Abstract
The Death Domain Fold superfamily of evolutionarily conserved protein-protein interaction domains consists of 4 subfamilies: the death domain, the death effector domain, the caspase recruitment domain, and the PYRIN domain. Interaction of Death Domain Fold containing proteins modulates the activity of several downstream effectors, such as caspases and transcription factors. Recent studies provide evidence for not only homotypic-, but also heterotypic interactions among different sub-families, and even unconventional non-death domain fold interactions. As the number of potential protein associations among Death Domain Fold containing proteins expands and their influence on cellular responses increases, a challenging field for new investigations opens up. This review will focus on PYRIN domain-containing proteins and discuss the recent advances that provide strong evidence that PYRIN domain-mediated signal transduction has broad implications on cellular functions, including innate immunity, inflammation, differentiation, apoptosis, and cancer.
Keywords: apoptosis, cancer, CARD domain, caspase activation, death domain fold, inflammation, interleukin-1, leucine rich region, NACHT domain, NF-κB, pathogen associated molecular pattern, pathogen recognition receptor, PYRIN domain, signal transduction
THE DEATH DOMAIN FOLD SUPERFAMILY OF PROTEIN INTERACTION DOMAINS
The death domain fold (DDF) is a highly conserved protein interaction domain spanning approximately 90 amino acid residues. This superfamily consists of four subfamilies: The death domain (DD), the death effector domain (DED), the caspase recruitment domain (CARD), and the pyrin N-terminal homology domain (PYRIN domain, PYD). Subfamilies are mainly defined by sequence homology, and display a common three-dimensional structure. However, the sequence identity between the subfamilies is usually lower than 25%, and even within one family mainly restricted to conserved residues of the hydrophobic core.
DDF containing proteins have been linked to the activation of caspases and subsequent initiation of apoptosis by the induced proximity mechanism. However, several additional downstream effectors link these domains not only to apoptosis, but also immunity, inflammation, differentiation and cancer. DDFs had originally been implicated to form exclusively homotypic interactions (e.g. DD-DD, DED-DED, CARD-CARD, PYD-PYD). Yet, recent evidence demonstrated their capability to also form heterotypic interactions among family members, and even unconventional, non-DDF interactions, thereby increasing the number of potential protein associations and cellular responses. The first evidence for unconventional, non-DDF interactions came from the observation that the DED of PEA-15 (PED) interacts with the C-terminal domain of ERK [1]. Also, the DD of Fas (Apo-1, CD95, TNFRSF6) was suggested to associate with calmodulin in a Fas ligand and Ca2+-dependent manner [2]. The apoptosis repressor with a CARD (ARC CARD2, Myp) can disrupt the intrinsic, mitochondrial apoptosis pathway by association of its CARD with the C-terminal domain of Bax. By heterotypic interaction, ARC disrupts the extrinsic, receptor mediated apoptosis pathway by interacting via its CARD with the DD of Fas and FADD, thereby preventing formation of the caspase-8 activating death-inducing signaling complex (DISC) [3]. The PYD of the apoptotic speck-like protein containing a CARD (ASC, TMS1, PYCARD, CARD5) also associates with Bax following DNA damage, which results in cytochrome c release and activation of caspase-9 [4]. The PYD of ASC further associates with pro-caspase-8, which results in activation of the caspase-8 zymogen [5]. Both, ARC and ASC also participate in classical, homotypic interactions [6, 7].
DDF interactions usually result in the formation of inducible, high molecular weight protein complexes. Homotypic interaction of CARD and DED-containing proteins with inactive caspase zymogens leads to the formation of multi-protein complexes, including the caspase-9 activating apoptosome [8], the caspase-1 and caspase-5 activating inflammasomes [9], the caspase 8 activating DISC [10], or the caspase-2 activating PIDDosome [11]. Trans-activation of these caspases in these complexes occurs through the induced proximity-, or the enhanced dimerization mechanism [12, 13].
The PYRIN domain - the most recent death domain family member
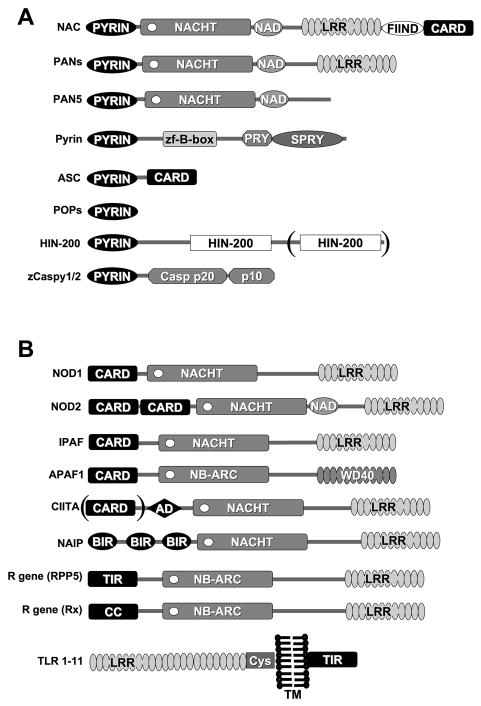
DD, DED, and CARD interactions are established mechanisms to transmit signals in apoptosis, inflammation, and immunity. More recently, a fourth domain was predicted to have a related fold and is known by several synonyms: PYRIN domain (PYD), pyrin, absent-in-melanoma, ASC and DD-like (PAAD), and domain in apoptosis and interferon response (DAPIN) [14–18]. At least 23 human proteins have been identified that contain a PYD. Table (1) and Fig (1A): PYD-containing proteins can be divided into at least 4 groups of proteins:
Table 1. PYRIN domain containing proteins.
(A) Members of the human PAN protein family; (B) Human PYRIN domain-containing adapter proteins; (C) Human regulatory or PYRIN domain-containing proteins; (D) Human HIN-200 family proteins; Activation (↑, +); Inhibition (↓, −); both is reported (+/−).
| A. Receptor proteins: | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Name | Synonym | Chromosome | Effector Domain | LRRs | Length (kDa) | Ligand | Binding to ASC | Apoptosis | Inflammation Immunity | Inducer |
| cryopyrin | CIAS1, PYPAF1, NALP3, CLR1.1 | 1q44 | PYD | 9 | 118 | MDP | + | + | +/− | (LPS, TNFα, LTA, dsRNA)↑ |
| Nac | CARD7, DEFCAP, NALP1, CLR17.1 | 17p12.13 | PYD, CARD | 5 | 161 | PGN | + | + | + | (LPS, Ionomycin, PMA)↑ |
| PAN1 | PYPAF2, NALP2, NBS1, CLR19.9 | 19q13.4 | PYD | 8 | 121 | + | −/+ | (IFNβ, IFN γ, LPS)↑ | ||
| PAN2 | PYPAF4, NALP4, CLR19.5 | 19q13.42 | PYD | 8 | 113 | − | − | |||
| PAN3 | PYPAF5, NALP6, CLR11.4 | 11p15 | PYD | 5 | 99 | + | + | |||
| PAN4 | NALP8, NOD16, CLR19.2 | 19q13.4 | PYD | 7 | 117 | |||||
| PAN5 | NALP10, PYNOD, NOD8, CLR11.1 | 11p15 | PYD | 0 | 75 | + | − | |||
| PAN6 | PYPAF7, NALP12, Monarch-1, CLR19.3 | 19q13.4 | PYD | 11 | 120 | + | + | NO↑, (TNFα, IFNγ)↓ | ||
| PAN7 | PYPAF3, NALP7, NOD12, CLR19.4 | 11q13.42 | PYD | 9 | 112 | − | (LPS, IL-1β)↑ | |||
| PAN8 | NALP14, NOD5, CLR11.2 | 11p15.4 | PYD | 12 | 125 | |||||
| PAN9 | 19q13.4 | PYD | 9 | 115 | ||||||
| PAN10 | PYPAF6, NALP11, NOD17, CLR19.6 | 19q13.42 | PYD | 9 | 118 | − | ||||
| PAN11 | PYPAF8, NALP5, (Mater1), CLR19.8 | 11q13.42 | PYD | 13 | 134 | |||||
| PAN12 | NALP9, NOD6, CLR19.1 | 19q13.4 | PYD | 10 | 113 | |||||
| PAN13 | NALP13, NOD14, CLR19.7 | 19q13.4 | PYD | 9 | 119 | |||||
| PAN14 | NOD27, CLR16.1 | 16q13 | PYD | 20 | 205 | LPS↓ | ||||
| B. Adaptor proteins: | ||||||||
|---|---|---|---|---|---|---|---|---|
| Name | Synonym | Chromosome | Effector Domain | Length (kDa) | Binding to ASC | Apoptosis | Inflammation Immunity | Inducer |
| ASC | TMS1, PYCARD, CARD5 | 16p12.1 | PYD, CARD | 22 | + | + | + | (TNF α, LPS, IL-1α, IL-1β, IFNα, IFNγ, p53)↑ |
| C. Regulatory proteins: | ||||||||
|---|---|---|---|---|---|---|---|---|
| Name | Synonym | Chromosome | Effector Domain | Length (kDa) | Binding to ASC | Apoptosis | Inflammation Immunity | Inducer |
| cPOP1 | ASC2, ASCI, ASCL | 16.p12.1 | PYD | 10 | + | + | − | |
| cPOP2 | 3q28 | PYD | 11 | + | − | |||
| pyrin | Marenostrin | 16p13.3 | PYD | 87 | + | − | − | (LPS, TNFα, IL-1β, IL-2, IL-4, IL-6, IL-10, IL-12)↑ |
| D. HIN-200 proteins: | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Name | Chromosome | Effector Domain | HIN-200 repeats | Length (kDa) | Apoptosis | Proliferation | Differentiation | Tumorigenesis | Inflammation Immunity | Inducer |
| IFI16 | 1q22 | HIN-200 | 2 | 80 | + | − | + | + | IFNγ↑ | |
| AIM2 | 1q22 | HIN-200 | 1 | 49 | − | + | − | IFNγ↑ | ||
| MNDA | 1q22 | HIN-200 | 1 | 46 | + | − | + | − | IFNα↑ | |
| IFIX | 1q21-23 | HIN-200 | 1 | 56 | − | − | IFNγ↑ | |||
Mater is the mouse orthologue, but an undefined effector domain replaces the PYRIN domain.
Figure 1. Domain organization of PYRIN domain containing proteins.
(A) PYRIN domain containing proteins and (B) PAN-related proteins. Please note the alternative names in Tables 1 and 2. APAF-1 contains a NACHT-related nucleotide binding NB-ARC domain and WD-repeats, which is also involved in ligand binding, similar to LRRs.
Receptor proteins: Table (1A): The most prominent members are potential PYD-containing intracellular pathogen recognition receptors (PRRs), known as PAN, NALP, PYPAF, Nod, Caterpiller and NLR proteins [19–24]. These proteins recognize pathogen associated molecular patterns (PAMPs). Which are products of the microbial metabolism that are conserved, essential, and unique to pathogens [25]. PAN-family proteins are composed of an N-terminal PYD, a central nucleotide binding NACHT domain, followed by a variable number of leucine rich regions (LRRs). 16 family members have been identified in the human genome. NAC (CARD7, DEFCAP, NALP1, CLR17.1) is one of only two human proteins that combine a PYD (N-terminal) and a CARD (C-terminal).
Adaptor protein: Table (1B): ASC, the apoptotic speck-like protein containing a CARD (TMS1, CARD5, PYCARD), also known as target of methylation induced silencing (TMS1), is a 22 kDa protein which encodes an N-terminal PYD followed by a short linker region and a C-terminal CARD [26, 27].
Regulatory proteins: Table (1C): This group consists of pyrin (marenostrin) and the cellular PYD-only proteins (cPOPs) cPOP1 and cPOP2 (Stehlik, C, unpublished) [28]. Pyrin, the founding member of the PYD protein family, is a protein of approximately 87 kDa, consisting of an N-terminal PYD, a central B-box zinc finger, followed by a PRY/SPRY (B30.1) domain. Two nuclear localization signals are encoded between the B-box and the PRY/SPRY region. Pyrin was initially predicted to function as a nuclear transcription factor of the RoRet protein family, but now appears to function as a regulator of the PYD signaling pathway [29–32]. cPOP1 (ASC2, ASCI, ASCL) is highly homologous (64% identical, 88% similar) to the PYD of ASC, and localizes just 14 kbps downstream of the ASC gene on human chromosome 16p12.1, suggesting that the genes encoding cPOP1 and ASC originally arose by gene duplication [28]. cPOP1 prevents binding of the PYD-containing adaptor protein ASC to PAN-family proteins, thereby blocking signal transduction [28]. A similar mechanism of regulation of signal transduction is found for some other DDFs, such as the CARD and the DED. DDF-only proteins include the CARD-only proteins Iceberg, COP (Pseudo-ICE), and INCA, or the DED-only proteins PEA-15 (PED), c-FLIP-s, and v-FLIP [33–38]. These DDF-only proteins usually function as endogenous dominant negative proteins that block specific DDF interactions.
HIN-200 proteins: Table (1D): The hematopoietic interferon-inducible nuclear proteins with the 200-amino-acid repeat (HIN-200) proteins form a distinct family [39]. Besides the presence of a PYD, these proteins are characterized by one or two copies of a 200 amino acid residue signature domain, termed domain A and B. [40, 41]. Members include human MNDA, AIM2, and several splice variants of IFI16 and IFIX, which are all encoded on chromosome 1q21-23, suggesting that these genes originally arose by gene duplication [40–42].
CARDs and DEDs are found in the pro-domain of caspases and mediate oligomerization and recruitment of adaptor proteins. Humans do not encode a caspase zymogen containing a PYD, whereas a PYD-containing pro-apoptotic caspase (caspase-15) is encoded in other mammalian species [43]. Zebrafish encodes two PYD-containing caspases, zCaspy1 and zCaspy2, supporting once more the close resemblance of the PYD with other DDFs [44].
PAN-family proteins are detected mainly in two clusters in the human genome, suggesting that these proteins evolved by gene duplication. One PAN-family protein cluster exists on the short arm on chromosome 11, whereas a second cluster localizes to the long arm on chromosome 19. The adaptor proteins ASC and pyrin, as well as the ASC inhibitor cPOP1, are clustered on chromosome 16, and HIN-200 members are found in close proximity on the long arm on chromosome 1.
SIGNAL TRANSDUCTION MEDIATED BY PYRIN DOMAIN INTERACTIONS
PYD-PYD interaction-dependent signaling is linked to inflammation, immunity, differentiation, apoptosis and cancer. The first indications on the potential function of the PYD came from studies of PAN-related proteins. PAN-family proteins contain an N-terminal PYD, a central nucleotide binding NACHT domain, a NACHT-associated domain (NAD), followed by a varying number of LRRs. Fig (1A). The C-terminal LRRs have been found in some PAN and PAN-related proteins to function as the ligand-binding domain, and are responsible for activating these proteins. The PYD is considered the effector domain, which recruits adaptor proteins and downstream effectors upon activation of PAN-family proteins. The NACHT domain is an NB-ARC-related predicted nucleoside triphosphatase (NTPase) domain, which consists of seven distinct motifs [45]. It is hypothesized that this domain mediates oligomerization of PAN-family proteins upon NTP binding and hydrolysis, and this domain is followed by the NAD, a domain of unknown function.
PAN-related proteins in inflammation and immunity
A number of structurally related proteins are well-established effectors of the host immune response to pathogens and mediators of apoptosis. Fig (1B) and Table (2). PAN-family proteins closely resemble the CARD containing proteins Nod1 (CARD4), Nod2 (CARD15), CLAN (IPAF, CARD12), APAF-1, the CARD and activation domain containing CIITA, the BIR domain containing NAIP, and plant resistance (R) genes [19, 21, 23, 24, 46, 47]. Common to these proteins are the NACHT and LRR domain structure, but they differ in their N-terminal effector domains. APAF-1 was identified as a CED4-like protein that mediates cytochrome c/ATP-dependent activation of caspase-9 [48]. The WD repeats are involved in ligand (cytochrome c) binding, and deletion of this domain converts APAF-1 to a constitutively active caspase-9 activator [49].
Table 2.
PAN-related proteins.
(A) The proteins listed show similar domain organization as PAN-family proteins, but contain a different effector domain (e.g. CARD). (B) PAN-related proteins are listed, which contain an effector domain unrelated to the death domain fold. Note that only the dendritic cell specific CIITA encodes a CARD. Activation (↑, +); Inhibition (↓, −); both is reported (+/−).
| A. Related death domain fold proteins: | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Name | Synonym | Chromosome | Effector Domain | LRRs | Length (kDa) | Ligand | Apoptosis | Inflammation Immunity | Inducer |
| Apaf1 | 12q23 | CARD | 12–13 WD40 | 130 | cytochrome c | + | p53↑ | ||
| Nod1 | CARD4 | 7p14.3 | CARD | 11 | 108 | iE-DAP | + | + | (Salmonellae, Staphylococcus, IFNγ)↑ |
| Nod2 | CARD15 | 16q12.1 | CARD, CARD | 12 | 115 | MDP | +/− | (IFNγ, TNFα, Salmonellae, Staphylococcus)↑ | |
| IPAF | CLAN, CARD12 | 2p22.3 | CARD | 14 | 116 | PGN, LTA, LPS | + | + | (p53, LPS, TNFα)↑ |
| CIITA | 16p13.13 | AD | 4 | 124 | + | IFNg↑ | |||
| B. Related non death domain fold proteins: | ||||||||
|---|---|---|---|---|---|---|---|---|
| Name | Synonym | Chromosome | Effector Domain | LRRs | Ligand | Apoptosis | Inflammation Immunity | Inducer |
| NAIP | BIRC1 | 5q13.2 | BIR, BIR, BIR | 14 | - | + | ||
| TLR1-11 | TIR | PAMPs | + | + | inflammation | |||
| Resistance (R) proteins | TIR, CC | PAMPs | + | |||||
The neuronal apoptosis inhibitory protein (NAIP) was identified in a functional screen for genes that are involved in spinal muscular atrophy. The encoded protein causes inappropriate motor neuron apoptosis, and its expression further correlates with disease severity [50, 51]. One function attributed to NAIP is its potential to inhibit apoptotic caspases via the BIR domains. NAIP association with, and inhibition of caspase-9 is dependent on ATP binding [52]. Of note is that recent evidence also connects NAIP to host defense. The NAIP-encoding genomic locus mediates resistance of inbred mice-derived macrophages to infection by Legionella pneumophila [53].
Bare lymphocyte syndrome type II is a hereditary MHC class II deficiency, and the class II transactivator (CIITA) is capable of restoring expression of MHC class II genes [54]. The transactivation potential depends on the binding and hydrolysis of GTP, and CIITA functions as a scaffold for several transcription factors that modulate MHC class II gene expression [55, 56]. The dendritic cell-specific CIITA gene also encodes a CARD, which enhances the transactivation potential of CIITA [57]. A polymorphism in one of the CIITA promoters, which results in a decreased expression of CIITA, was recently linked to susceptibility to several inflammatory disorders, including rheumatoid arthritis, multiple sclerosis and myocardial infarction [58].
The LRRs of the Toll like receptors (TLRs), a family of at least 11 conserved type I transmembrane receptors, function as PRRs that sense a number of different microbial PAMPs. Signal transduction occurs via their TIR (Toll/IL-1 receptor) domain [59, 60]. Plant disease resistance (R) genes also encode proteins with a related structure, and about 150 R genes are encoded in Arabidopsis thaliana. These proteins encode either a coiled coil (CC), or TIR effector domain, which is required to initiate the hypersensitive response, a form of cell death following PAMP recognition. This response supports the innate basal defense to prevent pathogen spread [47].
Nod1 and Nod2 function as PRRs that recognize proteoglycan (PGN), and both initiate similar downstream effector pathways. Nod1 and Nod2 recruit the CARD containing adaptor protein Cardiak (Rip2, Rick), but not ASC [61]. Nod1 recognizes bacterial diaminopimelate-containing N-acetylglucosamine-N-acetylmuramic acid (GlcNAc-MurNAc) tripeptide, which is unique to Gram-negative bacteria [62, 63]. Nod2 is characterized by the presence of two N-terminal CARDs, and recognizes muramyl dipeptide, (MDP) a moiety found in PGN from both Gram-negative and Gram-positive bacteria [64–66]. Evidence for an anti-bacterial function of the LRRs of Nod2 came from a recent study, where Nod2 expressing intestinal epithelial cells showed a smaller number of viable Salmonella typhimurium by invasion assay, when compared to control cells [67]. However, a common Crohn’s disease associated Nod2 mutation (3020insC), which causes truncation of the LRR domain, had no effect [67]. In addition, Nod2 was also reported to negatively regulate inflammation and immunity by supporting anti-inflammatory signals in response to the TLR2 activated TH1 response [68, 69]. Mutations associated with Crohn’s disease prevent the anti-inflammatory response mediated by the cytokines IL-10 and TGFβ, which might explain the excessive inflammatory response of these patients [68, 69].
CLAN functions as a direct activator of pro-caspase-1 in response to several PAMPs, including PGN, LTA and LPS [70–72]. Similarly to Nod2, CLAN expression also protects against viable Salmonella enteritidis and S. typhimurium by invasion assay in a caspase-independent manner, and induces caspase-1-independent apoptosis in response to Salmonella infection [70–72]. Fig (1B).
PYRIN domain containing proteins in inflammation and immunity
Activation of PAN-family proteins requires the leucine-rich region-dependent recognition of intracellular pathogens, which leads to an association with, and oligomerization of, the central PYD-containing adaptor protein ASC. This oligomerization is required for the formation of inflammasomes, which mediate activation of the pro-inflammatory caspases-1 and -5. ASC is the only identified adaptor protein, and is recruited to inflammasomes by PYD-PYD interaction. ASC then links pathogen recognition to the activation of downstream effectors of the innate immune response, which include activation of caspase-1-mediated processing of bioactive IL-1β IL-18 and probably IL-33, and activation of the transcription factor NF-κB. ASC−/− mice are defective in caspase-1 activation and the subsequent maturation of IL-1β and IL-18 in response to Gram negative and Gram positive pathogens, and are resistant to endotoxic shock [71, 73–76]. These results suggest that ASC is recruited to inflammasomes that are activated by PAMPs derived from Gram negative and Gram-positive pathogens. Recent progress has been made in defining the ligands recognized by PAN-family proteins. The NAC-containing inflammasome assembles and mediates activation of pro-caspase-1 and pro-caspase-5 in response to PGN, indicating that NAC might participate in PGN recognition [9, 77]. Pro-caspase-5 is directly recruited by the CARD of NAC, whereas pro-caspase-1 is recruited to the inflammasome via ASC, and both caspases are necessary to fully initiate IL-1β processing and secretion [9]. A similar observation has been made previously for murine caspase-1 and caspase-11 (the caspase-5 orthologue), which can form hetero-oligomers that are required for IL-1β and IL-18 processing and secretion [78–81].
Cryopyrin (CIAS-1, NALP-3, PYPAF-1) and PAN1 (PYPAF2, NALP2, NBS1, CLR19.9) also form inflammasomes upon recruitment of ASC and TUCAN (CARD8, Cardinal, NDPP1, KIAA0955), which results in the activation of pro-caspase-1, but not pro-caspase-5 [82]. The Ligand for PAN 1 is still elusive, but several ligands have been identified that are recognized by cryopyrin [73, 75, 83, 84]. Macrophages from cryopyrin−/− mice are deficient in the recognition of gout associated monosodium urate (MSU) or calcium pyrophosphate dihydrate (CPPD) crystals [84], bacterial RNA, CpG DNA, and the imidazoquinoline compounds R837 and R848 [73, 83], as well as LPS, Pam3CSK4, HKLM, PGN, and LTA in the presence of ATP [73, 75]. In contrast to macrophages from ASC−/− mice, macrophages from cryopyrin−/− mice show impaired caspase-1 activation only in response to infection by Gram positive bacteria [73, 74].
Besides activation of inflammatory caspases, PAN-family proteins are also indicated in the regulation of the pro-inflammatory transcription factor NF-κB. However, this has not yet been confirmed in vivo. Activation of this pathway depends upon recruitment of ASC to the inflammasome, which then bridges to the NF-κB activating IκB kinase (IKK) complex, and expression of dominant negative IKKβ (IKK2), or IKKγ (Nemo, FIP3) abolishes activation of NF-κB by PAN-family proteins [5, 85, 86]. However, the exact pathway leading from ASC to the IKK complex is still elusive, but might include caspase-8 [87].
Several PYD containing proteins, including ASC, cPOP1, PAN1, PAN2, and cryopyrin, have also been shown to block activation of NF-κB, which is mediated by the PYD [28, 88–91]. The NF-κB inhibitory role of PYDs has also been mapped to the IKK complex, suggesting that the PYD is capable of either activating or blocking the IKK complex. The effect of the PYD on the IKK complex, and subsequently on NF-κB activation, might be cell type-specific and might depend on which signals initiate activation of NF-κB. It appears that in situations, where NF-κB is activated by pro-inflammatory mediators, PYDs are able to block activation, whereas when signals are initiated by PAN-family proteins, PYDs mediate activation of NF-κB [22, 89, 91]. This observation is reminiscent of Nod2, which can activate NF-κB in response to MDP, but blocks NF-κB activating signals originating from TLR2 [68, 69].
PAN-family proteins together with ASC assemble inflammasomes, and several other PYD-containing proteins potentially function in the regulation of this complex. PAN1, PAN5, and the PYD-only protein cPOP1 function as inhibitors of PAN-family-induced activation of NF-κB, most likely by competing with the recruitment of the essential adaptor ASC to activated PAN-family proteins [92, 93]. This mechanism enables cells to fine-tune their responses. Expression of most PYD proteins is regulated by inflammatory or apoptotic mediators, and a well-balanced expression of these proteins appear to be critical to determine the cellular response.
Pyrin was first linked to the inflammatory response by the initial observation that it is expressed as a pro-inflammatory immediate-early response gene in monocytes in response to interferon (IFN)α, IFNγ, TNFα, and LPS, but expression is suppressed by the anti-inflammatory cytokines IL-4, IL-10, and TGFβ [94]. However, a contradictory analysis of pyrin expression in primary mouse macrophages identified pyrin as a protein induced in an NF-κB-dependent manner primarily by anti-inflammatory cytokines, and this finding is more consistent with its proposed function as a negative regulator of PYD signaling pathways [32, 95]. Furthermore, macrophages from pyrin−/− mice display increased caspase-1 activity and IL-1β secretion, and are more susceptible to septic shock [32, 95]. The adaptor ASC is also inducibly expressed in monocytes, macrophages and neutrophils in response to pro-inflammatory stimuli, and ASC−/− mice show impaired responses to experimental-induced septic shock [71, 76, 96, 97]. Inducible expression in response to inflammatory stimulation of cells was also observed for the PAN-family proteins cryopyrin, PAN1, PAN6, and PAN7 [90–92, 98].
Viral (v) homologs of the cellular PYD-only protein cPOP1 are encoded by several poxvirus strains, including the myxoma virus (MV, M013L), the Shope fibroma virus (SFV, gp013L), the swinepox virus (SPV, SPV14L), the Yaba-like disease virus (YLDV, 18L), and the Mule deer poxvirus (DpV, DPV83gp024) [18, 99, 100]. cPOP1 controls recruitment of the essential adapter ASC to PAN-family proteins, but also directly blocks IKK activation, and represents an endogenous dominant negative regulator [28, 101]. Poxviruses are known to hijack cellular proteins that are linked to inflammation, immunity and apoptosis, for immune evasive strategies [102]. The observations that poxviruses encode immune evasive proteins that target the PYD-signal transduction pathway, implies that viral PAMPs likely function as ligands for some PAN-family proteins, and emphasize the importance of the PYD signal transduction pathway for host defense.
Taken together, these findings suggest that PAMPs that are recognized by TLRs can also activate PAN-family proteins. However, activation of TLRs results in recruitment of TIR-containing adaptor proteins that link preferentially to the activation of MAPK and NF-κB, whereas PAN-family proteins recruit the adaptor protein ASC, which result mainly in activation of inflammatory pro-caspases-1 and -5, and PAMP recognition by both systems is required to mount an efficient immune response [73, 75].
TUCAN, an inflammasome-associated protein
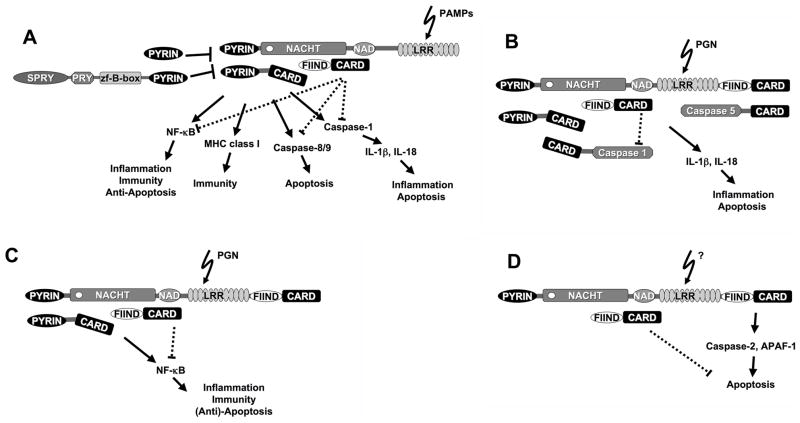
TUCAN contains a FIIND and CARD domain that resembles the C-terminal region of NAC, and is a component of some inflammasomes. ASC is recruited to inflammasomes by PYD-PYD interaction, whereas TUCAN associates with the NACHT domain of NAC, PAN1, and cryopyrin. Recruitment of TUCAN to NAC is not mediated via FIIND-FIIND or CARD-CARD association, as one would expect. However, this may enable the recruitment of TUCAN to inflammasomes containing cryopyrin, PAN1, and perhaps other PAN-family proteins, which do not encode these domains. In fact, the CARD of TUCAN cannot functionally compensate for the lack of the CARD in most of the PAN-family proteins. Fig (1A). The CARD in NAC recruits pro-caspase-5, but inflammasomes that contain cryopyrin or PAN1, although they contain TUCAN, do not recruit and activate pro-caspase-5 [77, 82]. TUCAN appears to be dispensable for caspase-1 activation by the inflammasome, and has been reported to suppress activation of this caspase [82, 103].
TUCAN associates with IKKγ and recruitment of TUCAN could provide a link of the inflammasome to the activation of NF-κB [103, 104]. Although TUCAN has been reported to block activation of the IKK complex, and subsequently activation of NF-κB, recruitment to the activated inflammasome might convert TUCAN from an inhibitor to an activator of NF-κB, reminiscent of ASC, but no direct evidence yet supports this hypothesis. A similar effect on NF-κB activation has been observed for DRAL, a TUCAN-associated protein [105]. On the other hand, recruitment of TUCAN could also be a mechanism to fine-tune the activity of inflammasomes. Fig (2). Because TUCAN can also bind the CARD of pro-caspase-9, it could potentially also link inflammasomes to the apoptotic machinery [103, 106, 107].
Figure 2. Models for the function of PAN-family proteins.
Signaling pathways involving PAN-family proteins. (A) In response to certain PAMPs, PAN-family proteins are activated (see Fig. 3 below), which results in the recruitment of adapter proteins by PYRIN domain interaction. To date ASC is the only identified adapter protein that is recruited to these PAN-family protein complexes. Activated downstream effector pathways include NF-κB, MHC class I gene expression, and caspases that are either linked to apoptosis or inflammation. Some PAN-family proteins also recruit TUCAN (CARD8, Cardinal, NDPP1, KIAA0955), a protein implicated in the negative regulation of NF-κB, caspase-1, and caspase-9. However, it is feasible that in response to ligand-dependent PAN-family protein activation, TUCAN might be converted from an inhibitor to an activator of these pathways. Alternatively, TUCAN might provide a mechanism to fine-tune responses elicited by activated PAN-family proteins. (B) NAC (CARD7, DEFCAP, NALP1, CLR17.1) represents an exception, as it also contains a CARD, which allows direct recruitment of pro-caspase-5 by CARD-CARD interaction (D) or links to the apoptotic pathway via pro-caspase-2 and APAF-1. (C) Recruitment of ASC could also link NAC to NF-κB activation.
The PYRIN domain in Apoptosis
Many of the PAN-family-related proteins are also implicated in the regulation of apoptosis, including APAF-1, Nod1, Nod2, Clan, NAIP, CIITA, and R genes [48, 50, 70, 108–115]. Also activated TLRs are capable of linking to the apoptotic machinery [116]. The PAN-family member NAC has initially been characterized as a pro-apoptotic protein that interacts with APAF-1 by CARD-CARD association, and is required for cytochrome c-dependent recruitment of pro-caspase-9 to the apoptosome [117]. Expression of NAC triggers caspase activation and subsequent initiation of apoptosis. This process can be abrogated by co-expressing either the CARD or the NACHT of NAC, while CARD-LRR represents a constitutively active apoptosis-inducing mutant [118]. NAC expression also induces cell death in cerebellar granule neurons, and was identified as a caspase-2 activator [118, 119]. These observations predict an interesting model, where PAN-family members are involved in several large protein complexes that are linked to the activation of CARD-containing caspase zymogens. These include the caspase-1 and caspase-5 activating inflammasomes, the caspase-2 activating PIDDosome, and the caspase-9 activating apoptosome [8, 9, 11, 48, 108, 117, 118]. As a result activation of one protein could result in the initiation of an inflammatory response, and if sustained, even provide a mechanism to directly eliminate infected cells. Whether other PAN-family members are also able to modulate inflammatory and apoptotic signals will need further investigation, but initial evidence suggests that cryopyrin is also involved in apoptosis [120].
ASC, the other essential component of the inflammasome, has also been implicated in regulation of apoptosis by interfering with either proteins that are crucial for activation of caspases, or directly with caspase zymogens. Caspase-1 is linked to apoptosis by activating effector caspase zymogens, preferentially under pathological conditions [121]. Therefore, ASC also contributes to caspase-1-mediated apoptosis in activated macrophages, and macrophages from ASC−/− mice display impaired caspase-1-mediated apoptosis in response to Salmonella typhimurium infection [71, 122–126]. The activation of pro-caspase-1 by ASC in mammals is dependent on the CARD. In Danio rerio (zebrafish), zASC is also directly involved in the activation of zcaspy, a pro-apoptotic zebrafish caspase. Both, zcaspy and zcaspy2, encode a PYD in their prodomain, only zcaspy interacts with ASC, and activation is dependent on ASC-mediated oligomerization and speck recruitment [44]. Both caspases show distinct substrate specificity. Zcaspy shows highest activity towards YVAD (a caspase-1 substrate) and zcaspy2 towards WEHD (a caspase-5 substrate), which are also the closest human orthologues [44].
ASC was originally identified as a pro-apoptotic, Triton X-100 insoluble protein in HL60 cells, and simultaneously as target of methylation-induced silencing (TMS1), and mediator of caspase-9-dependent apoptosis [26Conway, 2000 #795, 127]. In response to DNA-damaging anti-cancer drug-treatment of HL60 cells, ASC aggregates into specks, which are characteristic perinuclear protein aggregates. Furthermore, anti-sense-mediated knock-down of ASC expression impairs DNA-damage-induced apoptosis. ASC is inducibly expressed as a p53-response gene following DNA damage and p53 activation, and facilitates Bax translocation to mitochondrial membranes [4]. An unconventional, non DDF-interaction between the PYD of ASC and Bax is responsible and required for cytochrome c release, loss in membrane potential and activation of caspases-2, -3, and -9, suggesting that ASC represents an essential adaptor in the p53-mediated DNA damage response [4]. ASC, in collaboration with Clan, has also been implicated in the activation of pro-caspase-8, which requires a heterophilic interaction between the PYD of ASC and the DED of pro-caspase-8, an interaction that can be disrupted by pyrin [5]. ASC also associates with cryopyrin to induce apoptosis, and this complex formation can also be prevented by pyrin [120]. Pyrin prevents ASC-induced apoptosis by increasing the number of speck-positive cells, indicating that speck-formation does not predispose cells to undergo apoptosis [26, 128]. However, pyrin−/− macrophages are characterized by an impaired, rather than exaggerated apoptotic response, though the mechanism is still elusive. However, these are not pyrin null macrophages, and the pyrin gene was only disrupted. Therefore the observed effect could have resulted from a dominant negative effect of the PYD of pyrin, which is still expressed in these macrophages. The PYD could interfere with the interaction between ASC and caspase-8 or Bax [4, 5]. cPOP1 has also been implicated in the induction of apoptosis, which provides the most direct link between the PYD and apoptosis, because cPOP1 is only composed of a PYD [28, 129].
Signaling by HIN-200 proteins
HIN-200 proteins respond to interferons (IFNs) with an increase in expression [39]. These proteins are primarily nuclear proteins that are involved in transcriptional regulation of genes linked to cell cycle regulation, differentiation, tumorigenesis, and apoptosis, including NF-κB, AP-1, p53, E2F-1/DP-1, E2F-4/DP-1, MyoD, YY1, UBF1 [40, 41]. HIN-200 proteins also bind to members of the retinoblastoma tumor suppressor, resulting in decrease of cell growth via reduced G1-S transition. Some family members oligomerize, and this association is dependent on their PYD [130, 131]. IFI16, which contains two HIN-200 repeats and its murine homologs p202 and p204, appear to be involved in the terminal differentiation of myeloid cells. Expression of IFI16 induces growth arrest, inhibits colony formation and is associated with senescence [132]. IFI16 associates with p53 and impairs the transcriptional activity and stability of p53 [133, 134]. IFI16 dimerizes via PYD association, and has the ability to bind to DNA and function as a transcriptional repressor [135]. Absent in melanoma 2 (AIM2) contains a single HIN-200 repeat, localizes primarily to the cytoplasm, and is involved in suppression of tumorigenicity in melanoma, by inhibiting cell proliferation and induction of apoptosis [136, 137]. This observation correlates with its down-regulation in cancers [138, 139]. The most recently identified protein, IFIX, functions as a tumor suppressor in mammary epithelial cells by reducing cell proliferation and tumorigenicity in nude mice [42]. IFIX activates the cyclin-dependent kinase inhibitor p21CIP1, and is silenced in mammary tumors [42]. Expression of myeloid cell nuclear differentiation antigen (MNDA), which only contains one HIN-200 domain, and its murine homolog p205, is restricted to maturing myeloid cells and correlates with the differentiation status [140]. It stimulates the DNA binding activity of the transcription factor YY1, but also displays growth suppressive potential [141]. No direct evidence links the HIN-200 family members to inflammasomes or other innate immune responses.
THE PYRIN DOMAIN AND DISEASE
Hereditary mutations of PYRIN domain containing proteins and the link to autoinflammatory disorders
The physiological importance of PYD signal transduction pathways is emphasized by the close link of hereditary mutations in several PYD-containing proteins to autoinflammatory disorders. Autoinflammatory syndromes are systemic disorders, which are characterized by unprovoked and self-limited inflammation in the absence of high titer autoantibodies or antigen-specific T-cells [142]. Hereditary mutations in PYD containing proteins cause periodic fever syndromes, a type of autoinflammatory disorders that is defined by self-resolving and recurrent attacks of generalized inflammatory reactions, though the clinical symptoms are not based on infections or autoimmune reactions [143].
Hereditary mutations in the pyrin gene (marenostrin, MEFV) are linked to Familial Mediterranean Fever (FMF), a recessively inherited disease that was originally referred to as fever of unknown origin [144]. FMF is characterized by episodic fever peaks, which last up to three days and are usually followed by attack-free periods of several years. The FMF causing gene was mapped to the short arm of chromosome 16 and cloned in 1997 [30, 31]. More recently, pyrin was found to interact with the proline serine threonine phosphatase-interacting protein 1 (PSTPIP1), a cytoskeletal organizing protein, which when mutated, causes pyogenic arthritis, pyoderma gangrenosum, and acne (PAPA) syndrome [145, 146]. This observation identifies a common pathway as a cause for both disorders. Most FMF-associated mutations localize to the PRY/SPRY domain of pyrin, whose function is still elusive. Mutated PSTPIP1 shows increased association with pyrin, which correlates with the phosphorylation status of PSTPIP1. PBLs from PAPA patients secrete increased levels of IL-1β, suggesting that the enhanced interaction between mutated PSTPIP1 and pyrin prevents pyrin from properly regulating PYD signal transduction pathways [146]. It was suggested that pyrin-PSTPIP1 complexes might regulate inflammasome-cytoskeletal organization, eventually affecting activity of inflammasomes [147].
Cryopyrin (cold-induced autoinflammatory syndrome 1, CIAS1) is another PYD-containing gene, with which hereditary mutations are linked to three distinct periodic fever syndromes: familial cold autoinflammatory syndrome (FCAS), which is also known as familial cold urticaria (FCU), Muckle-Wells syndrome (MWS), and chronic infantile neurologic cutaneous and articular syndrome (CINCA), also referred to as neonatal-onset multisystem inflammatory disease (NOMID) [148–151]. Hereditary mutations in the PAN-related Nod2 gene are also linked to autoinflammatory disorders, including Crohn’s disease, a form of inflammatory bowel disease (IBD) and Blau syndrome (BS) [152–154].
The majority of mutations affect amino acid residues inside the NACHT domain of cryopyrin, resulting in a constitutively active protein, which spontaneously associates with ASC and initiates activation of downstream effectors [155]. This hyper-activity might explain the inflammatory component of these periodic fever syndromes. Mutations in pyrin that are associated with FMF, such as the common M694V mutation, might prevent pyrin from negatively regulating cryopyrin signaling, which also promotes aberrant inflammation in homozygous or compound heterozygous patients [30, 31]. BS-linked Nod2 mutations also affect residues in the NACHT domain, resulting in uncontrolled signaling, and perhaps association with the downstream adaptor Cardiak [154, 156]. In contrast, IBD-linked Nod2 mutations directly affect the LRRs, or residues close to the LRR domain, resulting in a defective response to muramyl dipeptide and impaired activation of downstream effectors [66, 152, 153, 157]. IBD patients suffer from inflammation, which might be caused by the impaired ability of Nod2 to sense infections and initiate secondary inflammatory responses.
A common mechanism of periodic fever syndromes appears to be aberrant activation of inflammatory effector pathways. This model is supported by the observation that disease-linked hereditary mutations in cryopyrin spontaneously associate with ASC and constitutively activate downstream pathways, without requirement for ligand recognition [82, 155]. Most hereditary mutations in PYD proteins affect activation of pro-caspase-1. Aberrant processing of pro-IL-1β appears to be the cause for many of these hereditary fever syndromes, though the cause of the periodic fever episodes is still unknown. Yet, this observation finally provided an efficient treatment for affected patients [158]. Application of a recombinant human IL-1 receptor antagonist, Kineret® (Anakinra), which is widely administered in patients with rheumatoid arthritis, proved to also be successful in patients with periodic fever syndromes [146, 159–165]. Phase II clinical trials are underway to test the efficiency of a recombinant fusion protein that contains the extracellular chains of both IL-1 receptors, IL-1RI and IL-1RacP, to mediate high-affinity IL-1β binding (IL-1 Trap) in patients with autoinflammatory disorders [166, 167].
A model for PAN activation in immunity and inflammation
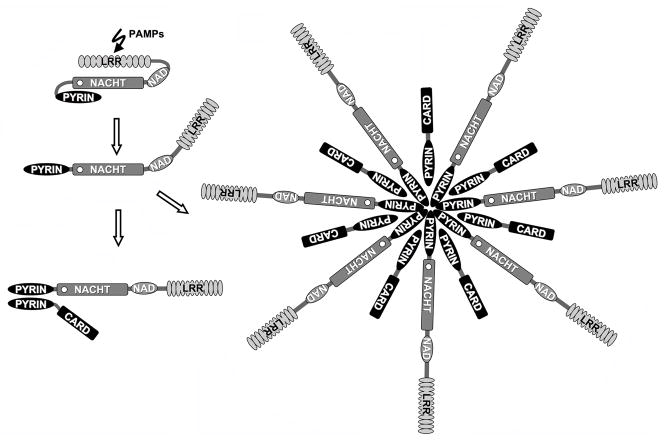
Little is known about the mechanism by which PAN-family proteins recognize PAMPs to initiate inflammasome assembly and activation of downstream effectors. Activation of PAN-family proteins might occur reminiscent of APAF-1 [168]. In this model, the LRRs could function as an auto-inhibitory domain similar to the WD40 repeats of APAF-1, where cytochrome c triggers a conformational change which results in exchange of the constitutively bound ADP for ATP. Hydrolysis of ATP induces another change in conformation that favors oligomerization and releases the CARD, which is required for recruitment of pro-caspase-9 [169]. Potential NTP-dependent oligomerization mediated by the NACHT domain of PAN-family proteins could require dislodging the LRRs from the NACHT domain in a ligand-dependent manner. Fig (3A). Oligomerization might then provide a scaffold for the recruitment of other PYD-containing proteins, such as ASC by PYD-PYD interaction to connect to downstream effectors. In an alternative mechanism, PAN-family proteins might function similarly to the monomeric R-gene product Rx of Solanaceae, in which the NB-ARC domain interacts simultaneously with the LRR and coiled coil (CC) domains [47, 170]. Fig (3B). This model is supported by the observation that the LRRs of NAC and cryopyrin appear to keep the inflammasome inactive, because overexpression of NAC or cryopyrin lacking LRRs, but not full-length proteins, induce activation of effectors [9, 92, 155]. Also several other PAN-family proteins are more potent in activating effector pathways in the absence of their LRR domain [5, 86, 120]. Ligands might be directly recognized and bind to the LRRs, or indirectly release LRR-mediated NACHT domain inhibition. Although LPS is not sensed by Nod1, but it has rather been the LPS-contaminating PGN, a study of Nunez and colleagues demonstrated direct binding of [3H]-labeled LPS (PGN?) to Nod1, providing first evidence for a potential direct interaction [171]. There is also evidence that at least some R proteins interact directly with PAMPs [172, 173].
Figure 3. A proposed model for the activation of PAN proteins.
PAN proteins probably exist as inactive proteins. This status is most likely mediated by the LRR domain, which presumably blocks access to the NACHT domain. This conformation might resemble APAF-1 and R proteins. The effector domain (PYRIN domain) is tightly packed against the NACHT domain, and is therefore inaccessible for binding partners. Ligand binding could release inhibition by the LRR domain, and results in unfolding of these domains. A requirement for NTP binding and hydrolysis is anticipated, but has not been demonstrated. Once unfolded, the PYRIN domain is able to recruit adapter proteins, such as ASC. Activation of downstream effectors is initiated by the recruitment of ASC-linked effectors, as described for the R-gene product Rx of Solanaceae [47, 170]. However, an alternative mechanism could be the oligomerization of PAN-family proteins, which may activate effectors by the enhanced dimerization mechanism, as described for APAF-1 [169].
The recruitment of ASC to PAN-family proteins appears to be dependent on their prior activation by PAMPs. Common hereditary mutations in cryopyrin that are linked to autoinflammatory disorders, spontaneously associate with ASC and activate effector pathways, even in the absence of an inducing ligand, and TUCAN is also efficiently recruited to the active inflammasome [82, 155]. Direct interaction of TUCAN with the NACHT domain of these PAN-family proteins can be detected following deletion of either the LRR, the PYD, or both domains of PAN proteins [82]. Increasing evidence also supports NACHT domain-dependent oligomerization, because the NACHT of CLAN is capable of interacting with other NACHT domains, including domains from cryopyrin, NAC, PAN2, NAIP, Nod1, and Nod2 [174]. In addition, CLAN interacts with Nod2 following treatment of monocytes with PGN, suggesting that NACHT domains form signaling networks that modulate activation of NF-κB and pro-caspase-1 [174]. Several lines of evidence suggest that PAN-family proteins are recruited to high molecular weight complexes that might support activation according to model A. However, NTP binding and hydrolysis by PAN-family proteins has not yet been demonstrated.
In agreement with the observation that deletion of the LRRs from PAN-family proteins renders them constitutively active, PAN5 (PYNOD, NALP10, NOD8, CLR11.1), the only PAN-family protein that lacks LRRs, associates with ASC and displays a constitutively active phenotype [93]. Fig (1). However, this protein does not activate downstream effectors, but rather functions as an inhibitor of the PYD signal transduction pathway. Association with ASC blocks activation of NF-κB and caspase-1 by cryopyrin, reminiscent of pyrin, suggesting that PAN5 competes with cryopyrin and perhaps other PAN-family proteins for binding to the adaptor ASC. Pyrin has a different domain organization than PAN-family proteins, but the observation that PAN5 blocks PYD signaling, indicates that not all PAN-family proteins are necessarily activators of innate immune effector pathways, and that their balance might ultimately determine activation of downstream effectors. PAN2 was the first identified family member that displays a dominant negative effect, and the PYD of this protein is responsible for inhibiting the kinase activity of the IKK complex and subsequent activation of NF-κB [89]. Also cryopyrin and PAN1 block activation of NF-κB, while PAN7 blocks activation of caspase-1 [90–92].
One likely explanation for this effect could be that these proteins block activation of downstream effectors, unless they are activated by their respective ligands. Ligand recognition might convert these proteins from inhibitors to activators. However, PAN5 is expected to retain the inhibitory function, because of the lack of LRRs that could sense PAMPs. Further support for this theory is provided by the indication that enforced expression of PAN-family proteins together with ASC triggers activation of downstream effectors, even in the absence of a ligand, indicating the potential of these proteins to activate pathways. Cryopyrin and PAN1 collaborate with ASC to activate pro-caspase-1, however, only cryopyrin, but not PAN1 mediates NF-κB activation in collaboration with ASC, suggesting the existence of additional regulatory mechanisms, and that sole recruitment of ASC might not be sufficient for effector activation [86, 88, 90, 97].
PAN2, as well as other PAN-family proteins that do not recruit the adaptor ASC, have not been demonstrated to activate downstream signal transduction pathways. However, ASC recruitment could depend upon activation of the PAN-family protein by their respective ligand. The important question, whether there exist additional proteins that can function as an adaptor for PAN-family proteins, is still not solved. Alternative adaptors could link to known pathways, or could potentially connect activation of PAN-family proteins to novel effectors. So far, the inability of at least PAN2, PAN7, and PAN10 to recruit ASC have been reported, but not all PAN-family proteins have been tested [89, 175]. Therefore, at least these PAN-family proteins can be expected to require another signaling adaptor, reminiscent on the different adaptors that are recruited to TLRs and R proteins [176–178]. In R proteins, the recruited adaptor protein is dependent on the effector domain, such as coiled coil or TIR (Toll/interleukin-1 receptor domain), and TLRs recruit at least five adaptor proteins that all depend on the TIR [176]. Grim-19 has been recently identified as an adaptor that can be recruited to Nod2, in addition to Cardiak, suggesting that also CARD containing Nod proteins utilize different adaptor proteins to transmit signals [179]. Also, the possibility of structural aspects regulating PYD-PYD interactions and their relevance in ASC recruitment will be discussed below. There is no other gene encoded in the human genome having a structure comparable to ASC. The only other gene with a PYD-CARD combination is NAC, but this protein appears to function as a PAN-family protein, and not as an adaptor protein [9]. These observations support a complex regulation of PYD-containing proteins and recruitment of ASC does not necessarily automatically determine activation of all known downstream effector pathways.
Silencing of PYRIN domain-containing proteins in cancer
ASC was originally identified as a gene silenced by promoter methylation in breast cancers [27]. Subsequently, silencing of ASC was also identified in lung-, ovarian-, hepatocellular-, and colorectal cancers, melanomas, and glioblastomas, suggesting ASC gene silencing as a common mechanism of carcinomas [27, 129, 180–184]. Significantly, one report exists that states a decrease in ASC expression correlates with the progression from astrocytic astrocytoma (stage III) to glioblastoma (stage IV) [181]. Silencing of tumor suppressor genes by methylation is a frequent mechanism of cancer cells to escape growth control and apoptosis. Also the promoter of cPOP1 was found to be methylated in hepatocellular carcinomas, and cPOP1 expression was demonstrated to have growth suppressive and pro-apoptotic effects [129]. This observation strongly suggests that the PYD of ASC might be the domain responsible for ASC silencing in carcinomas.
ASC and cPOP1 might be silenced to prevent their effects on signal transduction pathways, and both have been implicated to promote apoptosis [26, 27, 129]. Because ASC is important for DNA damage-induced apoptosis, loss of its expression might also affect efficiency of anti-tumor therapy. However, both are also capable of blocking the kinase activity of IKK complex-associated kinases to activate NF-κB [28, 88]. This might contribute to the observation that tumor cells frequently display high and constitutive NF-κB activity, which promotes proliferation, invasiveness and inhibition of apoptosis [185].
HIN-200 proteins are also linked to cancer, because chromosomal instability in their genomic loci is tightly linked to tumorigenesis, and these proteins areinvolved in regulation of cell growth and differentiation [39]. AIM2 is silenced by promoter methylation in response to immortalization of fibroblasts, whereas IFIX and IFI16 expression is lost in most breast cancers, and restored expression reduces tumorigenesis [42, 138, 186]. MNDA silencing correlates with the progression of metastatic prostate cancer [187]. The growth retarding potential of the HIN-200 domains appear to be related to their interaction with multiple transcription factors and their association with p53 and Rb [39].
STRUCTURE OF THE PYRIN DOMAIN
The Death Domain Fold
DDFs are usually characterized by an antiparallel arrangement of six-α-helical bundles with Greek-key topology and internal pseudo-twofold symmetry, but the length of the helices, the angles between individual helices, and the length of the linker region vary between subfamilies [188–190]. The X-ray crystallography structure of two DDF complexes, the CARD-CARD complex of APAF-1 and pro-caspase-9 and the DD-DD complex consisting of Pelle and Tube has been solved and is used to predict other DDF associations [191, 192]. Both dimers are based on different modes of interaction. The CARD-CARD interaction is strongly electrostatic, but also involves van der Waals interactions between hydrophobic residues of the core. A 50-degree bend in helix 1 of APAF-1 and pro-caspase-9 separates this helix into two smaller helices H1a and H1b. The strong dipolar surface charge is critical for this interaction. Helices 2 and 3 of APAF-1 contain several negatively charged residues that form a convex surface, which is recognized by the concave surface formed by helices 1a/b and 4 of pro-caspase-9 [191, 193]. This interaction is referred to as type 1, as opposed to the type 2 association observed for the DD-DD interaction of Pelle and Tube [194]. These DDs do not display the dipolar surface and association is mediated by a combination of charged, polar, and hydrophobic interactions involving two contact sites. Helix 4 of Pelle interacts with a groove formed by helices 1, 2, and 6, and the C-terminus of the DD of Tube binds to a groove of Pelle formed by helices 2, 3 and 4, 5 [192]. The DED of FADD on the other hand, is proposed to mediate interaction mainly via a hydrophobic interface [195]. Based on this model, other DDF complexes have been generated by homology modeling, including pro-caspase-2 and RAIDD and pro-caspase-1, Iceberg, and Rip2, which occur by electrostatic interaction between the dipolar surfaces [34, 196].
Although experimental proof exists only for DDF dimers, computer modeling predicted a possible trimeric complex. This type 3 interface was identified by superpositioning the APAF-1/pro-caspase-9 type 1 and the Pelle/Tube type 2 complexes [194]. This trimeric arrangement is also supported by mutagenesis data derived from Fas/FADD and TNFR/TRADD DD interactions [197–200]. Modeling NMR data available for the DD of Fas and FADD, predicted all three possible associations that would assemble a heterotrimeric complex, which was further predicted to result in the formation of a higher-order hexameric DISC complex [201, 202]. The formation of so called death filaments, resulting from over-expression of DEDs, is thought to occur by favored oligomerization via type 1 or type 2 interfaces, resulting in an extended homo oligomer, rather than a hetero oligomeric globular structure derived from a type 3 interface [194, 203].
The PYRIN Domain Fold
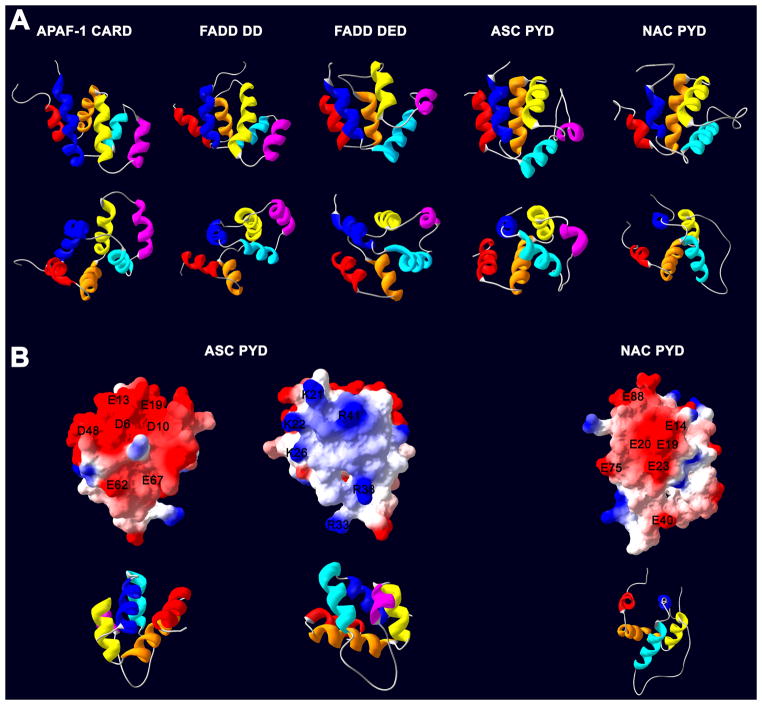
The NMR structure of two PYDs has been determined recently, and the backbone NMR assignment has been reported for another PYD [204–206]. All structures revealed close similarity to other DDFs, as predicted previously [14–18]. Fig (4A). The PYD of ASC displays a rather short helix 3 preceded by an unusually long loop region, and also the NMR assignment of cPOP1 predicts a similar fold [206]. Most surprisingly, the solved structure of the PYD of NALP1 suggests that helix 3 is not structured, but contains instead an unstructured, flexible loop [204]. The co-crystal structure for a PYD-PYD interaction has still not been obtained, which would be required to determine the heterodimeric structures of PYD complexes to identify the exact mode of oligomerization. The strong dipole present in the PYD has been predicted to form type 1 interactions [207]. Fig (4B). Acidic residues in helices 1 and 2 of NALP1 form a convex surface that can be predicted to fit onto the concave, basic surface of ASC, which is formed by helices 2 and 3. Fig (4B). However, since helix 3 is critically involved in a type 1 interaction, simple homology modeling, based on other DDFs, might not be possible.
Figure 4. Structure of the PYRIN domain.
(A) Schematic ribbon drawing of the average structure of the PYRIN domains of ASC (PDB 1UCP) [205], and NAC (PDB 1PN5) [204]. As comparison, structures of the CARD domain of APAF-1 (PDP 1C15) [193], the death domain of FADD (PDB 1FAD) [210], and the death effector domain of FADD (PDB 1A1Z) [195] are included. The lower panel represents a 90-degree rotation about the horizontal axis. Helix 1 is colored dark blue, helix 2 in light blue, helix 3 in cyan, helix 4 in yellow, helix 5 in orange and helix 6 in red. Note that also helix 3 in NAC is replaced by a flexible loop, and also helix 3 in ASC is rather short and contains some unstructured regions. (B) Representation of the solvent-accessible surface of the PYRIN domain of human ASC (left) [205] and human NAC (right) [204]. The electrostatic potential is colored in red (negative) and blue (positive), and the amino acid residues are indicated. The orientation of the α-helices is depicted below in a ribbon representation. Note, that the positively charged surface of ASC could potentially fit the negatively charged surface of NAC by charge and shape. Figures were produced using the published PDP codes with the program Swiss-PDB Viewer.
Structural implications for PYRIN domain-mediated signal transduction
The solved PYD structures provided valuable mechanistic explanations for the biological relevance of helix 3. A structured helix 3 is ultimately required for the known type 1 and type 2 interactions, and the observation of an unstructured or partially structured, but short region corresponding to helix 3, is highly significant. Unstructured regions in globular proteins are often involved in a conformational switch, and it could be speculated that local folding and unfolding of this region might regulate PYD-PYD interactions. Initial contact at other sites could induce structuring of helix 3, which might then be required for high-affinity interactions. Another possibility is that phosphorylation of specific residues in the PYD could be a mechanism to increase the helical propensity in this region, resulting in structuring of helix 3 and induction of specific PYD interactions, and could represent a mechanism to fine-tune these interactions and subsequently modulate signaling [28]. Furthermore, a potential type 3 interaction, where dimer formation could induce helix 3 intrinsic helical propensity, could result in a trimeric complex. One explanation might come from studies of the DD of Fas [208]. The V238N mutation of Fas, which is linked to the autoimmune lymphoproliferative syndrome type IA (ALPS), results in destabilization and local unfolding of helix 3, which impairs the Fas-FADD interaction and DISC formation [208]. Disruption of the van der Waals-mediated interaction between V238 of the hydrophobic core and hydrophobic residues of helix 3, lowers the already low intrinsic helical propensity in this region. This finding is significant in light of the observation that the common FMF mutation in the PYD of pyrin, R42W, localizes to the unstructured loop region corresponding to helix 3, next to a conserved basic surface patch [204]. Fig (5). It is therefore conceivable that W42 increases the intrinsic helical propensity sufficiently to induce local folding of helix 3, subsequently allowing association with other PYDs [209].
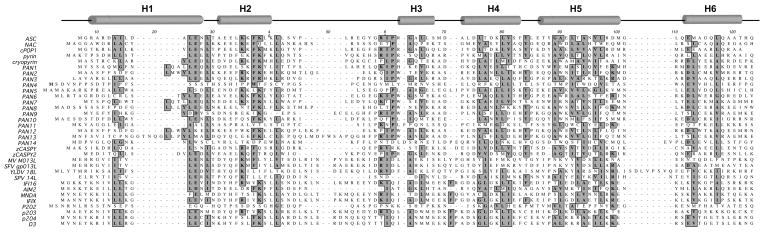
Figure 5. Amino acid sequence alignment of PYRIN domains.
Clustal W alignment of PYRIN domains, including viral POPs, murine HIN-200 family members p202, p203, p204, and D3, as well as zebrafish caspases. The α-helices, as defined for ASC, are marked on top (H1-6) [205]. Conserved residues are highlighted. Mainly residues of the hydrophobic core are conserved, but the linker distance is variable.
CONCLUSIONS AND FUTURE PERSPECTIVES
The recent demonstration that PAN proteins function as PRRs, thus complementing the TLR and Nod system is an exciting development which will increase our understanding of innate immune regulation and host defense. ASC is the central adaptor protein that is recruited to several PAN-family proteins to form the inflammasome to activate immune effectors, a step that is not well understood. More studies will be required to address some of the many remaining questions, such as, how inflammasome assembly is regulated, the exact mechanism of PYD-PYD interactions, or whether there exist other signaling adaptors for PAN-family proteins, similar to the TLR system. Most of the PAN-family proteins are still orphan receptors, and it can be expected that mutations could also be identified in other family members that could link to inflammatory and immune disorders. One of the big surprises has been the identification that mutations in different PYD-containing proteins affect the function of a single downstream effector, namely pro-caspase-1, which provided a molecular mechanism for periodic fever syndromes and treatment option for affected patients. Overall, in spite of the tremendous progress over the last years to decipher PYD-mediated signaling events, there remain many unanswered questions needed to sufficiently understand the physiological significance and complexity of PYD-containing proteins.
Table 3.
ASC-associated proteins.
Numerous proteins associate with ASC and are listed in this table. The method employed to detect this interaction is noted in column 3, as is the domain that was identified to mediate this interaction, and the resulting cellular response. co-immunoprecipitation assay (IP); in vitro pull-down assay (PD); yeast/mammalian two hybrid assay (2H); co-localization assay (L); (↑ increase; ↓ decrease; ↑ ↓ both is reported).
| Protein | Synonym | Domain | Method | Effect | Reference |
|---|---|---|---|---|---|
| ASC | TMS1, CARD5, PYCARD | PYRIN-PYRIN; CARD- CARD; PYRIN-CARD | IP, PD | NF-κB↓; apoptosis↑ | [26, 82, 97, 127, 211] |
| Bax | PYRIN-? | IP, PD, L | apoptosis↑ | [4] | |
| Cardiak | Rip2, Rick | CARD-CARD | IP, PD | Caspase-1↑ | [97] |
| caspase-1 | ICE | CARD-CARD | IP, PD, 2H, L | Caspase-1↑↓; | [85, 97, 212] |
| caspase-8 | MACH, FLICE, Mch5 | PYRIN-DED | IP, L | NF-κB↑; apoptosis↑ | [5, 87] |
| CLAN | IPAF, CARD12 | CARD-CARD | IP, 2H | NF-κB↑, Caspase-1↑ | [5, 85, 213] |
| cryopyrin | CIAS1, PYPAF1, NALP3, CLR1.1 | PYRIN-PYRIN | IP, L | NF-κB↑; Caspase-1↑; apoptosis↑ | [82, 86, 120] |
| IKK complex | PYRIN-? | IP, L | NF-κB↑↓ | [88] | |
| NAC | DEFCAP, CARD7, NALP1, CLR17.1 | PYRIN-PYRIN | IP, | Caspase-1↑ | [9] |
| cPOP1 | ASC2, ASCI | PYRIN-PYRIN | IP, PD, L | NF-κB↓; Caspase-1↑ | [28] |
| cPOP2 | PYRIN-PYRIN | IP, 2H | Caspase-1↓ | [C. Stehlik, unpublished] | |
| PAN1 | PYPAF2, NALP2, NBS1, CLR19.9 | PYRIN-PYRIN | IP, L | Caspase-1↑ | [82, 90] |
| PAN3 | PYPAF5, NALP6, CLR19.2 | PYRIN-PYRIN | L | NF-κB↑; Caspase-1↑ | [175] |
| PAN6 | PYPAF7, NALP12, Monarch-1, CLR19.3 | PYRIN-PYRIN | L | NF-κB↑; Caspase-1↑; MHC-I↑ | [85] |
| pyrin | marenostrin | PYRIN-PYRIN | IP, L | NF-κB↑↓; Caspase-1↓↑; apoptosis↓; specks ↑ | [5, 32, 120, 128] |
| zCaspy | PYRIN-PYRIN | IP, L | apoptosis↑ | [44] |
Acknowledgments
The author apologizes to those, whose work could not be cited due to space limitations, and recognizes the support provided by the National Institutes of Health (5P20RR016440, 1R21AI067680, 1R03AI067806), the Concern Foundation, the Art Ehrman Cancer Fund of the Fraternal Order of Eagles, and the Alexander Bland Osborn Cancer Center Endowment.
Abbreviations
- ALPS
autoimmune lymphoproliferative syndrome
- ASC
apoptotic speck-like protein containing a CARD
- BIR
baculovirus IAP repeat
- CARD
caspase recruitment domain
- Caterpiller (CLR)
CARD, transcription enhancer, R (purine)-binding, pyrin, lots of leucine repeats
- CIAS
cold-induced autoinflammatory syndrome
- CINCA
chronic infantile neurologic cutaneous articular
- CIITA
class II transactivator
- DAPIN
domain in apoptosis and interferon response
- DD
death domain
- DDF
death domain fold
- DED
death effector domain
- DISC
death-inducing signaling complex
- EST
expressed sequence tag
- FCAS
familial cold autoinflammatory syndrome
- GlcNAc-MurNAc
diaminopimelate-containing N-acetylglucosamine-N-acetylmuramic acid
- IAP
inhibitor of apoptosis
- FCU
familial cold urticaria
- FMF
familial mediterranean fever
- HIN-200
hematopoietic interferon-inducible nuclear proteins with the 200-amino-acid repeat
- IκB
inhibitor of kappa light polypeptide gene enhancer in B cells
- IKK
inhibitor of kappa light polypeptide gene enhancer in B cells kinase
- LPS
bacterial lipopolysaccharide
- LRR
leucine-rich repeat
- MWS
Muckle-Wells syndrome
- NACHT
NAIP, CIITA, HETE and TP1
- NAIP
neuronal apoptosis inhibitor protein
- NALP
NACHT leucine rich domain and pyrin-containing protein
- NBARC
nucleotide-binding, Apaf-1, plant R gene product
- NF-κB
nuclear factor kappa light polypeptide gene enhancer in B cells
- NOD
nucleotide-binding oligomerization domain
- NOMID
neonatal-onset multi-system inflammatory disease
- NTP
nucleotide triphosphate
- PAAD
pyrin, AIM (absent-in-melanoma), ASC and death domain-like
- PAMP
pathogen associated molecular pattern
- PAN
PAAD and NACHT
- PAPA syndrome
pyogenic arthritis, pyoderma gangrenosum, and acne syndrome
- PGN
peptidoglycan
- POP
PYD-only protein
- PRR
pathogen recognition receptor
- PSTPIP1
proline serine threonine phosphatase-interacting protein 1
- PYD
PYRIN domain
- PYPAF
pyrin-containing Apaf-1-like protein
- TIR
Toll/Interleukin-1 receptor domain
- TLR
Toll like receptor
References
- 1.Hill JM, Vaidyanathan H, Ramos JW, Ginsberg MH, Werner MH. Embo J. 2002;21:6494–6504. doi: 10.1093/emboj/cdf641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahn EY, Lim ST, Cook WJ, McDonald JM. J Biol Chem. 2004;279:5661–5666. doi: 10.1074/jbc.M311040200. [DOI] [PubMed] [Google Scholar]
- 3.Nam YJ, Mani K, Ashton AW, Peng CF, Krishnamurthy B, Hayakawa Y, Lee P, Korsmeyer SJ, Kitsis RN. Mol Cell. 2004;15:901–912. doi: 10.1016/j.molcel.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 4.Ohtsuka T, Ryu H, Minamishima YA, Macip S, Sagara J, Nakayama KI, Aaronson SA, Lee SW. Nat Cell Biol. 2004;6:121–128. doi: 10.1038/ncb1087. [DOI] [PubMed] [Google Scholar]
- 5.Masumoto J, Dowds TA, Schaner P, Chen FF, Ogura Y, Li M, Zhu L, Katsuyama T, Sagara J, Taniguchi S, Gumucio DL, Nunez G, Inohara N. Biochem Biophys Res Commun. 2003;303:69–73. doi: 10.1016/s0006-291x(03)00309-7. [DOI] [PubMed] [Google Scholar]
- 6.Koseki T, Inohara N, Chen S, Nunez G. Proc Natl Acad Sci USA. 1998;95:5156–5160. doi: 10.1073/pnas.95.9.5156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McConnell BB, Vertino PM. Apoptosis. 2004;9:5–18. doi: 10.1023/B:APPT.0000012117.32430.0c. [DOI] [PubMed] [Google Scholar]
- 8.Adams JM, Cory S. Curr Opin Cell Biol. 2002;14:715–720. doi: 10.1016/s0955-0674(02)00381-2. [DOI] [PubMed] [Google Scholar]
- 9.Martinon F, Burns K, Tschopp J. Mol Cell. 2002;10:417–426. doi: 10.1016/s1097-2765(02)00599-3. [DOI] [PubMed] [Google Scholar]
- 10.Peter ME, Krammer PH. Cell Death Differ. 2003;10:26–35. doi: 10.1038/sj.cdd.4401186. [DOI] [PubMed] [Google Scholar]
- 11.Tinel A, Tschopp J. Science 2004 [Google Scholar]
- 12.Salvesen GS, Dixit VM. Proc Natl Acad Sci USA. 1999;96:10964–10967. doi: 10.1073/pnas.96.20.10964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shi Y. Structure. 2002;10:285–288. doi: 10.1016/s0969-2126(02)00732-3. [DOI] [PubMed] [Google Scholar]
- 14.Bertin J, DiStefano PS. Cell Death Differ. 2000;7:1273–1274. doi: 10.1038/sj.cdd.4400774. [DOI] [PubMed] [Google Scholar]
- 15.Fairbrother WJ, Gordon NC, Humke EW, O’Rourke KM, Starovasnik MA, Yin JP, Dixit VM. Protein Sci. 2001;10:1911–1918. doi: 10.1110/ps.13801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martinon F, Hofmann K, Tschopp J. Curr Biol. 2001;11:R118–R120. doi: 10.1016/s0960-9822(01)00056-2. [DOI] [PubMed] [Google Scholar]
- 17.Pawlowski K, Pio F, Chu ZL, Reed JC, Godzik A. Trends Biochem Sci. 2001;26:85–87. doi: 10.1016/s0968-0004(00)01729-1. [DOI] [PubMed] [Google Scholar]
- 18.Staub E, Dahl E, Rosenthal A. Trends Biochem Sci. 2001;26:83–85. doi: 10.1016/s0968-0004(00)01717-5. [DOI] [PubMed] [Google Scholar]
- 19.Chamaillard M, Girardin SE, Viala J, Philpott DJ. Cell Microbiol. 2003;5:581–592. doi: 10.1046/j.1462-5822.2003.00304.x. [DOI] [PubMed] [Google Scholar]
- 20.Harton JA, Linhoff MW, Zhang J, Ting JP. J Immunol. 2002;169:4088–4093. doi: 10.4049/jimmunol.169.8.4088. [DOI] [PubMed] [Google Scholar]
- 21.Inohara N, Nunez G. Nat Rev Immunol. 2003;3:371–381. doi: 10.1038/nri1086. [DOI] [PubMed] [Google Scholar]
- 22.Stehlik C, Reed JC. J Exp Med. 2004;200:551–558. doi: 10.1084/jem.20032234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ting JP, Davis BK. Annu Rev Immunol. 2004;23:387–414. doi: 10.1146/annurev.immunol.23.021704.115616. [DOI] [PubMed] [Google Scholar]
- 24.Tschopp J, Martinon F, Burns K. Nat Rev Mol Cell Biol. 2003;4:95–104. doi: 10.1038/nrm1019. [DOI] [PubMed] [Google Scholar]
- 25.Janeway CA., Jr Cold Spring Harb Symp Quant Biol. 1989;54(Pt 1):1–13. [PubMed] [Google Scholar]
- 26.Masumoto J, Taniguchi S, Ayukawa K, Sarvotham H, Kishino T, Niikawa N, Hidaka E, Katsuyama T, Higuchi T, Sagara J. J Biol Chem. 1999;274:33835–33838. doi: 10.1074/jbc.274.48.33835. [DOI] [PubMed] [Google Scholar]
- 27.Conway KE, McConnell BB, Bowring CE, Donald CD, Warren ST, Vertino PM. Cancer Res. 2000;60:6236–6242. [PubMed] [Google Scholar]
- 28.Stehlik C, Krajewska M, Welsh K, Krajewski S, Godzik A, Reed JC. Biochem J. 2003;373:101–113. doi: 10.1042/BJ20030304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Centola M, Aksentijevich I, Kastner DL. Hum Mol Genet. 1998;7:1581–1588. doi: 10.1093/hmg/7.10.1581. [DOI] [PubMed] [Google Scholar]
- 30.Consortium TFF. Nat Genet. 1997;17:25–31. [Google Scholar]
- 31.Consortium TIF. Cell. 1997;90:797–807. doi: 10.1016/s0092-8674(00)80539-5. [DOI] [PubMed] [Google Scholar]
- 32.Chae JJ, Komarow HD, Cheng J, Wood G, Raben N, Liu PP, Kastner DL. Mol Cell. 2003;11:591–604. doi: 10.1016/s1097-2765(03)00056-x. [DOI] [PubMed] [Google Scholar]
- 33.Druilhe A, Srinivasula SM, Razmara M, Ahmad M, Alnemri ES. Cell Death Differ. 2001;8:649–657. doi: 10.1038/sj.cdd.4400881. [DOI] [PubMed] [Google Scholar]
- 34.Humke EW, Shriver SK, Starovasnik MA, Fairbrother WJ, Dixit VM. Cell. 2000;103:99–111. doi: 10.1016/s0092-8674(00)00108-2. [DOI] [PubMed] [Google Scholar]
- 35.Lamkanfi M, Denecker G, Kalai M, D’Hondt K, Meeus A, Declercq W, Saelens X, Vandenabeele P. J Biol Chem. 2004;279:51729–51738. doi: 10.1074/jbc.M407891200. [DOI] [PubMed] [Google Scholar]
- 36.Lee SH, Stehlik C, Reed JC. J Biol Chem. 2001;276:34495–34500. doi: 10.1074/jbc.M101415200. [DOI] [PubMed] [Google Scholar]
- 37.Condorelli G, Vigliotta G, Cafieri A, Trencia A, Andalo P, Oriente F, Miele C, Caruso M, Formisano P, Beguinot F. Oncogene. 1999;18:4409–4415. doi: 10.1038/sj.onc.1202831. [DOI] [PubMed] [Google Scholar]
- 38.Krueger A, Schmitz I, Baumann S, Krammer PH, Kirchhoff S. J Biol Chem. 2001;276:20633–20640. doi: 10.1074/jbc.M101780200. [DOI] [PubMed] [Google Scholar]
- 39.Asefa B, Klarmann KD, Copeland NG, Gilbert DJ, Jenkins NA, Keller JR. Blood Cells Mol Dis. 2004;32:155–167. doi: 10.1016/j.bcmd.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 40.Johnstone RW, Trapani JA. Mol Cell Biol. 1999;19:5833–5838. doi: 10.1128/mcb.19.9.5833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Landolfo S, Gariglio M, Gribaudo G, Lembo D. Biochimie. 1998;80:721–728. doi: 10.1016/s0300-9084(99)80025-x. [DOI] [PubMed] [Google Scholar]
- 42.Ding Y, Wang L, Su LK, Frey JA, Shao R, Hunt KK, Yan DH. Oncogene. 2004;23:4556–4566. doi: 10.1038/sj.onc.1207592. [DOI] [PubMed] [Google Scholar]
- 43.Eckhart L, Ballaun C, Uthman A, Kittel C, Stichenwirth M, Buchberger M, Fischer H, Sipos W, Tschachler E. J Biol Chem. 2005;280:35077–35080. doi: 10.1074/jbc.C500282200. [DOI] [PubMed] [Google Scholar]
- 44.Masumoto J, Zhou W, Chen FF, Su F, Kuwada JY, Hidaka E, Katsuyama T, Sagara J, Taniguchi S, Ngo-Hazelett P, Postlethwait JH, Nunez G, Inohara N. J Biol Chem. 2003;278:4268–4276. doi: 10.1074/jbc.M203944200. [DOI] [PubMed] [Google Scholar]
- 45.Koonin EV, Aravind L. Trends Biol Sci. 2000;25:223–224. doi: 10.1016/s0968-0004(00)01577-2. [DOI] [PubMed] [Google Scholar]
- 46.Gumucio DL, Diaz A, Schaner P, Richards N, Babcock C, Schaller M, Cesena T. Clin Exp Rheumatol. 2002;20:S45–53. [PubMed] [Google Scholar]
- 47.Belkhadir Y, Subramaniam R, Dangl JL. Curr Opin Plant Biol. 2004;7:391–399. doi: 10.1016/j.pbi.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 48.Zou H, Henzel WJ, Liu X, Lutschg A, Wang X. Cell. 1997;90:405–413. doi: 10.1016/s0092-8674(00)80501-2. [DOI] [PubMed] [Google Scholar]
- 49.Srinivasula SM, Ahmad M, Fernandes-Alnemri T, Alnemri ES. Mol Cell. 1998;1:949–957. doi: 10.1016/s1097-2765(00)80095-7. [DOI] [PubMed] [Google Scholar]
- 50.Roy N, Mahadevan MS, McLean M, Shutler G, Yaraghi Z, Farahani R, Baird S, Besner-Johnston A, Lefebvre C, Kang X, et al. Cell. 1995;80:167–178. doi: 10.1016/0092-8674(95)90461-1. [DOI] [PubMed] [Google Scholar]
- 51.Haider MZ, Moosa A, Dalal H, Habib Y, Reynold L. J Biomed Sci. 2001;8:191–196. doi: 10.1007/BF02256412. [DOI] [PubMed] [Google Scholar]
- 52.Davoodi J, Lin L, Kelly J, Liston P, MacKenzie AE. J Biol Chem. 2004;279:40622–40628. doi: 10.1074/jbc.M405963200. [DOI] [PubMed] [Google Scholar]
- 53.Diez E, Lee SH, Gauthier S, Yaraghi Z, Tremblay M, Vidal S, Gros P. Nat Genet. 2003;33:55–60. doi: 10.1038/ng1065. [DOI] [PubMed] [Google Scholar]
- 54.Steimle V, Otten LA, Zufferey M, Mach B. Cell. 1993;75:135–146. [PubMed] [Google Scholar]
- 55.Harton JA, Cressman DE, Chin KC, Der CJ, Ting JP. Science. 1999;285:1402–1405. doi: 10.1126/science.285.5432.1402. [DOI] [PubMed] [Google Scholar]
- 56.LeibundGut-Landmann S, Waldburger JM, Krawczyk M, Otten LA, Suter T, Fontana A, Acha-Orbea H, Reith W. Eur J Immunol. 2004;34:1513–1525. doi: 10.1002/eji.200424964. [DOI] [PubMed] [Google Scholar]
- 57.Nickerson K, Sisk TJ, Inohara N, Yee CS, Kennell J, Cho MC, Yannie PJ, 2nd, Nunez G, Chang CH. J Biol Chem. 2001;276:19089–19093. doi: 10.1074/jbc.M101295200. [DOI] [PubMed] [Google Scholar]
- 58.Swanberg M, Lidman O, Padyukov L, Eriksson P, Akesson E, Jagodic M, Lobell A, Khademi M, Borjesson O, Lindgren CM, Lundman P, Brookes AJ, Kere J, Luthman H, Alfredsson L, Hillert J, Klareskog L, Hamsten A, Piehl F, Olsson T. Nat Genet. 2005;37:486–494. doi: 10.1038/ng1544. [DOI] [PubMed] [Google Scholar]
- 59.Medzhitov R. Nat Rev Immunol. 2001;1:135–145. doi: 10.1038/35100529. [DOI] [PubMed] [Google Scholar]
- 60.Takeda K, Kaisho T, Akira S. Annu Rev Immunol. 2003;21:335–376. doi: 10.1146/annurev.immunol.21.120601.141126. [DOI] [PubMed] [Google Scholar]
- 61.Kobayashi K, Inohara N, Hernandez LD, Galan JE, Nunez G, Janeway CA, Medzhitov R, Flavell RA. Nature. 2002;416:194–199. doi: 10.1038/416194a. [DOI] [PubMed] [Google Scholar]
- 62.Girardin SE, Boneca IG, Carneiro LA, Antignac A, Jehanno M, Viala J, Tedin K, Taha MK, Labigne A, Zahringer U, Coyle AJ, DiStefano PS, Bertin J, Sansonetti PJ, Philpott DJ. Science. 2003;300:1584–1587. doi: 10.1126/science.1084677. [DOI] [PubMed] [Google Scholar]
- 63.Chamaillard M, Hashimoto M, Horie Y, Masumoto J, Qiu S, Saab L, Ogura Y, Kawasaki A, Fukase K, Kusumoto S, Valvano MA, Foster SJ, Mak TW, Nunez G, Inohara N. Nat Immunol. 2003;4:702–707. doi: 10.1038/ni945. [DOI] [PubMed] [Google Scholar]
- 64.Girardin SE, Boneca IG, Viala J, Chamaillard M, Labigne A, Thomas G, Philpott DJ, Sansonetti PJ. J Biol Chem. 2003;278:8869–8872. doi: 10.1074/jbc.C200651200. [DOI] [PubMed] [Google Scholar]
- 65.Girardin SE, Travassos LH, Herve M, Blanot D, Boneca IG, Philpott DJ, Sansonetti PJ, Mengin-Lecreulx D. J Biol Chem. 2003;278:41702–41708. doi: 10.1074/jbc.M307198200. [DOI] [PubMed] [Google Scholar]
- 66.Inohara N, Ogura Y, Fontalba A, Gutierrez O, Pons F, Crespo J, Fukase K, Inamura S, Kusumoto S, Hashimoto M, Foster SJ, Moran AP, Fernandez-Luna JL, Nunez G. J Biol Chem. 2003;278:5509–5512. doi: 10.1074/jbc.C200673200. [DOI] [PubMed] [Google Scholar]
- 67.Hisamatsu T, Suzuki M, Reinecker HC, Nadeau WJ, McCormick BA, Podolsky DK. Gastroenterology. 2003;124:993–1000. doi: 10.1053/gast.2003.50153. [DOI] [PubMed] [Google Scholar]
- 68.Watanabe T, Kitani A, Murray PJ, Strober W. Nat Immunol. 2004;5:800–808. doi: 10.1038/ni1092. [DOI] [PubMed] [Google Scholar]
- 69.Netea MG, Kullberg BJ, de Jong DJ, Franke B, Sprong T, Naber TH, Drenth JP, Van der Meer JW. Eur J Immunol. 2004;34:2052–2059. doi: 10.1002/eji.200425229. [DOI] [PubMed] [Google Scholar]
- 70.Damiano JS, Newman RM, Reed JC. J Immunol. 2004;173:6338–6345. doi: 10.4049/jimmunol.173.10.6338. [DOI] [PubMed] [Google Scholar]
- 71.Mariathasan S, Newton K, Monack DM, Vucic D, French DM, Lee WP, Roose-Girma M, Erickson S, Dixit VM. Nature. 2004;430:213–218. doi: 10.1038/nature02664. [DOI] [PubMed] [Google Scholar]
- 72.Franchi L, Amer A, Body-Malapel M, Kanneganti TD, Ozoren N, Jagirdar R, Inohara N, Vandenabeele P, Bertin J, Coyle A, Grant EP, Nunez G. Nat Immunol. 2006;7:576–582. doi: 10.1038/ni1346. [DOI] [PubMed] [Google Scholar]
- 73.Mariathasan S, Weiss DS, Newton K, McBride J, O’Rourke K, Roose-Girma M, Lee WP, Weinrauch Y, Monack DM, Dixit VM. Nature. 2006;440:228–232. doi: 10.1038/nature04515. [DOI] [PubMed] [Google Scholar]
- 74.Ozoren N, Masumoto J, Franchi L, Kanneganti TD, Body-Malapel M, Erturk I, Jagirdar R, Zhu L, Inohara N, Bertin J, Coyle A, Grant EP, Nunez G. J Immunol. 2006;176:4337–4342. doi: 10.4049/jimmunol.176.7.4337. [DOI] [PubMed] [Google Scholar]
- 75.Sutterwala FS, Ogura Y, Szczepanik M, Lara-Tejero M, Lichtenberger GS, Grant EP, Bertin J, Coyle AJ, Galan JE, Askenase PW, Flavell RA. Immunity. 2006;24:317–327. doi: 10.1016/j.immuni.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 76.Yamamoto M, Yaginuma K, Tsutsui H, Sagara J, Guan X, Seki E, Yasuda K, Yamamoto M, Akira S, Nakanishi K, Noda T, Taniguchi S. Genes Cells. 2004;9:1055–1067. doi: 10.1111/j.1365-2443.2004.00789.x. [DOI] [PubMed] [Google Scholar]
- 77.Martinon F, Agostini L, Meylan E, Tschopp J. Curr Biol. 2004;14:1929–1934. doi: 10.1016/j.cub.2004.10.027. [DOI] [PubMed] [Google Scholar]
- 78.Thornberry NA, Bull HG, Calaycay JR, Chapman KT, Howard AD, Kostura MJ, Miller DK, Molineaux SM, Weidner JR, Aunins J, et al. Nature. 1992;356:768–774. doi: 10.1038/356768a0. [DOI] [PubMed] [Google Scholar]
- 79.Li P, Allen H, Banerjee S, Franklin S, Herzog L, Johnston C, McDowell J, Paskind M, Rodman L, Salfeld J, Towne E, Tracey D, Wardwell S, Wei FY, Wong W, Kamen R, Seshadri T. Cell. 1995;80:401–411. doi: 10.1016/0092-8674(95)90490-5. [DOI] [PubMed] [Google Scholar]
- 80.Kuida K, Lippke JA, Ku G, Harding MW, Livingston DJ, Su MS, Flavell RA. Science. 1995;267:2000–2003. doi: 10.1126/science.7535475. [DOI] [PubMed] [Google Scholar]
- 81.Wang S, Miura M, Jung YK, Zhu H, Li E, Yuan J. Cell. 1998;92:501–509. doi: 10.1016/s0092-8674(00)80943-5. [DOI] [PubMed] [Google Scholar]
- 82.Agostini L, Martinon F, Burns K, McDermott MF, Hawkins PN, Tschopp J. Immunity. 2004;20:319–325. doi: 10.1016/s1074-7613(04)00046-9. [DOI] [PubMed] [Google Scholar]
- 83.Kanneganti TD, Ozoren N, Body-Malapel M, Amer A, Park JH, Franchi L, Whitfield J, Barchet W, Colonna M, Vandenabeele P, Bertin J, Coyle A, Grant EP, Akira S, Nunez G. Nature. 2006;440:232–236. doi: 10.1038/nature04517. [DOI] [PubMed] [Google Scholar]
- 84.Martinon F, Petrilli V, Mayor A, Tardivel A, Tschopp J. Nature. 2006;440:237–241. doi: 10.1038/nature04516. [DOI] [PubMed] [Google Scholar]
- 85.Wang L, Manji GA, Grenier JM, Al-Garawi A, Merriam S, Lora JM, Geddes BJ, Briskin M, DiStefano PS, Bertin J. J Biol Chem. 2002;277:29874–29880. doi: 10.1074/jbc.M203915200. [DOI] [PubMed] [Google Scholar]
- 86.Manji GA, Wang L, Geddes BJ, Brown M, Merriam S, Al-Garawi A, Mak S, Lora JM, Briskin M, Jurman M, Cao J, DiStefano PS, Bertin J. J Biol Chem. 2002;277:11570–11575. doi: 10.1074/jbc.M112208200. [DOI] [PubMed] [Google Scholar]
- 87.Hasegawa M, Imamura R, Kinoshita T, Matsumoto N, Masumoto J, Inohara N, Suda T. J Biol Chem. 2005;280:15122–15130. doi: 10.1074/jbc.M412284200. [DOI] [PubMed] [Google Scholar]
- 88.Stehlik C, Fiorentino L, Dorfleutner A, Bruey JM, Ariza EM, Sagara J, Reed JC. J Exp Med. 2002;196:1605–1615. doi: 10.1084/jem.20021552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fiorentino L, Stehlik C, Oliveira V, Ariza ME, Godzik A, Reed JC. J Biol Chem. 2002;277:35333–35340. doi: 10.1074/jbc.M200446200. [DOI] [PubMed] [Google Scholar]
- 90.Bruey JM, Bruey-Sedano N, Newman R, Chandler S, Stehlik C, Reed JC. J Biol Chem. 2004;279:51897–51907. doi: 10.1074/jbc.M406741200. [DOI] [PubMed] [Google Scholar]
- 91.O’Connor W, Jr, Harton JA, Zhu X, Linhoff MW, Ting JP. J Immunol. 2003;171:6329–6333. doi: 10.4049/jimmunol.171.12.6329. [DOI] [PubMed] [Google Scholar]
- 92.Kinoshita T, Wang Y, Hasegawa M, Imamura R, Suda T. J Biol Chem. 2005;280:21720–21725. doi: 10.1074/jbc.M410057200. [DOI] [PubMed] [Google Scholar]
- 93.Wang Y, Hasegawa M, Imamura R, Kinoshita T, Kondo C, Konaka K, Suda T. Int Immunol. 2004;16:777–786. doi: 10.1093/intimm/dxh081. [DOI] [PubMed] [Google Scholar]
- 94.Centola M, Wood G, Frucht DM, Galon J, Aringer M, Farrell C, Kingma DW, Horwitz ME, Mansfield E, Holland SM, O’Shea JJ, Rosenberg HF, Malech HL, Kastner DL. Blood. 2000;95:3223–3231. [PubMed] [Google Scholar]
- 95.Papin S, Cazeneuve C, Duquesnoy P, Jeru I, Sahali D, Amselem S. J Biol Chem. 2003;278:48839–48847. doi: 10.1074/jbc.M305166200. [DOI] [PubMed] [Google Scholar]
- 96.Shiohara M, Taniguchi S, Masumoto J, Yasui K, Koike K, Komiyama A, Sagara J. Biochem Biophys Res Commun. 2002;293:1314–1318. doi: 10.1016/S0006-291X(02)00384-4. [DOI] [PubMed] [Google Scholar]
- 97.Stehlik C, Lee SH, Dorfleutner A, Stassinopoulos A, Sagara J, Reed JC. J Immunol. 2003;171:6154–6163. doi: 10.4049/jimmunol.171.11.6154. [DOI] [PubMed] [Google Scholar]
- 98.Williams LK, Taxman JD, Linhoff WM, Reed W, Ting JP. J Immunol. 2003;170:5354–5358. doi: 10.4049/jimmunol.170.11.5354. [DOI] [PubMed] [Google Scholar]
- 99.Dorfleutner A, McDonald S, Bryan N, Funya K, Reed JC, Shi X, Flynn DC, Rojanasakul Y, Stehlik C. doi: 10.1007/s11262-007-0141-9. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Johnston JB, Barrett JW, Nazarian SH, Goodwin M, Ricuttio D, Wang G, McFadden G. Immunity. 2005;23:587–598. doi: 10.1016/j.immuni.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 101.Mariathasan S, Vucic D. Biochem J. 2003;373:e1–2. doi: 10.1042/BJ20030304COM. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Seet BT, Johnston JB, Brunetti CR, Barrett JW, Everett H, Cameron C, Sypula J, Nazarian SH, Lucas A, McFadden G. Annu Rev Immunol. 2003;21:377–423. doi: 10.1146/annurev.immunol.21.120601.141049. [DOI] [PubMed] [Google Scholar]
- 103.Razmara M, Srinivasula SM, Wang L, Poyet JL, Geddes BJ, DiStefano PS, Bertin J, Alnemri ES. J Biol Chem. 2002;277:13952–13958. doi: 10.1074/jbc.M107811200. [DOI] [PubMed] [Google Scholar]
- 104.Bouchier-Hayes L, Conroy H, Egan H, Adrain C, Creagh EM, MacFarlane M, Martin SJ. J Biol Chem. 2001;276:44069–44077. doi: 10.1074/jbc.M107373200. [DOI] [PubMed] [Google Scholar]
- 105.Stilo R, Leonardi A, Formisano L, Di Jeso B, Vito P, Liguoro D. FEBS Lett. 2002;521:165–169. doi: 10.1016/s0014-5793(02)02869-7. [DOI] [PubMed] [Google Scholar]
- 106.Pathan N, Marusawa H, Krajewska M, Matsuzawa SI, Kim H, Okada K, Torii S, Kitada S, Krajewski S, Welsh K, Pio F, Godzik A, Reed JC. J Biol Chem. 2001;276:32220–32229. doi: 10.1074/jbc.M100433200. [DOI] [PubMed] [Google Scholar]
- 107.Zhang H, Fu W. Int J Oncol. 2002;20:1035–1040. [PubMed] [Google Scholar]
- 108.Li P, Nijhawan D, Budihardjo I, Srinivasula SM, Ahmad M, Alnemri ES, Wang X. Cell. 1997;91:479–489. doi: 10.1016/s0092-8674(00)80434-1. [DOI] [PubMed] [Google Scholar]
- 109.Inohara N, Koseki T, del Peso L, Hu Y, Yee C, Chen S, Carrio R, Merino J, Liu D, Ni J, Nunez G. J Biol Chem. 1999;274:14560–14567. doi: 10.1074/jbc.274.21.14560. [DOI] [PubMed] [Google Scholar]
- 110.Bertin J, Nir WJ, Fischer CM, Tayber OV, Errada PR, Grant JR, Keilty JJ, Gosselin ML, Robison KE, Wong GH, Glucksmann MA, DiStefano PS. J Biol Chem. 1999;274:12955–12958. doi: 10.1074/jbc.274.19.12955. [DOI] [PubMed] [Google Scholar]
- 111.Poyet JL, Srinivasula SM, Tnani M, Razmara M, Fernandes-Alnemri T, Alnemri ES. J Biol Chem. 2001;276:28309–28313. doi: 10.1074/jbc.C100250200. [DOI] [PubMed] [Google Scholar]
- 112.Gotz R, Karch C, Digby MR, Troppmair J, Rapp UR, Sendtner M. Hum Mol Genet. 2000;9:2479–2489. doi: 10.1093/hmg/9.17.2479. [DOI] [PubMed] [Google Scholar]
- 113.Gourley TS, Chang CH. J Immunol. 2001;166:2917–2921. doi: 10.4049/jimmunol.166.5.2917. [DOI] [PubMed] [Google Scholar]
- 114.Gourley TS, Patel DR, Nickerson K, Hong SC, Chang CH. J Immunol. 2002;168:4414–4419. doi: 10.4049/jimmunol.168.9.4414. [DOI] [PubMed] [Google Scholar]
- 115.Shirasu K, Schulze-Lefert P. Plant Mol Biol. 2000;44:371–385. doi: 10.1023/a:1026552827716. [DOI] [PubMed] [Google Scholar]
- 116.Aliprantis AO, Yang RB, Mark MR, Suggett S, Devaux B, Radolf JD, Klimpel GR, Godowski P, Zychlinsky A. Science. 1999;285:736–739. doi: 10.1126/science.285.5428.736. [DOI] [PubMed] [Google Scholar]
- 117.Chu ZL, Pio F, Xie Z, Welsh K, Krajewska M, Krajewski S, Godzik A, Reed JC. J Biol Chem. 2001;276:9239–9245. doi: 10.1074/jbc.M006309200. [DOI] [PubMed] [Google Scholar]
- 118.Hlaing T, Guo RF, Dilley KA, Loussia JM, Morrish TA, Shi MM, Vincenz C, Ward PA. J Biol Chem. 2001;276:9230–9238. doi: 10.1074/jbc.M009853200. [DOI] [PubMed] [Google Scholar]
- 119.Liu F, Lo CF, Ning X, Kajkowski EM, Jin M, Chiriac C, Gonzales C, Naureckiene S, Lock YW, Pong K, Zaleska MM, Jacobsen JS, Silverman S, Ozenberger BA. Cell Signal. 2004;16:1013–1021. doi: 10.1016/j.cellsig.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 120.Dowds TA, Masumoto J, Chen FF, Ogura Y, Inohara N, Nunez G. Biochem Biophys Res Commun. 2003;302:575–580. doi: 10.1016/s0006-291x(03)00221-3. [DOI] [PubMed] [Google Scholar]
- 121.Marsden VS, O’Connor L, O’Reilly LA, Silke J, Metcalf D, Ekert PG, Huang DC, Cecconi F, Kuida K, Tomaselli KJ, Roy S, Nicholson DW, Vaux DL, Bouillet P, Adams JM, Strasser A. Nature. 2002;419:634–637. doi: 10.1038/nature01101. [DOI] [PubMed] [Google Scholar]
- 122.Mariathasan S, Weiss DS, Dixit VM, Monack DM. J Exp Med. 2005;202:1043–1049. doi: 10.1084/jem.20050977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Chen Y, Smith MR, Thirumalai K, Zychlinsky A. Embo J. 1996;15:3853–3860. [PMC free article] [PubMed] [Google Scholar]
- 124.Hersh D, Monack DM, Smith MR, Ghori N, Falkow S, Zychlinsky A. Proc Natl Acad Sci USA. 1999;96:2396–2401. doi: 10.1073/pnas.96.5.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Monack DM, Hersh D, Ghori N, Bouley D, Zychlinsky A, Falkow S. J Exp Med. 2000;192:249–258. doi: 10.1084/jem.192.2.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Monack DM, Navarre WW, Falkow S. Microbes Infect. 2001;3:1201–1212. doi: 10.1016/s1286-4579(01)01480-0. [DOI] [PubMed] [Google Scholar]
- 127.McConnell BB, Vertino PM. Cancer Res. 2000;60:6243–6247. [PubMed] [Google Scholar]
- 128.Richards N, Schaner P, Diaz A, Stcukey J, Shelden E, Wadhwa A, Gumucio DL. J Biol Chem. 2001;276:39320–39329. doi: 10.1074/jbc.M104730200. [DOI] [PubMed] [Google Scholar]
- 129.Kubo T, Yamamoto J, Shikauchi Y, Niwa Y, Matsubara K, Yoshikawa H. Cancer Res. 2004;64:5172–5177. doi: 10.1158/0008-5472.CAN-03-3314. [DOI] [PubMed] [Google Scholar]
- 130.Johnstone RW, Kershaw MH, Trapani JA. Biochemistry. 1998;37:11924–11931. doi: 10.1021/bi981069a. [DOI] [PubMed] [Google Scholar]
- 131.Xie J, Briggs JA, Briggs RC. FEBS Lett. 1997;408:151–155. doi: 10.1016/s0014-5793(97)00404-3. [DOI] [PubMed] [Google Scholar]
- 132.Xin H, Curry J, Johnstone RW, Nickoloff BJ, Choubey D. Oncogene. 2003;22:4831–4840. doi: 10.1038/sj.onc.1206754. [DOI] [PubMed] [Google Scholar]
- 133.Johnstone RW, Wei W, Greenway A, Trapani JA. Oncogene. 2000;19:6033–6042. doi: 10.1038/sj.onc.1204005. [DOI] [PubMed] [Google Scholar]
- 134.Kwak JC, Ongusaha PP, Ouchi T, Lee SW. J Biol Chem. 2003;278:40899–40904. doi: 10.1074/jbc.M308012200. [DOI] [PubMed] [Google Scholar]
- 135.Johnstone RW, Kerry JA, Trapani JA. J Biol Chem. 1998;273:17172–17177. doi: 10.1074/jbc.273.27.17172. [DOI] [PubMed] [Google Scholar]
- 136.DeYoung KL, Ray ME, Su YA, Anzick SL, Johnstone RW, Trapani JA, Meltzer PS, Trent JM. Oncogene. 1997;15:453–457. doi: 10.1038/sj.onc.1201206. [DOI] [PubMed] [Google Scholar]
- 137.Choubey D, Walter S, Geng Y, Xin H. FEBS Lett. 2000;474:38–42. doi: 10.1016/s0014-5793(00)01571-4. [DOI] [PubMed] [Google Scholar]
- 138.Kulaeva OI, Draghici S, Tang L, Kraniak JM, Land SJ, Tainsky MA. Oncogene. 2003;22:4118–4127. doi: 10.1038/sj.onc.1206594. [DOI] [PubMed] [Google Scholar]
- 139.Mori Y, Yin J, Rashid A, Leggett BA, Young J, Simms L, Kuehl PM, Langenberg P, Meltzer SJ, Stine OC. Cancer Res. 2001;61:6046–6049. [PubMed] [Google Scholar]
- 140.Cousar JB, Briggs RC. Leuk Res. 1990;14:915–920. doi: 10.1016/0145-2126(90)90182-9. [DOI] [PubMed] [Google Scholar]
- 141.Xie J, Briggs JA, Briggs RC. J Cell Biochem. 1998;70:489–506. [PubMed] [Google Scholar]
- 142.McDermott MF, Aksentijevich I, Galon J, McDermott EM, Ogunkolade BW, Centola M, Mansfield E, Gadina M, Karenko L, Pettersson T, McCarthy J, Frucht DM, Aringer M, Torosyan Y, Teppo AM, Wilson M, Karaarslan HM, Wan Y, Todd I, Wood G, Schlimgen R, Kumarajeewa TR, Cooper SM, Vella JP, Kastner DL, et al. Cell. 1999;97:133–144. doi: 10.1016/s0092-8674(00)80721-7. [DOI] [PubMed] [Google Scholar]
- 143.Drenth JP, van der Meer JW. N Engl J Med. 2001;345:1748–1757. doi: 10.1056/NEJMra010200. [DOI] [PubMed] [Google Scholar]
- 144.Alt HL, Barker MH. JAMA. 1930;94:1457–1461. [Google Scholar]
- 145.Wise CA, Gillum JD, Seidman CE, Lindor NM, Veile R, Bashiardes S, Lovett M. Hum Mol Genet. 2002;11:961–969. doi: 10.1093/hmg/11.8.961. [DOI] [PubMed] [Google Scholar]
- 146.Shoham NG, Centola M, Mansfield E, Hull KM, Wood G, Wise CA, Kastner DL. Proc Natl Acad Sci USA. 2003;100:13501–13506. doi: 10.1073/pnas.2135380100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.McDermott MF. Trends Immunol. 2004;25:457–460. doi: 10.1016/j.it.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 148.Cuisset L, Drenth JP, Berthelot JM, Meyrier A, Vaudour G, Watts RA, Scott DG, Nicholls A, Pavek S, Vasseur C, Beckmann JS, Delpech M, Grateau G. Am J Hum Genet. 1999;65:1054–1059. doi: 10.1086/302589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Hoffman HM, Mueller JL, Broide DH, Wanderer AA, Kolodner RD. Nat Genet. 2001;29:301–305. doi: 10.1038/ng756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Aksentijevich I, Nowak M, Mallah M, Chae JJ, Watford WT, Hofmann SR, Stein L, Russo R, Goldsmith D, Dent P, Rosenberg HF, Austin F, Remmers EF, Balow JE, Jr, Rosenzweig S, Komarow H, Shoham NG, Wood G, Jones J, Mangra N, Carrero H, Adams BS, Moore TL, Schikler K, Hoffman H, Lovell DJ, Lipnick R, Barron K, O’Shea JJ, Kastner DL, Goldbach-Mansky R. Arthritis Rheum. 2002;46:3340–3348. doi: 10.1002/art.10688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Feldmann J, Prieur AM, Quartier P, Berquin P, Certain S, Cortis E, Teillac-Hamel D, Fischer A, de Saint Basile G. Am J Hum Genet. 2002;71:198–203. doi: 10.1086/341357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Ogura Y, Bonen DK, Inohara N, Nicolae DL, Chen FF, Ramos R, Britton H, Moran T, Karalluskas R, Duerr RH, Achkar JP, Brant SR, Bayless TM, Kirschner BS, Hanauer SB, Nunez G, Cho JN. Nature. 2001;411:603–606. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 153.Hugot JP, Chamaillard M, Zouali H, Lesage S, Cezard JP, Belaiche J, Almer S, Tysk C, O’Morain CA, Gassull M, Binder V, Finkel Y, Cortot A, Modigllani R, Laurent-Plug P, Gower-Rousseau C, Macry J, Colombel JF, Sahbatou M, Thomas G. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 154.Miceli-Richard C, Lesage S, Rybojad M, Prieur AM, Manouvrier-Hanu S, Hafner R, Chamaillard M, Zouali H, Thomas G, Hugot JP. Nat Genet. 2001;29:19–20. doi: 10.1038/ng720. [DOI] [PubMed] [Google Scholar]
- 155.Dowds TA, Masumoto J, Zhu L, Inohara N, Nunez G. J Biol Chem. 2004;279:21924–21928. doi: 10.1074/jbc.M401178200. [DOI] [PubMed] [Google Scholar]
- 156.Chamaillard M, Philpott D, Girardin SE, Zouali H, Lesage S, Chareyre F, Bui TH, Giovannini M, Zaehringer U, Penard-Lacronique V, Sansonetti PJ, Hugot JP, Thomas G. Proc Natl Acad Sci USA. 2003;100:3455–3460. doi: 10.1073/pnas.0530276100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Bonen DK, Ogura Y, Nicolae DL, Inohara N, Saab L, Tanabe T, Chen FF, Foster SJ, Duerr RH, Brant SR, Cho JH, Nunez G. Gastroenterology. 2003;124:140–146. doi: 10.1053/gast.2003.50019. [DOI] [PubMed] [Google Scholar]
- 158.Dinarello CA. J Exp Med. 2005;201:1355–1359. doi: 10.1084/jem.20050640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Furst DE. Clin Ther. 2004;26:1960–1975. doi: 10.1016/j.clinthera.2004.12.019. [DOI] [PubMed] [Google Scholar]
- 160.Hawkins PN, Lachmann HJ, Aganna E, McDermott MF. Arthritis Rheum. 2004;50:607–612. doi: 10.1002/art.20033. [DOI] [PubMed] [Google Scholar]
- 161.Granel B, Serratrice J, Disdier P, Weiller PJ. Rheumatology (Oxford) 2005;44:689–690. doi: 10.1093/rheumatology/keh547. [DOI] [PubMed] [Google Scholar]
- 162.Hoffman HM, Patel DD. Arthritis Rheum. 2004;50:345–349. doi: 10.1002/art.20032. [DOI] [PubMed] [Google Scholar]
- 163.Hoffman HM, Rosengren S, Boyle DL, Cho JY, Nayar J, Mueller JL, Anderson JP, Wanderer AA, Firestein GS. Lancet. 2004;364:1779–1785. doi: 10.1016/S0140-6736(04)17401-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Lovell DJ, Bowyer SL, Solinger AM. Arthritis Rheum. 2005;52:1283–1286. doi: 10.1002/art.20953. [DOI] [PubMed] [Google Scholar]
- 165.Ramos E, Arostegui JI, Campuzano S, Rius J, Bousono C, Yague J. Rheumatology (Oxford) 2005 doi: 10.1093/rheumatology/keh652. [DOI] [PubMed] [Google Scholar]
- 166.Dinarello CA. Nat Med. 2003;9:20–22. doi: 10.1038/nm0103-20. [DOI] [PubMed] [Google Scholar]
- 167.Economides AN, Carpenter LR, Rudge JS, Wong V, Koehler-Stec EM, Hartnett C, Pyles EA, Xu X, Daly TJ, Young MR, Fandl JP, Lee F, Carver S, McNay J, Bailey K, Ramakanth S, Hutabarat R, Huang TT, Radziejewski C, Yancopoulos GD, Stahl N. Nat Med. 2003;9:47–52. doi: 10.1038/nm811. [DOI] [PubMed] [Google Scholar]
- 168.Saleh A, Srinivasula SM, Acharya S, Fishel R, Alnemri ES. J Biol Chem. 1999;274:17941–17945. doi: 10.1074/jbc.274.25.17941. [DOI] [PubMed] [Google Scholar]
- 169.Riedl SJ, Li W, Chao Y, Schwarzenbacher R, Shi Y. Nature. 2005;434:926–933. doi: 10.1038/nature03465. [DOI] [PubMed] [Google Scholar]
- 170.Moffett P, Farnham G, Peart J, Baulcombe DC. EMBO J. 2002;21:4511–4519. doi: 10.1093/emboj/cdf453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Inohara N, Ogura Y, Chen FF, Muto A, Nunez G. J Biol Chem. 2001;276:2551–2554. doi: 10.1074/jbc.M009728200. [DOI] [PubMed] [Google Scholar]
- 172.Deslandes L, Olivier J, Peeters N, Feng DX, Khounlotham M, Boucher C, Somssich I, Genin S, Marco Y. Proc Natl Acad Sci USA. 2003;100:8024–8029. doi: 10.1073/pnas.1230660100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Jia Y, McAdams SA, Bryan GT, Hershey HP, Valent B. Embo J. 2000;19:4004–4014. doi: 10.1093/emboj/19.15.4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Damiano J, Oliveira V, Welsh K, Reed JC. Biochem J. 2004;381:213–219. doi: 10.1042/BJ20031506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Grenier JM, Wang L, Manji GA, Huang WJ, Al-Garawi A, Kelly R, Carlson A, Merriam S, Lora JM, Briskin M, DiStefano PS, Bertin J. FEBS Lett. 2002;530:73–78. doi: 10.1016/s0014-5793(02)03416-6. [DOI] [PubMed] [Google Scholar]
- 176.Dunne A, O’Neill L. FEBS Lett. 2005;579:3330–3335. doi: 10.1016/j.febslet.2005.04.024. [DOI] [PubMed] [Google Scholar]
- 177.Aarts N, Metz M, Holub E, Staskawicz BJ, Daniels MJ, Parker JE. Proc Natl Acad Sci USA. 1998;95:10306–10311. doi: 10.1073/pnas.95.17.10306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.McDowell JM, Cuzick A, Can C, Beynon J, Dangl JL, Holub EB. Plant J. 2000;22:523–529. doi: 10.1046/j.1365-313x.2000.00771.x. [DOI] [PubMed] [Google Scholar]
- 179.Barnich N, Hisamatsu T, Aguirre JE, Xavier R, Reinecker HC, Podolsky DK. J Biol Chem. 2005;280:19021–19026. doi: 10.1074/jbc.M413776200. [DOI] [PubMed] [Google Scholar]
- 180.Guan X, Sagara J, Yokoyama T, Koganehira Y, Oguchi M, Saida T, Taniguchi S. Int J Cancer. 2003;107:202–208. doi: 10.1002/ijc.11376. [DOI] [PubMed] [Google Scholar]
- 181.Stone AR, Bobo W, Brat DJ, Devi NS, Van Meir EG, Vertino PM. Am J Pathol. 2004;165:1151–1161. doi: 10.1016/S0002-9440(10)63376-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Terasawa K, Sagae S, Toyota M, Tsukada K, Ogi K, Satoh A, Mita H, Imai K, Tokino T, Kudo R. Clin Cancer Res. 2004;10:2000–2006. doi: 10.1158/1078-0432.ccr-0932-03. [DOI] [PubMed] [Google Scholar]
- 183.Virmani A, Rathi A, Sugio K, Sathyanarayana UG, Toyooka S, Kischel FC, Tonk V, Padar A, Takahashi T, Roth JA, Euhus DM, Minna JD, Gazdar AF. Int J Cancer. 2003;106:198–204. doi: 10.1002/ijc.11206. [DOI] [PubMed] [Google Scholar]
- 184.Yokoyama T, Sagara J, Guan X, Masumoto J, Takeoka M, Komiyama Y, Miyata K, Higuchi K, Taniguchi S. Cancer Lett. 2003;202:101–108. doi: 10.1016/j.canlet.2003.08.027. [DOI] [PubMed] [Google Scholar]
- 185.Karin M, Cao Y, Greten FR, Li ZW. Nat Rev Cancer. 2002;2:301–310. doi: 10.1038/nrc780. [DOI] [PubMed] [Google Scholar]
- 186.Fujiuchi N, Aglipay JA, Ohtsuka T, Maehara N, Sahin F, Su GH, Lee SW, Ouchi T. J Biol Chem. 2004;279:20339–20344. doi: 10.1074/jbc.M400344200. [DOI] [PubMed] [Google Scholar]
- 187.Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, Ghosh D, Pienta KJ, Sewalt RG, Otte AP, Rubin MA, Chinnaiyan AM. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 188.Aravind L, Dixit VM, Koonin EV. Trends Biochem Sci. 1999;24:47–53. doi: 10.1016/s0968-0004(98)01341-3. [DOI] [PubMed] [Google Scholar]
- 189.Hofmann K. Cell Mol Life Sci. 1999;55:1113–1128. doi: 10.1007/s000180050361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Vaughn DE, Rodriguez J, Lazebnik Y, Joshua-Tor L. J Mol Biol. 1999;293:439–447. doi: 10.1006/jmbi.1999.3177. [DOI] [PubMed] [Google Scholar]
- 191.Qin H, Srinivasula SM, Wu G, Fernandes-Alnemri T, Alnemri ES, Shi Y. Nature. 1999;399:549–557. doi: 10.1038/21124. [DOI] [PubMed] [Google Scholar]
- 192.Xiao T, Towb P, Wasserman SA, Sprang SR. Cell. 1999;99:545–555. doi: 10.1016/s0092-8674(00)81542-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Zhou P, Chou J, Olea RS, Yuan J, Wagner G. Proc Natl Acad Sci USA. 1999;96:11265–11270. doi: 10.1073/pnas.96.20.11265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Weber CH, Vincenz C. Trends Biochem Sci. 2001;26:475–481. doi: 10.1016/s0968-0004(01)01905-3. [DOI] [PubMed] [Google Scholar]
- 195.Eberstadt M, Huang B, Chen Z, Meadows RP, Ng SC, Zheng L, Lenardo MJ, Fesik SW. Nature. 1998;392:941–945. doi: 10.1038/31972. [DOI] [PubMed] [Google Scholar]
- 196.Chou JJ, Matsuo H, Duan H, Wagner G. Cell. 1998;94:171–180. doi: 10.1016/s0092-8674(00)81417-8. [DOI] [PubMed] [Google Scholar]
- 197.Hill JM, Morisawa G, Kim T, Huang T, Wei Y, Wei Y, Werner MH. J Biol Chem. 2004;279:1474–1481. doi: 10.1074/jbc.M304996200. [DOI] [PubMed] [Google Scholar]
- 198.Martin DA, Zheng L, Siegel RM, Huang B, Fisher GH, Wang J, Jackson CE, Puck JM, Dale J, Straus SE, Peter ME, Krammer PH, Fesik S, Lenardo MJ. Proc Natl Acad Sci USA. 1999;96:4552–4557. doi: 10.1073/pnas.96.8.4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Park A, Baichwal VR. J Biol Chem. 1996;271:9858–9862. doi: 10.1074/jbc.271.16.9858. [DOI] [PubMed] [Google Scholar]
- 200.Tartaglia LA, Ayres TM, Wong GH, Goeddel DV. Cell. 1993;74:845–853. doi: 10.1016/0092-8674(93)90464-2. [DOI] [PubMed] [Google Scholar]
- 201.Weber CH, Vincenz C. FEBS Lett. 2001;492:171–176. doi: 10.1016/s0014-5793(01)02162-7. [DOI] [PubMed] [Google Scholar]
- 202.Kaufmann M, Bozic D, Briand C, Bodmer JL, Zerbe O, Kohl A, Tschopp J, Grutter MG. FEBS Lett. 2002;527:250–254. doi: 10.1016/s0014-5793(02)03146-0. [DOI] [PubMed] [Google Scholar]
- 203.Siegel R, Martin D, Zheng L, Ng S, Bertin J, Cohen J, Lenardo M. J Cell Biol. 1998;141:1243–1253. doi: 10.1083/jcb.141.5.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Hiller S, Kohl A, Fiorito F, Herrmann T, Wider G, Tschopp J, Grutter MG, Wuthrich K. Structure. 2003;11:1199–1205. doi: 10.1016/j.str.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 205.Liepinsh E, Barbals R, Dahl E, Sharipo A, Staub E, Otting G. J Mol Biol. 2003;332:1155–1163. doi: 10.1016/j.jmb.2003.07.007. [DOI] [PubMed] [Google Scholar]
- 206.Espejo F, Green M, Preece NE, Assa-Munt N. J Biomol NMR. 2002;23:151–152. doi: 10.1023/a:1016398403157. [DOI] [PubMed] [Google Scholar]
- 207.Liu T, Rojas A, Ye Y, Godzik A. Protein Sci. 2003;12:1872–1881. doi: 10.1110/ps.0359603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Eberstadt M, Huang B, Olejniczak ET, Fesik SW. Nat Struct Biol. 1997;4:983–985. doi: 10.1038/nsb1297-983. [DOI] [PubMed] [Google Scholar]
- 209.Eliezer D. Structure. 2003;11:1190–1191. doi: 10.1016/j.str.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 210.Jeong EJ, Bang S, Lee TH, Park YI, Sim WS, Kim KS. J Biol Chem. 1999;274:16337–16342. doi: 10.1074/jbc.274.23.16337. [DOI] [PubMed] [Google Scholar]
- 211.Masumoto J, Taniguchi S, Sagara J. Biochem Biophysical Res Commun. 2001;280:652–655. doi: 10.1006/bbrc.2000.4190. [DOI] [PubMed] [Google Scholar]
- 212.Srinivasula SM, Poyet JL, Razmara M, Datta P, Zhang Z, Alnemri ES. J Biol Chem. 2002;277:21119–21122. doi: 10.1074/jbc.C200179200. [DOI] [PubMed] [Google Scholar]
- 213.Geddes BJ, Wang L, Huang WJ, Lavellee M, Manji GA, Brown M, Jurman M, Cao J, Morgenstern J, Merriam S, Gluckmann MA, DiStefano PS, Bertin J. Biochem Biophys Res Comm. 2001;284:77–82. doi: 10.1006/bbrc.2001.4928. [DOI] [PubMed] [Google Scholar]