the strength but not the mass of limb muscles. 2010;107:14327–14332. doi: 10.1073/pnas.1009536107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CS, Kokontis J, Liao ST. Molecular cloning of human and rat complementary DNA encoding androgen receptors. Science. 1988;240:324–326. doi: 10.1126/science.3353726. [DOI] [PubMed] [Google Scholar]
- Chen SY, Wang J, Yu GQ, Liu WH, Pearce D. Androgen and glucocorticoid receptor heterodimer formation - A possible mechanism for mutual inhibition of transcriptional activity. Androgen and glucocorticoid receptor heterodimer formation - A possible mechanism for mutual inhibition of transcriptional activity. 1997;272:14087–14092. doi: 10.1074/jbc.272.22.14087. [DOI] [PubMed] [Google Scholar]
- Christodoulides C, Lagathu C, Sethi JK, Vidal-Puig A. Adipogenesis and wnt signaling. Trends Endocrinol Metab. 2008;20:16–24. doi: 10.1016/j.tem.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes C, Vapnik V. Support vector networks. Machine Learning. 1995;20:273–297. [Google Scholar]
- Coviello AD, Legro RS, Dunaif A. Adolescent girls with polycystic ovary syndrome have an increased risk of the metabolic syndrome associated with increasing androgen levels independent of obesity and insulin resistance. J Clin Endocrinol Metab. 2006;91:492–497. doi: 10.1210/jc.2005-1666. [DOI] [PubMed] [Google Scholar]
- Dati E, Baroncelli GI, Mora S, Russo G, Baldinotti F, Parrini D, Erba P, Simi P, Bertelloni S. Body composition and metabolic profile in women with complete androgen insensitivity syndrome. Sex Dev. 2009;3:188–193. doi: 10.1159/000228719. [DOI] [PubMed] [Google Scholar]
- Davies P, Rushmere NK. Association of glucocorticoid receptors with prostate nuclear sites for androgen receptors and androgen receptor response elements. J Mol Endocrinol. 1990;5:117–127. doi: 10.1677/jme.0.0050117. [DOI] [PubMed] [Google Scholar]
- De Vos P, Lefebvre AM, Miller SG, Guerre-Millo M, Wong K, Saladin R, Hamann LG, Staels B, Briggs MR, Auwerx J. Thiazaolidinediones repress ob gene expression in rodents via activation of peroxisome proliferator-activated receptor gamma. J Clin Invest. 1996;98:1004–1009. doi: 10.1172/JCI118860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feige JN, Sage D, Wahli W, Desvergne B, Gelman L. PixFRET, an ImageJ plug-in fro FRET calculation that can accomodate variations in spectral bleed-throughs. Microsc Res Tech. 2005;68:51–58. doi: 10.1002/jemt.20215. [DOI] [PubMed] [Google Scholar]
- Fu M, Sun T, Bookout AL, Downes M, Yu RT, Evans RM, Mangelsdorf DJ. A nuclear receptor atlas: 3T3-L1 adipogenesis. Mol Endocinol. 2005;19:2437–2450. doi: 10.1210/me.2004-0539. [DOI] [PubMed] [Google Scholar]
- Fujikura J, Fujimoto M, Yasue S, Noguchi M, Masuzaki H, Hosoda K, Tachibana T, Sugihara H, Nakao K. Insulin-induced lipohypertrophy: Report of a case with histopathology. Insulin-induced lipohypertrophy: Report of a case with histopathology. 2005;52:623–628. doi: 10.1507/endocrj.52.623. [DOI] [PubMed] [Google Scholar]
- Furutani T, Watanabe T, Tanimoto K, Hashimoto T, Koutoku H, Kudoh M, Shimizu Y, Kato S, Shikama H. Stabilization of androgen receptor protein is induced by agonist, not by antagonists. Biochem Biophys Res Commun. 2002;294:779–784. doi: 10.1016/S0006-291X(02)00564-8. [DOI] [PubMed] [Google Scholar]
- Gould DC, Kirby RS, Amoroso P. Hypoandrogen-metabolic syndrome: a potentially common and underdiagnosed condition in men. Int J Clin Pract. 2007;61:341–344. doi: 10.1111/j.1742-1241.2006.01239.x. [DOI] [PubMed] [Google Scholar]
- Guilherme A, Virbasius JV, Puri V, Czech MP. Adipocyte dysfunctions linking obesity to insulin resistance and type 2 diabetes. Adipocyte dysfunctions linking obesity to insulin resistance and type 2 diabetes. 2008;9:367–377. doi: 10.1038/nrm2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta V, Bhasin S, Guo W, Singh R, Miki R, Chauhan P, Choong K, Tchkonia T, Lebrasseur NK, Flanagan JN, Hamilton JA, Viereck JC, Narula NS, Kirkland JL, Jasuja R. Effects of dihydrotestosterone on differentiation and proliferation of human mesenchymal stem cells and preadipocytes. Effects of dihydrotestosterone on differentiation and proliferation of human mesenchymal stem cells and preadipocytes. 2008;296:32–40. doi: 10.1016/j.mce.2008.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartig SM, He B, Long W, Buehrer BM, Mancini MA. Homeostatic levels of SRC-2 and SRC-3 promote early human adipogenesis. Homeostatic levels of SRC-2 and SRC-3 promote early human adipogenesis. 2011;192:55–72. doi: 10.1083/jcb.201004026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotamisligil GS, Johnson RS, Distel RJ, Ellis R, Papaioannou VE, Spiegelman BM. Uncoupling of obesity from insulin resistance through a targeted mutation in ap2, the adipocyte fatty acid binding protein. Science. 1996;274:1377–1379. doi: 10.1126/science.274.5291.1377. [DOI] [PubMed] [Google Scholar]
- Jennrich RI. Stepwise discriminant analysis. In: Enslein K, Ralston A, Wilf HS, editors. Statistical methods for digital computers. John Wiley & Sons; New York, NY: 1977. pp. 77–95. [Google Scholar]
- Jones JO, An WF, Diamond MI. AR inhibitors identified by high-throughput microscopy detection of conformational change and subcellular localization. ACS Chem Biol. 2009;4:199–208. doi: 10.1021/cb900024z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemppainen JA, Langley E, Wong CI, Bobseine K, Kelce WR, Wilson EM. Distinguishing androgen receptor agonists and antagonists: distinct mechanisms of activation by medroxyprogesterone acetate and dihydrotestosterone. Mol Endocrinol. 1999;13:440–454. doi: 10.1210/mend.13.3.0255. [DOI] [PubMed] [Google Scholar]
- Klokk TI, Kurys P, Elbi C, Nagaich AK, Hendarwanto A, Slagsvold T, Chang CY, Hager GL, Saatcioglu F. Ligand-specific dynamics of the androgen receptor at its response element in living cells. Ligand-specific dynamics of the androgen receptor at its response element in living cells. 2007;27:1823–1843. doi: 10.1128/MCB.01297-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahnalampi M, Heinaniemi M, Sinkkonen L, Wabitsch M, Carlberg C. Time-resolved expression profiling of the nuclear receptor superfamily in human adipogenesis. PLOS One. 2010;5:e12991. doi: 10.1371/journal.pone.0012991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langley E, Zhou ZX, Wilson EM. Evidence for an anti-parallel orientation of the ligand-activated human androgen receptor dimer. J Biol Chem. 1995;270:29983–29990. doi: 10.1074/jbc.270.50.29983. [DOI] [PubMed] [Google Scholar]
- Lee MJ, Fried SK, Mundt SS, Wang Y, Sullivan S, Steffani A, Daugherty BL, Hermanowski-Vosatka Depot-specific regulation of the conversion of cortisone to cortisol in human adipose tissue. Obesity. 2008;16:1178–1185. doi: 10.1038/oby.2008.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman BA, Bona BJ, Edwards DP, Nordeen SK. The constitution of a progesterone response element. Mol Endocinol. 1993;7:515–527. doi: 10.1210/mend.7.4.8388996. [DOI] [PubMed] [Google Scholar]
- Liu J, Farmer SR. Regulating the balance between peroxisome proliferator-activated receptor gamma and beta-catenin signaling during adipogenesis. A glycogen synthase 3beta phosphorylation-defective mutant of beta-catenin inhibits teh expression of a subset of adipogenic genes. J Biol Chem. 2004;279:45020–45027. doi: 10.1074/jbc.M407050200. [DOI] [PubMed] [Google Scholar]
- Liu S, Navarro G, Mauvais-Jarvis F. Androgen excess produces systemic oxidative stress and predisposes to beta-cell failure in female mice. PLoS ONE. 2010;5:e11302. doi: 10.1371/journal.pone.0011302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombes M, Binart N, Oblin ME, Joulin V, Baulieu EE. Characterization of the interaction of the human mineralocorticosteroid receptor with hormone response elements. Biochem J. 1993;292:577–583. doi: 10.1042/bj2920577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubahn DB, Joseph DR, Sullivan PM, Willard HF, French FS, Wilson EM. Cloning of human androgen receptor complementary DNA and localization to teh X chromosome. Science. 1988;240:327–330. doi: 10.1126/science.3353727. [DOI] [PubMed] [Google Scholar]
- Madsen L, Pedersen LM, Liaset B, Ma T, Petersen RK, van den Berg S, Pan J, Muller-Decker K, Dulsner ED, Kleemann R, Kooistra T, Doskeland SO, Kristiansen K. cAMP-dependent signaling regulates the adipogenic effect of n-6 polyunsaturated fatty acids. cAMP-dependent signaling regulates the adipogenic effect of n-6 polyunsaturated fatty acids. 2008;283:7196–7205. doi: 10.1074/jbc.M707775200. [DOI] [PubMed] [Google Scholar]
- Magee JA, Chang LW, Stormo GD, Milbrandt J. Direct, androgen receptor-mediated regulation of the FKBP5 gene via a distal enhancer element. Endocrinology. 2006;147:590–598. doi: 10.1210/en.2005-1001. [DOI] [PubMed] [Google Scholar]
- Massie CE, Lynch A, Ramos-Montoya A, Boren J, Stark R, Fazil L, Warren A, Scott H, Madhu B, Sharma N, Bon H, Zecchini V, Smith DM, Denicola GM, Matthews N, Osborne M, Hadfield J, Macarthur S, Adryan B, Lyons SK, Brindle KM, Griffiths J, Gleave ME, Rennie PS, Neal DE, Mills IG. The androgen receptor fuels prostate cancer by regulating central metabolism and biosynthesis. EMBO J. 2011;30:2719–2733. doi: 10.1038/emboj.2011.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuzaki H, Paterson J, Shinyama H, Morton NM, Mullins JJ, Seckl JR, Flier JS. A transgenic model of visceral obesity and the metabolic syndrome. A transgenic model of visceral obesity and the metabolic syndrome. 2001;294:2166–2170. doi: 10.1126/science.1066285. [DOI] [PubMed] [Google Scholar]
- Masuzaki H, Yamamoto H, Kenyon CJ, Elmquist JK, Morton NM, Paterson JM, Shinyama H, Sharp MGF, Fleming S, Mullins JJ, Seckl JR, Flier JS. Transgenic amplification of glucocorticoid action in adipose tissue causes high blood pressure in mice. Transgenic amplification of glucocorticoid action in adipose tissue causes high blood pressure in mice. 2003;112:83–90. doi: 10.1172/JCI17845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Xu Z, Zhang X, Wang L, Gimble JM, Lander ES, Rosen ED. Comparative epigenomic analysis of murine and human adipogenesis. Cell. 2010;143:156–169. doi: 10.1016/j.cell.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss PE, Lyles BE, Stewart LV. The PPAR gamma ligand ciglitazone regulates androgen receptor activation differently in androgen-dependent versus androgen-independent human prostate cancer cells. The PPAR gamma ligand ciglitazone regulates androgen receptor activation differently in androgen-dependent versus androgen-independent human prostate cancer cells. 2010;316:3478–3488. doi: 10.1016/j.yexcr.2010.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazareth LV, Stenoien DL, Bingman WEr, James AJ, Wu C, Zhang Y, Edwards DP, Mancini M, Marcelli M, Lamb DJ, Weigel NL. A C619Y mutation in the human androgen receptor causes inactivation and mislocalization of the receptor with concomitant sequestration of SRC-1 (steroid receptor coactivator 1) Mol Endocrinol. 1999;13:2065–2075. doi: 10.1210/mend.13.12.0382. [DOI] [PubMed] [Google Scholar]
- Nishino N, Tamori Y, Tateya S, Kawaguchi T, Shibakusa T, Mizunoya W, Inoue K, Kitazawa R, Matsuki Y, Hiramatsu R, Masubuchi S, Omachi A, Kimura K, Saito M, Amo T, Ohta S, Yamaguchi T, Osumi T, Cheng J, Fujimoto T, Nakao K, Aiba A, Okamura H, Fushiki T, Kasuga M. FSP27 contributes to efficient energy storage in murine white adipocytes by promoting the formation of unilocular lipid droplets. J Clin Invest. 2008;118:2808–2021. doi: 10.1172/JCI34090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordeen SK, Suh BJ, Kuhnel B, Hutchinson CAr. Structural determinants of a glucocorticoid receptor recognition element. Mol Endocrinol. 1990;4:1866–1873. doi: 10.1210/mend-4-12-1866. [DOI] [PubMed] [Google Scholar]
- Pantoja C, Huff JT, Yamamoto KR. Glucocorticoid Signaling Defines a Novel Commitment State during Adipogenesis In Vitro. Glucocorticoid Signaling Defines a Novel Commitment State during Adipogenesis In Vitro. 2008;19:4032–4041. doi: 10.1091/mbc.E08-04-0420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rariy CM, Ratcliffe SJ, Weinstein R, Bhasin S, Blackman MR, Cauley JA, Robbins J, Zmuda JM, Harris TB, Cappola AR. Higher serum free testosterone concentration in older women is associated with greater bone mineral density, lean body mass, and total fat mass: the cardiovascular health study. J Clin Endocrinol Metab. 2011;96:989–996. doi: 10.1210/jc.2010-0926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy TE, Pauli F, Sprouse RO, Neff NF, Newberry KM, Garabedian MJ, Myers RM. Genomic determination of glucocorticoid response reveals unexpected mechanisms of gene regulation. Genome Res. 2009;19:2163–2171. doi: 10.1101/gr.097022.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche PJ, Hoare SA, Parker MG. A consensus DNA-binding site for the androgen receptor. Mol Endocrinol. 1992;6:2229–2235. doi: 10.1210/mend.6.12.1491700. [DOI] [PubMed] [Google Scholar]
- Rosen ED, Hsu CH, Wang XZ, Sakai S, Freeman MW, Gonzalez FJ, Spiegelman BM. C/EBP alpha induces adipogenesis through PPAR gamma: a unified pathway. C/EBP alpha induces adipogenesis through PPAR gamma: a unified pathway. 2002;16:22–26. doi: 10.1101/gad.948702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen ED, Sarraf P, Troy AE, Bradwin G, Moore K, Milstone DS, Spiegelman BM, Mortensen RM. PPAR gamma is required for the differentiation of adipose tissue in vivo and in vitro. Mol Cell. 1999;4:611–617. doi: 10.1016/s1097-2765(00)80211-7. [DOI] [PubMed] [Google Scholar]
- Schaufele F, Carbonell X, Guerbadot M, Borngraeber S, Chapman MS, Ma AAK, Miner JN, Diamond MI. The structural basis of androgen receptor activation: Intramolecular and intermolecular amino-carboxy interactions. The structural basis of androgen receptor activation: Intramolecular and intermolecular amino-carboxy interactions. 2005;102:9802–9807. doi: 10.1073/pnas.0408819102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenmakers E, Verrijdt G, Peeters B, Verhoeven G, Rombauts W, Claessens F. Differences in DNA binding characteristics of the androgen and glucocorticoid receptors can determine hormone-specific responses. J Biol Chem. 2000;275:12290–12297. doi: 10.1074/jbc.275.16.12290. [DOI] [PubMed] [Google Scholar]
- Schupp M, Cristancho AG, Lefterova MI, Hanniman EA, Briggs ER, Steger DJ, Qatanani M, Curtin JC, Schug J, Ochsner SA, McKenna NJ, Lazar MA. Re-expression of GATA2 Cooperates with Peroxisome Proliferator-activated Receptor-gamma Depletion to Revert the Adipocyte Phenotype. Re-expression of GATA2 Cooperates with Peroxisome Proliferator-activated Receptor-gamma Depletion to Revert the Adipocyte Phenotype. 2009;284:9458–9464. doi: 10.1074/jbc.M809498200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Artaza JN, Taylor WE, Braga M, Yuan X, Gonzalez-Cadavid NF, Bhasin S. Testosterone inhibits adipogenic differentiation in 3T3-L1 cells: Nuclear translocation of androgen receptor complex with beta-catenin and T-cell factor 4 may bypass canonical Wnt signaling to down-regulate adipogenic transcription factors. Testosterone inhibits adipogenic differentiation in 3T3-L1 cells: Nuclear translocation of androgen receptor complex with beta-catenin and T-cell factor 4 may bypass canonical Wnt signaling to down-regulate adipogenic transcription factors. 2006;147:141–154. doi: 10.1210/en.2004-1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Artaza JN, Taylor WE, Gonzalez-Cadavid NF, Bhasin S. Androgens stimulate myogenic differentiation and inhibit adipogenesis in C3H 10T1/2 pluripotent cells through an androgen receptor-mediated pathway. Androgens stimulate myogenic differentiation and inhibit adipogenesis in C3H 10T1/2 pluripotent cells through an androgen receptor-mediated pathway. 2003;144:5081–5088. doi: 10.1210/en.2003-0741. [DOI] [PubMed] [Google Scholar]
- Soccio RE, Tuteja G, Everett LJ, Li Z, Lazar MA, Kaestner KH. Species-specific strategies underlying conserved functions of metabolic transcription factors. Mol Endocrinol. 2011;25:694–706. doi: 10.1210/me.2010-0454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanworth RD, Jones TH. Testosterone in Obesity, Metabolic Syndrome and Type 2 Diabetes. Advances in the Management of Testosterone Deficiency. 2009;37:74–90. doi: 10.1159/000176046. [DOI] [PubMed] [Google Scholar]
- Steger DJ, Grant GR, Schupp M, Tomaru T, Lefterova MI, Schug J, Manduchi E, Stoeckert CJ, Lazar MA. Propagation of adipogenic signals through an epigenomic transition state. Propagation of adipogenic signals through an epigenomic transition state. 2010;24:1035–1044. doi: 10.1101/gad.1907110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szafran AT, Szwarc M, Marcelli M, Mancini MA. Androgen receptor functional analyses by high throughput imaging: Determination of ligand, cell cycle, and mutation-specific effects. PLOS One. 2008;3:e6205. doi: 10.1371/journal.pone.0003605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenover JS. Effects of testosterone supplementation in the aging male. J Clin Endocrinol Metab. 1992;75:1092–1098. doi: 10.1210/jcem.75.4.1400877. [DOI] [PubMed] [Google Scholar]
- Tontonoz P, Hu E, Spiegelman BM. Stimulation of adipogenesis in fibroblasts by PPAR gamma 2, a lipid-activated transcription factor. Cell. 1994;79:1147–1156. doi: 10.1016/0092-8674(94)90006-x. [DOI] [PubMed] [Google Scholar]
- Veilleux A, Cote JA, Blouin K, Nadeau M, Pelletier M, Marceau P, Laberge PY, Luu-The V, Tchernof A. Glucocorticoid-induced androgen inactivation by aldo-keto reductase 1C2 promotes adipogenesis in human preadipocytes. Am J Physiol Endocrinol Metab. 2012;302:E941–949. doi: 10.1152/ajpendo.00069.2011. [DOI] [PubMed] [Google Scholar]
- Wake DJ, Strand M, Rask E, Westerbacka J, Livingstone DEW, Soderberg S, Andrew R, Yki-Jarvinen H, Olsson T, Walker BR. Intra-adipose sex steroid metabolism and body fat distribution in idiopathic human obesity. Clin Endocrinol. 2007;66:440–446. doi: 10.1111/j.1365-2265.2007.02755.x. [DOI] [PubMed] [Google Scholar]
- Waki H, Park KW, Mitro N, Pei L, Damoiseaux R, Wilpitz DC, Reue K, Saez E, Tontonoz P. The small molecule harmine is an antidiabetic cell-type specific regulator of PPARgamma expression. Cell Metab. 2007;5:357–370. doi: 10.1016/j.cmet.2007.03.010. [DOI] [PubMed] [Google Scholar]
- Wallace AD, Cidlowski JA. Proteasome-mediated glucocorticoid receptor degradation restricts transcriptional signaling by glucocorticoids. J Biol Chem. 2001;276:42714–42721. doi: 10.1074/jbc.M106033200. [DOI] [PubMed] [Google Scholar]
- Wilson EM, French FS. Binding properties of androgen receptors. Evidence for identical receptors in rat testis, epididymis, and prostate. J Biol Chem. 1976;251:5620–5629. [PubMed] [Google Scholar]
- Yang CC, Ku CY, Wei S, Shiau CW, Chen CS, Pinzone JJ, Ringel MD. Peroxisome proliferator-activated receptor gamma-independent repression of prostate-specific antigen expression by thiazolidinediones in prostate cancer cells. Peroxisome proliferator-activated receptor gamma-independent repression of prostate-specific antigen expression by thiazolidinediones in prostate cancer cells. 2006;69:1564–1570. doi: 10.1124/mol.105.018333. [DOI] [PubMed] [Google Scholar]
- Yen PM, Liu Y, Palvimo JJ, Trifiro M, Whang J, Pinsky L, Janne OA, Chin WW. Mutant and wild-type androgen receptors exhibit cross-talk on androgen-, glucocorticoid-, and progesterone-mediated transcription. Mutant and wild-type androgen receptors exhibit cross-talk on androgen-, glucocorticoid-, and progesterone-mediated transcription. 1997;11:162–171. doi: 10.1210/mend.11.2.9886. [DOI] [PubMed] [Google Scholar]
- Yu CY, Mayba O, Lee JV, Tran J, Harris C, Speed TP. Genome-wide analysis of glucocorticoid receptor binding regions in adipocytes reveal gene network involved in triglyceride homeostasis. PLoS One. 2010;5:e15188. doi: 10.1371/journal.pone.0015188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zitzmann M. Testosterone deficiency, insulin resistance and the metabolic syndrome. Testosterone deficiency, insulin resistance and the metabolic syndrome. 2009;5:673–681. doi: 10.1038/nrendo.2009.212. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.