Abstract
Reported concentrations for endocannabinoids and related lipids in biological tissues can vary greatly; therefore, methods used to quantify these compounds need to be validated. This report describes a method to quantify anandamide (AEA), oleoylethanolamide (OEA) and palmitoylethanolamide (PEA) from rodent brain tissue. Analytes were extracted using acetonitrile without further sample clean up, resolved on a C18 reverse-phase column using a gradient mobile phase and detected using electrospray ionization in positive selected ion monitoring mode on a single quadrupole mass spectrometer. The method produced high recovery rates for AEA, OEA and PEA, ranging from 98.1% to 106.2%, 98.5% to 102.2% and 85.4% to 89.5%, respectively. The method resulted in adequate sensitivity with a lower limit of quantification for AEA, OEA and PEA of 1.4 ng/mL, 0.6 ng/mL and 0.5 ng/mL, respectively. The method was reproducible as intraday and interday accuracies and precisions were under 15%. This method was suitable for quantifying AEA, OEA and PEA from rat brain following pharmacological inhibition of fatty acid amide hydrolase.
Keywords: Endocannabinoids, Acylethanolamides, Anandamide, OEA, PEA, LC–MS
1. Introduction
The discovery of the endocannabinoid (eCB) system in the early 1990s intensified research on cannabinoid pharmacology, physiology and pathophysiology. To date, two cannabinoid receptors have been identified, including the CB1 receptor [1] and the CB2 receptor [2]. Additionally, two primary endogenous ligands have been discovered, including N-arachidonylethanolamide (anandamide, AEA) [3] and 2-arachidonylglycerol (2-AG) [4]. Since these seminal discoveries, the complexities of the eCB system have become increasingly apparent. For example, it is now known that multiple receptors, including the transient receptor potential cation channel V1 (TRPV1) [5], G-protein receptor 55 (GPR55) [6] and peroxisome proliferator-activated receptor (PPAR) isoforms [7], have the capacity to bind cannabinoid ligands. Additionally, 2-AG and AEA, although the most widely studied, are not the only endogenous ligands with the capacity to bind cannabinoid receptors [8]. Moreover, other bioactive lipids, particularly the N-acylethanolamides (NAEs), acylglycerols, and acylamides have been shown to be important in cannabinoid physiology as they act as entourage compounds by enhancing the activity of AEA and/or 2-AG [9]. For example, both palmitoylethanolamide (PEA) and oleoylethanolamide (OEA) have been shown to reduce AEA degradation by competing for fatty acid amide hydrolase (FAAH) [10], [11] and PEA can decrease FAAH expression [12]. Interestingly, OEA and PEA have the capability to displace [3H]-CP55,940 and [3H]-WIN55,212-2 binding to CB1 and CB2 receptors expressed in CHO cells [11]. It has also been demonstrated that both PEA and OEA can increase the affinity of AEA for TRPV1 receptors [13], an effect which contributes to the crosstalk between eCBs and other signaling systems. Due to these complexities, understanding eCB pharmacology, physiology and pathophysiology requires investigation of these lipid classes as a whole, rather than in isolation. Therefore, it is critical to develop analytical techniques that can simultaneously measure several eCBs and entourage compounds from biological tissues under normal and pathological conditions. Furthermore, development of easy, accurate, and reproducible analytical techniques to monitor eCBs and related lipids will aid preclinical and clinical testing of agents that modulate the eCB system, such as FAAH inhibitors.
A variety of analytical methods have been developed for the measurement of eCBs and other related lipids since their discovery [14], [15]. Many of the initial methods were developed using gas chromatography–mass spectroscopy (GC–MS) instrumentation to measure AEA [16] and/or 2-AG [17] and had on-column sensitivity in the femtamolar (fmol) to picomolar (pmol) range. For example, Giuffrida et al. [18], using GC–electron ionization (EI)-MS, reported limits of detection (LOD) of 300, 200 and 100 fmol for AEA, OEA and PEA, respectively, while another report by Maccarrone et al. [19] achieved sensitivity of 20±10 pmol for a variety of eCBs and related lipids, including AEA, OEA and PEA. However, eCBs typically need to be derivatized for GC–MS to increase volatility and sensitivity, which is a complex and time consuming task. Therefore, more recent methods for the quantification of eCBs and related lipids take advantage of liquid chromatography–mass spectroscopy (LC–MS) [18], [20] and LC–MS–MS [21], [22], as additional derivatization procedures are not necessary for these methodologies. Additionally, LC–MS systems have fmol sensitivity and in many cases may have greater sensitivity than GC–MS for measuring eCB and related lipid species. For example, Chen et al. [20] reported a LC–electrospray ionization (ESI)-MS method with a lower limit of quantification (LLOQ) of 2.5 ng/mL (28.8 fmol on column) for AEA, and Koga et al. [23] reported a LOD of 200 fmol using a LC–atmospheric pressure chemical ionization (APCI)-MS instrument. Further, reports describing methods for eCB quantification using LC–MS–MS have also reported LLOQ and LODs in the low fmol range [21], [22]. Specifically, Richardson et al. [21] reported a LOD of 25 fmol for AEA, OEA and PEA. Although MS–MS systems have benefits over MS systems for the measurement of eCBs and related compounds, such as increased selectivity and signal to noise ratios [15], single quadrupole systems may be the only available option for many laboratories. Therefore, development of methods for single quadrupole systems is valuable and necessary.
Extraction and sample preparation procedures for eCB and related lipids differ widely among published analytical methods. A chloroform/methanol solvent mixture is commonly used for extracting eCBs and related lipids from biological matrices; however, this is typically followed by sample clean-up using solid phase extraction (SPE) or thin-layer chromatography (TLC) [14]. Moreover, organic solvents such as toluene [24], ethyl acetate/hexane [21], [25] or acetonitrile (ACN) [20], [26] have also been used for protein precipitation and lipid extraction; however, many of the reported methods using these solvents still require further sample clean-up using SPE or TLC. For many extraction methods, it is necessary to further purify analytes of interest from high levels of other lipid constituents that can cause ion supression and peak interference [15]. However, SPE and TLC procedures are time consuming and expensive, and not ideal for analysis of multiple biological samples and/or high throughput drug discovery. Therefore, combined extraction and chromatographic methods that can reduce contamination, without using SPE or TLC, and adequately resolve analytes of interest, will significantly reduce the burden of measuring eCBs and related lipids in brain tissue.
The endogenous nature of eCBs and related lipids renders method validation using biological matrices rather difficult. In fact, many reported methods fail to use an appropriate matrix during method validation; however, this approach can negatively impact accurate quantification. Validation using surrogate matrices does not account for either ion suppression or analyte specific recovery from biological matrices, which can result in inaccurate estimation of analyte concentration. For example, reports that include validation in biological matrices have demonstrated that extraction efficiencies for eCBs and related lipids can deviate greatly depending on the analyte of interest [15], [21]. Therefore, potential interactions between the biological matrix and analytes should be taken into consideration when developing analytical methods.
In this study, simultaneous measurement of AEA, OEA and PEA in rodent whole brain tissue is reported. This report describes the first validated method to quantify this combination of analytes with a simple protein precipitation procedure followed by single quadrupole LC–MS detection, using low milligrams of brain tissue. Notably, this method was validated in whole brain matrix while accounting for the endogenous nature of these eCB and related lipid species. Additionally, the developed method was successfully implemented to quantify AEA, OEA and PEA in the rat hippocampus and entorhinal cortex following pharmacological inhibition of FAAH.
2. Materials and methods
2.1. Chemicals
Methanol and ACN were of HPLC grade, while all other chemicals used were of analytical grade. AEA, OEA, PEA and URB597 were purchased from Cayman Chemicals (Ann Arbor, MI, USA) and had a purity of ≥98%. Methanol and ACN were purchased from VWR International (Batavia, IL, USA), acetic acid was purchased from Fisher Scientific (Fairlawn, NJ, USA), ammonium acetate was purchased from Mallinckrodt Chemicals (Phillipsburg, NJ, USA), and ethanol was purchased from Pharmco-AAPER (Brookfield, CT, USA). Water was obtained from a Milli-Q® Advantage A10 purification and filtration system (Millipore, Billerica, MA, USA). Mobile phase was filtered at 0.2 μm using a nylon membrane filter (Supelco, Bellefonte, PA, USA).
2.2. Calibration and quality control sample preparation
Stock solutions of AEA, OEA and PEA were prepared in ethanol at 2.0 mg/mL and stored at −20 °C. From these stock solutions, 50 µg/mL working stocks and subsequent working solutions of appropriate concentration were prepared for each compound in ACN. From the working solutions, calibration and quality control (QC) samples were prepared for each compound by adding 5 µL of working solution of appropriate concentration to 95 µL of ACN or tissue homogenate. Calibration curves were constructed for AEA using the following calibration concentrations: 2.5, 5, 15, 25, 50 and 100 ng/mL, while curves for OEA and PEA were constructed using the following concentrations: 5, 10, 25, 50, 100, 250 and 500 ng/mL. QC samples for AEA were prepared at three concentration levels including 7.5, 35 and 75 ng/mL, while QC samples for OEA and PEA were prepared at 15, 30 and 90 ng/mL. The concentration ranges used for calibration and QC samples were chosen to encompass basal concentrations of AEA, OEA and PEA and concentrations anticipated following FAAH inhibition by URB597 [27], [28].
2.3. Sample extraction
AEA, OEA and PEA were extracted from rat brain tissue using a protein precipitation protocol modified from Chen et al. [20]. Brain tissue was weighed and then homogenized with equal volumes of ice cold saline in a siliconized microcentrifuge tube by rapid sonication on ice using a Sonic Dismembrator (Fisher Scientific, Fairlawn, NJ, USA). Following sonication, 100 µL of homogenate was transferred to a fresh siliconized microcentrifuge tube containing 1 mL of ACN. Samples were then vortexed for approximately 30 s and centrifuged at 13,000g for 20 min at 4 °C. ACN extracts were then transferred to a 5 mL siliconized test tube and gently evaporated under nitrogen at 37 °C. Following evaporation, samples were reconstituted in 100 µL ACN. To ensure maximum reconstitution of analytes of interest from the dry residue, samples were vortexed for 30 s, sonicated in an ice cold water bath for 15 min and vortexed again for 30 s. Samples were then transferred to siliconized microcentrifuge tubes and centrifuged at 13,000g at 4 °C to remove precipitates following reconstitution. Finally, the reconstituted samples were transferred to HPLC vials fitted with siliconized low volume inserts and placed in a temperature regulated autosampler (4 °C) for analysis. A 20 μL aliquot of sample was injected for LC–MS quantification. To ensure maximal extraction of analytes of interest, the effect of one vs. three extraction cycles on analyte recovery was investigated. To that end, homogenates were created and extracted one or three times according to the procedures described above and the relative MS signal between one and three extractions was compared as percent of one extraction cycle. This preliminary experiment showed that multiple extraction cycles were necessary to achieve maximal analyte recovery (Table 1). Therefore, for all subsequent validation experiments, analytes were extracted a total of three times and extracts were pooled before reconstitution by evaporating in the same test tube.
Table 1.
Effect of multiple extraction cycles on analyte recovery.
| Analyte | 1 Cycle (n=3) |
3 Cycles (n=3) |
||
|---|---|---|---|---|
| Mean (% C1) | CV (%) | Mean (% C1) | CV (%) | |
| AEA | 100.0 | 14.1 | 121.4 | 11.4 |
| OEA | 100.0 | 8.9 | 163.7 | 3.7 |
| PEA | 100.0 | 8.0 | 218.4 | 3.3 |
2.4. LC–MS conditions
HPLC was performed using a Waters Alliance® 2695 LC pump (Waters, Milford, MA, USA) equipped with a Waters Alliance® 2695 autosampler and thermostatic column compartment which was maintained at 37 °C. Separation was achieved using a Waters Symmetry® C18 (2.1 mm×150 mm, 5 µm) column coupled with a Waters Symmetry® C18 guard column (2.1 mm×10 mm, 3.5 µm). A gradient elution protocol was adapted from Patel et al. [26]. Mobile phase A consisted of 1 mM ammonium acetate and 0.1% acetic acid (v/v) in methanol and mobile phase B consisted of 1 mM ammonium acetate, 0.1% acetic acid (v/v) and 5% methanol in water. Initial conditions were set at 70% A and 30% B. A was increased linearly to 85% over 25 min and maintained for 1 min, then increased linearly to 100% over 1 min and held at 100% for 5 min. Finally, A was returned linearly to 70% over 1 min and held for 10 min for column equilibration. Flow rate was maintained at 0.3 mL/min. The MS detector used was a Micromass® ZQ™ (Waters, Milford, MA, USA) with an ESI probe. The MS conditions were set according to Chen et al. [20]: nitrogen desolvation gas 450 L/h, nitrogen cone gas 50 L/h, source temperature 120 °C, desolvation temperature 250 °C, capillary voltage 3.5 kV, cone voltage 25 kV, extractor voltage 5.0 kV and RF lens voltage 0.5 kV. ESI was set to the positive mode and selective ion monitoring was set to the following protonated ions, m/z 348.28 [M+H]+ (AEA), m/z 326.6 [M+H]+ (OEA) and m/z 300.5 [M+H]+ (PEA) with dwell times of 0.3 s for each ion.
2.5. Validation
The method was validated by examining linearity of standard curves, LLOQ, intra- and interday accuracy, intra- and interday precision, process efficiency (PE) and short-term analyte stability in rat brain extract at 4 °C. AEA, OEA and PEA linearity was evaluated over concentrations ranging from 2.5 to 100 ng/mL for AEA and 5 to 500 ng/mL for OEA and PEA in ACN and brain tissue by performing linear regression analysis. The LLOQ for each analyte was defined as the lowest concentration producing a peak height (signal) 10× greater than the baseline height (noise). This concentration was back calculated from a linear regression analysis of signal:noise vs. analyte concentration at analyte levels ranging from 2.5 to 25 ng/mL. Accuracy and precision were determined for each analyte at three different QC levels. Intraday QC's were run in triplicate on two separate occasions and accuracies and precisions were reported for intraday 1 and intraday 2. Interday QC's (n=8) were run on separate days with each day ranging from 1 to 3 replications. Accuracy was calculated as the following, accuracy=(calculated concentration/nominal concentration) ×100, while precision (% coefficient of variation, CV) was calculated as, %CV=(standard deviation/mean calculated concentration)×100. Accuracy and precision were considered acceptable when within 15%. PE was calculated for each analyte at each QC level and was defined as the ratio between the relative MS signal of the QC extracted from brain homogenate and the relative MS signal of the QC in ACN; PE=(QC spiked/QC in ACN)×100. As endogenous AEA, OEA and PEA may interfere with standardization and validation, great care was taken to ensure uniformity of homogenates. To that end, bulk brain homogenate used for the preparation of calibration and QC samples was produced by sonication on ice, followed by vigorous vortexing. Consistent background levels of AEA, OEA and PEA were achieved with this method, as repeated background measurements (n=5–7) varied by only 8.1%, 6.7% and 3.3%, respectively. Background signal of each analyte was subtracted prior to calculation of the measured calibrators and QC concentrations. To study the short-term stability of AEA, OEA and PEA, extracts were stored in the autosampler at 4 °C and injected at T=0 h and T=18 h. Relative MS signals between the two time points were compared as percent of T=0 h.
2.6. Biological application
Adult male Sprague-Dawley rats weighing approximately 330 g (n=12; Charles River, Raleigh, NC) were used in this study. All treatment protocols followed the Guide for the Care and Use of Laboratory Animals by the National Research Council (1996) and were approved by the University of Kentucky Institutional Animal Care and Use Committee. Rats were singly housed in Plexiglas cages in a University of Kentucky vivarium on a 12 h light/dark cycle with access to rat chow and water ad libitum. Rats were allowed to acclimate to housing conditions for 5 days and were handled for at least 3 days. During experimentation, rat chow was removed from cages and rats were fed a nutritionally complete diet consisting of Vanilla Ensure Plus© (Abbott Laboratories, Columbus, OH) and dextrose in water. Rats received diet by oral gavage every 8 h for 24 h. Rats were treated with the FAAH inhibitor, URB597 (0.3 mg/kg) by intraperitoneal injection at a concentration of 1.0 mg/mL in dimethylsulfoxide (DMSO). Two hours after injection, rats were euthanized by rapid decapitation; brains were extracted; and hippocampi and entorhinal cortices were dissected, placed in microcentrifuge tubes and flash frozen using a slurry of dry ice and 70% ethanol. The entire process from decapitation to rapid freezing was kept under 6 min to minimize post-mortem eCB and NAE accumulation [15]. Tissue was stored at −80 °C until further processing. Fifty mg of tissue was sufficient to achieve quantifiable levels of AEA, OEA and PEA in the hippocampus and entorhinal cortex. QCs were run prior to batch analysis of samples and periodically during analysis to ensure fidelity of the analytical method.
2.7. Statistics
LC–MS data were analyzed using Quanlynx (Waters, Milford, MA, USA) and analyte concentrations and LLOQs were determined using peak area and peak height, respectively. Statistics were performed using Prism (GraphPad version 4.03, La Jolla, CA, USA). Linear regression was used to assess linearity of calibration curves for AEA, OEA and PEA. Student's t-tests were used to compare differences in AEA, OEA and PEA content between vehicle and URB597 treated rats for each brain region. Statistical significance was accepted at p<0.05. Values are given as mean±SD unless otherwise indicated.
3. Results and discussion
3.1. Method development
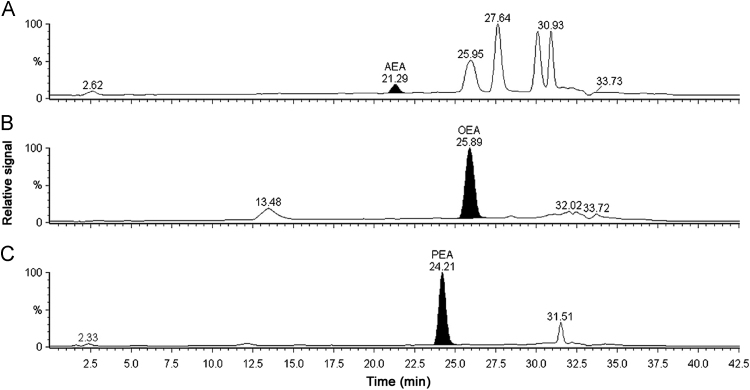
This report describes a LC–MS method for quantifying AEA, OEA and PEA in rat brain tissue. The LC protocol used in the current report was adapted from Chen et al. [20], with modification to prevent peak interference from co-eluting analytes. AEA, OEA and PEA eluted at 21.29 min, 25.89 min and 24.21 min, respectively, as shown in Fig. 1. Quantification of eCBs and related lipids using LC–MS typically requires long run times [18], [20]. Therefore, method optimization towards shorter elution times can provide significant advantages when analyzing biological samples such as decreasing the possibility of sample instability in the autosampler and increasing sample throughput. With the current protocol, run time was decreased by 7 min relative to Chen et al. [20]. Moreover, during method development, a final step involving washing with 100% methanol for 5 min was incorporated into the gradient protocol to ensure elution of analytes that would otherwise cause interference on subsequent runs.
Fig. 1.
Representative LC–MS chromatograms of analytes in whole brain tissue. (A) AEA, retention time (RT) 21.29 min, (B) OEA, RT 25.89 min and (C) PEA, RT 24.21 min.
Recovery of eCBs and related lipids from biological tissues is performed using liquid–liquid extraction/precipitation protocols [14], [15] and different solvent conditions can result in varying extraction efficiencies for specific analytes [29]. Thus, preliminary experiments were performed in order to determine the optimum solvent or solvent combination for simultaneously extracting AEA, OEA and PEA. These solvents included ACN [20], [26], [30], methanol [15] and a 9:1 ethyl acetate/hexane mixture [21], [25]. ACN and methanol extractions produced similar chromatograms and extraction efficiencies, while the ethyl acetate/hexane extraction was more time consuming and did not offer any significant benefit over the other two protocols (data not shown). Considering the well-documented superiority of ACN over methanol as a protein precipitant [31], ACN was selected as an extraction solvent. To further optimize analyte extraction, the effect of one vs. three extraction cycles was determined to ensure maximal analyte recovery from brain tissue homogenates. The effect of one and three extraction cycles is shown in Table 1. Repeated extractions enhanced the relative recovery of each analyte while the CV remained at acceptable levels (<15%). Interestingly, a correlation was observed between the relative endogenous abundance of each analyte and the magnitude of recovery following repeated extraction. The majority of AEA, which has low endogenous levels (low pmol range), was efficiently extracted following 1 extraction cycle, while recovery of PEA, found at higher endogenous levels (high pmol range), was greatly enhanced after 3 extractions (118.4% increase). Thus, this study suggests that multiple extractions with 1 mL of ACN are required to overcome a limited capacity of ACN to extract eCBs and related lipids from brain tissue.
3.2. Validation
Calibration curves were linear for AEA (R2=0.999) over a concentration range of 2.5–100 ng/mL and linear for OEA (R2=0.999) and PEA (R2=0.989) over a concentration range of 5–500 ng/mL. The concentration ranges for calibration curves were chosen to span across anticipated basal levels of endogenous AEA, OEA and PEA and levels expected following FAAH inhibition. The calculated LLOQs for AEA, OEA and PEA were 1.4, 0.6 and 0.5 ng/mL, respectively. This analytical method was found accurate, precise and reproducible for the simultaneous measurement of AEA, OEA and PEA in rodent brain tissue (Table 2). Intraday and interday accuracies were generally within 15% of the nominal concentration and CVs were also generally well below 15%. These results are acceptable for the developed method; however, the slight discrepancies reported in Table 2 are attributed to the endogenous nature of AEA, OEA and PEA in biological tissues, which needs to be accounted for while validating analytical methods. eCBs and related lipids are notoriously difficult to quantify accurately because their endogenous nature adds variability during method validation. Methods using biological matrices for validation have to account for endogenous analyte levels and correct for these basal levels to calculate calibration and QC concentrations. However, this can be a difficult task because basal levels can vary greatly in tissue homogenates. For example, it is well known that eCB and related lipids accumulate postmortem and during sample processing [15], [16]. To reduce variability, tissue needs to be rapidly dissected, flash frozen and kept ice cold during processing. However, tissue processing procedures are not standardized in the literature [21], [22], [25], which may partially account for varying estimations of eCB and NAE tissue content reported in the literature [20], [21]. In this report, bulk homogenates were created each day and great care was taken to ensure uniformity by vigorously vortexing the homogenate while ice cold. Additionally, new background measurements were acquired each day and for every batch of homogenate. Comparatively, in Richardson et al. [21], homogenates were prepared by freezing tissue samples in liquid nitrogen prior to being ground up using a mortar and pestle and aliquoted for validation experiments. It is possible that these differences in preparation could result in differences in homogenate quality and therefore account for seemingly different accuracies and precisions reported in the literature.
Table 2.
Linearity, intra- and interday accuracy and precision of analytical method for NAE measurement from brain matrix.
| Analyte | R2 | Nominal concentration (ng/mL) | Intraday 1 (n=3) |
Intraday 2 (n=3) |
Interday (n=8) |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean (ng/mL) | Accuracy (%) | CV (%) | Mean (ng/mL) | Accuracy (%) | CV (%) | Mean (ng/mL) | Accuracy (%) | CV (%) | |||
| AEA | 0.999 | 7.5 | 8.6 | 115.0 | 9.4 | 7.9 | 105.8 | 1.3 | 8.2 | 109.5 | 7.8 |
| 35 | 31.7 | 105.7 | 6.2 | 34.2 | 114.0 | 6.1 | 33.2 | 110.8 | 7.5 | ||
| 75 | 68.1 | 90.8 | 7.5 | 72.5 | 96.7 | 9.1 | 68.5 | 91.4 | 8.2 | ||
| OEA | 0.989 | 15 | 18.2 | 121.5 | 18.8 | 14.8 | 98.4 | 5.5 | 15.9 | 105.8 | 17.6 |
| 30 | 30.9 | 102.9 | 5.1 | 25.1 | 83.6 | 2.8 | 28.4 | 94.8 | 13.3 | ||
| 90 | 85.8 | 95.4 | 5.3 | 90.6 | 100.7 | 4.2 | 85.9 | 95.4 | 6.1 | ||
| PEA | 0.999 | 15 | 14.9 | 99.4 | 11.6 | 13.9 | 93.0 | 3.6 | 14.3 | 95.0 | 8.6 |
| 30 | 28.5 | 95.0 | 13.2 | 29.1 | 97.1 | 7.2 | 28.6 | 95.8 | 8.7 | ||
| 90 | 94.8 | 105.3 | 5.4 | 80.7 | 89.6 | 4.9 | 88.9 | 98.8 | 8.9 | ||
In order to circumvent the obstacles associated with the endogenous nature of eCBs and related lipids, some reports use surrogate matrices. However, opinions on whether or not the use of artificial matrices is appropriate while developing analytical methods vary in the literature. Some reports use alternative matrices commonly consisting of water and bovine serum albumin (BSA) for method validation [22], [30]. On the other hand, other reports use biological matrices and therefore can adjust for extraction efficiency and potential matrix effects for every analyte of interest [20], [21], [25]. Adjusting for extraction efficiency and matrix effects is particularly important when not using an internal standard or when only using a representative internal standard for multiple analytes during validation and sample quantification [20]. For the current study, the later approach was used for the reasons mentioned above.
PE was examined for the extraction of AEA, OEA and PEA from the whole brain homogenates (Table 3). Concentration levels to study PE were selected to reflect both reported levels of endogenous AEA, OEA and PEA [27] and results from preliminary analysis of analyte content. PE values for AEA and OEA ranged from 98.1% to 106.2% and 98.5% to 102.2%, respectively (Table 3). These high PE values indicate that the current extraction method was optimal and that the method did not result in a matrix effect. Moreover, PE for PEA ranged from 85.4% to 89.5%. Although a lower PE was consistently observed for PEA compared to AEA and OEA, the calculated CV was acceptable and accurate and precise quantification of PEA was achieved, as indicated in Table 2. These high process efficiencies achieved are consistent with previous literature using other extraction procedures [21], [22]. Noteworthy, although it appears that significant matrix–analyte interactions were not encountered, it is possible that this could occur when validating and quantifying other related lipid species using the present method.
Table 3.
Process efficiency.
| Analyte | Nominal concentration (ng/mL) | PE (%, n=3) | CV (%) |
|---|---|---|---|
| AEA | 7.5 | 106.2 | 2.4 |
| 35 | 99.2 | 2.3 | |
| 75 | 98.1 | 9.6 | |
| OEA | 15 | 102.2 | 2.2 |
| 30 | 99.5 | 16.5 | |
| 90 | 98.5 | 6.8 | |
| PEA | 15 | 89.5 | 10.4 |
| 30 | 85.4 | 17.1 | |
| 90 | 85.8 | 8.1 | |
Due to long sample run times (43 min), short-term sample stability was examined in the autosampler at 4 °C. The MS signals for AEA, OEA and PEA at T=18 h were 130%, 112.4% and 107% of T=0 h, respectively (Table 4). These data suggest that degradation processes are not occurring under the specified storage conditions. However, in order to reduce inflation of estimated analyte concentrations, samples were not allowed to remain in the autosampler longer than 12 h before analysis.
Table 4.
Short-term analyte stability at 4 °C.
| Analyte | 0 h (n=3) |
18 h (n=3) |
||
|---|---|---|---|---|
| Mean (% 0 h) | CV (%) | Mean (% 0 h) | CV (%) | |
| AEA | 100.0 | 13.1 | 130.5 | 2.3 |
| OEA | 100.0 | 3.3 | 112.4 | 3.5 |
| PEA | 100.0 | 3.7 | 107.0 | 4.0 |
3.3. Biological application
The developed method was used to quantify AEA, OEA and PEA in the rat hippocampus and entorhinal cortex following administration of the FAAH inhibitor, URB597. FAAH is the major enzyme responsible for the degradation of NAEs [32], is expressed throughout the central nervous system (CNS) [32], [33] and thus is implicated in a variety of physiological and behavioral processes. For example, pharmacological inhibition and/or genetic deletion of FAAH modulates depressive-like behavior [34], reduces inflammatory pain [35], alters drug reward [36] and affords neuroprotection [37]. Therefore, FAAH is under intense investigation for its therapeutic utility in a variety of CNS disorders. In the present study, a significant elevation in AEA, OEA and PEA was observed 2 h following intraperitoneal administration of URB597 (Table 5). In the hippocampus, AEA, OEA and PEA tissue content was increased by 57.5%, 475.6% and 986.6%, respectively. Contrastingly, in the entorhinal cortex, an increase in AEA content was not observed and much smaller increases of 250.2% and 435.0% were observed for OEA and PEA, respectively. This is the first study to demonstrate brain region and compound specific alterations in eCB and NAE content following a single dose of URB597. Comparatively, these data are consistent with studies examining NAE content following chronic URB597 administration. After weeks of URB597 administration, Bortolato et al. [34] observed elevations of AEA in the midbrain, thalamus and striatum; however, this effect was absent in the prefrontal cortex and hippocampus [32]. Although the original characterization of URB597 reported elevations of AEA, OEA and PEA in the whole brain tissue following a single dose [28], the aforementioned study and the findings of this report suggest that FAAH inhibition results in brain region specific regulation of eCBs and NAEs, providing insight into the neurochemical effects of FAAH inhibition.
Table 5.
Effect of URB596 on endogenous levels of AEA, OEA and PEA.
| Analyte | Brain region | Vehicle | URB597 (0.3 mg/kg) |
|---|---|---|---|
| AEA (nmol/g tissue) | Hippocampus | 37.9±20.5 | 59.7±9.4⁎ |
| Entorhinal cortex | 43.3±12.0 | 44.0±12.4 | |
| OEA (nmol/g tissue) | Hippocampus | 82.9±11.4 | 477.2±90.2⁎⁎ |
| Entorhinal cortex | 171.5±146.6 | 300.1±56.0⁎⁎ | |
| PEA (nmol/g tissue) | Hippocampus | 155.7±60.1 | 1691.8±377.6⁎⁎ |
| Entorhinal cortex | 85.7±50.0 | 917.6±195.8⁎⁎ | |
Values are given as mean±SD.
p<0.05.
p<0.001 compared to respective vehicle group.
4. Conclusions
The current study describes a novel method for quantifying eCBs and related lipids with acceptable accuracy and precision. This method has been developed to quantify AEA, OEA and PEA from rodent brain tissue and offers multiple advantages over other validated methods. A simple extraction protocol was used without time consuming and costly sample clean-up, analytes were measured on a single quadrupole MS with satisfactory sensitivity, the method was validated using appropriate biological matrices, which accurately accounts for analyte–matrix interactions, and low milligrams of brain tissue such as bilateral adult rat hippocampi was sufficient for the quantification of AEA, OEA and PEA. Finally, the method was proven effective in detecting elevations in AEA, OEA and PEA in rodent hippocampi and entorhinal cortices following administration of the FAAH inhibitor, URB597.
Acknowledgments
This work was supported by funding from the National Institute of Alcohol Abuse and Alcoholism AA016959 (KN), AA016499 (KN) and AA019853 (DJL) and the Kentucky Science and Technology Corporation (KSTC-184-512-07-029; ALS). Sponsors had no contribution to the execution, interpretation or preparation of the current report.
Footnotes
Peer review under responsibility of Xi'an Jiaotong University.
References
- 1.Matsuda L.A., Lolait S.J., Brownstein M.J. Structure of a cannabinoid receptor and functional expression of the cloned cDNA. Nature. 1990;346:561–564. doi: 10.1038/346561a0. [DOI] [PubMed] [Google Scholar]
- 2.Munro S., Thomas K.L., Abu-Shaar M. Molecular characterization of a peripheral receptor for cannabinoids. Nature. 1993;365:61–65. doi: 10.1038/365061a0. [DOI] [PubMed] [Google Scholar]
- 3.Devane W.A., Hanus L., Breuer A. Isolation and structure of a brain constituent that binds to the cannabinoid receptor. Science. 1992;258:1946–1949. doi: 10.1126/science.1470919. [DOI] [PubMed] [Google Scholar]
- 4.Mechoulam R., Ben-Shabat S., Hanus L. Identification of an endogenous 2-monoglyceride, present in canine gut, that binds to cannabinoid receptors. Biochem. Pharmacol. 1995;50:83–90. doi: 10.1016/0006-2952(95)00109-d. [DOI] [PubMed] [Google Scholar]
- 5.Ross R.A. Anandamide and vanilloid TRPV1 receptors. Br. J. Pharmacol. 1993;140:790–801. doi: 10.1038/sj.bjp.0705467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sharir H., Abood M.E. Pharmacological characterization of GPR55, a putative cannabinoid receptor. Pharmacol. Ther. 2010;126:301–313. doi: 10.1016/j.pharmthera.2010.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun Y., Alexander S.P., Garle M.J. Cannabinoid activation of PPAR alpha; a novel neuroprotective mechanism. Br. J. Pharmacol. 2007;152:734–743. doi: 10.1038/sj.bjp.0707478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kano M., Ohno-Shosaku T., Hashimotodani Y. Endocannabinoid-mediated control of synaptic transmission. Physiol. Rev. 2009;89:309–380. doi: 10.1152/physrev.00019.2008. [DOI] [PubMed] [Google Scholar]
- 9.Bradshaw H.B., Walker J.M. The expanding field of cannabimimetic and related lipid mediators. Br. J. Pharmacol. 2005;144:459–465. doi: 10.1038/sj.bjp.0706093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bisogno T., Maurelli S., Melck D. Biosynthesis, uptake, and degradation of anandamide and palmitoylethanolamide in leukocytes. J. Biol. Chem. 1997;272:3315–3323. doi: 10.1074/jbc.272.6.3315. [DOI] [PubMed] [Google Scholar]
- 11.Jonsson K.O., Vandevoorde S., Lambert D.M. Effects of homologues and analogues of palmitoylethanolamide upon the inactivation of the endocannabinoid anandamide. Br. J. Pharmacol. 2001;133:1263–1275. doi: 10.1038/sj.bjp.0704199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Di Marzo V., Melck D., Orlando P. Palmitoylethanolamide inhibits the expression of fatty acid amide hydrolase and enhances the anti-proliferative effect of anandamide in human breast cancer cells. Biochem. J. 2001;358:249–255. doi: 10.1042/0264-6021:3580249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Petrocellis L., Davis J.B., Di Marzo V. Palmitoylethanolamide enhances anandamide stimulation of human vanilloid VR1 receptors. FEBS Lett. 2001;506:253–256. doi: 10.1016/s0014-5793(01)02934-9. [DOI] [PubMed] [Google Scholar]
- 14.Zoerner A.A., Gutzki F.M., Batkai S. Quantification of endocannabinoids in biological systems by chromatography and mass spectrometry: a comprehensive review from an analytical and biological perspective. Biochem. Biophys. Acta. 2011;1811:706–723. doi: 10.1016/j.bbalip.2011.08.004. [DOI] [PubMed] [Google Scholar]
- 15.Buczynski M.W., Parsons L.H. Quantification of brain endocannabinoid levels: methods, interpretations and pitfalls. Br. J. Pharmacol. 2010;160:423–442. doi: 10.1111/j.1476-5381.2010.00787.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kemp K., Hsu F.F., Bohrer A. Isotope dilution mass spectrometric measurements indicate that arachidonylethanolamide, the proposed endogenous ligand of the cannabinoid receptor, accumulates in rat brain tissue post mortem but is contained at low levels in or is absent from fresh tissue. J. Biol. Chem. 1996;271:17287–17295. doi: 10.1074/jbc.271.29.17287. [DOI] [PubMed] [Google Scholar]
- 17.Kondo S., Kondo H., Nakane S. 2-Arachidonoylglycerol, an endogenous cannabinoid receptor agonist: identification as one of the major species of monoacylglycerols in various rat tissues, and evidence for its generation through Ca2+-dependent and -independent mechanisms. FEBS Lett. 1998;429:152–156. doi: 10.1016/s0014-5793(98)00581-x. [DOI] [PubMed] [Google Scholar]
- 18.Giuffrida A., Rodriguez de Fonseca F., Piomelli D. Quantification of bioactive acylethanolamides in rat plasma by electrospray mass spectrometry. Anal. Biochem. 2000;280:87–93. doi: 10.1006/abio.2000.4509. [DOI] [PubMed] [Google Scholar]
- 19.Maccarrone M., Attina M., Cartoni A. Gas chromatography–mass spectrometry analysis of endogenous cannabinoids in healthy and tumoral human brain and human cells in culture. J. Neurochem. 2001;76:594–601. doi: 10.1046/j.1471-4159.2001.00092.x. [DOI] [PubMed] [Google Scholar]
- 20.Chen J., Paudel K.S., Derbenev A.V. Simultaneous quantification of anandamide and other endocannabinoids in dorsal vagal complex of rat brainstem by LC–MS. Chromatographia. 2009;69:1–7. doi: 10.1365/s10337-008-0841-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Richardson D., Ortori C.A., Chapman V. Quantitative profiling of endocannabinoids and related compounds in rat brain using liquid chromatography–tandem electrospray ionization mass spectrometry. Anal. Biochem. 2007;360:216–226. doi: 10.1016/j.ab.2006.10.039. [DOI] [PubMed] [Google Scholar]
- 22.Williams J., Wood J., Pandarinathan L. Quantitative method for the profiling of the endocannabinoid metabolome by LC–atmospheric pressure chemical ionization-MS. Anal. Chem. 2007;79:5582–5593. doi: 10.1021/ac0624086. [DOI] [PubMed] [Google Scholar]
- 23.Koga D., Santa T., Fukushima T. Liquid chromatographic–atmospheric pressure chemical ionization mass spectrometric determination of anadamide and its analogs in rat brain and peripheral tissues. J. Chromatogr. B. 1997;690:7–13. doi: 10.1016/s0378-4347(96)00391-x. [DOI] [PubMed] [Google Scholar]
- 24.Felder C.C., Nielsen A., Briley E.M. Isolation and measurement of the endogenous cannabinoid receptor agonist, anandamide, in brain and peripheral tissues of human and rat. FEBS Lett. 1996;393:231–235. doi: 10.1016/0014-5793(96)00891-5. [DOI] [PubMed] [Google Scholar]
- 25.Kingsley P.J., Marnett L.J. Analysis of endocannabinoids by Ag+ coordination tandem mass spectrometry. Anal. Biochem. 2003;314:8–15. doi: 10.1016/s0003-2697(02)00643-7. [DOI] [PubMed] [Google Scholar]
- 26.Patel S., Wohlfeil E.R., Rademacher D.J. The general anesthetic propofol increases brain N-arachidonylethanolamine (anandamide) content and inhibits fatty acid amide hydrolase. Br. J. Pharmacol. 2003;139:1005–1013. doi: 10.1038/sj.bjp.0705334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmid P.C., Krebsbach R.J., Perry S.R. Occurrence and postmortem generation of anandamide and other long-chain N-acylethanolamines in mammalian brain. FEBS Lett. 1995;375:117–120. doi: 10.1016/0014-5793(95)01194-j. [DOI] [PubMed] [Google Scholar]
- 28.Fegley D., Gaetani S., Duranti A. Characterization of the fatty acid amide hydrolase inhibitor cyclohexyl carbamic acid 3′-carbamoyl-biphenyl-3-yl ester (URB597): effects on anandamide and oleoylethanolamide deactivation. J. Pharmacol. Exp. Ther. 2005;313:352–358. doi: 10.1124/jpet.104.078980. [DOI] [PubMed] [Google Scholar]
- 29.Zhang M.Y., Gao Y., Btesh J. Simultaneous determination of 2-arachidonoylglycerol, 1-arachidonoylglycerol and arachidonic acid in mouse brain tissue using liquid chromatography/tandem mass spectrometry. J. Mass Spectrom. 2010;45:167–177. doi: 10.1002/jms.1701. [DOI] [PubMed] [Google Scholar]
- 30.Bradshaw H.B., Rimmerman N., Krey J.F. Sex and hormonal cycle differences in rat brain levels of pain-related cannabimimetic lipid mediators. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2006;291:R349–358. doi: 10.1152/ajpregu.00933.2005. [DOI] [PubMed] [Google Scholar]
- 31.Polson C., Sarkar P., Inchedon B. Optimization of protein precipitation based upon effectiveness of protein removal and ionization effect in liquid chromatography–tandem mass spectrometry. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2003;785:263–275. doi: 10.1016/s1570-0232(02)00914-5. [DOI] [PubMed] [Google Scholar]
- 32.Cravatt B.F., Giang D.K., Mayfield S.P. Molecular characterization of an enzyme that degrades neuromodulatory fatty-acid amides. Nature. 1996;384:83–87. doi: 10.1038/384083a0. [DOI] [PubMed] [Google Scholar]
- 33.Thomas E.A., Cravatt B.F., Danielson P.E. Fatty acid amide hydrolase, the degradative enzyme for anandamide and oleamide, has selective distribution in neurons within the rat central nervous system. J. Neurosci. Res. 1997;50:1047–1052. doi: 10.1002/(SICI)1097-4547(19971215)50:6<1047::AID-JNR16>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 34.Bortolato M., Mangieri R.A., Fu J. Antidepressant-like activity of the fatty acid amide hydrolase inhibitor URB597 in a rat model of chronic mild stress. Biol. Psychiatry. 2007;62:1103–1110. doi: 10.1016/j.biopsych.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 35.Naidu P.S., Kinsey S.G., Guo T.L. Regulation of inflammatory pain by inhibition of fatty acid amide hydrolase. J. Pharmacol. Exp. Ther. 2010;334:182–190. doi: 10.1124/jpet.109.164806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scherma M., Panlilio L.V., Fadda P. Inhibition of anandamide hydrolysis by cyclohexyl carbamic acid 3′-carbamoyl-3-yl ester (URB597) reverses abuse-related behavioral and neurochemical effects of nicotine in rats. J. Pharmacol. Exp. Ther. 2008;327:482–490. doi: 10.1124/jpet.108.142224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hwang J., Adamson C., Butler D. Enhancement of endocannabinoid signaling by fatty acid amide hydrolase inhibition: a neuroprotective therapeutic modality. Life Sci. 2010;86:615–623. doi: 10.1016/j.lfs.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]