Abstract
PCSK9 is a promising target for the treatment of hyperlipidemia and cardiovascular disease. A Quantitative Systems Pharmacology model of the mechanisms of action of statin and anti-PCSK9 therapies was developed to predict low density lipoprotein (LDL) changes in response to anti-PCSK9 mAb for different treatment protocols and patient subpopulations. Mechanistic interactions and cross-regulation of LDL, LDL receptor, and PCSK9 were modeled, and numerous virtual subjects were developed and validated against clinical data. Simulations predict a slightly greater maximum percent reduction in LDL cholesterol (LDLc) when anti-PCSK9 is administered on statin background therapy compared to as a monotherapy. The difference results primarily from higher PCSK9 levels in patients on statin background. However, higher PCSK9 levels are also predicted to increase clearance of anti-PCSK9, resulting in a faster rebound of LDLc. Simulations of subjects with impaired LDL receptor (LDLR) function predict compromised anti-PCSK9 responses in patients such as homozygous familial hypercholesterolemics, whose functional LDLR is below 10% of normal.
Proprotein convertase subtilisin-like kexin type 9 (PCSK9) is a promising target for treatment of hyperlipidemia and cardiovascular disease.1 Extracellular PCSK9 protein binds to low density lipoprotein (LDL) receptors on hepatocytes, targeting the receptor for intracellular degradation and reducing the availability of receptors for removal of LDL from the circulation. The level of circulating LDL cholesterol (LDLc) is one of the strongest indicators of CV risk. High levels present a major risk factor for CV disease and events, and LDLc lowering therapies such as statins significantly reduce this risk. While statins can reduce LDLc levels up to 70%, statins alone are not always sufficient to achieve desired LDLc levels and some patients are intolerant to statins. Reducing the functional PCSK9 activity may offer an additional treatment option in these situations. Individuals with inactivating PCSK9 mutations have naturally low levels of circulating LDLc,2 and ongoing clinical trials have shown efficient LDL-lowering with anti-PCSK9 antibodies.3
PCSK9-targeted therapies will likely be administered on a statin background when possible. However, statin (HMG-CoA reductase inhibitor) inhibition of hepatic cholesterol synthesis leads to an upregulation of PCSK9 that might impact the efficacy of combination regimens. Furthermore, subjects with different mechanistic causes of hyperlipidemia might respond differently to anti-PCSK9. Familial hypercholesterolemic (FH) subjects commonly exhibit mutations to the LDL receptors (LDLR) that reduce LDLR functionality and increase systemic LDLc levels. Limited clinical data on the impact of background statin therapy and patient phenotype on anti-PCSK9 response have been reported.4,5,6,7,8,9 However, the impact of background statin therapy and altered LDLR-mediated cholesterol uptake and clearance in FH subjects on anti-PCSK9 efficacy is not well quantified. Finally, patient biomarkers predictive of anti-PCSK9 effects on LDLc would be valuable as a diagnostic. Baseline PCSK9 and LDLc levels are obvious candidate biomarkers but sufficient clinical data are not yet available to characterize the relationship of these biomarkers to anti-PCSK9 responsiveness.
To investigate these questions and predict the impact on LDLc of treatment with RG7652, a fully human monoclonal antibody antagonizing PSCK9 activity,10 we have developed a quantitative systems pharmacology (QSP) model of the mechanistic interaction and cross-regulation of LDLc, LDLR, and PCSK9 in health and dyslipidemic disease, including statin and anti-PCSK9 mechanisms of action and effects. QSP approaches are being increasingly applied to integrate mechanistic biology, preclinical and clinical data, and drug pharmacology to provide quantitative insight and guidance in pharmaceutical R&D.11 QSP models have been successfully developed and applied to a wide range of questions and therapeutic areas.12,13,14 In this work, we have adopted a QSP approach to integrate preclinical and mechanistic data as well as available clinical results to make predictions for scenarios for which clinical data are currently unavailable or insufficient. These predictions have helped set expectations for phase II results in advance of trial readout, inform early planning for subsequent development, increase understanding of the impact of statin background therapy, and support biomarker interpretation.
Results
Clinical calibration and validation: phase I virtual population response to statins and anti-PCSK9
All model-based research was conducted via simulation of clinical protocols in virtual populations that reproduce clinically observed variability as described in the Methods. Virtual subjects included in the phase I virtual population were selected to match RG7652 phase I clinical data on baseline levels of LDLc and PCSK9 and the correlation between the two (Supplementary Table S4). The implementations of statin and RG7652 therapies were calibrated to match the change in LDLc with atorvastatin15 and the phase I PK and change in LDLc of single-dose RG7652 in this population. The effect of atorvastatin on PCSK9 levels and the effect of multi-dose RG7652 as monotherapy or in combination with atorvastatin were used for validation of the virtual population. Details of this calibration and validation results are provided below.
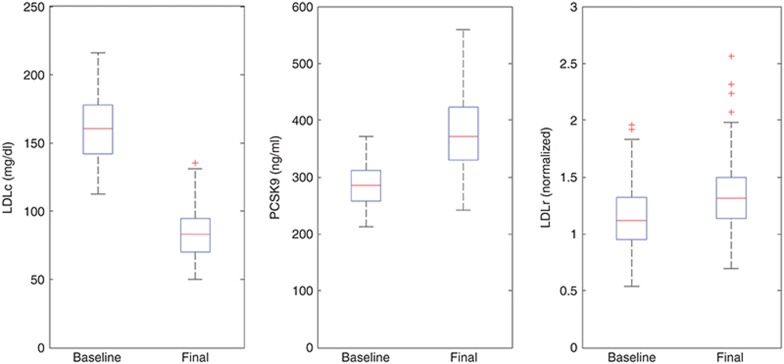
Statin response calibration and validation. Sustained inhibition of cholesterol synthesis by high dose (40–80 mg) atorvastatin was calibrated at 80% (CV = 25%) to capture a ~50% reduction in LDLc reported in the clinical literature.16 Figure 1 shows the corresponding LDLc distribution at baseline and after atorvastatin treatment for the phase I virtual population. In order to partially validate the statin implementation and test its ability to capture known effects of the drug on the modeled behaviors, we verified that simulations in the phase I population recapitulated the increase in PCSK9 reported clinically for statin treatment.17 Figure 1 confirms a simulated mean increase of 35% in PCSK9 levels as a result of high dose atorvastatin treatment, consistent with the reported range of 20–50%,17 offering validation that the regulation of PCSK9 synthesis and LDLR expression modeled based on preclinical data18,19,20 appropriately reproduces clinical behavior.
Figure 1.
Phase I virtual population selection and validation: LDLc, PCSK9, and LDLR for the virtual population at baseline and post high dose atorvastatin treatment. LDLc, low density lipoprotein cholesterol; LDLR, low density lipoprotein receptors.
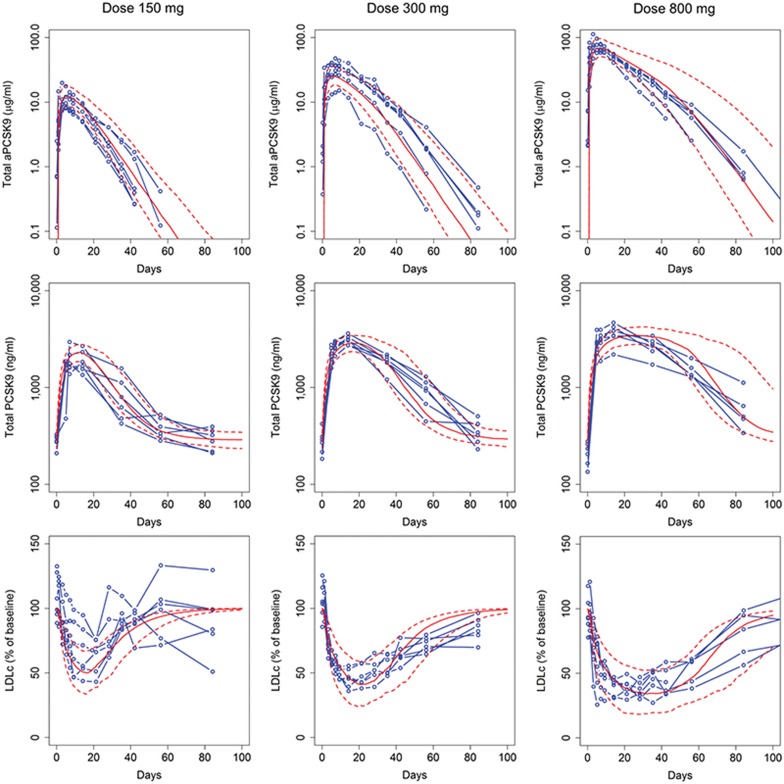
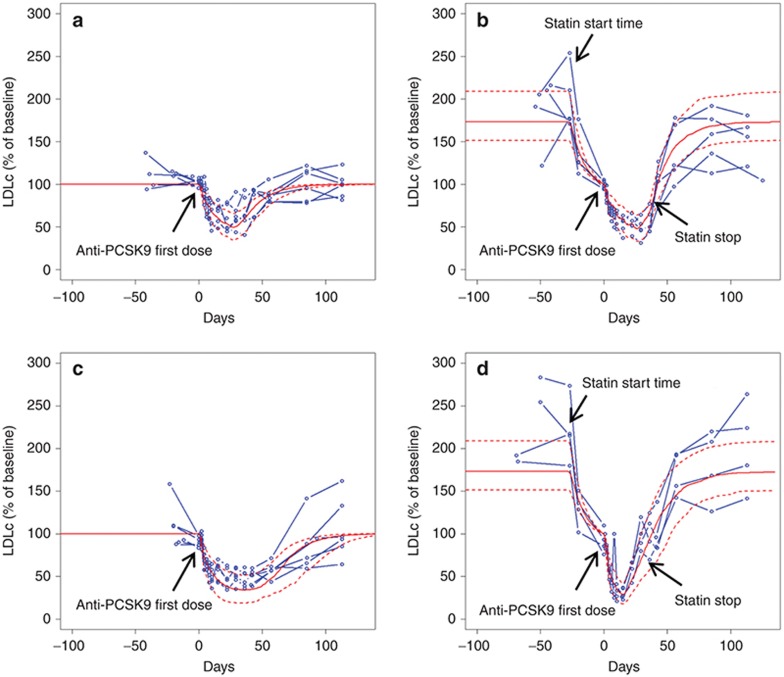
Anti-PCSK9 response calibration and validation. The TMDD PK model was calibrated to phase I single dose RG7652 (10–800 mg) data (N.R. Budha et al., unpublished data). This dataset included total anti-PCSK9 and total PCSK9 level time series profiles for six subjects per dose group. The model parameters representing LDLR degradation with PCSK9 were calibrated to capture the observed LDLc reduction profiles for the different doses of anti-PCSK9 (Figure 2). Three measurements were included in this calibration step: total anti-PCSK9 (free drug + drug-target complex), total PCSK9 (target + drug-target complex), and LDLc. The implementation was then validated by testing the population response to multi-dose anti-PCSK9 both as a monotherapy and in combination with atorvastatin. Figure 3 confirms that the virtual population simulations recapitulate the individual patient clinical data for the two doses of anti-PCS9 for which data were available (40 and 150 mg), providing validation for the implementation of anti-PCSK9 monotherapy and combination therapy with statins.
Figure 2.
Calibration of the model to match the total anti-PCSK9, total PCSK9, and LDLc reduction data from the phase I clinical study for RG7652 (results shown for three dose groups: 150, 300, and 800 mg); blue: individual clinical subjects; red: mean and 10–90 percentile range for virtual subjects. LDLc, low density lipoprotein cholesterol.
Figure 3.
Model validation against phase I clinical data. LDLc lowering (% of baseline: calculated relative to the LDLc at time of first anti-PCSK9 dose) in response to multiple dose anti-PCSK9 as monotherapy and in combination with atorvastatin; (a) 40 mg anti-PCSK9, (b) 40 mg anti-PCSK9 + atorvastatin, (c) 150 mg anti-PCSK9, (d) 150 mg anti-PCSK9 + atorvastatin. Blue: individual clinical subjects; red: mean and 10–90 percentile range for virtual subjects. LDLc, low density lipoprotein cholesterol.
Model-based predictions
LDLc lowering for phase II RG7652 dosing regimens. The phase II study of RG7652 includes the following five arms for anti-PCSK9 administration: 400 mg q4w; 200, 400, and 800 mg q8w; and 800 mg q12w. The trial is open to all-comers with LDLc >90 mg/dl, and subjects may be on statin background or statin intolerant.
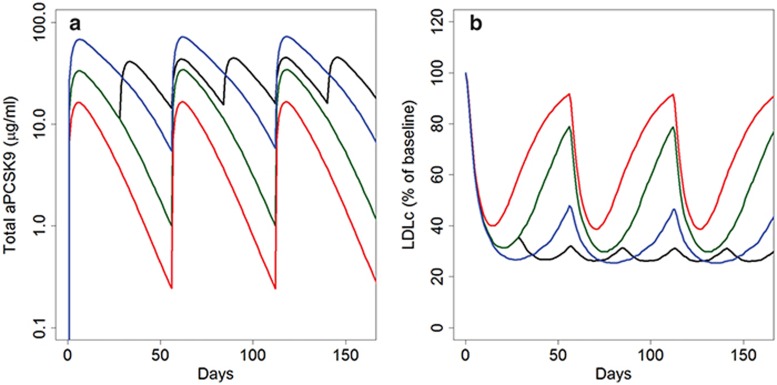
The anti-PCSK9 QSP model was used to simulate and predict results of the phase II trial to help anticipate likely results, predict the implications of mechanistic interactions between anti-PCSK9 and statin mediated regulation of cholesterol processing, and support planning for future clinical development. Two virtual patient populations were developed based on the anticipated LDLc and PCSK9 ranges of the phase II subjects: statin-background subjects (anticipated to be 80% of the trial cohort), statin-intolerant subjects (anticipated at 20% of the trial cohort). Median predictions for each of the phase II virtual populations for the different study arms are shown in Figure 4 (Supplementary Figure S5 shows population variability in response). Simulations for a regimen of 400 mg q4w show sustained reductions in LDLc without significant rebound in LDLc whereas a q8w schedule is predicted to require the higher dose of 800 mg to limit LDLc fluctuations. Doses of 400 and 200 mg are also predicted to decrease LDLc but reductions are not maintained due to inadequate drug concentrations towards the end of the dosing interval. Based on these predictions 400 mg q4w or 800 mg q8w are predicted to be the most efficacious regimens among those tested for sustained reduction of LDLc.
Figure 4.
Model predictions for RG7652 phase II study. (a) Median anti-PCSK9 and (b) median LDLc as a percent of baseline simulations for four different dose regimens shown; black: 400 mg q4w; blue: 800 mg q8w; dark green: 400 q8w; red: 200 q8w. LDLc, low density lipoprotein cholesterol.
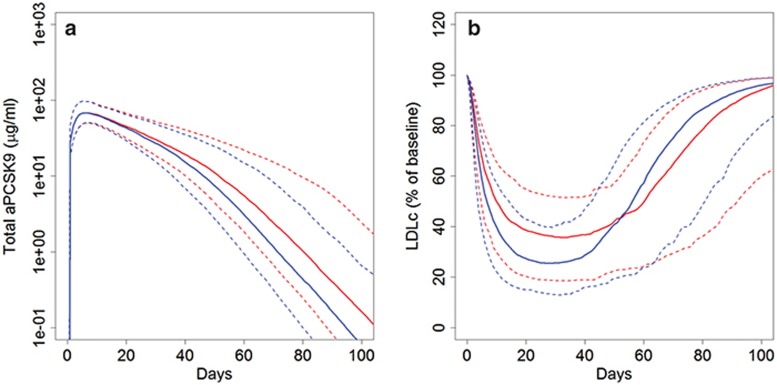
Statin background therapy causes a slight increase in maximum LDLc reduction but faster LDLc rebound. Statin therapy increases PCSK9 levels because the statin-induced reduction in hepatic cholesterol content leads to upregulation of the sterol regulatory element-binding protein 2 (SREBP2), which in turn increases synthesis of PCSK9 and LDLR. Thus, statin background therapy might influence the response to anti-PCSK9. Phase I data were insufficient to characterize this effect due to a small sample size (N = 12 patients on the combination of anti-PCSK9 and statins) and the short duration of statin treatment and limited statin doses in this trial. In order to predict and understand the impact of statin background therapy on the response to anti-PCSK9, we performed model-based investigations in the phase II virtual populations with and without statin background therapy. Based on trial entry criteria and enrollment expectations, statin-treated and statin-intolerant populations were developed with similar entry LDLc levels in the presence and absence of statin background, respectively. Despite comparable entry LDLc levels, statin background resulted in the anticipated higher simulated entry PCSK9 levels. Figure 5 shows the predicted response of the two virtual populations to an 800 mg single administration of anti-PCSK9. Median LDLc lowering is initially greater for subjects on a statin background (Figure 5b), due to neutralization of the greater LDL-increasing effects of higher levels of PCSK9. Higher PCSK9 levels also result in greater predicted target-mediated drug disposition, increasing clearance of anti-PCSK9 (Figure 5a) and resulting in a faster rebound of LDLc in the statin-treated subjects (Figure 5b). However, due to the modest (20–50%) statin-induced increase in PCSK9, the differences in predicted % LDLc lowering between statin-background and statin-untreated subjects are relatively small.
Figure 5.
Effect of statin background on response to anti-PCSK9. Response to single dose of 800 mg anti-PCSK9 for subjects on statin background (blue) and statin naïve subjects (red). (a) Total anti-PCSK9 and (b) LDLc. Solid lines show median predictions and dashed lines indicate the 10–90 percentile range. LDLc, low density lipoprotein cholesterol.
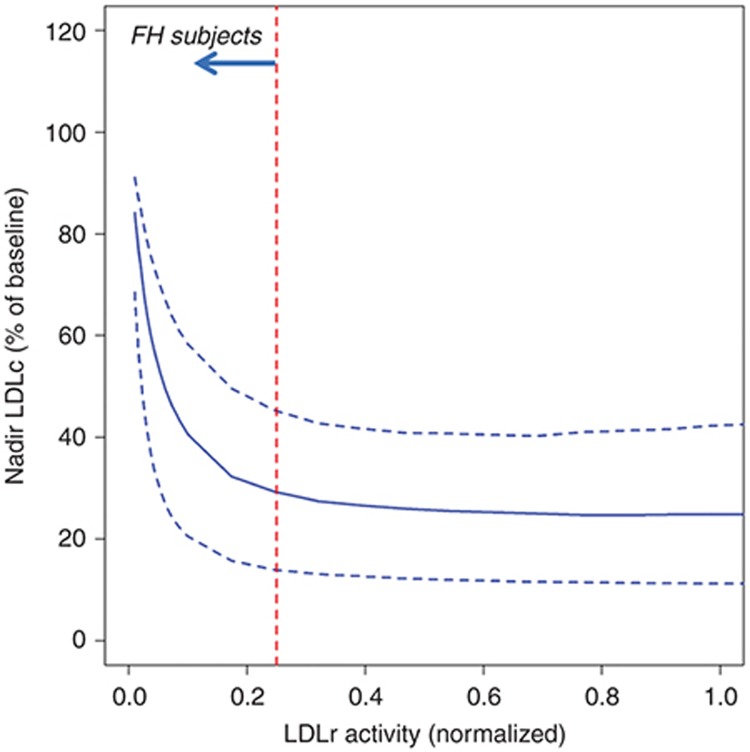
Response of FH subjects to anti-PCSK9 depends on functional LDLR activity. FH subjects have high LDLc levels, primarily due to an impaired LDLR functionality that reduces cellular LDL uptake. A virtual population consisting of both FH and dyslipidemics was utilized to evaluate the impact of LDLR activity on the response to anti-PCSK9. The FH subjects in the virtual population were calibrated with functional LDLR activity <25% of normal in keeping with clinical data21 as described in the Methods. The virtual population captures the correlation (R = 0.7) reported between PCSK9 levels and LDLc in FH subjects.22 Figure 6 shows the nadir LDLc levels as a function of LDLR activity for the virtual cohort in response to a simulated single 800 mg dose of anti-PCSK9 on statin background. For functional LDLR levels >10% of normal, LDLR activity does not appreciably impact LDLc lowering in response to anti-PCSK9. However, subjects with LDLR activity below 10% of normal are predicted to exhibit a weaker LDL response to anti-PCSK9 due to a greater relative importance of non-LDLR mediated LDLc clearance in these subjects. Variability in predicted response at a given LDLR activity is due to concurrent variability in the parameters associated with other biological mechanisms.
Figure 6.
Nadir LDLc (% of baseline) vs. functional LDLR level/activity. FH, familial hypercholesterolemic; LDLc, low density lipoprotein cholesterol; LDLR, low density lipoprotein receptors.
Minimal impact of baseline LDLc and PCSK9 on anti-PCSK9 driven LDLc % lowering. Phase I data did not reveal any significant correlation between response to anti-PCSK9 and baseline levels of either LDLc or PCSK9. We examined the impact of anti-PCSK9 on LDLc levels in the virtual subjects. The relative lowering in LDLc is predicted to be independent of baseline LDLc levels, with variability in absolute nadir LDLc levels over a wide range of baseline LDLc (see Supplementary Figure S1). Simulations suggested a weak but significant correlation between LDLc lowering and baseline PCSK9, with higher baseline PCSK9 levels resulting in lower nadir LDLc.
Discussion
We have developed a QSP model of the mechanistic interaction and cross-regulation of LDLc, LDLR, and PCSK9 in dyslipidemic disease and used it to predict anti-PCSK9 mAb induced reduction in LDLc in subjects with and without statin background therapy and in hypercholesterolemic subjects. Additionally, we have explored the relationship between baseline LDLc and PCSK9 biomarker levels and anti-PCSK9 induced changes in LDLc.
The model structure and numerous parameterizations representing different virtual subjects were developed based on preclinical mechanistic data and calibrated to clinical data on: untreated subjects; LDLc lowering by statins; and RG7652 PK and effects on total PCSK9 and LDLc in single-administration treatment in the phase I trial. Virtual populations comprising sets of virtual subjects were validated by verifying that changes in PCSK9 in response to atorvastatin and changes in LDLc and PCSK9 in response to multiple administration of RG7652 (as monotherapy and in combination with atorvastatin) quantitatively and statistically matched phase I clinical data. While somewhat limited by clinical data availability, these validation efforts confirm that the quantitative representation of critical elements of the model including PCSK9 and LDLc cross-regulation, statin and anti-PCSK9 effects, and interactions between the drugs, are consistent with available clinical data.
Exploration of alternate parameterizations via virtual subjects is an important aspect of research in mechanistic QSP models.23,24 Systems models typically include numerous parameters that cannot be uniquely identified. Furthermore, many of these parameters demonstrate inter-subject variability. However, the potential impact of this variability and uncertainty can be addressed by exploring alternate parameterizations that recapitulate higher level behaviors of interest. An analysis of numerous biological systems models found that the majority of parameters in such models are “sloppy parameters” whose values need not be accurately determined for confidence in model predictions.25 Rather, reproducing system level (phenotypic and clinical) outputs of interest was found to be more critical to ensuring subsequent predictability of the same outputs or measurements. Thus, recapitulation of LDLc and PCSK9 levels in response to statin and anti-PCSK9 treatment provides the validation needed to successfully apply the model and virtual populations to forward simulation and prediction of LDLc and PCSK9 levels under different conditions and protocols. However, because virtual population variability addresses not only inter-patient variability, but also alternate mechanistic hypotheses and parameter uncertainty, the virtual population variability does not correspond directly to clinical variability. The richer the clinical data on interventions modifying the pathways of interest, the stronger the constraints on critical parameters and the greater the correspondence between virtual population variability and clinical variability.
A key application of the model was the prediction of LDLc modulation for various regimens of RG7652 treatment in an ongoing phase II trial. The predictions helped set expectations for the outcomes of phase II results, including optimal dosing regimens, and increased confidence in simulations and dose recommendations obtained with a nonlinear mixed-effects population PKPD model (N.R. Budha et al., unpublished data). Population PKPD analysis is particularly useful for the characterization of variability and identification of covariates based on clinical data. However, due to lack of mechanistic detail, the PKPD modeling (N.R. Budha et al., unpublished data) was unable to select between two indirect response models with alternate explanations for the differential LDLc response to anti-PCSK9 in statin-untreated and treated arms in the phase I study of RG7652. Furthermore, clinical-data driven indirect response models cannot be confidently applied to predict responses in patient populations with fundamentally different underlying biology than those previously tested. However, because of its emphasis on mechanism rather than prior clinical data, a QSP approach is better suited to forward-prediction in scenarios for which there is minimal clinical experience, such as the impact of background statin therapy on anti-PCSK9 efficacy or the prediction of treatment response in FH or other untested populations. Simulation of anti-PCSK9 treatment with and without background statins predicted a small but potentially detectable impact of statin-treatment on anti-PCSK9 mediated LDLc lowering with statin treatment. Peak LDLc reduction was predicted to be slightly better on a statin background, due to neutralization of the statin-induced PCSK9 and its detrimental effect on LDLc. However, higher PCSK9 in statin-treated subjects was also predicted to lead to greater target-mediated clearance of anti-PCSK9 and rebound of LDLc. These trends are consistent with limited published clinical data for anti-PCSK9 efficacy with and without statins.4,5,6,7,8,9,26 However, the predicted differences in LDLc-profiles between anti-PCSK9 treated subjects with and without background statin are small, suggesting that anti-PCSK9 dose need not be adjusted to compensate for statin-mediated PCSK9 up-regulation and that statin background therapy is unlikely to cause significantly greater anti-PCSK9 induced LDLc lowering, a potential safety concern when combining two potent LDLc-lowering agents.
The model was also used to compare predicted responses in dyslipidemics vs. FH subjects. Results suggested that the reduced LDLR activity in heterozygous FH (functional LDLR activity of 10–25% of normal) is unlikely to strongly impact the anti-PCSK9 responsiveness whereas a reduction of functional LDLR levels to less than 10% of normal, which may be seen in homozygous FH subjects, is predicted to reduce the efficacy of anti-PCSK9, due to the greater relative importance LDLR-independent LDL uptake mechanisms. This effort would not have been easily conducted in an indirect-response model lacking the mechanistic detail of the QSP model.
Finally, the model was used to investigate potential predictive biomarkers. Although the relative lowering of LDLc was predicted to be independent of baseline LDLc levels, the variability in the absolute nadir levels in LDLc helps identify the anticipated variability in response of subjects over a wide range of baseline LDLc levels. We also predicted a weak but significant correlation between baseline PCSK9 and LDLc lowering on anti-PCSK9 that is unlikely to impact efficacy or dose requirements to meet desired LDLc levels.
One limitation of the work described here as well as of many shorter biomarker-focused early clinical trials in cardiovascular disease is a focus on lipid biomarkers as an endpoint, instead of the more clinically important outcome of cardiovascular (CV) risk reduction. For example, despite early promise of CETP inhibitors for treatment of atherosclerosis, recent phase III trials have failed to show improvement in CV risk despite increases in HDL cholesterol.27,28 However, unlike HDL elevation, there is strong evidence for LDL reduction as a predictive marker of CV risk reduction. A linear correlation of LDLc to CV risk derived from clinical trials of statin therapy supports a causative role of LDLc reduction on CV risk reduction. However, it remains to be determined whether this relationship applies to other LDL-lowering agents, as statins might have anti-inflammatory and additional effects that could contribute to CV risk reduction.29
Other biology not included in the model scope could also influence clinical results. The model does not consider LDL particle size distribution or HDL dynamics. Although total LDLc is a marker for CV risk, small, dense LDL particles are considered more atherogenic,30 and alteration of the LDL particle distribution by anti-PCSK9 may influence CV risk beyond what would be predicted based on LDLc alone. Similarly, any impact on HDL resulting from LDL modification could also influence inflammation, reverse cholesterol transport, plaque accumulation, and CV risk. These aspects of the biology can be considered in future development of the model as warranted by ongoing clinical research. The model can also be used to evaluate new or next generation drugs targeting the pathways or proteins represented.
In conclusion, the application of the QSP model of LDLc/PCSK9 interactions has led to an increased understanding of the effects of anti-PCSK9 treatment on different patient subtypes, thus enabling more robust planning for potential clinical scenarios where data are currently limited. Additionally, the model is adaptable and extendable to address questions related to future investigation of additional interventions and broader related biology.
Methods
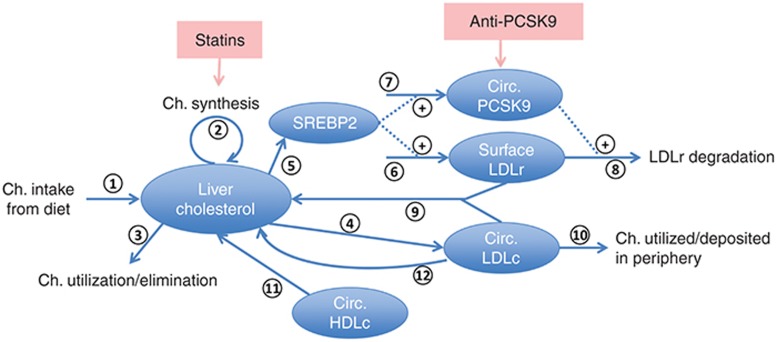
A mechanistic mathematical model of the response of LDLc to intervention with anti-PCSK9 and statins was developed in the Simbiology software package (Mathworks, Natick, MA). A schematic of the model is depicted in Figure 7 and details on model states, parameters and equations are included in the Supplementary Tables S1-S3 and Figures S2-4.
Figure 7.
Schematic of Quantitative Systems Pharmacology model for evaluation of LDLc lowering with anti-PCSK9 as monotherapy and combination with statins. Ch., cholesterol; Circ., circulating; LDLc, low density lipoprotein cholesterol.
Model structure
The model shown includes the following major processes in the systemic circulation and hepatic compartment:
1. Cholesterol consumption, excretion, and utilization: The primary sources of cholesterol are represented and include dietary cholesterol intake with ~50% absorption31 (arrow 1) and endogenous synthesis by hepatocytes32 (arrow 2). A sink from the hepatic cholesterol pool (arrow 3) corresponding primarily to bile formation completes the mass balance.
2. Hepatocyte cholesterol synthesis, uptake, and degradation: The model includes a mean representation of circulating LDL particles with a cholesterol content of 0.92 mg/nmol of particles.33 The formation of the LDL particles34 is represented in a single step with a rate proportional to the hepatic cholesterol content (arrow 4). Details of apoB-100 particle formation and metabolism are not included in the model. Circulating PCSK9 (ng/ml) and hepatocyte LDLR levels are modeled. Both PCSK9 and LDLR are transcriptionally upregulated by SREBP2,35 which is expressed in normalized units relative to an untreated, healthy subject level. SREBP2 is regulated by hepatic cholesterol content (arrow 5) and positively impacts LDLR and PCSK9 production (arrows 6 and 7). PCSK9 directs LDLR to lysosomal degradation rather than recycling in hepatocytes and thereby favors greater LDLc levels in plasma.36,37 Correspondingly, LDLR degradation is modeled as driven by PCSK9 (arrow 8). Free LDLR is modeled as binding LDLc, resulting in hepatocyte LDLc uptake and increased liver cholesterol levels (arrow 9). LDL particles are also consumed in the periphery (arrow 10). In addition to uptake of LDL particles, hepatocytes also take up cholesterol in the form of HDL particles (arrow 11). The model does not address HDL dynamics, and instead represents a fixed, invariant pool of HDL. A LDLR independent route of hepatocyte LDLc uptake (arrow 12) is also included.38 The cholesterol metabolism model developed here is focused on the key mechanisms and details relevant represent the impact of the statin and anti-PCSK9 combination therapy. A more detailed cholesterol metabolism model is presented in ref. 39; the scope of the current model can be expanded if other interventions such as ezetimibe or HDL modifying therapies are of interest.
3. Therapeutic modulation of cholesterol processing: Anti-PCSK9 and statin drugs are represented based on their mechanisms of action. Statin induced inhibition of HMG-CoA reductase and thus cholesterol synthesis is represented as a constant inhibition of the endogenous hepatic cholesterol synthesis rate. Given the daily treatment regimen for statins, temporal fluctuations due to statin PK can be neglected for predictions on the days-weeks time-scale. A full target mediated drug disposition (TMDD) model40 is included to capture the dynamics of anti-PCSK9 (drug) and its impact on PCSK9 (target) levels. Anti-PCSK9 is administered subcutaneously (SC) and absorption from the depot to the central plasma compartment is represented by a first order process with rate constant ka and bioavailability F. Reversible elementary binding kinetics of drug and target are modeled with forward and reverse rate constants kon and koff. The clearances of the free drug, target, and drug-target complex are represented as first order processes. Distribution of the free drug to a peripheral compartment is also included. Due to the highly perfused nature of the liver, extracellular hepatic drug and LDLc concentrations are assumed to be equivalent to serum concentrations.
Model calibration and development of virtual population
A reference untreated, healthy subject was developed using a corresponding reference parameterization (details on model equations and parameters are included in Supplementary Tables S1–3). Parameter values were primarily obtained from literature data on the pathways and proteins. Parameters for which specific data were not available were calibrated to reproduce phenotypes and measurements for a reference subject corresponding to a typical or average healthy subject. The subsequent methodology to develop and validate virtual subjects and each virtual population comprised three key steps:
1. Parameter exploration: Variability in key model parameters was explored to generate alternate parameterizations, each of which represents a potential virtual subject. Reference values for the parameters and the ranges of exploration were determined from available literature data. A list of the model parameters explored for generation of the virtual subjects is shown in Supplementary Table S2.
2. Patient selection: The baseline characteristics and selected responses to clinical interventions of each virtual subject was simulated. Supplementary Table S4 shows the clinical data used for virtual subject selection. A virtual subject was acceptable if simulated behaviors matched specified clinical data characterizing the patient phenotype(s) of interest. A subset of all valid virtual subjects was selected to match the desired distributions of clinical measurements. Each virtual subject's relative contribution to virtual population measurements is weighted by a virtual subject specific prevalence weight;24 a simple weighting scheme was applied by which each virtual subject was assigned a weight of either 1 or 0 to optimize the fit to clinical data distributions.
3. Patient validation: A subset of the clinical data was reserved for patient validation. The subjects selected in step (ii) were validated against this clinical data to increase confidence in the predictive ability of the model and of the virtual subjects selected for research. Supplementary Table S4 shows the clinical data used for virtual subject validation.
Four virtual populations were specified as collections of virtual subjects that together reproduce mean (SD) clinical trial entry criteria/data for baseline LDLc (mg/dl) and PCSK9 (ng/ml) for the corresponding clinical populations, as follows:
A phase I virtual population with untreated LDLc = 160(20), PCSK9 = 290(40)
A phase II statin-background virtual population with statin-treated LDLc = 125(20), PCSK9 = 360(40)
A phase II statin-intolerant virtual population with untreated LDLc = 125(20), PCSK9 = 275(40)
An FH virtual population
The baseline LDLc distribution in the phase II study is not known, and is assumed to be 125(20) (mean(SD)) for both the statin treated and statin intolerant populations to allow an unbiased comparison of anti-PCSK9 monotherapy and combination therapy with statins. Virtual subjects in the FH population were restricted to those with functional LDLR levels below 25% of normal.21 Only non-FH virtual subjects with functional LDLR levels of 25–100% of normal were included in phase I and phase II populations because of the low representation of FH subjects in the broader hypercholesterolemic population. The phase I virtual population was used for calibration and validation of the model to clinical observations. The phase II and FH virtual populations were used for model-based predictions.
Conflict of interest
All authors are full time employees at Genentech Inc., a subsidiary of Roche.
Author contributions
S.R. and K.G. wrote the manuscript. S.R., K.G., A.B., and P.F. designed the research. K.G. and N.B. performed the research. S.R., K.G., N.B., A.B., J.D.D., and P.F. analyzed the data. N.B. contributed new reagents/analytical tools.
Study Highlights
Acknowledgments
The authors thank Whit Tingley, Genentech, for technical discussions and review of the manuscript and Asawari Samant, Mathworks, for Simbiology and MATLAB software support.
Supplementary Material
References
- Duff C.J., Hooper N.M. PCSK9: an emerging target for treatment of hypercholesterolemia. Expert Opin. Ther. Targets. 2011;15:157–168. doi: 10.1517/14728222.2011.547480. [DOI] [PubMed] [Google Scholar]
- Cohen J., Pertsemlidis A., Kotowski I.K., Graham R., Garcia C.K., Hobbs H.H. Low LDL cholesterol in individuals of African descent resulting from frequent nonsense mutations in PCSK9. Nat. Genet. 2005;37:161–165. doi: 10.1038/ng1509. [DOI] [PubMed] [Google Scholar]
- Hooper A.J., Burnett J.R. Anti-PCSK9 therapies for the treatment of hypercholesterolemia. Expert Opin. Biol. Ther. 2013;13:429–435. doi: 10.1517/14712598.2012.748743. [DOI] [PubMed] [Google Scholar]
- Giugliano R.P., et al. LAPLACE-TIMI 57 Investigators Efficacy, safety, and tolerability of a monoclonal antibody to proprotein convertase subtilisin/kexin type 9 in combination with a statin in patients with hypercholesterolaemia (LAPLACE-TIMI 57): a randomised, placebo-controlled, dose-ranging, phase 2 study. Lancet. 2012;380:2007–2017. doi: 10.1016/S0140-6736(12)61770-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenney J.M., Koren M.J., Kereiakes D.J., Hanotin C., Ferrand A.C., Stein E.A. Safety and efficacy of a monoclonal antibody to proprotein convertase subtilisin/kexin type 9 serine protease, SAR236553/REGN727, in patients with primary hypercholesterolemia receiving ongoing stable atorvastatin therapy. J. Am. Coll. Cardiol. 2012;59:2344–2353. doi: 10.1016/j.jacc.2012.03.007. [DOI] [PubMed] [Google Scholar]
- Dias C.S., et al. Effects of AMG 145 on low-density lipoprotein cholesterol levels: results from 2 randomized, double-blind, placebo-controlled, ascending-dose phase 1 studies in healthy volunteers and hypercholesterolemic subjects on statins. J. Am. Coll. Cardiol. 2012;60:1888–1898. doi: 10.1016/j.jacc.2012.08.986. [DOI] [PubMed] [Google Scholar]
- Stein E.A., et al. Effect of a monoclonal antibody to PCSK9, REGN727/SAR236553, to reduce low-density lipoprotein cholesterol in patients with heterozygous familial hypercholesterolaemia on stable statin dose with or without ezetimibe therapy: a phase 2 randomised controlled trial. Lancet. 2012;380:29–36. doi: 10.1016/S0140-6736(12)60771-5. [DOI] [PubMed] [Google Scholar]
- Roth E.M., McKenney J.M., Hanotin C., Asset G., Stein E.A. Atorvastatin with or without an antibody to PCSK9 in primary hypercholesterolemia. N. Engl. J. Med. 2012;367:1891–1900. doi: 10.1056/NEJMoa1201832. [DOI] [PubMed] [Google Scholar]
- Raal F., et al. Low-density lipoprotein cholesterol-lowering effects of AMG 145, a monoclonal antibody to proprotein convertase subtilisin/kexin type 9 serine protease in patients with heterozygous familial hypercholesterolemia: the Reduction of LDL-C with PCSK9 Inhibition in Heterozygous Familial Hypercholesterolemia Disorder (RUTHERFORD) randomized trial. Circulation. 2012;126:2408–2417. doi: 10.1161/CIRCULATIONAHA.112.144055. [DOI] [PubMed] [Google Scholar]
- Tingley W. Effects of RG7652, a fully human mAb against proprotein convertase subtilisin/kexin type 9, on LDL-c: a Phase I, randomised, double-blind, placebo-controlled, single- and multiple-dose study. Eur Heart J. 2013;34 (suppl 1:P4183. [Google Scholar]
- van der Graaf P.H., Benson N. Systems pharmacology: bridging systems biology and pharmacokinetics-pharmacodynamics (PKPD) in drug discovery and development. Pharm. Res. 2011;28:1460–1464. doi: 10.1007/s11095-011-0467-9. [DOI] [PubMed] [Google Scholar]
- Peterson M.C., Riggs M.M. A physiologically based mathematical model of integrated calcium homeostasis and bone remodeling. Bone. 2010;46:49–63. doi: 10.1016/j.bone.2009.08.053. [DOI] [PubMed] [Google Scholar]
- Rullmann J.A., Struemper H., Defranoux N.A., Ramanujan S., Meeuwisse C.M., van Elsas A. Systems biology for battling rheumatoid arthritis: application of the Entelos PhysioLab platform. Syst. Biol. (Stevenage) 2005;152:256–262. doi: 10.1049/ip-syb:20050053. [DOI] [PubMed] [Google Scholar]
- Kirouac D.C., et al. Computational modeling of ERBB2-amplified breast cancer identifies combined ErbB2/3 blockade as superior to the combination of MEK and AKT inhibitors. Sci. Signal. 2013;6:ra68. doi: 10.1126/scisignal.2004008. [DOI] [PubMed] [Google Scholar]
- Nissen S.E., et al. REVERSAL Investigators Effect of intensive compared with moderate lipid-lowering therapy on progression of coronary atherosclerosis: a randomized controlled trial. JAMA. 2004;291:1071–1080. doi: 10.1001/jama.291.9.1071. [DOI] [PubMed] [Google Scholar]
- Stern R.H., Yang B.B., Hounslow N.J., MacMahon M., Abel R.B., Olson S.C. Pharmacodynamics and pharmacokinetic-pharmacodynamic relationships of atorvastatin, an HMG-CoA reductase inhibitor. J. Clin. Pharmacol. 2000;40:616–623. [PubMed] [Google Scholar]
- Careskey H.E., Davis R.A., Alborn W.E., Troutt J.S., Cao G., Konrad R.J. Atorvastatin increases human serum levels of proprotein convertase subtilisin/kexin type 9. J. Lipid Res. 2008;49:394–398. doi: 10.1194/jlr.M700437-JLR200. [DOI] [PubMed] [Google Scholar]
- Dubuc G., et al. Statins upregulate PCSK9, the gene encoding the proprotein convertase neural apoptosis-regulated convertase-1 implicated in familial hypercholesterolemia. Arterioscler. Thromb. Vasc. Biol. 2004;24:1454–1459. doi: 10.1161/01.ATV.0000134621.14315.43. [DOI] [PubMed] [Google Scholar]
- Mayne J., et al. Plasma PCSK9 levels are significantly modified by statins and fibrates in humans. Lipids Health Dis. 2008;7:22. doi: 10.1186/1476-511X-7-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., et al. An anti-PCSK9 antibody reduces LDL-cholesterol on top of a statin and suppresses hepatocyte SREBP-regulated genes. Int. J. Biol. Sci. 2012;8:310–327. doi: 10.7150/ijbs.3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh S.E., Foster A.H., Whittall R.A., Hubbart C.S., Humphries S.E. Update and analysis of the University College London low density lipoprotein receptor familial hypercholesterolemia database. Ann. Hum. Genet. 2008;72:485–498. doi: 10.1111/j.1469-1809.2008.00436.x. [DOI] [PubMed] [Google Scholar]
- Raal F., Panz V., Immelman A., Pilcher G. Elevated PCSK9 levels in untreated patients with heterozygous or homozygous familial hypercholesterolemia and the response to high-dose statin therapy. J. Am. Heart Assoc. 2013;2:e000028. doi: 10.1161/JAHA.112.000028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoda L., et al. The Type 1 Diabetes PhysioLab Platform: a validated physiologically based mathematical model of pathogenesis in the non-obese diabetic mouse. Clin. Exp. Immunol. 2010;161:250–267. doi: 10.1111/j.1365-2249.2010.04166.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt B.J., Casey F.P., Paterson T., Chan J.R. Alternate virtual populations elucidate the type I interferon signature predictive of the response to rituximab in rheumatoid arthritis. BMC Bioinformatics. 2013;14:221. doi: 10.1186/1471-2105-14-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutenkunst R.N., Waterfall J.J., Casey F.P., Brown K.S., Myers C.R., Sethna J.P. Universally sloppy parameter sensitivities in systems biology models. PLoS Comput. Biol. 2007;3:1871–1878. doi: 10.1371/journal.pcbi.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein E.A., et al. Effect of a monoclonal antibody to PCSK9 on LDL cholesterol. N. Engl. J. Med. 2012;366:1108–1118. doi: 10.1056/NEJMoa1105803. [DOI] [PubMed] [Google Scholar]
- Mohammadpour A.H., Akhlaghi F. Future of cholesteryl ester transfer protein (CETP) inhibitors: a pharmacological perspective. Clin. Pharmacokinet. 2013;52:615–626. doi: 10.1007/s40262-013-0071-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barter P., Rye K.A. Cholesteryl ester transfer protein inhibition to reduce cardiovascular risk: Where are we now. Trends Pharmacol. Sci. 2011;32:694–699. doi: 10.1016/j.tips.2011.07.004. [DOI] [PubMed] [Google Scholar]
- Lefer D.J. Statins as potent antiinflammatory drugs. Circulation. 2002;106:2041–2042. doi: 10.1161/01.cir.0000033635.42612.88. [DOI] [PubMed] [Google Scholar]
- Sacks F.M., Campos H. Clinical review 163: Cardiovascular endocrinology: Low-density lipoprotein size and cardiovascular disease: a reappraisal. J. Clin. Endocrinol. Metab. 2003;88:4525–4532. doi: 10.1210/jc.2003-030636. [DOI] [PubMed] [Google Scholar]
- Bosner M.S., Lange L.G., Stenson W.F., Ostlund R.E. Percent cholesterol absorption in normal women and men quantified with dual stable isotopic tracers and negative ion mass spectrometry. J. Lipid Res. 1999;40:302–308. [PubMed] [Google Scholar]
- Sudhop T., et al. Changes in cholesterol absorption and cholesterol synthesis caused by ezetimibe and/or simvastatin in men. J. Lipid Res. 2009;50:2117–2123. doi: 10.1194/jlr.P900004-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromwell W.C., et al. LDL Particle Number and Risk of Future Cardiovascular Disease in the Framingham Offspring Study - Implications for LDL Management. J. Clin. Lipidol. 2007;1:583–592. doi: 10.1016/j.jacl.2007.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havel R.J. The formation of LDL: mechanisms and regulation. J. Lipid Res. 1984;25:1570–1576. [PubMed] [Google Scholar]
- Horton J.D., et al. Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc. Natl. Acad. Sci. U.S.A. 2003;100:12027–12032. doi: 10.1073/pnas.1534923100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton J.D., Cohen J.C., Hobbs H.H. Molecular biology of PCSK9: its role in LDL metabolism. Trends Biochem. Sci. 2007;32:71–77. doi: 10.1016/j.tibs.2006.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan D.C., Lambert G., Barrett P.H., Rye K.A., Ooi E.M., Watts G.F. Plasma proprotein convertase subtilisin/kexin type 9: a marker of LDL apolipoprotein B-100 catabolism. Clin. Chem. 2009;55:2049–2052. doi: 10.1373/clinchem.2009.128645. [DOI] [PubMed] [Google Scholar]
- Osono Y., Woollett L.A., Herz J., Dietschy J.M. Role of the low density lipoprotein receptor in the flux of cholesterol through the plasma and across the tissues of the mouse. J. Clin. Invest. 1995;95:1124–1132. doi: 10.1172/JCI117760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mc Auley M.T., Wilkinson D.J., Jones J.J., Kirkwood T.B. A whole-body mathematical model of cholesterol metabolism and its age-associated dysregulation. BMC Syst. Biol. 2012;6:130. doi: 10.1186/1752-0509-6-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mager D.E., Jusko W.J. General pharmacokinetic model for drugs exhibiting target-mediated drug disposition. J. Pharmacokinet. Pharmacodyn. 2001;28:507–532. doi: 10.1023/a:1014414520282. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.