Dear Editor,
Recent discoveries suggest that N6-methyladenosine (m6A) modification, a prevalent internal modification in eukaryotic RNA, is an essential RNA regulatory mechanism. This modification is post-transcriptionally installed by m6A methyltransferases (METTL3-METTL14-WTAP complex)1,2,3,4 and oxidatively removed by m6A demethylases (FTO and ALKBH5)5,6. These 'writer' and 'eraser' enzymes are required for embryo development, energy homeostasis and fertility, suggesting fundamental regulatory roles of m6A1,2,5.
Very recently, the human YTH (YT521-B homology) domain family proteins 1-3 (YTHDF1-3) were shown to act as specific m6A 'readers' to regulate mRNA degradation7. YTHDF2 specifically binds to m6A-containing mRNA via its C-terminal YTH domain, and recruits the mRNA to cytoplasmic foci (P bodies) to control mRNA degradation via its P/Q/N-rich N terminus7. A further mechanistic understanding of the function of m6A modification requires the knowledge of the molecular basis of m6A-specific recognition by the YTH domain.
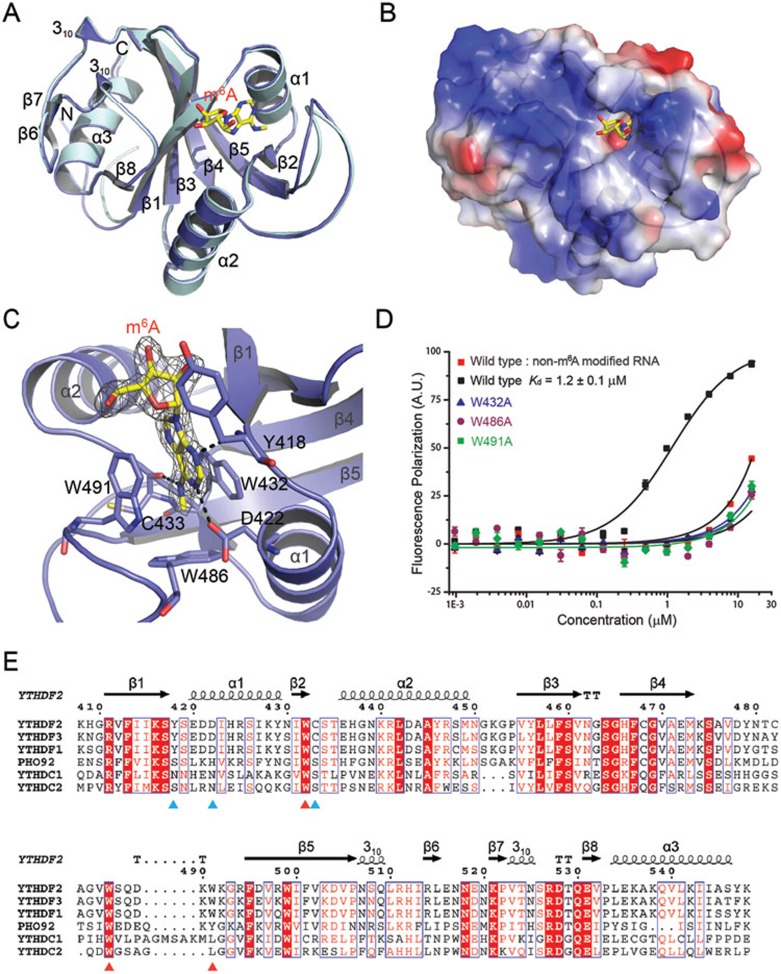
We first characterized the binding affinity of the YTH domain of human YTHDF2 (YTH-YTHDF2) to m6A-containing RNA using fluorescence polarization (FP) assays. YTH-YTHDF2 binds to an m6A-containing RNA probe (AUGG(m6A)CUCC, sequence derived from son mRNA which colocalized with YTHDF2 in vivo7) with a dissociation constant (Kd) of 1.2 ± 0.1 μM; the affinity is dramatically decreased when the RNA probe is unmethylated (Figure 1D).
Figure 1.
Structure of YTH-YTHDF2 in complex with an m6A mononucleotide. (A) Structural superimposition of free (colored in palecyan) and m6A-bound YTH-YTHDF2 (colored in blue) structures. The m6A mononucleotide is shown in yellow stick. (B) Electrostatic potential surface of YTH-YTHDF2 in complex with the m6A mononucleotide. (C) Detailed interactions between YTH-YTHDF2 and the m6A mononucleotide. The weighted 2Fo-Fc electron density map of m6A is shown. (D) FP assays of wild type and mutants of YTH-YTHDF2 incubated with m6A-containing RNA probe or unmethylated RNA probe. (E) Sequence alignment of the YTH domains of Homo sapiens YTHDF1-3, YTHDC1 and YTHDC2, and S. cerevisiae PHO92. Residues forming the aromatic cage are indicated with red triangles and residues involved in hydrogen bond interactions with m6A are indicated with cyan triangles. The structure figures were prepared with PyMOL.
In order to uncover how the specific recognition of m6A by YTH-YTHDF2 is achieved, we solved the crystal structures of the YTH domain of human YTHDF2 (NP_057342.2) in the free form and in complex with an m6A mononucleotide. The crystal structure of apo YTH-YTHDF2 was refined to 2.15 Å resolution (Supplementary information, Table S1). The asymmetric unit contains six YTH-YTHDF2 molecules with a backbone root-mean-square deviation (RMSD) of 0.2 Å. YTH-YTHDF2 adopts a mixed α-helix-β-sheet fold, which is composed of three α helices (α1-α3), eight β strands (β1-β8) and two 310 helices (Figure 1A). The six central β strands (β8-β1-β3-β4-β5-β2) are arranged in an atypical β-barrel fold and covered by the three α helices, constituting the hydrophobic core (Figure 1A). There are a small β-sheet formed by strands β6 and β7 and two 310 helices between strands β5 and β8, packing on the surface of α3 (Figure 1A).
The crystal structure of YTH-YTHDF2 in complex with an m6A mononucleotide was refined to 2.10 Å resolution (Supplementary information, Table S1). Each asymmetric unit contains two protein molecules and each molecule binds to an m6A mononucleotide in a similar manner. Structural superimposition of free and m6A-bound YTH-YTHDF2 structures reveals a local conformation adjustment of the loop between β4 and β5 (β4-β5 loop) (Figure 1A). The m6A mononucleotide is tightly locked in a hydrophobic pocket formed by residues from α1 helix, β2 strand and β4-β5 loop (Figure 1A and 1B). Specifically, the m6A mononucleotide is positioned in an aromatic cage of three aromatic amino acids, of which Trp486 from β4-β5 loop forms the base, and Trp432 from β2 strand and Trp491 from β4-β5 loop form the walls (Figure 1C). The aromatic rings of Trp432 and Trp491, which undergo an induced flipping coupled with m6A binding, are almost parallel to each other (Supplementary information, Figure S1A). The m6A adenine moiety is sandwiched by these two aromatic walls. The methyl group of the m6A mononucleotide is pointed toward the aromatic ring of Trp486 with a distance of about 3.8 Å, and is also sandwiched by the two aromatic ring walls with a distance of about 4 Å (Supplementary information, Figure S1B). Mutations of the aromatic cage residues (Trp432Ala, Trp486Ala and Trp491Ala) significantly reduced the binding affinity of YTH-YTHDF2 to the m6A RNA probe, confirming the importance of the aromatic cage for m6A binding (Figure 1D). The m6A-YTH-YTHDF2 interaction also involves the formation of intermolecular hydrogen bonds between the N1 nitrogen atom and the side chain carbonyl oxygen of Asp422, the N3 nitrogen atom and the backbone NH of Tyr418, and the N6 nitrogen atom and the backbone carbonyl oxygen of Cys433 (Figure 1C). Overall, the recognition of m6A is facilitated by the π-π interactions between the adenine base and the aromatic cage, the cation-π interactions between the N6-methyl moiety and the aromatic cage, and a set of hydrogen bonds, demonstrating at atomic resolution a well-organized pocket for m6A reading.
The arrangement of aromatic residues is reminiscent of a common molecular architecture for the recognition of methylated lysine and arginine, examples of which can be found in chromo, tudor and PHD domains8. Two of the three residues (Trp432 and Trp486) in YTHDF2 that form the aromatic cage are completely conserved in YTH domain-containing proteins (Figure 1E). The third aromatic cage residue (Trp491 in YTHDF2) is replaced by Tyr237 in PHO92, the only known YTH domain-containing protein in Saccharomyces cerevisiae (S. cerevisiae)9, and a leucine residue in YTHDC1/2 which have been reported to bind to a degenerate unmethylated RNA sequence10 (Figure 1E). During the preparation of this manuscript, structures of the YTH domains of human YTHDC111 and ZrMRB1 (the ortholog of S. cerevisiae PHO92 in Zygosaccharomyces rouxii)12 were reported. The three YTH domains show high structural similarity with all pairwise backbone RMSD values below 1 Å and possess a similar pocket for m6A binding despite the variance in the third aromatic cage residue (Supplementary information, Figure S1C and S1D).
In summary, our data presented here provide the molecular basis for the recognition of m6A by the YTH domain of YTHDF2. As the G nucleotide at the −1 position relative to the m6A site adopts different conformations in the two recently reported structures11,12, further studies are needed to illustrate the structural basis of preferential recognition of the conserved G(m6A)C motif by YTH-YTHDF27.
The atomic coordinates of YTH-YTHDF2 in the free state and in complex with an m6A mononucleotide are deposited in the Protein Data Bank under accession numbers 4RDO and 4RDN, respectively.
Acknowledgments
We thank Dr Chao He (Anhui University), Lijun Wang, Juncheng Wang, Yiyang Jiang, Hongyu Bao and Zhonghua Liu for helpful discussions about the experiments. We thank the staff at BL17U of Shanghai Synchrotron Radiation Facilities for their help in the X-ray data collection. This work was financially supported by the National Basic Research Program of China (973 Program; 2011CB966302, 2012CB917201 and 2011CB911104), the National Natural Science Foundation of China (31170693 and 31330018), the Chinese Academy of Sciences (KJZD-EW-L05), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB08010101 and XDB08030302) and China Postdoctoral Science Foundation (2013M540519).
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
(A) Structural superimposition of free (palecyan) and m6A bound YTH-YTHDF2 (blue).
Data collection, and refinement statistics
References
- Liu J, Yue Y, Han D, et al. Nat Chem Biol. 2014. pp. 93–95. [DOI] [PMC free article] [PubMed]
- Wang Y, Li Y, Toth JI, et al. Nat Cell Biol. 2014. pp. 191–198. [DOI] [PMC free article] [PubMed]
- Ping XL, Sun BF, Wang L, et al. Cell Res. 2014. pp. 177–189. [DOI] [PMC free article] [PubMed]
- Schwartz S, Mumbach MR, Jovanovic M, et al. Cell Rep. 2014. pp. 284–296. [DOI] [PMC free article] [PubMed]
- Zheng GQ, Dahl JA, Niu YM, et al. Mol Cell. 2013. pp. 18–29. [DOI] [PMC free article] [PubMed]
- Jia GF, Fu Y, Zhao X, et al. Nat Chem Biol. 2012. pp. 1008–1008.
- Wang X, Lu Z, Gomez A, et al. Nature. 2014. pp. 117–120. [DOI] [PMC free article] [PubMed]
- Taverna SD, Li H, Ruthenburg AJ, et al. Nat Struct Mol Biol. 2007. pp. 1025–1040. [DOI] [PMC free article] [PubMed]
- Kang HJ, Jeong SJ, Kim KN, et al. Biochem J. 2014. pp. 391–400. [DOI] [PubMed]
- Zhang ZY, Theler D, Kaminska KH, et al. J Biol Chem. 2010. pp. 14701–14710. [DOI] [PMC free article] [PubMed]
- Xu C, Wang X, Liu K, et al. Nat Chem Biol. 2014. pp. 927–929. [DOI] [PubMed]
- Luo S, Tong L. Proc Natl Acad Sci USA. 2014. pp. 13834–13839. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Structural superimposition of free (palecyan) and m6A bound YTH-YTHDF2 (blue).
Data collection, and refinement statistics