Abstract
During the past decade, research on c-di-GMP expanded greatly, uncovering several roles it plays among bacteria
Although my students might think that all major components and important systems in biology are known, I tell them that nothing could be further from the truth. The field of cyclic di-GMP (c-di-GMP) signaling is a case in point. Asking someone at an ASM General Meeting a decade or more ago about c-di-GMP would likely trigger blank stares, unless that person worked with Moshe Benziman at the Hebrew University of Jerusalem in Israel.
This year, however, I chaired a session covering second messenger signaling in bacteria during the 2012 ASM General Meeting in San Francisco. Now the vast majority of papers in this field focus on c-di-GMP. What is c-di-GMP, and why is interest in it sweeping through microbiology? What progress are we making in understanding c-di-GMP, and what challenges do we face? Researchers are now addressing many questions about the environmental cues that control enzymes to produce and degrade this molecule, about downstream effectors, and the many roles of c-di-GMP in bacteria (Table 1). Knowledge from this research will lead to a deeper understanding of how bacteria use this signaling molecule to respond and adapt to different environments. Moreover, deciphering c-di-GMP signaling may lead to new strategies for treating biofilm-based chronic infections.
TABLE 1.
New frontiers in c-di-GMP signaling
| New Frontier | Rationale |
|---|---|
| Identifying environmental signals that control c-di-GMP | Only a handful of environmental signals that control c-di-GMP synthesis or degradation have been identified, despite the fact that thousands of DGCs and PDEs are present in bacterial genomes. |
| Understanding c-di-GMP in other bacteria | C-di-GMP is widespread and remains to be intensively studied in many bacteria, including the Gram-positives |
| Elucidating c-di-GMP controlled phenotypes | The last few years have revealed many new phenotypes controlled by c-di-GMP. What other behaviors are impacted by c-di-GMP? |
| Discovering c-di-GMP effectors | Although a number of c-di-GMP binding effectors have been identified, it is my opinion that we have just found the tip of the iceberg. |
| Determining signaling specificity | Does c-di-GMP function via low- or high-specificity signaling pathways, or a mixture of both? What are the molecular mechanisms that maintain segregation of c-di-GMP in the cell? |
| Development of a single-cell c-di-GMP sensor | Additional biosensors to quantify c-di-GMP at single-cell resolution need to be developed to allow examination of this signal in challenging environments such as in vivo infection. |
| Targeting c-di-GMP | Disrupting c-di-GMP signaling is an attractive strategy to treat biofilm-based infections. |
Research on Cellulose Synthesis Leads to Discovery of c-di-GMP
Benziman and his collaborators discovered c-di-GMP, as part of a broader effort to study cellulose synthesis. Because of constraints studying this process in plants, Benziman studied cellulose synthesis in the bacterium Gluconacetobacter xylinus (formerly Acetobacterium xylinum), which produces floating cellulose mats to colonize air-liquid interfaces, where cells maximize their access to oxygen.
Although Benziman suspected that cellulose is made in the cell membrane, something was missing when he and his collaborators tried to follow this process biochemically in vitro. However, adding GTP to the reaction mixture stimulated activity comparable to that seen in living bacteria. He and his collaborators determined that a diguanylate cyclase (DGC) enzyme in the membrane fraction cyclized two GTP molecules into the chemical bis (3′→5′)-cyclic diguanylic acid (c-di-GMP), which binds to and stimulates the cellulose synthesis enzyme.
Benziman and his collaborators began to study c-di-GMP in the mid-1980s, but another 15 years passed before other researchers began to appreciate how widely active this molecule is within the microbial world. That realization depended on two critical developments: genomic sequencing, which revealed that approximately 85% of bacteria analyzed encode genes for making and degrading c-di-GMP, and growing interest in how biofilms form, a process in which c-di-GMP plays a critical role.
c-di-GMP Helps to Control Processes from Biofilms to Virulence
The commonly accepted function of c-di-GMP is to control the transition of gram-negative bacteria from free-living planktonic to sessile biofilms. In general, when c-di-GMP is reduced or abolished, biofilm formation is much lower, suggesting c-di-GMP is essential for this process. Relatively little is known about the function of c-di-GMP in gram-positive bacteria.
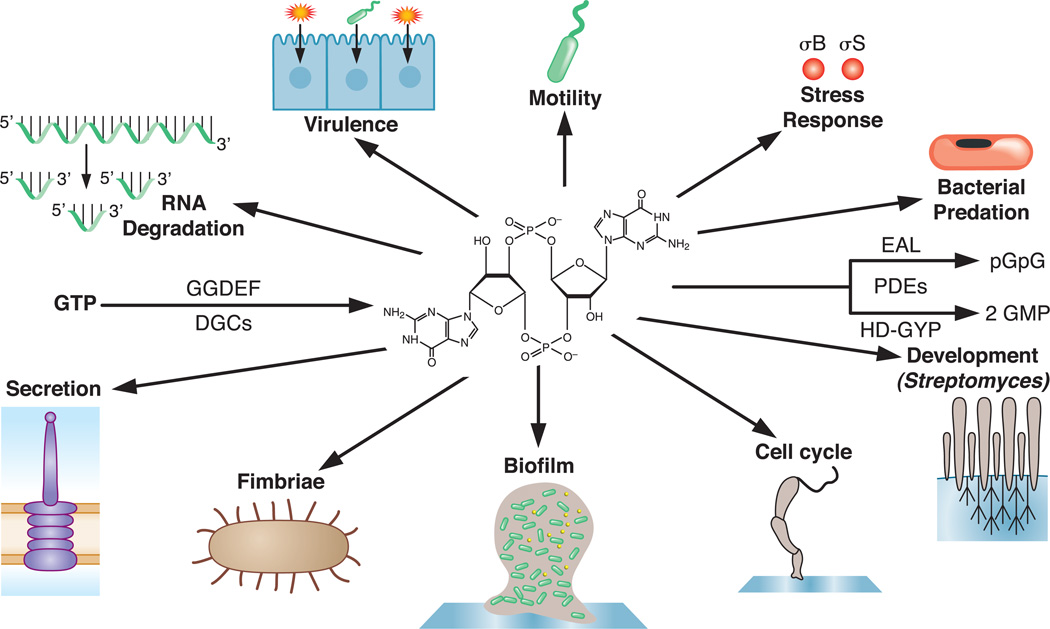
Although the most appreciated role for c-di-GMP is regulating biofilms and motility, this molecule affects many more bacterial processes. One exciting development is the realization that c-di-GMP controls a number of other phenotypes and behaviors, including Bdellovibrio predation, Streptomyces development, turnover of mRNA in Caulobacter crescentus, and RNA degradasomes in Escherichia coli (Fig. 1).
FIGURE 1.
C-di-GMP is synthesized by GGDEF enzymes possessing diguanylate cyclase (DGC) activity and degraded by EAL and HD-GYP enzymes possessing phosphodiesterase (PDE) activity. EAL enzymes cleave one bond of the cyclic structure generating the linear molecule pGpG while HD-GYP enzymes cleave both bonds to generate GMP. Many bacterial behaviors in addition to biofilm formation and motility are regulated, both positively and negatively, by c-di-GMP.
Of particular interest, c-di-GMP inhibits the virulence of pathogens that cause acute infections. For example, in Vibrio cholerae, high intracellular concentrations of c-di-GMP reduce virulence in two ways: by forming biofilms and by repressing virulence factor expression. Thus, levels of c-di-GMP in V. cholerae are high during environmental persistence and low upon infection, according to Andrew Camilli at Tufts Medical School in Boston, Mass. However, for Pseudomonas aeruginosa both c-di-GMP-degrading and -producing enzymes are important when this pathogen causes pneumonia in mice, according to Stephen Lory of Harvard University in Cambridge, Mass. Thus, its signaling role in virulence is complex, perhaps shifting during critical phases of infection.
Meanwhile, c-di-GMP positively influences chronic, biofilm-based infectious diseases. For example, when colonizing the lungs of cystic fibrosis patients, P. aeruginosa typically becomes a hyper-biofilm-forming mucoid strain or small-colony variant, both of which are associated with high levels of c-di-GMP. Because these robust biofilm-forming morphotypes persist in lungs and survive antibiotic therapy, their associations with c-di-GMP make them a prime target for development of antibiofilm intervention strategies.
The c-di-GMP System Includes Many DGCs and PDEs
DGC enzymes, typically encoding a GGDEF domain, synthesize c-di-GMP from two GTP molecules. Phosphodiesterase enzymes (PDE), typically containing either an EAL or HD-GYP domain, degrade c-di-GMP molecules (Fig. 1). These domains are named for the critical amino acids in the respective active sites of these two types of enzyme.
This seemingly simple situation proves more complicated because many bacteria encode enzymes that contain both DGC and PDE domains within the same polypeptide. In some cases, only one of these two domains may be active. However, GGDEF/EAL hybrid proteins are found, with the most studied being ScrC of Vibrio parahaemolyticus, in which signals lead these dual-domain enzymes to switch between DGC and PDE activity, according to Linda McCarter at the University of Iowa.
One of the most striking features of c-di-GMP signaling systems is the sheer number of DGCs and PDEs encoded by bacteria. For example V. cholerae encodes 41 GGDEF domains, 22 EAL domains, and 9 HD-GYP domains! Understanding why bacteria encode so many enzymes that modulate c-di-GMP is amajor unanswered question in this field. Researchers advanced two main hypotheses, which are not mutually exclusive, to explain this plenitude. The multitude of enzymes could be used to sample a variety of different environmental signals, or subsets of enzymes could be specifically linked to the control of specific behaviors.
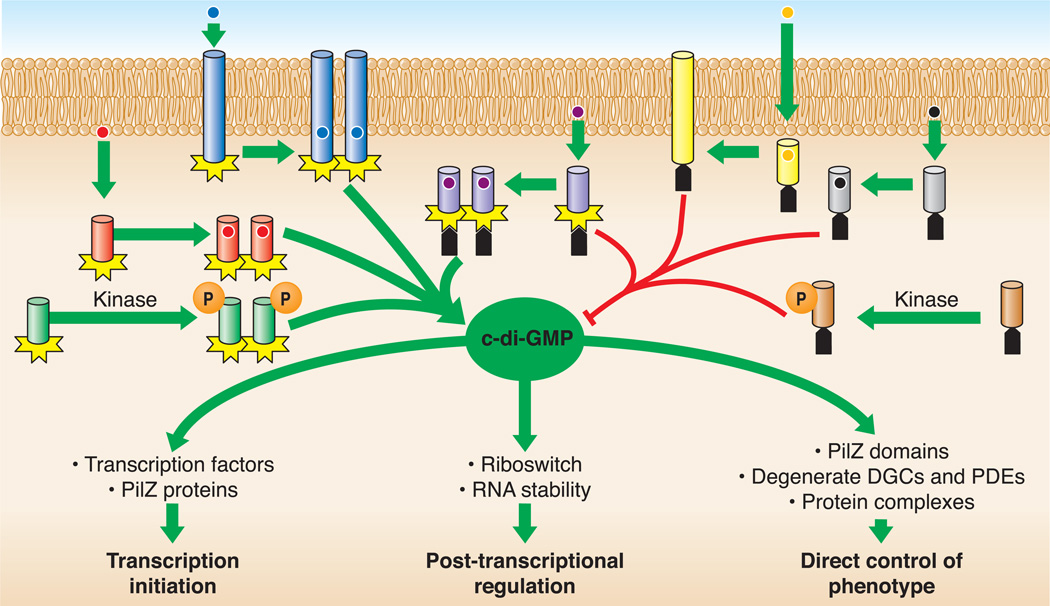
DGCs and PDEs, which reside in both the cytoplasm and inner membrane, are modular proteins that encode the catalytic domain in the C-terminus and signal reception domains in the N-terminus, reminiscent of two-component signaling systems (Fig. 2). The N-termini have divergent sequences that include a variety of signal reception domains such as PAS, GAF, or receiver domains. Generally, these N-terminal domains respond to specific environmental cues, controlling activity in the enzymatic domains. When signaling domains interact with stimulatory signals, the DGC enzymes form dimers, bringing the GTP-bearing GGDEF domains together to make c-di-GMP.
FIGURE 2.
Numerous environmental signals are translated into c-di-GMP to modulate downstream phenotypes. DGC domains (black plus) are modular in nature and are linked to various sensory domains. Recognition of stimulatory signals or modification by upstream signal transduction pathways, as indicated by phosphorylation (P) of a receiver domain by a kinase, drives dimerization to induce catalysis of c-di-GMP. These enzymes can be located either in the cytoplasm or inner membrane (the two lines refer to the inner and outer membrane of the cell). PDE domains (grey pentagon) are similar. A subclass of proteins contains both a DGC and PDE domain. C-di-GMP interacts with multiple effectors to regulate behaviors at the levels of transcription initiation, posttranscriptional control, and direct control of phenotypic output.
The activity cues are known for only a handful of GGDEF enzymes. These signals include nearby oxygen or small molecules, quorum sensing autoinducers, or physical components of the environment. Through these signal reception domains, these enzymes help cells adapt to specific environmental niches by controlling c-di-GMP-mediated behaviors, including the switch between the motile and sessile lifestyles. In support of this hypothesis, bacteria that commonly shift between different environments tend to encode more DGCs and PDEs, such as V. cholerae and P. aeruginosa, whereas those limited to more fixed environments, such as facultative intracellular pathogens, have few or no c-di-GMP signaling systems.
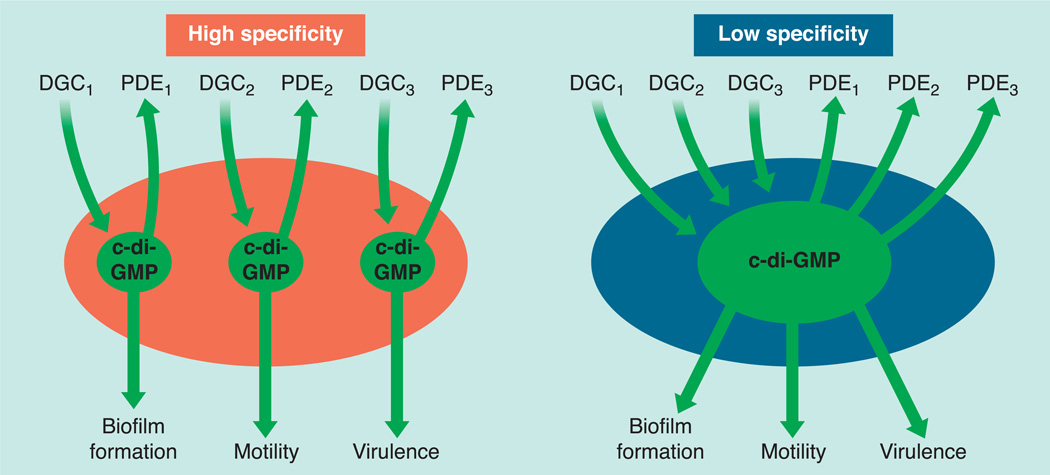
In some bacteria, specific DGC or PDE enzymes might preferentially control one or a subset of c-di-GMP regulated phenotypes (Fig. 3). Such high-specificity signaling entails parallel pathways with little crosstalk between distinct c-di-GMP “pools.” Alternatively, in low specificity systems, all DGCs and PDEs modulate the global c-di-GMP pool. In support of low-specificity signaling, DGC enzymes that show no homology apart from the conserved GGDEF domain can complement mutations in one another, suggesting that c-di-GMP synthesis is alone sufficient to restore activity. However, overproducing these enzymes may merely disrupt molecular mechanisms that segregate independent pools of c-di-GMP.
FIGURE 3.
A major question in the field of c-di-GMP signaling is the degree of signaling specificity. C-di-GMP may exist as a high specificity pathway in which segregated “pools” linked to specific DGCs or PDEs regulate downstream phenotypes via parallel signaling pathways (left). Alternatively, in a low specificity system all DGCs and PDEs modulate a common c-di-GMP hub that impacts all regulated phenotypes (right).
Meanwhile, for several species, including C. crescentus, Pseudomonas fluorescens, and V. cholerae, the intracellular concentrations of c-di-GMP do not directly correlate with phenotypic outputs, implying that DGCs or PDEs differentially contribute to regulating phenotypes. Thus, accumulating evidence suggests that c-di-GMP does not function via shared global intracellular signaling pools uniformly distributed throughout individual bacterial cells.
The c-di-GMP System Includes Many Kinds of Effectors
How are pools of specific signaling molecules segregated within bacterial cells? In C. crescentus, for example, differentially localized DGCs and PDEs could produce segregated pools of c-di-GMP. In addition, large protein complexes might segregate c-di-GMP molecules from the rest of the cell, enabling signaling specificity.
In another example, particular GGDEFs within the plant pathogen Xanthomonas campestris specifically associate with HD-GYP proteins in response to the cell density signal, called diffusible signal factor, according to Maxwell Dow at University College Cork in Ireland and his collaborators. This intriguing finding suggests that DGCs and PDEs themselves might form multicomponent protein complexes.
The control of output responses to changing c-di-GMP levels is also complex (Fig. 2). Unlike its cousin, the second messenger cyclic AMP, c-di-GMP binds to multiple effector molecules to control gene expression at all levels, including transcription initiation, posttranscriptional modulation, and allosteric regulation of protein activity. A number of factors directly bind c-di-GMP to regulate transcription, and this list is rapidly growing. These transcription factors fall into three general groups, namely, enhancer-binding proteins such as FleQ and VpsR; members of the FixJ, LuxR, and CsgD families, such as VpsT and CpsQ; and the cAMP activator protein (CAP) family, including Clp and Bcam1349. Moreover, c-di-GMP can bind to a PilZ-containing protein to control transcription.
In addition to regulating transcription, c-di-GMP binds to a widely distributed riboswitch motif, presumably altering other stages of gene expression. A riboswitch is an untranslated sequence of RNA that alters tertiary structure upon binding to a small ligand cofactor to control gene expression in cis. Two classes of c-di-GMP riboswitches, recognized as conserved DNA sequences, are found upstream of numerous genes in many species of bacteria. However, little is known about the impact of c-di-GMP riboswitches on c-di-GMP output responses.
C-di-GMP also binds to and modulates the activity of various proteins throughout the cell. The PilZ domain, first identified using bioinformatics analysis by Michael Galperin at the National Institutes of Health in Bethesda, Md., was the first protein shown to bind c-di-GMP. These domains, which are found throughout bacteria, associate in a modular fashion with other conserved domains. YcgR, the best-understood PilZ encoding protein, associates with the flagellar body and binds to c-di-GMP to inhibit motility by altering the rotation of the flagella or changing its speed.
Enzymatically inactive DGCs and PDEs also are c-di-GMP effectors. For example, in the Lap system of P. fluorescens, the inner membrane-associated protein LapD, which encodes an enzymatically inactive PDE enzyme, binds to c-di-GMP at its degenerate active site, according to George O’Toole and his collaborators at Dartmouth University. This binding leads to titration of the periplasmic protease LapG by LapD. At low levels of c-di-GMP, the conformation of LapD is altered, releasing LapG to cleave the outer-membrane anchored adhesion LapA, leading P. fluorescens to dissociate from biofilms.
The Exciting Future of c-di-GMP
With the explosion of genomic sequencing and interest in biofilms, the field of c-di-GMP signaling has grown from its birth and infancy in Benziman’s laboratory and continues to mature. As new organisms and new regulatory systems are found to be involved, c-di-GMP daily becomes more complex and difficult to understand. Unraveling this complexity and the role of c-di-GMP in both pathogenic and nonpathogenic behaviors is an exciting challenge. Important questions about the environmental cues that control DGC and PDE activity, signal integration, identification and characterization of downstream effectors, and the role of c-di-GMP in bacteria that have not yet been examined remain to be answered. This knowledge will provide a deeper understanding of how bacteria use this signaling molecule to respond and adapt to different environments. I am amazed that in under a decade this initially obscure field pioneered by Benziman has exploded into all facets of microbiology, and I am left to contemplate what new areas of microbiology remain hidden.
AUTHOR PROFILE.
Waters: Fascinated with the Natural World, and Studying Bacteria Fits His Personality
As a child, Chris Waters loved logic puzzles. “I could do those all day,” he says. “I think this desire to solve puzzles is one of the main reasons why I do what I do, and why I love doing it. Now, I’m just trying to solve nature’s puzzles, but the game is the same … I do it with things like genetic circuits, signaling pathways, the structure of communities, and the interactions of organisms.”
Waters, 37, in the Department of Microbiology and Molecular Genetics at Michigan State University (MSU), studies chemical signaling in bacteria, with the goal of better understanding how biofilms form and finding ways to disrupt them. “This is a really cool field to be in” he says. “There are many, many fundamental questions that remain to be answered.”
Waters and his older brother, a fisheries biologist, became the only scientists from a family of Methodist ministers. That religious background provides “an interesting perspective on the conflict between science and religion,” he says. “I am very grateful that I grew up in Kansas. It is such an open place with rolling prairies and a huge, wide, open sky that I think it taught me to not limit my ideas.” In his youth, Waters was active outdoors, including hunting, fishing, camping, canoeing, and hiking. “I’ve been very connected to and fascinated by the natural world,” he says. “I knew I wanted to do something in science, but I wasn’t sure what.”
Microbiology was not on his radar screen when he started his undergraduate studies at the University of Kansas, from which he graduated in 1997 with a B.S. in genetics. However, he found the enthusiasm of his first microbiology teacher, John Brown, contagious. “He had a passion for science and especially microbiology, which rubbed off on me,” he says. Brown also connected Waters with Eric Elsinghorst, who was studying pathogenic Escherichia coli, “and my work with him started my love affair with microbes,” Waters says. “Studying bacteria is a perfect fit for my personality, as I … try a lot of different experiments, and I’m not very patient.”
After completing his Ph.D. in microbiology at the University of Minnesota with Gary Dunny in 2002, Waters did postdoctoral research there and with Bonnie Bassler at Princeton University before moving in 2008 to MSU. “I’ve been lucky to have such fantastic mentors throughout my career,” he says. He describes three professional goals. “First, I want to be a good mentor and teacher, and help develop the next generation of scientists,” he says. “Second, I want my lab to discover fundamentally important new pieces of information. Third, I want to do something that benefits society. We have an obligation to apply our science towards improving the human condition.”
Waters is married to his high school sweetheart, Jennie, a stay-at-home mom, who also helps with administrative tasks in his lab. They have three children, Emma, 10, Bella, 8, and Luke, 5. When his oldest was in kindergarten, he visited her school’s science carnival to explain bioluminescent Vibrio harveyi to her classmates. ‘All of the kids could relate because of the glowing fish in “Finding Nemo,”’ he says. “They couldn’t believe that the flask of bacteria that I was holding had more bacteria in it than people on the planet.”
He enjoys athletics and plays basketball weekly, mostly with grad students. He also coaches his kids’ soccer and basketball teams. Despite living in Michigan, however, he continues to support the Kansas Jayhawks, he says. “My current Michigan State students are trying to convert me to be a full-blooded Spartan fan, but their efforts are in vain.”
Marlene Cimons
Marlene Cimons lives and writes in Bethesda, Md.
SUMMARY.
-
➤
Research on cyclic di-GMP (c-di-GMP) will lead to a deeper understanding of how bacteria use this signaling molecule and could yield new strategies for treating biofilm-based chronic infections.
-
➤
Moshe Benziman at the Hebrew University of Jerusalem and his collaborators identified c-di-GMP as a signal molecule that stimulates cellulose synthesis in bacteria.
-
➤
In general, c-di-GMP stimulates biofilm formation; however, it plays a complex role in disease, inhibiting acute infections and promoting chronic infections.
-
➤
A striking feature of c-di-GMP signaling systems is the sheer number of bacterial genes that encode enzymes to produce and degrade this signaling molecule.
-
➤
Those enzymes or other proteins might form complexes that effectively compartmentalize various c-di-GMP signaling systems within bacterial cells.
Acknowledgments
I am thankful to Brian Hammer, Michael Federle, and Jennie Waters for critical reading of the manuscript. Funding for my laboratory is provided by grants by NIH grants K22-AI-080937, the Region V ‘Great Lakes’ RCE NIH award U54-AI-057153, the NSF sponsored BEACON Science and Technology Center Cooperative Agreement No. DBI-0939454, and Michigan State University.
Suggested Reading
- Fang X, Gomelsky M. A post-translational, c-di-GMP-dependent mechanism regulating flagellar motility. Mol. Microbiol. 2010;76:1295–1305. doi: 10.1111/j.1365-2958.2010.07179.x. [DOI] [PubMed] [Google Scholar]
- Hickman JW, Harwood CS. Identification of FleQ from Pseudomonas aeruginosa as a c-di-GMP-responsive transcription factor. Mol. Microbiol. 2008;69:376–389. doi: 10.1111/j.1365-2958.2008.06281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasteva PV, Fong JC, Shikuma NJ, Beyhan S, Navarro MV, Yildiz FH, Sondermann H. Vibrio cholerae VpsT regulates matrix production and motility by directly sensing cyclic di-GMP. Science. 2010;327:866–868. doi: 10.1126/science.1181185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newell PD, Boyd CD, Sondermann H, O’Toole GA. A c-di-GMP effector system controls cell adhesion by inside-out signaling and surface protein cleavage. PLoS Biol. 2011;9:e1000587. doi: 10.1371/journal.pbio.1000587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul R, Weiser S, Amiot NC, Chan C, Schirmer T, Giese B, Jenal U. Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Genes Dev. 2004;18:715–727. doi: 10.1101/gad.289504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross P, Weinhouse H, Aloni Y, Michaeli D, Weinberger-Ohana P, Mayer R, Braun S, de Vroom E, van der Marel GA, van Boom JH, Benziman M. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature. 1987;325:279–281. doi: 10.1038/325279a0. [DOI] [PubMed] [Google Scholar]
- Sudarsan N, Lee ER, Weinberg Z, Moy RH, Kim JN, Link KH, Breaker RR. Riboswitches in eubacteria sense the second messenger cyclic di-GMP. Science. 2008;321:411–413. doi: 10.1126/science.1159519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischler AD, Camilli A. Cyclic diguanylate (c-di-GMP) regulates Vibrio cholerae biofilm formation. Mol. Microbiol. 2004;53:857–869. doi: 10.1111/j.1365-2958.2004.04155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimble MJ, McCarter LL. Bis-(3’-5’)-cyclic dimeric GMP-linked quorum sensing controls swarming in Vibrio parahaemolyticus. Proc. Natl. Acad. Sci. USA. 2011;108:18079–18084. doi: 10.1073/pnas.1113790108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe A, Visick K. The second messenger cyclic di-GMP. Washington, D.C.: ASM Press; 2010. [Google Scholar]