Abstract
Eukaryotic cells utilize various RNA quality control mechanisms to ensure high fidelity of gene expression, thus protecting against the accumulation of nonfunctional RNA and the subsequent production of abnormal peptides. Messenger RNAs (mRNAs) are largely responsible for protein production, and mRNA quality control is particularly important for protecting the cell against the downstream effects of genetic mutations. Nonsense-mediated decay (NMD) is an evolutionarily conserved mRNA quality control system in all eukaryotes that degrades transcripts containing premature termination codons (PTCs). By degrading these aberrant transcripts, NMD acts to prevent the production of truncated proteins that could otherwise harm the cell through various insults, such as dominant negative effects or the ER stress response. Although NMD functions to protect the cell against the deleterious effects of aberrant mRNA, there is a growing body of evidence that mutation-, codon-, gene-, cell-, and tissue-specific differences in NMD efficiency can alter the underlying pathology of genetic disease. In addition, the protective role that NMD plays in genetic disease can undermine current therapeutic strategies aimed at increasing the production of full-length functional protein from genes harboring nonsense mutations. Here, we review the normal function of this RNA surveillance pathway and how it is regulated, provide current evidence for the role that it plays in modulating genetic disease phenotypes, and how NMD can be used as a therapeutic target.
Keywords: Genetic disease, nonsense-mediated decay, nonsense suppression, read-through
1. Introduction
According to the National Organization for Rare Disorders (NORD), there are nearly 7,000 known rare genetic disorders. In the US, there are approximately 30 million people that have a genetic disorder, of which approximately 30% arise as a consequence of a nonsense or frameshift mutation that introduces a premature termination codon (PTC, also called a nonsense codon or premature stop codon) [1, 2]. Nonsense mutations in particular account for 20.3% of all disease-associated single-base pair mutations, and are three times more likely to come to clinical attention than missense mutations [3, 4]. These mutations commonly inactivate gene function due to the production of truncated protein products, as well as leading to a significant decrease in cytoplasmic mRNA abundance due to targeted degradation by nonsense-mediated mRNA decay (NMD).
NMD is an evolutionarily conserved translation-dependent mechanism in all eukaryotes that is responsible for recognizing and eliminating aberrant messenger RNA (mRNA) transcripts to prevent the production of truncated peptides that could have toxic and detrimental effects on the cell [5-9]. NMD plays a critical role in preventing the potential dominant-negative effect of non-functional protein within the cell, as well as the prevention of misfolded protein accumulation and subsequent initiation of the ER stress response. NMD primarily protects the cell against the deleterious effects of PTCs, but there is a growing body of evidence that mutation-, codon-, gene-, cell-, and tissue-specific differences in NMD efficiency can alter the underlying disease pathology [2, 10-12]. In fact, there is evidence that in certain genetic disorders, NMD can act to aggravate disease pathology and worsen the clinical phenotype, because degradation of the mutated mRNA prevents translation and accumulation of truncated peptides that retain residual activity [9-11]. In addition, there is evidence that inter-individual variability in NMD efficiency leads to differences in disease presentation and therapeutic outcome [13]. Therefore, NMD has emerged as a potent modulator of disease severity and clinical phenotype in light of its important role in recognizing and degrading mutated transcripts. The variable involvement of NMD in genetic disease has important implications for therapy, including decreased efficacy of read-through compounds intended to suppress the effects of nonsense mutations. In this review, we summarize current evidence that shows how NMD can modulate genetic disease pathology, why variability in NMD efficiency is important, and how NMD can be used as a therapeutic target.
2. Mechanism of Substrate Recognition and Degradation
Eukaryotic cells contain a number of RNA quality control mechanisms to ensure the high fidelity of genetic expression, which protects the cell against the accumulation of nonfunctional RNA and the production of abnormal peptides [14]. mRNAs are largely responsible for protein production, and mRNA quality control is particularly important for protecting the cell against the harmful effects of genetic mutations. The synthesis and processing of mRNA involves transcription, capping, splicing, polyadenylation, nuclear membrane transport, translation, and degradation [14]. An aberrant mRNA containing a PTC can arise as a consequence of a variety of events, and must be recognized before it is fully processed into a mature mRNA capable of producing truncated protein products. NMD is a critical RNA quality control mechanism that involves multiple cellular components that recognize the presence of a PTC within a mRNA transcript. During synthesis by RNA polymerase II, mRNA is cotranscriptionally processed into a messenger ribonucleoprotein complex (mRNP) that contains a 5’-m7GpppN cap structure at the 5’ end and a poly(A)-tail at the 3’ end, which protect the newly synthesized RNA transcript from premature degradation. In addition, the cap-binding protein (CBP) heterodimer CBP80-CBP20 complex (CBC) binds to and protects the 5’-cap, and the nuclear poly(A)-binding protein N1 (PABPN1) and the primarily cytoplasmic poly(A)-binding protein C1 (PABPC1) bind to the poly(A) tail. Another important factor, eIF4G, also binds to and stabilizes the poly(A)-tail. More than 90% of human mRNA transcripts undergo alternative splicing to remove introns and ligate the coding regions of the transcript (exons) together [15]. During this process, a complex of proteins (eIF4AIII, MAGOH, MLN51, and Y14) called the exon junction complex (EJC) is deposited 20-24 nucleotides upstream of every exon-exon splice site within the mRNP [16]. The CBC-bound mRNA then undergoes the pioneer round of translation, in which one or more ribosomes load onto the mRNP. During this process, the CBC is replaced by eukaryotic translation initiation factor 4E (eIF4E), the EJCs are displaced, PABPN1 is entirely replaced by PABPC1, and the mature mRNA is directed into steady-state rounds of bulk translation [17]. Within mammalian cells, mature transcripts are bound by eIF4E at their caps and PABPC1 at their poly(A) tails, lack EJCs, and are immune to NMD.
The processing of pre-mRNA into mature mRNA relies heavily on the correct placement of splicing factors during early biogenesis. These factors are functionally coupled to every step of mRNA synthesis, including 5’ capping, 3’ polyadenylation, splicing, regulation of transcript stability, nuclear export, cytoplasmic transport, and translation [18, 19]. The EJC is essential for the initiation of NMD for transcripts harboring PTCs and is crucial for the decay of most aberrant mRNA transcripts. This is exemplified by the fact that all transcripts from intronless genes studied to date are invulnerable to NMD [15, 20, 21].
There are several parallel mRNA decay pathways, including the UPF2-independent NMD branch, the EJC-independent NMD branch, the UPF3-independent NMD branch, and the classical NMD branch [22-24]. In addition, there are alternative mRNA decay pathways that involve NMD factors, such as the Stau1-mediated decay pathway, which recruits up-frameshift 1 (UPF1) [25]. For the purposes of this review, we will focus only to the classical branch of the NMD pathway.
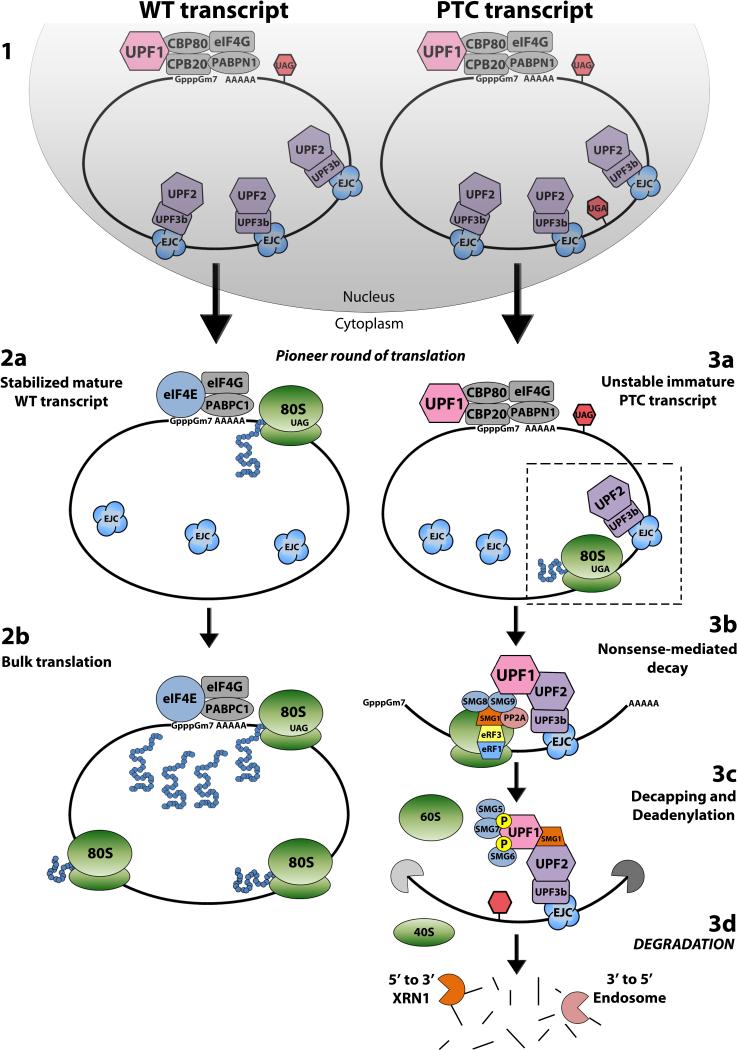
The classical NMD pathway (Figure 1) is thought to be triggered during the pioneer round of translation when the first ribosome translates the processed CBC-bound mRNA and stalls at a PTC that is located more than 50-55 nucleotides upstream of the last EJC-bearing exon-exon junction [5-9, 26]. During ribosomal stalling, the PTC moves into the A-site of the ribosome and is subsequently bound by eukaryotic release factor 1 (eRF1) and eRF3. CBP80 loosely associates with the key NMD factor, UPF1, prior to the pioneer round of translation. During NMD, CBP80 promotes recruitment of UPF1 by eRF1-bound eRF3 to the stalled ribosome. Next, the suppressor of morphogenetic effect on genitalia 1 (SMG1), a serine/threonine protein kinase, binds together with UPF1 to eRF1/3 to form the SMG1-UPF1-eRF1-eRF3 (SURF) complex. SMG8 and SMG9 also bind to the SURF complex, temporarily blocking phosphorylation of UPF1 by SMG1. This entire complex is then translocated from the ribosome to UPF2 and UPF3, which are bound to a downstream EJC. This releases SMG8 and SMG9 from SURF, allowing SMG1 to phosphorylate and subsequently activate UPF1. Phosphorylated UPF1 triggers the dissociation of the ribosome from the mRNP, blocks further translation initiation by targeting eIF3, and recruits SMG5, SMG6, and SMG7. This is followed by SMG5-SMG7-mediated decay of the transcript, which occurs by decapping and/or deadenylation, followed by degradation in the 5’-to-3’ direction from the decapped 5’ end by XRN1, and degradation in the 3’-to-5’ direction from the deadenylated 3’ end by the exosome complex [27]. UPF1 ATPase activity is required for stimulating the complete removal of NMD factors and complete mRNP disassembly, which allows for NMD factor recycling [28].
Figure 1. A Model of Nonsense-Mediated Decay.
(1) During synthesis by RNA polymerase II, wild type and PTC-bearing transcripts undergo cotranscriptional processing into messenger ribonucleoprotein complexes (mRNPs) that contain a 5’-m7GpppN cap and a poly(A)-tail. The cap-binding protein (CBP) heterodimer CBP80-CBP20 complex (CBC) binds to and protects the 5’-cap, and the nuclear poly(A)-binding protein N1 (PABPN1) and the primarily cytoplasmic PABPC1 bind to and protect the poly(A) tail along with eIF4G. In addition, the exon junction complex (EJC) is deposited 20-24 nucleotides upstream of every exon-exon splice site within the mRNP during synthesis. The CBC-bound mRNA is then exported from the nucleus(2a) Wild type transcripts are stabilized during the pioneer round of translation when the CBC is replaced by eukaryotic translation initiation factor 4E (eIF4E), the EJCs are displaced, and PABPN1 is entirely replaced by PABPC1. The mature mRNA is then directed into (2b) steady-state rounds of bulk translation. (3a) PTC-bearing transcripts trigger the classical NMD pathway during the pioneer round of translation when the first ribosome stalls at a PTC that is located more than 50-55 nucleotides upstream of the last EJC-bearing exon-exon junction. (3b) During ribosomal stalling, eukaryotic release factor 1 (eRF1) and eRF3 bind to the ribosomal A site and associate with UPF1 which recruits SMG1 to form the SMG1-UPF1-eRF1-eRF3 (SURF) complex. SMG8 and SMG9 are then recruited to the SURF complex. (3c) The SURF complex is then translocated from the ribosome to UPF2-UPF3 on a downstream EJC. This releases SMG8 and SMG9 from SURF, allowing SMG1 to phosphorylate, and subsequently activate UPF1. Phosphorylated UPF1 triggers the dissociation of the ribosome from the mRNP and recruits SMG5, SMG6, and SMG7. This is followed by SMG5-SMG7-mediated decay of the transcript, which occurs by decapping and/or deadenylation, followed by (3d) degradation by XRN1 and the exosome complex.
An alternative pathway, involving SMG6-mediated endonucleolytic degradation of the transcript can occur, resulting in a 5’ cleavage product and 3’ cleavage product. These cleavage products are rapidly degraded in a similar manner to the decapped/deadenylated transcript by XRN1 and the exosome complex. SMG5-SMG7 and/or SMG6 then promote dephosphorylation of UPF1 by protein phosphatase 2A (PP2A), which recycles UPF1 [5-7].
3. Regulation of Nonsense-Mediated Decay
In addition to RNA quality control, NMD acts as an RNA regulatory pathway that broadly controls approximately 10% of all cellular mRNAs in a wide variety of eukaryotes [24, 29, 30], and also acts as an important regulator of mRNA splicing [31, 32]. The dual role that NMD plays in RNA quality control and regulation of normal mRNA expression means NMD itself is under tight regulatory control. There are a number of examples in which NMD is inhibited by environmental insults, such as hypoxia, ultraviolet irradiation, DNA damage-inducing agents, amino acid starvation, and conditions that activate the unfolded protein response [33-35]. Genetic mutations in NMD factors have also been shown to downregulate or completely remove NMD activity. For example, mutations in UPF2, UPF3A, UPF3B, SMG6, EIF4A3 and RNPS1 are linked to various neurodevelopmental disorders and intellectual disability [13, 36]. NMD factors also function as important modulators in other metabolic pathways necessary for proper embryonic development. It has been shown in several species, including the nematode (C. elegans), fruitfly (D. melanogaster), zebrafish (D. rerio) and mouse (M. musculus), that disruption of NMD through the loss of various NMD factors can lead to growth abnormalities and embryonic lethality [37]. With this in mind, it is necessary to explore NMD homeostatic and regulatory mechanisms and their implications in the manifestation and clinical severity of different genetic diseases when considering therapies that target and/or disrupt this pathway. These variations in NMD efficiency and subsequent mRNA degradation due to differential regulation in a given cell or tissue type reveal an important mechanistic link between specific mutations and the manifestation of genetic disease [38, 39].
3.1 NMD factor regulatory mechanisms
Individual NMD factors can coordinately regulate NMD activity and control the abundance and/or availability of one or more NMD factors. Perhaps the best example of this type of regulation is the cycle of phosphorylation and dephosphorylation that occurs to UPF1 by SMG1 [40]. The phosphorylated form of UPF1 prevents additional rounds of translation, and recruits NMD factors required for exonucleolytic degradation or endonucleolytic degradation through interactions with SMG5-SMG7 or SMG6 [41, 42]. SMG5-SMG7 and/or SMG6 recruit protein phosphatase 2A (PP2A) to dephosphorylate UPF1 [43, 44]. These NMD factor interactions lead to UPF1 recycling, making it available for continued use in NMD.
Another well-characterized regulatory interaction that alters the abundance of NMD factors is the interaction between UPF3 (UPF3A) and UPF3X (UPF3B). UPF3X has a stronger affinity for the EJC than UPF3, and can effectively out-compete UPF3 for EJC binding, resulting in UPF3 destabilization. NMD activity is dependent upon UPF3X outcompeting the functionally less effective UPF3, which will bind UPF2 and inhibit NMD [7, 23].
NMD factors can act through coordinated autoregulatory branched feedback loops that control NMD factor activity and abundance [45, 46]. This pathway of autoregulation is based on the remarkably simple fact that the mRNAs that encode the NMD factors UPF1, UPF2, UPF3X, SMG1, SMG5, SMG6 and SMG7 are NMD substrates themselves. This type of feedback buffers against perturbations that could disrupt NMD factor abundance, thus acting to prevent changes in NMD activity in a particular cell type [45].
Another example of a regulatory mechanism that modulates the activity of NMD through NMD factor abundance is microRNA-mediated decay [47]. MicroRNAs (miRNAs) are small (~20-22-nt) non-protein-coding RNAs that bind and repress the expression of specific mRNA targets through translational repression and/or mRNA decay [48-53]. A microRNA/NMD regulatory pathway exists in the brain that has the ability to fine-tune gene expression in a developmentally regulated cell- and tissue-specific manner. This predominantly neural-specific miRNA, miR-128, represses NMD by targeting UPF1 and MLN1 [47]. Initial evidence shows that miR-128 is differentially expressed in different neurons in distinct regions of the brain during different time points of embryological development [47]. This suggests that neurons with high levels of miR-128 would have repressed NMD and increased levels of NMD-targeted proteins, and neurons with low levels of miR-128 would have increased NMD activity and decreased levels of NMD-targeted proteins [54]. In addition, there is increasing evidence that the dysregulation of miR-128 leads to autism [55], prion-induced neurodegeneration [56], Huntington's disease [57], Parkinson's disease [58], and Alzheimer's disease [59]. Taken together with the fact that mutations that disrupt NMD cause intellectual disability and psychological disorders [36], the homeostatic interplay between miR-128 and NMD increases the clinical relevance of this regulatory circuit and provides an additional feedback/regulatory system for NMD.
3.2 Cell type specific NMD
Variation in the efficiency of NMD between specific cell types is dependent on the expression of the targeted transcript, the relative abundance of NMD factors, as well as the inherent activity of NMD. There are various examples of cell-type-specific differences in NMD. Recent evidence suggests the involvement of the miR-128/NMD circuit in T cell development within the thymus. In addition, it was demonstrated that thymocyte development is perturbed in NMD-deficient mice [61]. This provides evidence that NMD-targeted degradation of particular transcripts is optimal in the cell type that normally expresses it. Another example of transcript-specific degradation within a specific cell type is the targeted degradation of collagen X gene (COL10A1) transcripts, which are degraded when they contain nonsense mutations in cartilage cells, where they are normally expressed, but not in noncartilage cells, such as lymphoblasts and bone cells, derived from a patient with Schmid metaphyseal chondrodysplasia [38, 62]. Additional evidence shows variability in NMD efficiency for ESCO2 transcripts carrying a PTC among different tissues [63], as well as variability in NMD efficiency of CYH2 transcripts carrying a PTC among different yeast strains [64].
Not only is NMD elicited in a transcript-specific manner in different cell types, but different cell types have variable inherent NMD activity in general. Linde et al. [65] found that CFTR and β-globin transcripts carrying a disease-causing PTC, as well as five physiologic NMD substrates, had differential expression due to variable NMD efficiency in different cell lines, including HeLa, CFP15a, CF15b, CFP22a, and MCF7. In contrast, there has been reported interindividual variation in NMD efficiency among patients carrying identical mutations, which caused variable reduction of PTC-bearing transcripts in the same cell type [66, 67].
Variation in the abundance of NMD factors can also lead to variation in NMD activity between cell types. For example, decreased abundance of the EJC accessory protein RNPS1 leads to low NMD efficiency in a HeLa cell model system [68]. In another study, siRNA-mediated knock-down of UPF1 and SMG1 caused DNA damage and suppression of cell growth in HeLa cells, but not in fibroblasts [69].
3.3 Tissue specific NMD
Tissue specific variations in NMD efficiency have been observed in different mouse tissues. In mice heterozygous for a frameshift mutation in the Men1 gene, there are significant differences in the expression ratio of the PTC-containing allele versus the wild type allele in 13 different tissues [39]. The first group of tissues, which included testis, ovary, brain and heart, displayed a strong decrease of the mutated PTC-bearing transcript (average ratio of 18% of mutant versus wild type Men1 transcripts). The second group of tissues, which included lung, intestine and thymus, displayed much lower NMD efficiency (average ratio of 35%). There was also significant variation in NMD factor gene expression. However, these variations in NMD factor gene expression could not account for the variations seen in NMD efficiency. In other words, some other tissue-specific NMD regulatory mechanism besides NMD factor abundance must be responsible for the differences seen in this mouse model.
Tissue-specific variations in NMD efficiency have also been observed in human tissues. Lamin A (LMNA) mRNA carrying a nonsense mutation from a family affected by sudden cardiac death, dilated cardiomyopathy, and rhythm disturbances was significantly downregulated compared to wild type in explanted myocardial tissue from the left and right ventricle, but was not downregulated in cultured fibroblasts [70]. This is an example of tissue-specific NMD efficiency modulating the manifestation of a genetic disease in a transcript-specific manner. Variations in NMD efficiency due to heritable polymorphisms could help explain the differences in the clinical severity seen in many patients with the same genetic mutation. One potential contributing factor to the inter-individual differences seen in NMD efficiency are polymorphisms found in proteins involved in NMD. For example, A large-scale study using data obtained from the International HapMap project found several heritable polymorphisms, including a single-nucleotide polymorphism (SNP) in the DCP1A gene, which forms part of the mRNA decapping complex [71]. The mutant form of this decapping enzyme is less efficient, possibly resulting in a greater proportion of PTC-bearing transcripts. Using expression quantitative trait loci (eQTL) mapping, several SNPs associated with the expression and efficiency of NMD have been found in SMG7, UPF3B, RBM8A and MAGOH [13]. While genetic polymorphisms could explain the variations seen between patients with genetic disease, these differences could also be due to epigenetic modifications.
4. Nonsense-Mediated Decay and Genetic Disease
Variable regulation of NMD factors leads to transcript-, cell-, and tissue-specific differences in NMD efficiency and modulates the manifestation and clinical severity of a number of genetic disorders in at least three major ways: 1) altering the pattern of inheritance, 2) causing distinct traits to manifest from mutations in the same gene, and 3) modifying the specific clinical phenotype [10]. There are many examples where NMD plays a significant role in the manifestation of human genetic disease.
4.1 NMD Alters the Pattern of Inheritance
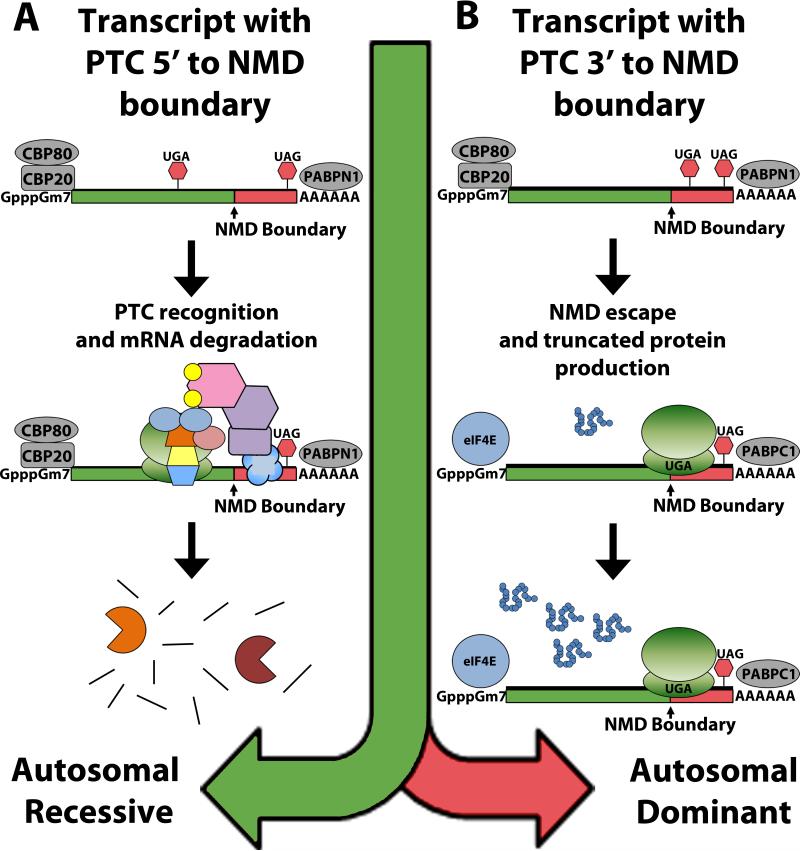
The first way in which NMD modulates the manifestation of genetic disease is by altering the pattern of inheritance. Differential recognition and degradation of mutated transcripts can occur due to the location of the PTC either 5’ to the NMD boundary (NMD-competent) or 3’ to the NMD boundary (NMD-incompetent) [11]. These position-dependent effects of NMD alter the pattern of inheritance for that specific mutant allele (Figure 2). If located within an NMD-competent region of the transcript, NMD will recognize the mutation and degrade that allele, depleting the truncated peptide product from the cell, and preventing its toxic effects. This means that a heterozygous carrier of the mutated gene can still rely on the wild type allele for proper function, leading to an autosomal recessive pattern of inheritance. However, if the mutation is located within an NMD-incompetent region of the transcript, NMD will not recognize nor degrade that allele, allowing the truncated peptide product to accumulate and cause catastrophic damage to the cell, leading to an autosomal dominant pattern of inheritance.
Figure 2. Nonsense-Mediated Decay Alters the Pattern of Inheritance.
Differential recognition and degradation of mutated transcripts can occur due to the location of the PTC either 5’ to the NMD boundary (NMD-competent) or 3’ to the NMD boundary (NMD-incompetent). (A) If the mutation is located within an NMD-competent region of the transcript, NMD will degrade the transcript, depleting the truncated peptide product from the cell, and preventing its toxic effects. This means that a heterozygous carrier of the mutated gene can still rely on the wild type allele for proper function, leading to an autosomal recessive pattern of inheritance. (B) If the mutation is located within an NMD-incompetent region of the transcript, it is not recognized by NMD, allowing the truncated peptide product to accumulate, causing catastrophic damage to the cell, which leads to an autosomal dominant pattern of inheritance.
For example, PTC mutations in the β-globin gene (HBB) located 5’ to the NMD boundary cause the recessively inherited form of β-thalassemia (heterozygotes are healthy), whereas PTC mutations located 3’ to the NMD boundary cause the dominantly inherited form of β-thalassemia [72-75]. Other conditions that display this type of mutation-specific relationship with inheritance include susceptibility to mycobacterial infection (interferon gamma receptor 1; IFNGR1) [76, 77], Robinow syndrome and brachydactyly type B (receptor tyrosine kinase-like orphan receptor 2; ROR2) [78, 79], von Willebrand disease (von Willebrand factor; VWF) [80], factor X deficiency (factor X; F10) [81], retinitis pigmentosa and blindness (rhodopsin; RHO) [82, 83], Becker disease and Thomsen disease (chloride channel 1; CLCN1) [84], Leber congenital amaurosis (cone-rod homeobox-containing gene; CRX) [85], and pseudoxanthoma elasticum (ATP-binding cassette, subfamily C member 6; ABCC6) [86] (Table 1).
Table 1.
NMD Alters the Pattern of Inheritance
| Gene Symbol | Gene Name | OMIM reference no. | 5’ PTC AR 3’ PTC AD | References |
|---|---|---|---|---|
| HBB | β-globin gene | 141900 | β-thalassemia | [72-75] |
| IFNGR1 | Interferon gamma receptor 1 | 107470 | Mycobacterial infection | [76, 77] |
| ROR2 | Receptor tyrosine kinase-like orphan receptor 2 | 602337 | 5’ PTC Robinow syndrome 3’ PTC Brachydactyly type B | [78, 79] |
| VWF | von Willebrand factor | 613160 | von Willebrand disease | [80] |
| F10 | Factor X | 613872 | Factor X deficiency | [81] |
| RHO | Rhodopsin | 180380 | Retinitis pigmentosa | [82, 83] |
| CLCN1 | Chloride channel 1 | 118425 | 5’ PTC Becker disease 3’ PTC Thomsen disease | [84] |
| CRX | Cone-rod homeobox-containing gene | 602225 | Leber congenital amaurosis 5’ PTC heterozygous normal 3’ PTC AD | [85] |
| ABCC6 | ATP-binding cassette, subfamily C member 6 | 603234 | Pseudoxanthoma elasticum | [86] |
AD: autosomal dominant; AR: autosomal recessive; PCWH: peripheral demyelinating neuropathy, central dysmyelinating leukodystrophy, Waardenburg syndrome and Hirschsprung disease; PTC: premature termination codon
The role that NMD plays in altering the pattern of inheritance for many genetic disorders lends support to the hypothesis that heterozygous carriers of genes with PTC mutations are protected from manifesting a more severe dominant disease phenotype that would result from the toxic effects of truncated peptides. It is difficult to estimate the number of genetic disorders in which NMD plays a protective role in disease pathology. Although the number of mutations leading to PTCs can be accurately determined, at this point it is not possible to determine the number of conditions where NMD alters the pattern of inheritance.
4.2 NMD Causes Distinct Traits to Manifest from Mutations in the Same Gene
Although most PTCs resulting from nonsense and frameshift mutations lead to a complete loss-of-function of the truncated peptide, the position-dependent recognition of the PTC within the gene (NMD-sensitive versus NMD-insensitive) can cause distinct traits to manifest due to the variable involvement of NMD and subsequent effects on the expression of dominant-negative traits (Table 2).
Table 2.
NMD Causes Distinct Traits to Manifest from the Same Gene
| Gene Symbol | Gene Name | OMIMreference no. | 5’ haploinsufficiency 3’ PTC dominant-negative or gain-of-function | References |
|---|---|---|---|---|
| SOX10 | SRY-box 10 | 602229 | 5’ PTC Waardenburg syndrome 3’ PTC PCWH | [87, 88] |
| MPZ | Myelin protein zero | 159440 | 5’ PTC Charcot-Marie-Tooth disease type 1B 3’ PTC Congenital hypomyelinating neuropathy | [89] |
| ELN | Elastin | 130160 | 5’ PTC Supravalvular aortic stenosis 3’ PTC Congenital cutis laxa | [90] |
| COL1A2 | Collagen type 1 alpha 2 | 120160 | 5’ PTC Ehlers-Danlos syndrome (EDS) 3’ PTC Osteogenesis imperfecta | [91] |
| COL4A1 | Collagen type 4 alpha 1 | 120130 | 5’ PTC Small vessel brain disease 3’ PTC HANAC | [92, 93] |
| NDUFS4 | NADH-ubiquinone oxidoreductase Fe-S protein 4 | 602694 | 5’ PTC Leigh syndrome 3’ PTC Respiratory complex I deficiency | [94] |
| SLC4A1 | Solute carrier family four member 1 | 109270 | 5’ PTC Spherocytosis 3’ PTC Renal tubular acidosis | [95, 96] |
| ALS2 | Alsin | 606352 | 5’ PTC Amyotrophic lateral sclerosis 3’ PTC Infantile-onset ascending spastic paralysis | [97, 98] |
| GLI3 | Gli-Kruppel family member 3 | 165240 | 5’ PTC Greig cephalopolysyndactyly syndrome 3’ PTC Pallister-Hall syndrome & acrocallosal syndrome | [99, 100] |
HANAC: hereditary angiopathy with nephropathy, aneurysm, and muscle cramps syndrome; PCWH: peripheral demyelinating neuropathy, central dysmyelinating leukodystrophy, Waardenburg syndrome and Hirschsprung disease; PTC: premature termination codon
For example, nonsense mutations 3’ to the NMD boundary in SOX10 produce truncated proteins with potent dominant-negative activity that lead to the severe and complex neurological phenotype of peripheral demyelinating neuropathy, central dysmyelinating leukodystrophy, Waardenburg syndrome and Hirschsprung disease (PCWH). Whereas, nonsense mutations 5’ to the NMD boundary in SOX10 are recognized by NMD, which reduces the dominant-negative expression, leads to a relatively mild phenotype that only combines Waardenburg and Hirschsprung diseases (WS4) [87, 88]. Much like SOX10, different mutations in the myelin protein zero gene (MPZ) cause distinct neurological diseases, each affecting the myelin of the peripheral nervous system [89]. Nonsense mutations 3’ to the NMD boundary in MPZ lead to the more severe congenital hypomyelinating neuropathy (CHN) and Dejerine-Sottas neuropathy (DNS), whereas nonsense mutations located 5’ to the NMD boundary cause the less severe, adult onset Charcot-Marie-Tooth disease type 1B (CMT1B) [87].
Other conditions that display this type of mutation-specific relationship with disease manifestation include supravalvular aortic stenosis (SVAC) and congential cutis laxa (elastin; ELN) [90], Ehlers-Danlos syndrome (EDS) and osteogenesis imperfecta (OI) (collagen type I alpha 2; COL1A2) [91], small vessel brain disease and Hereditary angiopathy with nephropathy, aneurysms, and muscle cramps (HANAC) syndrome (collagen type 4 alpha 1; COL4A1) [92, 93], Leigh syndrome and respiratory complex I deficiency (NADH-ubiquinone oxidoreductase Fe-S protein 4; NDUFS4) [94], spherocytosis and renal tubular acidosis (solute carrier family four member 1; SLC4A1) [95, 96], amyotrophic lateral sclerosis (ALS) and infantile-onset ascending spastic paralysis (alsin, ALS2) [97, 98], and Greig Cephalopolysyndactyly syndrome (GCS), Pallister-Hall syndrome (PHS) and acrocallosal syndrome (Gli-Kruppel family member 3; GLI3) [10, 99, 100]. For the examples given, NMD alters the manifestation of disease traits by converting the dominant-negative expression of the mutated protein to haploinsufficiency, therefore leading to milder disease pathology.
4.3 NMD Modulates the Specific Clinical Phenotype
NMD generally plays an important beneficial role in the expression of recessive traits by preventing the dominant-negative toxicity of mutant proteins. In addition to altering inheritance patterns and disease trait manifestation, NMD also plays a more subtle role in modulating disease pathology. Much like the position-dependent effects on inheritance and trait manifestation, the position of the PTC within the gene can modulate the severity of the specific clinical phenotype (Table 3).
Table 3.
NMD Modulates the Clinical Phenotype
| Gene Symbol | Gene Name | OMIM reference no. | Phenotype | References |
|---|---|---|---|---|
| COL1A1 | Collagen type I, al | 120150 | 5’ PTC osteogenesis imperfecta type I 3’ PTC osteogenesis imperfect type II-IV | [101, 102] |
| COL2A1 | Collagen type II, al | 120140 | 5’ PTC Stickler syndrome 3’ PTC spondyloepiphyseal dysplasia | [101, 103] |
| DMD | Dystrophin | 300377 | 5’ PTC Duchenne muscular dystrophy 3’ PTC Becker muscular dystrophy | [104, 105] |
| FBN1 | Fibrillin-1 | 134797 | Marfan syndrome and type 1 fibrillinopathies 5’ PTC more severe | [106, 107] |
| SALL1 | Sal-like 1 | 107480 | 5’ PTC milder phenotype 3’ PTC Townes-Brocks syndrome | [100] |
| GHR | Growth hormone receptor | 265500 | 5’ PTC Growth hormone insensitivity syndrome | [108] |
| FGD4 | Frabin | 611104 | Charcot Marie Tooth disease type 4H 5’ PTC less severe | [109, 110] |
| HEXA | Hexosaminidase A | 606869 | Tay-Sachs disease 5’ PTC infantile (severe) | [111] |
| RB1 | Retinoblastoma | 180200 | Retinoblastoma 5’ PTC early onset | [112] |
| ATM | Ataxia-telangiectasia mutated | 607585 | 5’ PTC mild 3’ PTC severe | [113] |
PTC: premature termination codon
For example, missense mutations in the collagen type I alpha 1 (COL1A1) and collagen type II alpha 1 (COL2A1) genes cause the severe disease phenotypes of osteogenesis imperfecta (OI) type II-IV and spondyloepiphyseal dysplasia, respectively. Whereas, all nonsense mutations 5’ to the NMD boundary in both genes cause haploinsuffiency, leading to the milder clinical phenotypes of OI type I (COL1A1) [101, 102] and Stickler syndrome (COL2A1) [103]. In contrast, there are genetic disorders where truncated peptides retain function, such as Duchenne muscular dystrophy (DMD), and NMD acts to worsen the disease phenotype by degrading NMD-sensitive transcripts [104, 105]. Dystrophin gene (DMD) transcripts carrying mutations that are NMD-insensitive produce truncated peptides with residual activity that can yield Becker muscular dystrophy (BMD), a milder form of DMD.
Other conditions that display this mutation-specific expression of disease severity include the clinical phenotypes of Marfan syndrome (MFS) (fibrillin-1; FBN1) [106, 107], Townes-Brocks syndrome (TBS) (sal-like 1; SALL1) [100], growth hormone insensitivity syndrome (GHIS) (growth hormone receptor; GHR) [108], Charcot Marie Tooth type 4H (CMT4H) (Frabin; FGD4) [109, 110], Tay-Sachs disease (hexosaminidase A; HEXA) [111], retinoblastoma (retinoblastoma; RB1) [112], and ataxia-telangiectasia (ataxia-telangiaectasia mutated; ATM) [113]. In addition to the examples given above, we have shown previously that NMD recognizes numerous mutations from a variety of genes that cause neuronal ceroid lipofuscinosis (NCL), a group of fatal neurodegenerative disorders, leading to variable gene expression and subsequent differences in protein function and clinical severity [114-116].
Multiple biochemical mechanisms govern the mutation-, transcript-, cell-, and tissue-specific variability of NMD efficiency that underlies the pattern of inheritance, expression of distinct traits, and modulation of clinical severity in genetic disease. The involvement of NMD in the pathology of genetic disease provides a therapeutic target for the treatment of almost all genetic disorders with in-frame PTCs. This has led to the discovery and development of many compounds that target the NMD pathway, suppress the effects of nonsense mutations, and rescue the disease phenotype by producing full-length and often fully functional proteins.
5. Current Therapeutic Approaches That Target Nonsense Mutations
5.1 Nonsense suppression
The recognition of sense codons during translation is facilitated when aminoacyl-tRNAs enter the A site of the ribosome and sample the codon until a cognate aminoacyl-tRNA can bind. A cognate aminoacyl-tRNA has an anticodon sequence that can correctly pair with all three nucleotides of the A site codon. A proofreading step then occurs to confirm the correct codonanticodon pairing and the cognate aminoacyl-tRNA becomes fully accommodated into the A site. This is followed by peptide bond formation, which occurs through addition of the amino acid carried by the cognate aminoacyl-tRNA onto the C-terminus of the polypeptide chain. The peptidyl-tRNA is then translocated into the ribosomal P site and aminoacyl-tRNA selection continues at the next codon until reaching a stop codon.
There are three stop codons (UAA, UAG and UGA) that are not recognized by aminoacyl-tRNAs, but instead, are recognized by eukaryotic release factors (eRFs). The termination of eukaryotic translation occurs when a stop codon enters the ribosomal A site and is bound by the class I eukaryotic release factor (eRF1). eRF1 has a similar three-dimensional structure as an aminoacyl-tRNA and forms a complex with the class II release factor (eRF3). eRF3 is a GTPase that assists eRF1 with stop codon recognition and entrance into the peptidyl transferase center (P site) to release the polypeptide [1, 117, 118]. Normally, stop codon recognition is mediated by the same sampling process that occurs at a sense codon. Near-cognate aminoacyl-tRNAs with anticodons that have two out of three complementary nucleotides of the stop codon can bind at the A site. Usually, the stop codon is recognized by the eRF1/3 complex, which out-competes near-cognate aminoacyl-tRNAs, leading to translation termination and polypeptide chain release. However, near-cognate aminoacyl-tRNAs can occasionally become accommodated into the A site and their amino acid is incorporated into the growing polypeptide chain, which recodes the stop codon into a sense codon [1]. When this occurs at an in-frame PTC, one of several possible amino acids is inserted, and the synthesis of the full-length protein occurs. This process is called ‘stop codon suppression’ (nonsense suppression) or ‘read-through’. Under normal conditions, natural read-through occurs at a rate of 0.01-1% at PTCs and 0.001-0.1% at normal stop codons [118].
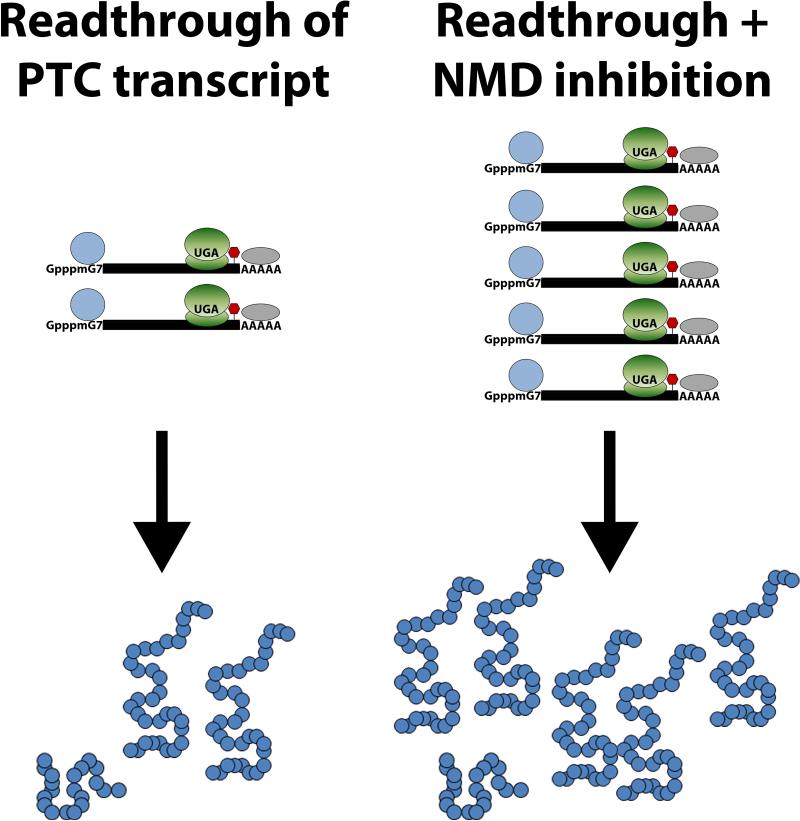
The goal of nonsense suppression therapy is to exploit this natural process and enhance read-through by allowing near-cognate aminoacyl-tRNAs to out-compete the release factor complex and enter the ribosomal A site. By recoding the PTC into a sense codon, enough full-length and possibly functional protein may be produced to provide a therapeutic benefit to patients with genetic disease (Figure 3).
Figure 3. Nonsense Suppression Therapy.
PTC-bearing mRNA transcripts that are not recognized and degraded by NMD will mature and produce truncated proteins that have deleterious effects on the cell. Read-through compounds will bind to either the 40S or 60S subunit of the ribosome and decrease the fidelity of the PTC. The purpose of nonsense suppression therapy is to trick the ribosome into accepting near-cognate aminoacyl-tRNAs into the A-site, therefore enhancing natural PTC read-through and increasing the abundance of full-length protein.
There are several compounds that can induce read-through of PTCs, such as aminoglycosides, modified aminoglycosides (NB30, NB54, NB84), ataluren (PTC124) and RTC13. These compounds have been shown in both in vitro and in vivo models to alleviate disease pathogenesis by enhancing PTC read-through [119]. Examples include cystic fibrosis (CF) [120-131], Becker and Duchenne muscular dystrophy (BMD/DMD) [128, 129, 132-140], ataxia telangiectasia [139, 141, 142], Rett syndrome (RTT) [143-146], Usher syndrome type I (USH1) [147-149], Hurler syndrome (MPS1) [150-154], Maroteaux-Lamy syndrome (MPSVI) [155], carnitine palmitoyltransferase 1A (CPT1A) [156], hemophilia [157, 158], methylmalonic acidura (MMA) [159], neuronal ceroid lipofuscinosis (NCL) [114, 160, 161], spinal muscular atrophy (SMA) [162], peroxisome biogenesis disorder (PBD) [163, 164], obesity [165], poor drug metabolism [166], and cancer [151].
A few of these disorders have been heavily studied using nonsense suppression therapy and have proceeded into clinical trials. These diseases include CF, BMD/DMD, factor VII deficiency, Hailey-Hailey disease, hemophilia A and hemophilia B, leucocyte adhesion deficiency 1 (LAD1) and McArdle disease [119].
5.2 Limitations to nonsense suppression
Although there is a potential benefit from using nonsense suppression in patients with genetic disorders (as seen with in vitro and in vivo disease models), read-through compounds have had few reported successes in human subjects.
The efficacy of each read-through compound depends on the innate ability of each compound to trick the ribosome into allowing near-cognate aminoacyl-tRNAs into the A site and also the innate fidelity of each of the three different stop codons. The termination fidelity of each stop codon is inversely related to read-through capacity. UGA has the lowest termination fidelity and highest read-through potential, followed by UAG, with an intermediate fidelity, and UAA with the highest termination fidelity [167]. In addition, the 5’ and 3’ sequence adjacent to each stop codon within the mRNA transcript can modulate termination fidelity [168, 169]. It is also important to point out that cell- and tissue-specific changes in NMD efficiency will also alter read-through capacity. Therefore, it is important to consider the transcript- and codon-specific context of every mutation as well as the cell- and tissue-specific differences in NMD efficiency when evaluating the potential therapeutic outcome of using read-through compounds for nonsense suppression to effectively treat genetic disease. Another often overlooked variable affecting read-through is the relative bioavailability of each compound in the tissue being targeted. For example, it is not currently known how many compounds can effectively cross the blood brain barrier (BBB) when treating diseases of the central nervous system.
Another possible limitation to the efficacy of read-through is the incorporation of a nonfunctional amino acid at the site of the PTC. This is further complicated by the fact that the three stop codons (UAG, UGA, UAA) have a limited number of near-cognate aminoacyl-tRNAs with which they can associate within the A site of the ribosome. UGA is most commonly recoded as tryptophan, arginine and cysteine during read-through, while UAG is recoded as either glutamine or tryptophan, and UAA is recoded as glutamine [119]. Therefore, nonsense suppression can result in the production of a full-length protein with a missense mutation, which may not yield functional protein activity.
Although some read-through compounds, such as ataluren, have very low toxicity, many of the aminoglycosides that are used for nonsense suppression pose significant risks, including nephrotoxicity and ototoxicity. In fact, approximately 2-25% of patients treated with aminoglycosides show signs of nephrotoxicity and/or ototoxicity [118]. The kidney damage caused by aminoglycoside treatment is reversible, but damage to the inner ear and subsequent hearing loss is permanent.
PTCs typically lead to a complete absence of full-length protein. In certain cases, there is evidence that suggests the involvement of a T-cell mediated immune response against full-length or nearly full-length protein produced during nonsense suppression [170]. Immune-mediated degradation of full-length protein may not occur in all cases, but must be taken into consideration as a potential limitation to read-through therapy in human subjects.
Although there are many potential factors limiting the efficacy of nonsense suppression, many steps are being taken to thwart these factors and enhance read-through therapy. For example, many researchers are currently developing and testing potentially safer and more effective read-through compounds with decreased toxicity, such as the compounds RTC13 and RTC14 [139, 140]. In addition, modifications made to the aminoglycosides paromomycin, amikacin and G418 have produced less toxic derivatives with better efficacy, such as NB30, NB54 and NB84 [118]. Other techniques, such as the coadministration of daptomycin and poly-L-aspartic acid (PAA) alongside aminoglycoside treatment can alleviate nephrotoxicity and ototoxicity. Interestingly, coadministration of PAA with aminoglycosides has been shown to enhance nonsense suppression in human MPS-I fibroblasts and a CF mouse model through increased intracellular aminoglycoside concentrations [171].
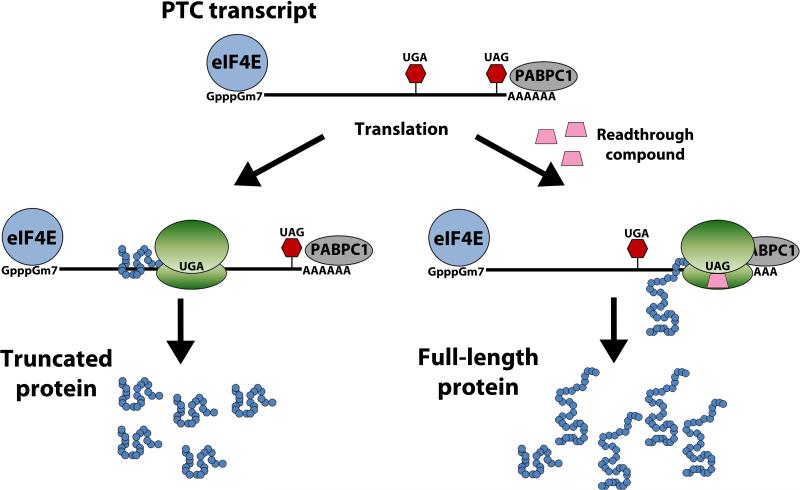
Although NMD protects the cell against the potentially toxic effects of truncated protein products, the targeted degradation of mRNA transcripts limits the efficacy of read-through therapies and dampens the potential rescue of the disease phenotype by limiting the amount of mRNA from which full-length protein is produced. Therefore, an obvious way to increase nonsense suppression efficacy is to partially inhibit NMD to increase the available pool of PTC-containing mRNA for translation and read-through. Phosphatidylinositol-3-kinase inhibitors such as caffeine, wortmannin, and LY294002SMG1 inhibit SMG1, the protein responsible for phosphorylating UPF1. In addition, another NMD inhibitor called NMD-I was recently discovered. NMD-I inhibits UPF1 dephosphorylation by blocking the interaction between UPF1 and SMG5, thus preventing the recruitment of PP2A and dephosphorylation of UPF1 [172, 173]. For the treatment of many genetic disorders, the use of any single therapy may not surpass the therapeutic threshold needed to produce enough functional protein to effectively alter disease outcome. To enhance the overall therapeutic benefit of nonsense suppression therapy, a combination of approaches could be used to further enhance the production of functional protein. Combining NMD inhibition with nonsense suppression could provide additional PTC-bearing transcripts during read-through, thus allowing for increased levels of full-length functional protein (Figure 4).
Figure 4. Combining NMD Inhibition with Nonsense Suppression to Enhance Therapeutic Outcome.
Nonsense-mediated decay targets PTC-bearing transcripts in the cytoplasm, leading to a decrease in the pool of mRNA, which produces little to no full-length protein. The goal of nonsense suppression therapy is to produce full-length functional protein to alleviate disease pathology. However, nonsense-mediated degradation of PTC-bearing transcripts decreases the abundance of mRNA, limiting the efficacy of read-through therapy and decreasing the chance of surpassing the therapeutic threshold. By combining NMD inhibition with nonsense suppression there is an increased abundance of PTC-bearing mRNA with which read-through drugs can act on, leading to an increase in the production of full-length protein.
6. Conclusion and Future Outlook
Although NMD primarily acts to protect the cell from the downstream effects of nonsense mutations within the genome, there are many variables that act to undermine the potential benefit of this evolutionarily conserved mechanism. The dynamics of NMD regulation and the mutation-, codon-, gene-, cell-, and tissue-specific differences in NMD efficiency can alter the underlying disease pathology, which leads to different patterns of inheritance (autosomal recessive vs. autosomal dominant), as well as causing distinct traits to manifest from mutations in the same gene, and modulating the specific clinical phenotype. In addition, NMD limits the efficacy of current nonsense suppression therapies intended to enhance read-through and increase levels of full-length protein. Therefore, there are two sides to the NMD coin: the beneficial action of targeting PTC-bearing mRNA transcripts for degradation to prevent the toxic side effects of truncated protein and the unintended consequence of limiting the potential benefit of read-through therapy. With this in mind, it is important to understand the role of NMD in genetic disease pathology on a case-by-case basis, which will lead to better techniques enhancing the production of full-length functional protein. Utilizing combination therapies, such as nonsense suppression alongside NMD inhibition, may move us closer to finding treatments that break the therapeutic threshold for the more than 7,000 rare genetic disorders affecting more than 30 million people in the US.
Acknowledgments
Funding
Partially supported by NIH R01NS43310 and the Beat Batten Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest Statement
The authors declare no conflict of interest.
References
- 1.Keeling KM, Bedwell DM. Suppression of nonsense mutations as a therapeutic approach to treat genetic diseases. Wiley Interdiscip Rev RNA. 2011;2:837–852. doi: 10.1002/wrna.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frischmeyer PA, Dietz HC. Nonsense-mediated mRNA decay in health and disease. Hum Mol Genet. 1999;8:1893–1900. doi: 10.1093/hmg/8.10.1893. [DOI] [PubMed] [Google Scholar]
- 3.Krawczak M, Ball EV, Cooper DN. Neighboring-nucleotide effects on the rates of germ-line single-base-pair substitution in human genes. Am J Hum Genet. 1998;63:474–488. doi: 10.1086/301965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mort M, et al. A meta-analysis of nonsense mutations causing human genetic disease. Hum Mutat. 2008;29:1037–1047. doi: 10.1002/humu.20763. [DOI] [PubMed] [Google Scholar]
- 5.Chang YF, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- 6.Kervestin S, Jacobson A. NMD: a multifaceted response to premature translational termination. Nat Rev Mol Cell Biol. 2012;13:700–712. doi: 10.1038/nrm3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schoenberg DR, Maquat LE. Regulation of cytoplasmic mRNA decay. Nat Rev Genet. 2012;13:246–259. doi: 10.1038/nrg3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Balagopal V, Fluch L, Nissan T. Ways and means of eukaryotic mRNA decay. Biochim Biophys Acta. 2012;1819:593–603. doi: 10.1016/j.bbagrm.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 9.Muhlemann O, et al. Recognition and elimination of nonsense mRNA. Biochim Biophys Acta. 2008;1779:538–549. doi: 10.1016/j.bbagrm.2008.06.012. [DOI] [PubMed] [Google Scholar]
- 10.Khajavi M, Inoue K, Lupski JR. Nonsense-mediated mRNA decay modulates clinical outcome of genetic disease. Eur J Hum Genet. 2006;14:1074–1081. doi: 10.1038/sj.ejhg.5201649. [DOI] [PubMed] [Google Scholar]
- 11.Holbrook JA, et al. Nonsense-mediated decay approaches the clinic. Nat Genet. 2004;36:801–808. doi: 10.1038/ng1403. [DOI] [PubMed] [Google Scholar]
- 12.Bashyam MD. Nonsense-mediated decay: linking a basic cellular process to human disease. Expert Rev Mol Diagn. 2009;9:299–303. doi: 10.1586/erm.09.18. [DOI] [PubMed] [Google Scholar]
- 13.Doma MK, Parker R. RNA quality control in eukaryotes. Cell. 2007;131:660–668. doi: 10.1016/j.cell.2007.10.041. [DOI] [PubMed] [Google Scholar]
- 14.Grzybowska EA. Human intronless genes: functional groups, associated diseases, evolution, and mRNA processing in absence of splicing. Biochem Biophys Res Commun. 2012;424:1–6. doi: 10.1016/j.bbrc.2012.06.092. [DOI] [PubMed] [Google Scholar]
- 15.Lee HC, et al. Nonsense-mediated translational repression involves exon junction complex downstream of premature translation termination codon. FEBS Lett. 2010;584:795–800. doi: 10.1016/j.febslet.2010.01.003. [DOI] [PubMed] [Google Scholar]
- 16.Maquat LE, Tarn WY, Isken O. The pioneer round of translation: features and functions. Cell. 2010;142:368–374. doi: 10.1016/j.cell.2010.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maniatis T, Reed R. An extensive network of coupling among gene expression machines. Nature. 2002;416:499–506. doi: 10.1038/416499a. [DOI] [PubMed] [Google Scholar]
- 18.Moore MJ, Proudfoot NJ. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 2009;136:688–700. doi: 10.1016/j.cell.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 19.Brocke KS, et al. The human intronless melanocortin 4-receptor gene is NMD insensitive. Hum Mol Genet. 2002;11:331–335. doi: 10.1093/hmg/11.3.331. [DOI] [PubMed] [Google Scholar]
- 20.Maquat LE, Li X. Mammalian heat shock p70 and histone H4 transcripts, which derive from naturally intronless genes, are immune to nonsense-mediated decay. RNA. 2001;7:445–456. doi: 10.1017/s1355838201002229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chan WK, et al. An alternative branch of the nonsense-mediated decay pathway. EMBO J. 2007;26:1820–1830. doi: 10.1038/sj.emboj.7601628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chan WK, et al. A UPF3-mediated regulatory switch that maintains RNA surveillance. Nat Struct Mol Biol. 2009;16:747–753. doi: 10.1038/nsmb.1612. [DOI] [PubMed] [Google Scholar]
- 23.Huang L, Wilkinson MF. Regulation of nonsense-mediated mRNA decay. Wiley Interdiscip Rev RNA. 2012;3:807–828. doi: 10.1002/wrna.1137. [DOI] [PubMed] [Google Scholar]
- 24.Kim YK, et al. Mammalian Staufen1 recruits Upf1 to specific mRNA 3'UTRs so as to elicit mRNA decay. Cell. 2005;120:195–208. doi: 10.1016/j.cell.2004.11.050. [DOI] [PubMed] [Google Scholar]
- 25.Nagy E, Maquat LE. A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem Sci. 1998;23:198–199. doi: 10.1016/s0968-0004(98)01208-0. [DOI] [PubMed] [Google Scholar]
- 26.Houseley J, LaCava J, Tollervey D. RNA-quality control by the exosome. Nat Rev Mol Cell Biol. 2006;7:529–539. doi: 10.1038/nrm1964. [DOI] [PubMed] [Google Scholar]
- 27.Franks TM, Singh G, Lykke-Andersen J. Upf1 ATPase-dependent mRNP disassembly is required for completion of nonsense- mediated mRNA decay. Cell. 2010;143:938–950. doi: 10.1016/j.cell.2010.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mendell JT, et al. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat Genet. 2004;36:1073–1078. doi: 10.1038/ng1429. [DOI] [PubMed] [Google Scholar]
- 29.Wilkinson MF. A new function for nonsense-mediated mRNA-decay factors. Trends Genet. 2005;21:143–148. doi: 10.1016/j.tig.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 30.Lejeune F, Maquat LE. Mechanistic links between nonsense-mediated mRNA decay and pre-mRNA splicing in mammalian cells. Curr Opin Cell Biol. 2005;17:309–315. doi: 10.1016/j.ceb.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 31.Weischenfeldt J, et al. Mammalian tissues defective in nonsense-mediated mRNA decay display highly aberrant splicing patterns. Genome Biol. 2012;13:R35. doi: 10.1186/gb-2012-13-5-r35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sharifi NA, Dietz HC. Nonsense-Mediated mRNA Decay. Landes Bioscience; Austin, TX: 2006. Physiologic substrates and functions for mammalian NMD. [Google Scholar]
- 33.Gardner LB. Hypoxic inhibition of nonsense-mediated RNA decay regulates gene expression and the integrated stress response. Mol Cell Biol. 2008;28:3729–3741. doi: 10.1128/MCB.02284-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gardner LB, Corn PG. Hypoxic regulation of mRNA expression. Cell Cycle. 2008;7:1916–1924. doi: 10.4161/cc.7.13.6203. [DOI] [PubMed] [Google Scholar]
- 35.Tarpey PS, et al. Mutations in UPF3B, a member of the nonsense-mediated mRNA decay complex, cause syndromic and nonsyndromic mental retardation. Nat Genet. 2007;39:1127–1133. doi: 10.1038/ng2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nguyen LS, et al. Contribution of copy number variants involving nonsense-mediated mRNA decay pathway genes to neuro-developmental disorders. Hum Mol Genet. 2013;22:1816–1825. doi: 10.1093/hmg/ddt035. [DOI] [PubMed] [Google Scholar]
- 37.Hwang J, Maquat LE. Nonsense-mediated mRNA decay (NMD) in animal embryogenesis: to die or not to die, that is the question. Curr Opin Genet Dev. 2011;21:422–430. doi: 10.1016/j.gde.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bateman JF, et al. Tissue-specific RNA surveillance? Nonsense-mediated mRNA decay causes collagen X haploinsufficiency in Schmid metaphyseal chondrodysplasia cartilage. Hum Mol Genet. 2003;12:217–225. doi: 10.1093/hmg/ddg054. [DOI] [PubMed] [Google Scholar]
- 39.Zetoune AB, et al. Comparison of nonsense-mediated mRNA decay efficiency in various murine tissues. BMC Genet. 2008;9:83. doi: 10.1186/1471-2156-9-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hwang J, et al. UPF1 association with the cap-binding protein, CBP80, promotes nonsense-mediated mRNA decay at two distinct steps. Mol Cell. 2010;39:396–409. doi: 10.1016/j.molcel.2010.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Isken O, et al. Upf1 phosphorylation triggers translational repression during nonsense-mediated mRNA decay. Cell. 2008;133:314–327. doi: 10.1016/j.cell.2008.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Okada-Katsuhata Y, et al. N- and C-terminal Upf1 phosphorylations create binding platforms for SMG-6 and SMG-5:SMG-7 during NMD. Nucleic Acids Res. 2012;40:1251–1266. doi: 10.1093/nar/gkr791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kashima I, et al. Binding of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev. 2006;20:355–367. doi: 10.1101/gad.1389006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ohnishi T, et al. Phosphorylation of hUPF1 induces formation of mRNA surveillance complexes containing hSMG-5 and hSMG-7. Mol Cell. 2003;12:1187–1200. doi: 10.1016/s1097-2765(03)00443-x. [DOI] [PubMed] [Google Scholar]
- 45.Huang L, et al. RNA homeostasis governed by cell type-specific and branched feedback loops acting on NMD. Mol Cell. 2011;43:950–961. doi: 10.1016/j.molcel.2011.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yepiskoposyan H, et al. Autoregulation of the nonsense-mediated mRNA decay pathway in human cells. RNA. 2011;17:2108–2118. doi: 10.1261/rna.030247.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bruno IG, et al. Identification of a microRNA that activates gene expression by repressing nonsense-mediated RNA decay. Mol Cell. 2011;42:500–510. doi: 10.1016/j.molcel.2011.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 49.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 50.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 51.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Baek D, et al. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Selbach M, et al. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 54.Karam R, Wilkinson M. A conserved microRNA/NMD regulatory circuit controls gene expression. RNA Biol. 2012;9:22–26. doi: 10.4161/rna.9.1.18010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Abu-Elneel K, et al. Heterogeneous dysregulation of microRNAs across the autism spectrum. Neurogenetics. 2008;9:153–161. doi: 10.1007/s10048-008-0133-5. [DOI] [PubMed] [Google Scholar]
- 56.Saba R, et al. A miRNA signature of prion induced neurodegeneration. PLoS One. 2008;3:e3652. doi: 10.1371/journal.pone.0003652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee ST, et al. Altered microRNA regulation in Huntington's disease models. Exp Neurol. 2011;227:172–179. doi: 10.1016/j.expneurol.2010.10.012. [DOI] [PubMed] [Google Scholar]
- 58.Kim J, et al. A MicroRNA feedback circuit in midbrain dopamine neurons. Science. 2007;317:1220–1224. doi: 10.1126/science.1140481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lukiw WJ. Micro-RNA speciation in fetal, adult and Alzheimer's disease hippocampus. Neuroreport. 2007;18:297–300. doi: 10.1097/WNR.0b013e3280148e8b. [DOI] [PubMed] [Google Scholar]
- 60.Carter MS, Li S, Wilkinson MF. A splicing-dependent regulatory mechanism that detects translation signals. EMBO J. 1996;15:5965–5975. [PMC free article] [PubMed] [Google Scholar]
- 61.Frischmeyer-Guerrerio PA, et al. Perturbation of thymocyte development in nonsense-mediated decay (NMD)-deficient mice. Proc Natl Acad Sci U S A. 2011;108:10638–10643. doi: 10.1073/pnas.1019352108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tan JT, et al. Competency for nonsense-mediated reduction in collagen X mRNA is specified by the 3'UTR and corresponds to the position of mutations in Schmid metaphyseal chondrodysplasia. Am J Hum Genet. 2008;82:786–793. doi: 10.1016/j.ajhg.2008.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Resta N, et al. A homozygous frameshift mutation in the ESCO2 gene: evidence of intertissue and interindividual variation in Nmd efficiency. J Cell Physiol. 2006;209:67–73. doi: 10.1002/jcp.20708. [DOI] [PubMed] [Google Scholar]
- 64.Kebaara B, et al. Genetic background affects relative nonsense mRNA accumulation in wild-type and upf mutant yeast strains. Curr Genet. 2003;43:171–177. doi: 10.1007/s00294-003-0386-3. [DOI] [PubMed] [Google Scholar]
- 65.Linde L, et al. The efficiency of nonsense-mediated mRNA decay is an inherent character and varies among different cells. Eur J Hum Genet. 2007;15:1156–1162. doi: 10.1038/sj.ejhg.5201889. [DOI] [PubMed] [Google Scholar]
- 66.Perrin-Vidoz L, et al. The nonsense-mediated mRNA decay pathway triggers degradation of most BRCA1 mRNAs bearing premature termination codons. Hum Mol Genet. 2002;11:2805–2814. doi: 10.1093/hmg/11.23.2805. [DOI] [PubMed] [Google Scholar]
- 67.Linde L, et al. Nonsense-mediated mRNA decay affects nonsense transcript levels and governs response of cystic fibrosis patients to gentamicin. J Clin Invest. 2007;117:683–692. doi: 10.1172/JCI28523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Viegas MH, et al. The abundance of RNPS1, a protein component of the exon junction complex, can determine the variability in efficiency of the Nonsense Mediated Decay pathway. Nucleic Acids Res. 2007;35:4542–4551. doi: 10.1093/nar/gkm461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Usuki F, et al. Specific inhibition of nonsense-mediated mRNA decay components, SMG-1 or Upf1, rescues the phenotype of Ullrich disease fibroblasts. Mol Ther. 2006;14:351–360. doi: 10.1016/j.ymthe.2006.04.011. [DOI] [PubMed] [Google Scholar]
- 70.Geiger SK, et al. Incomplete nonsense-mediated decay of mutant lamin A/C mRNA provokes dilated cardiomyopathy and ventricular tachycardia. J Mol Med (Berl) 2008;86:281–289. doi: 10.1007/s00109-007-0275-1. [DOI] [PubMed] [Google Scholar]
- 71.Seoighe C, Gehring C. Heritability in the efficiency of nonsense-mediated mRNA decay in humans. PLoS One. 2010;5:e11657. doi: 10.1371/journal.pone.0011657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hall GW, Thein S. Nonsense codon mutations in the terminal exon of the beta-globin gene are not associated with a reduction in beta-mRNA accumulation: a mechanism for the phenotype of dominant beta-thalassemia. Blood. 1994;83:2031–2037. [PubMed] [Google Scholar]
- 73.Thein SL, et al. Molecular basis for dominantly inherited inclusion body beta-thalassemia. Proc Natl Acad Sci U S A. 1990;87:3924–3928. doi: 10.1073/pnas.87.10.3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Baserga SJ, Benz EJ., Jr. Nonsense mutations in the human beta-globin gene affect mRNA metabolism. Proc Natl Acad Sci U S A. 1988;85:2056–2060. doi: 10.1073/pnas.85.7.2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Forget BG, et al. Absence of messenger RNA for beta globin chain in beta(0) thalassaemia. Nature. 1974;247:379–381. doi: 10.1038/247379a0. [DOI] [PubMed] [Google Scholar]
- 76.Jouanguy E, et al. Interferon-gamma-receptor deficiency in an infant with fatal bacille Calmette-Guerin infection. N Engl J Med. 1996;335:1956–1961. doi: 10.1056/NEJM199612263352604. [DOI] [PubMed] [Google Scholar]
- 77.Jouanguy E, et al. A human IFNGR1 small deletion hotspot associated with dominant susceptibility to mycobacterial infection. Nat Genet. 1999;21:370–378. doi: 10.1038/7701. [DOI] [PubMed] [Google Scholar]
- 78.Schwabe GC, et al. Distinct mutations in the receptor tyrosine kinase gene ROR2 cause brachydactyly type B. Am J Hum Genet. 2000;67:822–831. doi: 10.1086/303084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Patton MA, Afzal AR. Robinow syndrome. J Med Genet. 2002;39:305–310. doi: 10.1136/jmg.39.5.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schneppenheim R, et al. Expression and characterization of von Willebrand factor dimerization defects in different types of von Willebrand disease. Blood. 2001;97:2059–2066. doi: 10.1182/blood.v97.7.2059. [DOI] [PubMed] [Google Scholar]
- 81.Millar DS, et al. Molecular analysis of the genotype-phenotype relationship in factor X deficiency. Hum Genet. 2000;106:249–257. doi: 10.1007/s004390051035. [DOI] [PubMed] [Google Scholar]
- 82.Rosenfeld PJ, et al. A null mutation in the rhodopsin gene causes rod photoreceptor dysfunction and autosomal recessive retinitis pigmentosa. Nat Genet. 1992;1:209–213. doi: 10.1038/ng0692-209. [DOI] [PubMed] [Google Scholar]
- 83.Sung CH, et al. Rhodopsin mutations in autosomal dominant retinitis pigmentosa. Proc Natl Acad Sci U S A. 1991;88:6481–6485. doi: 10.1073/pnas.88.15.6481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pusch M. Myotonia caused by mutations in the muscle chloride channel gene CLCN1. Hum Mutat. 2002;19:423–434. doi: 10.1002/humu.10063. [DOI] [PubMed] [Google Scholar]
- 85.Rivolta C, Berson EL, Dryja TP. Dominant Leber congenital amaurosis, cone-rod degeneration, and retinitis pigmentosa caused by mutant versions of the transcription factor CRX. Hum Mutat. 2001;18:488–498. doi: 10.1002/humu.1226. [DOI] [PubMed] [Google Scholar]
- 86.Plomp AS, et al. Does autosomal dominant pseudoxanthoma elasticum exist? Am J Med Genet A. 2004;126A:403–412. doi: 10.1002/ajmg.a.20632. [DOI] [PubMed] [Google Scholar]
- 87.Inoue K, et al. Molecular mechanism for distinct neurological phenotypes conveyed by allelic truncating mutations. Nat Genet. 2004;36:361–369. doi: 10.1038/ng1322. [DOI] [PubMed] [Google Scholar]
- 88.Inoue K, et al. Translation of SOX10 3′ untranslated region causes a complex severe neurocristopathy by generation of a deleterious functional domain. Hum Mol Genet. 2007;16:3037–3046. doi: 10.1093/hmg/ddm262. [DOI] [PubMed] [Google Scholar]
- 89.Warner LE, et al. Clinical phenotypes of different MPZ (P0) mutations may include Charcot-Marie-Tooth type 1B, Dejerine-Sottas, and congenital hypomyelination. Neuron. 1996;17:451–460. doi: 10.1016/s0896-6273(00)80177-4. [DOI] [PubMed] [Google Scholar]
- 90.Tassabehji M, et al. Elastin: genomic structure and point mutations in patients with supravalvular aortic stenosis. Hum Mol Genet. 1997;6:1029–1036. doi: 10.1093/hmg/6.7.1029. [DOI] [PubMed] [Google Scholar]
- 91.Schwarze U, et al. Rare autosomal recessive cardiac valvular form of Ehlers-Danlos syndrome results from mutations in the COL1A2 gene that activate the nonsense-mediated RNA decay pathway. Am J Hum Genet. 2004;74:917–930. doi: 10.1086/420794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Plaisier E, et al. COL4A1 mutations and hereditary angiopathy, nephropathy, aneurysms, and muscle cramps. N Engl J Med. 2007;357:2687–2695. doi: 10.1056/NEJMoa071906. [DOI] [PubMed] [Google Scholar]
- 93.Lemmens R, et al. Novel COL4A1 mutations cause cerebral small vessel disease by haploinsufficiency. Hum Mol Genet. 2013;22:391–397. doi: 10.1093/hmg/dds436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Petruzzella V, et al. Mutations in the NDUFS4 gene of mitochondrial complex I alter stability of the splice variants. FEBS Lett. 2005;579:3770–3776. doi: 10.1016/j.febslet.2005.05.035. [DOI] [PubMed] [Google Scholar]
- 95.Jenkins PB, et al. A nonsense mutation in the erythrocyte band 3 gene associated with decreased mRNA accumulation in a kindred with dominant hereditary spherocytosis. J Clin Invest. 1996;97:373–380. doi: 10.1172/JCI118425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Toye AM, et al. Band 3 Walton, a C-terminal deletion associated with distal renal tubular acidosis, is expressed in the red cell membrane but retained internally in kidney cells. Blood. 2002;99:342–347. doi: 10.1182/blood.v99.1.342. [DOI] [PubMed] [Google Scholar]
- 97.Yang Y, et al. The gene encoding alsin, a protein with three guanine-nucleotide exchange factor domains, is mutated in a form of recessive amyotrophic lateral sclerosis. Nat Genet. 2001;29:160–165. doi: 10.1038/ng1001-160. [DOI] [PubMed] [Google Scholar]
- 98.Gros-Louis F, et al. An ALS2 gene mutation causes hereditary spastic paraplegia in a Pakistani kindred. Ann Neurol. 2003;53:144–145. doi: 10.1002/ana.10422. [DOI] [PubMed] [Google Scholar]
- 99.Johnston JJ, et al. Molecular and clinical analyses of Greig cephalopolysyndactyly and Pallister-Hall syndromes: robust phenotype prediction from the type and position of GLI3 mutations. Am J Hum Genet. 2005;76:609–622. doi: 10.1086/429346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Furniss D, et al. Nonsense-mediated decay and the molecular pathogenesis of mutations in SALL1 and GLI3. Am J Med Genet A. 2007;143A:3150–3160. doi: 10.1002/ajmg.a.32097. [DOI] [PubMed] [Google Scholar]
- 101.Korkko J, et al. Analysis of the COL1A1 and COL1A2 genes by PCR amplification and scanning by conformation-sensitive gel electrophoresis identifies only COL1A1 mutations in 15 patients with osteogenesis imperfecta type I: identification of common sequences of null-allele mutations. Am J Hum Genet. 1998;62:98–110. doi: 10.1086/301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Willing MC, et al. Premature chain termination is a unifying mechanism for COL1A1 null alleles in osteogenesis imperfecta type I cell strains. Am J Hum Genet. 1996;59:799–809. [PMC free article] [PubMed] [Google Scholar]
- 103.Snead MP, Yates JR. Clinical and Molecular genetics of Stickler syndrome. J Med Genet. 1999;36:353–359. [PMC free article] [PubMed] [Google Scholar]
- 104.Kerr TP, et al. Long mutant dystrophins and variable phenotypes: evasion of nonsense-mediated decay? Hum Genet. 2001;109:402–407. doi: 10.1007/s004390100598. [DOI] [PubMed] [Google Scholar]
- 105.Pillers DA, et al. Duchenne/Becker muscular dystrophy: correlation of phenotype by electroretinography with sites of dystrophin mutations. Hum Genet. 1999;105:2–9. doi: 10.1007/s004399900111. [DOI] [PubMed] [Google Scholar]
- 106.Schrijver I, et al. Premature termination mutations in FBN1: distinct effects on differential allelic expression and on protein and clinical phenotypes. Am J Hum Genet. 2002;71:223–237. doi: 10.1086/341581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Faivre L, et al. Effect of mutation type and location on clinical outcome in 1,013 probands with Marfan syndrome or related phenotypes and FBN1 mutations: an international study. Am J Hum Genet. 2007;81:454–466. doi: 10.1086/520125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gorbenko del Blanco D, et al. Growth hormone insensitivity syndrome caused by a heterozygous GHR mutation: phenotypic variability owing to moderation by nonsense-mediated decay. Clin Endocrinol (Oxf) 2012;76:706–712. doi: 10.1111/j.1365-2265.2011.04304.x. [DOI] [PubMed] [Google Scholar]
- 109.Houlden H, et al. A novel Frabin (FGD4) nonsense mutation p.R275X associated with phenotypic variability in CMT4H. Neurology. 2009;72:617–620. doi: 10.1212/01.wnl.0000342463.35089.cc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Stendel C, et al. Peripheral nerve demyelination caused by a mutant Rho GTPase guanine nucleotide exchange factor, frabin/FGD4. Am J Hum Genet. 2007;81:158–164. doi: 10.1086/518770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Myerowitz R. Tay-Sachs disease-causing mutations and neutral polymorphisms in the Hex A gene. Hum Mutat. 1997;9:195–208. doi: 10.1002/(SICI)1098-1004(1997)9:3<195::AID-HUMU1>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 112.Alonso J, et al. Spectrum of germline RB1 gene mutations in Spanish retinoblastoma patients: Phenotypic and molecular epidemiological implications. Hum Mutat. 2001;17:412–422. doi: 10.1002/humu.1117. [DOI] [PubMed] [Google Scholar]
- 113.Li A, Swift M. Mutations at the ataxia-telangiectasia locus and clinical phenotypes of A-T patients. Am J Med Genet. 2000;92:170–177. doi: 10.1002/(sici)1096-8628(20000529)92:3<170::aid-ajmg3>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 114.Miller JN, Chan CH, Pearce DA. The role of nonsense-mediated decay in neuronal ceroid lipofuscinosis. Hum Mol Genet. 2013;22:2723–2734. doi: 10.1093/hmg/ddt120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Drack AV, Miller JN, Pearce DA. A Novel c.1135_1138delCTGT Mutation in CLN3 Leads to Juvenile Neuronal Ceroid Lipofuscinosis. J Child Neurol. 2013 doi: 10.1177/0883073813494812. [DOI] [PubMed] [Google Scholar]
- 116.Miller JN, Pearce DA. A Novel c.776_777insA Mutation in CLN1 Leads to Infantile Neuronal Ceroid Lipofuscinosis. J Child Neurol. 2013 doi: 10.1177/0883073813494267. [DOI] [PubMed] [Google Scholar]
- 117.Jackson RJ, Hellen CU, Pestova TV. Termination and post-termination events in eukaryotic translation. Adv Protein Chem Struct Biol. 2012;86:45–93. doi: 10.1016/B978-0-12-386497-0.00002-5. [DOI] [PubMed] [Google Scholar]
- 118.Keeling KM, et al. Suppression of premature termination codons as a therapeutic approach. Crit Rev Biochem Mol Biol. 2012;47:444–463. doi: 10.3109/10409238.2012.694846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lee HL, Dougherty JP. Pharmaceutical therapies to recode nonsense mutations in inherited diseases. Pharmacol Ther. 2012;136:227–266. doi: 10.1016/j.pharmthera.2012.07.007. [DOI] [PubMed] [Google Scholar]
- 120.Howard M, Frizzell RA, Bedwell DM. Aminoglycoside antibiotics restore CFTR function by overcoming premature stop mutations. Nat Med. 1996;2:467–469. doi: 10.1038/nm0496-467. [DOI] [PubMed] [Google Scholar]
- 121.Bedwell DM, et al. Suppression of a CFTR premature stop mutation in a bronchial epithelial cell line. Nat Med. 1997;3:1280–1284. doi: 10.1038/nm1197-1280. [DOI] [PubMed] [Google Scholar]
- 122.Clancy JP, et al. Evidence that systemic gentamicin suppresses premature stop mutations in patients with cystic fibrosis. Am J Respir Crit Care Med. 2001;163:1683–1692. doi: 10.1164/ajrccm.163.7.2004001. [DOI] [PubMed] [Google Scholar]
- 123.Zsembery A, et al. Correction of CFTR malfunction and stimulation of Ca-activated Cl channels restore HCO3- secretion in cystic fibrosis bile ductular cells. Hepatology. 2002;35:95–104. doi: 10.1053/jhep.2002.30423. [DOI] [PubMed] [Google Scholar]
- 124.Rowe SM, et al. Restoration of W1282X CFTR activity by enhanced expression. Am J Respir Cell Mol Biol. 2007;37:347–356. doi: 10.1165/rcmb.2006-0176OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Du M, et al. Aminoglycoside suppression of a premature stop mutation in a Cftr−/− mouse carrying a human CFTR-G542X transgene. J Mol Med (Berl) 2002;80:595–604. doi: 10.1007/s00109-002-0363-1. [DOI] [PubMed] [Google Scholar]
- 126.Du M, et al. Clinical doses of amikacin provide more effective suppression of the human CFTR-G542X stop mutation than gentamicin in a transgenic CF mouse model. J Mol Med (Berl) 2006;84:573–582. doi: 10.1007/s00109-006-0045-5. [DOI] [PubMed] [Google Scholar]
- 127.Du M, et al. PTC124 is an orally bioavailable compound that promotes suppression of the human CFTR G542X nonsense allele in a CF mouse model. Proc Natl Acad Sci U S A. 2008;105:2064–2069. doi: 10.1073/pnas.0711795105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Nudelman I, et al. Development of novel aminoglycoside (NB54) with reduced toxicity and enhanced suppression of disease-causing premature stop mutations. J Med Chem. 2009;52:2836–2845. doi: 10.1021/jm801640k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Nudelman I, et al. Repairing faulty genes by aminoglycosides: development of new derivatives of geneticin (G418) with enhanced suppression of diseases-causing nonsense mutations. Bioorg Med Chem. 2010;18:3735–3746. doi: 10.1016/j.bmc.2010.03.060. [DOI] [PubMed] [Google Scholar]
- 130.Sermet-Gaudelus I, et al. Ataluren (PTC124) induces cystic fibrosis transmembrane conductance regulator protein expression and activity in children with nonsense mutation cystic fibrosis. Am J Respir Crit Care Med. 2010;182:1262–1272. doi: 10.1164/rccm.201001-0137OC. [DOI] [PubMed] [Google Scholar]
- 131.Rowe SM, et al. Suppression of CFTR premature termination codons and rescue of CFTR protein and function by the synthetic aminoglycoside NB54. J Mol Med (Berl) 2011;89:1149–1161. doi: 10.1007/s00109-011-0787-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Barton-Davis ER, et al. Aminoglycoside antibiotics restore dystrophin function to skeletal muscles of mdx mice. J Clin Invest. 1999;104:375–381. doi: 10.1172/JCI7866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Howard MT, et al. Sequence specificity of aminoglycoside-induced stop condon readthrough: potential implications for treatment of Duchenne muscular dystrophy. Ann Neurol. 2000;48:164–169. [PubMed] [Google Scholar]
- 134.Bidou L, et al. Premature stop codons involved in muscular dystrophies show a broad spectrum of readthrough efficiencies in response to gentamicin treatment. Gene Ther. 2004;11:619–627. doi: 10.1038/sj.gt.3302211. [DOI] [PubMed] [Google Scholar]
- 135.Howard MT, et al. Readthrough of dystrophin stop codon mutations induced by aminoglycosides. Ann Neurol. 2004;55:422–426. doi: 10.1002/ana.20052. [DOI] [PubMed] [Google Scholar]
- 136.Loufrani L, et al. Absence of dystrophin in mice reduces NO-dependent vascular function and vascular density: total recovery after a treatment with the aminoglycoside gentamicin. Arterioscler Thromb Vasc Biol. 2004;24:671–676. doi: 10.1161/01.ATV.0000118683.99628.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Welch EM, et al. PTC124 targets genetic disorders caused by nonsense mutations. Nature. 2007;447:87–91. doi: 10.1038/nature05756. [DOI] [PubMed] [Google Scholar]
- 138.Allamand V, et al. Drug-induced readthrough of premature stop codons leads to the stabilization of laminin alpha2 chain mRNA in CMD myotubes. J Gene Med. 2008;10:217–224. doi: 10.1002/jgm.1140. [DOI] [PubMed] [Google Scholar]
- 139.Du L, et al. Nonaminoglycoside compounds induce readthrough of nonsense mutations. J Exp Med. 2009;206:2285–2297. doi: 10.1084/jem.20081940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kayali R, et al. Read-through compound 13 restores dystrophin expression and improves muscle function in the mdx mouse model for Duchenne muscular dystrophy. Hum Mol Genet. 2012;21:4007–4020. doi: 10.1093/hmg/dds223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Lai CH, et al. Correction of ATM gene function by aminoglycoside-induced read-through of premature termination codons. Proc Natl Acad Sci U S A. 2004;101:15676–15681. doi: 10.1073/pnas.0405155101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Nakamura K, et al. Functional characterization and targeted correction of ATM mutations identified in Japanese patients with ataxia-telangiectasia. Hum Mutat. 2012;33:198–208. doi: 10.1002/humu.21632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Brendel C, et al. Suppression of nonsense mutations in Rett syndrome by aminoglycoside antibiotics. Pediatr Res. 2009;65:520–523. doi: 10.1203/PDR.0b013e31819d9ebc. [DOI] [PubMed] [Google Scholar]
- 144.Popescu AC, et al. Aminoglycoside-mediated partial suppression of MECP2 nonsense mutations responsible for Rett syndrome in vitro. J Neurosci Res. 2010;88:2316–2324. doi: 10.1002/jnr.22409. [DOI] [PubMed] [Google Scholar]
- 145.Brendel C, et al. Readthrough of nonsense mutations in Rett syndrome: evaluation of novel aminoglycosides and generation of a new mouse model. J Mol Med (Berl) 2011;89:389–398. doi: 10.1007/s00109-010-0704-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Vecsler M, et al. Ex vivo treatment with a novel synthetic aminoglycoside NB54 in primary fibroblasts from Rett syndrome patients suppresses MECP2 nonsense mutations. PLoS One. 2011;6:e20733. doi: 10.1371/journal.pone.0020733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Rebibo-Sabbah A, et al. In vitro and ex vivo suppression by aminoglycosides of PCDH15 nonsense mutations underlying type 1 Usher syndrome. Hum Genet. 2007;122:373–381. doi: 10.1007/s00439-007-0410-7. [DOI] [PubMed] [Google Scholar]
- 148.Nudelman I, et al. Redesign of aminoglycosides for treatment of human genetic diseases caused by premature stop mutations. Bioorg Med Chem Lett. 2006;16:6310–6315. doi: 10.1016/j.bmcl.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 149.Goldmann T, et al. PTC124-mediated translational readthrough of a nonsense mutation causing Usher syndrome type 1C. Hum Gene Ther. 2011;22:537–547. doi: 10.1089/hum.2010.067. [DOI] [PubMed] [Google Scholar]
- 150.Keeling KM, et al. Gentamicin-mediated suppression of Hurler syndrome stop mutations restores a low level of alpha-L-iduronidase activity and reduces lysosomal glycosaminoglycan accumulation. Hum Mol Genet. 2001;10:291–299. doi: 10.1093/hmg/10.3.291. [DOI] [PubMed] [Google Scholar]
- 151.Keeling KM, Bedwell DM. Clinically relevant aminoglycosides can suppress disease-associated premature stop mutations in the IDUA and P53 cDNAs in a mammalian translation system. J Mol Med (Berl) 2002;80:367–376. doi: 10.1007/s00109-001-0317-z. [DOI] [PubMed] [Google Scholar]
- 152.Hein LK, et al. alpha-L-iduronidase premature stop codons and potential read-through in mucopolysaccharidosis type I patients. J Mol Biol. 2004;338:453–462. doi: 10.1016/j.jmb.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 153.Brooks DA, Muller VJ, Hopwood JJ. Stop-codon read-through for patients affected by a lysosomal storage disorder. Trends Mol Med. 2006;12:367–373. doi: 10.1016/j.molmed.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 154.Wang D, et al. The designer aminoglycoside NB84 significantly reduces glycosaminoglycan accumulation associated with MPS I-H in the Idua-W392X mouse. Mol Genet Metab. 2012;105:116–125. doi: 10.1016/j.ymgme.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Bartolomeo R, et al. Pharmacological read-through of nonsense ARSB mutations as a potential therapeutic approach for mucopolysaccharidosis VI. J Inherit Metab Dis. 2013;36:363–371. doi: 10.1007/s10545-012-9521-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Tan L, et al. PTC124 improves readthrough and increases enzymatic activity of the CPT1A R160X nonsense mutation. J Inherit Metab Dis. 2011;34:443–447. doi: 10.1007/s10545-010-9265-5. [DOI] [PubMed] [Google Scholar]
- 157.James PD, et al. Aminoglycoside suppression of nonsense mutations in severe hemophilia. Blood. 2005;106:3043–3048. doi: 10.1182/blood-2005-03-1307. [DOI] [PubMed] [Google Scholar]
- 158.Yang C, et al. A mouse model for nonsense mutation bypass therapy shows a dramatic multiday response to geneticin. Proc Natl Acad Sci U S A. 2007;104:15394–15399. doi: 10.1073/pnas.0610878104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Buck NE, et al. Stop codon read-through of a methylmalonic aciduria mutation. Mol Genet Metab. 2009;97:244–249. doi: 10.1016/j.ymgme.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 160.Sleat DE, et al. Aminoglycoside-mediated suppression of nonsense mutations in late infantile neuronal ceroid lipofuscinosis. Eur J Paediatr Neurol. 2001;5(Suppl A):57–62. doi: 10.1053/ejpn.2000.0436. [DOI] [PubMed] [Google Scholar]
- 161.Sarkar C, Zhang Z, Mukherjee AB. Stop codon read-through with PTC124 induces palmitoyl-protein thioesterase-1 activity, reduces thioester load and suppresses apoptosis in cultured cells from INCL patients. Mol Genet Metab. 2011;104:338–345. doi: 10.1016/j.ymgme.2011.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Wolstencroft EC, et al. A non-sequence-specific requirement for SMN protein activity: the role of aminoglycosides in inducing elevated SMN protein levels. Hum Mol Genet. 2005;14:1199–1210. doi: 10.1093/hmg/ddi131. [DOI] [PubMed] [Google Scholar]
- 163.Allen LA, Raetz CR. Partial phenotypic suppression of a peroxisome-deficient animal cell mutant treated with aminoglycoside G418. J Biol Chem. 1992;267:13191–13199. [PubMed] [Google Scholar]
- 164.Dranchak PK, et al. Nonsense suppressor therapies rescue peroxisome lipid metabolism and assembly in cells from patients with specific PEX gene mutations. J Cell Biochem. 2011;112:1250–1258. doi: 10.1002/jcb.22979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Brumm H, et al. Rescue of melanocortin 4 receptor (MC4R) nonsense mutations by aminoglycoside-mediated read-through. Obesity (Silver Spring) 2012;20:1074–1081. doi: 10.1038/oby.2011.202. [DOI] [PubMed] [Google Scholar]
- 166.Fuchshuber-Moraes M, et al. Aminoglycoside-induced suppression of CYP2C19*3 premature stop codon. Pharmacogenet Genomics. 2011;21:694–700. doi: 10.1097/FPC.0b013e328349daba. [DOI] [PubMed] [Google Scholar]
- 167.Sun J, et al. Relationships among stop codon usage bias, its context, isochores, and gene expression level in various eukaryotes. J Mol Evol. 2005;61:437–444. doi: 10.1007/s00239-004-0277-3. [DOI] [PubMed] [Google Scholar]
- 168.Tate WP, et al. Translational termination efficiency in both bacteria and mammals is regulated by the base following the stop codon. Biochem Cell Biol. 1995;73:1095–1103. doi: 10.1139/o95-118. [DOI] [PubMed] [Google Scholar]
- 169.Tork S, et al. The major 5′ determinant in stop codon read-through involves two adjacent adenines. Nucleic Acids Res. 2004;32:415–421. doi: 10.1093/nar/gkh201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Malik V, et al. Gentamicin-induced readthrough of stop codons in Duchenne muscular dystrophy. Ann Neurol. 2010;67:771–780. doi: 10.1002/ana.22024. [DOI] [PubMed] [Google Scholar]
- 171.Du M, et al. Poly-L-aspartic acid enhances and prolongs gentamicin-mediated suppression of the CFTR G542X mutation in a cystic fibrosis mouse model. J Biol Chem. 2009;284:6885–6892. doi: 10.1074/jbc.M806728200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Durand S, et al. Inhibition of nonsense-mediated mRNA decay (NMD) by a new chemical molecule reveals the dynamic of NMD factors in P-bodies. J Cell Biol. 2007;178:1145–1160. doi: 10.1083/jcb.200611086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Keeling KM, et al. Attenuation of nonsense-mediated mRNA decay enhances in vivo nonsense suppression. PLoS One. 2013;8:e60478. doi: 10.1371/journal.pone.0060478. [DOI] [PMC free article] [PubMed] [Google Scholar]