Abstract
Glyphosate is a widely applied broad-spectrum systemic herbicide that inhibits competitively the penultimate enzyme 5-enolpyruvylshikimate 3-phosphate synthase (EPSPS) from the shikimate pathway, thereby causing deleterious effects. A glyphosate-resistant Arabidopsis mutant (gre1) was isolated and genetic analyses indicated that a dysfunctional red (R) and far-red (FR) light receptor, phytochrome B (phyB), caused this phenotype. This finding is consistent with increased glyphosate sensitivity and glyphosate-induced shikimate accumulation in low R:FR light, and the induction of genes encoding enzymes of the shikimate pathway in high R:FR light. Expression of the shikimate pathway genes exhibited diurnal oscillation and this oscillation was altered in the phyB mutant. Furthermore, transcript analysis suggested that this diurnal oscillation was not only dependent on phyB but was also due to circadian regulatory mechanisms. Our data offer an explanation of the well documented observation that glyphosate treatment at various times throughout the day, with their specific composition of light quality and intensity, results in different efficiencies of the herbicide.
Keywords: Arabidopsis thaliana, glyphosate, herbicide resistance, shikimate pathway, phyB, circadian rhythm
INTRODUCTION
Glyphosate (N-phosphonomethyl glycine) is a highly effective, post-emergence, broad-spectrum, systemic herbicide that mobilizes to the chloroplasts of actively growing sink tissues and inhibits irreversibly a shikimate pathway component EPSPS (EC 2.5.1.19) (Figure S1), causing subsequent plant death due to the disturbance of the carbon metabolism required to produce aromatic amino acids and their derivatives in plants (Amrhein et al., 1980; Herrmann, 1995b; Bromilow and Chamberlain, 2000). Since 1996, glyphosate-resistant (GR) crops have been in wide commercial use partly due to the apparently low toxicity (Franz et al., 1997; Williams et al., 2000) and fast immobilization and degradation characteristics of glyphosate in soils (Rüppel et al., 1977; Zhou et al., 2004). The glyphosate application range has increased dramatically mainly because of their cost-effective, superior weed control technology and no-till monoculture systems (Gianessi, 2008; Duke and Powles, 2009). Widespread glyphosate use causes high selection pressure on weeds that has resulted in weed population shifts and the evolution of over 20 GR weed species (Benbrook, 2012; Heap, 2013). The mechanism(s) of resistance of most of these species, while not well characterized, are believed to be due mainly to alterations of the target enzyme. In the cases of goosegrass (Eleusine indica) and ryegrass species (Lolium rigidum, and Lolium multiflorum) a mutation at Pro106 in EPSPS confers glyphosate resistance (Baerson et al., 2002; Wakelin and Preston, 2006; Jasieniuk et al., 2008; Kaundun et al., 2011), whereas in populations of Palmer's amaranth (Amaranthus palmeri) and in Italian ryegrass (Lolium multiflorum), an amplification of EPSPS underlies the increased glyphosate resistance (Gaines et al., 2010; Salas et al., 2012). In addition, high pressure liquid chromatography (HPLC) and nuclear magnetic resonance (NMR) analyses have revealed tissue-specific translocation rates and vacuolar sequestration in ryegrass (Lolium rigidum) and horseweed (Conyza canadensis) populations (Lorraine-Colwill et al., 2002; Ge et al., 2010). Furthermore, several reports have suggested that glyphosate efficiency is affected by the time of day at which it is sprayed (Martinson et al., 2005) and have attributed this effect to factors such as leaf angle changes (Mohr et al., 2007).
In order to gain a better understanding of glyphosate-resistance mechanisms, we performed a forward genetic screen using a T-DNA tagged (pGPTV-Bar) Arabidopsis thaliana mutant population, ecotype Col0, and obtained a number of mutants that are either hypersensitive or hyposensitive to glyphosate. One mutant, the glyphosate response 1 (gre1), showed an elevated resistant phenotype in shoot and root growth caused by a loss of function of the red and far-red (FR) light receptor, phytochrome B (phyB). Here we demonstrate the role of phyB on glyphosate sensitivity and present a model that links phyB signaling and circadian clock associated control mechanism to the complex regulation of the shikimate pathway.
RESULTS
Characterization of the GR mutant gre1
A glyphosate response mutation screen was performed on agar plates under 16 h white light at a fluence rate of 22 ± 2 μmol m−2 s−1 (Figure S2) and this screen led to the isolation of the gre1 mutant. The gre1 plant showed GR traits such as vigorous shoot growth and new leaf formation, development of lateral root branches and nodules, and high chlorophyll content and reduced shikimate levels in the shoot in response to 15–50 μm glyphosate treatments over 12 days (Figure1). Glyphosate treatments ≤5 μm did not affect shoot growth, however they did cause induction of lateral root development in both genotypes (Figure1a). Glyphosate concentrations ≥15 and ≥30 μm inhibited lateral root and primary leaf formation in the wild-type (wt) only, whereas, in gre1 lateral roots, root nodules and primary rosette leaves developed on glyphosate concentrations up to 50 μm (Figure1a). We determined that 15 and 30 μm glyphosate caused marginal and complete chlorosis respectively in newly formed primary leaves in the wt but not in gre1 (Figure1a). The glyphosate-response phenotype was confirmed by fresh weight comparisons, chlorophyll content analyses and shikimate accumulation assays, and it was shown that gre1 plants gain more fresh weight, contain more chlorophyll and less shikimate in response to glyphosate treatment with the largest differences noticed at an exposure to 15 μm glyphosate (Figure1b–d).
Figure 1.
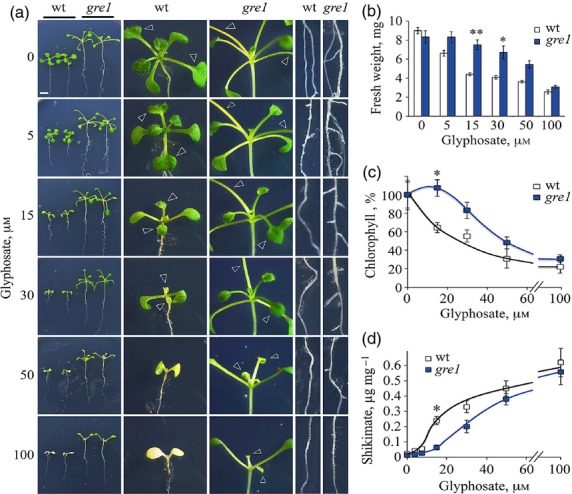
Gre1 plants show glyphosate resistant phenotypes.(a) Phenotypes of wt and gre1 plants screened on 15 or 30 μm glyphosate medium for 12 days under LI = 22 ± 2 μmol m−2 sec−1, 16 h light regime. Close-up pictures show shoot and root phenotypes of wt and gre1 plants. White scale bars represent 5 mm, and empty arrowheads point to the primary leaves.(b–d) Fresh weight, relative chlorophyll content and shikimate level of wt and gre1 plants in glyphosate dose–response studies. Fresh weight data were taken at day 12 and chlorophyll content and shikimate analyses were done at day 9. Each data point represents mean ± standard errors (SEs), with 15 plants analysed for fresh weight, and five plants for chlorophyll and shikimate content analyses. Double and single asterisks indicate P-values < 0.05 and < 0.1 respectively.
The gre1 phenotype at 30 μm glyphosate treatment is strictly fluence rate-dependent, with the most pronounced characteristics in light intensities between 15 and 25 μmol m−2 s−1 (Figure S3). Glyphosate resistance in gre1 is lost when plants are exposed to light intensities outside this range. Gre1 also exhibits morphological differences from the wt, such as longer hypocotyls and shorter roots, earlier flowering, paler green leaves and bright yellow colored seeds (Figure S4).
In addition to the phenotypic characterization of gre1 plants grown on agar plates, we also determined glyphosate resistance of soil grown plants (Figure S5) under a higher fluence rate (75 ± 5 μmol m−2 s−1). The higher fluence rate in soil was required to avoid elongated hypocotyl and petiole due to the Shade Avoidance Syndrome (SAS). In contrast with wt plants, gre1 plants continued to grow after treatment with 200 μm glyphosate (0.2535 mg m−2) and produced seeds even after the death of older leaves (Figure S5). These phenotypic and physiological characteristics of gre1 clearly point to the presence of a glyphosate-resistance mechanism that prevents the irreversible inhibitory effects on the target enzyme and/or a compensating activity on the shikimate pathway.
The glyphosate resistant phenotype of gre1 is caused by dysfunctional phyB
A back-cross analysis (wt × gre1) was performed on gre1 plants to confirm the inheritance of glyphosate resistance (Table S1). In the analysis, all tested 134 F1 plants inherited both glyphosate- and bialaphos-resistant phenotypes from the gre1 parental line and these phenotypes segregated 1:3 (sensitive:resistant) in the F2 progenies, indicating that resistance is a dominant trait. We performed a genetic co-segregation analysis in the F3 GR homozygote progeny using both glyphosate and bialaphos selection procedures and all the 380 lines tested were resistant to both herbicides and confirmed the tight linkage between T-DNA insertion and the glyphosate-response locus.
A thermal asymmetric interlaced polymerase chain reaction (TAIL-PCR) analysis of the mutant identified a T-DNA border sequence in the first exon of PHYB (At2g18790) that encodes an apoprotein of the red (R) and far-red (FR) light receptor phyB (Figure2a). We then performed a PHYB expression analysis (Figure2b) and showed the absence of the PHYB amplicon in gre1. The GR phenotypes of gre1 lines were confirmed by testing another allelic phyB loss-of-function mutant line (phyB-9) that also confers resistance to glyphosate (Figure2c). This result further supported the conclusion that a dysfunctional phyB can cause glyphosate resistance, and suggested that over-expression of PHYB may increase sensitivity to glyphosate. When PHYB was over-expressed on wt and gre1 genetic backgrounds (pbc1 and gpbc1), hypersensitive phenotypes were observed in response to glyphosate with traits that included arrested shoot and root development, twisted cotyledons and shoot anthocyanin accumulation (Figure2c).
Figure 2.
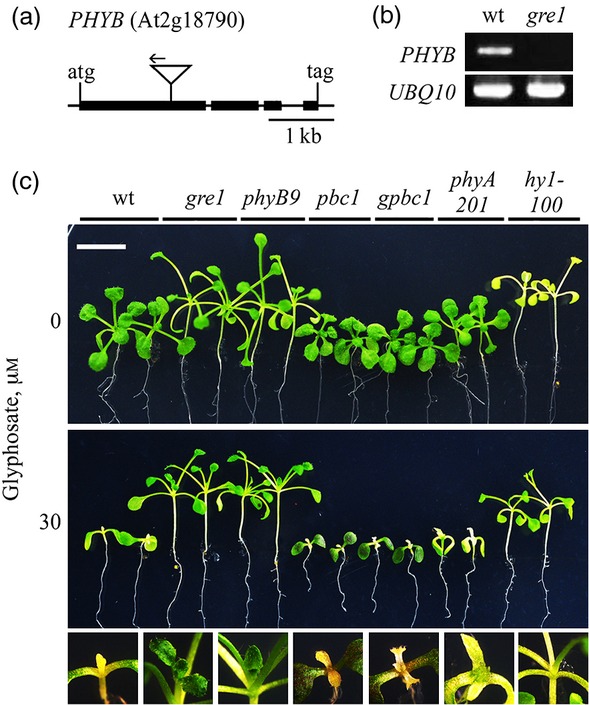
The glyphosate resistant phenotypes of gre1 are conferred by the knock-out mutation of PHYB gene. (a) TAIL-PCR revealed a T-DNA insertion (triangle) at the first exon of PHYB gene (At2g18790) in the gre1 plant. Arrow indicates the left border of T-DNA at the 1.45 kb downstream of the start codon.(b) RT-PCR analysis of PHYB expression in total RNA extracted from wt and gre1 plants. Expression of the housekeeping gene, UBIQUITIN10 (UBQ10) was used to normalize band intensities.(c) Comparison of glyphosate hypersensitive and resistance phenotypes of wt and gre1 plants with an allelic phyB mutant line (phyB-9), PHYB over-expressed lines in wt (pbc1) and in gre1 (gpbc1), a phyA deficient line (phyA-201), and plastid heme-oxygenase loss-of-function mutant (hy1-100), in 30 μm glyphosate treatments under a 16 h, 22 ± 2 μmol m−2 sec−1 light regime. Close-up pictures depicting shoot growth are placed below each corresponding line. Pictures were taken at day 12 of the treatment. Scale bars = 10 mm.
We also tested the response of phyA (the light labile phytochrome) deficient (phyA-201) and phytochromobilin (the light absorbing unit of phytochromes) deficient (hy1-100) plants on agar containing 30 μm glyphosate (Figure2c). The analysis revealed that phyA deficiency does not lead to glyphosate resistance, whereas phytochromobilin deficiency conferred glyphosate resistance and that the glyphosate response is similar to that of gre1 and phyB-9 plants (Figure2c).
Arabidopsis glyphosate hypersensitive phenotypes are far-red-light specific
Phytochrome exists in two distinct photo-reversible isoforms, Pr and Pfr in FR (near 730 nm) and in R (near 660 nm) respectively (Andel et al., 1996; Rockwell et al., 2006). It is generally accepted that the physiologically active form of phytochrome is Pfr (Rüdiger et al., 1983), as it localizes in the nucleus where it interacts with transcription factors (TFs) to regulate gene expression causing photomorphogenesis that in turn enables adaptation of the plant to changing environmental conditions (Nagatani, 2004). In order to examine the physiological relevance of the Pfr in the glyphosate response, wt and gre1 plants were treated with 20 μm glyphosate and grown under high R:FR and low R:FR conditions (Figure3a). Both wt and gre1 showed the same resistance response under high R:FR light conditions, whereas only the wt was hypersensitive under low R:FR. This FR light-specific (low R:FR) glyphosate hypersensitivity of wt plants is reflected in the glyphosate-induced shikimate accumulation. Glyphosate treatment caused a seven-fold increase in shikimate accumulation in shoots at low R:FR as compared with the control plants, but only a three-fold increase at high R:FR (Figure3b). In gre1, both high and low R:FR light caused significantly low amounts of shikimate accumulation in the glyphosate treatment if compared with wt (Figure3b).
Figure 3.
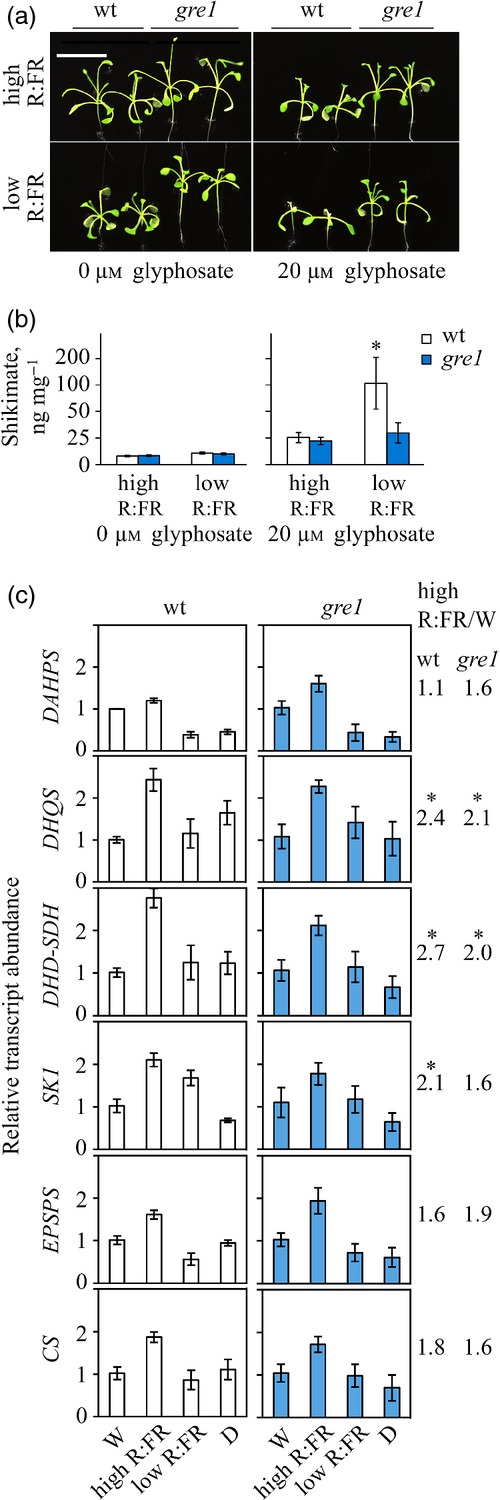
Low R:FR light causes glyphosate hypersensitivity. (a) Four-day-old wt and gre1 plants were tested under high (3.8 μmol m−2 sec−1) and low (0.09 μmol m−2 sec−1) R:FR light enriched with white light (total LI = 22 ± 2 μmol m−2 sec−1) in a 16 h light regime with (20 μm) or without (control) glyphosate treatments for 9 days. White scale bars are 10 mm.(b) Shikimate assay was done on glyphosate treated plants treated under the same conditions described above. Data represent the mean of five plants with ± standard error (SE). Asterisk indicates P-values < 0.05.(c) Shikimate pathway regulatory gene expression was performed by qPCR on 12-day-old, agar plate grown plants treated under white (W), high or low R:FR light conditions, or dark (D) condition for 2 h after the 8 h dark period. Relative transcript abundance was calculated using W light treated wt mRNA as the calibrator and normalized with respect to βACTIN7 gene transcript level. Error bars represent ± SE of the mean of three samples. Ratio of relative transcript numbers in high R:FR/W of both genotype are written next to the corresponding gene. Asterisks indicate a significant increase.
We also performed transcript analyses of all six genes that encoded enzymes of the shikimate pathway (Figure S1) under dark (D), white light (W) and high and low R:FR light conditions 2 h after the end of the night cycle (Figure3c) and observed a hyper-accumulation in the response to high R:FR light. As compared with W light, high R:FR light caused a significant increase in DeHydro Quianate Synthase (DHQS), 3-DeHydroquiante Dehydratase–Shikimate DeHydrogenase (DHD-SDH) and Shikimare Kinase 1 (SK1) transcripts in the wt. In the mutant this increase was slightly reduced. The high R:FR light-dependent transcript level of Deoxy d-Arbino Heptulsonate 7-Phosphate Synthase 1 (DAHPS1) was more pronounced in gre1 than in the wt. Except for DAHPS1, there was hardly any difference between the expression values during the 2 h exposure to D and W in the wt as well as in gre1 (Figure3c).
Roles of phyB and the circadian clock in the regulation of the shikimate pathway
Given the phyB-dependent glyphosate hypersensitive response (Figures2 and 3), we looked for light-regulated transcription factor-binding sites (TFSs) in the promoters of the shikimate pathway genes. We found that the first and the fourth genes of the pathway, DAHPS1 and SK1, harbored circadian clock-related TFS motifs such as the Evening Element (EE) motif (AAAATATCT), and circadian clock associated 1 (CCA1) binding site (AAMAATCT), while the second and fifth genes, DHQS and Enol Pyruvyl Shikimate Phosphate Synthase 1 (EPSPS1), harbored the Phytochrome-Interacting Factor (PIF) binding G-motif promoter elements (CACGTG) (Table S2). The presence of both phytochrome and circadian clock associated TFSs suggested an expression analysis on shikimate pathway genes and genes that encode components of the circadian clock (EarLy Flowering 4 - ELF4 and CCA1) and phyB-dependent signaling molecules (Phytochrome-Interacting Factor 4 and 5 - PIF4 and PIF5). These analyses were performed on plants that were grown on both agar plates under 22 ± 2 μmol m−2 s−1 white light and in soil under 75 ± 5 μmol m−2 s−1 white light. Under both growth conditions the gre1 phenotype was obtained in response to glyphosate (Figures1 and S5). Quantitative PCR (qPCR) revealed a diurnal expression pattern of the shikimate pathway genes under both conditions with only slight differences (Figures4a and S6a).
Figure 4.
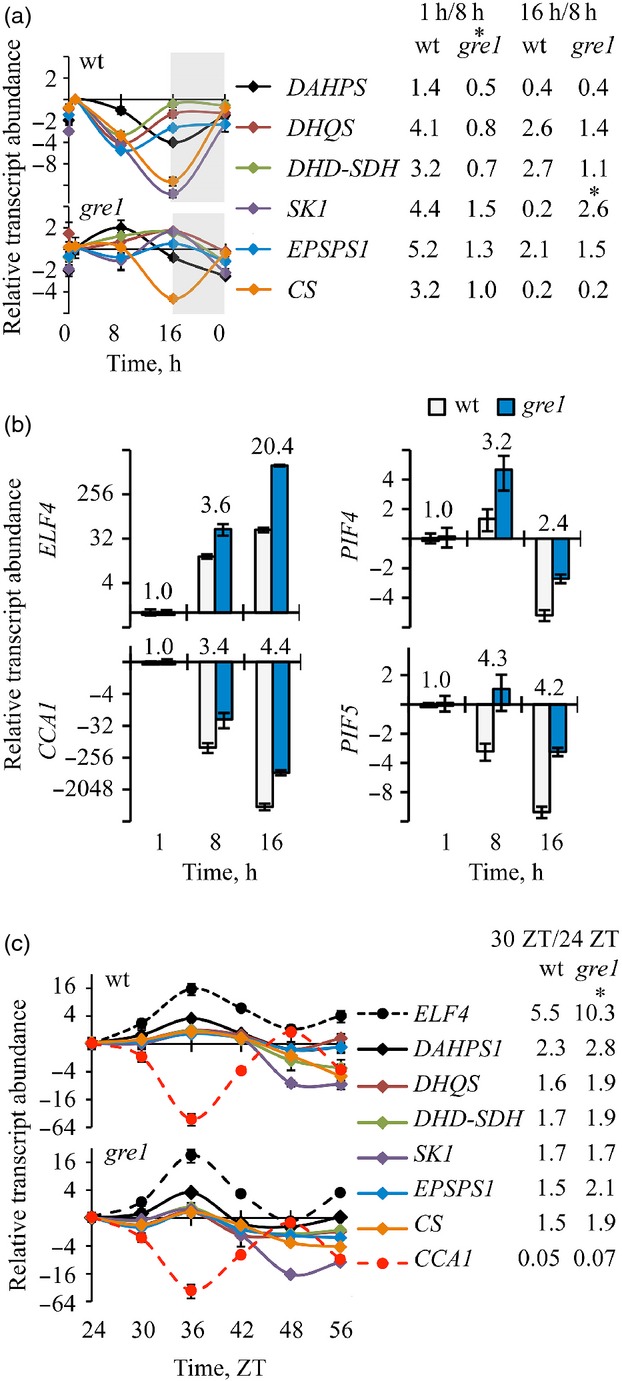
The shikimate pathway is affected by the circadian clock and phyB signaling.Twelve-day-old agar plate grown plants under LI = 22 ± 2 μmol m−2 sec−1 were used in these experiments. Values are means ± standard error (SE) of three replicates and times start from 0 h as day light begins. Shaded areas indicate the night cycle. Data are expressed as log2 and asterisks indicate significant differences.(a) Transcript accumulation of shikimate pathway components under regular day-night regime. Ratio of relative transcript numbers at 1 h/8 h and 16 h/8 h of both genotypes are written next to the corresponding gene. (b) Transcript abundance of circadian clock and phyB downstream signaling elements. Ratios of transcript levels of gre1/wt are written above the bars of the corresponding genes. (c) Transcript level of shikimate pathway and circadian clock components under continuous light condition with the ratios of 30 ZT/24 ZT plotted next to the corresponding genes.
When we studied the agar plate grown plants, in the wt, the peak level of DAHPS1, SK1 and Chorismate Synthase (CS) transcript accumulation occurred after 1 h light exposure and the lowest levels occurred at the onset of dark. In contrast, transcript accumulation of DHQS, DHD-SDH and EPSPS1 reached a peak at the onset of dark and the lowest level was observed at midday (8 h). In gre1 plants, the oscillations of DAHPS1 and SK1 were altered, with DAHPS1 peaking at midday (8 h) and SK1 at the onset of the dark period (16 h), the lowest transcript levels of these genes shifted to the onset of the light period. The pronounced reduction of transcripts seen in the wt plants at midday did not occur in gre1 (Figure4a).
In soil, all genes of the pathway were highly transcribed during the dark period, and immediately down-regulated at the onset of light period (Figure S6a). Transcript accumulation of wt genes, except for DAHPS1, appeared to increase continuously from the second half of the day and reached a peak at the onset of the light period. DAHPS1 transcripts started accumulating after 1 h light. In gre1 plants, the maximum transcript accumulation of these genes appears to be shifted to the end of the light period (16 h) and extended into the dark period in gre1. This shift in relative transcript amounts in gre1 is seen clearly between 8 and 16 h. In comparison with the wt, all mRNA ratios of 16 h/8 h in gre1 were higher, significantly (≥2-fold) in DAHPS1 and EPSPS1. The transcript peak times for both DAHPS1 and SK1 in wt were at the end of the light period and the peak times were not altered in gre1 (Figure S6a). The different transcript accumulation patterns of DAHPS1, SK1 and CS genes under different fluence-rates implicated light intensity as a factor in the regulation of the shikimate pathway (Figures4a and S6a).
Furthermore, we have analysed transcript accumulation of the circadian clock regulatory elements: CCA1, and ELF4, and phyB downstream signaling components, PIF4 and PIF5 under the same conditions as described above. qPCR revealed that the expression profiles of these TFs were similar in both genotypes, however, transcript accumulation of each gene was more pronounced in gre1 (Figures4b and S6b).
We also checked the maintenance of diurnal patterns of the pathway under continuous light in order to detect whether this oscillation is controlled by the internal time keeping mechanism. qPCR revealed that the transcript accumulation pattern of all shikimate pathway genes still oscillated in a rhythm similar to the one of the circadian clock component ELF4 under continuous light in both wt and gre1 (Figures4c and S6c). The peak value of ELF4 was two times more pronounced in agar and six times more in soil for gre1 as compared with wt. The oscillation patterns during the light period – ZT (Zeitgeber Time) – were identical in both genotypes on the agar plate but more pronounced in gre1 than in wt in soil grown plants. The gre1 mRNA ratios (32 ZT/24 ZT) were 4–11.4 times bigger than those of the corresponding wt ratios (Figure S6c). As light has a strong inhibitory effect on shikimate pathway gene expression (Figures4a and S6a), it is conceivable that continuous dark might increase the amplitude of the circadian oscillation of these genes. To test this hypothesis, the circadian oscillation effect was studied under continuously dark conditions in the soil grown plants and revealed a response that was similar to that under continuous light. In the dark, transcript abundance of all pathway genes in gre1 was more pronounced (1.7–3.6 times) at 32 ZT when compared with wt and also follows ELF4 oscillation (Figure S7).
Glyphosate effects depend on shikimate pathway activity
Our previous results revealed that both the phyB-dependent mechanism and the circadian clock regulatory components are essential parts of the transcriptional regulation of shikimate pathway genes, and that defective phyB causes transcript accumulation in major components of the shikimate pathway in gre1 (Figures4 and S6). Thus, enhanced transcript accumulation is the most likely reason for the development of glyphosate resistance in gre1. In order to test this hypothesis, we performed glyphosate experiments on greenhouse grown plants that had been sprayed at different times. The growth inhibitory potential of the herbicide varied according to the time of day the spray was applied. The plant survival rates were determined based on the development of stem elongation (Figure5). In comparison with gre1 plants, growth of wt plants was arrested completely when they were sprayed 200 μm glyphosate (0.2535 mg m−2) during the first half of the day, whereas glyphosate spraying in late afternoon caused only a partial inhibition (Figure5a, b). When glyphosate was applied either at 5 or 9 o'clock (am) it was lethal to wt plants, however, all wt plants survived if the application occurred later. All gre1 plants survived regardless of the time of spray, however, the growth rate was still dependent upon the spray time; the least growth occurred if sprayed in the morning and better growth occurred at the later applications (Figure5a, b).
Figure 5.
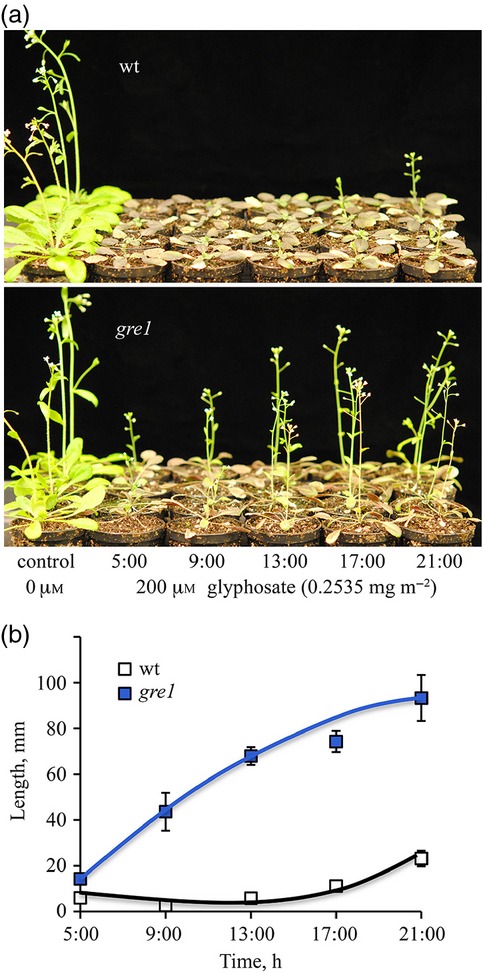
Glyphosate effects are dependent on the time of day at which spraying occurs. (a) Comparison of plant survival rates after 200 μm glyphosate (0.2535 mg m−2) application at 5:00, 9:00, 13:00, 17:00 and 21:00 h on 20-day-old greenhouse grown plants and the control was a spray without glyphosate. Pictures were taken 12 days after the treatment.(b) Shoot length of glyphosate treated plants. Measurement was done at day 12 after the application (n = 10).
DISCUSSION
PhyB mode of action determines the response to glyphosate
To-date more than 20 GR weed species have been reported, but only in goosegrass (Eleusine indica) and ryegrass species (Lolium rigidum, and Lolium multiflorum) can resistance be attributed to alterations at the binding site of EPSPS (Baerson et al., 2002; Wakelin and Preston, 2006; Jasieniuk et al., 2008; Kaundun et al., 2011; Heap, 2013). In the case of Palmer's amaranth (Amaranthus palmeri) and ryegrass (Lolium multiflorum) resistance has been attributed to EPSPS amplification (Gaines et al., 2010; Salas et al., 2012). However, the molecular mechanisms that underlie resistance remain to be elucidated and gre1 may turn out to be a valuable tool that can contribute to the understanding of how different environmental conditions affect EPSPS levels and hence sensitivity to the herbicide.
Genetic analyses revealed that GR phenotypes of gre1 are due to a dominant knock-out mutation of the R and FR receptor phyB (Figure2 and Table S1). This conclusion is supported by the glyphosate resistance response of a second allelic mutant (phyB-9) and glyphosate hypersensitivity of phyB over-expressing lines (Figure2c). The phenotypic characteristics of the phyB mutation are consistent with previous reports of dominant (semi-dominant), loss-of-function phenotypes and can be explained by haplo insufficiency (Veitia, 2002; Exner et al., 2010; Wang et al., 2011; Meinke, 2013). Arabidopsis contains five phytochromes (phyA, B, C, D and E) classified as type I or type II based on their light stability (Sharrock and Quail, 1989; Furuya, 1993). PhyA is the only light labile type I and is predominant in dark-grown seedlings (Furuya, 1993; Smith, 2000). That the phyA-deficient mutant phyA-201 does not show a GR phenotype further confirms that the glyphosate response phenotypes of gre1 are due specifically to a dysfunction of phyB. Chromophore deficiency could also have a role in the glyphosate response as all phytochrome apoproteins gain functionality only when assembled covalently with phytochromobilin, a light-absorbing linear tetrapyrrole chromophore, by forming 250 kDa dimers in the cytosol (Muramoto et al., 1999; Quail, 2002). A heme-oxygenase, which catalyzes an oxygenation reaction for the formation of most of the phytochromobilins, is encoded by the HY1 locus in Arabidopsis (Muramoto et al., 1999). Our glyphosate response analysis suggested that HY1 locus was also capable of conferring glyphosate resistance as the chromophore biosynthesis-deficient hy1-100 mutant exhibits glyphosate resistance that is similar to phytochrome-deficient mutants (Figure2c). However, chromophore-related glyphosate responses might be attributed to a dysfunctionality of all phytochromes, while we propose that it is caused specifically by phyB deficiency as phyB is the most abundant phytochrome in light-grown plants (Sharrock and Clack, 2002).
Based on their specific sensitivities to light intensity, three types of modes of action of phytochromes have been suggested: (1) a very-low-fluence response (VLFR); (2) a low-fluence response (LFR); and (3) a high-irradiance response (HIR). PhyB is considered to mediate FR light reversible LFR in R light (Casal et al., 1998). It is therefore reasonable to conclude that phyB-dependent glyphosate sensitivity must be linked to the LFR and this is consistent with the observation that, firstly, the most pronounced phenotype of gre1 in agar plate assays could be seen at light intensities between 15–25 μmol m−2 s−1 (Figure S3), and secondly, that the wt phenotype under these LFR conditions is reversed by high R:FR light (Figure3a). Given this dependence of the phenotype on the light regime, the agar plate experiments presented and discussed here were performed at the fluence rate of 22 ± 2 μmol m−2 s−1 (Figures2c, S2, 3 and 4). In addition, gre1 plants that are grown in soil under a fluence rate of 75 ± 5 μmol m−2 s−1 where no SAS occurs, also exhibited a resistant phenotype after spray application of glyphosate (Figure S5). The fluence rate-dependent glyphosate effect on phyB signaling under different environmental and developmental conditions will need to be elucidated further. Nevertheless, one of our glyphosate spray experiments in the greenhouse demonstrated that the wt and gre1 plants showed increased resistance to glyphosate application when treated in the afternoon or in the late evening (Figure5). This finding is consistent with the evening LFR response mediated by phyB. There are numerous reports that link glyphosate efficiency to the time of day at which it is applied, and it was observed that greatest annual weed control effect was obtained when glyphosate was applied between 9 a.m. and 6 p.m. rather than at 6 a.m. or between 9 p.m. and 12 a.m. (Martinson et al., 2005). A diurnal effect on leaf angle change, a phytochrome-dependent process, has also been recognized as one of the major reasons of greater biomass accumulation in broadleaf weeds when glyphosate was applied in the early morning rather than in late afternoon (Mohr et al., 2007).
PhyB signaling and the circadian clock affect shikimate pathway activity
The shikimate pathway is considered to be a key link between carbohydrate metabolism and the biosynthesis of aromatic amino acids and many secondary metabolites (Singh et al., 1991; Herrmann, 1995a) and consists of a sequence of six metabolic steps that lead to the synthesis of chorismate (Figure S1). The fact that gre1 is a light receptor mutant that affects chorismate biosynthesis makes it a potentially highly useful tool for the study of light effects on the pathway. Our results suggested that the glyphosate resistance of gre1 is not related to any modification of the target enzyme but is the consequence of increased activity of the entire shikimate pathway due to altered regulation of the pathway caused by dysfunctional phyB (Figures4 and S6). It is noteworthy that the gre1 mutant shows less shikimate accumulation in response to glyphosate under low R:FR light condition and this result is indicative of an active shikimate pathway (Figure3b). Accumulation of shikimate, an intermediate product of the shikimate pathway, is considered one of the immediate physiological responses to glyphosate inhibition (Amrhein et al., 1980; Smart et al., 1985). These findings, together with our studies on circadian regulatory mechanisms, give rise to a more detailed proposed model of the regulation of the pathway that can, at least in part, explain the link between glyphosate resistance and light quality (Figure6).
Figure 6.
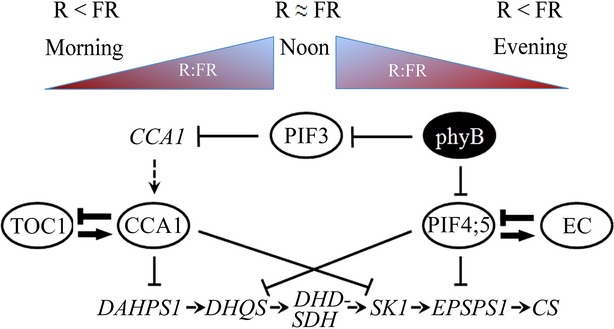
A model proposing controls of the shikimate pathway.PhyB exerts control over the pathway by changing its active and non-active, Pfr and Pr, conformations according to the light quality (R:FR). The R:FR increases gradually from the onset of light till midday and decreases towards evening. Expression of DAHPS1 and SK1 is repressed by the circadian component CCA1 during the day, while phyB (Pfr) promotes the accumulation of DHQS and EPSPS1 in response to high R:FR through the interaction with PIF4 and PIF5. During the evening, EC elements have a role in stabilizing PIF4 and PIF5. Arrows and blunt end lines indicate stimulatory and inhibitory effects respectively, and dashed arrows indicate transcriptional activation. The proposed feed-back regulation of the circadian clock and EC-dependent regulation are shown as embossed arrows.
The phyB-dependent regulatory mechanism of the shikimate pathway is likely mediated through PIFs given that the promoters of both DHQS and EPSPS1 harbor PIF-binding G-motif promoter elements (CACGTG) (Table S2). Among the Arabidopsis basic helix–loop–helix (bHLH) TFs, PIF4 or PIF5 are the most likely candidates as these TFs tend to accumulate in the dark e.g. in dense vegetation where R:FR ratios are lowest thereby causing the SAS, and this response is limited by their interaction with the Pfr isomer of phyB in high R:FR (Huq and Quail, 2002; Nozue et al., 2007; Shen et al., 2007; Lorrain et al., 2008). Under the low R:FR, the wt plants showed glyphosate hypersensitive phenotypes that were reversible by high R:FR (Figure3a). This response is consistent with the high R:FR induced accumulation of the physiologically active Pfr isoform of phyB in the nucleus where it selectively binds to PIF4 and PIF5, targeting them for proteasomal degradation (Huq and Quail, 2002; Shen et al., 2007; Lorrain et al., 2008). This conclusion in turn is consistent with a positive regulatory role of phyB and a negative regulatory role of PIFs on shikimate pathway activity (Figure6). Indeed, the shikimate pathway tends to be activated in response to high R:FR, as transcript levels of the pathway genes are higher in high R:FR than in low R:FR (Figure3c).
The regulatory effect of phyB on the shikimate pathway activity cannot account for all transcriptional responses in the regular day–night regime and in continuous light conditions, because the genes responsible for the pathway are also under a diurnal rhythm (Figures4a, c and S6a, c). Among these, the first and the fourth gene of the pathway, DAHPS1 and SK1, showed the most pronounced and stable oscillation in both the regular day–night regime and continuous light conditions. The distinct expression patterns of these two genes suggested the presence of an additional regulatory mechanism that is under circadian control. Given that both DAHPS1 and SK1 genes contain EE- and CCA1-binding TFSs (Table S2), we propose that the regulation is most likely operated through negative regulation of CCA1 (Figure6). CCA1 is the central component of circadian clock and it recognizes both EE and CCA1 motifs that are conserved in the promoter regions of light-regulated genes to induce peak expression in the evening (Harmer et al., 2000), or activate R light-dependent transcription (Wang et al., 1997). This explanation is consistent with the up- and down-regulation of the first and fourth genes, DAHPS1 and SK1, of the pathway and constant diurnal oscillation of the shikimate pathway that is inverse to that of the CCA1 expression pattern (Figures4a, c and S6a, c). The plant circadian clock is composed of many regulatory factors that operate interlinked transcriptional and post-translational positive and negative feed-back loops to enable day and night anticipation (Harmer, 2009; Más and Yanovsky, 2009). It has been proven that CCA1 and its close homologue LHY (Late Elongated Hypocotyl) suppress expression of evening-phased genes TOC1 and ELF4 through binding EEs present in their promoter regions. Conversely, both TOC1 and ELF4 promote expression of CCA1 and LHY genes and form a transcriptional feed-back loop, thereby enabling circadian oscillation (Alabadi et al., 2001; Kikis et al., 2005; Pruneda-Paz et al., 2009) (Figure6). In addition, positive phyB regulation of CCA1 and LHY expression through PIF3 bound to G-boxes in their promoter regions has also been suggested (Martıńnez-Garcıńa et al., 2000) (Figure6). It is this regulatory link that is disturbed in gre1 because of its dysfunctional phyB. However, the overriding system of the circadian clock may, at least in part, compensate this defect in CCA1 (Figure6).
Another circadian clock related regulatory mechanism that involve EC elements (‘evening complex’ (ELF4-ELF3-LUX)) has been proposed for PIF4 and PIF5 expression in the early evening (Nusinow et al., 2011) as one of the major components of EC, LUX, interacts with the promoter regions of both PIFs in vivo (Nusinow et al., 2011). Dysfunctional phyB is likely to lead to overproduction of EC, because regulation of PIF4 and PIF5 is crucial for the complex function (Nusinow et al., 2011). Therefore, our model proposes a transcriptional feed-back loop in which PIFs play a positive regulatory role in the transcription of LUX in the afternoon (Figure6). LUX might play a ‘housekeeping’ role in the absence of phyB during the night to control proper transcription of PIFs. This idea might explain the persisting high levels of the shikimate pathway genes at midday and the extended expression of DHQS and EPSPS1 in gre1 (Figures4a and S6a). Transcript analyses of ELF4, CCA1, PIF4 and PIF5 support the idea that the circadian rhythm and phyB affect the shikimate pathway as the ELF4 appears over-expressed and CCA1 and the two PIFs show reduced transcript accumulation during the afternoon in the gre1 (Figures4b and S6b). Furthermore, oscillation of all shikimate pathway genes under continuous light indicated that this pathway is under the control of the circadian clock (Figures4c and S6c). These data therefore link the circadian clock to phyB signaling and also enable integrated regulation of the shikimate pathway.
Based on our transcript accumulation studies of the major components of the shikimate pathway, we propose a regulatory network that involves both the circadian clock and phyB signaling. The circadian clock assures internal time keeping, whereas, phyB plays a stabilizing role and tunes the response to specific environmental conditions. In the case of gre1, hyper-activation of the shikimate pathway affects the entire pathway and overrides the functions normally under the control of circadian clock. The resulting increase of EPSPS1 contributes to an increase in the glyphosate target enzyme, which in turn causes increased resistance to glyphosate. This conclusion is consistent with the phenotype that shows reduced glyphosate sensitivity, increased fresh weight, reduced chlorophyll damage and reduced shikimate accumulation in the presence of glyphosate. Thus, downstream signaling of phyB may shed light on the regulation of the shikimate pathway and increased levels of EPSPS in particular, and this in turn may explain glyphosate resistance in some weed species.
EXPERIMENTAL PROCEDURES
Plant material and growth condition
T-DNA tagged (pGPTV-Bar (http://biotech.unl.edu/pgptv-bar)) mutant population of Arabidopsis thaliana, ecotype Col0, were used in the screen. To select for positive transformed lines, seedlings of each generation were sprayed with 0.01% Basta (bialaphos) solution (Aventis Crop Science Deutschland GmbH, Hattersheim, Germany) repeatedly at 3-day intervals for 2 weeks. Seeds of the bialaphos resistant 10 T2 (transformant 2) plants were pooled and screened for glyphosate resistance by sowing directly on 50 μm glyphosate (N–phosphonomethyl glycine, Sigma Aldrich, www.sigmaaldrich.com) that contained Murashige and Skoog (MS) medium with addition of agar (1.1%) and 20 g L−1 sucrose. Phenotypes of the selected mutants (T3 progenies) were confirmed in glyphosate dose–response studies, in which 4-day old plantlets were transferred on the same medium that contained 1, 5, 10, 20, 40, 80 or 100 μm glyphosate. Mutants obtained from each generation were recovered on bovine serum albumin (0.2%) supplemented MS – agar (8 g L−1), glyphosate free, 20 g L−1 sucrose-containing media. Homozygote gre1 plants were obtained from F2 segregating populations of the second time back crossed population. Screening was performed in a growth room equipped with fluorescent lights (Light intensity (LI) = 22 ± 2 μmol m−2 s−1) under a 16 h light regime. The PhyB over-expressed line – pbc1 (Salk Institute, www.salk.edu), and gpbc1 – was obtained by crossing gre1 × pbc1. PhyA-201 and hy1-100 lines were received from the Salk Institute. Light quality and gene expression studies were done in an LED light chamber that contains white, red and FR-light sources (FloraLED, CLF PlantClimatics GmbH, Munich, Germany). Light conditions were: high R:FR = 3.8 (R = 14.6 μmol m−2 sec−1; FR = 3.8 μmol m−2 sec−1) and low R:FR = 0.09 (R = 0.36 μmol m−2 sec−1; FR = 3.8 μmol m−2 sec−1). Total light intensities were adjusted to 22 ± 2 μmol m−2 sec−1 by white light for agar plate studies. For plants grown in Metro-Mix (Sun Gro Horticulture, Bellevue, WA, USA) soil, the light intensities were augmented to 75 ± 5 μmol m−2 sec−1 in order to avoid interference from the SAS.
Determination of chlorophyll content
Chlorophyll content was measured by placing plantlets in 95% ethanol and then boiling in a water bath for 5–10 min. Chlorophyll A and B contents in the ethanol extract were determined as described previously (Lichtenthaler and Wellburn, 1983), and total chlorophyll was calculated by adding both chlorophyll A and B values.
Shikimate content analysis
Shikimic acid was extracted from plantlets by floating them in 100 μL of 0.25 N HCl solution followed by incubation at 60°C for 15 min. Shikimate content was determined using a 96-well microtiter plate assay following a method detailed elsewhere (Shaner et al., 2005).
PCR conditions and gene expression analyses
Sequences of all oligo-nucleotides used for PCRs are described in Table S3. TAIL-PCR was done using three nested primers with the combination of two degenerative primers. For the PHYB gene expression analyses (RT-PCR) expression of UBIQUITIN 10 gene was used for normalizing the band intensity. Total RNA was extracted using the RNeasy plant mini kit (Qiagen, www.qiagen.com) and superscript RT-PCR first strand synthesis kit with oligo(dT) (Invitrogen, www.lifetechnologies.com) was used for cDNA synthesis. qPCRs were performed using fast SYBR green master mix (Applied Biosystems, www.appliedbiosystems.com) in the AB-7900HT Fast Real-Time PCR machine. In the QPCR, CT values of mRNA expression in the W light, at 1 h light treatment, and at 24 h ZT point were used as calibrators for gene expression analyses in the light quality studies, light and dark period and circadian oscillation studies respectively. Twelve-day-old plants were used for qPCR analyses with three replications. All qPCR data were normalized to the value of mRNA expression of βACTIN7.
Glyphosate spraying of plants in the greenhouse
Plants were grown in the greenhouse under natural light conditions. A 200 μm glyphosate solution (N-phosphonomethyl glycine (Sigma Aldrich) in 10 mm (NH4)2SO4 and 0.01% w/v Tween-20) was applied to 20-day-old soil grown plants at 5:00, 9:00, 13:00, 17:00 and 21:00 h by using a herbicide spray booth ‘Generation III’, (De Vries, Hollandale, MN, USA). The sprayed 200 μm glyphosate solution resulted in an application of 0.2535 mg m−2 as determined by spray speed, length and spray volume under 240 kpa pressure. The control spray contained 10 mm (NH4)2SO4 and 0.01% w/v Tween-20.
Acknowledgments
We thank Joanne Chory (the Salk Institute) for providing the pbc1 mutant strain. A.S. is grateful to Mike Hasegawa (Purdue University) for most helpful discussions and encouragement.
Supporting Information
Additional Supporting Information may be found in the online version of this article.
Plant shikimate pathway.
Figure S2 Glyphosate response mutation screen.
Figure S3 Glyphosate resistant phenotypes depend on light fluence rate.
Figure S4. Morphological comparison of wt and gre1.
Figure S5. Analysis of glyphosate sprayed plants grown on soil under 75 μmol m−2 sec−1 light.
Figure S6. Transcript analyses of shikimate pathway genes and circadian clock and phyB signaling components.
Figure S7. Transcript accumulation of shikimate pathway and circadian clock components in continues dark condition.
Back-cross analysis of gre1 line.
Table S2. Transcription factor binding sites on the promoter regions of shikimate pathway genes.
Table S3 Oligo-nucleotides used in PCRs.
REFERENCES
- Alabadi D, Oyama T, Yanovsky MJ, Harmon FG, Mas P, Kay SA. Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science. 2001;293:880–883. doi: 10.1126/science.1061320. [DOI] [PubMed] [Google Scholar]
- Amrhein N, Deus B, Gehrke P, Steinrücken HC. The site of the inhibition of the shikimate pathway by glyphosate: II. Interference of glyphosate with chorismate formation in vivo and in vitro. Plant Physiol. 1980;66:830–834. doi: 10.1104/pp.66.5.830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andel F, 3rd, Lagarias JC, Mathies RA. Resonance raman analysis of chromophore structure in the lumi-R photoproduct of phytochrome. Biochemistry. 1996;35:15997–16008. doi: 10.1021/bi962175k. [DOI] [PubMed] [Google Scholar]
- Baerson SR, Rodriguez DJ, Tran M, Feng Y, Biest NA, Dill GM. Glyphosate-resistant goosegrass. Identification of a mutation in the target enzyme 5-enolpyruvylshikimate-3-phosphate synthase. Plant Physiol. 2002;129:1265–1275. doi: 10.1104/pp.001560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benbrook CM. Impacts of genetically engineered crops on pesticide use in the U.S., the first sixteen years. Environ. Sci. Eur. 2012;24:1–13. [Google Scholar]
- Bromilow RH, Chamberlain K. The herbicide glyphosate and related molecules: physicochemical and structural factors determining their mobility in phloem. Pest Manag. Sci. 2000;56:368–373. [Google Scholar]
- Casal JJ, Sánchez RA, Botto JF. Modes of action of phytochromes. J. Exp. Bot. 1998;49:127–138. [Google Scholar]
- Duke SO, Powles SB. Glyphosate-resistant crops and weeds: now and in the future. J. Agrobio. Manag. Econ. 2009;12:346–357. [Google Scholar]
- Exner V, Alexandre C, Rosenfeldt G, Alfarano P, Nater M, Caflisch A, Gruissem W, Batschauer A, Hennig L. A gain-of-function mutation of Arabidopsis CRYPTOCHROME1 promotes flowering. Plant Physiol. 2010;154:1633–1645. doi: 10.1104/pp.110.160895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz JE, Mao MK, Sikorski JA. Glyphosate: A Unique Global Herbicide. Washington, DC: American Chemical Society; 1997. [Google Scholar]
- Furuya M. Phytochromes: their molecular species, gene families, and functions. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1993;44:617–645. [Google Scholar]
- Gaines TA, Zhang W, Wang D, et al. Gene amplification confers glyphosate resistance in Amaranthus palmeri. Proc. Natl Acad. Sci. USA. 2010;107:1029–1034. doi: 10.1073/pnas.0906649107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X, d'Avignon DA, Ackerman JJH, Sammons RD. Rapid vacuolar sequestration: the horseweed glyphosate resistance mechanism. Pest Manag. Sci. 2010;66:345–348. doi: 10.1002/ps.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gianessi LP. Economic impacts of glyphosate-resistant crops. Pest Manag. Sci. 2008;64:346–352. doi: 10.1002/ps.1490. [DOI] [PubMed] [Google Scholar]
- Harmer SL. The circadian system in higher plants. Annu. Rev. Plant Biol. 2009;60:357–377. doi: 10.1146/annurev.arplant.043008.092054. [DOI] [PubMed] [Google Scholar]
- Harmer SL, Hogenesch JB, Straume M, Chang H-S, Han B, Zhu T, Wang X, Kreps JA, Kay SA. Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science. 2000;290:2110–2113. doi: 10.1126/science.290.5499.2110. [DOI] [PubMed] [Google Scholar]
- Heap I. The international survey of herbicide resistant weeds: Weed Science Society of America, Weeds resistant to glycines. 2013. www.weedscience.org.
- Herrmann KM. The shikimate pathway as an entry to aromatic secondary metabolism. Plant Physiol. 1995a;107:7–12. doi: 10.1104/pp.107.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann KM. The Shikimate pathway: early steps in the biosynthesis of aromatic compounds. Plant Cell. 1995b;7:907–919. doi: 10.1105/tpc.7.7.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huq E, Quail PH. PIF4, a phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. EMBO J. 2002;21:2441–2450. doi: 10.1093/emboj/21.10.2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasieniuk M, Ahmad R, Sherwood AM, Firestone JL, Perez-Jones A, Lanini WT, Mallory-Smith C, Stednick Z. Glyphosate-resistant Italian ryegrass (Lolium multiflorum) in California: distribution, response to glyphosate, and molecular evidence for an altered target enzyme. Weed Sci. 2008;56:496–502. [Google Scholar]
- Kaundun SS, Dale RP, Zelaya IA, Dinelli G, Marotti I, McIndoe E, Cairns A. A novel P106L mutation in EPSPS and an unknown mechanism(s) act additively to confer resistance to glyphosate in a South African Lolium rigidum population. J. Agric. Food Chem. 2011;59:3227–3233. doi: 10.1021/jf104934j. [DOI] [PubMed] [Google Scholar]
- Kikis EA, Khanna R, Quail PH. ELF4 is a phytochrome-regulated component of a negative-feedback loop involving the central oscillator components CCA1 and LHY. Plant J. 2005;44:300–313. doi: 10.1111/j.1365-313X.2005.02531.x. [DOI] [PubMed] [Google Scholar]
- Lichtenthaler HK, Wellburn AR. Determinations of total carotenoids and chlorophylls a and b of leaf extracts in different solvents. Biochem. Soc. Trans. 1983;11:591–592. [Google Scholar]
- Lorrain S, Allen T, Duek PD, Whitelam GC, Fankhauser C. Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J. 2008;53:312–323. doi: 10.1111/j.1365-313X.2007.03341.x. [DOI] [PubMed] [Google Scholar]
- Lorraine-Colwill DF, Powles SB, Hawkes TR, Hollinshead PH, Warner SAJ, Preston C. Investigations into the mechanism of glyphosate resistance in Lolium rigidum. Pest Biochem. Physiol. 2002;74:62–72. [Google Scholar]
- Martıńnez-Garcıńa JF, Huq E, Quail PH. Direct targeting of light signals to a promoter element-bound transcription factor. Science. 2000;288:859–863. doi: 10.1126/science.288.5467.859. [DOI] [PubMed] [Google Scholar]
- Martinson KB, Durgan BR, Gunsolus JL, Sothern RB. Time of day of application effect on glyphosate and glufosinate efficacy. 2005. www.plantmanagementnetwork.org.
- Más P, Yanovsky MJ. Time for circadian rhythms: plants get synchronized. Curr. Opin. Plant Biol. 2009;12:574–579. doi: 10.1016/j.pbi.2009.07.010. [DOI] [PubMed] [Google Scholar]
- Meinke DW. A survey of dominant mutations in Arabidopsis thaliana. Trends Plant Sci. 2013;18:84–91. doi: 10.1016/j.tplants.2012.08.006. [DOI] [PubMed] [Google Scholar]
- Mohr K, Sellers BA, Smeda RJ. Application time of day influences glyphosate efficacy. Weed Tech. 2007;21:7–13. [Google Scholar]
- Muramoto T, Kohchi T, Yokota A, Hwang I, Goodman HM. The Arabidopsis photomorphogenic mutant hy1 is deficient in phytochrome chromophore biosynthesis as a result of a mutation in a plastid heme oxygenase. Plant Cell. 1999;11:335–347. doi: 10.1105/tpc.11.3.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagatani A. Light-regulated nuclear localization of phytochromes. Curr. Opin. Plant Biol. 2004;7:708–711. doi: 10.1016/j.pbi.2004.09.010. [DOI] [PubMed] [Google Scholar]
- Nozue K, Covington MF, Duek PD, Lorrain S, Fankhauser C, Harmer SL, Maloof JN. Rhythmic growth explained by coincidence between internal and external cues. Nature. 2007;448:358–361. doi: 10.1038/nature05946. [DOI] [PubMed] [Google Scholar]
- Nusinow DA, Helfer A, Hamilton EE, King JJ, Imaizumi T, Schultz TF, Farre EM, Kay SA. The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature. 2011;475:398–402. doi: 10.1038/nature10182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruneda-Paz JL, Breton G, Para A, Kay SA. A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science. 2009;323:1481–1485. doi: 10.1126/science.1167206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail PH. Phytochrome photosensory signalling networks. Nat. Rev. Mol. Cell Biol. 2002;3:85–93. doi: 10.1038/nrm728. [DOI] [PubMed] [Google Scholar]
- Rockwell NC, Su Y-S, Lagarias JC. Phytochrome structure and signaling mechanisms. Annu. Rev. Plant Biol. 2006;57:837–858. doi: 10.1146/annurev.arplant.56.032604.144208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rüdiger W, Thümmler F, Cmiel E, Schneider S. Chromophore structure of the physiologically active form (Pfr) of phytochrome. Proc. Natl Acad. Sci. USA. 1983;80:6244–6248. doi: 10.1073/pnas.80.20.6244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rüppel MI, Brightwell BB, Schaefer J, Marvel JT. Metabolism and degradation of glyphosate in soil and water. J. Agric. Food Chem. 1977;25:517–528. doi: 10.1021/jf60211a018. [DOI] [PubMed] [Google Scholar]
- Salas RA, Dayan FE, Pan Z, Watson SB, Dickson JW, Scott RC, Burgos NR. EPSPS gene amplification in glyphosate-resistant Italian ryegrass (Lolium perenne ssp. multiflorum) from Arkansas. Pest Manag. Sci. 2012;68:1223–1230. doi: 10.1002/ps.3342. [DOI] [PubMed] [Google Scholar]
- Shaner DL, Nadler-Hassar T, Henry WB, Koger CH. A rapid in vivo shikimate accumulation assay with excised leaf discs. Weed Sci. 2005;53:769–774. [Google Scholar]
- Sharrock RA, Clack T. Patterns of expression and normalized levels of the five Arabidopsis phytochromes. Plant Physiol. 2002;130:442–456. doi: 10.1104/pp.005389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharrock RA, Quail PH. Novel phytochrome sequences in Arabidopsis thaliana: structure, evolution, and differential expression of a plant regulatory photoreceptor family. Genes Dev. 1989;3:1745–1757. doi: 10.1101/gad.3.11.1745. [DOI] [PubMed] [Google Scholar]
- Shen Y, Khanna R, Carle CM, Quail PH. Phytochrome induces rapid PIF5 phosphorylation and degradation in response to red-light activation. Plant Physiol. 2007;145:1043–1051. doi: 10.1104/pp.107.105601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh BK, Siehl DL, Connelly JA. Shikimate pathway: why does it mean so much to so many? Oxford Surv. Plant Mol. Cell Biol. 1991;7:143–185. [Google Scholar]
- Smart CC, Johanning D, Muller G, Amrhein N. Selective overproduction of 5-enolpyruvyl-shikimic acid 3-phosphate synthase in a plant cell culture which tolerates high doses of the herbicide glyphosate. J. Biol. Chem. 1985;260:4724–4728. [PubMed] [Google Scholar]
- Smith H. Phytochromes and light signal perception by plants - an emerging synthesis. Nature. 2000;407:585–591. doi: 10.1038/35036500. [DOI] [PubMed] [Google Scholar]
- Veitia RA. Exploring the etiology of haploinsufficiency. BioEssays. 2002;24:175–184. doi: 10.1002/bies.10023. [DOI] [PubMed] [Google Scholar]
- Wakelin AM, Preston C. A target-site mutation is present in a glyphosate-resistant Lolium rigidum population. Weed Res. 2006;46:432–440. [Google Scholar]
- Wang ZY, Kenigsbuch D, Sun L, Harel E, Ong MS, Tobin EM. A Myb-related transcription factor is involved in the phytochrome regulation of an Arabidopsis Lhcb gene. Plant Cell. 1997;9:491–507. doi: 10.1105/tpc.9.4.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Roig-Villanova I, Khan S, Shanahan H, Quail PH, Martinez-Garcia JF, Devlin PF. A novel high-throughput in vivo molecular screen for shade avoidance mutants identifies a novel phyA mutation. J. Exp. Bot. 2011;62:2973–2987. doi: 10.1093/jxb/err062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams GM, Kroes R, Munro IC. Safety evaluation and risk assessment of the herbicide Roundup and its active ingredient, glyphosate, for humans. Regul. Toxicol. Pharma. 2000;31:117–165. doi: 10.1006/rtph.1999.1371. [DOI] [PubMed] [Google Scholar]
- Zhou D-M, Wang Y-J, Cang L, Hao X-Z, Luo X-S. Adsorption and cosorption of cadmium and glyphosate on two soils with different characteristics. Chemosphere. 2004;57:1237–1244. doi: 10.1016/j.chemosphere.2004.08.043. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Plant shikimate pathway.
Figure S2 Glyphosate response mutation screen.
Figure S3 Glyphosate resistant phenotypes depend on light fluence rate.
Figure S4. Morphological comparison of wt and gre1.
Figure S5. Analysis of glyphosate sprayed plants grown on soil under 75 μmol m−2 sec−1 light.
Figure S6. Transcript analyses of shikimate pathway genes and circadian clock and phyB signaling components.
Figure S7. Transcript accumulation of shikimate pathway and circadian clock components in continues dark condition.
Back-cross analysis of gre1 line.
Table S2. Transcription factor binding sites on the promoter regions of shikimate pathway genes.
Table S3 Oligo-nucleotides used in PCRs.


