Key Points
A new bone marrow DC culture method with FLT3L and GM-CSF efficiently generates functional Batf3-dependent CD103+ DCs.
GM-CSF promotes DC precursor polarization and CD103+ DC proliferation.
Abstract
Multiple subsets of FMS-like tyrosine kinase 3 ligand (FLT3L)-dependent dendritic cells (DCs) control T-cell tolerance and immunity. In mice, Batf3-dependent CD103+ DCs efficiently enter lymph nodes and cross-present antigens, rendering this conserved DC subset a promising target for tolerance induction or vaccination. However, only limited numbers of CD103+ DCs can be isolated with current methods. Established bone marrow culture protocols efficiently generate monocyte-derived DCs or produce a mixture of FLT3L-dependent DC subsets. We show that CD103+ DC development requires prolonged culture time and continuous action of both FLT3L and granulocyte macrophage colony-stimulating factor (GM-CSF), explained by a dual effect of GM-CSF on DC precursors and differentiating CD103+ DCs. Accordingly, we established a novel method to generate large numbers of CD103+ DCs (iCD103-DCs) with limited presence of other DC subsets. iCD103-DCs develop in a Batf3- and Irf8-dependent fashion, express a CD8α/CD103 DC gene signature, cross-present cell-associated antigens, and respond to TLR3 stimulation. Thus, iCD103-DCs reflect key features of tissue CD103+ DCs. Importantly, iCD103-DCs express high levels of CCR7 upon maturation and migrate to lymph nodes more efficiently than classical monocyte-derived DCs. Finally, iCD103-DCs induce T cell–mediated protective immunity in vivo. Our study provides insights into CD103+ DC development and function.
Introduction
Dendritic cells (DCs) form a complex network of bone marrow (BM)-derived immune cells, which populate lymphoid and nonlymphoid tissues and balance tolerance and immunity.1,2 Two major types of FMS-like tyrosine kinase 3 ligand (FLT3L)-dependent DCs have been identified: conventional DCs (cDCs) and plasmacytoid DCs (pDCs). In mice, cDCs can be further subdivided into the related lymphoid tissue-resident CD8α+ DCs and nonlymphoid tissue CD103+ cDCs that lack CD8α and have low CD11b expression.3 These cDC subsets have recently been unified across tissues and even species as CD8α-like cDCs with conserved developmental and functional properties.4-7 Similarly, a second, yet more heterogeneous, cDC group is formed by CD4+ DCs and CD11b+ DCs, which express high levels of signal regulatory protein α (SIRPα).1 In analogy to CD8α-like cDCs, we hereafter call them CD11b-like cDCs.
CD8α-like cDCs are excellent cross-presenters of cell-associated antigens.8-10 Whereas CD8α+ cDCs reside in lymphoid organs, CD103+ cDCs survey nonlymphoid tissues and migrate efficiently to lymph nodes during steady state and inflammation. Thus, CD103+ cDCs represent an attractive target for vaccination against intracellular pathogens and tumors, or for tolerance induction. Unfortunately, only very few CD103+ DCs can be isolated from tissues to investigate their function.
Large numbers of DCs can be differentiated from BM precursors (BM-derived DCs [BMDCs]). The culture of BM cells with granulocyte macrophage colony-stimulating factor (GM-CSF) DCs (GM-DCs) generates DCs primarily resembling monocyte-derived DCs (Mo-DCs).11 In contrast, culture with FLT3L (FL-DC) best reflects physiologic DC development, yet gives rise to a complex mixture of pDCs and cDCs, the latter including equivalents of CD8α-like cDCs and CD11b-like cDCs with poor enrichment for CD103+ DCs.1,12 CD8α-like FL-DCs lack CD8α and SIRPα but are identified by CD24 and Clec9A expression.10,12 Notably, only a fraction of CD8α-like FL-DCs expresses CD103 at variable levels.13,14 CD103 expression can be enhanced by supplementing GM-CSF during the last 2 days of FL-DC culture.13,14 However, these CD103+ DCs develop independent of Batf3.13,15 This transcription factor is nevertheless critical for CD8α-like cDC development in vivo.16 Furthermore, the in vivo role of GM-CSF for CD103+ DC development is controversial.13-15
Migration to lymph nodes is an important metric for determining effective vaccination. Only about 1% of DCs migrate to lymph nodes after subcutaneous injection.17 Around 104 DCs populate mouse lymph nodes at steady state, representing about 0.25% of total cells.18 Taken together, injection of >106 DCs is required to replace or significantly supplement resident lymph node DCs, approaching the limit for generating or isolating Batf3-dependent CD103+ DCs from one mouse with current methods.
We describe a new method for the selective and efficient in vitro generation of Batf3-dependent CD103+ DCs. This will enable insights into the development, function, and therapeutic potential of this DC subset.
Methods
All animal experiments were performed in accordance with institutional and state guidelines of Lower Saxony. Detailed methods are described in the supplemental material, available on the Blood Web site.
BMDCs
Briefly, BM cells were incubated with GM-CSF (GM-DCs) or FLT3L (FL-DCs) supernatants in RPMI1640 medium with 10% heat-inactivated fetal calf serum (Biochrom, Cambridge, UK), penicillin/streptomycin, and 50 µM β-mercaptoethanol. For iCD103-DCs, BM cells were incubated in 10 mL complete medium with FLT3L combined with GM-CSF for 9 days and subsequently replated with the same combination of cytokines and harvested at days 15 to 16.
Results
Generation of CD103+ DCs with current methods
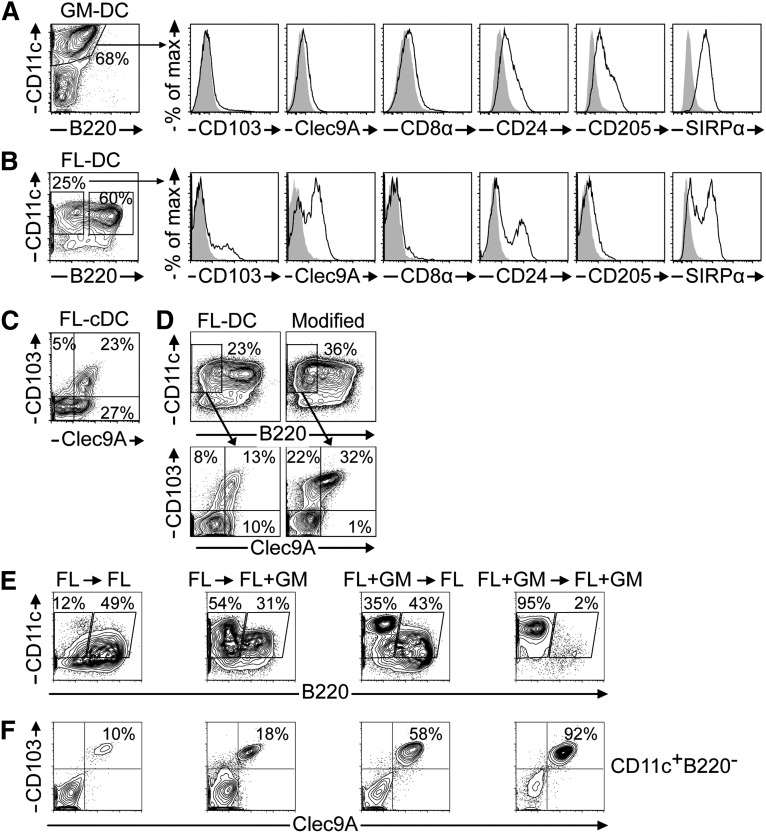
First, we investigated CD103+ DCs using established protocols. GM-DCs predominantly contain CD11c+B220– cells lacking CD103 expression (Figure 1A). Moreover, GM-DCs lack the CD8α-like cDC markers Clec9A and CD8α, but express SIRPα. Notably, GM-DCs weakly express CD24 and CD205, markers of CD8α-like cDCs that can also be expressed by other DC subsets.10 GM-DCs have been previously reported to resemble monocyte-derived inflammatory DCs.19 Thus, GM-CSF alone is insufficient for CD103+ cDC development.
Figure 1.
Generation of CD103+ DCs with established BMDC protocols. (A) GM-DCs and (B) FL-DCs were analyzed by flow cytometry. Histograms show CD8α-like DC marker expression (solid black line) of live-gated CD11c+B220– DCs compared with isotype controls (gray overlay). (C) Clec9A vs CD103 expression of live-gated CD11c+B220– FL-DCs. (D) FL-DCs and modified FL-DCs14 were compared. (E-F) BMDC cultures were established with FLT3L (FL) or a combination of FLT3L and GM-CSF (FL+GM). After 9 days, cells were harvested and replated with either FL or FL+GM. DCs were analyzed by flow cytometry on day 15. (E) Composition of DC subsets under different culture conditions. (F) Clec9A vs CD103 expression among live-gated CD11c+B220– DCs. Results are representative of 3 to 4 (A-B), 3 (C-D), or 2 (E-F) independent experiments.
FL-DCs are heterogeneous mixtures of CD11c+B220+ pDCs, which may also include cDC precursors,20 and CD11c+B220– cDCs (Figure 1B). cDCs split into SIRPα+ CD11b-like cDCs and SIRPα– CD8α-like cDCs (Figure 1B).12 We found variable expression of Clec9A, CD24, and CD103 among cDCs (Figure 1B). CD8α is absent and CD205 is only marginally expressed (Figure 1B). The discrepant expression of CD8α-like DC markers is further illustrated by co-staining of Clec9A and CD103. Many Clec9A+ FL-cDCs lack CD103 expression, whereas a second fraction co-expresses both markers (Figure 1C). This previously noted variability can be reduced by adding GM-CSF during the last 2 days of culture, which enhances CD103 expression (Figure 1D).14 However, these CD103+ DCs are Batf3-independent13,15 and are still a fraction among several DC subsets (Figure 1D). We speculated that d9 FL-DCs contain developmentally immature precursors of CD8α-like cDCs, consistent with the notion that CD103 is expressed late during the ontogeny of this cDC subset.15
CD103hi DC development follows delayed kinetics and requires the concerted action of FLT3L and GM-CSF
Because GM-CSF was implicated in the homeostasis of CD103+ DCs in vivo,21,22 we hypothesized that GM-CSF could differentiate and maintain CD103+ DCs from day 9 immature FL-DC precursors. BM cells were cultured for 9 days in the presence of supernatants containing FLT3L or FLT3L/GM-CSF. Cells were then harvested and replated for 7 days in the presence of refreshed FLT3L or FLT3L/GM-CSF. The combination of FLT3L and GM-CSF supernatants, especially during the first step of differentiation, results in an enhanced generation of CD11chiB220– cDCs (Figure 1E). Interestingly, CD11chiB220– cDCs predominate in cultures after the continuous presence of FLT3L/GM-CSF, whereas CD11cintB220+ pDCs are largely reduced (Figure 1E). The discrimination of pDCs and cDCs by different CD11c expression levels was previously only observed in vivo.12 Although CD103hi cDCs emerge in all conditions tested by day 16, the combination of FLT3L and GM-CSF throughout the culture period is necessary for an optimal yield (Figure 1F). This is not merely caused by the control of CD103 expression by GM-CSF, because complete co-expression of Clec9A and CD103 was also observed when GM-CSF was omitted at certain time points (Figure 1F). Thus, GM-CSF supernatant acts throughout the 16-day culture by promoting the differentiation and/or homeostasis of CD103hi DCs. The use of recombinant FLT3L and GM-CSF instead of in-house–produced supernatants confirmed the highly biased differentiation of CD103+Clec9A+ cDCs, despite a lower absolute yield (supplemental Figure 1). We term this novel BMDC protocol induced CD103-DCs (iCD103-DCs), which predominantly gives rise to CD11chiB220–CD103hi cDCs (Figure 1E-F).
iCD103-DC cultures efficiently and preferentially generate CD8α-like cDCs
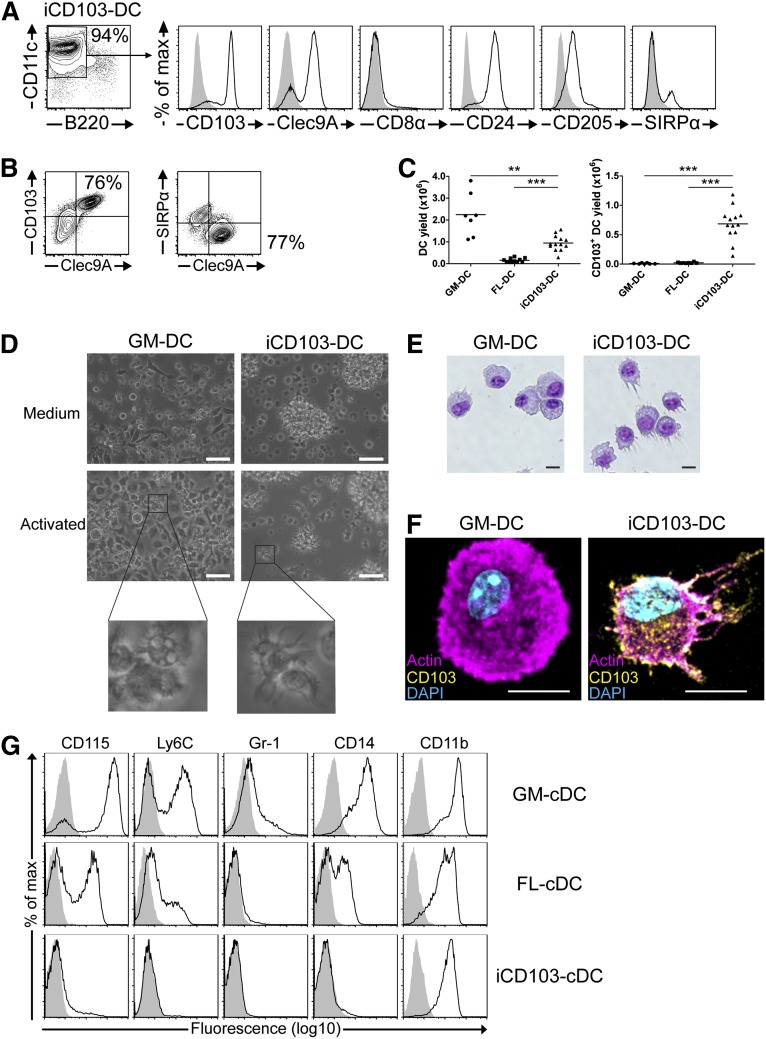
iCD103-DCs display a high expression of Clec9A, CD24, and CD205, whereas the majority of cells lacks SIRPα (Figure 2A), indicative of a CD8α-like cDC phenotype. CD8α is not expressed (Figure 2A), similar to FL-DCs.12 Co-stainings revealed that iCD103-DCs mainly consist of CD103hiClec9A+SIRPα– CD8α-like cDCs and a small proportion of CD103–/lowClec9A–SIRPα+ CD11b-like cDCs (Figure 2B).
Figure 2.
A novel culture method (iCD103) preferentially and efficiently generates CD103hi CD8α-like DCs. BM cells were cultured to generate GM-DCs, FL-DCs, or iCD103-DCs. (A) iCD103-DCs were analyzed by flow cytometry. Histograms show CD8α-like DC marker expression (solid black line) of live-gated CD11c+B220– DCs compared with isotype controls (gray overlay). (B) Clec9A vs CD103 (left) and Clec9A vs SIRPα (right) expression of live-gated CD11c+B220– iCD103-DCs. (C) Yield comparison per 1 × 106 input BM cells of CD11c+B220– DCs (left) and CD103+CD11c+B220– DCs (right) using GM-DC, FL-DC, and iCD103-DC culture methods. (D) Typical light micrographs of GM-DC and iCD103-DC cultures during steady state (Medium, upper panel) or after overnight CpG stimulation (Activated, lower panel). Enlarged views of activated DCs are depicted. Scale bars represent 50 µm. (E) Bright-field micrographs of cytospins of sorted CD11c+ GM-DCs and CD11c+B220–CD103+ iCD103-DCs after Giemsa staining. (F) Fluorescence micrographs of CpG-activated GM-DCs and iCD103-DCs stained for actin (magenta), CD103 (yellow), and nuclei (DAPI; cyan). (E-F) Scale bars represent 10 µm. (G) Myeloid lineage marker expression among live-gated CD11c+ GM-DCs or CD11c+B220– cDCs (FL-cDCs and iCD103-cDCs). Specific (solid black lines) and isotype control stainings (gray overlay) are depicted. Results are representative of 3 to 4 (A-B) or 3 (D-G) independent experiments. Data for (C) were combined from 7 (GM-DC), 3 (FL-DC), and 6 (iCD103-DC) independent experiments, each involving cultures from 1 to 6 mice. **P < .01, ***P < .001; 2-tailed Mann-Whitney U test.
We next compared the efficiencies of the different BMDC methods to generate CD11c+B220– DCs and CD103+CD11c+B220– DCs in terms of absolute cell numbers. We analyzed GM-DCs and FL-DCs at day 9 and iCD103-DCs at day 16 to compare the established protocols to our new method. GM-DCs exhibit high DC yields because they proliferate vigorously, and a low number of BM cells are initially plated (Figure 2C, left). In contrast, FL-DCs generate comparably fewer DCs relative to the input of BM cells, because only a fraction of them are cDCs and a high number of BM cells are seeded. Interestingly, iCD103-DCs generate significantly higher numbers of cDCs compared with FL-DCs (Figure 2C, left). When comparing the absolute yields of CD103+CD11c+B220– DCs, iCD103-DC cultures are the only method that efficiently generates CD103+ DCs (Figure 2C, right; supplemental Figure 1B).
D16 iCD103-DCs tend to cluster into floating cell aggregates, whereas GM-DCs and FL-DCs predominantly form single-cell suspensions, with GM-DCs exhibiting many adherent cells (Figure 2D and supplemental Figure 1C). Both GM-DCs and iCD103-DCs have a dendritic morphology, especially upon activation (Figure 2D). GM-DCs become highly adherent after activation or prolonged culture (not shown) and contain phagocytic inclusions, whereas activated iCD103-DCs enhance the formation of floating clusters and no inclusions are seen (Figure 2D). GM-DCs have a pronounced cytoplasm and polarized nuclei, whereas iCD103-DCs possess less cytoplasm and a more irregular and dendritic morphology (Figure 2E). Morphology and CD103 expression were substantiated by confocal microscopy of CpG-activated BMDCs (Figure 2F).
DC differentiation is associated with a downregulation of monocyte/macrophage lineage markers. Because GM-CSF differentiates monocytes in vitro and because FL-DCs seemed developmentally immature, we compared the expression of myeloid markers among the various BMDC types. GM-DCs express high levels of CD115 and CD14, whereas a subpopulation expresses Ly6C, in line with a monocytic relationship (Figure 2G). Some Gr-1+ cells were also detected (Figure 2G). Notably, a substantial fraction of FL-cDCs express CD115 and CD14, albeit at lower levels compared with GM-DCs (Figure 2G). Ly6C is expressed at low levels and Gr-1 is absent from FL-DCs (Figure 2G). The expression of monocytic markers underlines that d9 FL-DCs may not have fully developed into cDCs. Importantly, iCD103-DCs are largely devoid of CD115, CD14, Gr-1, and Ly6C expression, confirming full differentiation into cDCs (Figure 2G). CD11b is expressed by all types of BMDCs, which is frequently observed after in vitro culture.12 We calculated slightly higher CD11b MFI ratios for GM-DCs (54 ± 5.9) compared with FL-DCs (26 ± 5.2) and iCD103-DCs (23.8 ± 4.0) from 4 independent experiments. In summary, iCD103-DC cultures efficiently and preferentially generate developmentally mature CD8α-like cDCs.
CD103hi iCD103-DCs are Batf3/Irf8-dependent and develop independent of monocytes
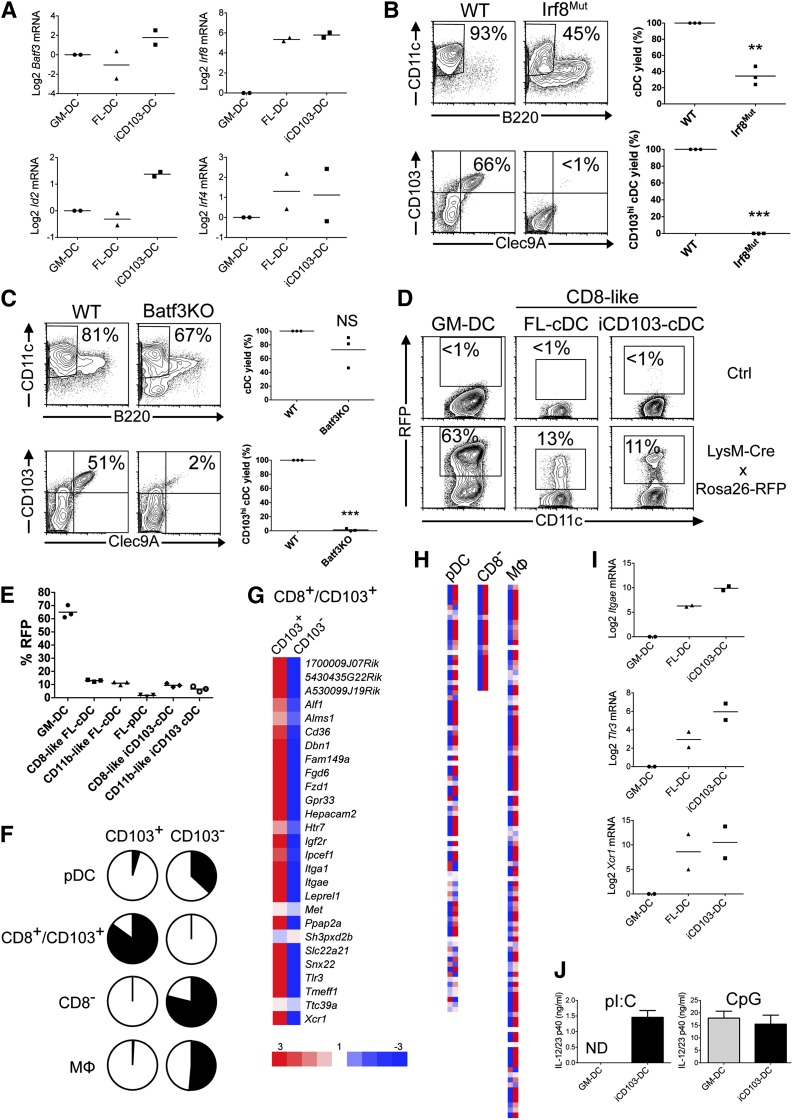
Mouse CD8α/CD103 cDCs develop from pre-DCs in a Batf3-, Id2-, and Irf8-dependent manner, whereas pre–DC-derived CD103+CD11b+ intestinal cDCs develop independent of these transcription factors and resemble CD11b+ cDCs.7 Because iCD103-DCs express CD11b, their developmental origin was unclear. Moreover, previous studies suggest that monocytes contribute to CD103+ DC development.23,24 The expression of Batf3 and Id2 is enriched in iCD103-DCs (Figure 3A). Irf8 is highly expressed in both FL-DCs and iCD103-DCs compared with GM-DCs (Figure 3A), in line with its more upstream role in FLT3L-dependent DC development. In contrast, Irf4 expression, which is required for the development of CD11b+ cDCs,25 is not consistently enriched in any BMDC type (Figure 3A). Importantly, when BM from Irf8-mutant and Batf3-deficient mice is differentiated into iCD103-DCs, impaired cDC development and the virtual absence of CD103hiClec9A+ cDCs are noted (Figure 3B-C). In contrast, GM-DCs develop normally in the absence of Irf8 and Batf3 (data not shown).
Figure 3.
CD103hi iCD103-DCs develop independent of monocytes through Batf3- and Irf8-sensitive pathways and express CD8α/CD103 DC signature genes. (A) Relative expression of transcription factors by GM-DCs, FL-DCs, and iCD103-DCs. (B-C) iCD103-DCs generated from wild-type (WT) and (B) BXH2 (Irf8mut) or (C) Batf3−/− (Batf3KO) mice were analyzed by flow cytometry. The upper left plots show frequencies of CD11c+B220– cDCs and the lower left plots show frequencies of CD103hiClec9A+ DCs among cDCs (representative of 3 independent experiments). Quantifications of DC yields relative to WT cells are shown on the right for 3 independent experiments. NS, not statistically significant; **P < .01, ***P < .0001; paired Student t test. (D-E) GM-DCs, FL-DCs, and iCD103-DCs generated from LysM-Cre × Rosa26-RFP or Rosa26-RFP control mice (Ctrl) were analyzed for RFP expression by flow cytometry. One of 2 independent experiments, each involving 3 mice per genotype, is shown. (D) Representative plots compare CD11c vs RFP expression among CD11c+B220– BMDC subsets. (E) Summarized percentages of RFP+ DCs among CD11c+ GM-DCs, CD11c+B220–SIRPα– CD8α-like FL-DCs, CD11c+B220–SIRPα+ CD11b-like FL-DCs, CD11c+B220+ FL-pDCs, CD11c+B220–SIRPα+CD103– CD11b-like iCD103-DCs, and CD11c+B220–SIRPα–CD103hi CD8α-like iCD103-DCs. (F-H) Relative mRNA expression from FACS-sorted CD103hi and CD103– iCD103-DCs by Agilent microarray analysis. The expression of published gene signatures for pDCs, CD8+/CD103+ DCs, CD8– DCs, and macrophages was compared. See also supplemental Table 1. (F) Fraction of signature genes (black) upregulated ≥threefold in CD103+ vs CD103– iCD103-DCs (left; pDC: 4.3%, CD8+/CD103+: 82.1%, CD8–: 0%, MF: 0.9%) or upregulated ≥threefold in CD103– vs CD103+ iCD103-DCs (right; pDC: 32.3%, CD8+/CD103+: 0%, CD8–: 71.4%, MF: 47.4%). (G) Degree of overexpression (red) or underexpression (blue) for all genes of the CD8+/CD103+ DC signature in CD103+ vs CD103– iCD103-DCs (equal expression in white). (H) Signatures of pDCs, CD8– DCs, and macrophages as in (G). (I) Relative expression levels of Itgae, Tlr3, and Xcr1 among BMDCs. (J) GM-DCs or iCD103-DCs were stimulated for 24 hours with poly(inosine:cytidine) (pI:C) or CpG. IL-12/23p40 was quantified in the supernatant by enzyme-linked immunosorbent assay. Mean + SD (n = 3) is shown for 1 of 2 independent experiments. ND, not detectable. Data from (A) and (I) are combined from 2 independent experiments. Values are normalized to Hprt and GM-DCs.
We next used LysM-Cre × Rosa26-RFP mice to irreversibly label LysM+ cells (including monocytes and their progeny) by RFP expression.21 More than 60% of GM-DCs express RFP (Figure 3D-E), compatible with their development from monocytes.21,26 In contrast, cDC and pDC subsets from FL-DC cultures express RFP at a frequency <20%, which can be attributed to the ∼15% targeting of their precursors (MDPs) in LysM-Cre mice.27 Similarly, only ∼10% of both CD11b-like and CD8α-like iCD103-DC subsets express RFP (Figure 3D-E). Thus, CD103hi iCD103-DCs develop from pre-cDCs in an Irf8- and Batf3-dependent fashion, independent of monocytes.
iCD103-DCs express CD8α/CD103 DC-specific genes and respond to TLR3 stimulation
Our data suggest that CD103hi iCD103-DCs are CD8α-like cDCs. Recent transcriptional network analysis identified a signature of 28 genes specific to or enriched in the CD8α+/CD103+ (CD8α-like) cDC subset, including Tlr3 and Xcr1.28 Similarly, signatures for CD8– (CD11b-like) cDCs, pDCs, and macrophages were previously identified.28,29 We performed Agilent microarray analysis and calculated the coverage of genes overexpressed more than threefold in CD103+ vs CD103– or in CD103– vs CD103+ iCD103-DCs within each signature (Figure 3F and supplemental Table 1). Importantly, CD103+ iCD103-DCs are strongly and selectively enriched in the CD8α-like signature genes including Itgae (CD103), Tlr3, and Xcr1, but not in other signatures (Figure 3F-H). In contrast, CD103– iCD103-DCs predominantly express the CD11b-like DC signature, yet also display enrichment in pDC and macrophage genes (Figure 3F,H). Similar results were obtained independently using Affymetrix microarrays (data not shown).
We next validated these data by quantitative polymerase chain reaction. Itgae (CD103) and Tlr3 expression is lowest in GM-DCs, moderate in FL-DCs, and highly enriched in iCD103-DCs (Figure 3I). Xcr1 is highly overexpressed in FLT3L-dependent DCs, with slight enrichment in iCD103-DCs (Figure 3I). TLR3 drives cytokine production upon poly(I:C) stimulation of CD8α-like cDCs.30,31 Indeed, poly(I:C) stimulation results in IL-12p40 production by iCD103-DCs but not GM-DCs, whereas both DC types respond to TLR9-dependent CpG stimulation (Figure 3J). Based on these analyses we propose that CD103+ iCD103-DCs share key developmental and functional features with in vivo CD103+ CD8α-like DCs.
GM-CSF supernatant selectively promotes the differentiation and homeostasis of CD8α-like cDCs
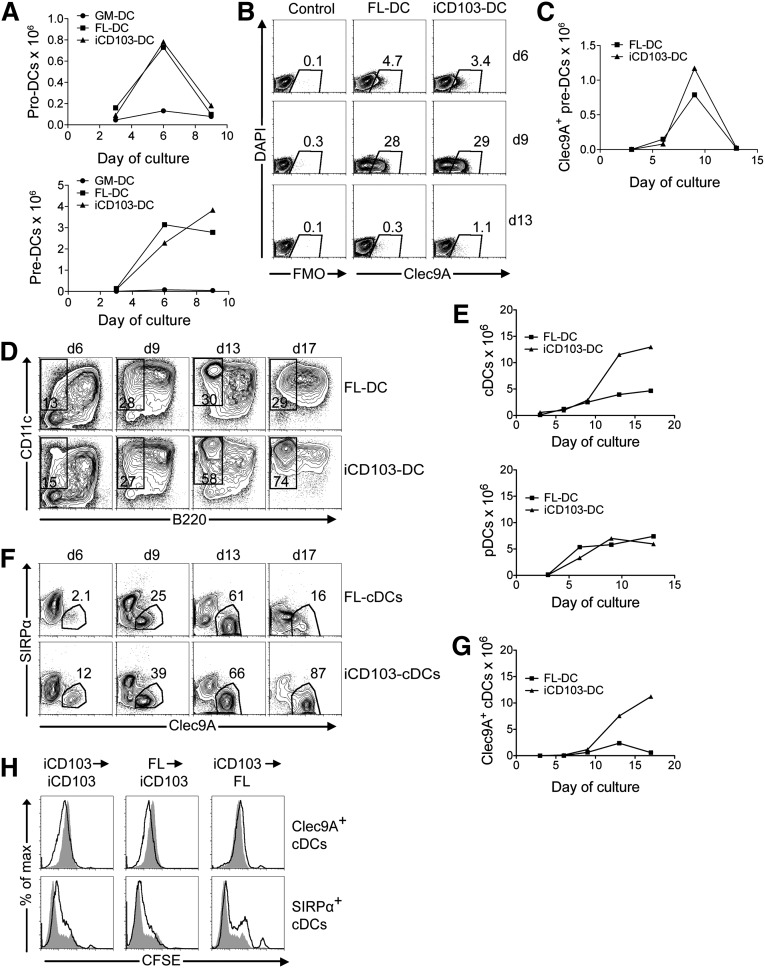
To understand the mechanisms through which GM-CSF supernatant is essential for iCD103-DC development, a detailed kinetics analysis of the precursor-product relationship was performed. DC precursors can be identified in FLT3L cultures as pro-DCs (corresponding to common DC progenitors [CDPs]) and the more downstream pre-DCs, which may be further subdivided into B220+ and B220– subsets.20,32,33 Pro-DCs peak at day 6 of FLT3L culture and decline sharply by day 9 (Figure 4A, upper panel). Pre-DCs also accumulate at day 6 but are maintained at high levels until day 9 (Figure 4A, lower panel). GM-CSF alone is insufficient to expand DC precursors (Figure 4A). Furthermore, the combination of FLT3L and GM-CSF (iCD103-DC) has little effect on DC precursor numbers when compared with FL-DC cultures (Figure 4A).
Figure 4.
GM-CSF supernatant selectively promotes the differentiation and homeostasis of CD103+ DCs. Equal numbers of CFSE-labeled BM cells were plated to establish GM-DC, FL-DC, and iCD103-DC cultures. Cells were harvested after 3, 6, 9, 13, or 17 days for flow cytometric analysis. On day 9, cells were harvested, CFSE-labeled, and replated at equal numbers. (A) Kinetics for total pro-DC numbers and pre-DC numbers are shown. (B) Representative plots display Clec9A expression among pre-DCs of FL-DC and iCD103-DC cultures over time. (C) Total numbers of Clec9A+ pre-DCs. (D) The percentages of CD11c+B220– cDCs are shown. (E) Total numbers of CD11c+B220– cDCs (upper graph) and CD11c+B220+ pDCs (lower graph) over time. (F) Clec9A vs SIRPα expression among CD11c+B220– cDCs is shown. (G) Absolute numbers of CD11c+B220–Clec9A+ cDCs. (H) FL-DCs and iCD103-DCs were harvested on day 9 and labeled with CFSE before replating for 4 days with either FLT3L (FL) or FLT3L and GM-CSF (iCD103). The upper panel shows CFSE signals of CD11c+B220–Clec9A+ cDCs, and the lower panel indicates CFSE signals of CD11c+B220–SIRPα+ cDCs within the same cultures (black lines). Cells cultured only in presence of FLT3L on both day 0 and day 9 are overlaid as gray histograms. Representative results show 1 of 2 independent experiments (A-H).
CD24hi pre-DCs preferentially develop into CD8α-like cDCs.34 We speculated that this also holds true for Clec9A, a more specific marker of the CD8α-like cDC lineage, recently shown to be already expressed by DC precursors.35,36 Interestingly, pre-DCs start to express Clec9A on day 6 and Clec9A expression peaks on day 9, with about 30% of pre-DCs being Clec9A+ in both FL-DC and iCD103-DC cultures (Figure 4B-C). Thus, by comparing FL-DC and iCD103-DC cultures, GM-CSF has little effect on Clec9A expression (Figure 4B) and Clec9A+ pre-DC numbers (Figure 4C). Hence, Clec9A+ pre-DCs accumulate at day 9 (Figure 4C) when FL-DCs are routinely harvested.12 This further underlines the immature character of FL-DCs (Figure 1B-C and Figure 2G). Indeed, a sharp drop in Clec9A+ pre-DCs occurs from day 9 to day 13 after reculture with DC growth factors (Figure 4C). To address whether this is explained by their differentiation into the CD8α-like cDC lineage, we compared DC precursor kinetics to those of the mature DC subpopulations. CD11c+B220+ cells dominate day 6 and day 9 cultures independent of GM-CSF (Figure 4D). CD11c+B220– cDCs already split into Clec9A+ and SIRPα+ subsets, and GM-CSF increases the proportion of Clec9A+ CD8α-like cDCs at these time points (Figure 4F). However, the effect of GM-CSF on cDC and Clec9A+ cDC numbers is negligible until day 9 (Figure 4E,G). Interestingly, after day 9, GM-CSF supernatant massively promotes the generation of CD11chiClec9A+ cDCs in iCD103-DC cultures, resulting in the predominance of this subset on day 17 (Figure 4D-G), as shown before (Figure 1E-F). In contrast, cDCs, and specifically Clec9A+ cDCs, decline in FL-DC cultures in the absence of GM-CSF by day 17 (Figure 4D-G). Because FLT3L supernatant alone is sufficient to differentiate Clec9A+ cDCs initially (Figure 4F, day 13), GM-CSF supernatant likely promotes the proliferation and/or survival of CD8α-like cDCs.
To evaluate proliferation specifically, we carboxyfluorescein succinimidyl ester (CFSE)-labeled FL-DCs and iCD103-DCs on day 9 and compared the CFSE dilution of cDC subsets on day 13. GM-CSF enhanced the CFSE dilution of Clec9A+ DCs in iCD103-DC cultures when compared with cells cultured with FLT3L alone (Figure 4H; iCD103→iCD103). This was mainly mediated by the late action of GM-CSF from day 9, because enhanced CFSE dilution of Clec9A+ cDCs was also observed when GM-CSF was initially absent on day 0, but not in the reciprocal experiment (Figure 4H; FL→iCD103 and iCD103→FL). Notably, this effect is selective for Clec9A+ cDCs because the kinetics of pDC numbers is not altered by GM-CSF (Figure 4E, lower graph). Moreover, CFSE dilution of SIRPα+ cDCs within the same cultures is not enhanced by GM-CSF (Figure 4H). In summary, GM-CSF supernatant acts by promoting the differentiation and homeostasis of CD8α-like cDCs after day 9 of FLT3L culture, but also has early effects on DC precursors without markedly affecting their numbers.
iCD103-DCs cross-present cell-associated antigens
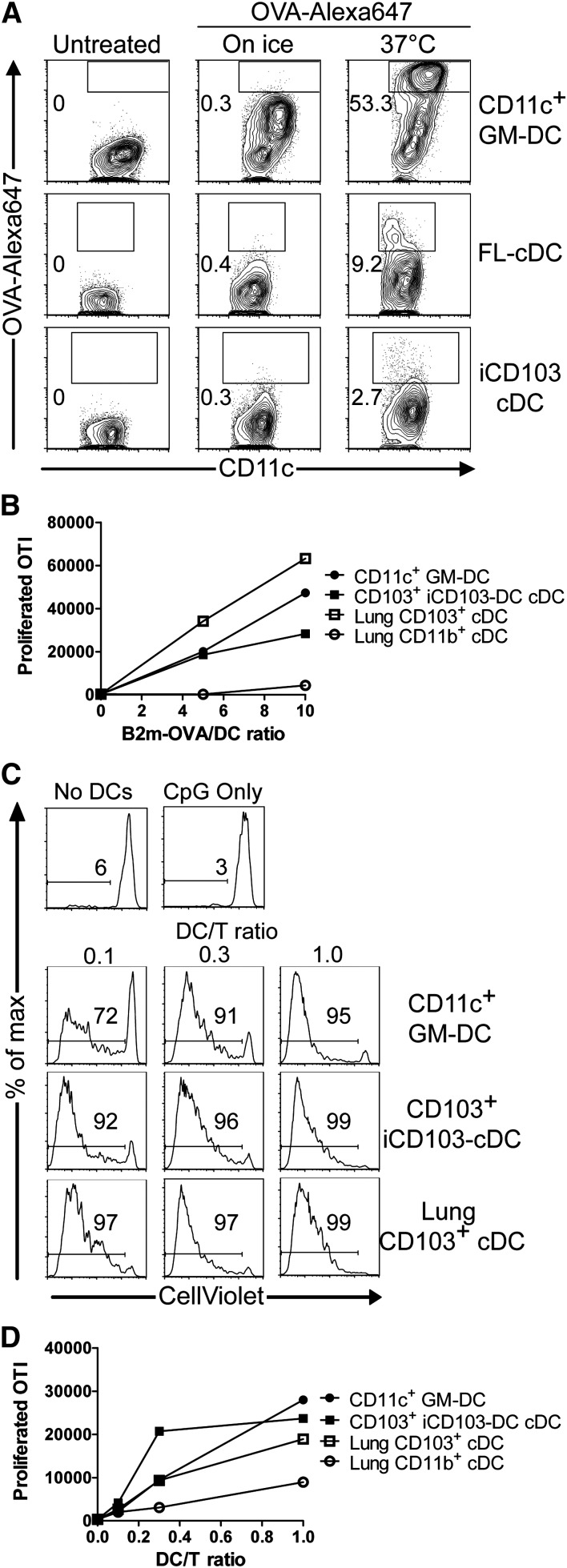
CD8α-like cDCs efficiently cross-present cell-associated antigens.8,9,37 Antigen uptake represents an important factor that can modulate antigen presentation. CD103+ DCs take up antigens less efficiently than CD103– DCs.38 Consistently, >50% of GM-DCs incorporate high levels of ovalbumin (OVA), compared with <10% of FL-DCs and iCD103-DCs (Figure 5A).
Figure 5.
CD103hi iCD103-DCs cross-present cell-associated antigens. (A) GM-DCs, FL-DCs, and iCD103-DCs were seeded at equal numbers. OVA-Alexa647 was added for 10 minutes at 37°C. Controls were kept on ice or were untreated. The uptake of OVA was quantified by flow cytometry. Plots display CD11c expression vs OVA-Alex647 signal intensity among CD11c+ GM-DCs (top), CD11c+B220– FL-cDCs (middle), and CD11c+B220– iCD103-cDCs (bottom). Numbers indicate percentages of DCs that have actively internalized OVA-Alexa647. (B-D) CD11c+ GM-DCs, CD11c+B220–CD103hi iCD103-DCs, lung CD103+ DCs, and lung CD11b+ DCs were FACS-sorted. DCs were co-cultured at varying ratios with CD8+CellViolet+ .OTI T cells in the presence of (B) irradiated OVA-coated B2m−/− splenocytes or (C-D) irradiated B16-OVA melanoma cells and CpG. Proliferation was quantified by flow cytometry after 5 days of culture. (B,D) The graphs show the numbers of proliferated CD8β+ OTI cells (mean values of duplicate wells) in response to (B) splenocytes or (D) B16-OVA cells. (C) Representative histograms show CellViolet dilution of CD8β+ OTI cells in response to B16-OVA. Wells without DCs (No DCs) or without DCs and B16-OVA cells (CpG only) served as controls. Results are representative of 2 independent experiments (A-D).
We next compared the capacity of fluorescence-activated cell (FACS)-sorted iCD103-DCs, GM-DCs, and lung DC subsets to cross-present cell-associated antigen. Lung CD103+ DCs efficiently induce OTI T-cell proliferation in contrast to lung CD11b+ DCs (Figure 5B), as expected.37 Notably, iCD103-DCs and GM-DCs also cross-present cell-associated OVA, albeit with slightly reduced efficiency compared with lung CD103+ DCs (Figure 5B). Cross-presentation was proposed to be promoted by GM-CSF or toll-like receptor (TLR) signaling.30,39 Conversely, tumor cells can inhibit DC functions and render them tolerogenic.40,41 We established a cross-presentation assay of irradiated B16-OVA melanoma cells. Neither BMDCs nor CD103+ lung DCs are able to induce OTI T-cell proliferation under noninflammatory conditions in this assay (data not shown). However, using CpG as an inflammatory TLR9 stimulus,42 iCD103-DCs potently cross-present melanoma-derived antigen at even slightly higher efficiency when compared with lung CD103+ DCs (Figure 5C-D). Thus, iCD103-DCs can efficiently cross-present cell-associated antigens.
iCD103-DCs are not inherently tolerogenic
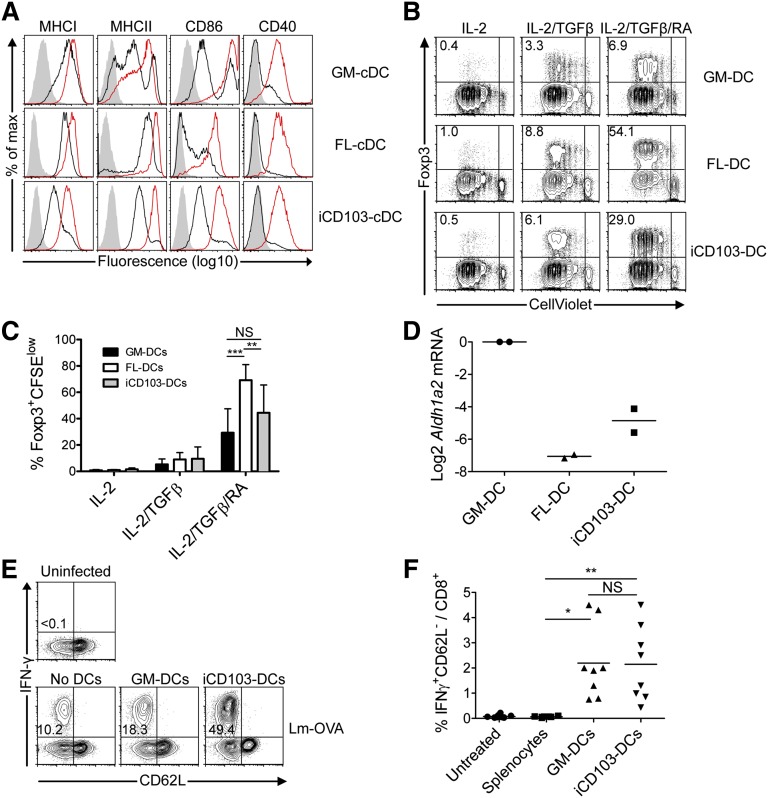
The maturation status of DCs can influence their effector functions. GM-DCs have slightly higher levels of MHC class I, CD86, and CD40 at steady state compared with iCD103-DCs (Figure 6A). In addition, iCD103-cDCs are mainly MHC-class-IIint, whereas GM-DCs contain a higher proportion of both MHC-class-IIhi and MHC-class-IIlow DCs (Figure 6A). In contrast, FL-cDCs display higher levels of MHC-class-I and MHC-class-II molecules than do GM-DCs, yet low levels of co-stimulatory molecules (Figure 6A). Upon CpG activation, all BMDCs upregulate the maturation markers tested (Figure 6A).
Figure 6.
iCD103-DCs are not specialized to induce tolerance. (A) DC maturation marker expression among CD11c+ GM-DCs or CD11c+B220– FL-cDCs and CD11c+B220– iCD103-cDCs in the presence or absence of overnight CpG stimulation. Unstimulated (solid black lines), CpG-stimulated (solid red lines), and isotype control stainings (gray overlay) are shown. (B-C) CellViolet-labeled CD4+ Rag1−/− × OTII T cells were co-cultured with GM-DCs (upper panel), FL-DCs (middle panel), or iCD103-DCs (lower panel) in the presence of OVA323-339 peptide, IL-2, and combinations of TGFβ and RA. After 5 days, cells were analyzed by flow cytometry. (B) Plots display CellViolet dilution vs Foxp3 expression among CD4+ cells. (C) Quantification of proliferated Foxp3+ cells in different culture conditions. Mean + SD is shown. **P < .01, ***P < .001; NS, not significant; 2-way analysis of variance with Bonferroni’s post-test. (D) Relative Aldh1a2 expression among GM-DCs, FL-DCs, and iCD103-DCs is shown normalized to Hprt and GM-DCs. Results show 2 independent cDNA preparations. (E-F) Naïve WT mice were injected intravenously (IV) with equal numbers of OVA257-264-pulsed FACS-sorted CD11c+ GM-DCs or CD103hiCD11c+B220– iCD103-DCs. Mice were infected 8 days later IV with 5 × 103 colony-forming units (CFU) Lm-OVA. Seven days post infection, splenocytes were restimulated with OVA257-264 to determine the antigen-specific CD8+ T-cell response. (E) Representative plots display CD62L vs interferon (IFN)-γ expression among restimulated CD8+ T cells. (F) Quantification of IFN-γ–secreting CD8+ T cells. Results are representative of 2 to 5 (A), 4 (B), or 2 (D-F) independent experiments. Data were combined from 4 independent experiments with individual culture preparations (C) or 2 independent experiments with 4 to 5 mice per experimental group (F). *P < .05, **P < .005; NS, not significant; 2-tailed Mann-Whitney U test.
Intestinal CD103+ DCs and splenic CD8α+ DCs induce Foxp3+ regulatory T cells (Tregs) via transforming growth factor (TGF)-β and retinoic acid (RA).43,44 Moreover, given that iCD103-DCs are slightly less mature than GM-DCs, we next compared their tolerogenic properties. Interestingly, all types of BMDCs lack the ability to induce Foxp3+ Tregs, whereas Treg induction occurs comparably after exogenous addition of TGF-β (Figure 6B-C). The addition of RA-enhanced Treg induction and FL-DCs were significantly more efficient under these conditions than GM-DCs and iCD103-DCs (Figure 6B-C). The increased proficiency of FL-DCs to induce Foxp3+ Tregs correlates with lower co-stimulatory molecule expression (Figure 6A). Aldh1a2, encoding the enzyme necessary for RA production in DCs,43 was expressed at much lower levels in iCD103-DCs and FL-DCs compared with GM-DCs (Figure 6D), potentially explaining why FL-DCs exhibit stronger Foxp3+ Treg induction after exogenous RA supplementation. Thus, the ability to induce Foxp3+ Tregs is acquired independent of the development of CD103+ DCs.
Finally, we assessed whether iCD103-DCs induce T-cell tolerance in vivo. Interestingly, the adoptive transfer of OVA257-264-pulsed BMDCs does not result in CD8+ T-cell tolerance, but rather enhances the OVA-specific immune response against OVA-expressing Listeria monocytogenes (LmOVA; Figure 6E-F). Thus, iCD103-DCs do not have inherent tolerogenic properties but rather promote adaptive immunity.
Mature iCD103-DCs express high levels of CCR7 and induce protective T-cell immunity in vivo
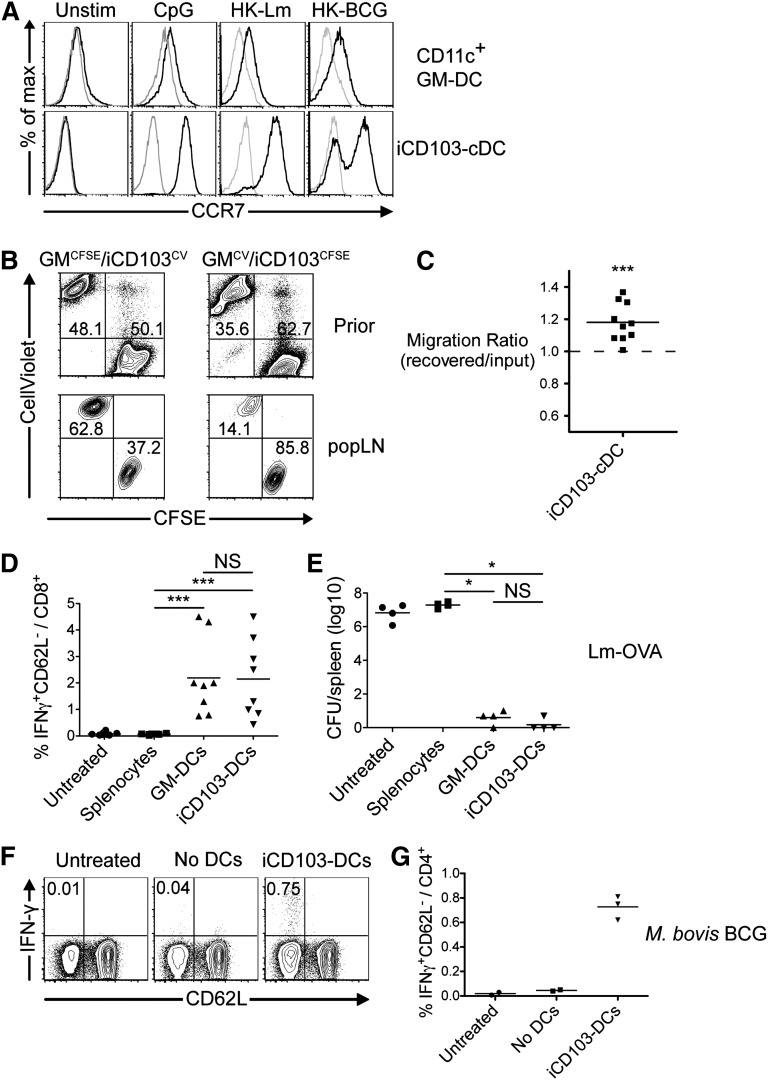
We next tested matured iCD103-DCs in vaccination experiments. An important requirement for successful DC vaccines is their efficient migration to draining lymph nodes after subcutaneous application. Lymph node homing is controlled by CCR7 expression on migrating DCs.45 Unstimulated BMDCs are largely devoid of CCR7 expression, yet this receptor is slightly upregulated on GM-DCs after stimulation with CpG or inactivated pathogens (Figure 7A, upper panel). In contrast, virtually all CpG- and Listeria-activated iCD103-DCs express high levels of CCR7, and CCR7 is also upregulated after Mycobacterium bovis bacillus Calmette-Guérin (BCG) stimulation (Figure 7A, lower panel). GM-DCs and iCD103-DCs were differentially labeled, mixed, and injected together with CpG into the footpad. iCD103-DCs more efficiently reach the draining LN 2 days after injection compared with GM-DCs (Figure 7B-C), consistent with higher CCR7 expression.
Figure 7.
iCD103-DCs upregulate CCR7, efficiently enter lymph nodes, and induce protective T-cell immunity. (A) GM-DCs and iCD103-DCs were kept untreated (unstim) or were stimulated for 16 hours with CpG, heat-killed Listeria monocytogenes (HK-Lm), or heat-killed M bovis (HK-BCG). CCR7 expression is shown for live-gated CD11c+B220–CD103hi iCD103-cDCs and CD11c+ GM-DCs (black lines). Overlays indicate control stainings (gray lines). (B-C) GM-DCs were labeled with CFSE and mixed with CellViolet-labeled iCD103-DCs (GMCFSE/iCD103CV) or vice versa (GMCV/iCD103CFSE). (B) Flow cytometric analysis of DC mixtures before injection (upper panel) and recovered DCs from popliteal lymph nodes (popLN) 48 hours after subcutaneous injection in combination with CpG. (C) Migration ratio calculated as the frequency of labeled iCD103-DCs recovered from popLN (2 per mouse) after 48 hours over the initial frequency in the injected DC mixtures. A ratio of 1 indicates equivalent migration capacity of GM-DCs and iCD103-DCs. ***P = .001; 1-sample Student t test compared with value of 1.0. (D) iCD103-DCs and GM-DCs were stimulated and FACS-sorted as CD11c+B220–CD103hi or CD11c+ cells. DCs were then pulsed with OVA257-264 and OVA323-339 peptides, washed, and injected subcutaneously. Pulsed splenocytes served as a control. After 7 days, mice were bled and leukocytes were restimulated with OVA257-264 in the presence of Brefeldin A followed by flow cytometric analysis. The frequencies of CD62L–IFN-γ+ cells among CD8+ T cells are quantified. (E) Vaccinated mice were challenged on day 8 with a lethal dose of Lm-OVA (1 × 106 CFU) IV. CFUs in the spleen were determined 3 days later. (F-G) iCD103-DCs were stimulated with M bovis BCG and transferred subcutaneously into mice. Identically treated samples without DCs (No DCs) were injected as a control. Seven days later, splenocytes were restimulated with Ag85B240-254 and analyzed by flow cytometry. (F) Representative plots show CD62L vs IFN-γ expression among CD4+ live-gated cells. (G) Quantification of Ag85B240-254-specific IFN-γ–producing cells. Results are representative of or quantify 2 to 3 independent experiments (A), 2 independent experiments with 2 to 3 (B-C,F-G), 2 independent experiments with 4 to 5 (D), or 2 independent experiments with 1 to 4 (E) mice per group. **P < .01, **P < .001; NS, not significant; 2-tailed Mann-Whitney U test.
We next assessed whether iCD103-DC vaccination is sufficient to induce protective T-cell immunity. Restimulation of blood leukocytes confirms that OVA257-264-specific CD8+ T-cell responses are mounted after GM-DC and iCD103-DC vaccination, but not after splenocyte vaccination (Figure 7D). Eight days after vaccination, mice were challenged intravenously with a lethal dose of Lm-OVA. Untreated and splenocyte-vaccinated mice exhibited a high bacterial burden (Figure 7E). In contrast, vaccination of mice with iCD103-DCs or GM-DCs effectively controlled the lethal Lm-OVA dose (Figure 7E). We next vaccinated mice with M bovis BCG-pulsed iCD103-DCs to investigate whether iCD103-DCs also elicit immunity against a natural pathogen–derived antigen. Indeed, an Ag85B-specific T-cell response was elicited after iCD103-DC vaccination (Figure 7F-G). These results collectively demonstrate that iCD103-DCs are functional in vivo and induce T-cell immunity.
Discussion
Here we describe a novel BMDC protocol, termed iCD103-DC. From one mouse, 30 to 50 million CD103+ iCD103-DCs can be easily obtained, compared with <0.5 million CD103+ DCs that must be isolated with much effort from tissues by digestion and cell sorting. Because iCD103-DCs preferentially are CD103+ cDCs, even unseparated cultures can be useful for many experimental questions. In contrast, short-term FL-DC cultures generate all FLT3L-dependent DC subsets simultaneously. This is advantageous when pDCs or CD11b-like cDCs are the subject of investigation, yet it necessitates subset isolation with a very low yield of CD103+ DCs.
It is controversial whether GM-CSF is required for CD103+ DC development in vivo, because this cytokine was shown to control CD103 expression.13-15 Furthermore, the putative effects of GM-CSF on the development, survival, or migration of CD103+ DCs were previously difficult to distinguish. Using 15- to 16-day iCD103-DC cultures, where migration can be excluded, and by using detailed kinetics analysis of the precursor/product relationship, we established that GM-CSF supernatant is essential for the efficient generation of CD103+ DCs along with FLT3L supernatant. This is independent of the mere regulation of CD103 expression, because concomitant staining for SIRPα and Clec9A allows for the faithful identification of CD8α-like cDCs. Moreover, prolonging GM-DC (data not shown) or FL-DC culture (Figure 1E-F and Figure 4F-G) to days 15 to 16 fails to efficiently induce CD8α-like cDCs. Conversely, omission of GM-CSF at either day 0 or day 9 of the iCD103-DC method lowers the yield of CD8α-like cDCs. Thus, CD103+ DC development is not just a matter of kinetics of established methods. Instead, we found that GM-CSF supernatant selectively promotes CD8α-like cDC development via a dual mechanism throughout the culture period, as is further described next.
First, the early presence of GM-CSF was necessary for the optimal development of CD103+ DCs at later time points. Indeed, CDPs are known to express Csf2ra mRNA at levels comparable with splenic cDCs.46 Moreover, pre-DCs express surface Csf2rb similar to CDPs.22 However, GM-CSF did not substantially alter the numbers of DC precursors in iCD103-DC cultures, in line with previous in vivo data showing that GM-CSF–unresponsive BM cells normally develop into MDPs, CDPs, and pre-DCs in mixed chimeras.22 Similarly, Csf2−/− mice (Csf2 encodes for GM-CSF) have normal numbers of MDPs and CDPs in the bone marrow, yet the combined loss of GM-CSF and FLT3L results in a synergistic loss of DC precursors.46 Whether this is a cell-intrinsic phenomenon has not yet been addressed. It is tempting to speculate that GM-CSF signaling modulates DC precursors in a way that facilitates development into the CD103+ DC lineage. This requires further investigation. Notably, CD11c+B220+ cells preceded the development of CD11c+B220– cDCs in both FL-DCs and iCD103-DCs, consistent with their potential overlap with early, uncommitted DC precursors.20
Second, we showed that GM-CSF was specifically essential to promote the proliferation and persistence of CD11chiClec9A+ cDCs during the second differentiation step. Given that the numbers of Clec9A+ DC precursors peak at day 9 and sharply drop by day 13, this is likely explained by their terminal differentiation into the CD8α-like cDC lineage. Our results are in agreement with the strong reduction of nonlymphoid tissue CD8α-like cDCs in the absence of GM-CSF signaling that has been attributed in part to defective survival.22 Our finding that CD11chi cDC development requires prolonged culture could resolve a previous discrepancy between lymphoid organs and FL-DC cultures: in the latter, CD11c expression among pDCs and cDCs is identically intermediate, whereas in lymphoid organs12 and iCD103-DC cultures, high CD11c expression discriminates cDCs from the few emerging pDCs (Figure 1E, Figure 4D, and supplemental Figure 1A).
The human counterpart to mouse CD8α-like DCs are the CD141+CLEC9A+ DCs.5,6 Notably, human CD8α-like DCs can be generated from CD34+ stem cells,6,47 yet the efficiency and purity is low compared with the mouse iCD103-DC protocol. However, it can be noted that the reported method is also 2-step–based, similar to iCD103-DCs, including an expansion of DC precursors and their differentiation.6 Thus, CD8α-like DC differentiation being a late event6,14,15 seems to be conserved between mice and humans. In addition, FLT3L and GM-CSF were shown to be essential for human CD8α-like DC differentiation,6 as we describe here for mouse CD103+ DCs. Furthermore, the human counterparts of iCD103-DCs were rather immature regarding CD40 and CCR7 expression, a finding that we also confirmed. However, this does not translate directly to tolerogenic DC functions: iCD103-DCs were poorly phagocytic, lacked the abilities to endogenously produce retinoic or to generate Foxp3+ Tregs, and also did not induce tolerance in vivo. In contrast, antibody-mediated antigen targeting to steady state CD103+ DCs in vivo promotes tolerance in the absence of adjuvants.48 Thus, tolerogenic functions can be acquired independent of DC development and maturation. Several factors that imprint tolerogenic activities in DCs have been recently identified (reviewed in Mayer et al2). The iCD103-DC culture, apparently devoid of such factors, might be a powerful tool in exploring DC tolerogenicity.
Maturation and migratory behavior of iCD103-DCs can, however, be readily induced by TLR-mediated activation. Matured iCD103-DCs express higher levels of CCR7 and migrate more efficiently to draining lymph nodes when compared with GM-DCs after subcutaneous injection. Given the marked difference in CCR7 expression, it is surprising that the migratory activity of iCD103-DCs is not more pronounced. However, it is possible that the migratory activity is counter-regulated. For example, CD103/β7 binds E-cadherin on epithelial cells and is critical for the retention of CD103+ cells at peripheral tissues.49 Because iCD103-DCs express high levels of CD103 in contrast to GM-DCs, they may be preferentially trapped after subcutaneous injection. However, CD103 can be downregulated by anti-CD40 stimulation.50 Thus, the right combination of maturation signals and different application routes could potentially further enhance the migratory activity of iCD103-DCs compared with classic GM-DCs.
There are also some differences between the human and the mouse culture settings. For example, IL-4 was essential in the human protocol for the development of CD8α-like DCs.6 Thus, human CD141+CLEC9A+ DC differentiation may on rely on additional or distinct factors than the corresponding mouse DC subset. Conversely, we could not formally rule out a contribution of endogenous IL-4 in iCD103-DC cultures. We found that recombinant FLT3L and GM-CSF are sufficient to replicate the highly biased differentiation of CD103+ DCs, yet in-house–produced supernatants yielded higher cell numbers and are thus the currently favored protocol (supplemental Figure 1). This could relate to different biological properties of the recombinant cytokines or to accessory factors in the FLT3L/GM-CSF supernatants that might increase CD103+ DC numbers by regulating the proliferation or survival of DCs or their precursors. Future investigation of these interesting aspects will be useful for the further optimization of protocols generating human DC subsets.
In summary, we describe a simple BMDC culture method combining GM-CSF and FLT3L supernatants to generate large numbers of CD103+ DCs that resemble tissue CD8α-like cDCs. iCD103-DC cultures open many future possibilities for studying CD103+ DC development and function.
Acknowledgments
The authors thank Dr. Robert Geffers (Helmholtz Center for Infection Research, Braunschweig) for help with microarray analyses and the Cell Sorting Core Facility of the Hannover Medical School. The study benefitted from data assembled by the ImmGen Consortium.
This study was supported in part by the Braukmann-Wittenberg-Herz-Stiftung and Deutsche Forschungsgemeinschaft (DFG), the German National Academic Foundation (C.T.M.), a stipend from the German Academic Exchange Service (A.N.), a stipend from the Hannover Biomedical Research School and the Zentrum für Infektionsbiologie (P.G.), the International Research Training Group 1273 funded by the DFG (M.D), the DFG (SFB900), and the EuroTransBio initiative.
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: C.T.M., L.B., C.A.-S., F.P., and T.S. planned experiments; C.T.M., P.G., A.N., M.D., C.H., L.B., P.S., C.A.-S., and F.P. performed experiments and analyzed data; M.M. provided critical reagents and advice; C.T.M., P.G., and T.S. wrote the manuscript; and C.T.M. and T.S. conceived the project.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for C.T.M. is Laboratory of Molecular Immunology, The Rockefeller University, New York, New York.
Correspondence: Tim Sparwasser, Institute of Infection Immunology, TWINCORE/Centre for Experimental and Clinical Infection Research, Feodor-Lynen-Straße 7, 30625 Hannover, Germany; e-mail: tim.sparwasser@twincore.de.
References
- 1.Liu K, Nussenzweig MC. Origin and development of dendritic cells. Immunol Rev. 2010;234(1):45–54. doi: 10.1111/j.0105-2896.2009.00879.x. [DOI] [PubMed] [Google Scholar]
- 2.Mayer CT, Berod L, Sparwasser T. Layers of dendritic cell-mediated T cell tolerance, their regulation and the prevention of autoimmunity. Front Immunol. 2012;3:183. doi: 10.3389/fimmu.2012.00183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ginhoux F, Liu K, Helft J, et al. The origin and development of nonlymphoid tissue CD103+ DCs. J Exp Med. 2009;206(13):3115–3130. doi: 10.1084/jem.20091756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Contreras V, Urien C, Guiton R, et al. Existence of CD8α-like dendritic cells with a conserved functional specialization and a common molecular signature in distant mammalian species. J Immunol. 2010;185(6):3313–3325. doi: 10.4049/jimmunol.1000824. [DOI] [PubMed] [Google Scholar]
- 5.Crozat K, Guiton R, Contreras V, et al. The XC chemokine receptor 1 is a conserved selective marker of mammalian cells homologous to mouse CD8alpha+ dendritic cells. J Exp Med. 2010;207(6):1283–1292. doi: 10.1084/jem.20100223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poulin LF, Salio M, Griessinger E, et al. Characterization of human DNGR-1+ BDCA3+ leukocytes as putative equivalents of mouse CD8alpha+ dendritic cells. J Exp Med. 2010;207(6):1261–1271. doi: 10.1084/jem.20092618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Satpathy AT, Wu X, Albring JC, Murphy KM. Re(de)fining the dendritic cell lineage. Nat Immunol. 2012;13(12):1145–1154. doi: 10.1038/ni.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bedoui S, Whitney PG, Waithman J, et al. Cross-presentation of viral and self antigens by skin-derived CD103+ dendritic cells. Nat Immunol. 2009;10(5):488–495. doi: 10.1038/ni.1724. [DOI] [PubMed] [Google Scholar]
- 9.Henri S, Poulin LF, Tamoutounour S, et al. CD207+ CD103+ dermal dendritic cells cross-present keratinocyte-derived antigens irrespective of the presence of Langerhans cells. J Exp Med. 2010;207(1) doi: 10.1084/jem.20091964. 189-206, S181-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Merad M, Sathe P, Helft J, Miller J, Mortha A. The dendritic cell lineage: ontogeny and function of dendritic cells and their subsets in the steady state and the inflamed setting. Annu Rev Immunol. 2013;31:563–604. doi: 10.1146/annurev-immunol-020711-074950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lutz MB, Kukutsch N, Ogilvie AL, et al. An advanced culture method for generating large quantities of highly pure dendritic cells from mouse bone marrow. J Immunol Methods. 1999;223(1):77–92. doi: 10.1016/s0022-1759(98)00204-x. [DOI] [PubMed] [Google Scholar]
- 12.Naik SH, Proietto AI, Wilson NS, et al. Cutting edge: generation of splenic CD8+ and CD8- dendritic cell equivalents in Fms-like tyrosine kinase 3 ligand bone marrow cultures. J Immunol. 2005;174(11):6592–6597. doi: 10.4049/jimmunol.174.11.6592. [DOI] [PubMed] [Google Scholar]
- 13.Edelson BT, Bradstreet TR, Kc W, et al. Batf3-dependent CD11b(low/-) peripheral dendritic cells are GM-CSF-independent and are not required for Th cell priming after subcutaneous immunization. PLoS ONE. 2011;6(10):e25660. doi: 10.1371/journal.pone.0025660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sathe P, Pooley J, Vremec D, et al. The acquisition of antigen cross-presentation function by newly formed dendritic cells. J Immunol. 2011;186(9):5184–5192. doi: 10.4049/jimmunol.1002683. [DOI] [PubMed] [Google Scholar]
- 15.Jackson JT, Hu Y, Liu R, et al. Id2 expression delineates differential checkpoints in the genetic program of CD8α+ and CD103+ dendritic cell lineages. EMBO J. 2011;30(13):2690–2704. doi: 10.1038/emboj.2011.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hildner K, Edelson BT, Purtha WE, et al. Batf3 deficiency reveals a critical role for CD8alpha+ dendritic cells in cytotoxic T cell immunity. Science. 2008;322(5904):1097–1100. doi: 10.1126/science.1164206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.MartIn-Fontecha A, Sebastiani S, Höpken UE, et al. Regulation of dendritic cell migration to the draining lymph node: impact on T lymphocyte traffic and priming. J Exp Med. 2003;198(4):615–621. doi: 10.1084/jem.20030448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dawicki W, Jawdat DW, Xu N, Marshall JS. Mast cells, histamine, and IL-6 regulate the selective influx of dendritic cell subsets into an inflamed lymph node. J Immunol. 2010;184(4):2116–2123. doi: 10.4049/jimmunol.0803894. [DOI] [PubMed] [Google Scholar]
- 19.Shortman K, Naik SH. Steady-state and inflammatory dendritic-cell development. Nat Rev Immunol. 2007;7(1):19–30. doi: 10.1038/nri1996. [DOI] [PubMed] [Google Scholar]
- 20.Diao J, Winter E, Chen W, Cantin C, Cattral MS. Characterization of distinct conventional and plasmacytoid dendritic cell-committed precursors in murine bone marrow. J Immunol. 2004;173(3):1826–1833. doi: 10.4049/jimmunol.173.3.1826. [DOI] [PubMed] [Google Scholar]
- 21.Bogunovic M, Ginhoux F, Helft J, et al. Origin of the lamina propria dendritic cell network. Immunity. 2009;31(3):513–525. doi: 10.1016/j.immuni.2009.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Greter M, Helft J, Chow A, et al. GM-CSF controls nonlymphoid tissue dendritic cell homeostasis but is dispensable for the differentiation of inflammatory dendritic cells. Immunity. 2012;36(6):1031–1046. doi: 10.1016/j.immuni.2012.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jakubzick C, Tacke F, Ginhoux F, et al. Blood monocyte subsets differentially give rise to CD103+ and CD103- pulmonary dendritic cell populations. J Immunol. 2008;180(5):3019–3027. doi: 10.4049/jimmunol.180.5.3019. [DOI] [PubMed] [Google Scholar]
- 24.Landsman L, Varol C, Jung S. Distinct differentiation potential of blood monocyte subsets in the lung. J Immunol. 2007;178(4):2000–2007. doi: 10.4049/jimmunol.178.4.2000. [DOI] [PubMed] [Google Scholar]
- 25.Suzuki S, Honma K, Matsuyama T, et al. Critical roles of interferon regulatory factor 4 in CD11bhighCD8alpha- dendritic cell development. Proc Natl Acad Sci USA. 2004;101(24):8981–8986. doi: 10.1073/pnas.0402139101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Merad M, Manz MG. Dendritic cell homeostasis. Blood. 2009;113(15):3418–3427. doi: 10.1182/blood-2008-12-180646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jakubzick C, Bogunovic M, Bonito AJ, Kuan EL, Merad M, Randolph GJ. Lymph-migrating, tissue-derived dendritic cells are minor constituents within steady-state lymph nodes. J Exp Med. 2008;205(12):2839–2850. doi: 10.1084/jem.20081430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller JC, Brown BD, Shay T, et al. Immunological Genome Consortium. Deciphering the transcriptional network of the dendritic cell lineage. Nat Immunol. 2012;13(9):888–899. doi: 10.1038/ni.2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gautier EL, Shay T, Miller J, et al. Immunological Genome Consortium. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat Immunol. 2012;13(11):1118–1128. doi: 10.1038/ni.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jelinek I, Leonard JN, Price GE, et al. TLR3-specific double-stranded RNA oligonucleotide adjuvants induce dendritic cell cross-presentation, CTL responses, and antiviral protection. J Immunol. 2011;186(4):2422–2429. doi: 10.4049/jimmunol.1002845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hochrein H, Shortman K, Vremec D, Scott B, Hertzog P, O’Keeffe M. Differential production of IL-12, IFN-alpha, and IFN-gamma by mouse dendritic cell subsets. J Immunol. 2001;166(9):5448–5455. doi: 10.4049/jimmunol.166.9.5448. [DOI] [PubMed] [Google Scholar]
- 32.Naik SH. Generation of large numbers of pro-DCs and pre-DCs in vitro. Methods Mol Biol. 2010;595:177–186. doi: 10.1007/978-1-60761-421-0_11. [DOI] [PubMed] [Google Scholar]
- 33.Segura E, Wong J, Villadangos JA. Cutting edge: B220+CCR9- dendritic cells are not plasmacytoid dendritic cells but are precursors of conventional dendritic cells. J Immunol. 2009;183(3):1514–1517. doi: 10.4049/jimmunol.0901524. [DOI] [PubMed] [Google Scholar]
- 34.Naik SH, Metcalf D, van Nieuwenhuijze A, et al. Intrasplenic steady-state dendritic cell precursors that are distinct from monocytes. Nat Immunol. 2006;7(6):663–671. doi: 10.1038/ni1340. [DOI] [PubMed] [Google Scholar]
- 35.Poulin LF, Reyal Y, Uronen-Hansson H, et al. DNGR-1 is a specific and universal marker of mouse and human Batf3-dependent dendritic cells in lymphoid and nonlymphoid tissues. Blood. 2012;119(25):6052–6062. doi: 10.1182/blood-2012-01-406967. [DOI] [PubMed] [Google Scholar]
- 36.Schraml BU, van Blijswijk J, Zelenay S, et al. Genetic tracing via DNGR-1 expression history defines dendritic cells as a hematopoietic lineage. Cell. 2013;154(4):843–858. doi: 10.1016/j.cell.2013.07.014. [DOI] [PubMed] [Google Scholar]
- 37.Helft J, Manicassamy B, Guermonprez P, et al. Cross-presenting CD103+ dendritic cells are protected from influenza virus infection. J Clin Invest. 2012;122(11):4037–4047. doi: 10.1172/JCI60659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.del Rio ML, Rodriguez-Barbosa JI, Bölter J, et al. CX3CR1+ c-kit+ bone marrow cells give rise to CD103+ and CD103- dendritic cells with distinct functional properties. J Immunol. 2008;181(9):6178–6188. doi: 10.4049/jimmunol.181.9.6178. [DOI] [PubMed] [Google Scholar]
- 39.Zhan Y, Carrington EM, van Nieuwenhuijze A, et al. GM-CSF increases cross-presentation and CD103 expression by mouse CD8⁺ spleen dendritic cells. Eur J Immunol. 2011;41(9):2585–2595. doi: 10.1002/eji.201141540. [DOI] [PubMed] [Google Scholar]
- 40.Zou W. Regulatory T cells, tumour immunity and immunotherapy. Nat Rev Immunol. 2006;6(4):295–307. doi: 10.1038/nri1806. [DOI] [PubMed] [Google Scholar]
- 41.Palucka K, Banchereau J. Cancer immunotherapy via dendritic cells. Nat Rev Cancer. 2012;12(4):265–277. doi: 10.1038/nrc3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sparwasser T, Koch ES, Vabulas RM, et al. Bacterial DNA and immunostimulatory CpG oligonucleotides trigger maturation and activation of murine dendritic cells. Eur J Immunol. 1998;28(6):2045–2054. doi: 10.1002/(SICI)1521-4141(199806)28:06<2045::AID-IMMU2045>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 43.Coombes JL, Siddiqui KR, Arancibia-Cárcamo CV, et al. A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF-beta and retinoic acid-dependent mechanism. J Exp Med. 2007;204(8):1757–1764. doi: 10.1084/jem.20070590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamazaki S, Dudziak D, Heidkamp GF, et al. CD8+ CD205+ splenic dendritic cells are specialized to induce Foxp3+ regulatory T cells. J Immunol. 2008;181(10):6923–6933. doi: 10.4049/jimmunol.181.10.6923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ohl L, Mohaupt M, Czeloth N, et al. CCR7 governs skin dendritic cell migration under inflammatory and steady-state conditions. Immunity. 2004;21(2):279–288. doi: 10.1016/j.immuni.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 46.Kingston D, Schmid MA, Onai N, Obata-Onai A, Baumjohann D, Manz MG. The concerted action of GM-CSF and Flt3-ligand on in vivo dendritic cell homeostasis. Blood. 2009;114(4):835–843. doi: 10.1182/blood-2009-02-206318. [DOI] [PubMed] [Google Scholar]
- 47.Proietto AI, Mittag D, Roberts AW, Sprigg N, Wu L. The equivalents of human blood and spleen dendritic cell subtypes can be generated in vitro from human CD34(+) stem cells in the presence of fms-like tyrosine kinase 3 ligand and thrombopoietin. Cell Mol Immunol. 2012;9(6):446–454. doi: 10.1038/cmi.2012.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Semmrich M, Plantinga M, Svensson-Frej M, et al. Directed antigen targeting in vivo identifies a role for CD103+ dendritic cells in both tolerogenic and immunogenic T-cell responses. Mucosal Immunol. 2012;5(2):150–160. doi: 10.1038/mi.2011.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Suffia I, Reckling SK, Salay G, Belkaid Y. A role for CD103 in the retention of CD4+CD25+ Treg and control of Leishmania major infection. J Immunol. 2005;174(9):5444–5455. doi: 10.4049/jimmunol.174.9.5444. [DOI] [PubMed] [Google Scholar]
- 50.Annacker O, Coombes JL, Malmstrom V, et al. Essential role for CD103 in the T cell-mediated regulation of experimental colitis. J Exp Med. 2005;202(8):1051–1061. doi: 10.1084/jem.20040662. [DOI] [PMC free article] [PubMed] [Google Scholar]