Dear Editor,
SWEETs represent a novel family of membrane sugar transporters that have been identified in plants, worms, and mammals. They selectively transport mono- or disaccharides across plasma or intracellular membranes, and are involved in a number of essential physiological processes1. The functions of SWEETs are best characterized in plants. In Arabidopsis thaliana, AtSWEET1/4/5/7/8/13 mediate glucose efflux1, AtSWEET11/12 function as sucrose transporters2, and AtSWEET17 permeates fructose3,4. These SWEETs are important for the growth and development of plants, and some are hijacked by pathogens or symbionts for their own sugar supply.
SWEETs belong to the MtN3 family in plants and SLC50 sugar efflux transporter family in human5. It was predicted that SWEETs comprise seven transmembrane (TM) helices that are folded into two parallel three-helix bundles connected by one central TM1,5. Homologues of SWEETs were recently identified in bacteria5. Each bacterial SWEET monomer consists of three TMs, reminiscent of one three-helix bundle in the eukaryotic SWEETs. Therefore they are named SemiSWEETs. A representative homologue from B. japonicum USDA 110, BjSemiSWEET1, exhibited sucrose transport activity5.
In an attempt to understand the molecular basis underlying substrate selectivity and transport mechanism of sugar transporters, we sought to determine the crystal structure of SemiSWEETs. We have focused on BjSemiSWEET1 and its close homologue from T. yellowstonii DSM 11347 (TySemiSWEET). Both homologues gave rise to high-yield and well-behaved recombinant proteins. We were able to crystalize both proteins purified in detergent micelles using hanging-drop diffusion method. However, these crystals never diffracted X-rays beyond 10 Å resolution. Finally, crystals of TySemiSWEET were obtained in the space group P212121 using lipidic cubic phase approach and diffracted X-rays beyond 2.4 Å resolution at BL32XU, SPring-8. The structure of TySemiSWEET was determined by molecular replacement using the recently reported structure of a SemiSWEET protein from L. biflexa (LbSemiSWEET)6 as search model and refined to 2.4 Å resolution (Supplementary information, Figure S1 and Table S1).
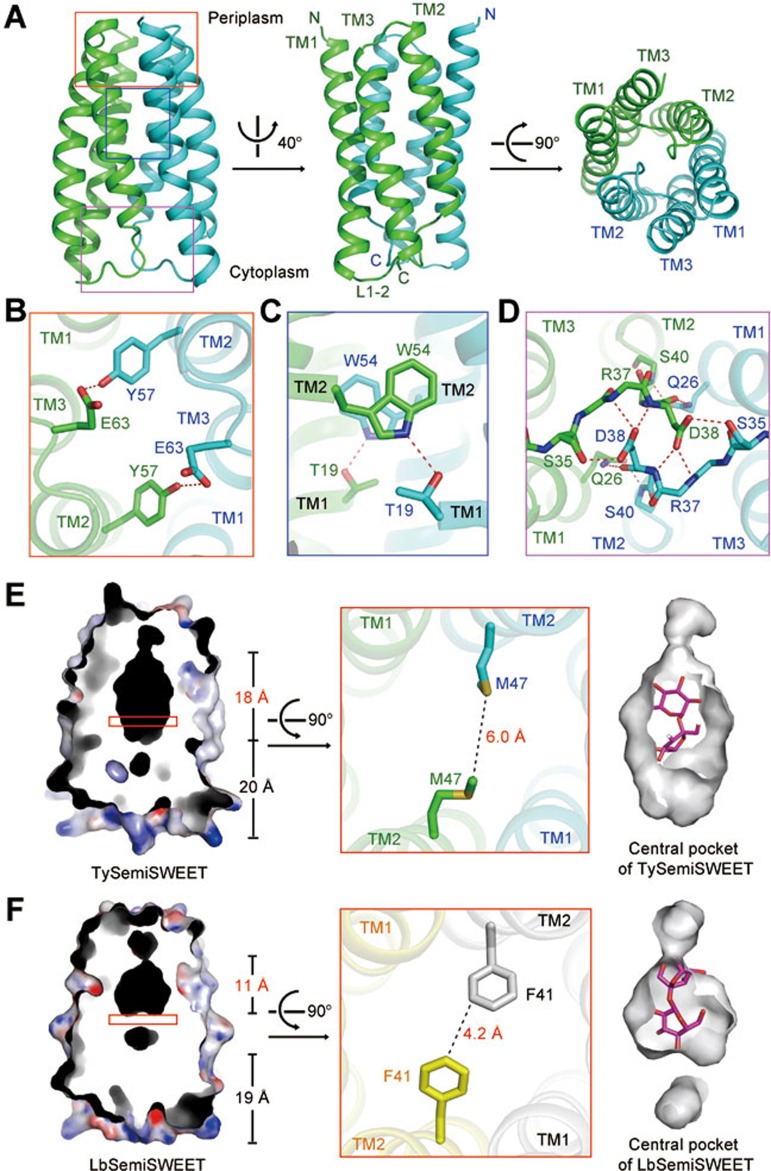
Six TySemiSWEET molecules that are arranged into three dimers were found in each asymmetric unit (Figure 1A and Supplementary information, Figure S1). While two dimers are arranged in a parallel fashion, the third one is positioned in the opposite orientation, further supporting the dimeric assembly of SemiSWEETs6. Within each dimer, the two parallel protomers that exhibit almost identical conformations are related by 180° rotation around an axis perpendicular to the membrane plane. Within each protomer, TM1 and TM2 are connected by an extended linker (L1-2), and TM3 is positioned between TM1 and TM2 (Figure 1A). Note that the L1-2 linker is enriched with positively charged residues (Supplementary information, Figure S2). According to the “positive-inside rule” and the topological analysis of AtSWEET11, the L1-2 should be located on the cytoplasmic side, which leaves the N-terminus of each protomer on the periplasmic side of the membrane (Figure 1A)6,7.
Figure 1.
Crystal structure of the SemiSWEET from T. yellowstonii (TySemiSWEET) in an occluded conformation. (A) Overall structure of the dimeric TySemiSWEET. The two protomers are colored green and cyan. (B-D) The dimer interface of TySemiSWEET consists of three clusters of H-bonds between residues on TM1 of one protomer and TM2 of the other, including a pair of H-bonds at the extracellular side (B), a pair close to the center of the membrane (C), and an extensive H-bond network on the cytoplasmic side (D). The H-bonds, together with extensive van der Waals contacts between the two protomers, sealed the dimer in an occluded conformation. (E-F) The central cavity of TySemiSWEET is considerably larger than that of LbSemiSWEET. Residues Phe41 from the two protomers of LbSemiSWEET close the central pocket at approximately the midway of the membrane, whereas the corresponding Met47 residues in TySemiSWEET leave enough space for an elongated cavity. A sucrose molecule can be accommodated by TySemiSWEET, but not LbSemiSWEET (right panels). All structure figures were prepared with PyMol10.
It is noteworthy that the basic structural and functional unit of the major facilitator superfamily (MFS) transporters is also the 3-helix bundle8. Interestingly, when TM1 and TM2 in each 3-helix bundle are aligned on the same plane, the position of TM3 in SemiSWEETs is on the opposite side to that of the MFS 3-helix bundle (Supplementary information, Figure S3). The difference in the organization of the 3-helix bundles may argue against a common ancestor between SWEET and MFS transporters.
The two protomers in each TySemiSWEET dimer enclose a central cavity that is sealed from both sides of the membrane. Therefore, the structure represents an occluded conformation. The dimer interface is mediated through three clusters of hydrogen bonds (H-bonds) between TM1 of one protomer and TM2 of the other (Figure 1B-1D) as well as extensive van der Waals contacts. Because of the 2-fold symmetry, described here are the interactions on one side of the dimer and the two protomers are named Mol A and Mol B. On the extracellular side, the hydroxyl group of Tyr57 of Mol A-TM1 donates a H-bond to the carboxyl oxygen of Glu63 which demarcates the beginning of TM2 in Mol B (Figure 1B). One helical turn below, Trp54 of Mol A is H-bonded to the side chain of Thr19 on Mol B-TM2 (Figure 1C). On the cytosolic side, an extensive H-bond network exists between the two L1-2 linkers, which appear to serve as the inner gate (Figure 1D). The side chain of Asp38 of Mol A accepts one hydrogen bond from the side chain hydroxyl group of Ser35 and donates two hydrogen bonds to the main chain amides of Arg37 and Asp38 of Mol B. The side chain of Gln26 of Mol A is H-bonded to both the main chain oxygen of Asp38 and the main chain amide of Ser40 of Mol B.
TySemiSWEET shares sequence identities of 44.2% and 40.2% with BjSemiSWEET1 and LbSemiSWEET, respectively (Supplementary information, Figure S4). The structure of LbSemiSWEET was also determined in an occluded conformation6. The dimeric structures of TySemiSWEET and LbSemiSWEET can be superimposed with a root mean squared deviation (r.m.s.d.) of 0.64 Å over 144 Cα atoms. However, evident difference can be observed in the central pockets of the two highly similar structures (Figure 1E and 1F). The central pocket of TySemiSWEET is 18 Å long, with an overall surface of 463 Å2 and volume of 613 Å3. In contrast, the central pocket of LbSemiSWEET is 11 Å long, with an overall surface of 327 Å2 and volume of 424 Å3. The difference is caused mainly by variation of one amino acid, Met47 in TySemiSWEET versus the corresponding Phe41 in LbSemiSWEET. The bulky side groups of the two Phe41 in LbSemiSWEET dimer close the central pocket in the midway of the membrane, whereas Met47 residues in TySemiSWEET leave enough space for an elongated central pocket (Figure 1E, 1F and Supplementary information, Figure S4).
During structural refinement of TySemiSWEET, an extended electron density was seen in the central cavity. The quality of the density is insufficient for accurate identification and assignment of a ligand. Nevertheless, the crystals of TySemiSWEET were obtained in the presence of 20 mM sucrose, and the contour of the electron density is reminiscent of a disaccharide molecule (Supplementary information, Figure S5A). We therefore tentatively built a sucrose molecule into the density. The disaccharide molecule can be accommodated by the surrounding residues in the central pocket (Figure 1E, and Supplementary information, Figure S5B). Notably, all the 16 residues in each protomer that constitute the central pocket are highly conserved, with 10 invariants, between TySemiSWEET and BjSemiSWEET1 (Supplementary information, Figure S5C), indicating a similar central pocket in BjSemiSWEET1, the sucrose transporter. In contrast, the pocket in LbSemiSWEET is too small to accommodate a disaccharide molecule, consistent with its function of being a glucose transporter (Figure 1F, right panel).
Structural comparison of TySemiSWEET and LbSemiSWEET provides important clue to understanding substrate selectivity of SemiSWEETs. If the central cavities observed in TySemiSWEET and LbSemiSWEET represent the primary binding site for ligands in SemiSWEETs, two questions immediately stand out. First, is there a second substrate binding site along the transport path? Second, what is the transport mechanism?
All the known transporters utilize a general alternating access mechanism which requires the structural shifts of a transporter between at least two conformations, the outward-open and inward-open, to upload and release of the substrate(s) on the two sides of the membrane. The positions of the cavity in TySemiSWEET and LbSemiSWEET, both located closer to the periplasmic side, deviate from those in the known structures of sugar transporters, in which a primary substrate binding site is usually placed in the midway of the membrane8,9. Examination of the transport path of TySemiSWEET and the other two SemiSWEET structures reveals that the cytoplasmic half of the putative transport path is highly hydrophobic, and may not constitute a stable binding site for the highly polar substrate (Supplementary information, Figure S6)6. Notably, the hydrophobic half-path is also observed in the transmembrane domains of the ABC (ATP-binding cassette) maltose transporter MalFG. Structures of the maltose transporter were obtained in multiple conformations and a single binding site in the transmembrane domain is identified9. It is possible that TySemiSWEET only contains one substrate binding site as observed in the structure. The enriched hydrophobic residues along the transport path may facilitate the passage of the polar ligand by lowering potential resistance during substrate penetration.
The structures of the outward-open and occluded states have been obtained for the SemiSWEETs6. An inward-open structure is yet to be captured to elucidate the alternating access cycle of SemiSWEETs. On top of that, an intriguing and critical question in the study of SemiSWEETs and SWEETs is the driving force for their conformational changes. It remains to be characterized whether SemiSWEETs and SWEETs are facilitative uniporters or secondary active co-transporters. The structures reported here and previously lay out the foundation to address these important questions.
The atomic coordinates have been deposited in the Protein Data Bank with the accession code 4RNG.
Acknowledgments
We thank Meng Ke, Ruobing Ren, Wenlin Ren, and Xiangyu Liu for technical assistance. This work was supported by funds from the Ministry of Science and Technology of China (2011CB910501 and 2015CB910101). The research of Nieng Yan was supported in part by an International Early Career Scientist grant from the Howard Hughes Medical Institute and an endowed professorship from Bayer Healthcare.
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
Structure determination of TySemiSWEET.
References
- Chen LQ, Hou BH, Lalonde S, et al. Nature. 2010. pp. 527–532. [DOI] [PMC free article] [PubMed]
- Chen LQ, Qu XQ, Hou BH, et al. Science. 2012. pp. 207–211. [DOI] [PubMed]
- Chardon F, Bedu M, Calenge F, et al. Curr Biol. 2013. pp. 697–702. [DOI] [PubMed]
- Guo WJ, Nagy R, Chen HY, et al. Plant Physiol. 2014. pp. 777–789. [DOI] [PMC free article] [PubMed]
- Xuan YH, Hu YB, Chen LQ, et al. Proc Natl Acad Sci USA. 2013. pp. E3685–E3694. [DOI] [PMC free article] [PubMed]
- Xu Y, Tao Y, Cheung LS, et al. Nature 2014. Sep 3. doi: 10.1038/nature13670 [DOI]
- von Heijne G, Gavel Y. Eur J Biochem. 1988. pp. 671–678. [DOI] [PubMed]
- Yan N. Trends Biochem Sci. 2013. pp. 151–159. [DOI] [PubMed]
- Chen J. Curr Opin Struct Biol. 2013. pp. 492–498. [DOI] [PMC free article] [PubMed]
- DeLano WL . http://www.pymol.org 2002
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Structure determination of TySemiSWEET.