Abstract
Effects of exosomes present in human plasma on immune cells have not been examined in detail. Immunological studies with plasma-derived exosomes require their isolation by procedures involving ultracentrifugation. These procedures were largely developed using supernatants of cultured cells. To test biologic activities of plasma-derived exosomes, methods are necessary that ensure adequate recovery of exosome fractions free of contaminating larger vesicles, cell fragments and protein/nucleic acid aggregates. Here, an optimized method for exosome isolation from human plasma/serum specimens of normal controls (NC) or cancer patients and its advantages and pitfalls are described. To remove undesirable plasma-contaminating components, ultrafiltration of differentially-centrifuged plasma/serum followed by size-exclusion chromatography prior to ultracentrifugation facilitated the removal of contaminants. Plasma or serum was equally acceptable as a source of exosomes based on the recovered protein levels (in μg protein/mL plasma) and TEM image quality. Centrifugation on sucrose density gradients led to large exosome losses. Fresh plasma was the best source of morphologically-intact exosomes, while the use of frozen/thawed plasma decreased exosome purity but not their biologic activity. Treatments of frozen plasma with DNAse, RNAse or hyaluronidase did not improve exosome purity and are not recommended. Cancer patients’ plasma consistently yielded more isolated exosomes than did NCs’ plasma. Cancer patients’ exosomes also mediated higher immune suppression as evidenced by decreased CD69 expression on responder CD4+ T effector cells. Thus, the described procedure yields biologically-active, morphologically-intact exosomes that have reasonably good purity without large protein losses and can be used for immunological, biomarker and other studies.
Keywords: Exosome isolation, human plasma, size-exclusion chromatography, immunological studies, exosome characteristics
Introduction
Exosomes are virus-sized vesicles, ranging from 20–100nm in size, and enveloped by a phospholipid membrane (Thery et al., 2002). Formed within the endocytic compartments, exosomes are released into the extracellular space via fusion of multivesicular bodies with the cell surface membrane (Thery et al., 2002). They are produced by normal as well as malignant cells and are present in all human body fluids, including blood (Hawari et al., 2004; Graner et al., 2009), urine (Pisitkun et al., 2004), cerebrospinal fluid (Harrington et al., 2009; Street et al., 2012) and ascites (Runz et al., 2007). Exosomes carry multiple membrane-tethered, biologically-active molecules, and their molecular cargo mimics the surface molecular profile of the mother cell (Skog et al., 2008; Mathivanan et al., 2010). Vesicular content of exosomes includes nucleic acids, enzymes, soluble factors and a variety of molecules derived from the cytosol of mother cells (Mathivanan and Simpson, 2009). Tumor cells are avid exosome producers, and we have previously shown that these tumor-derived exosomes, in contrast to exosomes produced by normal cells, exert immunosuppressive effects, impairing survival and functions of adaptive and innate immune cells (Taylor et al., 2003; Kim et al., 2005).
The nomenclature adopted for exosomes is based on their size and a mode of release from mother cells, with vesicles ranging in diameter from 30–150nm and formed as intraluminal vesicles by budding into multivesicular endosomes (MVEs) considered to be exosomes. In contrast, larger (200–500nm) vesicles which bud directly from the plasma membrane are referred to as “microvesicles” (MVs) (Raposo and Stoorvogel, 2014). There is also some evidence that microvesicles might have properties and functions distinct from exosomes (Raposo and Stoorvogel, 2014).
Studies of exosomes have been largely performed using supernatants of cultured cells. This is because in a cell culture, the origin of exosomes can be determined and because relatively simple chemical composition of most culture media facilitates isolation of exosomes devoid of ‘contaminating’ proteins, lipids and sugars. Human or animal serum used as a supplement for cell cultures is generally ultracentrifuged prior to its use to remove “contaminating” extracellular vesicles. Isolation of exosomes from plasma or other body fluids is more complex for several reasons. First, the source of exosomes present in plasma is unknown, as plasma contains a mix of exosomes derived from many different cells in varying proportions. Second, plasma exosomes are ‘coated’ with proteins, glycoproteins or glycolipids likely to cause their aggregation and a potential loss upon subsequent centrifugation. Third, separation of exosomes derived from a specific cell type is a considerable problem, as the vast majority of plasma exosomes originate from erythrocytes and platelets, and the desired exosomal fraction may represent only a small fraction of the total (Vlassov et al., 2012; Witwer et al., 2013; Gyorgy et al., 2014; Van der Meel et al., 2014).
Few attempts have been made to optimize exosome isolation from human plasma (Witwer et al., 2013). The most commonly used technique to isolate exosomes relies on their physical properties: because they are virus-size, they can only be isolated by ultracentrifugation and have a characteristic buoyancy of 1.10 to 1.19 g/mL on continuous sucrose density gradients (Thery et al., 2006; Keller et al., 2007). However, human body fluids represent complex mixtures of many components, including vesicles of several different sizes, protein complexes, protein-nucleic acid aggregates and subcellular fragments. Thus, isolation of well-defined exosomal fractions free of “contaminating” vesicular and non-vesicular components from human plasma for biologic studies is a challenging task. The tendency of exosomes to form aggregates of varying sizes results in losses, which further complicate their recovery.
We have previously used size-exclusion chromatography originally described by Taylor et al. (Taylor et al., 2002) prior to ultracentrifugation to isolate purified exosomes from cell supernatants or human plasma (Taylor et al., 2003; Kim et al., 2005). In this manuscript, we show that application of this methodology following differential centrifugation of human plasma and ultrafiltration results in increased purity and greater recovery of morphologically and functionally intact exosomes. In view of increasing recognition of exosomes as potentially useful biomarkers of disease, our goal was to obtain pure exosomal fractions and to optimize their quantitative recovery from human fresh or banked plasma specimens.
Materials and Methods
1.1. Peripheral blood specimens
Buffy coats obtained from healthy volunteers were purchased from the Central Blood Bank of Pittsburgh. Under an IRB-approved protocol (IRB #991206) venous blood samples were obtained from cancer patients or normal controls (NC). All subjects signed an informed consent prior to blood draws. Venous blood was collected into tubes with or without added heparin. Heparinized blood was centrifuged on Ficoll-Hypaque gradients (GE Healthcare Bioscience). PBMC were recovered, washed in AIM-V medium (Invitrogen, Grand Island, NY, USA) and were immediately used for experiments. Plasma or serum was harvested and either immediately used or aliquoted into 2mL vials and banked in liquid N2 until future use.
1.2. Exosome isolation
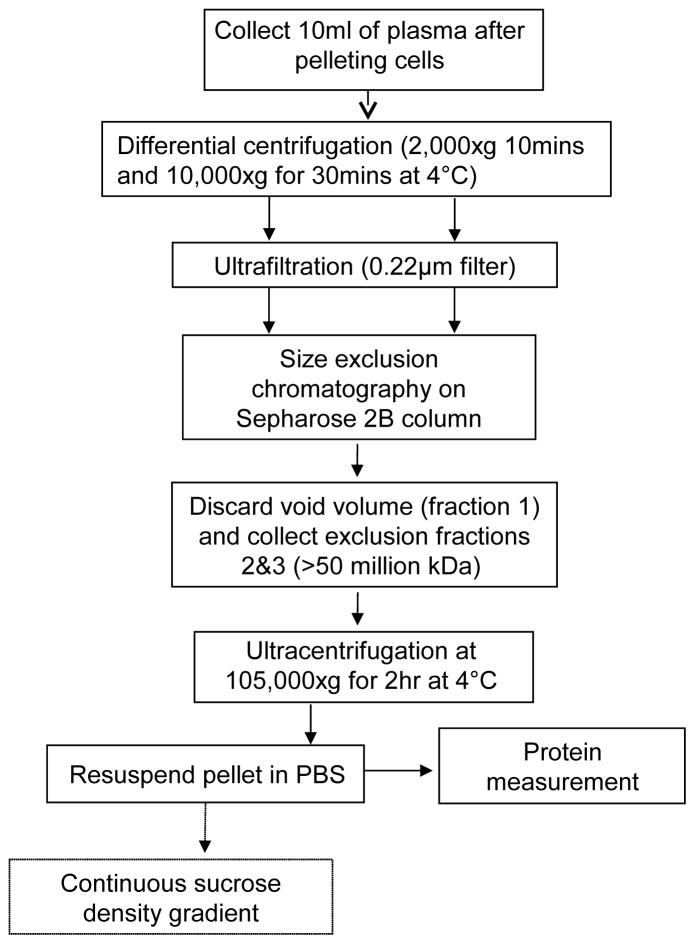
Exosomes were isolated from human plasma as previously described (Kim et al., 2005) with the modifications that included differential centrifugation of plasma (1,000 x g for 10 min at 4°C and 10,000 x g for 30 min at 4°C) followed by ultrafiltration (0.22μm filter; Millipore, Billicera, MA, USA) and modified size-exclusion chromatography. If high lipid content was present after the low-speed centrifugation (as evident by color), plasma was incubated for 2h at 4°C, and the precipitated fat was removed by centrifugation at 1000 x g for 10min at 4°C. A 9ml aliquot of plasma was applied to an A50m column (Bio-Rad Laboratories, Hercules, Ca, USA) packed with Sepharose 2B (Sigma-Aldrich, St. Louis, MO, USA), and the exclusion volume fractions (#2 and #3, each 9mL) were retained, while fraction #1 was discarded. Following ultracentrifugation (Figure 1), the exosome pellet was resuspended in PBS and protein concentration was measured. Isolated exosomes were immediately used for experiments or stored at 4°C. For a long-period storage, exosomes were frozen at −80°C.
Figure 1.
Schema of the isolation procedure for human plasma-derived exosomes.
1.3. Plasma specimens
Plasma obtained from cancer patients or NC was used fresh or subjected to one freeze/thaw cycle (rapid freezing at −80°C for 1h and slow thawing at 4°C). In some experiments, after a freeze/thaw cycle and prior to any exosome processing, deoxyribonuclease I (DN25, Sigma, St. Louis, MO, USA) and ribonuclease A (R6513, Sigma) and/or heparinase ((75 Sigma units; H2519) were added. In some experiments, ultrafiltration or size exclusion chromatography were omitted.
1.4. Protein measurements
Aliquots (5–10uL) of isolated exosomes were dispensed into wells of a 96-well plate, and the assay was performed as recommended by the manufacturer (Pierce BCA Protein Assay Kit, Thermo Scientific, Rockford, lL-61105, USA). Total protein concentrations were determined using a linear standard curve established with bovine serum albumin (BSA).
1.5. NanoSight
An aliquot (1uL) of isolated exosomes was diluted in PBS (at least 1:1000) to achieve a uniform particle distribution, and 6 sequential measurements (1 min each) at 23°C (viscosity 0.89–0.92 cp) were performed according to the manufacturer’s directions (NanoSight, Malvern Instruments Ltd, Malvern, UK). The instrument settings were: camera shutter 25–32 ms, 24.98–24.99 frames/sec; drift velocity 5011 to 6970 nm/sec; analysis: blur auto, detection threshold 5–6 multi, min track length auto and min expected size auto. At least 900 tracks were recorded per measurement.
1.6. Transmission Electron Microscopy (TEM)
TEM was performed at the Center for Biologic Imaging at the University of Pittsburgh. Two different methods were used to visualize exosomes: (a) freshly-isolated exosomes were put on a copper grid coated with 0.125% Formvar in chloroform. The grids were stained with 1% v/v uranyl acetate in ddH2O and the samples were examined immediately. (b) The isolated exosomes were centrifuged using an airfuge at 100.000xg with 25 PSI for 45mins. The pellet was fixed with cold 2.5% v/v glutaraldehyde in 0.1M PBS, rinsed in PBS, dehydrated through a graded series of ethanol and embedded in Epon. Ultra-thin sections (65nm) were stained with uranyl aetate and Reynold’s lead citrate. A JEOL 1011 transmission electron microscope was used for imaging.
1.7. Western Blots
Western blots were performed as described previously (Wieckowski et al., 2009). Exosome samples were adjusted to the same protein content and boiled for 5mins in 5x Laemmli buffer. Proteins were then separated by SDS-PAGE. The following antibodies were used to visualize the exosome cargo: Ab specific for the glycoprotein IIb/IIIa that is found in activated platelets (sc-53417, Santa Cruz, Biotechnologies, Santa Cruz, CA, USA); Abs specific for exosomal markers TSG101 (ab30871, Santa Cruz Biotechnologies), CD81 (PA5-13582, Thermo Scientific, Pittsburgh, PA, USA); and Ab specific for the housekeeping protein GAPDH (sc-25778, Santa Cruz Biotechnologies).
1.8. Flow cytometry
Isolated exosomes were captured on aldehyde sulfate-coated latex beads (Invitrogen, Grand Island, NY, USA). Blocking of the beads with 0.5% w/v BSA was performed prior to capture. The non-specifically captured exosomes were detected with anti-CD9-FITC Ab (eBioSN4, eBioscience, San Diego, CA, USA) and mouse IgG1-FITC isotype (11-4714-42, eBioscience).
Cells to be used for flow cytometry were washed and incubated in the dark for 20 min with anti-CD4 (A07752, Beckman Coulter, Brea, CA, USA), anti-CD69 (555530, BD Biosciences, San Jose, CA, USA) or anti-CD154 (12-1548, eBioscience) Abs at room temperature. Isotypes were used for each experiment (mouse IgG1-κ-FITC, 555909, BD Biosciences and IgG1-κ-PE, 12-4714, eBioscience). Cells were again washed, re-suspended in flow buffer and analyzed using an EPICS XL-MCL or a Gallios flow cytometer (Beckman Coulter). At least 5x104 events were collected, and the data were analyzed using the Kaluza software (Beckman Coulter).
1.9. Density gradients
Isolated exosomes were placed on a continuous sucrose density gradient (0.25–2.5M). An overnight ultracentrifugation was performed at 4°C as previously described (Montecalvo et al., 2013). 1ml fractions with an increasing density were collected. The refractive index of each fraction was measured (Fisher Scientific bench-type refractometer, Pittsburgh, PA, USA). Next, each fraction was diluted in PBS and subjected to ultracentrifugation at 100,000 x g for 2h at 4°C. The pellets were re-suspended in a low volume of PBS and Laemmli buffer was added prior to western blots.
1.10. Functional Studies
T-cell subsets were isolated via an immunoaffinity-based capture procedure using Miltenyi beads as previously described (Saze et al., 2013). Negative selection to isolate CD4+ T cells was followed by the separation of CD4+CD39+ and CD4+CD39neg T cells using anti-CD39 Ab-coated Miltenyi beads by AutoMACS. The purity of the isolated cells was determined by flow cytometry. The isolated T-cell subsets were activated (or not) by anti-CD3/anti-CD28 antibody (Ab)-coated beads and IL-2 (150U/ml) for 4h and co-incubated with 50ug of isolated exosomes for 16h. Cells were harvested for flow cytometry, and the frequency of CD69+ cells as well as CD69 expression (MFI) on the cell surface were determined.
1.11. Statistics
The data are presented as mean values +/− SD or in form of boxplots. To determine significance, a one-sided student t-test for homoscedastic values or a Wilcoxon signed rank test were used. A p-value of 0.05 was considered to be significant. The software used was R (R Development Core Team (2008). R: A language and environment for statistical computing. Foundation for Statistical Computing, Vienna, Austria. ISBN 3-900051-07-0, http://www.R-project.org).
Results
2.1. Quality of exosomes recovered from human plasma
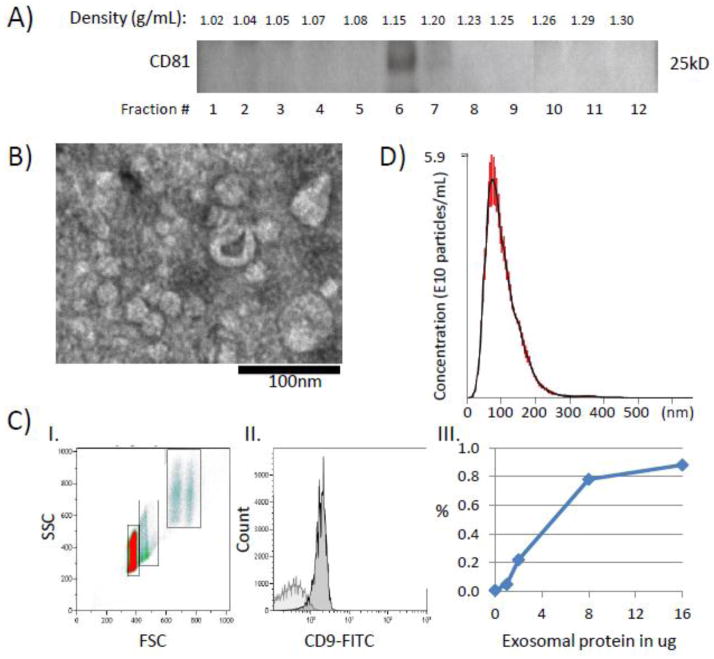
Exosomes isolated from freshly-harvested NC’s or cancer patients’ plasma, as illustrated in Figure 1, were evaluated by western blots, flow cytometry, NanoSight and TEM (Figure 2). Initially, to confirm exosome buoyancy, differential centrifugation, column chromatography and ultracentrifugation of plasma exosomes were followed by their separation on a continuous sucrose density gradient. This yielded distinct bands containing CD81 at the densities of 1.15 to 1.20 g/ml (fractions 6–7, Figure 2A), as expected (Thery et al., 2006). When sucrose gradients were omitted, the exosome recovery was considerably greater (e.g., 360 vs 274 μg protein/mL plasma in a representative experiment) than that in fractions 6–7 recovered from the gradient. This loss of proteins upon sucrose gradient centrifugation likely reflects the presence of protein aggregates and aggregated exosomes in ultracentrifuged exosome fractions, as also seen by TEM (data not shown). To avoid exosome losses and simplify exosome isolation, sucrose gradients were omitted in the subsequent experiments.
Figure 2.
Characterization of isolated exosomes. A) Western blot of isolated cancer plasma-derived exosomes floated on a continuous sucrose gradient (0.25–2.5M). Exosomes expressing CD81 are located in fractions 6–7. B) A representative TEM image of isolated exosomes. Exosomes range in size from 20–80nm. C) Flow cytometry of exosomes captured on beads. I. Ungated forward and side scatter of single and aggregated exosome-carrying beads. II. A representative histogram of exosomes (16μg). Note a clear right-shift of the CD9-FITC signal compared to the isotype control (light gray). The gate is set on single beads. III. A sigmoidal curve defines the relationship of exosome input (μg protein) and MIF values for the CD9-FITC signal. D) Exosomes isolated from plasma of a cancer patient were evaluated by NanoSight. The plot shows a broad size distribution (mean 102nm, SD ±46) and the concentration of cancer exosomes (5.6x1010/ml of plasma). The data are means of 6 measurements ± SE.
The ultracentrifuged, resuspended exosomes appear in TEM as well defined membrane-bound vesicles ranging in size from 20–100 nm (Figure 2B). Flow cytometry of the isolated exosomes captured on latex beads discriminated single beads from aggregated beads (Figure 2CI). Upon gating on single beads (Figure 2CII), it was possible to quantitate exosomal protein from the linear part of the curve generated by adding increasing concentrations of PE-labeled anti-CD9 Ab to single beads carrying captured exosomes, as shown in Figure 2CIII). The sigmoid curve reaches a plateau at the input of 4ug total exosomal protein/10uL aliquot of beads. Examination of exosomes isolated from plasma of cancer patients in the NanoSight instrument gave a broad peak corresponding to the mean particle size of 100nm, with the range of 15–360 nm, and the total particle count of 5.6 x 1010/mL of plasma (Figure 2D).
2.2. Exosome recovery
Exosome recovery from fresh or frozen/thawed plasma specimens after sequential isolation steps (Figure 1) was measured in ug protein normalized to 1ml of plasma. Tables 1–3 summarize the results for exosome recovery from plasma specimens of NC or patients with cancer. Whenever ultrafiltration or size-exclusion chromatography was omitted, the total protein levels in exosome fractions were significantly increased. Upon freezing/thawing NC plasma protein concentrations were significantly higher than those in paired fresh specimens. These protein levels were reduced after enzymatic treatments (see below). Overall, there was no difference in protein levels of exosomes recovered from NC’s or patients’ plasma compared to serum.
Table 1.
Protein levels in exosome fractions isolated from normal donors’ plasma ± ultrafiltration/chromatography a
| Isolation Protocol | Protein b (μg/mL plasma) |
|---|---|
| + ultrafiltration | 86 ± 7 |
| No Chromatography | |
| No ultrafiltration | 14 ± 3 * |
| + chromatography | |
| + ultrafiltration | 4 ± 4 * |
| + chromatography |
The exosome isolation procedure is shown in Figure 1. Fresh plasma specimens were processed with or without ultrafiltration/chromatography steps.
The data are means ± SD from 3 independent experiments with plasma obtained from different normal donors. Asterisks indicate significant differences (p < 0.01) in exosomal protein levels from those in exosomes isolated without size-exclusion chromatography.
Table 3.
Protein levels, numbers of particles and particle size ranges following ultrafiltration and chromatography.a
| Method used for isolation | Particles x109 (NanoSight) | Protein level (μg/mL) | Size Range (nm) (NanoSight) | Mode Size (nm) (NanoSight) | Mean Size (nm) NanoSight) |
|---|---|---|---|---|---|
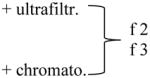
|
499 ± 47 | 5 ± 7 | 10 – 390 | 80±22 | 114±2 |
| 2,155 ± 530 | 34 ± 17 | 10 – 790 | 66±5 | 90±6 | |
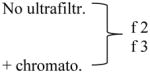
|
600 ± 31 | 2 ± 2 | 10 – 450 | 110±6 | 141±21 |
| 3,160 ± 735 | 68 ± 40 | 10 – 310 | 78±8 | 97±7 | |

|
9,460 ± 1,047 | 166 ± 50 | 10 – 410 | 52±7 | 77±1 |
Plasma specimens obtained from two patients with HNSCC were processed using differential centrifugation and evaluated ± ultrafiltration and size-exclusion chromatography. Fractions 2 and 3 collected from the column were evaluated by NanoSight for numbers and size of particles and by the BioRad assay for protein levels.
Analyzing different ultracentrifuged exosome fractions collected after size-exclusion chromatography columns by NanoSight, we confirmed that proteins with small molecular weight eluting in fraction 4 can be removed with this technique. As shown in Table 3, NanoSight measurements indicated that the particle count and protein concentrations were considerably higher when sample ultrafiltration or column chromatography were omitted. The limited size range in the last row of Table 3 is probably biased by the high dilution needed for Nanosight-based analysis.
2.3. Differential centrifugation and ultrafiltration of plasma
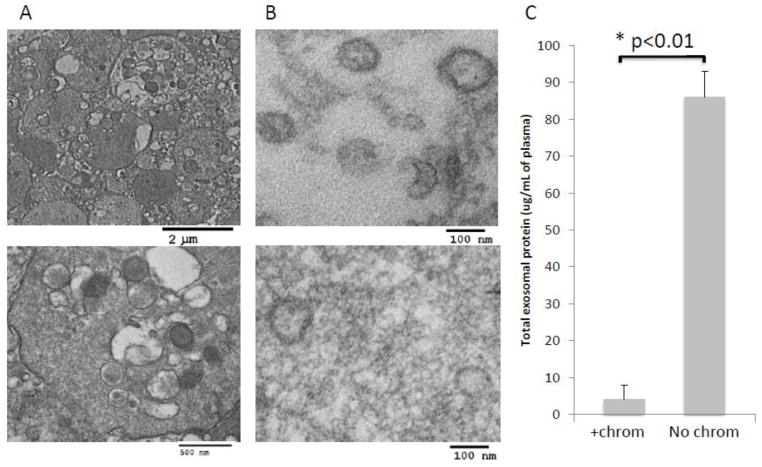
Differential centrifugation at a low (2,000rpm) then at a high speed (10,000rpm) is necessary to remove platelets, subcellular fragments and larger microvesicles from plasma as previously emphasized (Raposo et al., 1996; Thery et al., 2006). In addition, ultrafiltration using 0.2μm bacterial filter was found to be necessary for removal of larger vesicles (above 200nm) and thrombocytes (~1–2μm), which remain in plasma even after differential centrifugation (Figure 3A). Apart from thrombocytes and microvesicles, other “contaminating” elements such as lysosomes, mitochondria, nucleic acid-protein aggregates, centrioles and even bacteria may be present in differentially-centrifuged plasma as seen by TEM (data not shown). Ultrafiltration removes the majority of these “contaminants.”
Figure 3.
Effects of ultrafiltration and size-exclusion chromatography on isolated exosomes. A) TEM images of plasma-derived exosomes from a representative NC of 5 examined. Note the presence of thrombocytes in non-filtered plasma (upper image: low mag. and lower image: high mag.) B) TEM of epon-embedded and sectioned exosomes from plasma of a representative cancer patient isolated by size-exclusion chromatography (upper image) or omitting chromatography (lower image). Note increased purity of the exosomal preparation obtained after chromatography. C) Protein levels in exosomes isolated from plasma of patients with cancer (n=3) ± size-exclusion chromatography. The data are means ± SE.
2.4. Size-exclusion chromatography
Size exclusion chromatography was previously reported to improve the purity of exosomes isolated from supernatants of cell lines (Taylor et al., 2003; Kim et al., 2005). We have confirmed these earlier data and showed that the recovery and quality of plasma-derived exosomes is also greatly improved by the use of size-exclusion chromatography. When size-exclusion chromatography was omitted, exosomes appeared to be “dirty” by TEM relative to “clean” exosomes preparations obtained after column chromatography (Figure 3B). The “clean” exosomal preparations yielded significantly lower total protein concentrations after ultracentrifugation than those seen in the “dirty” exosomal fractions (Figure 3C and Tables 1–3). These data show that size exclusion chromatography prior to ultracentrifugation is an essential step for removing “contaminating” plasma protein and other small molecules from preparations of plasma-derived exosomes.
2.5. Exosome isolation from serum vs. plasma
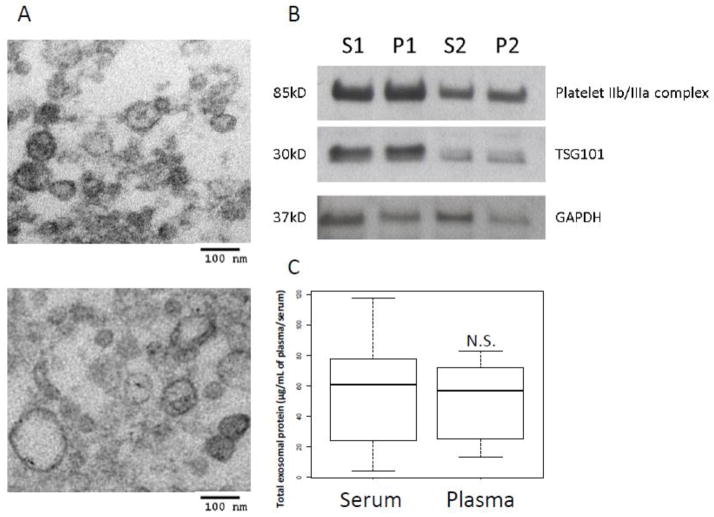
Freshly-harvested plasma vs. serum samples were compared as a source of exosomes. A concern existed that a loss of exosomes trapped within the clot can occur when serum is used. On the other hand, heparin in plasma could facilitate the formation of exosome-heparin complexes and aggregation of exosomes as previously reported (Atai et al., 2013). However no significant differences were observed in exosome recovery from paired serum vs. plasma specimens obtained from 6 patients with cancer. Exosome microscopic appearance was also comparable (Figure 4A). Since activation of the clotting cascade could lead to platelets activation and release of “contaminating” granules, western blots were performed to determine the presence of glycoproteins IIb/IIIa (markers of platelet activation) in isolated exosomes (Figure 4B). The levels of glycoproteins IIb/IIIa relative to the exosomal GAPDH levels were somewhat higher in plasma than serum specimens. This was not seen when the ratio of glycoproteins IIb/IIIa to TSG101 was considered, possibly because TSG101 levels vary widely in exosomes isolated from plasma or sera of different donors. Also, protein levels of exosome fractions recovered from serum vs plasma were not significantly different (Figure 4C), although a higher SD for the serum values suggests that clotting introduces substantial variability in exosome protein levels. In aggregate, based on our data, we concluded that either serum or plasma were comparable sources of exosomes. To avoid the variability due to a clot formation, we selected to utilize plasma specimens for all further studies. To address a concern that heparin present in plasma might interfere with the action of polymerases and thus impair the recovery of exosomal nucleic acids, we compared the mRNA recovery from plasma- or serum-derived exosomes and found no significant differences (data not shown).
Figure 4.
Comparison of serum-derived with plasma-derived exosomes. A) TEM of serum-derived exosomes (upper image) or plasma-derived exosomes (lower image) obtained from the same cancer patient. Note the similar morphology of the isolated exosomes. B) The presence of platelet IIb/IIIa complex in exosomes (10ug) derived from serum (S1, S2) versus plasma (P1, P2) of two cancer patients as seen in a Western blot. An exosomal marker TSG101 and a housekeeping protein GAPDH were used as loading controls. C) A box plot shows no significant (N.S.) difference in total proteins of exosomal fractions isolated from paired serum or plasma specimens of the same 6 patients with cancer. Note a considerably larger SE in values for serum-derived exosomes than those isolated from plasma. The bar inside the box indicates the median, the box shows interquartile range (25–75%) and whiskers extend to 1.5 x the interquartile range.
2.6. Exosome isolation from fresh vs. frozen plasma
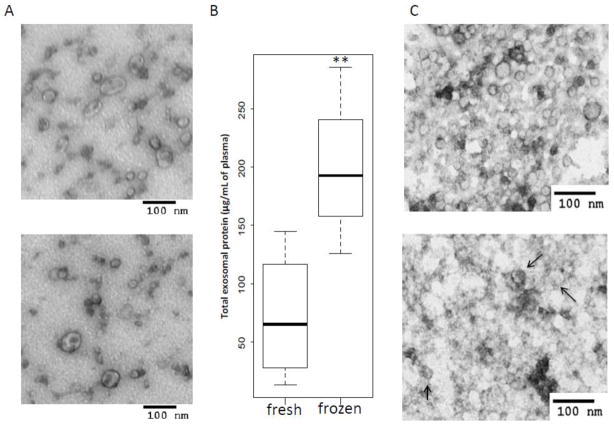
To determine whether banking, storage and freezing/thawing of plasma negatively affects exosome isolation, we measured total protein recovery and evaluated exosome image quality prior to and after freezing. Freshly-harvested exosomes were quite resistant to freeze/thaw cycles. Once purified, no noticeable difference of their morphology by TEM was evident after a single freeze/thaw (Figure 5A). On the other hand, the use of frozen plasma (i.e., plasma frozen after low-speed centrifugation) as a source of exosomes, yielded exosomes containing numerous “contaminants”, which were difficult to remove completely. Long-term storage (e.g., up to 7 yrs) of plasma at −80°C did not alter exosome morphology, although it substantially increased protein/nucleic acid aggregation as indicated by high background levels seen by TEM (see below). When total exosomal protein recovered from the same volume of plasma before and after a single freeze/thaw was measured, lower protein values were consistently seen in exosomes isolated from fresh, unfrozen plasma (Figure 5B). This observation suggested that freezing of plasma leads to significant enrichment of free proteins in exosomal fractions. TEM images of these exosomes (Figure 5C) suggest that freezing significantly increases the background, probably due to a loss of disrupted vesicles or protein/nucleic acid leakage from damaged vesicles, without a noticeable change in their morphology.
Figure 5.
Exosome quality and recovery before and after freezing of plasma. A) TEM of isolated exosomes before (upper image) and after a single freeze/thaw cycle (lower image). B) The increase in total protein levels of exosome fractions are higher when fresh plasma is frozen after low-speed centrifugation than in plasma frozen after differential centrifugation. The values are from 3 independent experiments with plasma of different NC. The data are presented as box plots (see the legend to Figure 4). C) Differences in morphology of exosomes in TEM (negative staining) from fresh (upper image) as compared to exosomes from frozen plasma samples examined after differential centrifugation (lower image). Plasma was obtained from a representative cancer patient. The arrows indicate exosomes surrounded by exogenous material.
2.7. Removal of nucleic acid-protein or heparin-protein aggregates
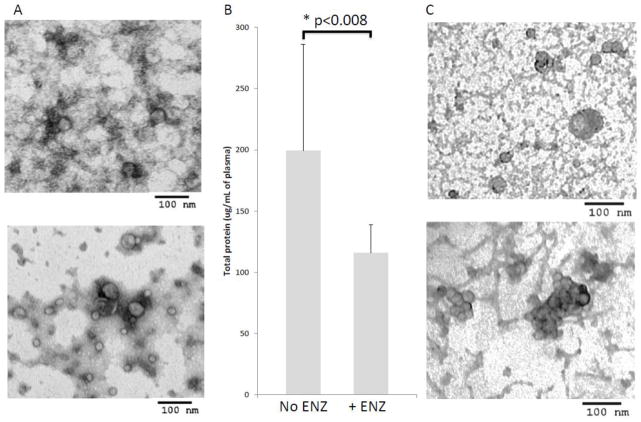
Because plasma-derived exosomes, especially those isolated from frozen/thawed plasma specimens, contained what appeared to be protein-nucleic acid aggregates by TEM, we considered using DNAses and RNAses to remove these aggregates. Treatments of frozen/thawed plasma after differential centrifugation with these enzymes as described in M&M in part removed the background material, giving a clearer image of exosomes as seen in the EM images of negatively stained preparations (Figure 6A) and concomitantly decreased total protein levels (Figure 6B). Finally, treatments of plasma with hyaluronidase also reduced the background staining and gave cleaner exosome images, but induced exosome aggregation (Figure 6C). These experiments suggested that enzymatic treatments of plasma designed to improve the quality and recovery of plasma-derived exosomes were only partially effective, and because of the considerable cost of the enzymatic treatment/sample were not applicable to routine exosome isolation from plasma.
Figure 6.
Enzymatic treatments of frozen/thawed plasma. A) TEM (negative staining) of a representative frozen sample examined before (upper image) and after (lower image) treatment with DNAse and RNAse. Note decrease in the background and also a considerable loss of exosomes. B) Change in total exosomal protein levels ± enzyme treatment. A representative experiment of 3 independent experiments performed with samples of different NC. C) TEM (negative staining) of exosomes isolated from cancer patients’ plasma stored frozen for 7 years: without enzymatic treatment (upper image) and after treatment with h DNAse/RNAse or Heparinase (lower image).
2.8. Functions of plasma-derived exosomes
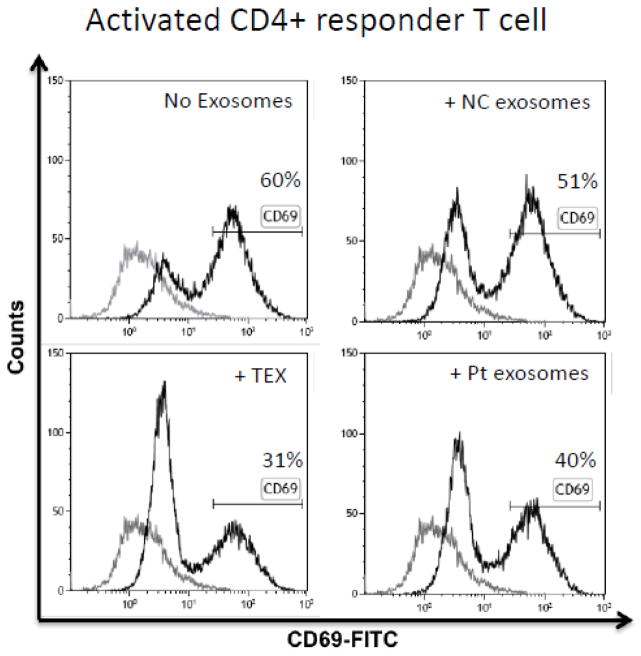
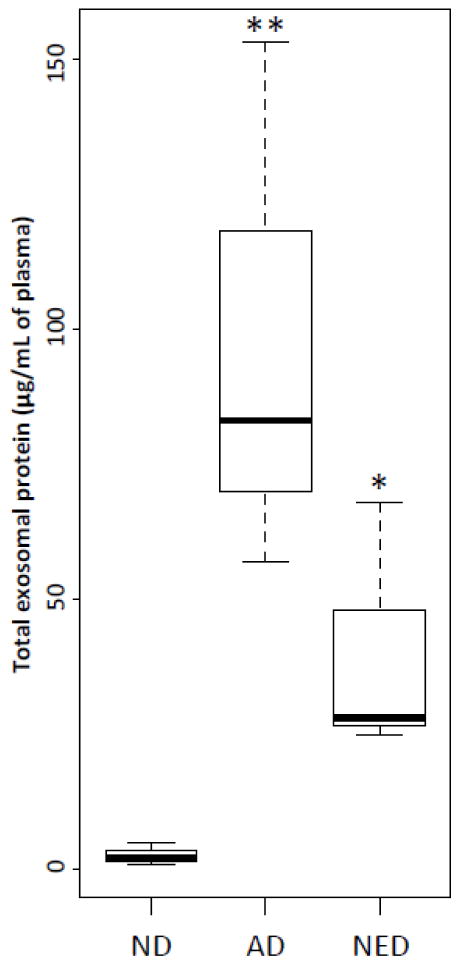
The protocol for exosome isolation shown in Figure 1 was developed to obtain purified, biologically-active exosomes from human plasma. To evaluate their functional competence, exosomes isolated from fresh plasma of NC or patients with head and neck squamous cell carcinoma (HNSCC) were co-incubated with activated CD4+ conventional T cells for 7h to measure exosome immunosuppressive ability (Wieckowski et al., 2009; Whiteside, 2013). These CD4+ T cells had a purity of at least 95%, and the frequency of CD69+ cells after activation was 60%. Following co-incubation with exosomes, the frequency of CD4+CD69+ T-responder cells was measured by flow cytometry (Ruitenberg et al., 2011; Canavan et al., 2012). As expected, cancer patients’ plasma-derived exosomes inhibited CD69 expression in activated CD4+ responder T cells (40% remained CD69+). Exosomes isolated from NC’s plasma were less suppressive, with 51% CD4+ responder T cells remaining CD69+ (Figure 7). TEX isolated from supernatants of a HNSCC cell line were most suppressive (30% CD69+ T cells). These data show that the isolation procedure we describe allows for isolation of functionally-competent exosomes from plasma of NC or patients with cancer. In all instances, cancer patients’ specimens contained significantly more exosomal proteins than those of NC (Figure 8).
Figure 7.
Functional activity of isolated exosomes co-incubated with activated T effector cells. Flow cytometry showing the differential suppressive effects of TEX, cancer patient’s plasma-derived or NC exosomes on T effector cells (CD4+) represented by CD69-expression. Note the higher suppressive effect of cancer plasma-derived exosomes vs. NC exosomes. The curve in light gray represents the isotype control.
Figure 8.
Levels of total protein in exosomes isolated as shown in Figure 1 from fresh plasma specimens obtained from NC or HNSCC patients with active disease (AD) or no evident disease after therapy (NED). n=3 for each group, * p<0.05 and ** p<0.01. These data are presented as box plots (see the legend to Figure 4).
Discussion
To be able to use exosomes from body fluids of humans for immunologic studies or as potential biomarkers of prognosis, prediction of responses to therapies or survival, it is first necessary to isolate them without losses as purified, biologically-active vesicles. A variety of commercially-developed methods are currently available for exosome enrichment and isolation (Witwer et al., 2013; Gyorgy et al., 2014; Van der Meel et al., 2014). These methods largely aim at isolation of exosomal nucleic acid, especially miRNA. Other approaches use immunocapture of exosomes on beads coated with Abs specific for various exosome markers (Clayton et al., 2001; Kim et al., 2012; Yoo et al., 2012). These methods aim at a simplified, one-step method of exosome isolation that could be applied to large-scale, high throughput biomarker-type studies. In contrast, studies of exosome biologic activities such as, e.g., effects on functions of immune or non-immune cells, require isolation from human plasma of exosomes recovered without substantial losses, free of plasma proteins and nucleic acids, morphologically intact and functionally active. We have evaluated and attempted to optimize such an isolation procedure and describe here its merits and potential pitfalls. Importantly, human plasma or serum served as equally good sources of exosomes based on recovery, purity, morphology and function.
Exosome isolation from fresh and especially from frozen human plasma (after low-speed centrifugation) faces considerable difficulties, largely related to the potential “contamination” with larger microvesicles, subcellular fractions, protein aggregates, protein-nucleic acid aggregates or plasma proteins. Placing exosomes isolated by ultracentrifugation on continuous sucrose gradients to take advantage of their characteristic buoyant density, as conventionally done (Raposo et al., 1996; Thery et al., 2006), not only results in substantial exosomal losses but also is labor-intensive. Differential centrifugation of plasma alone is not adequate to remove various “contaminants.” A much “cleaner” plasma is obtained after ultrafiltration, and the subsequent application of such plasma to a size-exclusion chromatography column is critical for removal of plasma proteins and other soluble components. The void volume fractions collected by column chromatography are enriched in relatively “clean” exosomes, at least when fresh plasma specimens serve as a source of exosomes. The recovery, quality and functionality of exosomes purified from such plasma or serum specimens were found to be adequate for immunological studies.
Most exosome studies performed today are retrospective and utilize specimens frozen/thawed for months if not years (Witwer et al., 2013). Few if any prospective exosome-related immunologic studies have been conducted so far. This is because of a need for clinical results to be available for correlations with exosome molecular or functional profiles. However, the use of banked frozen/thawed plasma or serum specimens for exosome isolation presents some problems. When plasma is frozen immediately after low-speed centrifugation, larger vesicles that are left behind as well as some exosomes are disrupted or damaged during freezing and release nucleic acids and proteins. We showed by TEM that even the differentially-centrifuged frozen/thawed plasma contains numerous string-like aggregates, which surround exosomes. Removal of these aggregates by enzymatic treatments with DNAse, RNAse or hyaluronidase in part reduced their presence but tended to aggregate exosomes and did not significantly decrease the overall protein content. Thus, attempts at improving exosome recovery from frozen/thawed plasma specimens by enzymatic treatments were neither productive nor cost effective and are not recommended. Based on our findings, we recommend the use of fresh plasma as a source of exosomes whenever possible. If frozen plasma has to be used, it should be centrifugated at low-speed and 10,000 x g, as well as ultrafiltrated before freezing. After thawing of plasma, a second ultrafiltration and processing as indicated in Figure 1 greatly reduces “contaminants” and allows for a recovery of immunologically-active exosomes.
Freezing of freshly isolated exosomes (i.e., after their recovery by ultracentrifugation) does not alter exosomal morphology and does not seem to impair their function, as also previously reported (Sokolova et al., 2011; Jayachandran et al., 2012). The exosome resistance to freezing is probably due to the more balanced surface to volume ratio, as compared to much larger cells or larger vesicles. We, therefore, recommend that if isolated exosomes cannot be used within a few days, they are aliquoted and stored at −80°C for future use.
Importantly, exosomes isolated from fresh or frozen-thawed plasma by the procedure shown in Figure 1, have a characteristic morphology in TEM and range in diameter from 20 to 100nml. They also mediate biologic activity, e.g., immune suppression as shown by their abilty to down-regulate CD69 expression on human activated CD4+ T cells. Consistently, exosomes isolated from plasma of patients with cancer had higher protein levels/mL of plasma than those isolated from plasma of normal donors (Figure 3). Exosomes isolated from cancer patients’ plasma also induced higher suppression of CD69 expression than those isolated from ND’s plasma.
In aggregate, it appears that biologically-competent, morphologically intact exosomes can be successfully purified from fresh or frozen human plasma by the method we have described. The modifications introduced to the conventional exosome purification method as previously described (Raposo et al., 1996; Thery et al., 2006; Bianco et al., 2007) are necessary for processing human plasma, and potentially other body fluids, to remove non-relevant plasma proteins without incurring losses in exosome recovery.
Table 2.
Protein levels in exosome fractions isolated from fresh or frozen plasma of cancer patients a
| Protein (μg/mL plasma) | ||
|---|---|---|
| Plasma Before Freezing | 72 ± 40 |
|
| Plasma After Freezing | 199 ± 63 | |
| Plasma After Freezing + enzymatic treatments | 116 ± 23 | |
Exosomes were isolated as shown in Figure 1. Plasma obtained from cancer patients was processed fresh or was frozen after differential centrifugation and thawed prior to processing.
The data are means ± SD obtained in four independent experiments.
p< 0.01
Highlights.
Isolation of immunologically-active exosomes from human plasma
Advantages of plasma ultrafiltration and size-exclusion chromatography
Plasma or serum is an equally acceptable source of exosomes
Fresh plasma yields “cleaner” exosomes than frozen/thawed plasma
Minimized losses due to exosome aggregates with nucleic acids or plasma proteins
Abbreviations
- Ab
antibody
- AD
active disease
- EXO
exosomes
- HNSCC
head and neck squamous cell carcinoma
- MF
mean fluorescence intensity
- NC
normal control
- PBS
phosphate buffered saline
- Pt
patient
- TEM
transmission electron microscopy
- TEX
tumor-derived exosomes
Footnotes
Financial Disclosure
This manuscript is supported in part by NIH grant, RO1 CA168628 (TLW), and by the Swiss National Foundation, grant # PBSKP3_140119/1 (LM). Flow cytometry facility was supported by the NIH CSSG grant 5P30 CA047904.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Atai NA, Balaj L, van Veen H, Breakefield XO, Jarzyna PA, Van Noorden CJ, Skog J, Maguire CA. Heparin blocks transfer of extracellular vesicles between donor and recipient cells. Journal of Neuro-Oncology. 2013;115:343–51. doi: 10.1007/s11060-013-1235-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco NR, Kim SH, Morelli AE, Robbins PD. Modulation of the immune response using dendritic cell-derived exosomes. Methods in Molecular Biology. 2007;380:443–55. doi: 10.1007/978-1-59745-395-0_28. [DOI] [PubMed] [Google Scholar]
- Canavan JB, Afzali B, Scotta C, Fazekasova H, Edozie FC, Macdonald TT, Hernandez-Fuentes MP, Lombardi G, Lord GM. A rapid diagnostic test for human regulatory T-cell function to enable regulatory T-cell therapy. Blood. 2012;119:e57–66. doi: 10.1182/blood-2011-09-380048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton A, Court J, Navabi H, Adams M, Mason MD, Hobot JA, Newman GR, Jasani B. Analysis of antigen presenting cell derived exosomes, based on immuno-magnetic isolation and flow cytometry. Journal of Immunological Methods. 2001;247:163–74. doi: 10.1016/s0022-1759(00)00321-5. [DOI] [PubMed] [Google Scholar]
- Graner MW, Alzate O, Dechkovskaia AM, Keene JD, Sampson JH, Mitchell DA, Bigner DD. Proteomic and immunologic analyses of brain tumor exosomes. FASEB Journal : official publication of the Federation of American Societies for Experimental Biology. 2009;23:1541–57. doi: 10.1096/fj.08-122184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyorgy B, Paloczi K, Kovacs A, Barabas E, Beko G, Varnai K, Pallinger E, Szabo-Taylor K, Szabo TG, Kiss AA, Falus A, Buzas EI. Improved circulating microparticle analysis in acid-citrate dextrose (ACD) anticoagulant tube. Thrombosis Resaerch. 2014;133:285–92. doi: 10.1016/j.thromres.2013.11.010. [DOI] [PubMed] [Google Scholar]
- Harrington MG, Fonteh AN, Oborina E, Liao P, Cowan RP, McComb G, Chavez JN, Rush J, Biringer RG, Huhmer AF. The morphology and biochemistry of nanostructures provide evidence for synthesis and signaling functions in human cerebrospinal fluid. Cerebrospinal Fluid Research. 2009;6:10. doi: 10.1186/1743-8454-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawari FI, Rouhani FN, Cui X, Yu ZX, Buckley C, Kaler M, Levine SJ. Release of full-length 55-kDa TNF receptor 1 in exosome-like vesicles: a mechanism for generation of soluble cytokine receptors. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:1297–302. doi: 10.1073/pnas.0307981100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayachandran M, Miller VM, Heit JA, Owen WG. Methodology for isolation, identification and characterization of microvesicles in peripheral blood. Journal of Immunological Methods. 2012;375:207–14. doi: 10.1016/j.jim.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller S, Rupp C, Stoeck A, Runz S, Fogel M, Lugert S, Hager HD, Abdel-Bakky MS, Gutwein P, Altevogt P. CD24 is a marker of exosomes secreted into urine and amniotic fluid. Kidney International. 2007;72:1095–102. doi: 10.1038/sj.ki.5002486. [DOI] [PubMed] [Google Scholar]
- Kim G, Yoo CE, Kim M, Kang HJ, Park D, Lee M, Huh N. Noble polymeric surface conjugated with zwitterionic moieties and antibodies for the isolation of exosomes from human serum. Bioconjugate Chemistry. 2012;23:2114–20. doi: 10.1021/bc300339b. [DOI] [PubMed] [Google Scholar]
- Kim JW, Wieckowski E, Taylor DD, Reichert TE, Watkins S, Whiteside TL. Fas ligand-positive membranous vesicles isolated from sera of patients with oral cancer induce apoptosis of activated T lymphocytes. Clinical Cancer Research : an Official Journal of the American Association for Cancer Research. 2005;11:1010–20. [PubMed] [Google Scholar]
- Mathivanan S, Lim JW, Tauro BJ, Ji H, Moritz RL, Simpson RJ. Proteomics analysis of A33 immunoaffinity-purified exosomes released from the human colon tumor cell line LIM1215 reveals a tissue-specific protein signature. Molecular & Cellular Proteomics : MCP. 2010;9:197–208. doi: 10.1074/mcp.M900152-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathivanan S, Simpson RJ. ExoCarta: A compendium of exosomal proteins and RNA. Proteomics. 2009;9:4997–5000. doi: 10.1002/pmic.200900351. [DOI] [PubMed] [Google Scholar]
- Montecalvo A, Larregina AT, Morelli AE. Methods of analysis of dendritic cell-derived exosome-shuttle microRNA and its horizontal propagation between dendritic cells. Methods in Molecular Biology. 2013;1024:19–40. doi: 10.1007/978-1-62703-453-1_3. [DOI] [PubMed] [Google Scholar]
- Pisitkun T, Shen RF, Knepper MA. Identification and proteomic profiling of exosomes in human urine. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:13368–73. doi: 10.1073/pnas.0403453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raposo G, Nijman HW, Stoorvogel W, Liejendekker R, Harding CV, Melief CJ, Geuze HJ. B lymphocytes secrete antigen-presenting vesicles. The Journal of Experimental Medicine. 1996;183:1161–72. doi: 10.1084/jem.183.3.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raposo G, Stoorvogel W. Extracellular vesicles: Exosomes, microvesicles and friends. Journal of Cell Biology. 2013;200:373–83. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruitenberg JJ, Boyce C, Hingorani R, Putnam A, Ghanekar SA. Rapid assessment of in vitro expanded human regulatory T cell function. Journal of Immunological Methods. 2011;372:95–106. doi: 10.1016/j.jim.2011.07.001. [DOI] [PubMed] [Google Scholar]
- Runz S, Keller S, Rupp C, Stoeck A, Issa Y, Koensgen D, Mustea A, Sehouli J, Kristiansen G, Altevogt P. Malignant ascites-derived exosomes of ovarian carcinoma patients contain CD24 and EpCAM. Gynecologic Oncology. 2007;107:563–71. doi: 10.1016/j.ygyno.2007.08.064. [DOI] [PubMed] [Google Scholar]
- Skog J, Wurdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, Curry WT, Jr, Carter BS, Krichevsky AM, Breakefield XO. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nature Cell Biology. 2008;10:1470–6. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolova V, Ludwig AK, Hornung S, Rotan O, Horn PA, Epple M, Giebel B. Characterisation of exosomes derived from human cells by nanoparticle tracking analysis and scanning electron microscopy. Colloids and Surfaces B, Biointerfaces. 2011;87:146–50. doi: 10.1016/j.colsurfb.2011.05.013. [DOI] [PubMed] [Google Scholar]
- Street JM, Barran PE, Mackay CL, Weidt S, Balmforth C, Walsh TS, Chalmers RT, Webb DJ, Dear JW. Identification and proteomic profiling of exosomes in human cerebrospinal fluid. Journal of Translational Medicine. 2012;10:5. doi: 10.1186/1479-5876-10-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor DD, Gercel-Taylor C, Lyons KS, Stanson J, Whiteside TL. T-cell apoptosis and suppression of T-cell receptor/CD3-zeta by Fas ligand-containing membrane vesicles shed from ovarian tumors. Clinical Cancer Research : an Official Journal of the American Association for Cancer Research. 2003;9:5113–9. [PubMed] [Google Scholar]
- Taylor DD, Lyons KS, Gercel-Taylor C. Shed membrane fragment-associated markers for endometrial and ovarian cancers. Gynecologic Oncology. 2002;84:443–8. doi: 10.1006/gyno.2001.6551. [DOI] [PubMed] [Google Scholar]
- Thery C, Amigorena S, Raposo G, Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. In: Bonifacino Juan S, et al., editors. Current Protocols in Cell Biology. Unit 3. Chapter 3. 2006. p. 22. [DOI] [PubMed] [Google Scholar]
- Thery C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nature Reviews Immunology. 2002;2:569–79. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- Van der Meel R, Krawczyk-Durka M, Van Solinge WW, Schiffelers RM. Toward routine detection of extracellular vesicles in clinical samples. International Journal of Laboratory Hematology. 2014;36:244–253. doi: 10.1111/ijlh.12247. [DOI] [PubMed] [Google Scholar]
- Vlassov AV, Magdaleno S, Setterquist R, Conrad R. Exosomes: Current knowledge of their composition, biological functions and diagnostic and therapeutic potentials. Biochimica et Biophysica Acta. 2012;1820:940–948. doi: 10.1016/j.bbagen.2012.03.017. [DOI] [PubMed] [Google Scholar]
- Whiteside TL. Immune modulation of T-cell and NK (natural killer) cell activities by TEXs (tumour-derived exosomes) Biochemical Society Transactions. 2013;41:245–51. doi: 10.1042/BST20120265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieckowski EU, Visus C, Szajnik M, Szczepanski MJ, Storkus WJ, Whiteside TL. Tumor-derived microvesicles promote regulatory T cell expansion and induce apoptosis in tumor-reactive activated CD8+ T lymphocytes. Journal of Immunology. 2009;183:3720–30. doi: 10.4049/jimmunol.0900970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witwer KW, Buzas EI, Bemis LT, Bora A, Lasser C, Lotvall J, Nolte-‘t Hoen EN, Piper MG, Sivaraman S, Skog J, Thery C, Wauben MH, Hochberg F. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. Journal of Extracellular Vesicles. 2013;2 doi: 10.3402/jev.v2i0.20360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo CE, Kim G, Kim M, Park D, Kang HJ, Lee M, Huh N. A direct extraction method for microRNAs from exosomes captured by immunoaffinity beads. Analytical Biochemistry. 2012;431:96–8. doi: 10.1016/j.ab.2012.09.008. [DOI] [PubMed] [Google Scholar]