Abstract
The role of Fat Mass and Obesity-associated protein (FTO) and its substrate N6-methyladenosine (m6A) in mRNA processing and adipogenesis remains largely unknown. We show that FTO expression and m6A levels are inversely correlated during adipogenesis. FTO depletion blocks differentiation and only catalytically active FTO restores adipogenesis. Transcriptome analyses in combination with m6A-seq revealed that gene expression and mRNA splicing of grouped genes are regulated by FTO. M6A is enriched in exonic regions flanking 5′- and 3′-splice sites, spatially overlapping with mRNA splicing regulatory serine/arginine-rich (SR) protein exonic splicing enhancer binding regions. Enhanced levels of m6A in response to FTO depletion promotes the RNA binding ability of SRSF2 protein, leading to increased inclusion of target exons. FTO controls exonic splicing of adipogenic regulatory factor RUNX1T1 by regulating m6A levels around splice sites and thereby modulates differentiation. These findings provide compelling evidence that FTO-dependent m6A demethylation functions as a novel regulatory mechanism of RNA processing and plays a critical role in the regulation of adipogenesis.
Keywords: N6-methyladenosine (m6A), METTL3, FTO, mRNA splicing, adipogenesis
Introduction
The Fat Mass and Obesity-associated (FTO) protein belongs to the AlkB family of non-heme Fe (II)/dioxygenases1. FTO is the first gene shown to contribute to non-syndromic human obesity2,3,4. In vivo studies using FTO overexpression or knockout mouse models have revealed abnormal adipose tissues and body mass, suggesting a pivotal role of FTO in adipogenesis and energy homeostasis5,6,7,8. In vitro FTO demethylates various methylated nucleic acids1,9,10. However, only demethylation of N6-methyladenosine (m6A) in RNA has been confirmed in vivo9. First identified in mammalian mRNA in the 1970s, m6A has been proposed to regulate mRNA processing including alternative splicing, RNA degradation and translation11,12,13,14,15. In both mammals and yeast, m6A modification of RNA preferentially occurs within the consensus sequence RRACH (R = G or A; H = A, C or U) in gene coding regions and 3′UTRs, implicating its fundamental roles in RNA processing and translational control16,17,18,19. There is a dramatic increase in m6A levels during embryonic development and neuronal maturation in mammals and recent studies implicate that m6A destabilizes developmental regulators in embryonic stem cells, and that the speed of the circadian clock is also regulated by m6A -dependent RNA processing17,18,20,21. In addition, in neuron-specific FTO-deficient mice, a subset of mRNAs involved in dopaminergic (DA) signaling shows increased m6A levels22. FTO is most highly expressed in fetal tissues and in adult brain, adipose and muscle tissues. FTO deficiency results in a lean phenotype in mice5,23. However, the exact role of FTO in energy homeostasis and body weight control remains unclear24. Recently YTHDF2 was identified as an m6A-binding protein involved in the regulation of translation and stability of m6A-containing mRNAs25. The recent identification and characterization of the m6A methyltransferase complex (METTL3-METTL14-WTAP) and its role in RNA splicing further highlight the biological significance of m6A modification26,27,28.
Splicing of mRNA precursors (pre-mRNA) to mature mRNAs is a highly dynamic and complex process influencing most biological functions in eukaryotes. RNA splicing is controlled by both cis-regulatory sequences within the RNA and trans-regulatory proteins. Two major classes of trans-acting factors, serine/arginine-rich (SR) proteins and heterogeneous ribonucleoproteins (hnRNPs), impact splice site recognition29,30. Splice site selection is determined by cis-acting RNA sequences including intronic dinucleotides GU and AG (with variations used by the minor spliceosome) and the surrounding sequence context such as exonic/intronic splicing enhancers (E/ISEs) and silencers (E/ISSs)31. Other factors including factors modulating the chromatin and transcription complexes also affect RNA splicing32. Chemical inhibition of m6A formation causes changes in the ratio between precursor and mature mRNAs33, and m6A has been observed to be significantly enriched in both multi-isoform genes and alternatively spliced exons18. Consistently, binding clusters of the m6A methyltransferase components WTAP and METTL3 are preferentially present in genes with multiple isoforms27. Depletion of either METTL3 or ALKBH5 causes abnormal mRNA splicing as evident in genes in the p53 signaling pathways18,34. Speckle-localization of the m6A methyltransferase WMM complex components as well as demethylases ALKBH5 and FTO further supports the role of m6A in RNA processing9,27,34. ALKBH5 deficiency causes diminished SRSF1 (ASF/SF2) signal in the speckle34. Taken together, all these observations suggest a cis-regulatory element-dependent role of m6A in RNA splicing.
The reported association of FTO activity with obesity prompted us to investigate how FTO activity influences RNA processing through regulation of m6A abundance during adipogenesis. By combining transcriptome and m6A-seq analyses, we demonstrate that m6A is enriched in exons adjacent to intronic 5′/3′splice sites flanking spliced exons. In addition, we observe a striking spatial overlap of m6A sites with mRNA exonic splicing enhancers recognized by SRSF2. Furthermore our results suggest that FTO controls mRNA splicing by regulating the RNA binding ability of SRSF2, and that FTO-regulated m6A demethylation acts as a novel regulatory mechanism of RNA splicing, explaining its important role during adipogenesis.
Results
FTO-dependent m6A demethylation regulates adipogenesis
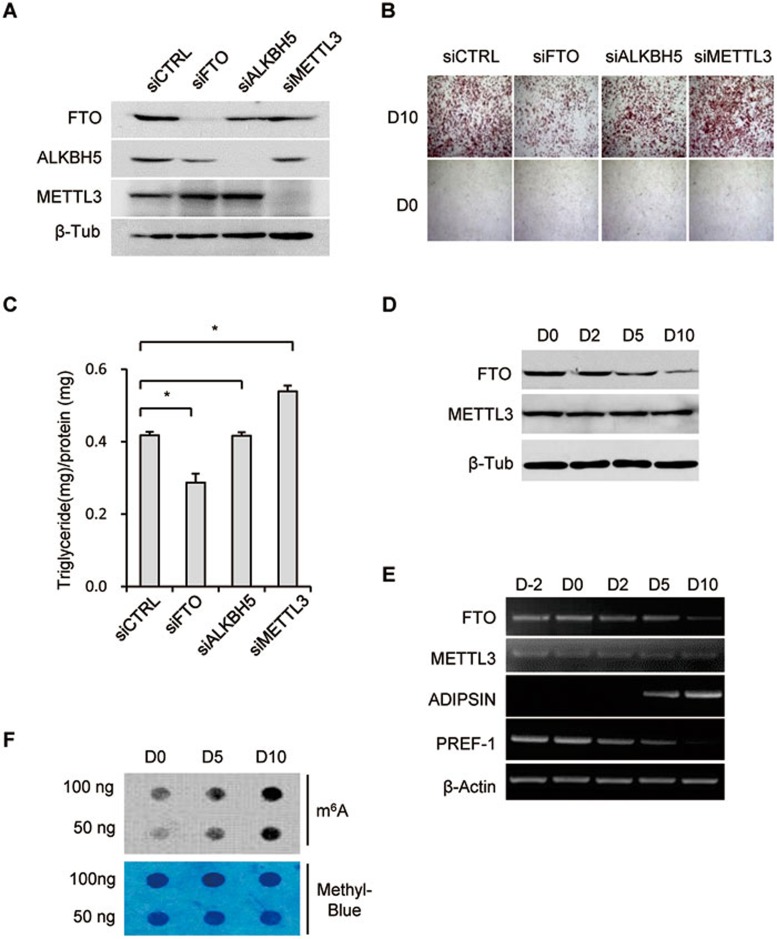
To investigate the function of m6A modification in adipogenesis, we took advantage of the well-characterized mouse 3T3-L1 pre-adipocyte cell line (Supplementary information, Figure S1A). FTO, the other m6A demethylase ALKBH5 or methyltransferase METTL3 was depleted individually by siRNA and then the pre-adipocytes were induced to differentiate (Figure 1A). Interestingly, FTO depletion severely impaired differentiation as demonstrated by Oil Red O staining (Figure 1B) and triglyceride content measurements (Figure 1C). This is consistent with decreased fat tissue accumulation observed in FTO-null mouse5,8,35. While METTL3 deficiency appeared to promote pre-adipocyte differentiation (Figure 1A-1C), ALKBH5 which is important for spermatogenesis34, had no obvious effect on adipogenesis (Figure 1A-1C). Since FTO and METTL3 have opposing catalytic activities, their different effects on differentiation may be caused by changes in m6A dynamics when either of them is depleted.
Figure 1.
FTO depletion interferes with adipogenesis. (A-C) 3T3-L1 pre-adipocytes (Day −2) were treated with FTO, ALKBH5, METTL3 or control siRNA. Forty-eight hours later, cells were lysed and subjected to immunoblotting with the indicated antibodies (A). Forty-eight hours following siRNA treatment differentiation was induced by incubation with the differentiation cocktail (IBMX/DEX/Insulin) on Day 0. Differentiation status was determined by Oil Red O staining (B) and triglyceride assay (C) on Day 0 and 10. (C) Triglyceride content was quantified and normalized to protein content. *P < 0.05 is considered significant. Results are shown as mean ± SD. (D) 3T3-L1 cells collected at different time points (D0/2/5/10) during adipogenesis were lysed and subjected to immunoblotting with the indicated antibodies. β-tublin was used as loading control. (E) RT-PCR detected the expression levels of FTO, METTL3, as well as adipogenic markers, including ADIPSIN and PREF-1, during adipocyte differentiation. β-Actin was used as loading control. (F) mRNA was isolated from multiple stages (D0/5/10) of adipogenesis and used in dot blot analyses with m6A antibody. mRNA was loaded by two-fold serial dilution. The m6A contents are shown in the upper panel. Equal loading of mRNA was verified by methylene blue staining (lower panel). See also Supplementary information, Figure S1.
We next measured the expression of FTO and METTL3 during 3T3-L1 pre-adipocyte differentiation. Interestingly, the expression of FTO decreased during differentiation and reached the lowest level in mature fat cells (D10) (Figure 1D, 1E and Supplementary information, Figure S1A), while the global m6A levels were increased during the course of differentiation and reached the highest level in mature adipocytes (Figure 1F). METTL3 expression remained unchanged (Figure 1D and 1E). Successful differentiation was evidenced by the characteristic increase in expression of the adipocyte marker, complement factor D (ADIPSIN), and decrease in expression of preadipocyte factor-1 (PREF-1)36,37,38 (Figure 1E).
Differentiation of FTO-depleted pre-adipocytes could be fully restored by complementation with wild-type (WT) FTO but not by empty vector control or a FTO mutant protein in which three residues critical for FTO catalytic activity had been mutated1 (H231A/D233A/H307A, referred to as HDH; Supplementary information, Figure S1B-S1D). These results indicate that FTO regulates adipogenesis by modulating m6A levels.
We next performed RNA-seq and m6A-seq analyses in control (siCTRL) and FTO-deficient (siFTO) 3T3-L1 pre-adipocytes, as well as in 3T3-L1 pre-adipocytes at five stages (D–2/0/2/5/10) of adipogenesis. RNA-seq analysis was also performed in METTL3-depleted 3T3-L1 cells. All of the cDNA libraries were sequenced using Hiseq 2000 (Illumina, 101 bp) and at least two biological replicates were performed for each experiment except for the experiments where we analyzed changes in five stages of adipogenesis. The deep sequencing data were mapped to mouse genome version mm10 without any gap and a maximum of two mismatches were allowed. The clean reads, mapped reads, uniquely mapped reads and mapping rates for all 22 samples are listed in Supplementary information, Table S1. Overall, 22-69 million uniquely mapped reads with mapping quality higher than or equal to 20 were obtained for these samples and their mapping rates ranged from 72% to 90%. The m6A regions (m6A peaks) were identified by comparing the read abundance between m6A-Seq and RNA-seq samples of the same locus according to a method previously described17. In total, 8 710 and 10 137 m6A-modified genes in control (siCTRL) and FTO-depleted (siFTO) cells with significant m6A enrichment were identified, respectively (Supplementary information, Table S2). This is consistent with the overall increase of m6A levels in FTO-depleted cells compared with control cells as observed by dot blot analysis (Supplementary information, Figure S2A). The m6A distribution profiles and motifs are similar to previous observations17,18, verifying the successful enrichment of m6A-mRNAs by m6A-IP (Supplementary information, Figure S2B and S2C). Recent reports demonstrate that the METTL3-METTL14-WTAP complex is responsible for N6-methylation of adenosine in mRNA26,27,28. Therefore, we speculated that FTO and METTL3 might dynamically regulate mRNA m6A levels and gene expression. We found that the expression of 5 071 genes was affected by FTO depletion, while 6 065 genes were altered by METTL3 depletion. By overlapping differentially expressed genes (DEGs) upon FTO and METTL3 knockdown, we found that 524/553 genes were up/downregulated by FTO depletion while down/upregulated by METTL3 depletion (Supplementary information, Figure S2D). 428 of these 1 077 (524 + 553) genes showed increased m6A levels upon FTO depletion, suggesting that m6A modification in this subset of genes is co-regulated by FTO and METTL3 (Supplementary information, Figure S2E and S2F). To investigate the m6A distribution patterns within these 428 genes in more detail we divided each transcript into 5′-UTR, CDS and 3′-UTR regions. 174 genes showed no changes in overall m6A distribution pattern but higher m6A levels within individual peaks while in the other 254 genes, 337 new m6A peaks were identified. New m6A sites were mainly located in CDS (179) and 3′-UTR (135) regions and ∼90% of the modifications were located in RRACH consensus motifs (Supplementary information, Figure S2G). As expected, only 74 m6A peaks disappeared upon FTO depletion and 63 of them were located in 5′-UTR regions.
Gene ontology (GO) analysis revealed that the 428 co-regulated genes were enriched in lipid synthesis, protein localization and transport, cellular movement and translation-related pathways, suggesting that FTO and METTL3 might co-regulate a group of functionally diverse genes (Supplementary information, Figure S2H).
Next we characterized the m6A dynamics during adipogenesis by performing m6A-seq at five stages (D–2/0/2/5/10) of adipogenesis. 11 427 genes with m6A were observed at the five stages and 6 313 of them were found to contain m6A modification throughout the five stages (Supplementary information, Figure S3A). The GGAC (A/U) motif with m6A modification was highly enriched at all stages (Supplementary information, Figure S3B). Overall, m6A level was inversely associated with gene expression as illustrated in the heatmap (Supplementary information, Figure S3C). Genes highly expressed at early stages of differentiation were associated with regulation of transcription, cellular metabolic processes and cell growth, while highly expressed genes in fat cells were involved in the regulation of adipocyte differentiation and oxidation reduction processes (Supplementary information, Figure S3C).
FTO-dependent demethylation of m6A controls RNA splicing
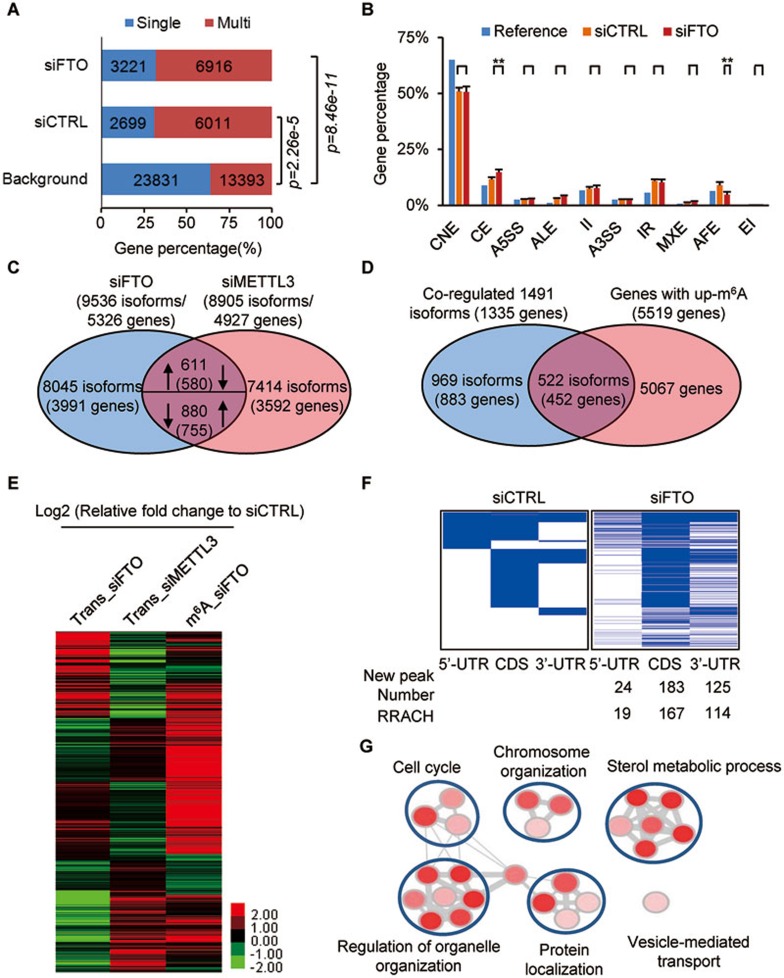
Previous findings have shown that m6A demethylases and members of the m6A methyltransferase complex are localized in nuclear speckles, suggesting a potential role of m6A in RNA splicing9,27,34,39. Analyses by Cufflink showed that 279 out of 694 genes without obvious changes in gene expression levels (fold change < 2) but with significant changes in the main isoforms (fold change > 2), were m6A-modified (P = 2.2e-10, hypergeometric test). Scripture analysis showed that 6 329 of 7 374 genes with differently spliced isoforms were m6A-modified (P < 1e-305, hypergeometric test). When comparing single- and multi-isoform genes with the Ensembl-annotated reference (background), it was evident that m6A is more common in multi-isoform genes compared to single-isoform genes (Figure 2A). In addition, the average number of m6A peaks per gene was also higher in multi-isoform genes (4.52 peaks per gene) compared to single-isoform genes (3.65 peaks per gene). At the exon/intron level, 376 of 798 differentially expressed exons (P = 3.9e-27, hypergeometric test) and 1 613 of 3 521 differentially expressed introns (P = 2.6e-105, hypergeometric test) bare m6A. Comparing the m6A-modified exons with Ensembl-annotated genomic exons, we found that 51% of the constitutive exons (CNE) were m6A-modified, which is 14% less than the 65% predicted by the Ensembl-annotation (P < 2.2e-16, Fisher's exact test). In contrast, m6A was overrepresented in both alternative cassette (CE) exons and intron retention (IR) splicing events, and m6A peaks within CE exons increased upon FTO depletion (P < 0.01, t-test) (Figure 2B), suggesting that FTO directly regulates m6A levels during this type of splicing.
Figure 2.
FTO affects RNA splicing via modulating m6A. (A) The percentage of m6A-mRNAs in control (6 011 multi-isoform/2 699 single-isoform, P = 2.26e-5, Fisher test) and FTO-knockdown cells (6 916 multi-isoform/3 221 single-isoform, P = 8.46e-11, Fisher test) derived from single-isoform or multi-isoform genes compared to the distribution predicted by the Ensembl-annotation reference (background). (B) The distribution of m6A-containing exons across different categories of splicing events in both control (orange) and FTO-depleted samples (red) compared to that Ensembl-annotation reference (Blue). P value for each category was calculated between siCTRL and siFTO samples. **P < 0.01 (Student's t-test) is considered significant (n = 2). Results are shown as mean ± SD. CNE, constitutive exon; CE, cassette exon; A5SS, alternative 5′ splice site; ALE, alternative last exon; II, intron isoform; A3SS, alternative 3′ splice site; IR, intron retention; MXE, mutually exclusive exons; AFE, alternative first exon; EI, exon isoform. (C) 1 491 isoforms (1 335 genes) are co-regulated by FTO and METTL3. Arrows pointing up/down indicates up/down-regulation. (D) FTO depletion resulted in increased m6A levels in 522 isoforms (452 genes) of the 1 491 isoforms (1 335 genes) co-regulated by FTO and METTL3 (shown in C). (E) The heatmap shows the expression levels of 522 reverse-regulated isoforms by FTO and METTL3, as well as the m6A modification in FTO-depleted cells. (F) The heatmap shows m6A peaks in 5′-UTR, CDS, and 3′-UTR (522 isoforms co-regulated by FTO and METTL3) in control and FTO-deficient cells. Blue lines represent m6A peaks. Each horizontal line represents one gene. The number of new m6A peaks upon FTO depletion and new m6A peaks within a RRACH motif are shown below the heatmap. (G) Function enrichment analysis of 522 isoforms (452 genes) based on the DAVID GO analysis result. The cutoff parameters for enrichment analysis with Cytoscape software are: P < 0.005, FDR q < 0.1, overlap cutoff > 0.5. See also Supplementary information, Figures S2-S4.
We next used Cufflink to calculate the m6A level of each isoform using the FPKM (fragments per kilobase of transcript per million mapped reads) method to make the estimate. When comparing the isoform changes upon FTO and METTL3 depletion, an inverse expression pattern was observed in 1 491 (611 + 880) isoforms of 1 335 genes (580 + 755) (Figure 2C). Of these, 452 genes (522 isoforms) showed increased m6A levels (in either peak number or peak enrichment) following FTO knockdown (Figure 2D and 2E), suggesting that m6A may directly influence isoform expression in this subset of genes. Analysis of m6A distribution in 5′-UTR, CDS and 3′-UTR segments revealed that 214 of the 452 co-regulated genes contained conserved m6A sites, while 332 new m6A peaks had occurred in the other 238 genes upon FTO depletion (Figure 2F). GO analysis showed enrichment of genes with functions related to sterol metabolic processes and organelle organization (Figure 2G). During the course of adipogenesis, we identified 17 637 significantly differentially expressed isoforms. M6A levels at different stages were illustrated in a heatmap (Supplementary information, Figure S3D). A representative gene FBXO9 displays increased m6A levels in FTO-depleted 3T3-L1 cells and progressively rising m6A levels during adipocyte differentiation (Supplementary information, Figure S4).
M6A sites spatially overlap with 5′ and 3′ exonic flanking sequences
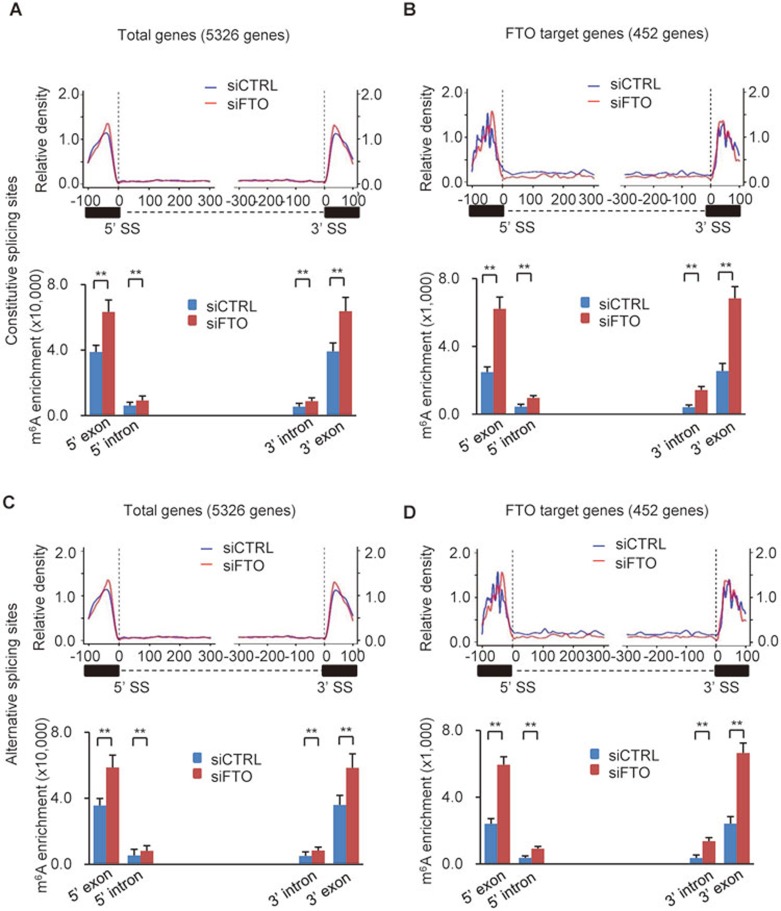
Cis-elements found within exons, such as exonic splicing enhancers (ESEs) and exonic splicing silencers (ESSs) in the vicinity of splice sites (both 5′ and 3′ splice sites), play important roles in regulating inclusion or skipping of exons40. In addition, in human and mouse the conserved intronic sequences flanking both sides of an alternatively spliced exon have also been proposed to be involved in the regulation of alternative splicing41,42. The findings presented so far in combination with previous reports suggest the possible involvement of m6A in the regulation of RNA splicing18,27. To further test this, we examined the distribution of m6A sites in the vicinity of splice sites. Using procedures reported previously43, we analyzed m6A peak distributions in mRNAs within 100-300 nt upstream or downstream of the constitutive splice sites. Meanwhile, to analyze the effect of FTO depletion on m6A peak number and enrichment, total m6A enrichments for m6A peaks located within 100 nt up- and downstream of 5′ and 3′ splice sites were calculated, respectively. For 5 326 genes showing changes in isoform expression upon FTO depletion, a significant overrepresentation of m6A sites in 5′ and 3′ exonic sequences flanking the constitutive splice site was observed when compared to intronic sequences adjacent to the splice site (Figure 3A and Supplementary information, Table S3). FTO deficiency further increased the m6A enrichment in these regions (Figure 3A, lower panel and Supplementary information, Table S3). For the 522 isoforms (452 genes) that were inversely affected in expression levels by FTO and METTL3 depletion and showed increased m6A levels upon FTO depletion, the effect of FTO depletion on m6A levels in regions surrounding constitutive splice sites was even more pronounced (5′ exon, 2.50-fold; 3′ exon, 2.68-fold; Figure 3B, lower panel). Similar results regarding m6A distribution pattern and enrichment were obtained around alternative splice sites (Figure 3C and 3D). On the whole, although FTO depletion barely affected the overall distribution pattern of m6A peaks, it increased the m6A levels at individual m6A peaks (Figure 3A-3D). We next analyzed previously published m6A-Seq/MeRIP-Seq datasets17,18. Consistently, in these datasets the majority of m6A peaks in exons were also mainly located in regions adjacent to constitutive and alternative splice sites (Supplementary information, Figure S5A).
Figure 3.
m6A is enriched at exonic splice sites. (A-D) Schematic analysis of the distribution of m6A peaks (upper panel in each figure; 100 nt upstream and 300 nt downstream from 5′SS; 300 nt upstream and 100 nt downstream from 3′SS) and m6A enrichment (lower panel in each figure; 100 nt up/downstream of the 5′ SS or 3′ SS) along 5′ exon-intron and 3′ intron-exon boundaries in control (blue) and FTO-deficient (red) cells. (A, C) m6A density in the vicinity of constitutive SS (A) or alternative SS (C) in the 5 326 genes showing changes in isoform expression upon FTO depletion. (B, D) Similar to A and C but here the m6A density was determined in the 452 FTO target genes showing increased m6A levels after FTO knockdown. The m6A levels in each region between control and FTO depletion are shown as mean ± SD and statistical analysis on their difference were performed by Student's t-test. **P < 0.01. See also Supplementary information, Figures S4 and S5.
The spatial overlap of m6A with constitutive/alternative splice sites was observed at all stages of adipogenesis (Supplementary information, Figure S5B-S5E). Consistent with our above findings that the global m6A levels increased while FTO expression levels decreased during the course of adipocyte differentiation (Figure 1D and 1E), a minor increase in m6A levels around splice sites was evident in more differentiated cells (Supplementary information, Figure S5B-S5E). Taken together, these observations strongly support a regulatory role of m6A in RNA splicing.
M6A sites show a spatial overlap with SRSF1- and 2-binding clusters
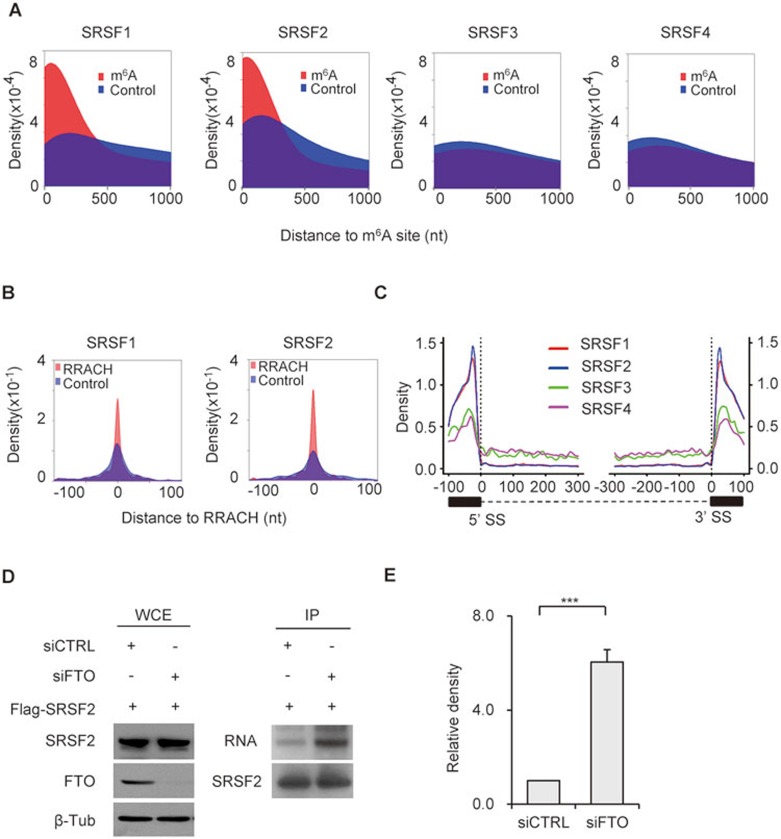
SR proteins are important for splice site recognition and intron processing. They recognize the pre-mRNA cis-acting elements, ESEs, and thereby participate in the selection of splice sites. ESEs recognized by SRSF1 and 2, like m6A, are highly enriched at the edges of exons44, suggesting a potential cooperative role between SRSF1/2 and m6A in RNA splicing. To test this, we calculated the distances between SRSF1-4-binding clusters based on the previously published CLIP-seq datasets45,46 and the nearest m6A sites. As a control, randomly selected sites with the same length as the m6A peak within the same exons were generated by BEDTools' shuffleBed. There was a significant spatial overlap between SRSF1- and 2-binding clusters and m6A sites (Figure 4A). In contrast, no overlap between SRSF3, 4 or ribonucleoproteins hnRNPC/hnRNRH47,48 and m6A sites was detected (Figure 4A and Supplementary information, Figure S6A-S6D). PhyloP scores across 60 vertebrates49 generated by comparing the specific sites with random exonic regions of the same size indicated a significant conservation of the overlapping m6A and SRSF1- and 2-binding sites (P = 2.2e-16, Kolmogorov-Smirnov test; Supplementary information, Figure S7A). Furthermore, the potential SRSF1- or 2-recognized ESEs identified from m6A-modified exons by ESEfinder50,51 significantly overlapped with m6A sites. These predicted ESEs were also enriched with the RRACH motif found in m6A sites (Figure 4B). In details, 60%-80% of the predicted ESEs overlapped with m6A sites (P = 2.2e-16, Fisher test) and 20%-30% overlapped with the RRACH motif (P = 2.2e-16, Fisher test). Collectively, these results suggest that m6A might specifically influence the binding of SRSF1 and 2 to ESEs.
Figure 4.
m6A modification influences RNA binding ability of SRSF2. (A) Density plots showing the relative distance (X-axis), calculated by BEDTools' closestBed, between m6A sites and SRSF1-4-binding sites. Randomly selected sites (blue color) within the same co-regulated genes were used as control. (B) The distance between the top-ranking SRSF1 and 2 ESEs predicted by ESEfinder and the nearest RRACH in identified m6A peak (red color) were calculated by BEDTools' closestBed (X axis). The distances of m6A-modified RRACH sites to the ESEs predicted by ESEfinder were calculated and their relative density distributions were shown as density plot. Randomly selected sites (blue color) within the same co-regulated genes were used as control. (C) Analysis of the distribution of SRSF1-4-binding clusters along 5′ exon-intron and 3′ intron-exon boundaries (100 nt upstream and 300 nt downstream of 5′SS; 300 nt upstream and 100 nt downstream of 3′SS). (D) PAR-CLIP of SRSF2 in control and FTO-depleted 3T3-L1 cells transfected with the pCS2-Flag-SRSF2 plasmid. SRSF2 protein-RNA complex was pulled down with Flag M2 Affinity Gel (mouse), and then RNA was labeled and detected following instruction of biotin labeling kit. The expression levels of SRSF2 protein in whole cell lysate (WCE) was detected by western blotting with Flag antibody (rabbit). Knockdown efficiency of FTO was detected with its specific antibody. β-tubulin was used as loading control. (E) RNA binding ability of SRSF2 was calculated by normalizing binding RNAs to the corresponding pull-downed proteins. Statistical analysis shows the relative binding ability of SRSF2 in FTO-depleted cells (P < 0.0001) to that in control cells (siCTRL). See also Supplementary information, Figures S6-S7.
Consistently, like m6A, we found that SRSF1- and 2-binding clusters were strongly enriched around splice sites, while SRSF3 and 4 binding patterns were much less pronounced (Figure 4C). The mRNA binding of both hnRNPC and hnRNRH was decreased at the splice sites (Supplementary information, Figure S6A-S6D). Interestingly, in addition to the known GGAGA motif for SRSF1 and GA-rich motif for SRSF245 binding, we found that m6A motifs can also serve as SRSF1- (UGGAC, P = 1e-547; AGGACCU, P = 1e-99) and 2- (UGGAC, P = 1e-1207; AAGGACC, P = 1e-182) binding sites, but not SRSF3- or 4-binding sites (Supplementary information, Figure S7B).
Using SRSF2 as an example, we next tested the effects of FTO depletion on its RNA binding ability. Interestingly, FTO knockdown promoted the RNA binding ability of SRSF2 (Figure 4D and 4E), but had no obvious effect on that of SRSF4 (Supplementary information, Figure S7C and S7D). As FTO does not interact with SRSF2 (data not shown), changes in RNA binding ability of SRSF2 cannot be explained by potential changes in direct protein-protein recruitment of SRSF2 by FTO. Taken together, these results suggest that m6A may serve as a new exonic cis-regulatory signal for RNA splicing, which together with ESEs co-regulates SRSF2 protein recruitment.
M6A modification regulates splicing of SRSF2 target genes
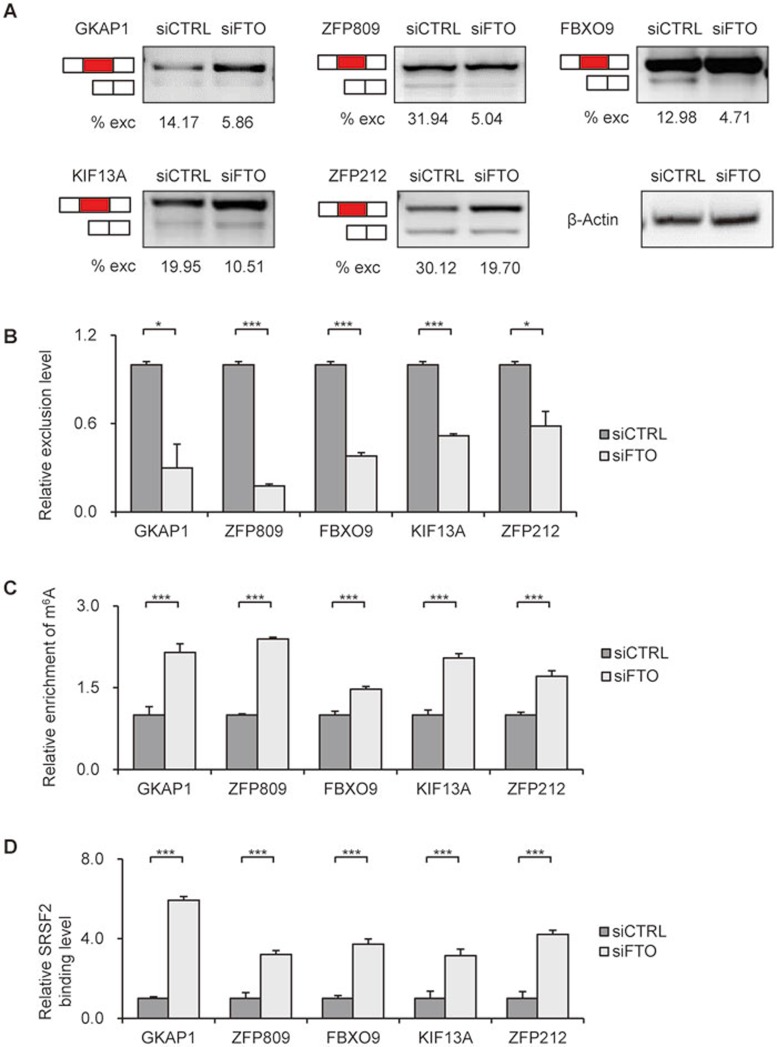
To test our above hypothesis, we chose alternatively spliced genes that contain spatially overlapped SRSF2-binding sites and m6A sites around spliced exons (including 5′ and 3′ flanking exons and internal alternative exons), for alternative splicing analysis by RT-PCR. A representative gene FBXO9 showed increased m6A levels around splicing site upon FTO depletion (Supplementary information, Figure S4). We found that FTO depletion inhibited exon skipping and promoted inclusion of the alternative exons of GKAP1, ZFP809, FBXO9, KIF13A and ZFP212 genes, as shown by the decreased exclusion levels (Figure 5A, 5B and Supplementary information, Figure S6E). Supporting our findings, previously published CLIP-seq data showed that SRSF2 binds to exons around alternative splice sites of these genes45.
Figure 5.
FTO regulates splicing in a subset of SRSF2 target genes. (A) Splicing analysis of SRSF2 target genes by RT-PCR in FTO-depleted cells. Red boxes indicate the spliced internal exons. % exc (exclusion) indicates the exclusion level of the internal exons. The expression of Actin was detected to confirm the equal loading of PCR products. (B) Statistical analysis of the relative exclusion level of spliced exons in FTO-depleted 3T3-L1 cells to that in control cells. ***P < 0.0001, *P < 0.05. Results are shown as mean ± SD. (C) Detection of m6A levels around splicing sites of FTO target genes by m6A-IP and RT-qPCR. ***P < 0.0001. Results are shown as mean ± SD. (D) Effects of FTO depletion on SRSF2 binding ability to its target genes validated by PAR-CLIP and RT-qPCR. ***P < 0.0001. Results are shown as mean ± SD. See also Supplementary information, Figure S8.
We performed m6A-IP and PAR-CLIP in control and FTO-depleted 3T3-L1 cells. All of the tested spliced genes showed increased m6A levels upon FTO depletion, supporting the notion that they are direct targets of FTO (Figure 5C). Consistent with the global effect of FTO depletion on the binding ability of SRSF2, binding of SRSF2 to above tested genes increased when FTO was knocked down (Figure 5D). SRSF2 is a splicing regulator reported to influence RNA splicing. When SRSF2 binding exceeds a certain level, inclusion of the alternative exons is ensured45. Hence, the increased binding ability of SRSF2 to RNA supported by increased m6A levels may promote exon inclusion in alternative splicing of FTO target genes.
We further confirmed the splicing events in METTL3-depleted 3T3-L1 cells. Exon exclusion levels of FBXO9, KIF13A and ZFP212 increased upon METTL3 knockdown (Supplementary information, Figure S8A-S8C). The opposite responses of the splicing events in FTO- and METTL3-depleted cells indicate that they are under the direct influence of m6A dynamics. Next, we asked whether these splicing events were SRSF2-dependent. We examined this hypothesis in SRSF2-depleted 3T3-L1 cells. Exon exclusion levels of FBXO9 and KIF13A were increased in response to SRSF2 deletion, opposite to the effects of FTO depletion (Supplementary information, Figure S8D and S8E).
FTO regulates alternative splicing of RUNX1T1 which modulates preadipocyte diffentiation.
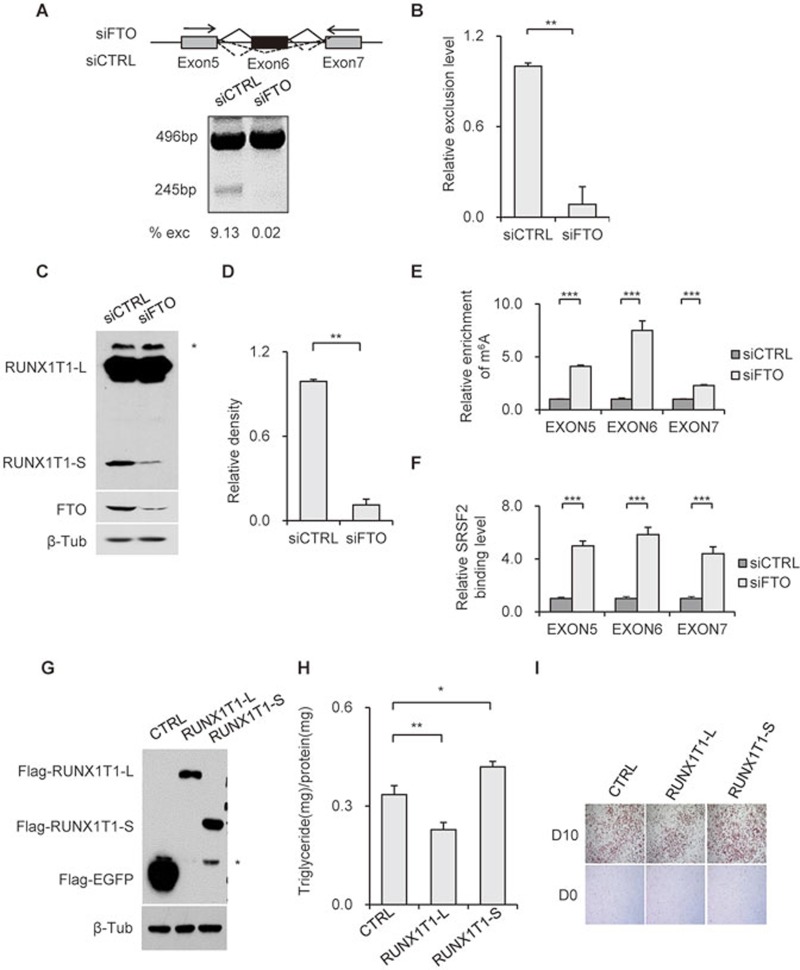
To validate the biological relevance of m6A-dependent regulation of SRSF2 binding and mRNA splicing, we chose to thoroughly examine the expression and function of an adipogenesis-related transcription factor, Runt-related transcription factor 1 (RUNX1T1)52. RUNX1T1 can be expressed in two isoforms. The PCR product of the constitutive isoform RUNX1T1-L is 496 bp, while that of the exon 6-skipped isoform RUNX1T1-S is 245 bp (Figure 6A). The PCR products were verified by sequencing analysis (Supplementary information, Figure S9A). In FTO-depleted 3T3-L1 cells, the exclusion level of exon 6 was decreased (Figure 6A and 6B). To the contrary, the exclusion level was increased in METTL3-depleted 3T3-L1 cells (Supplementary information, Figure S9B and S9C).
Figure 6.
FTO regulates alternative splicing of adipogenensis-related factor RUNX1T1. (A) Alternative splicing of RUNX1T1 was identified by RT-PCR using the indicated PCR primer set (arrows). Depending on isoform, bands of either 496 bp or 245 bp can be detected. (B) Statistical analysis of the relative exclusion level of RUNX1T1 in FTO-depleted 3T3-L1 cells to that in control cells. **P < 0.01. Results are shown as mean ± SD. (C) RUNX1T1 detection in 3T3-L1 pre-adipocyte lysates shows the endogenous protein products of the two RUNX1T1 isoforms in control and FTO-deficient 3T3-L1 pre-adipocytes. An unspecific band was labeled by *. (D) Statistical analysis of RUNX1T1-S protein expression levels in FTO-depleted 3T3-L1 cells to that in control cells. **P < 0.01. Results are shown as mean ± SD. (E) Detection of m6A levels around splicing sites of RUNX1T1 gene by m6A-IP and RT-qPCR. ***P < 0.0001. Results are shown as mean ± SD. (F) Effect of FTO depletion on SRSF2 binding ability to exons around splicing sites validated by PAR-CLIP and RT-qPCR. ***P < 0.0001. Results are shown as mean ± SD. (G-I) 3T3-L1 pre-adipocytes were transfected with pEGFP-C1b, pEGFP-C1b-Runx1T1-L or pEGFP-C1b-Runx1T1-S. Fory-eight hours later, transfection cells were lysed and immunoblotted with the indicated antibodies (G). An unspecific band or degradation band was labeled by *. Transfected cells were induced to differentiation and subjected to triglyceride analysis (H) and Oil Red O staining (I) on D10. Triglyceride content was quantified and normalized to protein content. *P < 0.05 is considered significant. **P < 0.01. Results are shown as mean ± SD. See also Supplementary information, Figure S9.
We applied a commercially available antibody recognizing both RUNX1T1 isoforms to verify that the mRNA expression pattern corresponded to what could be observed at the protein level. The RUNX1T1-L protein (68 kDa) was equally expressed in both control and FTO-depleted cells, while only very little amount of RUNX1T1-S (27 kDa) could be detected in FTO-depleted cells compared with control cells (Figure 6C and 6D). FTO depletion resulted in increased m6A levels in exons 5-7 (Figure 6E), which was accompanied by significant increase in SRSF2 binding (Figure 6F), suggesting that FTO may control the alternative splicing of RUNX1T1 by regulating m6A levels and thus SRSF2 accumulation around the splice sites. The finding that RUNX1T1 isoform expression is regulated by FTO prompted us to explore the potential role of RUNX1T1-L and -S in adipogenesis.
During adipocyte differentiation, RUNX1T1-S expression displays a progressive decrease as that of FTO (Supplementary information, Figure S9D and S9E). This result is in consistent with the observed splicing pattern of RUNX1T1 upon FTO knockdown. Oil Red O staining and triglyceride assays showed a significant inhibition of differentiation by RUNX1T1-L, whereas RUNX1T1-S appeared to promote differentiation (Figure 6G-6I), suggesting that FTO via its regulation of m6A levels and SRSF2 binding controls RUNX1T1 isoform expression, which in turn contributes to the regulation of adipogenesis. The significantly reduced expression of RUNX1T1-S might partially explain the differentiation defect observed in FTO-deficient cells.
Discussion
In this study we demonstrate that FTO contributes to the regulation of mRNA alternative splicing by modulating m6A levels and hence SRSF2 binding at splice sites. We show a novel FTO-dependent function of m6A in the regulation of mRNA splicing in adipocytes and in the process of adipogenesis. There are several pieces of evidence suggesting a role of m6A in RNA splicing: m6A are preferentially found in exons and UTR regions in multi-isoform genes and alternatively spliced exons18, binding of the m6A methyltransferase complex is especially enriched in genes with multiple isoforms and alternatively spliced exon junctions27, and finally, depletion of either METTL3 or ALKBH5 causes mRNA splicing alterations18,34. However, it has remained unclear how m6A may contribute to the regulation of mRNA alternative splicing. The pronounced enrichment of m6A in exonic sequences spatially overlapping with ESEs and binding clusters of specific SR proteins, together with the finding that m6A regulates RNA binding of SRSF2, suggests that m6A might influence the choice of exon inclusion/exclusion by regulating SRSF2 access to its targets. The accurate recognition of exon/intron boundaries is essential for correct splicing and generation of mature mRNA. In higher eukaryotes, exon-intron junctions are defined by conserved intronic cis-elements, including the 5′ splice site and 3′ splice site. Exonic cis-elements such as exonic splicing enhancers (ESEs) or silencers (ESSs) are important for correct splice-site identification and prevalent in most exons, including constitutive ones. ESEs serve as binding sites for specific serine/arginine-rich (SR) proteins. ESEs and their binding SR proteins frequently exist in protein-coding exons, compared with flanking intronic regions53. Similarly, in CDS regions, m6A is highly enriched at the edges of exons adjacent to splice sites relative to intron regions. Knockdown of FTO specifically affects the RNA binding ability of SRSF2, but not that of SRSF4. This might not be surprising since SRSF2 readily binds to exons in contrast to other SR proteins such as SRSF3 and SRSF4 which have been reported to preferentially bind to intronless genes, ncRNAs and introns or 3′-UTRs of intron-containing genes45,46,54. Hence m6A dynamics appears to specifically affect the RNA binding ability of SRSF2, thus specifically influencing the splicing outcome of genes under the regulation of SRSF2 (Figures 5 and 6), which can promote both inclusion and skipping of exons45. However, in the case of m6A-dependent SRSF2 regulation of mRNA splicing, SRSF2 appears to exclusively promote constitutive splicing (inclusion of exons) while inhibiting alternative splicing (exclusion of exons). Thus m6A seems to bring another layer of regulation to the mRNA splicing process (Figure 7). A thorough mechanistic understanding of the interplay between m6A, ESEs and SR proteins will be a challenge for future studies.
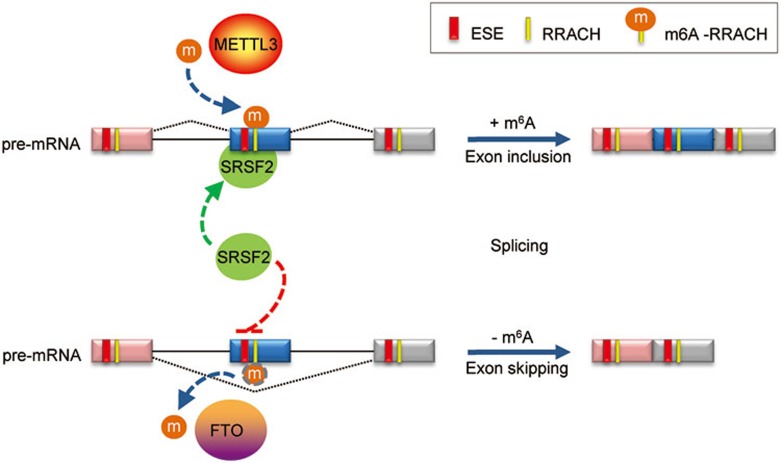
Figure 7.
Cooperative role of m6A in regulating SRSF2 function at splice sites. The splicing factor SRSF2 can recognize exon splicing enhancer (ESE), inducing exon inclusion. METTL3-mediated m6A modification enhances the recruitment of SRSF2 to its target ESE, promoting exon inclusion. On the other hand demethylation of m6A-RRACHs near ESEs by FTO prevents recognition of the ESEs by SRSF2, thereby inhibiting inclusion and instead promoting exon skipping of the specific exon. M6A co-regulated by methyltransferases and demthylases serves as a new RNA splicing exonic cis-regulatory element that in combination with ESEs regulates SRSF2 protein recruitment.
FTO has for a long time been implicated in obesity. Our findings show a clear connection between FTO-dependent m6A regulation and the ability of pre-adipocytes to differentiate. It appears, at least in part, to be due to the apparently very different regulatory functions of the two alternative RUNX1T1 isoforms in adipogenesis. Our finding that FTO regulates the splicing of a subset of genes, such as sterol metabolic process-related genes, via controlling m6A levels (Figure 2G), may help explain why FTO appears to play a role in obesity55. We believe it is reasonable to imagine that changes in the ratio of specific isoforms might have dramatic impact on not only adipogenesis and obesity but also a wide range of biological functions. The finding of a specific role of FTO but not ALKBH5 suggests that these two demethylases function in different physiological processes likely by regulating different subsets of mRNAs.
In summary, we demonstrate a novel FTO-dependent regulatory role of m6A and SRSF2 in mRNA splicing and adipocyte differentiation. The dynamic regulation of m6A by FTO in adipocytes is important in the determination of splicing and gene expression patterns in a subset of genes contributing to the regulation of adipogenesis. The spatial overlap of m6A sites with splice sites and cis-regulatory elements as well as the apparent regulation of SRSF2 binding by m6A call for a further elucidation of the role of m6A in RNA splicing. The direct connection between isoform expression of the adipogenic regulatory factor RUNX1T1 and its m6A status in the context of adipocyte differentiation sheds new light on our understanding of the biological significance of RNA m6A modification.
Materials and Methods
Cell line, plasmids and antibodies
3T3-L1 pre-adipocytes were obtained from ATCC (Manassas, VA). The human FTO and mouse RUNX1T1 genes were cloned into pEGFP-C1B56. Human SR plasmids were bought from OriGene Technologies (Rockville, MD). The expression constructs were generated using PCR and subcloned into pCS2+ vectors with an N-terminal FLAG tag57. Polyclonal rabbit anti-FTO antibody was affinity-purified from rabbits immunized with 6×His-tagged full-length human FTO protein as previously reported9. Polyclonal rabbit anti-ALKBH5 antibody was generated against synthesized peptide (RKYQEDSDERSD, 58-70 amino acids of human ALKBH5) by CWBio (Beijing) as previously reported34. The primary antibodies were purchased from commercial sources: rabbit anti-METTL3 (MT-A70; 15073-1-AP, Proteintech Group); rabbit anti-RUNX1T1 (15494-1-AP, Proteintech); mouse anti-β-tubulin (t-5293, Sigma); rabbit anti-m6A (202003; Synaptic Systems); mouse anti-HA (cw0092B, CWbiotech); rabbit anti-Flag (F7425, Sigma); mouse anti-Flag (F1804, Sigma). The secondary antibodies used for immunoblotting and dot-blotting: anti-Rabbit IgG-HRP (p0448, dakocytomation); anti-Mouse IgG-HRP (p0161, dakocytomation).
Cell culture and adipocyte differentiation
3T3-L1 pre-adipocytes were grown in DMEM Dulbecco's Modified Eagle Medium (DMEM) containing 10% newborn calf serum (SH30401.01, Hyclone) and 1% antibiotics until confluence and induced to differentiation as previously described. In brief, two days' post-confluence (day 0, D0), cells were exposed to differentiation medium containing 0.5 mmol/L isobutylmethylxanthine (I5879, Sigma), 1 μmol/L dexamethasone (D2915, Sigma), 10 μg/mL insulin (I6634, Sigma), 2 μmol/L rosiglitazone (R2408, Sigma) and 10% fetal bovine serum (FBS; A15-151, PAA) for three days. At the end of day 3, culture medium was replaced with DMEM supplemented only with 10 μg/mL insulin and 10% FBS, and replenished every other day. After the differentiation process, at least 90% of the cells had accumulated lipid droplets at day 10, and were used as mature adipocytes.
Gene expression and splicing analyses
Total RNA was isolated from cells with TRI Reagent (Sigma). cDNA library was constructed using TruSeq. RNA Sample Prep Kit and then sequenced with HiSeq 2000 system (Illumina Inc.). RNA-seq reads were mapped to the mouse reference genome (mm10) by Tophat (V2.0.4)58,59. Number of reads mapped to each Ensembl gene were counted using the HTSeq python package and the DEGs in different samples were determined by R-package DEGseq60. The expressions of transcripts were quantified as Reads Per Kilobase of exon model per Million mapped reads (RPKM).
For splicing analysis, spliced events were identified and compared with Cufflink and Scripture softwares61,62. Cuffdiff, one part of the Cufflinks package were applied to calculate the reads count and FPKM for each isoform of each gene. The differentially expressed isoforms were identified using R-package DEGseq. Scripture was used to detect splicing events within each gene62. Differentially expressed exons analysis was done by the R package DEXSeq63. To differentially expressed introns, the number of reads falling in each intron were counted using RSeQC64, and differential expression analysis was done using the DEGseq60. The exon-exon junctions approach applied in previous study was also used to analyze alternative splicing event65.
M6A-seq analyses
mRNA was extracted using biotinylated poly(dT) oligo, followed by further removing of contaminated rRNA using RiboMinus Transcriptome Isolation Kit (K1550-02, Invitrogen). The mRNA quality was analyzed by NanoDrop. mRNA was fragmented and subjected to immunoprecipitation with m6A specific antibody as previously described approaches17,18. M6A-enriched RNA was used to generate the cDNA library in parallel with the input RNA, and then sequenced using the HiSeq 2000 system (Illumina Inc.). For m6A analysis, m6A regions (m6A peaks) were identified by comparing the read abundance between m6A-seq and RNA-seq samples of the same loci according to a method previously described17. Briefly, the entire mm10 genome was divided into 25 nt bins and the numbers of both m6A-seq reads and RNA-seq reads (used as control) mapped to each bin were counted and compared. Bins with statistically enriched m6A-seq reads compared with the RNA-seq reads (adjusted P < 1e-5, Fisher's exact test together with Benjamini-Hocberg procedure) were identified and concatenated adjacently. Using the same selection criteria, regions with significantly enriched RNA-seq reads were selected as control peaks. Sequence motifs enriched in m6A peaks were identified by HOMER with m6A peaks as the target sequences and control peaks as the background66. The spatial correlations between m6A sites (RRACH) and ESEs or binding clusters of SR proteins were analyzed by BEDTools software67.
PAR-CLIP
3T3-L1 cells were co-transfected with pCS2-SR plamids and control siRNAs and FTO siRNAs, respectively. Thirty-two hours later, cells were labeled with 200 μM 4-SU (Sigma T4509) for 16 h, then to induce crosslinking treated with 0.4 J/cm2 of 365 nm UV light in a BLX-E365 crosslinker BLX-E365 (Vilber). Irradiated cells were then lysed with NP-40 lysis buffer (50 mM pH 7.5 Tris-Cl, 150 mM NaCl, 2 mM EDTA, 0.5% (v/v) NP40) on ice for 30 min. Cell debris was removed by centrifugation and the crude lysates were incubated with Flag M2 Affinity Gel (A2220, Sigma) for 2 h at 4 °C. The immunoprecipitates were treated with CIP (NEB M0290) and subjected to the biotin labelling, following the instructions of the RNA 3′ end biotinylation kit (20160, Thermo). After separation on 4%-12% NuPAGE gels (NP0321B0X, Invitrogen), the protein-RNA complexes were transferred to PVDF membranes. Biotin-labelled RNA were detected and visualized according to manufactors description (chemiluminescent nuclei acid kit, Thermo 89880). SR proteins were identified by rabbit Flag antibody (F7425, Sigma).
Accession numbers
M6 A-seq and RNA-seq data have been uploaded to GEO database and can be accessed via accession number GSE53244 and GSE53249.
Acknowledgments
This work was supported by the National Basic Research Program of China (973 Program; 2011CB510103 and 2014CB964902), the National Natural Science Foundation of China (91319308, 31430022, and 31400672), and the Chinese Academy of Sciences (CAS) Strategic Priority Research Program (XDB14030300).
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
Wild-type but not catalytically inactive FTO can rescue adipogenesis in FTO-deficient preadipocytes. Related to Figure 1.
FTO and METTL3 inversely regulate isoform expression and m6A in a subset of genes. Related to Figure 2.
Characterization of the expression of spliced isoforms and m6A enrichment. Related to Figure 2.
m6A enrichment and exonic expression of representative gene FBXO9 upon FTO/METTL3 deletion and during adipocyte differentiation.
Spatial relationship between splice sites and m6A sites during adipogenesis. Related to Figure 3.
Spatial relationship between splice sites and m6A sites for hnRNPC and H. Related to Figure 4.
SRSF1-4 binding sequences and SRSF4 RNA binding. Related to Figure 4.
Splicing events upon METTL3 or SRSF2 deficiency. Related to Figure 5.
Verification of RUNX1T1 gene splicing pattern. Related to Figure 6.
Summary of the sequencing samples
Statistical analysis of m6A peak number and total enrichment in FTO proficient and deficient samples. p<0.05 in K-S test indicated as significant.
Statistical analysis of m6A peak number and enrichment locating in exons and introns adjacent to 5′ or 3′ splicing sites of the constitutive splicing and the alternative splicing patterns for global genes as well as FTO target genes.
Extended Experimental Procedures
References
- Gerken T, Girard CA, Tung YC, et al. The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science. 2007;318:1469–1472. doi: 10.1126/science.1151710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loos RJ, Bouchard C. FTO: the first gene contributing to common forms of human obesity. Obes Rev. 2008;9:246–250. doi: 10.1111/j.1467-789X.2008.00481.x. [DOI] [PubMed] [Google Scholar]
- Zabena C, Gonzalez-Sanchez JL, Martinez-Larrad MT, et al. The FTO obesity gene. Genotyping and gene expression analysis in morbidly obese patients. Obes Surg. 2009;19:87–95. doi: 10.1007/s11695-008-9727-0. [DOI] [PubMed] [Google Scholar]
- Loos RJ, Yeo GS. The bigger picture of FTO—the first GWAS-identified obesity gene. Nat Rev Endocrinol. 2014;10:51–61. doi: 10.1038/nrendo.2013.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer J, Koch L, Emmerling C, et al. Inactivation of the Fto gene protects from obesity. Nature. 2009;458:894–898. doi: 10.1038/nature07848. [DOI] [PubMed] [Google Scholar]
- Gao X, Shin YH, Li M, Wang F, Tong Q, Zhang P. The fat mass and obesity associated gene FTO functions in the brain to regulate postnatal growth in mice. PLoS One. 2010;5:e14005. doi: 10.1371/journal.pone.0014005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church C, Moir L, McMurray F, et al. Overexpression of Fto leads to increased food intake and results in obesity. Nat Genet. 2010;42:1086–1092. doi: 10.1038/ng.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurray F, Church CD, Larder R, et al. Adult onset global loss of the fto gene alters body composition and metabolism in the mouse. PLoS Genet. 2013;9:e1003166. doi: 10.1371/journal.pgen.1003166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia GF, Fu Y, Zhao X, et al. N6-Methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7:885–887. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia G, Yang CG, Yang S, et al. Oxidative demethylation of 3-methylthymine and 3-methyluracil in single-stranded DNA and RNA by mouse and human FTO. FEBS Lett. 2008;582:3313–3319. doi: 10.1016/j.febslet.2008.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan T. N6-methyl-adenosine modification in messenger and long non-coding RNA. Trends Biochem Sci. 2013;38:204–209. doi: 10.1016/j.tibs.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu Y, Zhao X, Wu YS, Li MM, Wang XJ, Yang YG. N6-methyl-adenosine (m6A) in RNA: an old modification with a novel epigenetic function. Genomics Proteomics Bioinformatics. 2013;11:8–17. doi: 10.1016/j.gpb.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, He C. Nucleic acid modifications with epigenetic significance. Curr Opin Chem Biol. 2012;16:516–524. doi: 10.1016/j.cbpa.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KD, Jaffrey SR. The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat Rev Mol Cell Biol. 2014;15:313–326. doi: 10.1038/nrm3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Dominissini D, Rechavi G, He C. Gene expression regulation mediated through reversible (m6A) RNA methylation. Nat Rev Genet. 2014;15:293–306. doi: 10.1038/nrg3724. [DOI] [PubMed] [Google Scholar]
- Bodi Z, Button JD, Grierson D, Fray RG. Yeast targets for mRNA methylation. Nucl Acids Res. 2010;38:5327–5335. doi: 10.1093/nar/gkq266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012;149:1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominissini D, Moshitch-Moshkovitz S, Schwartz S, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- Schwartz S, Agarwala SD, Mumbach MR, et al. High-resolution mapping reveals a conserved, widespread, dynamic mRNA methylation program in yeast meiosis. Cell. 2013;155:1409–1421. doi: 10.1016/j.cell.2013.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Li Y, Toth JI, Petroski MD, Zhang ZL, Zhao JC. N-6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat Cell Biol. 2014;16:191–198. doi: 10.1038/ncb2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fustin JM, Doi M, Yamaguchi Y, et al. RNA-Methylation-dependent RNA processing controls the speed of the circadian clock. Cell. 2013;155:793–806. doi: 10.1016/j.cell.2013.10.026. [DOI] [PubMed] [Google Scholar]
- Hess ME, Hess S, Meyer KD, et al. The fat mass and obesity associated gene (Fto) regulates activity of the dopaminergic midbrain circuitry. Nat Neurosci. 2013;16:1042–1048. doi: 10.1038/nn.3449. [DOI] [PubMed] [Google Scholar]
- Frayling TM, Timpson NJ, Weedon MN, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Yang Y, Sun BF, Zhao YL, Yang YG. FTO and obesity: mechanisms of association. Curr Diab Rep. 2014;14:486. doi: 10.1007/s11892-014-0486-0. [DOI] [PubMed] [Google Scholar]
- Wang X, Lu Z, Gomez A, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Yue Y, Han D, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol. 2014;10:93–95. doi: 10.1038/nchembio.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ping XL, Sun BF, Wang L, et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014;24:177–189. doi: 10.1038/cr.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, Mumbach MR, Jovanovic M, et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5 sites. Cell Rep. 2014;8:1–13. doi: 10.1016/j.celrep.2014.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JC, Caceres JF. The SR protein family of splicing factors: master regulators of gene expression. Biochem J. 2009;417:15–27. doi: 10.1042/BJ20081501. [DOI] [PubMed] [Google Scholar]
- Martinez-Contreras R, Cloutier P, Shkreta L, Fisette JF, Revil T, Chabot B. hnRNP proteins and splicing control. Advan Experiment Med Biol. 2007;623:123–147. doi: 10.1007/978-0-387-77374-2_8. [DOI] [PubMed] [Google Scholar]
- Matlin AJ, Clark F, Smith CW. Understanding alternative splicing: towards a cellular code. Nat Rev Mol Cell Biol. 2005;6:386–398. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- Braunschweig U, Gueroussov S, Plocik AM, Graveley BR, Blencowe BJ. Dynamic integration of splicing within gene regulatory pathways. Cell. 2013;152:1252–1269. doi: 10.1016/j.cell.2013.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoltzfus CM, Dane RW. Accumulation of spliced avian retrovirus mRNA is inhibited in S-adenosylmethionine-depleted chicken embryo fibroblasts. J Virol. 1982;42:918–931. doi: 10.1128/jvi.42.3.918-931.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G, Dahl JA, Niu Y, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49:18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tews D, Fischer-Posovszky P, Fromme T, et al. FTO deficiency induces UCP-1 expression and mitochondrial uncoupling in adipocytes. Endocrinology. 2013;154:3141–3151. doi: 10.1210/en.2012-1873. [DOI] [PubMed] [Google Scholar]
- Mitterberger MC, Lechner S, Mattesich M, et al. DLK1 (PREF1) is a negative regulator of adipogenesis in CD105(+)/CD90(+)/CD34(+)/CD31(−)/FABP4(−) adipose-derived stromal cells from subcutaneous abdominal fat pats of adult women. Stem Cell Res. 2012;9:35–48. doi: 10.1016/j.scr.2012.04.001. [DOI] [PubMed] [Google Scholar]
- Wang Y, Sul HS. Pref-1 regulates mesenchymal cell commitment and differentiation through Sox9. Cell Metab. 2009;9:287–302. doi: 10.1016/j.cmet.2009.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min HY, Spiegelman BM. Adipsin, the adipocyte serine protease: gene structure and control of expression by tumor necrosis factor. Nucl Acids Res. 1986;14:8879–8892. doi: 10.1093/nar/14.22.8879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berulava T, Ziehe M, Klein-Hitpass L, et al. FTO levels affect RNA modification and the transcriptome. Eur J Hum Genet. 2013;21:317–323. doi: 10.1038/ejhg.2012.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caceres JF, Kornblihtt AR. Alternative splicing: multiple control mechanisms and involvement in human disease. Trends Genet. 2002;18:186–193. doi: 10.1016/s0168-9525(01)02626-9. [DOI] [PubMed] [Google Scholar]
- Sorek R, Ast G. Intronic sequences flanking alternatively spliced exons are conserved between human and mouse. Genome Res. 2003;13:1631–1637. doi: 10.1101/gr.1208803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barash Y, Calarco JA, Gao W, et al. Deciphering the splicing code. Nature. 2010;465:53–59. doi: 10.1038/nature09000. [DOI] [PubMed] [Google Scholar]
- Wang Y, Gogol-Doring A, Hu H, et al. Integrative analysis revealed the molecular mechanism underlying RBM10-mediated splicing regulation. Embo Mol Med. 2013;5:1431–1442. doi: 10.1002/emmm.201302663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanford JR, Coutinho P, Hackett JA, Wang X, Ranahan W, Caceres JF. Identification of nuclear and cytoplasmic mRNA targets for the shuttling protein SF2/ASF. PLoS One. 2008;3:e3369. doi: 10.1371/journal.pone.0003369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandit S, Zhou Y, Shiue L, et al. Genome-wide analysis reveals SR protein cooperation and competition in regulated splicing. Mol Cell. 2013;50:223–235. doi: 10.1016/j.molcel.2013.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anko ML, Muller-McNicoll M, Brandl H, et al. The RNA-binding landscapes of two SR proteins reveal unique functions and binding to diverse RNA classes. Genome Biol. 2012;13:R17. doi: 10.1186/gb-2012-13-3-r17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konig J, Zarnack K, Rot G, et al. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat Struct Mol Biol. 2010;17:909–915. doi: 10.1038/nsmb.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelga SC, Vu AQ, Arnold JD, et al. Integrative genome-wide analysis reveals cooperative regulation of alternative splicing by hnRNP proteins. Cell Rep. 2012;1:167–178. doi: 10.1016/j.celrep.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010;20:110–121. doi: 10.1101/gr.097857.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith PJ, Zhang C, Wang J, Chew SL, Zhang MQ, Krainer AR. An increased specificity score matrix for the prediction of SF2/ASF-specific exonic splicing enhancers. Hum Mol Genet. 2006;15:2490–2508. doi: 10.1093/hmg/ddl171. [DOI] [PubMed] [Google Scholar]
- Cartegni L, Wang J, Zhu Z, Zhang MQ, Krainer AR. ESEfinder: A web resource to identify exonic splicing enhancers. Nucl Acids Res. 2003;31:3568–3571. doi: 10.1093/nar/gkg616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochford JJ, Semple RK, Laudes M, et al. ETO/MTG8 is an inhibitor of C/EBPbeta activity and a regulator of early adipogenesis. Mol Cell Biol. 2004;24:9863–9872. doi: 10.1128/MCB.24.22.9863-9872.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Smith PJ, Krainer AR, Zhang MQ. Distribution of SR protein exonic splicing enhancer motifs in human protein-coding genes. Nucl Acids Res. 2005;33:5053–5062. doi: 10.1093/nar/gki810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anko ML, Morales L, Henry I, Beyer A, Neugebauer KM. Global analysis reveals SRp20- and SRp75-specific mRNPs in cycling and neural cells. Nat Struct Mol Biol. 2010;17:962–970. doi: 10.1038/nsmb.1862. [DOI] [PubMed] [Google Scholar]
- Dai HJ, Wu JC, Tsai RT, Pan WH, Hsu WL. T-HOD: a literature-based candidate gene database for hypertension, obesity and diabetes. Database (Oxford) 2013;2013:bas061. doi: 10.1093/database/bas061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YG, Lindahl T, Barnes DE. Trex1 exonuclease degrades ssDNA to prevent chronic checkpoint activation and autoimmune disease. Cell. 2007;131:873–886. doi: 10.1016/j.cell.2007.10.017. [DOI] [PubMed] [Google Scholar]
- Fu Y, Huang B, Shi Z, et al. SRSF1 and SRSF9 RNA binding proteins promote Wnt signalling-mediated tumorigenesis by enhancing beta-catenin biosynthesis. Embo Mol Med. 2013;5:737–750. doi: 10.1002/emmm.201202218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Feng Z, Wang X, Zhang X. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 2010;26:136–138. doi: 10.1093/bioinformatics/btp612. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Garber M, Levin JZ, et al. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat Biotechnol. 2010;28:503–510. doi: 10.1038/nbt.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Reyes A, Huber W. Detecting differential usage of exons from RNA-seq data. Genome Res. 2012;22:2008–2017. doi: 10.1101/gr.133744.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wang S, Li W. RSeQC: quality control of RNA-seq experiments. Bioinformatics. 2012;28:2184–2185. doi: 10.1093/bioinformatics/bts356. [DOI] [PubMed] [Google Scholar]
- Han H, Irimia M, Ross PJ, et al. MBNL proteins repress ES-cell-specific alternative splicing and reprogramming. Nature. 2013;498:241–245. doi: 10.1038/nature12270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Wild-type but not catalytically inactive FTO can rescue adipogenesis in FTO-deficient preadipocytes. Related to Figure 1.
FTO and METTL3 inversely regulate isoform expression and m6A in a subset of genes. Related to Figure 2.
Characterization of the expression of spliced isoforms and m6A enrichment. Related to Figure 2.
m6A enrichment and exonic expression of representative gene FBXO9 upon FTO/METTL3 deletion and during adipocyte differentiation.
Spatial relationship between splice sites and m6A sites during adipogenesis. Related to Figure 3.
Spatial relationship between splice sites and m6A sites for hnRNPC and H. Related to Figure 4.
SRSF1-4 binding sequences and SRSF4 RNA binding. Related to Figure 4.
Splicing events upon METTL3 or SRSF2 deficiency. Related to Figure 5.
Verification of RUNX1T1 gene splicing pattern. Related to Figure 6.
Summary of the sequencing samples
Statistical analysis of m6A peak number and total enrichment in FTO proficient and deficient samples. p<0.05 in K-S test indicated as significant.
Statistical analysis of m6A peak number and enrichment locating in exons and introns adjacent to 5′ or 3′ splicing sites of the constitutive splicing and the alternative splicing patterns for global genes as well as FTO target genes.
Extended Experimental Procedures