Abstract
Studies of mechanisms of resistance to glyphosate have increased current understanding of herbicide resistance mechanisms. Thus far, single-codon non-synonymous mutations of EPSPS (5-enolypyruvylshikimate-3-phosphate synthase) have been rare and, relative to other herbicide mode of action target-site mutations, unconventionally weak in magnitude for resistance to glyphosate. However, it is possible that weeds will emerge with non-synonymous mutations of two codons of EPSPS to produce an enzyme endowing greater resistance to glyphosate. Today, target-gene duplication is a common glyphosate resistance mechanism and could become a fundamental process for developing any resistance trait. Based on competition and substrate selectivity studies in several species, rapid vacuole sequestration of glyphosate occurs via a transporter mechanism. Conversely, as the chloroplast requires transporters for uptake of important metabolites, transporters associated with the two plastid membranes may separately, or together, successfully block glyphosate delivery. A model based on finite glyphosate dose and limiting time required for chloroplast loading sets the stage for understanding how uniquely different mechanisms can contribute to overall glyphosate resistance.
Keywords: vacuole sequestration, gene duplication, gene amplification, EPSPS (5-enolypyruvylshikimate-3-phosphate synthase), 31P NMR, glyphosate, restricted translocation, reduced translocation, group G herbicides, herbicide resistance
1 INTRODUCTION
Glyphosate stands alone in many categories.1,2 This N-phosphonomethyl-modified derivative of glycine is able to form a stabile dead-end complex with EPSPS (5-enolypyruvylshikimate-3-phosphate synthase, EC 2.5.1.19)3 as a transition-state analogue competitive with phosphoenolpyruvate (PEP).4 The inhibition of the shikimate pathway located in the chloroplast was attributed to glyphosate's mode of action (MOA) 8 years after commercialization.5,6 Some thought resistance to glyphosate was unlikely to evolve owing to its transition-state mimicry, lack of plant metabolism and absence of known active transporters for herbicides in plants,7 and, indeed, during the first 15 years of glyphosate use, primarily as a non-selective treatment before planting, glyphosate-resistant (GR) weeds were not found (1972–1997). However, in the last 15 years (1998–2013), glyphosate resistance in 24 species on six continents has been documented.8,9
The basic herbicide resistance mechanisms can be catalogued as follows: (1) target-site resistance, typically represented by amino acid substitutions that affect herbicide interactions at the target enzyme; (2) metabolism, a chemical modification of the herbicide either by conjugation or degradation; (3) exclusion of the herbicide from the target, either physically with enhanced cuticular and other structural barriers or physiologically with active transporters.10 A fourth mechanism could be added – avoidance, pertaining to biochemical ability to handle the toxic agent produced by the pesticide and thereby avoid a toxic result. An example of avoidance in herbicides is the overexpression of superoxide dismutase as a rescue for paraquat.11 Intriguingly, the known glyphosate resistance mechanisms now exceed those described for any other herbicide. Mechanisms include target-site mutation, target-site gene duplication, active vacuole sequestration, limited cellular uptake and a rapid necrosis response. Notably, glyphosate resistance is generally lower in magnitude and less frequently involves target-site and metabolic mechanisms than in the case of herbicides that inhibit acetolactate synthase (ALS) or acetyl-CoA carboxylase (ACCase).8
2 EXCLUSION MECHANISMS
The first investigations of glyphosate resistance in Lolium rigidum found no biochemical differences between GR and glyphosate-susceptible (GS) populations, indicating that quite subtle differences in physiology could result in glyphosate resistance.12,13 Early 14C glyphosate translocation studies revealed restricted translocation in GR L. rigidum.14 Similarly, 14C glyphosate showed restricted systemic delivery in Conyza canadensis when comparing GR and GS populations.15 The curiosity was that target-site resistance (a Pro106Ser substitution, see Section 4) afforded approximately twofold resistance in Eleusine indica,16 whereas the very slight differences (<2×) in restricted translocation in C. canadensis and L. rigidum afforded resistance magnitudes 6–8× the recommended field rates (1× = 860 g ha−1). Something substantial was happening to the glyphosate beyond the usual target-site and metabolic resistance mechanisms observed for other herbicides. The observations of restricted translocation quickly highlighted a resistance mechanism that became widely used in discussions of glyphosate resistance. But really, in the words of a famous bard, it is necessary to ‘Find out the cause of this effect, Or rather say, the cause of this defect, For this effect defective comes by cause.’ (Hamlet 2:2, Shakespeare).
2.1 Vacuolar sequestration
Experiments using 31P nuclear magnetic resonance (NMR) to observe glyphosate in vivo entering cells and cellular compartments have identified at least two new resistance mechanisms that result in restricted translocation correlated with the glyphosate resistance observed.17 The observation18 of rapid sequestration of glyphosate in C. canadensis by 31P NMR demonstrated the application of an old technique19–21 to the biochemistry of resistance in weedy species. The pH versus chemical shift (31P NMR signal) changes of the glyphosate phosphonate have been well documented.22 Hence, after dosing a plant with glyphosate prior to or during NMR observation, the pH changes make it possible to follow the passage of glyphosate from the apoplast buffer (pH 5) into the cell cytoplasm (pH 6.8) and further into the vacuole (pH 5.5). The overlay of the apoplast glyphosate chemical shift in the spectrum with the vacuole glyphosate signal was resolved using a pulse chase method, by first using the leaf-sample circulating stabilization buffer to challenge with glyphosate. Spectra were collected while observing initial cellular glyphosate uptake for 10 h, and then removing glyphosate from the circulating buffer solution to eliminate the apoplast glyphosate signal and reveal the underlying glyphosate vacuole signal. The difference in glyphosate vacuole sequestration rates between GR and GS C. canadensis was about 10×. The range of concentrations for the glyphosate challenge allowed the rates of cellular uptake and even vacuole sequestration to be measured.17 These NMR results on the Delaware GR C. canadensis population23 showed that the resistant trait is likely a singular biochemical activity (sequestration) and corroborated inheritance data showing a nuclear inherited single gene.24
To characterize the specificity of glyphosate uptake, other solutes were added such as the glyphosate degradation product aminomethyl phosphonate (AMPA) and the structurally similar glycine. Both showed significant competitive inhibition of glyphosate uptake. This competitive nature of the glyphosate uptake is an expected property of an active transport system.17 Varying the glyphosate challenge concentration revealed cytoplasm concentrations accumulating with a linear dependence to >20 mM glyphosate, indicating that the cellular plasmalemma uptake transporter could not be saturated. During the chase period, the glyphosate vacuole accumulation observations indicated the rate of vacuole uptake. This vacuole uptake rate did saturate with increasing cytoplasmic glyphosate concentration. (Sammons RD, personal communication) Substrate saturation behavior (Km, Vmax) is a key characteristic of active transporters. Finally, another major observation in the pulse chase is that glyphosate does not leach out of the cytoplasm, nor out of the vacuole to the cytoplasm, unless there is evidence of cell death. That is, the transport of glyphosate across membranes is observed to be unidirectional in all species studied to date (23 in total).17 These observations also support the hypothesis that glyphosate cellular uptake and vacuole sequestration are mediated by active transport. Previously, glyphosate transport systems have been characterized insufficiently to explain observed uptake and translocation values.25,26 Given that these transporters are present, the realization that glyphosate may require assistance to cross any active bilayer further suggests that many transporters are working to allow glyphosate cell uptake, whole-plant translocation and subcellular distribution.
2.1.1 Temperature effects on vacuolar sequestration
C. canadensis survives freezing very well, which is consistent with its biennial lifecycle,27 making it a good candidate for low-temperature 31P NMR to determine how cold temperature affects the sequestration process. Pulse chase 31P NMR and whole-plant treatments of glyphosate in the cold (8 °C) demonstrated that vacuole sequestration was inhibited and that GR C. canadensis individuals were susceptible to glyphosate at cold temperatures.28 This provided an unequivocal demonstration that the resistance was due solely to the sequestration process and showed that the physiological properties of the resistance mechanism could be utilized to reverse the resistance to the herbicide. Interestingly, vacuolar sequestration has also been hypothesized as a mechanism of paraquat resistance,29 with a similar temperature dependence for paraquat resistance reported.30 Lipid bilayers are well known to respond to temperature differently, based on their lipid saturation levels, as these affect the membrane fluidity.31
The new exclusion mechanism of rapid vacuole sequestration was also demonstrated to be a major part of the glyphosate resistance in Lolium spp., with the sequestration rate correlating with the LD50.32 Glyphosate-resistant Lolium spp. from three continents were shown to sequester glyphosate in the vacuole, suggesting that the GR trait is present within diverse Lolium spp. and can be selected independently wherever Lolium is found. Glyphosate-resistant Sorghum halepense and Lolium spp. populations having reduced translocation were also made sensitive at suboptimal temperatures, further confirming that populations dependent on membrane mechanisms could be made sensitive in the cold.33 A comparison of GR L. multiflorum populations from Italy34 in cold acclimation and glyphosate treatments showed that the most resistant population, also shown to contain a EPSPS Pro106Ser mutation (see Section 4), was not made as sensitive by the cold acclimation. This supports the idea that multiple resistance mechanisms can contribute to the overall resistance magnitude, and not all mechanisms are temperature sensitive.
2.1.2 Variation among species in sensitivity to glyphosate
The demonstration of rapid vacuole sequestration then seemed to be the most reasonable explanation for the variation in species sensitivity to glyphosate. A hypothesis was developed that those species not using the vacuole would be more sensitive and those utilizing the vacuole to sequester glyphosate would be more tolerant and potential candidates for selection of resistance. Consequently, a survey of 23 species was carried out to compare glyphosate-sensitive [e.g. barley (Hordeum vulgare)] and hard-to-control weeds (e.g. Ipomoea lacunosa, Desmanthus illinoensis).17 The plants were all track sprayed with a 4× label rate of glyphosate and then examined 24 h later. No correlation of vacuole sequestration and glyphosate sensitivity was found, and in fact several different categories could be defined. Quite unexpectedly, it was observed that some species accumulated very little glyphosate in the cytoplasm. In one case, barley allowed very little glyphosate inside the cell, but a strong shikimate-3-phosphate (S3P) signal suggested a high degree of toxicity with the small amount of glyphosate, which is consistent with its sensitive categorization. However, Convolvulus arvensis also took up very little glyphosate, and there was no evidence of an S3P peak, which again is consistent with its hard-to-control classification. In another situation, Chenopodium album and I. lacunosa took up ∼10× more glyphosate than C. arvensis and did not sequester glyphosate in the vacuole, and yet no S3P was observed; therefore, glyphosate appeared to be less toxic, even though much more was entering the cytoplasm. Echinochloa colona uptake was similar to that of I. lacunosa, but 83% of the glyphosate was moved to the vacuole (the same as GR Conyza canadensis). Overall, 11 species took up very little glyphosate, including the following: Ambrosia artemisiifolia, E. colona, C. album, I. lacunosa, Xanthium strumarium, Camelina microcarpa, Setaria viridis, H. vulgare, Avena fatua, E. crus-galli and C. arvensis. Only E. colona sequestered glyphosate in the vacuole together with minimal uptake. A caveat would be that this lack of vacuole signal in these minimal uptake species was generally due to such low cytoplasm concentrations. The 13 species that took up moderate amounts of glyphosate included nine that used the vacuole, including Senna obtusifolia, Sida spinosa, Polygonum convolvulus, Ambrosia trifida, Sorghum bicolor, Urochloa platyphylla, L. rigidum and C. canadensis. P. convolvulus rivaled GR C. canadensis in its efficiency to sequester glyphosate.
For the remaining four species that did not use the vacuole but had good cellular uptake, GR populations of three species were able to restrict cellular uptake when compared with GS populations, including Amaranthus palmeri, A. tuberculatus and S. halepense. One particular case demonstrates this observation. In a survey of GR A. palmeri35 for various EPSPS copy numbers, two female plants (R20 and R25) were found to have very similar genomic EPSPS copy numbers (∼35 by qRT-PCR, see Section 3). Upon vegetative cloning of R20 and R25 and glyphosate titration (five replicates, WeatherMax®, 1/2× to 16× by factors of 2), the R20 plants survived 16× the field rate (13.8 kg ha−1), whereas the R25 plants only survived 4× the rate. Owing to this surprising result, both plants were then vegetatively cloned and examined by 31P NMR pulse chase with a 10 mM glyphosate challenge (according to the methods described).17 The R25 cloned individuals had a glyphosate cellular uptake rate 4× faster than the R20 cloned individuals, so that after 10 h the ratio of glyphosate/internal standard in R25 was 0.4 and for R20 the ratio was 0.1. Therefore, the reduced cellular uptake of glyphosate in the R20 individuals was shown to contribute >4× the label rate in resistance magnitude. These data showing limitations to glyphosate cellular uptake support a resistance mechanism of restricted cytoplasm entry. The 31P NMR data also indicated that glyphosate is not pumped out of the cell after the challenge is removed, so the limitation to glyphosate uptake is due to restricted entry rather than a shifted equilibrium due to the increased activity of an export transporter. Previously, Hetherington et al.36 studied 14C glyphosate efflux in tissue culture using transgenic GR soybean (Glycine max) and described slow rates of efflux correlating with cytoplasmic and vacuole pools measurable over long times because GR plant tissue was used. The time course of A. palmeri NMR experiments was not long enough to measure such slow efflux rates. Export transporters are very commonly found in microbial systems, and so multidrug resistance pumps would have been a reasonable hypothesis.37 Similar restricted entry results were observed for GR A. tuberculatus (Sammons RD, unpublished) and for GR S. halepense from Arkansas.17 Restricted entry into the source-leaf cells has been proposed in studies examining L. rigidum with 14C glyphosate translocation and offered as the origin of the restricted translocation observed for glyphosate systemic movement in the resistant populations.14 The translocation of 14C glyphosate in S. halepense was also observed to be restricted,38,39 and so a similar explanation can be offered. Specifically, translocation of glyphosate requires source-leaf cell uptake to access the phloem for systemic translocation. The restricted cellular entry of the R20 A. palmeri line and the GR A. tuberculatus described above does not result in the reduced translocation phenotype (Sammons RD, unpublished).
In this last group of four species studied in the survey, D. illinoensis was very unique, taking up 100× more glyphosate than C. arvensis, not using the vacuole and showing no S3P signal. This latter case prompts a new hypothesis whereby glyphosate has restricted entry into the chloroplast, and this last physiological barrier could well be the most potent of all glyphosate resistance mechanisms. This hypothesis allows explanations for the reduced toxicity of glyphosate even while present in the cell.
2.1.3 Molecular characterization of vacuolar sequestration
Some molecular evidence suggests that proteins associated with the vacuolar membrane may have roles in the glyphosate vacuolar sequestration resistance mechanism in C. canadensis, including ATP-binding cassette (ABC) transporters and a tonoplast intrinsic protein (TIP).40–42 Using a transcriptome profiling technique called iGentifier, a TIP was identified that was generally higher in GR than GS individuals from several populations and in GR individuals from segregating F2 populations.42 The TIP was also highly expressed in one GS population. A limitation of the iGentifier approach was that only 72 sequence tags were linked to previously obtained EST sequences, and only 52 sequence tags were able to be annotated. Using an Arabidopsis heterologous microarray for hybridization of untreated and glyphosate-treated RNA samples from one GR population, several genes were induced by glyphosate treatment, including four ABC transporters.42 To date, genes identified from transcriptome analysis have not been shown to confer glyphosate resistance. Genetic marker evidence (SSRs) suggested that resistance had evolved several times independently in C. canadensis, and clustering of GR populations based on iGentifier expression patterns suggested that similar mechanisms had evolved in several GR populations.42
The successful application of next-generation sequencing technology to GR C. canadensis identified ABC-transporter unigenes M10 and M11.41 M11 is an ortholog of Arabidopsis gene At3g13080 (an ATP-dependent multidrug-resistant protein-like transporter able to transport glutathione conjugates), which was also identified in the microarray study with approximately 30-fold increased expression following glyphosate treatment in a TN GR C. canadensis population.42 The M11 Arabidopsis ortholog is associated with the vacuolar membrane, making this gene an especially interesting candidate given the vacuole sequestration mechanism hypothesis.18 Using qRT-PCR to measure the expression relative to an actin gene, both M10 and M11 were constitutively more expressed in GR C. canadensis than in GS; both were upregulated in response to glyphosate treatment, and expression in GR remained higher than in GS.41 Additional ABC-transporter unigenes M6, M7 and P3 were also constitutively higher in GR than in GS, were induced in GS by glyphosate treatment and were unaffected by glyphosate treatment in GR.
Additional confirmatory results suggest a possible role for M10 and M11 in C. canadensis glyphosate resistance, when gene expression was compared using qRT-PCR in two GR and one GS Mediterranean C. canadensis populations.40 Constitutive expression of both M10 and M11 was higher in the GS population than in both GR populations. However, both M10 and M11 were highly (11–23-fold) induced in both GR populations by glyphosate treatment, and significant induction did not occur in the GS population.
Based on results available to date, it is not clear whether M10, M11 or other ABC transporters have a direct role in the C. canadensis vacuolar sequestration glyphosate resistance mechanism. Observed ABC-transporter expression has been generally constitutively higher in GR populations, and often induced to higher levels by glyphosate treatment.40–42 Additional research is necessary to establish the possible role(s) of ABC transporters in glyphosate resistance, including M10 and M11.
2.2 Other reduced translocation mechanisms
Some GR A. trifida populations have shown an interesting response to glyphosate application. Within 12 h after treatment, mature leaves show rapid necrosis and cell death.43 Glyphosate translocation is substantially reduced in GR individuals, presumably owing to the rapid necrosis and the inability of glyphosate to be exported from mature leaves. The molecular and genetic basis of the rapid necrosis response remains unknown, but future research will very likely provide important insights into cell signaling responses that occur after glyphosate treatment.
3 GENE DUPLICATION
Gene duplication is defined as the heritable replication of a DNA segment, resulting in one or more additional gene copies within the genome of an organism.44 Gene amplification is sometimes used synonymously with gene duplication, but gene amplification can also have a contrasting definition, such as the non-heritable replication of a DNA segment.44 In other fields, gene amplification is defined as the heritable replication of multiple copies of a DNA segment, while gene duplication indicates the replication of only a single copy.45 Gene duplication is vital for generating genomic diversity and is a common process in the evolutionary history of plants.46 Duplication can occur through unequal crossing-over events, chromosome duplication or transposable element activity.47 When a duplicated gene sequence contains transcription signals including promoters, or when a duplicated gene is inserted behind a promoter, the immediate results are increased expression of mRNA and protein. The extra gene copies can accumulate mutations without potentially negative consequences (e.g. loss of function), thereby increasing opportunities to select more advantageous mutations. Given the fitness advantages of improved mutations, duplicated gene copies can be deleted over time while retaining the improved gene copies, an important evolutionary process. Of particular relevance when considering glyphosate resistance mechanisms, examples have been reported of positive selection for gene dosage, leading to adaptive gene duplication in response to stressful environmental conditions.48
3.1 EPSPS gene duplication
Cell culture selection studies using glyphosate provided the first examples of EPSPS gene duplication in selected GR cell lines.49–53 In several cases, duplicated EPSPS copies were not maintained when glyphosate selection was removed, and not inherited when plantlets were cultured from selected cells, suggesting that the duplicated EPSPS genes were unstable or carried a fitness penalty.54
The first reported occurrence of EPSPS gene duplication in a field-evolved weed species was a GR A. palmeri population from Georgia, United States.55 Both EPSPS and ALS were measured using quantitative real-time PCR on genomic DNA, and EPSPS relative genomic copy number was calculated using ALS as an internal standard. Using this technique, the EPSPS gene was duplicated from two- to over 100-fold in comparison with a GS population. Both EPSPS mRNA and EPSPS protein expression correlated positively with genomic EPSPS copy number in the Georgia GR population55 and in a different GR population from Mississippi.56 In the Georgia A. palmeri population, the EPSPS gene was present on all chromosomes. Subsequently, a North Carolina GR A. palmeri population was reported with EPSPS gene duplication from 22- to 63-fold.57 A New Mexico GR A. palmeri population was sevenfold more resistant than a GS population and had EPSPS gene duplication ranging from two- to tenfold, and individuals with EPSPS gene duplication were able to survive label-rate glyphosate applications (0.82 kg ha−1).58,59
Additional species have recently been reported with EPSPS gene duplication in GR populations, including L. multiflorum,60 Kochia scoparia,61 A. tuberculatus62 and A. spinosus63 (summarized in Table 1). The reported number of duplicated EPSPS gene copies in K. scoparia and A. tuberculatus populations is lower (from four- to tenfold) than the Georgia, North Carolina and Mississippi55–57 A. palmeri populations (Table 1), and similar to the New Mexico A. palmeri population.58 EPSPS mRNA expression and EPSPS protein levels were found to have a linear correlation with EPSPS genomic copy number in L. multiflorum populations,60 as well as in K. scoparia populations.61
Table 1.
EPSPS gene duplication reported in glyphosate-resistant weed species
| Species | Population origin | EPSPS relative genomic copy number range | Reference |
|---|---|---|---|
| Amaranthus palmeri | Georgia | 40–100 | 55 |
| Amaranthus palmeri | North Carolina | 20–60 | 57 |
| Amaranthus palmeri | New Mexico | 2–10 | 58 |
| Amaranthus palmeri | Mississippi | 33–59 | 56 |
| Amaranthus tuberculatus | Missouri, Illinois | 4 | 62 |
| Lolium multiflorum | Arkansas | 15–25 | 60 |
| Kochia scoparia | Kansas, Colorado | 3–9 | 61 |
| Amaranthus spinosus | Mississippi | 26–37 | 63 |
A. spinosus and A. palmeri have been shown to be closely related using both molecular markers and gene sequence phylogeny.64,65 As glyphosate resistance can transfer from A. palmeri to A. spinosus through interspecific hybridization and the hybrid progeny are self-fertile,66 an important question is whether A. spinosus with EPSPS gene duplication63 has evolved independently, or whether the trait has transferred via interspecific hybridization.
The glyphosate resistance level appears to increase with higher EPSPS genomic copy number in several examples. In L. multiflorum, the dose required to achieve 50% reduction in plant growth increased in a linear relationship with EPSPS protein expression.60 Higher EPSPS copy number correlated with higher-level resistance in A. palmeri populations from both Georgia and Mississippi.56,67 Shikimate accumulation following glyphosate treatment decreased as EPSPS cDNA expression level increased in a New Mexico A. palmeri population.58 An open question is whether EPSPS genomic copy number in species such as K. scoparia and A. tuberculatus may respond to continuing glyphosate selection pressure and increase to higher levels than previously observed. The number of species using gene duplication begs the question as to how common the mechanism may be and why it has not been observed for other herbicide mechanisms of action? Gene duplication could be a possible explanation for the increased gene expression associated with rapid vacuole sequestration, or any trait that could be conferred by increasing gene expression through increased gene copy number.
3.2 Molecular mechanism and inheritance of EPSPS gene duplication
The A. palmeri genome likely contains two original EPSPS loci, and one of the loci appears to have been duplicated in GR populations.68,69 Genomic sequencing suggested that the gene duplication proceeded via a DNA-mediated mechanism, and the duplicated section of DNA including the 10 kb EPSPS gene is at least 30 kb long.68 In that study, various mobile genetic elements were identified within the duplicated sequence bordering EPSPS gene copies, including an unclassified DNA transposon, an Activator transposase and sequences with homology to miniature inverted-repeat transposable elements (MITEs). However, it remains unclear whether the gene duplication mechanism involves MITEs or any other mobile genetic elements.
EPSPS gene duplication inheritance appears to be more complex than inheritance of resistance-endowing single-gene mutations. The EPSPS gene duplication in several A. palmeri populations is inherited on the nuclear genome.56–58,67 Inheritance in a North Carolina A. palmeri population was generally consistent with polygenic inheritance and did not fit a single-gene model.57 EPSPS gene duplication inheritance in a New Mexico A. palmeri population was likewise not consistent with a single-gene model.58 Inheritance of high EPSPS copy number into F1 and F2 generations from a Georgia population was quantitative and not always stable, including transgressive segregation and non-Mendelian inheritance in at least one F2 family.69 This variable gene duplication transmission has parallels with the instability of duplicated EPSPS copies from glyphosate-selected cell cultures.54 EPSPS gene duplication inheritance in a Mississippi A. palmeri population showed maternal influence, as a higher copy number was inherited in progeny with a GR maternal parent than in progeny from a GS maternal parent.56 Evidence for apomixis was shown by the production of seeds on maternal A. palmeri individuals in the complete absence of pollination.56 Such apomixis may increase the frequency of the maternal genotype in the progeny, in part explaining the maternal influence on EPSPS copy number observed in the inheritance studies. Apomixis could be very advantageous for the evolution of gene duplication, ensuring that the selected trait is maintained in the population. Additional factors that may contribute to the observed unstable inheritance include ongoing activity of the duplication mechanism, protective genomic mechanisms that remove duplicated EPSPS gene copies and/or other unknown mechanisms. Notably, a lack of fitness cost due to EPSPS gene duplication has been reported, accounting for variations in genetic background and measuring multiple phenotypic parameters.70,71 Clearly, the inheritance and population genetics of EPSPS gene duplication are more complex than most known herbicide resistance mechanisms.
4 EPSPS TARGET-SITE MUTATIONS
The EPSPS gene in plants contains coding sequence for a transit peptide used to direct the protein to the chloroplast.72,73 The transit peptide, variable in sequence and length among different species, is cleaved upon chloroplast delivery, producing the mature EPSPS protein. For consistency and ease of comparison, most publications number the amino acid positions relative to the start codon of the mature plant EPSPS protein, without the chloroplast transit peptide leader sequence.
Reported mutations conferring glyphosate resistance in weeds change the hydrophobic Pro106 amino acid of EPSPS to the hydrophobic amino acids Ala or Leu, or the hydrophilic amino acids Ser and Thr (summarized in Table 2). The Pro106 codon (CCx) can mutate to the commonly reported Ser, Ala or Thr through substitutions at the first base of the codon (TCx, GCx and ACx respectively). Substitutions at the second base of the codon can produce Leu (CTx, the most infrequently reported mutation), Arg (CGx), Gln (CAA/G) or His (CAC/T). Pro106Arg, Gln and His mutations have not been reported to date. While Gln is hydrophilic, both Arg and His are positively charged and may be disruptive to the active site. EPSPS target-site mutations have been reported in six species, most frequently in the genus Lolium, including L. rigidum and L. multiflorum (Table 2). The first dicotyledonous species with a Pro106 mutation is A. tuberculatus, with two populations recently reported.74,75
Table 2.
Reported Pro106 target-site mutations in EPSPS endowing glyphosate resistance in weed species
| Species | Reference | Pro106a to | Fold resistance | Other mechanisms detected? |
|---|---|---|---|---|
| Eleusine indica | 16 | Ser | 2–4 | No |
| Amaranthus tuberculatus | 74 | Ser | 5 | Yes, reduced translocation |
| Amaranthus tuberculatus | 75 | Ser | 5 | No, Ser mutation did not fully account for resistance |
| Echinochloa colona | 108 | Ser | 6.6 | No |
| Lolium multiflorum | 109 | Ser | 2–5 | No |
| Ala | 5–15 | |||
| Eleusine indica | 110, 111 | Ser | 3 | No |
| Thr | 3 | |||
| Digitaria insularis | 88 | Thr | 4 | Yes, reduced absorption and reduced translocation |
| Eleusine indica | 112 | Ser | 2 | No |
| Lolium rigidum | 107 | Leu | 1.7 | Yes, unknown mechanism |
| Lolium rigidum | 106 | Ser | 6–8 | Yes, reduced translocation |
| Thr | 8–11 | Yes, reduced translocation | ||
| Lolium multiflorum | 113 | Ser | 5 | Yes, reduced translocation |
| Lolium multiflorum | 114 | Ser | 5 | No, reduced translocation detected in different population |
| Lolium rigidum | 115 | Thr | 2–3 | No |
| Lolium rigidum | 34 | Ser & Leu | 16–21 | No, but other mechanisms suspected |
| Lolium rigidum | 116 | Ser | — | No |
| Lolium rigidum | 117 | Ala | 14 | Yes, reduced translocation |
Numbered relative to the start of the mature EPSPS enzyme in plants; for example, the first ten amino acids of the mature petunia EPSPS (GenBank M21084.1) are KPSEIVLQPI, and for the Arabidopsis EPSPS gene AT2G45300 the first ten are KASEIVLQPI.
The Pro106 is not directly involved in molecular interactions with either glyphosate or the substrate PEP, but changing the Pro106 to a different amino acid causes a structural change in the active site and shifts other amino acids towards the inhibitor, reducing the available space.76 The Pro106Ser mutation has previously been shown to increase the inhibitory constant Ki(app) for glyphosate from 0.4 μM in the wild-type petunia (Petunia × hybrida) enzyme to 3 μM in the mutant, but also to increase the Km(app) for PEP from 5 to 44 μM.72 The ratio Ki/Km, considered to be a selectivity factor for PEP over glyphosate binding,72 showed little change due to the Pro106Ser mutation, from 0.08 in wild type to 0.07 in the mutant. Subsequent research using a Pro106Ser mutant cloned from E. indica showed a wild-type Ki/Km ratio of 0.013 and a Pro106Ser mutant ratio of 0.12, a 9.2-fold increase.16 The expected loss of substrate affinity was less severe than reported in petunia,72 suggesting that in the E. indica EPSPS the Pro106Ser mutation confers glyphosate resistance sufficient to confer survival in the field while maintaining adequate PEP affinity.16
Additional mutations in the EPSPS gene able to confer glyphosate resistance have been identified using directed mutagenesis and expression in E. coli and transgenic plants, but have not yet been reported in GR weeds (Table 3). These mutations should be considered as candidate glyphosate resistance mechanisms when considering EPSPS target-site resistance by sequencing the entire EPSPS gene. Several reports involve double mutations that provide high-level glyphosate resistance while maintaining sufficient affinity for PEP (Table 3). The double mutants Gly101Ala + Gly144Asp77 and Gly101Ala + Ala192Thr78 have been shown to confer glyphosate resistance in transgenic maize (Zea mays) and canola (Brassica napus) respectively. The Thr102Ile + Pro106Ser (TIPS) double mutation has been described in transgenic crops (GA21 event used in first-generation Roundup Ready maize) as providing high-level glyphosate resistance,79,80 and the enzyme kinetics shows improved glyphosate resistance and relatively acceptable PEP affinity in comparison with the Pro106Ser mutation alone.81 Recently, Jalaludin et al.82 reported a GR E. indica population that had the Thr102Ile mutation in combination with the Pro106Ser mutation, which had previously evolved. This first report of a double EPSPS mutation in a field-evolved GR weed population demonstrates the evolutionary process by which mutations can accumulate to confer increasingly efficient resistance when selection pressure is persistent. Consistent with empirical and theoretical predictions for evolution of fitter proteins,83,84 the Thr102Ile mutation would be unlikely to occur first or independently owing to its significant reduction in PEP Km (Table 3). However, as a second mutation, Thr102Ile can improve the Pro106Ser mutation that has evolved first in many GR species.
Table 3.
Additional target-site mutations in EPSPS shown to confer glyphosate resistance (increased Ki for glyphosate) with variable effects on PEP affinity (Km) when expressed in E. coli. Numbering system relative to the start of the mature EPSPS enzyme in plants, including petunia and Arabidopsis. Positions Gly101, Thr102, Pro106, Gly144 and Ala192 in plant mature EPSPS consensus correspond to Gly96, Thr97, Pro101, Gly137 and Ala183 in E. coli
| Mutation(s) | Ki(app) for glyphosate (μM) | Km (app) for PEP (μM) | Ki/Km | Reference |
|---|---|---|---|---|
| Petunia wild-type EPSPS | 0.4 | 5.2 | 0.08 | 118 |
| Gly101Ala | 2000 | 200 | 10.0 | 118 |
| Gly101Ala, Ala192Thr | 683 | 54 | 12.6 | 118 |
| Gly101Ala, Gly144Asp | 348 | 40 | 7.7 | 119 |
| Gly101Ala, Gly144Asn | 960 | 91 | 10.5 | 119 |
| Maize wild type EPSPS | 0.5 | 27 | 0.02 | 120 |
| Maize Thr102Ile | 148.6 | 233.0 | 0.6 | 120 |
| Maize Pro106Ser | 1.0 | 17.1 | 0.06 | 120 |
| Maize Thr102Ile, Pro106Ser | 58.0 | 10.6 | 5.5 | 120 |
| Maize Thr102Ile, Pro106Thr | 101.3 | 11.2 | 9.0 | 120 |
| Maize Thr102Ile, Pro106Gly | 38.6 | 23.0 | 1.68 | 120 |
| Maize Thr102Ile, Pro106Cys | 818.2 | 47.0 | 17.4 | 120 |
| Maize Thr102Ile, Pro106Ala | 148.3 | 10.2 | 14.5 | 120 |
| Maize Thr102Ile, Pro106Ile | 2500 | 60.3 | 41.5 | 120 |
| Maize Thr102Ile, Pro106Val | 1600 | 109.3 | 14.6 | 120 |
| Maize Thr102Ile, Pro106Met | 37 200 | 143.3 | 260 | 120 |
| Maize Thr102Ile, Pro106Leu | 2100 | 99.5 | 21.1 | 120 |
| Maize Pro106Thr | 4.0 | 24.6 | 0.16 | 120 |
| Maize Pro106Leu | 28.6 | 86.7 | 0.33 | 120 |
| Agrobacterium spp. CP4 | 5100 | 14.4 | 354 | 120 |
5 OTHER MECHANISMS
Because very high (e.g. >50× the label rate) resistance levels have not been reported in GR weeds as has been observed in some cases for herbicides that inhibit ALS or ACCase, it is hypothesized that low dose selection has been effective for glyphosate resistance mechanisms. Low dose recurrent selection is a major scientific methodology for investigating pesticide resistance evolution, including herbicides,85–87 primarily because weak resistance traits are successfully selected with a low dose. Cross-pollination among survivors of a low dose followed by recurrent selection combines additive contributions from genes of small effect to evolve commercially significant resistance.86 Such traits involving any mechanism that contributes to improved survival under glyphosate selection have appeared in the literature. Reduced glyphosate absorption has been reported in some GR populations.88,89 In the case of Chilean L. multiflorum, the leaf angle in the GR population was different from that in the GS population, enabling reduced foliar spray interception.89 In addition, the GR population absorbed less glyphosate and had lower glyphosate translocation from treated leaves.
The 31P NMR survey of species (Section 2.1.1) in part chose D. illinoensis and S. obtusifolia because of the report of approximately 50% metabolism of glyphosate to AMPA in 7 days.90 AMPA also has a phosphonate, making it visible by 31P NMR, and the signal is easily distinguished from glyphosate. However, in the short time of these 31P NMR experiments, no AMPA could be observed (Sammons RD, unpublished). The metabolism of glyphosate in plants at least is rare and is only documented in a few cases, such as Equisetum arvense91 and soybeans.92 Examination of glyphosate metabolism in multiple species found no relationship between conversion to AMPA and glyphosate resistance level.90,93 Some recent reports employing a new methodology claim that two metabolic pathways for glyphosate are present in C. canadensis and Digitaria insularis.88,94 The detection of both glyoxylate and sarcosine in a glyphosate-dependent manner, when these compounds can also be readily metabolized in plants, requires confirmation by other methods. The unequivocal observations that weedy legumes90 can oxidize glyphosate to AMPA will spur on others to reinvestigate this long-overlooked mechanism and suggest that glyphosate degradation mechanisms may contribute to resistance eventually in selected species, particularly legumes.
6 INTEGRATIVE MECHANISTIC MODEL FOR GLYPHOSATE RESISTANCE
The concept starts with the realization that a finite dose of glyphosate is delivered to a plant, more particularly to the source leaves on the plant. Previous research has shown that sink leaves do not take up appreciable glyphosate and are incapable of transporting it systemically.95 The idea that the dose is finite builds on the loading that eventually ends. Many different formulations were evaluated,96 and while the kinetics of uptake varied among formulations, no formulation continued to complete glyphosate loading, indicating that there is a time period for loading and it does end. The model consequently has each source cell presented with a finite dose of glyphosate, and the fate of the glyphosate in the cell would depend on the pathways available once inside. Figure 1A shows theoretical glyphosate uptake and fate within a normal source-leaf cell. The y-axis is a measure of the glyphosate concentration in a relative sense, and the x-axis is time limited to 1 day. The accumulation of glyphosate in the cytoplasm presents glyphosate to organelles and most particularly the chloroplast, which must accumulate glyphosate. Chloroplast accumulation rates depend on the concentration in the cytoplasm (whether uptake is passive or active transport). Because most source cells supply assimilate, usually sucrose, to the phloem by way of cell-to-cell connectivity via plasmodesmata, glyphosate phloem mobilization (translocation) is dependent on the glyphosate concentration in the cytoplasm.97 The model needs some leniency from the reader for the ‘mobilized’ curve, as it really represents the rate of glyphosate being translocated out of the cell rather than a concentration in the phloem. The applied glyphosate dose is finite, limited by the concentration-volume of the application solution and the loading competency of the surfactant system.96 The amount of glyphosate entering the source cell slows and stops at about 10 h (Fig. 1A). The cell then continues to disperse the cytoplasmic glyphosate to the organelles and the plasmodesmata-phloem until some equilibrium is reached or the cytoplasm is drained of glyphosate. The concentration of glyphosate in the chloroplast determines the herbicidal activity, as partial EPSPS inhibition results in varying degrees of stunting and sublethal effects on the cell and the leaf. A concentration higher than the minimum inhibitory concentration sustained for an adequate period of time inhibits the EPSPS and blocks the shikimate pathway sufficiently to destroy the chloroplast (Fig. 1A). Because chloroplast destruction is fast98 and appears as swollen plastids, the idea that chloroplasts have lost their osmotic integrity seems likely. The potential phytotoxic action of rapid chloroplast destruction differs from aromatic amino acid starvation often attributed to the glyphosate mechanism of action (slow death due to lack of aromatic amino acids). The actual cellular toxic mechanism is not clear, although it could be hyperaccumulation of shikimate faster than it can be exported99 or some other indirectly linked process interrupted.100 However, it should be pointed out that killing a cell or a leaf, or a root tip, etc., is not sufficient to kill a plant, as this is only part of the consequential biochemical activity, and so the model does not mean to explain overall plant death.
Figure 1.
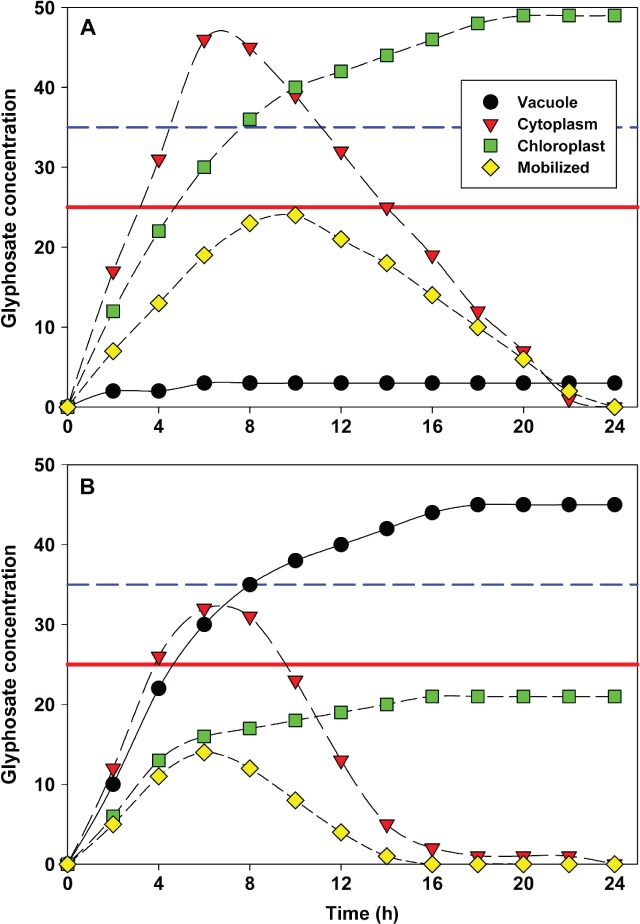
A theoretical cellular-level model of glyphosate uptake and distribution for (A) a normal, glyphosate-susceptible source cell and (B) a glyphosate-resistant source cell using the vacuole to sequester glyphosate. The units for glyphosate are relative concentration, with a theoretical chloroplast minimum inhibitory concentration (25) indicated by a red line, and a chloroplast glyphosate concentration (35) consistent with saturated inhibition indicated by a dashed blue line.
Figure 1B represents a GR cell where, in addition, the vacuole is used to sequester glyphosate. The concentration of glyphosate in the cytoplasm of the GR source-leaf cell (Fig. 1B) is somewhat less at any given time than in the cytoplasm of the GS source-leaf cell (Fig. 1A) because there is now an extra pathway competing for glyphosate. The cytoplasm might be considered to be a conduit for the other destinations, so the total flux (loading and dispersal) is the same. The vacuole captures most of the glyphosate entering the cell, thereby decreasing the amount available to the chloroplast and phloem. The three pathways together decrease the amount of glyphosate in the GR cytoplasm faster than in the GS case, resulting in glyphosate concentrations lower than the minimum inhibitory concentration in the chloroplast and less glyphosate being mobilized (Fig. 1B). Resistance results, then, from a lower cytoplasm glyphosate concentration and less time to load the chloroplast. Therefore, Fig. 1B demonstrates the case where sequestration would provide a shield to the plant by decreasing the net available dose of glyphosate. The glyphosate trapped in the vacuole might be considered to be a Trojan horse, but this never seems to be realized in surviving herbaceous plants.
The model also works for sink tissue. In this case, glyphosate is delivered to the cytoplasm symplastically via the phloem, as long as the phloem is delivering assimilate.101 With decreased vacuole size in sink tissue, the volume of the cytoplasm is a much larger relative proportion, holding more glyphosate and allowing for an increased dose delivered to the chloroplasts. Sink tissues seem to be more reliant on their chloroplasts, as each unit of glyphosate is more toxic in sink tissues,98,102 but this could be explained by less organelle competition for glyphosate, resulting in much higher concentrations in the developing sink-leaf chloroplasts. One argument could be that the increased sensitivity of apical tissue would stop phloem delivery due to toxicity (blocked respiration of sucrose), leaving a high concentration of glyphosate in the source-leaf cytoplasm that could further load the chloroplasts there with increased time. This seems possible, but still it remains a fact that source leaves are generally the last green tissues in a lethally dosed glyphosate plant.
The model works very well for explaining the data observed by 31P NMR, 14C glyphosate and shikimate determinations, so the consequence of other potential resistance mechanisms can be examined. Firstly, an assumption illustrated in the model needs to be highlighted because of its critical impact on the predictions. The 31P NMR experiments demonstrate that glyphosate only crosses a lipid bilayer in one direction in the time course of 2–3 days (for living cells).17 This means that, once entering the cell, glyphosate cannot diffuse back to the apoplast, and similarly, once in the vacuole, glyphosate does not go back to the cytoplasm. For this reason it has been assumed that glyphosate entering the chloroplast also cannot go back to the cytoplasm. This makes the modeling somewhat easier, as a reversible equilibrium need not be included, but it can be accommodated if necessary when data are produced that support the efflux.
Why is the very weak mutation Pro106 to Ser or Thr found so often in so many species? The mutation itself on purified enzyme makes very slight changes to the Km of PEP and Ki for glyphosate. The experimentally determined EPSPS I50 values will be used primarily because the interactions of Ki and Km for mutants is beyond the scope of the present discussion.103 If the glyphosate concentration in the chloroplast is well below or well above the I50, then the mutant EPSPS may be of very little consequence, but small differences leading to life or death could be observed at chloroplast glyphosate concentrations near the EPSPS I50.104 Mutant EPSPS would effectively raise the minimum inhibitory concentration shown in Fig. 1. Because the Pro106 mutation is consistently selected in field situations, chloroplast glyphosate concentrations must be near the EPSPS I50. While the Pro106 mutation is weak, the addition of any other mechanism that lowers the glyphosate cytoplasm concentration, and therefore reduces the chloroplast glyphosate concentration below the mutant EPSPS I50, would have an additive interaction beneficial for survival. A good example described is when this weak Pro106Ser mutation was combined with sequestration as observed in L. multiflorum, R336.32,34 Such Pro106 mutations are frequently reported, which may suggest the presence of other contributing mechanisms. Obviously, a potent mutation that significantly raises the Ki will easily be selected, as the chloroplast is unlikely to accumulate such very high levels of glyphosate in the time allowed (Powles S, private communication). Higher EPSPS concentrations in the chloroplast would increase the total units of enzyme activity, requiring more glyphosate to obtain sufficient EPSPS inhibition. The I50 remains the same, but enough EPSPS activity remains for plant survival even when EPSPS is 95% inhibited. The EPSPS gene duplication system recently characterized in several species provides more enzyme, which requires more glyphosate in the chloroplast for sufficient inhibition of EPSPS.55
Other barriers in the model can easily be envisioned. If glyphosate has reduced uptake by a change in plasmalemma transport, then the cytoplasm concentration is decreased, limiting access to the chloroplast as measured for S. halepense.17,38 Similarly, if glyphosate uptake into the chloroplast is reduced, then the effective concentration of glyphosate could be drastically reduced as proposed for D. illinoensis.17 The combination of reduced cellular uptake and reduced chloroplast uptake together with vacuole sequestration would create a series of impressive barriers to delivering glyphosate.
This model has two unique perspectives built in. Firstly, there is a finite dose that must be dealt with by the plant at the cellular level. Secondly, there is a time component, as the limited amount of glyphosate available must reach the minimum inhibitory concentration (shown in Fig. 1) within a particular timeframe. Very many sequential barriers could decrease the glyphosate available to reach the chloroplast, and even physical cuticle barriers could play a role.89 Therefore, a number of specific physical and biochemical barriers can reduce the effective glyphosate available to inhibit EPSPS. The NMR data for a survey of cellular uptake suggests that there are three barriers used in very different ways by plants.17 Some reduce the cellular uptake, some sequester glyphosate using the vacuole and others seem to be able to restrict the entry of glyphosate into the chloroplast.
7 SUMMARY
The study of resistance mechanisms in GR weeds has provided fascinating insights into the evolutionary processes utilized by genetically diverse weed species in response to intense selection pressure. Previously unknown resistance mechanisms have been discovered in GR weed species, including vacuolar glyphosate sequestration,18 gene duplication55 and rapid mature leaf necrosis resulting in reduced translocation.43 Target-site mutations for glyphosate resistance have been reported at one codon, are apparently rare within populations and endow lower levels of resistance than target-site mutations for some other herbicide mechanisms of action. A common trend is for glyphosate resistance mechanisms to combine within populations and within individuals.105 Several examples have been reported of Pro106 target-site mutations acting additively with reduced translocation, or other unknown mechanisms, to provide higher-level resistance than either mechanism alone.74,106,107 In other cases, gene duplication or target-site mutations were detected but did not seem fully to explain the higher than expected levels of resistance.34,75 Accumulation of multiple mechanisms is a common occurrence, particularly in cross-pollinated species, producing higher resistance levels in response to continuing glyphosate selection pressure. The recruitment of a variety of biochemical processes to establish resistance to glyphosate in so many species suggests that weed management will have to diversify and use additional methods to control glyphosate-resistant weeds.
Acknowledgments
T.G. was supported by Bayer CropScience (Weed Resistance Research, Frankfurt am Main, Germany) and the Australian Herbicide Resistance Initiative at the University of Western Australia.
REFERENCES
- 1.Duke SO, Powles SB. Glyphosate: a once-in-a-century herbicide. Pest Manag Sci. 2008;64:319–325. doi: 10.1002/ps.1518. [DOI] [PubMed] [Google Scholar]
- 2.Dill G, Sammons RD, Feng P, Kohn F, Mehrsheikh A, Bleeke M, et al., editors. Glyphosate Resistance in Crops and Weeds. Hoboken, NJ: Wiley; 2010. Glyphosate: discovery, development, applications and properties; pp. 1–34. [Google Scholar]
- 3.Anderson KS, Sammons RD, Leo GC, Sikorski JA, Benesi AJ, Johnson KA. Observation by carbon-13 NMR of the EPSP synthase tetrahedral intermediate bound to the enzyme active site. Biochemistry. 1990;29:1460–1465. doi: 10.1021/bi00458a017. [DOI] [PubMed] [Google Scholar]
- 4.Schonbrunn E, Eschenburg S, Shuttleworth WA, Schloss JV, Amrhein N, Evans JNS, et al. Interaction of the herbicide glyphosate with its target enzyme 5-enolpyvuvylshikimate-3-phosphate synthase in atomic detail. Proc Natl Acad Sci USA. 2001;98:1376–1380. doi: 10.1073/pnas.98.4.1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Steinrucken HC, Amrhein N. The herbicide glyphosate is a potent inhibitor of 5-enolpyruvylshikimic acid-3-phosphate synthase. Biochem Biophys Res Comm. 1980;94:1207–1212. doi: 10.1016/0006-291x(80)90547-1. [DOI] [PubMed] [Google Scholar]
- 6.Baird DD, Upchurch RP, Homesley WB, Franz JE, editors. Introduction of a new broad spectrum post emergence herbicide of a new broad spectrum post emergence herbicide class with utility for herbaceous perennial weed control. Proceedings 26th North Central Weed Control Conf, Dec. 7–9; Kansas City, MO. 1971. pp. 64–68. [Google Scholar]
- 7.Bradshaw LD, Padgette SR, Kimball SL, Wells BH. Perspectives on glyphosate resistance. Weed Technol. 1997;11:189–198. [Google Scholar]
- 8.Heap I. International Survey of Herbicide Resistant Weeds. Online]. Available: weedscience.org [1 October 2013] [Google Scholar]
- 9.Powles SB. Evolved glyphosate-resistant weeds around the world: lessons to be learnt. Pest Manag Sci. 2008;64:360–365. doi: 10.1002/ps.1525. [DOI] [PubMed] [Google Scholar]
- 10.Sammons R, Heering D, DiNicola N, Glick H, Elmore G. Sustainability and stewardship of glyphosate and glyphosate-resistant crops. Weed Technol. 2007;21:347–354. [Google Scholar]
- 11.Bowler C, Montagu MV, Inze D. Superoxide dismutase and stress tolerance. Annu Rev Plant Phys Plant Mol Biol. 1992;43:83–116. [Google Scholar]
- 12.Lorraine-Colwill DF, Hawkes TR, Williams PH, Warner SAJ, Sutton PB, Powles SB, et al. Resistance to glyphosate in Lolium rigidum. Pestic Sci. 1999;55:489–491. [Google Scholar]
- 13.Feng PCC, Pratley JE, Bohn JA. Resistance to glyphosate in Lolium rigidum. II. Uptake, translocation, and metabolism. Weed Sci. 1999;47:412–415. [Google Scholar]
- 14.Lorraine-Colwill DF, Powles SB, Hawkes TR, Hollinshead PH, Warner SAJ, Preston C. Investigations into the mechanism of glyphosate resistance in Lolium rigidum. Pestic Biochem Phys. 2002;74:62–72. [Google Scholar]
- 15.Feng PCC, Tran M, Chiu T, Sammons RD, Heck GR, CaJacob CA. Investigations into glyphosate-resistant horseweed (Conyza canadensis): retention, uptake, translocation, and metabolism. Weed Sci. 2004;52:498–505. [Google Scholar]
- 16.Baerson SR, Rodriguez DJ, Tran M, Feng YM, Biest NA, Dill GM. Glyphosate-resistant goosegrass. Identification of a mutation in the target enzyme 5-enolpyruvylshikimate-3-phosphate synthase. Plant Physiol. 2002;129:1265–1275. doi: 10.1104/pp.001560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ge X, d'Avignon DA, Ackerman J, Ostrander E, Sammons RD. Herbicides: Biological Activity, Classification and Health and Environmental Implications. Hauppauge, NY: Nova Science; 2013. Applications of 31P NMR spectroscopy to glyphosate studies in plants: insights into cellular uptake and vacuolar sequestration correlated to herbicide resistance. [Google Scholar]
- 18.Ge X, d'Avignon DA, Ackerman JJH, Sammons RD. Rapid vacuolar sequestration: the horseweed glyphosate resistance mechanism. Pest Manag Sci. 2010;66:345–348. doi: 10.1002/ps.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rueppel ML, Marvel JT. 1H and 31P NMR spectra of substituted methylphosphonic acids with indirect determination of 31P shifts. Organic Magnetic Resonance. 1976;8:19–20. [Google Scholar]
- 20.Rueppel ML, Brightwell BB, Schaefer J, Marvel JT. Metabolism and degradation of glyphosate in soil and water. Journal of Agricultural and Food Chemistry. 1977;25:517–528. doi: 10.1021/jf60211a018. [DOI] [PubMed] [Google Scholar]
- 21.Roberts JKM, Ray PM, Wade-Jardetsky N, Jardetsky O. Estimation of cytoplasmic and vacuolar pH in higher plant cells by 31P NMR. Nature. 1980;283:870–872. [Google Scholar]
- 22.Castellino S, Leo GC, Sammons RD, Sikorski JA. Phosphorus-31, nitrogen-15 and carbon-13 NMR of glyphosate: comparison of pH titrations to the herbicidal dead-end complex with 5-enolpyruvylshikimate-3-phosphate synthase. Biochemistry. 1989;28:3856–3868. [Google Scholar]
- 23.VanGessel MJ. Glyphosate-resistant horseweed from Delaware. Weed Sci. 2001;49:703–705. [Google Scholar]
- 24.Zelaya IA, Owen MDK, VanGessel MJ. Inheritance of evolved glyphosate resistance in Conyza canadensis (L.) Cronq. Theor Appl Genet. 2004;110:58–70. doi: 10.1007/s00122-004-1804-8. [DOI] [PubMed] [Google Scholar]
- 25.Denis MH, Delrot S. Carrier-mediated uptake of glyphosate in broad bean (Vicia faba) via a phosphate transporter. Physiol Plantarum. 1993;87:569–575. [Google Scholar]
- 26.Anthelme F, Marigo G. Glyphosate uptake in Catharanthus roseus cells: involvement of a plasma membrane redox system? Pest Biochem Phys. 1998;62:73–86. [Google Scholar]
- 27.Weaver SE. The biology of Canadian weeds. 115. Conyza canadensis. Can J Plant Sci. 2001;81:867–875. [Google Scholar]
- 28.Ge X, d'Avignon DA, Ackerman JJH, Duncan B, Spaur MB, Sammons RD. Glyphosate-resistant horseweed made sensitive to glyphosate: low-temperature suppression of glyphosate vacuolar sequestration revealed by 31P NMR. Pest Manag Sci. 2011;67:1215–1221. doi: 10.1002/ps.2169. [DOI] [PubMed] [Google Scholar]
- 29.Yu Q, Huang S, Powles S. Direct measurement of paraquat in leaf protoplasts indicates vacuolar paraquat sequestration as a resistance mechanism in Lolium rigidum. Pest Biochem Phys. 2010;98:104–109. [Google Scholar]
- 30.Purba E, Preston C, Powles SB. The mechanism of resistance to paraquat is strongly temperature-dependent in resistant Hordeum leporinum Link and H. Glaucum Steud. Planta. 1995;196:464–468. [Google Scholar]
- 31.Lyons JM, Graham D, Raison JK, editors. The Role of the Membrane. New York, NY: Academic Press; 1979. Low temperature stress in crop plants; p. 565. [Google Scholar]
- 32.Ge X, d'Avignon DA, Ackerman JJH, Collavo A, Sattin M, Ostrander EL, et al. Vacuolar glyphosate-sequestration correlates with glyphosate resistance in ryegrass (Lolium spp.) from Australia, South America, and Europe: a 31P NMR investigation. J Agric Food Chem. 2012;60:1243–1250. doi: 10.1021/jf203472s. [DOI] [PubMed] [Google Scholar]
- 33.Vila-Aiub MM, Gundel PE. Yu Q and Powles SB. Glyphosate resistance in Sorghum halepense and Lolium rigidum is reduced at suboptimal growing temperatures. Pest Manag Sci. 2013;69:228–232. doi: 10.1002/ps.3464. [DOI] [PubMed] [Google Scholar]
- 34.Collavo A, Sattin M. Resistance to glyphosate in Lolium rigidum selected in Italian perennial crops: bioevaluation, management and molecular bases of target-site resistance. Weed Res. 2012;52:16–24. [Google Scholar]
- 35.Culpepper AS, Grey TL, Vencill WK, Kichler JM, Webster TM, Brown SM, et al. Glyphosate-resistant Palmer amaranth (Amaranthus palmeri) confirmed in Georgia. Weed Sci. 2006;54:620–626. [Google Scholar]
- 36.Hetherington PR, Marshall G, Kirkwood RC, Warner JM. Absorption and efflux of glyphosate by cell suspensions. J Exp Bot. 1998;49:527–533. [Google Scholar]
- 37.Kamicker BJ, Sweeney MT, Kaczmarek F, Dib-Hajj F, Shang W, Crimin K. Bacterial efflux pump inhibitors. In: Champney WS, et al., editors. Methods in Molecular Medicine. Totowa, NJ: Humana Press; 2008. pp. 187–204. [DOI] [PubMed] [Google Scholar]
- 38.Riar DS, Norsworthy JK, Johnson DB, Scott RC, Bagavathiannan M. Glyphosate resistance in a Johnsongrass (Sorghum halepense) biotype from Arkansas. Weed Sci. 2011;59:299–304. [Google Scholar]
- 39.Vila-Aiub MM, Balbi MC, Distefano AJ, Fernandez L, Hopp E, Yu Q, et al. Glyphosate resistance in perennial Sorghum halepense (Johnsongrass), endowed by reduced glyphosate translocation and leaf uptake. Pest Manag Sci. 2012;68:430–436. doi: 10.1002/ps.2286. [DOI] [PubMed] [Google Scholar]
- 40.Nol N, Tsikou D, Eid M, Livieratos IC, Giannopolitis CN. Shikimate leaf disc assay for early detection of glyphosate resistance in Conyza canadensis and relative transcript levels of EPSPS and ABC transporter genes. Weed Res. 2012;52:233–241. [Google Scholar]
- 41.Peng Y, Abercrombie LLG, Yuan JS, Riggins CW, Sammons RD, Tranel PJ, et al. Characterization of the horseweed (Conyza canadensis) transcriptome using GS-FLX 454 pyrosequencing and its application for expression analysis of candidate non-target herbicide resistance genes. Pest Manag Sci. 2010;66:1053–1062. doi: 10.1002/ps.2004. [DOI] [PubMed] [Google Scholar]
- 42.Yuan JS, Abercrombie LLG, Cao Y, Halfhill MD, Zhou X, Peng Y, et al. Functional genomics analysis of horseweed (Conyza canadensis) with special reference to the evolution of non-target-site glyphosate resistance. Weed Sci. 2010;58:109–117. [Google Scholar]
- 43.Robertson RR. Department of Horticulture and Landscape Architecture. West Lafayette, IN: Purdue University; 2010. Physiological and biochemical characterization of glyphosate resistant Ambrosia trifida L. MS Thesis. [Google Scholar]
- 44.Innan H, Kondrashov F. The evolution of gene duplications: classifying and distinguishing between models. Nat Rev Genet. 2010;11:97–108. doi: 10.1038/nrg2689. [DOI] [PubMed] [Google Scholar]
- 45.Bass C, Field LM. Gene amplification and insecticide resistance. Pest Manag Sci. 2011;67:886–890. doi: 10.1002/ps.2189. [DOI] [PubMed] [Google Scholar]
- 46.Flagel LE, Wendel JF. Gene duplication and evolutionary novelty in plants. New Phytol. 2009;183:557–564. doi: 10.1111/j.1469-8137.2009.02923.x. [DOI] [PubMed] [Google Scholar]
- 47.Zhang J. Evolution by gene duplication: an update. Trends Ecol Evol. 2003;18:292–298. [Google Scholar]
- 48.Kondrashov FA. Gene duplication as a mechanism of genomic adaptation to a changing environment. Proc R Soc B Biol Sci. 2012;279:5048–5057. doi: 10.1098/rspb.2012.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boerboom CM, Wyse DL, Somers DA. Mechanism of glyphosate tolerance in birdsfoot-trefoil (Lotus corniculatus) Weed Sci. 1990;38:463–467. [Google Scholar]
- 50.Jones JD, Goldsbrough PB, Weller SC. Stability and expression of amplified EPSPS genes in glyphosate resistant tobacco cells and plantlets. Plant Cell Rep. 1996;15:431–436. doi: 10.1007/BF00232070. [DOI] [PubMed] [Google Scholar]
- 51.Shah DM, Horsch RB, Klee HJ, Kishore GM, Winter JA, Tumer NE, et al. Engineering herbicide tolerance in transgenic plants. Science. 1986;233:478–481. doi: 10.1126/science.233.4762.478. [DOI] [PubMed] [Google Scholar]
- 52.Suh H, Hepburn AG, Kriz AL, Widholm JM. Structure of the amplified 5-enolpyruvylshikimate-3-phosphate synthase gene in glyphosate-resistant carrot cells. Plant Mol Biol. 1993;22:195–205. doi: 10.1007/BF00014928. [DOI] [PubMed] [Google Scholar]
- 53.Widholm JM, Chinnala AR, Ryu JH, Song HS, Eggett T, Brotherton JE. Glyphosate selection of gene amplification in suspension cultures of 3 plant species. Physiol Plantarum. 2001;112:540–545. doi: 10.1034/j.1399-3054.2001.1120411.x. [DOI] [PubMed] [Google Scholar]
- 54.Pline-Srnic W. Physiological mechanisms of glyphosate resistance. Weed Technol. 2006;20:290–300. [Google Scholar]
- 55.Gaines TA, Zhang W, Wang D, Bukun B, Chisholm ST, Shaner DL, et al. Gene amplification confers glyphosate resistance in Amaranthus palmeri. Proc Natl Acad Sci USA. 2010;107:1029–1034. doi: 10.1073/pnas.0906649107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ribeiro DN, Pan Z, Duke SO, Nandula VK, Baldwin BS, Shaw DR, et al. Involvement of facultative apomixis in inheritance of EPSPS gene amplification in glyphosate-resistant Amaranthus palmeri. Planta. 2014;239:199–212. doi: 10.1007/s00425-013-1972-3. [DOI] [PubMed] [Google Scholar]
- 57.Chandi A, Milla-Lewis SR, Giacomini D, Westra P, Preston C, Jordan DL, et al. Int J Agron: 2012. Inheritance of evolved glyphosate resistance in a North Carolina Palmer amaranth (Amaranthus palmeri) biotype. DOI: 10.1155/2012/176108. [Google Scholar]
- 58.Mohseni-Moghadam M, Schroeder J, Ashigh J. Mechanism of resistance and inheritance in glyphosate resistant Palmer amaranth (Amaranthus palmeri) populations from New Mexico. USA. Weed Sci. 2013;61:517–525. [Google Scholar]
- 59.Mohseni-Moghadam M, Schroeder J, Heerema R, Ashigh J. Resistance to glyphosate in Palmer Amaranth (Amaranthus palmeri) populations from New Mexico pecan orchards. Weed Technol. 2013;27:85–91. [Google Scholar]
- 60.Salas RA, Dayan FE, Pan Z, Watson SB, Dickson JW, Scott RC, et al. EPSPS gene amplification in glyphosate-resistant Italian ryegrass (Lolium perenne ssp. multiflorum) from Arkansas. Pest Manag Sci. 2012;68:1223–1230. doi: 10.1002/ps.3342. [DOI] [PubMed] [Google Scholar]
- 61.Wiersma AT. Department of Bioagricultural Sciences and Pest Management. Fort Collins, CO: Colorado State University; 2012. Regional whole plant and molecular response of Kochia scoparia to glyphosate. MS Thesis. [Google Scholar]
- 62.Tranel PJ, Riggins CW, Bell MS, Hager AG. Herbicide resistances in Amaranthus tuberculatus: a call for new options. J Agric Food Chem. 2011;59:5808–5812. doi: 10.1021/jf103797n. [DOI] [PubMed] [Google Scholar]
- 63.Nandula VK, Wright A, Bond J, Eubank T, Molin W. Glyphosate resistance in spiny amaranth. Weed Sci Soc Am Proc. 2013;53:115. [Google Scholar]
- 64.Riggins CW, Peng YH, Stewart CN, Tranel PJ. Characterization of de novo transcriptome for waterhemp (Amaranthus tuberculatus) using GS-FLX 454 pyrosequencing and its application for studies of herbicide target-site genes. Pest Manag Sci. 2010;66:1042–1052. doi: 10.1002/ps.2006. [DOI] [PubMed] [Google Scholar]
- 65.Wassom JJ, Tranel PJ. Amplified fragment length polymorphism-based genetic relationships among weedy Amaranthus species. J Heredity. 2005;96:410–416. doi: 10.1093/jhered/esi065. [DOI] [PubMed] [Google Scholar]
- 66.Gaines TA, Ward SM, Bukun B, Preston C, Leach JE, Westra P. Interspecific hybridization transfers, a previously unknown glyphosate resistance mechanism in Amaranthus species. Evol Appl. 2012;5:29–38. doi: 10.1111/j.1752-4571.2011.00204.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gaines TA, Shaner DL, Ward SM, Leach JE, Preston C, Westra P. Mechanism of resistance of evolved glyphosate-resistant Palmer amaranth (Amaranthus palmeri) J Agric Food Chem. 2011;59:5886–5889. doi: 10.1021/jf104719k. [DOI] [PubMed] [Google Scholar]
- 68.Gaines TA, Wright AA, Molin WM, Lorentz L, Riggins CW, Tranel PJ, et al. Identification of genetic elements associated with EPSPS gene amplification. PloS ONE. 2013;8:e65819. doi: 10.1371/journal.pone.0065819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Giacomini DA, Westra P, Ward SM, Sammons RD. The inheritance of amplified EPSPS gene copies in Palmer amaranth (Amaranthus palmeri) Weed Sci Soc Am Proc. 2013;53:311. [Google Scholar]
- 70.Giacomini D, Westra P, Ward SM. Impact of genetic background in fitness cost studies: an example from glyphosate-resistant Palmer amaranth. Weed Sci. 2014;62:29–37. [Google Scholar]
- 71.Vila-Aiub MM, Goh SS, Gaines TA, Han H, Busi R, Yu Q, et al. No fitness cost of glyphosate resistance endowed by massive EPSPS gene amplification in Amaranthus palmeri. Planta. 2014 doi: 10.1007/s00425-013-2022-x. DOI: 10.1007/s00425-00013-02022-x. [DOI] [PubMed] [Google Scholar]
- 72.Padgette SR, Re DB, Gasser CS, Eichholtz DA, Frazier RB, Hironaka CM, et al. Site-directed mutagenesis of a conserved region of the 5-enolpyruvylshikimate-3-phosphate synthase active site. J Biol Chem. 1991;266:22 364–22 369. [PubMed] [Google Scholar]
- 73.Gasser CS, Winter JA, Hironaka CM, Shah DM. Structure, expression, and evolution of the 5-enolpyruvylshikimate-3-phosphate synthase genes of petunia and tomato. J Biol Chem. 1988;263:4280–4289. [PubMed] [Google Scholar]
- 74.Nandula VK, Ray JD, Ribeiro DN, Pan Z, Reddy KN. Glyphosate resistance in tall waterhemp (Amaranthus tuberculatus) from Mississippi is due to both altered target-site and nontarget-site mechanisms. Weed Sci. 2013;61:374–383. [Google Scholar]
- 75.Bell MS, Hager AG, Tranel PJ. Multiple resistance to herbicides from four site-of-action groups in waterhemp (Amaranthus tuberculatus) Weed Sci. 2013;61:460–468. [Google Scholar]
- 76.Healy-Fried ML, Funke T, Priestman MA, Han H, Schonbrunn E. Structural basis of glyphosate tolerance resulting from mutations of Pro(101) in Escherichia coli 5-enolpyruvylshikimate-3-phosphate synthase. J Biol Chem. 2007;282:32 949–32 955. doi: 10.1074/jbc.M705624200. [DOI] [PubMed] [Google Scholar]
- 77.Howe AR, Gasser CS, Brown SM, Padgette SR, Hart J, Parker GB, et al. Glyphosate as a selective agent for the production of fertile transgenic maize (Zea mays L.) plants. Mol Breed. 2002;10:153–164. [Google Scholar]
- 78.Kahrizi D, Salmanian AH, Afshari A, Moieni A, Mousavi A. Simultaneous substitution of Gly96 to Ala and Ala183 to Thr in 5-enolpyruvylshikimate-3-phosphate synthase gene of E. coli (k12) and transformation of rapeseed (Brassica napus L.) in order to make tolerance to glyphosate. Plant Cell Rep. 2007;26:95–104. doi: 10.1007/s00299-006-0208-4. [DOI] [PubMed] [Google Scholar]
- 79.Lebrun M, Sailland A, Freyssinet G, Degryse E. Mutated 5-enolpyruvylshikimate-3-phosphate synthase, gene coding for said protein and transformed plants containing said gene. US 6,566,587, Bayer CropScience SA; 2003. pp. 1–17. [Google Scholar]
- 80.Spencer M, Mumm R, Gwyn J. Glyphosate resistant maize lines. 2000. pp. 1–59. US Patent 6040497, Dekalb Genetics Corporation.
- 81.Funke T, Yang Y, Han H, Healy-Fried M, Olesen S, Becker A, et al. Structural basis of glyphosate resistance resulting from the double mutation Thr(97) → Ile and Pro(101) → Ser in 5-enolpyruvylshikimate-3-phosphate synthase from Escherichia coli. J Biol Chem. 2009;284:9854–9860. doi: 10.1074/jbc.M809771200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jalaludin A, Han H, Yu Q, Powles S. Proceedings of Global Herbicide Resistance Challenge. Australia: Freemantle; Evolution in action: a double amino acid substitution in the EPSPS gene endows high-level glyphosate resistance. Feb. 24, [Online]. (2013). Available: http://www.herbicideresistanceconference.com.au/Program. [Google Scholar]
- 83.Weinreich DM, Delaney NF, DePristo MA, Hartl DL. Darwinian evolution can follow only very few mutational paths to fitter proteins. Science. 2006;312:111–114. doi: 10.1126/science.1123539. [DOI] [PubMed] [Google Scholar]
- 84.McCandlish DM. On the findability of genotypes. Evolution. 2013;67:2592–2603. doi: 10.1111/evo.12128. [DOI] [PubMed] [Google Scholar]
- 85.Busi R, Gaines TA, Walsh MJ, Powles SB. Understanding the potential for resistance evolution to the new herbicide pyroxasulfone: field selection at high doses versus recurrent selection at low doses. Weed Res. 2012;52:489–499. [Google Scholar]
- 86.Busi R, Powles SB. Evolution of glyphosate resistance in a Lolium rigidum population by glyphosate selection at sublethal doses. Heredity. 2009;103:318–325. doi: 10.1038/hdy.2009.64. [DOI] [PubMed] [Google Scholar]
- 87.Neve P, Powles S. Recurrent selection with reduced herbicide rates results in the rapid evolution of herbicide resistance in Lolium rigidum. Theor Appl Genet. 2005;110:1154–1166. doi: 10.1007/s00122-005-1947-2. [DOI] [PubMed] [Google Scholar]
- 88.de Carvalho LB, Alves P, Gonzalez-Torralva F, Cruz-Hipolito HE, Rojano-Delgado AM, De Prado R, et al. Pool of resistance mechanisms to glyphosate in Digitaria insularis. J Agric Food Chem. 2012;60:615–622. doi: 10.1021/jf204089d. [DOI] [PubMed] [Google Scholar]
- 89.Michitte P, De Prado R, Espinoza N, Ruiz-Santaella JP, Gauvrit C. Mechanisms of resistance to glyphosate in a ryegrass (Lolium multiflorum) biotype from Chile. Weed Sci. 2007;55:435–440. [Google Scholar]
- 90.Reddy KN, Rimando AM, Duke SO, Nandula VK. Aminomethylphosphonic acid accumulation in plant species treated with glyphosate. J Agric Food Chem. 2008;56:2125–2130. doi: 10.1021/jf072954f. [DOI] [PubMed] [Google Scholar]
- 91.Marshall G, Kirkwood RC, Martin DJ. Studies on the mode of action of asulam, aminotriazole and glyphosate in Equisetum arvense L. (field horsetail). II. The metabolism of carbon-14 asulam, carbon-14 aminotriazole and carbon-14 glyphosate. Pestic Sci. 1987;18:65–78. [Google Scholar]
- 92.Reddy KN, Rimando AM, Duke SO. Aminomethylphosphonic acid, a metabolite of glyphosate, causes injury in glyphosate-treated, glyphosate-resistant soybean. J Agric Food Chem. 2004;52:5139–5143. doi: 10.1021/jf049605v. [DOI] [PubMed] [Google Scholar]
- 93.Simarmata M, Kaufmann JE, Penner D. Potential basis of glyphosate resistance in California rigid ryegrass (Lolium rigidum) Weed Sci. 2003;51:678–682. [Google Scholar]
- 94.Gonzalez-Torralva F, Rojano-Delgado AM, Luque de Castro MD, Muelleder N, De Prado R. Two non-target mechanisms are involved in glyphosate-resistant horseweed (Conyza canadensis L. Cronq.) biotypes. J Plant Phys. 2012;169:1673–1679. doi: 10.1016/j.jplph.2012.06.014. [DOI] [PubMed] [Google Scholar]
- 95.Feng PCC, Sandbrink JJ, Sammons RD. Retention, uptake, and translocation of 14C-glyphosate from track-spray applications and correlation to rainfastness in velvetleaf (Abutilon theophrasti) Weed Technol. 2000;14:127–132. [Google Scholar]
- 96.Ryerse JS, Downer RA, Sammons RD, Feng PC. Effect of glyphosate spray droplets on leaf cytology in velvetleaf (Abutilon theophrasti) Weed Sci. 2004;52:302–309. [Google Scholar]
- 97.Gougler JA, Geiger DR. Uptake and distribution of N-phosphonomethylglycine in sugar beet plants. Plant Physiol. 1981;68:668–672. doi: 10.1104/pp.68.3.668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mollenhauer C, Smart CC, Amrhein N. Glyphosate toxicity in the shoot apical region of the tomato plant. 1. Plastid swelling is the initial ultrastructural feature following in-vivo inhibition of 5-enolpyruvylshikimic acid-3-phosphate synthase. Pestic Biochem Phys. 1987;29:55–65. [Google Scholar]
- 99.Hollander-Czytko H, Amrhein N. Sub cellular compartmentation of shikimic-acid and phenylalanine in buckwheat (Fagopyrum esculentum) cell suspension cultures grown in the presence of shikimate pathway inhibitors. Plant Sci Lett. 1983;29:89–96. [Google Scholar]
- 100.Ge X, d'Avignon DA, Ackerman JJH, Sammons RD. Observation and identification of 2-C-methyl-D-erythritol-2,4-cyclopyrophosphate in horseweed and ryegrass treated with glyphosate. Pestic Biochem Phys. 2012;104:187–191. [Google Scholar]
- 101.Geiger DR, Bestman HD. Self-limitation of herbicide mobility by phytotoxic action. Weed Sci. 1990;38:324–329. [Google Scholar]
- 102.Feng PCC, Chiu T, Sammons RD. Glyphosate efficacy is contributed by its tissue concentration and sensitivity in velvetleaf (Abutilon theophrasti) Pestic Biochem Phys. 77:83–91. [Google Scholar]
- Phillips MA, Kaplan AP, Rutter WJ, Bartlett PA. Transition-state characterization: a new approach combining inhibitor analogues and variation in enzyme structure. Biochemistry. 1992;31:959–963. doi: 10.1021/bi00119a003. [DOI] [PubMed] [Google Scholar]
- 104.Cornish-Bowden A. Why is uncompetitive inhibition so rare? A possible explanation, with implications for the design of drugs and pesticides. FEBS Lett. 1986;203:3–6. doi: 10.1016/0014-5793(86)81424-7. [DOI] [PubMed] [Google Scholar]
- 105.Preston C, Wakelin AM, Dolman FC, Bostamam Y, Boutsalis P. A decade of glyphosate-resistant Lolium around the world: mechanisms, genes, fitness, and agronomic management. Weed Sci. 2009;57:435–441. [Google Scholar]
- 106.Bostamam Y, Malone JM, Dolman FC, Boutsalis P, Preston C. Rigid ryegrass (Lolium rigidum) populations containing a target site mutation in EPSPS and reduced glyphosate translocation are more resistant to glyphosate. Weed Sci. 2012;60:474–479. [Google Scholar]
- 107.Kaundun SS, Dale RP, Zelaya IA, Dinelli G, Marotti I, McIndoe E, et al. A novel P106L mutation in EPSPS and an unknown mechanism(s) act additively to confer resistance to glyphosate in a South African Lolium rigidum population. J Agric Food Chem. 2011;59:3227–3233. doi: 10.1021/jf104934j. [DOI] [PubMed] [Google Scholar]
- 108.Alarcón-Reverte R, García A, Urzúa J, Fischer AJ. Resistance to glyphosate in junglerice (Echinochloa colona) from California. Weed Sci. 2012;61:48–54. [Google Scholar]
- 109.Jasieniuk M, Ahmad R, Sherwood AM, Firestone JL, Perez-Jones A, Lanini WT, et al. Glyphosate-resistant Italian ryegrass (Lolium multiflorum) in California: distribution, response to glyphosate, and molecular evidence for an altered target enzyme. Weed Sci. 2008;56:496–502. [Google Scholar]
- 110.Ng CH, Wickneswari R, Salmijah S, Teng YT, Ismail BS. Gene polymorphisms in glyphosate-resistant and -susceptible biotypes of Eleusine indica from Malaysia. Weed Res. 2003;43:108–115. [Google Scholar]
- 111.Ng CH, Wickneswary R, Salmijah S, Teng YT, Ismail BS. Glyphosate resistance in Eleusine indica (L.) Gaertn. from different origins and polymerase chain reaction amplification of specific alleles. Aust J Agric Res. 2004;55:407–414. [Google Scholar]
- 112.Kaundun SS, Zelaya IA, Dale RP, Lycett AJ, Carter P, Sharpies KR, et al. Importance of the P106S target-site mutation in conferring resistance to glyphosate in a goosegrass (Eleusine indica) population from the Philippines. Weed Sci. 2008;56:637–646. [Google Scholar]
- 113.Gonzalez-Torralva F, Gil-Humanes J, Barro F, Brants I, De Prado R. Target site mutation and reduced translocation are present in a glyphosate-resistant Lolium multiflorum Lam. biotype from Spain. Plant Physiol Biochem. 2012;58:16–22. doi: 10.1016/j.plaphy.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 114.Perez-Jones A, Park KW, Polge N, Colquhoun J, Mallory-Smith CA. Investigating the mechanisms of glyphosate resistance in Lolium multiflorum. Planta. 2007;226:395–404. doi: 10.1007/s00425-007-0490-6. [DOI] [PubMed] [Google Scholar]
- 115.Wakelin AM, Preston C. A target-site mutation is present in a glyphosate-resistant Lolium rigidum population. Weed Res. 2006;46:432–440. [Google Scholar]
- 116.Simarmata M, Penner D. The basis for glyphosate resistance in rigid ryegrass (Lolium rigidum) from California. Weed Sci. 2008;56:181–188. [Google Scholar]
- 117.Yu Q, Cairns A, Powles S. Glyphosate, paraquat and ACCase multiple herbicide resistance evolved in a Lolium rigidum biotype. Planta. 2007;225:499–513. doi: 10.1007/s00425-006-0364-3. [DOI] [PubMed] [Google Scholar]
- 118.Eichholtz DA, Gasser CS, Kishore GM. Modified gene encoding glyphosate-tolerant 5-enolpyruvyl-3-phosphoshikimate synthase. US Patent 6225114, Monsanto Company, pp. 1–39 (2001)
- 119.Eichholtz DA, Gasser CS, Kishore GM. Glyphosate-tolerant 3-enolpyruvyl-3-phosphoshikimate synthases. 1994. pp. 1–31. US Patent 5310667, Monsanto Company.
- 120.Alibhai MF, CaJacob C, Feng PCC, Heck GR, Qi Y, Flasinski S, et al. Glyphosate resistant Class I 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS) 2010. pp. 1–49. US Patent 7723575 B2, Monsanto Technology.


