Abstract
The microbial mechanisms and key metabolites that shape the composition of the human gut microbiota are largely unknown, impeding efforts to manipulate dysbiotic microbial communities towards stability and health. Vitamins, which by definition are not synthesized in sufficient quantities by the host and can mediate fundamental biological processes in microbes, represent an attractive target for reshaping microbial communities. Here, we discuss how vitamin B12 (cobalamin) impacts diverse host-microbe symbioses. Although cobalamin is synthesized by some human gut microbes, it is a precious resource in the gut and is likely not provisioned to the host in significant quantities. However, this vitamin may make an unrecognized contribution in shaping the structure and function of human gut microbial communities.
Introduction
The human gut harbors trillions of microbes, dominated by two broad phylogenetic groups (Bacteroidetes and Firmicutes) that collectively encode over one hundred fold more genes than the human genome (Lozupone et al., 2012; Qin et al., 2010). Furthermore, genes encoded by an individual’s gut microbial community also exhibit substantially greater interpersonal variation than their host. For example, individual humans have been estimated to be heterozygous for ~100 loss-of-function gene variants in their own genomes (MacArthur et al., 2012), whereas their microbiomes generally encode hundreds of thousands of genes that are not present in the majority of sampled individuals: over 2 million microbiome-encoded genes are observed in less than 20% of individuals (Qin et al., 2010). These metagenomic studies suggest that the microbiota may represent an important factor that contributes to disease predisposition and response to treatment. However, the underlying factors and key metabolites that shape microbial community variation are largely unknown, and as a result methods to rationally alter the composition of the gut microbiota for therapeutic purposes remain elusive.
Biologists have long appreciated the contributions of the gut microbiota to health. In fact, Pasteur proposed in 1885 that animals would not be viable without their associated microbes (Gordon and Pesti, 1971). Although the establishment of germfree mice, rats, and other mammals 60 years later disproved this bold hypothesis, gnotobiotic (known life) studies comparing germfree and conventional animals have revealed a tremendous and wide-ranging impact of resident microbes on host development, metabolism, immune responses, and behavior (Dantas et al., 2013; Diaz Heijtz et al., 2011; Faith et al., 2010; Goodman and Gordon, 2010; Smith et al., 2007). These experiments quickly indicated that one fundamental service of the gut microbiota is production of vitamins for the host. Indeed, early studies emphasized the requirement for increased dietary vitamin K, B1 (thiamin), B6 (pyridoxine), B7 (biotin), B9 (folic acid) and B12 (cobalamin) for the health of germfree animals (Ikeda et al., 1979; Sumi et al., 1977; Wostmann, 1981; Wostmann and Knight, 1965). Members of the genera Bifidobacterium and Lactobacillus may play important roles in this regard (Lozupone et al., 2012).
Because vitamins (by definition) are not produced in sufficient quantities by the host, are safely consumed by humans, and often mediate fundamental biological processes in most cells, including bacteria, these compounds represent attractive targets for manipulating gut microbial community composition. Here, we discuss important nuances in evaluating the potential role of one vitamin, cobalamin, in mediating both host-microbe and microbe-microbe interactions in the gut. This Perspective has two goals: first, we review evidence for gut microbial contribution to host cobalamin levels in diverse microbe-host symbioses and present evidence that in the case of humans, gut microbial communities are likely competitors for dietary cobalamin, rather than contributors of this essential vitamin to the host. Second, we propose a potential role for cobalamin and related cofactors (corrinoids) in shaping the composition of human gut microbial communities and their expressed functions.
Role of microbial cobalamin in non-human systems
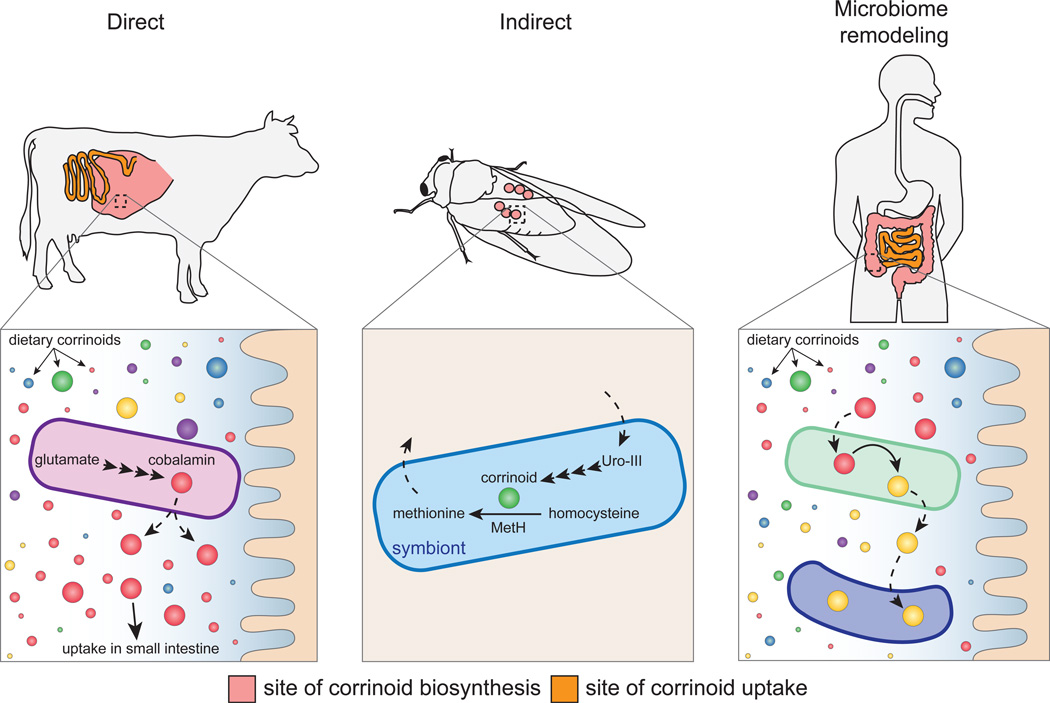
Cobalamin is an unusual vitamin in that it is not made by plants and instead is synthesized exclusively by bacteria and archaea (Roth et al., 1996). However, eukaryotes can have both direct and indirect requirements for cobalamin and other corrinoids, and can potentially be further influenced by corrinoid-dependent metabolism by their gut microbiota (Figure 2). As with both bacteria and archaea, only some eukaryotic lineages encode corrinoid-dependent enzymes (e.g., vertebrates, some protists and invertebrates, but not plants, insects or fungi) (Roth et al., 1996). Strikingly, like humans the majority of eukaryotes that require corrinoids have retained genes encoding corrinoid-dependent methylmalonyl CoA mutase (MCM, EC 5.4.99.2) and/or corrinoid-dependent methionine synthase (MetH, EC 2.1.1.13). These corrinoid-dependent genes are retained in these eukaryotes despite the existence of corrinoid-independent analogs or pathways that utilize different intermediates. MCM catalyzes the interconversion of R-methylmalonyl-CoA and succinyl-CoA, a step in the metabolism of several branched chain amino acids, fatty acids, cholesterol, and sugars to propionate (Banerjee and Chowdhury, 1999). In eukaryotes, this process primarily occurs in the mitochondria (Frenkel and Kitchens, 1975). MetH catalyzes the final step in the biosynthesis of methionine. This reaction involves the transfer of a methyl group from another cofactor, methyltetrahydrofolate, to the substrate, homocysteine (Matthews, 1999). In addition to MCM and MetH, some eukaryotes also encode a corrinoid-dependent ribonucleotide reductase (NrdJ/RNR II, EC 1.17.4.1) (Torrents et al., 2006). This enzyme is critical for DNA synthesis as it is responsible for the synthesis of deoxynucleotides (dNDP or dNTP) from ribonucleotides (NDP or NTP) (Roth et al., 1996). The essential nature of these pathways makes many eukaryotes dependent on microbial corrinoids.
Figure 2. Microbial corrinoid metabolism in the gut.
In coprophagic and ruminant animals, gut microbes provide a direct source of cobalamin (direct effect) (Girard et al., 2009). In insects, corrinoids are essential cofactors for obligate symbionts that provide key nutrients to the host (indirect effect) (McCutcheon et al., 2009). Competition and exchange of corrinoids likely shape gut microbial community composition and expressed functions in humans and other animals (microbiome remodeling) (Allen and Stabler, 2008; Goodman et al., 2011; Seth and Taga, 2014).
Although most plants neither produce nor require cobalamin, nearly half of all algae require this vitamin (Croft et al., 2005; Helliwell et al., 2011). These algae use cobalamin as a cofactor for MCM, MetH and/or RNR II (Helliwell et al., 2011). Many species that do not require cobalamin have been found to encode MetE, the cobalamin-independent isozyme of MetH, facilitating growth in cobalamin deplete conditions (Helliwell et al., 2011). Average free cobalamin levels in marine systems are too low to support cobalamin dependent algal growth in vitro; however, co-culture with cobalamin producing bacteria can rescue growth (Croft et al., 2005). Some of these bacteria colonize the extracellular mucus layers surrounding algal cells. This close, physical association likely promotes exchange of algal photosynthates and bacterial cobalamin, forming a mutualism (Croft et al., 2005). Notably, this requirement is not neatly divided across the algal phylogenetic tree, and is likely the result of multiple independent losses of MetE or MetH from an ancestor that encoded both enzymes (Helliwell et al., 2011). Differences in cobalamin availability and cobalamin independent metabolic pathways are presumed to have contributed to the heterogeneous occurrences of cobalamin-requiring enzymes in algae (Helliwell et al., 2011; Sañudo-Wilhelmy et al., 2012). Similar mutualisms have been predicted in marine sponges and their associated microbes (Siegl et al., 2011; Thomas et al., 2010). It is unclear if algae or sponges can use corrinoids other than cobalamin.
In nematodes, including the model Caenorhabditis elegans, cobalamin limitation results in reduced fertility and longevity and recapitulates symptoms of cobalamin deficiency in humans (Bito et al., 2013). C. elegans feeds on soil and plant associated bacteria that can colonize the gut (Felix and Duveau, 2012). As a result, the bacterial diet determines cobalamin status in C. elegans: worms fed with bacteria that produce cobalamin radically alter gene expression and show accelerated development and reduced mortality (MacNeil et al., 2013; Watson et al., 2014). In this system, increased MCM activity prevents the toxic buildup of propionic acid, reducing mortality. Further, MetH activity enhances the production of S-adenosylmethionine, which accelerates development, leading to an early cessation of egg laying and consequently, reduced fertility (Watson et al., 2014). Resident gut microbes that produce cobalamin are similarly important in several freshwater fish species by obviating the need for dietary cobalamin (Sugita et al., 1991). One of these cobalamin producing microbes, Cetobacterium somerae, is ubiquitous in wild and laboratory-reared zebrafish, suggesting a possible metabolic importance (Roeselers et al., 2011).
Eukaryotes that do not encode cobalamin-dependent enzymes can maintain an indirect requirement for cobalamin if the vitamin is essential for microbial symbionts they carry. For example, neither plants nor insects directly require cobalamin (Dadd, 1973; Roth et al., 1996) but in each of these lineages symbioses with bacteria have evolved that rely on this essential cofactor. Several legume species including Medicago sativa (alfalfa) form intracellular associations with rhizobial bacteria such as Sinorhizobium meliloti. S. meliloti resides within specialized root structures called nodules, where it fixes atmospheric nitrogen and significantly contributes to plant growth under nitrogen limiting conditions (Jones et al., 2007). Successful colonization of these nodules by S. meliloti is dependent on cobalamin for ribonucleotide reductase (NrdJ) and methionine synthase (MetH) activity (Taga and Walker 2010; Campbell et al. 2006). Consequently, cobalamin deficiency severely inhibits plant growth under nitrogen limiting conditions due to the lack of nitrogen fixation by its bacterial partner (Campbell et al., 2006; Taga and Walker, 2010).
Numerous insects have forged intracellular, nutritional symbioses with bacteria (Moran et al., 2008). Cicadas, for example, feed on xylem sap, an incredibly poor nutritional source. To obtain the essential vitamins and amino acids (including methionine) missing from this diet, these insects maintain a co-symbiosis with two bacterial species, Sulcia muelleri (Bacteroidetes) and Hodgkinia cicadicola (Alphaproteobacteria) (McCutcheon et al., 2009). H. cicadicola synthesizes methionine via its cobalamin-dependent MetH enzyme; as a result, it maintains 13 genes for corrinoid biosynthesis in its diminutive 144-kbp genome (McCutcheon et al., 2009). Without these enzymes, all three members of this tripartite symbiosis would likely die out.
Gut microbes are unlikely to provide significant sources of cobalamin to humans
In humans, cobalamin limitation can result from both uptake disorders (including pernicious anemia, an autoimmune loss of intrinsic factor responsible for uptake of cobalamin that is fatal if untreated) as well as dietary deficiencies (Stabler and Allen, 2004). The physiologic and neurologic consequences of cobalamin limitation and cobalamin trafficking disorders have been discussed in numerous excellent reviews (Alpers and Russell-Jones, 2013; Gherasim et al., 2013; Rosenblatt and Fenton, 1999).
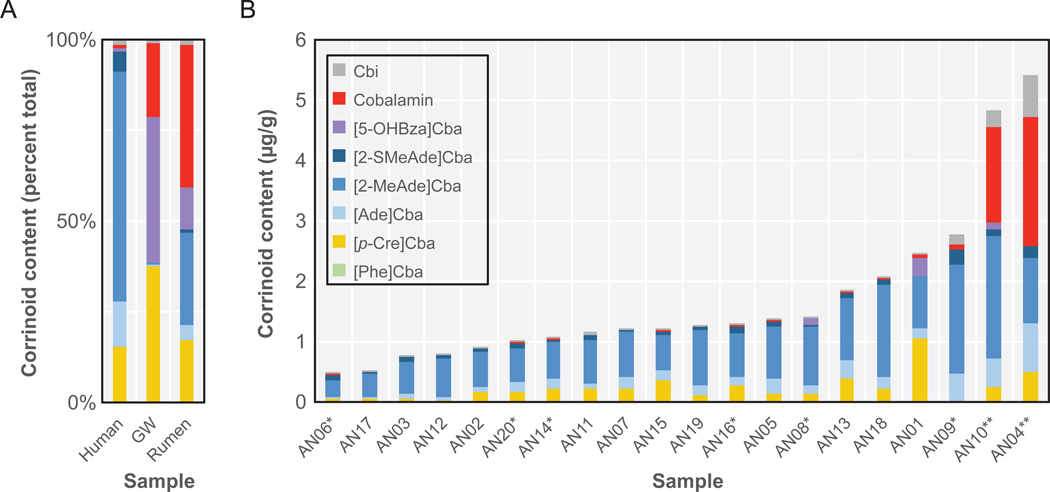
Although corrinoids are abundant in the large intestine due to the activity of the gut microbiota, multiple factors prevent humans from acquiring significant levels of cobalamin from this source. First, cobalamin produced by gut microbes represents less than 2% of the total corrinoid content in feces (Allen and Stabler, 2008). Further, cobalamin produced in the colon, where microbial numbers are highest, is not bioavailable because the receptors necessary for absorbing the vitamin are found in the small intestine, upstream of the site of corrinoid production (Seetharam and Alpers, 1982). A notable exception is found in mammals that practice coprophagy: these animals receive the benefit of microbial cobalamin production in the large intestine by consuming their feces which localizes microbial cobalamin (and other vitamins) to the upper part of the digestive tract where it can be absorbed (Mickelsen, 1956). Furthermore, ruminants are able to absorb cobalamin produced by microbes that reside in the rumen, a microbe-rich digestive organ located upstream of the small intestine (Girard et al., 2009). Perhaps coprophagic and ruminant animals might thus select for gut microbes that produce cobalamin rather than other corrinoids, while other species would not benefit from such enrichment. Indeed, the cobalamin content of bovine rumen (38% of total corrinoids) is substantially higher than that of human feces (Figure 3A) (Girard et al., 2009).
Figure 3. Corrinoid composition of microbial communities measured by LCMS.
A. The average corrinoid composition of fecal samples from 18 humans in the absence of cobalamin supplementation (Human); a groundwater enrichment community amended with trichloroethene (GW); and bovine rumen (Rumen) is shown (adapted with permission from (Allen and Stabler, 2008; Girard et al., 2009; Men et al., 2014b)). B. Corrinoid composition of fecal samples from 20 individuals (adapted with permission from (Allen and Stabler, 2008)). Asterisks indicate individuals supplemented daily with ≤25 μg cobalamin. Double asterisks indicate individuals supplemented daily with ≤1,000 μg cobalamin. Corrinoid abbreviations are defined in Box 1.
In humans, diet is instead more likely to constitute the primary source of cobalamin. Given that cobalamin is not found in plants, animal products are the main source of cobalamin for most individuals. It is for this reason that vegetarians, and especially vegans, are more commonly found to have low serum levels of cobalamin (Antony, 2003). The edible macroalga Porphyra yezoensis (commonly known as purple laver or nori) has been found to contain corrinoid levels sufficient to reverse the effects of cobalamin deficiency in rats (Takenaka et al., 2001). Cobalamin has also been reported to be present at lower levels in other non-animal foods including other edible algae, some mushrooms, and fermented foods such as tempe, kimchi, and tea (Watanabe et al., 2014).
Several other dietary items have previously been reported to contain cobalamin, but more thorough chemical analyses later revealed that other corrinoids were present. Much of the confusion about the availability of cobalamin in foods and probiotics is derived from ambiguity in the methods used to measure cobalamin. The classical method of measuring cobalamin levels is a microbiological bioassay in which the growth of a corrinoid-dependent organism such as Lactobacillus spp. or Euglena gracilis is used as a proxy (Raven et al., 1972). Bioassays are still commonly used today because of their sensitivity and convenience. However, it is clear that these assays cannot distinguish cobalamin from other corrinoids because several different corrinoids can be used as cofactors for the bioassay strains. Assays with higher specificity for cobalamin have also been developed that use purified intrinsic factor, the mammalian protein with the highest selectivity for cobalamin (Lau et al., 1965). However, to our knowledge, not all of the known naturally occurring corrinoids have been tested with this assay. The recent development of liquid chromatography-mass spectrometry methods for corrinoid measurements has enabled the identification of specific corrinoids in biological samples (Allen and Stabler, 2008; Men et al., 2014b). The use of such methods has revealed that food-associated bacteria previously thought to provide cobalamin, such as the probiotic bacterium Lactobacillus reuteri and the edible cyanobacterium Arthrospira (formerly Spirulina) platensis, actually produce only pseudo-cobalamin ([Ade]Cba), which cannot be used by humans (Santos et al., 2007; Tanioka et al., 2010).
Indeed, human gut microbes are likely to present direct competition with their host for cobalamin. The great majority of human gut microbial species are predicted to require exogenous corrinoids (Degnan et al., 2014). Notably, individuals with high bacterial loads in their small intestines tend to have low cobalamin status (Albert et al., 1980; Brandt et al., 1977; Murphy et al., 1986), possibly due to either competition for cobalamin between the host and microbes, or an overabundance of corrinoids other than cobalamin. This observation is particularly notable given that several bacteria isolated from the small intestine are capable of synthesizing corrinoids in vitro (Albert et al., 1980). Further, bacterial cells and purified intrinsic factor bind cobalamin with similar affinities (Giannella et al., 1971, 1972). Together these observations suggest that gut microbes likely compete for dietary cobalamin in the small intestine. Whether small intestinal microbial community configurations enriched in corrinoid scavengers contribute to cobalamin deficiencies in predisposed individuals (due to diet or genetic factors) is unknown.
A potential role for corrinoids in gut microbial ecology
There is great interest in developing ways of manipulating the microbiota to promote health. There are presently no effective methods of controlling the composition of microbes in the human gut, with the exception of antibiotic therapy, which targets broad taxonomic groups, or fecal transplantation, in which the entire microbiota is replaced with that of another individual (Dethlefsen et al., 2008; Seekatz et al., 2014). Treatment with probiotics, live bacteria ingested for the purpose of altering the composition or metabolism in the gut, can improve symptoms of gastrointestinal disease in some cases, though these probiotic organisms may not become integrated as members of the microbiota (Brigidi et al., 2000; McNulty et al., 2011).
Based on recent studies of the role of corrinoids in the human gut, we propose that targeted manipulations of the gut microbiota could be achieved by altering the levels of specific corrinoids. This hypothesis is based on the following observations. First, over 80% of sequenced human gut bacteria are predicted to use corrinoids, but less than 25% have the genetic capacity to synthesize these molecules (Degnan et al., 2014); Second, human fecal samples have been found to contain up to eight distinct corrinoids (Allen and Stabler, 2008). Third, only a fraction of the corrinoids available in a microbial community can be used by a given organism because corrinoid-dependent enzymes have native specificity for their preferred cofactors (Keller et al., 2013; Mok and Taga, 2013; Yi et al., 2012). Fourth, corrinoids other than cobalamin are poorly recognized by human intrinsic factor, the key protein required for transport of cobalamin from the lumen of the small intestine. Finally, corrinoids are synthesized exclusively by bacteria and archaea, unlike many other vitamins that are also prevalent in plant-based dietary components (Roth et al., 1996). Thus, because different bacteria require distinct groups of corrinoids, we hypothesize that microbial communities can be manipulated by altering the levels of particular corrinoids.
An analysis of the corrinoid composition of fecal samples of 20 individuals by Allen and Stabler revealed differences in both the total corrinoid levels and the proportion of each corrinoid (Figure 3B). It still remains to be tested whether these differences are correlated with distinct microbial signatures in these communities. For example, a corrinoid found almost exclusively in methanogens, 5-hydroxybenzimidazolylcobamide ([5-OHBza]Cba) (Pol et al., 1982; Ryzhkova and Brukhanov, 2009), was detected in only 30% of the samples analyzed. It would be interesting to test whether the presence or amount of this corrinoid correlates with the abundance or metabolic activity of methanogenic archaea, which are also found in approximately 30% of individuals (Hansen et al., 2011). Similarly, the levels of p-cresolylcobamide ([p-Cre]Cba), a corrinoid only known to be synthesized by Veillonellaceae, could be reflective of the abundance of this taxon (Chan and Escalante-Semerena, 2011; Men et al., 2014a; Men et al., 2014b). Despite the observed differences among these individuals, a much greater difference is seen between the human fecal samples and those from bovine rumen and a trichloroethene (TCE)-containing groundwater enrichment, suggesting that the human gut environment selects for microbes that produce a particular set of corrinoids (Figure 3A; (Girard et al., 2009; Men et al., 2014b)). However, it remains to be determined whether a broader diversity of corrinoid profiles can be found in individuals with greater differences in diet, lifestyle, or geographic location.
Allen and tabler’s landmark study of corrinoids in human fecal samples also revealed the effect of high doses of cobalamin on the corrinoid composition of the gut (Allen and Stabler, 2008). Individuals who ingested 1–2 mg cobalamin per day (nearly 1000-fold higher than the recommended daily allowance in the U.S.A.) had higher levels of this vitamin in the feces, as would be expected if some of the supplement was not absorbed in the small intestine, but also had high levels of nearly all other corrinoids (Figure 3B). Furthermore, when examined over time in a single individual, the increase in corrinoid levels was found to be transient, as each corrinoid returned to the original level after only ten days. The increase in total corrinoid levels following cobalamin supplementation is indicative of corrinoid remodeling, in which the lower ligand of a corrinoid is removed and another is attached (Gray and Escalante-Semerena, 2009). Corrinoid remodeling has been observed in archaea and bacteria grown in pure culture and is thought to be a mechanism of obtaining specific corrinoids needed for metabolism (Escalante-Semerena, 2007; Gray and Escalante-Semerena, 2009; Yi et al., 2012). The observation that corrinoid remodeling allows the gut microbiota to maintain a stable corrinoid composition points to the robustness of this community and an important challenge in modulating corrinoid levels by supplementation with a biologically active corrinoid. Alternative approaches could be to provide synthetic corrinoid analogs that are not recognized by the remodeling machinery, or lower ligand bases to be incorporated to form desired corrinoids (known as guided biosynthesis). It would be interesting to examine the microbial composition and gene expression profiles following cobalamin supplementation as a means to understand the short-term and long-term effects of perturbations of corrinoid metabolism.
Corrinoids play a role in the physiology of most human gut microbes
Corrinoids are best known for their roles as cofactors for enzymes that perform radical rearrangements or methyl transfer reactions. MCM and MetH, the only two cobalamin-dependent enzymes present in humans, are also among the most abundant corrinoid-dependent enzymes in the human gut microbiome (Degnan et al., 2014). The conversion of succinyl-CoA to methylmalonyl-CoA by MCM and the subsequent decarboxylation to propionyl CoA generates CO2, which has been hypothesized to facilitate respiration in anaerobic environments such as the gut that contain low levels of alternate electron acceptors (Fischbach and Sonnenburg, 2011; Macy et al., 1978). During this process, Na+ is pumped out of the cell, establishing a gradient that can be exploited for ATP synthesis (Dimroth, 1987). One third of the bacterial species that contain a MetH homolog apparently do not depend on corrinoids for MetH activity under at least some conditions because they also contain a homolog of MetE (Degnan et al., 2014). However, numerous additional genes encoding a variety of other corrinoid-dependent enzymes not found in human cells are also present in the human gut microbiome and contribute to specific metabolic processes (Table 1). For example, the corrinoid-dependent enzyme ethanolamine ammonia lyase, which is present in 17% of sequenced human gut commensals (Degnan et al., 2014), allows bacteria to use ethanolamine, a breakdown product of the membrane phospholipid phosphatidylethanolamine that is found in the gut, as a carbon and nitrogen source (Garsin, 2010). As described below, the large number of other genes in the gut whose expression is potentially regulated by corrinoids suggests that additional corrinoid-dependent processes remain to be discovered.
Table 1. Corrinoid dependent enzymes in humans and their gut microbiota.
| Ortholog | Humans | Gut microbes | Percent of gut microbes |
Protein description | EC No. |
|---|---|---|---|---|---|
| MetH | ✓ | ✓ | 67% | B12-dependent methionine synthase | 2.1.1.13 |
| MCM / ScpA / MutAB | ✓ | ✓ | 25% | Methylmalonyl-CoA mutase | 5.4.99.2 |
| RNR II / NrdJ | – | ✓ | 35% | Vitamin B12-dependent ribonucleotide reductase | 1.17.4.1 |
| GlmES | – | ✓ | 8% | Glutamate mutase, methylaspartate mutase | 5.4.99.1 |
| Mgm | – | ✓ | 1% | 2-methyleneglutarate mutase | 5.4.99.4 |
| IcmAB / IcmF / Hcm | – | ✓ | 4% | Isobutyryl-CoA mutase / 2-hydroxyisobutyryl-CoA mutase | 5.4.99.13 / 5.4.99.- |
| EutBC | – | ✓ | 17% | Ethanolamine ammonia lyase | 4.1.3.7 |
| PduCDE | – | ✓ | 11% | Propanediol dehydratase / Glycerol dehydratase | 4.2.1.28 / 4.2.1.30 |
| KamED | – | ✓ | 8% | Beta-lysine 5,6-aminomutase | 5.4.3.3 |
| OraSE | – | ✓ | 2% | D-ornithine 4,5-aminomutase | 5.4.3.5 |
| QueG | – | ✓ | 33% | Epoxyqueuosine reductase | no EC |
| PceA | – | ✓ | 1% | Tetrachloroethene reductive dehalogenase | 1.97.1.8 |
| AcsECD | – | ✓ | 4% | CO dehydrogenase corrinoid/iron-sulfur protein complex | no EC |
| Mta / Mtt | – | ✓ | 30% | Methanol/methylamine/trimethylamine methyltransferases | 2.1.1.247 |
| HpnR/Hyp. | – | ✓ | 43% | B12-binding domain/radical SAM domain proteins | no EC |
Corrinoid-dependent gene regulation in the gut microbiome
In addition to their roles as enzyme cofactors, corrinoids also act as regulators of gene expression in diverse bacteria including human gut species. Riboswitches are regulatory RNA elements commonly found in the 5’ untranslated region (UTR) of gene transcripts that modulate gene expression by binding specific small molecule metabolites such as vitamins or amino acids (Tucker and Breaker, 2005). The change in RNA secondary structure caused by binding the small molecule ligand affects transcription or translation of the attached mRNA (Nahvi et al., 2002; Waters and Storz, 2009). Corrinoid riboswitches (classified as cobalamin or adenosylcobalamin riboswitches) have been found to act by ligand induced transcriptional or translational attenuation in select model bacteria (Vitreschak et al., 2003). In these organisms, most of the genes regulated by riboswitches are involved in corrinoid biosynthesis or transport. Gut microbes that are dependent on corrinoids frequently encode multiple corrinoid riboswitches, with nearly 41% of species recently analyzed possessing ≥3 (106/260) and some with as many as 17 (Degnan et al., 2014). Apart from corrinoid biosynthesis and transport genes, these mRNA regulators in the human microbiome are found upstream of a variety of corrinoid-dependent genes and their corrinoid-independent isozyme counterparts such as metE and corrinoid-independent ribonucleotide reductases (Rodionov et al., 2003). Strikingly, known corrinoid biosynthesis, corrinoid transport, corrinoid-dependent enzymes and their corrinoid-independent isozymes constitute only half of the nearly 4,000 genes putatively regulated by corrinoid riboswitches in sequenced human gut microbial species (Degnan et al., 2014). As such, many additional genes are potentially subject to expression modulation in response to corrinoid availability. For example, more than half of the sequenced Bacteroidetes genomes encode a corrinoid riboswitch-regulated multisubunit carboxylase gene cluster (EC 6.4.1.-). Although the substrate is uncertain (possibly propionate (Rodionov et al., 2003)) the widespread regulation of this locus in response to corrinoids may be important for modulating flux through MCM. In addition, many putative transporters of unknown substrates are found downstream of corrinoid riboswitches and warrant further investigation (Degnan et al., 2014).
Corrinoid-dependent regulation in the gut microbiome may also extend beyond riboswitch-regulated genes. Nearly 100 corrinoid riboswitch-regulated ORFs identified in our recent survey of human gut microbial genomes encode predicted transcriptional regulators and DNA-binding proteins which themselves likely influence the expression of numerous downstream genes (Degnan et al., 2014). Indeed, genome-wide transcriptional profiling studies of corrinoid response in the organohalide respiring bacterium Dehalococcoides mccartyi (formerly D. ethenogenes) reveal that many corrinoid-responsive genes in this organism do not possess corrinoid riboswitches (Johnson et al., 2009). Multiple bacterial species also use cobalamin to directly control the activity of transcriptional regulators (Klug, 2014). In the best-studied examples, these regulators leverage the photo-labile nature of the upper ligand of the molecule to respond to light (Cheng et al., 2014; Ortiz-Guerrero et al., 2011). It is not known whether similar processes exist in microbes that transition between host-associated and external habitats, or whether these or other transcription factors respond to the extensive corrinoid structural diversity present in polymicrobial environments.
Selective acquisition of corrinoids
Unlike other essential small molecules and metals, a single type of transport mechanism for cobalamin (and other corrinoids) predominates among bacteria: the ABC-type transporter BtuFCD in conjunction with the outer membrane receptor BtuB in Gram-negative bacteria (but see (Gopinath et al., 2013)). Although microbes and their corrinoid-dependent enzymes have been shown to require corrinoids with specific lower ligands (Mok and Taga, 2013; Reinhold et al., 2012; Yi et al., 2012), the few BtuBFCD systems that have been characterized are capable of transporting a variety of corrinoids and their precursors (Bradbeer et al., 1978; Degnan et al., 2014). This promiscuity likely allows for effective corrinoid salvaging and remodeling. However, for microbes without the necessary enzymes to carry out these processes, developing transporter specificity for compatible corrinoids could be advantageous. We recently showed that unlike Proteobacteria (e.g., Escherichia coli), gut Bacteroidetes (which constitute 20–80% of the human gut microbiome) often encode multiple copies of BtuB (Degnan et al., 2014). In Bacteroides thetaiotaomicron, the three copies exhibit a corrinoid-specific functional hierarchy. One copy, BtuB2, readily transports cobalamin and 6 additional corrinoids tested. Strains lacking this single transporter (with the two additional transporters BtuB1 and BtuB3 intact) are unable to compete with wild type bacteria in gnotobiotic mice with cobalamin as the only available corrinoid (Degnan et al., 2014; Goodman et al., 2009). These experiments, and in vitro studies, suggest that BtuB1 and BtuB3 have a lower affinity or specificity for cobalamin compared to BtuB2. Further, the remaining transporters BtuB1 and BtuB3 demonstrate preferences in vitro and in vivo for distinct corrinoids common in the gut. Strains encoding only BtuB1 outcompete strains encoding BtuB3 when [Ade]Cba and [Bza]Cba are provided but are outcompeted in the presence of cobalamin, [2-MeAde]Cba, [5-OMeBza]Cba and [5-MeBza]Cba. Our computational analyses suggest that these three B. thetaiotaomicron transporters represent only 3% of known BtuB sequence and functional diversity in human gut bacteria. As such, their relative transport preferences represent a small portion of the complexity of corrinoid recognition in the human gut microbiome.
Conclusion
Human-associated commensal microbes engage in elegant and complex chemical communication and metabolic interactions (Dorrestein et al., 2014). Manipulation of these pathways provides an attractive strategy for maximizing the contribution of the microbiota to health, but which metabolites play fundamental roles in shaping these communities remains unknown. This challenge is further compounded by the observation that the enormous diversity of microbial metabolites remains largely unexplored (Cimermancic et al., 2014). A surge in genome sequencing efforts, coupled with new approaches for surveying, clustering, and annotating chemical metabolites, make this an exciting era in small molecule discovery (Bouslimani et al., 2014; Charlop-Powers et al., 2014). However, known small molecules with well-established, fundamental roles in bacterial metabolism and gene regulation, including vitamins, should not be overlooked in this process and may represent promising opportunities to translate decades of studies of microbial metabolism into effective methods to manipulate the microbiota.
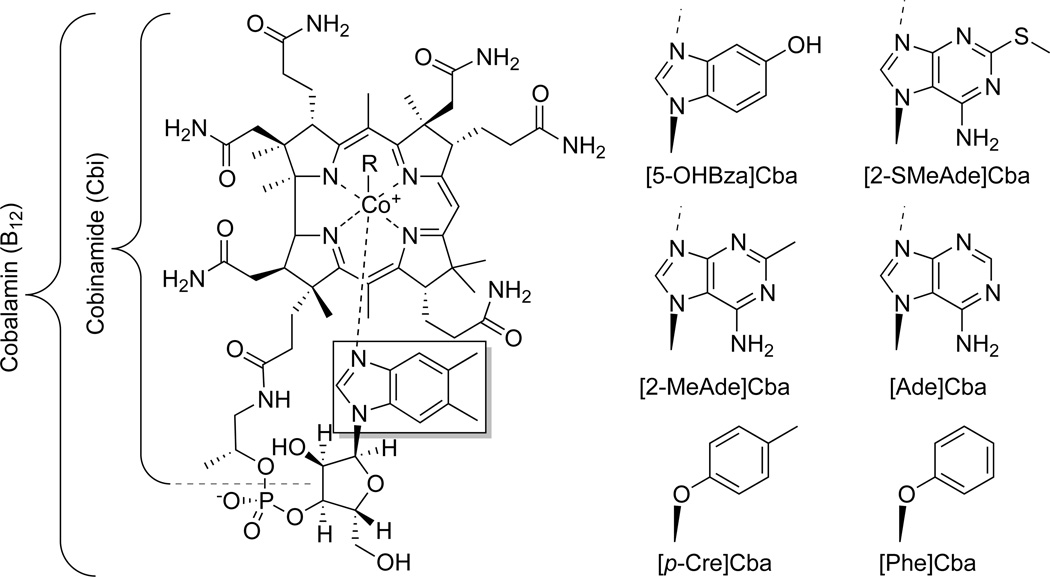
Figure 1. Representative examples of corrinoid structural diversity.
Box 1: Corrinoid diversity in the human gut.
Over a dozen corrinoids have been described that differ in the structure of the lower ligand, a portion of the molecule that plays an indirect role in the reactions catalyzed by corrinoid-dependent enzymes (Figure 1). Of these, eight have been found in the human gut in varying proportions (Allen and Stabler, 2008). Although all corrinoids are formally capable of performing both radical-mediated reactions and methyl group transfers via chemistry in the upper ligand (labeled as R in Figure 1) (Kräutler, 2005), humans and many bacteria are restricted in the corrinoids they can use (Keller et al., 2013; Mok and Taga, 2013; Rosenblatt and Fenton, 1999; Yi et al., 2012). Elaborate mechanisms exist that allow cobalamin entry into human cells while blocking the absorption of other corrinoids, thereby shielding corrinoid-dependent enzymes from exposure to corrinoids that cannot be used as cofactors (Nielsen et al., 2012). Likewise, bacteria that rely on corrinoid import rather than de novo biosynthesis must have mechanisms of acquiring the specific corrinoids that function in their metabolism or the ability to remodel other corrinoids (Gray and Escalante-Semerena, 2009; Men et al., 2014a; Seth and Taga, 2014; Yi et al., 2012). Notably, gut microbes possibly remodel 80% of dietary cobalamin based on supplementation studies (Allen and Stabler, 2008; Brandt et al., 1977). Mechanisms for corrinoid export have not been identified in bacteria.
Acknowledgements
We thank members of the Degnan, Taga, and Goodman labs for helpful discussion. We appreciate support from NIH grants DK089121 and GM103574 (to A.L.G.) and NSF grant MCB1122046 and the Hellman Family Faculty Fund (to M.E.T.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Albert MJ, Mathan VI, Baker SJ. Vitamin B12 synthesis by human small intestinal bacteria. Nature. 1980;283:781–782. doi: 10.1038/283781a0. [DOI] [PubMed] [Google Scholar]
- Allen RH, Stabler SP. Identification and quantitation of cobalamin and cobalamin analogues in human feces. Am J Clin Nutr. 2008;87:1324–1335. doi: 10.1093/ajcn/87.5.1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alpers DH, Russell-Jones G. Gastric intrinsic factor: the gastric and small intestinal stages of cobalamin absorption. a personal journey. Biochimie. 2013;95:989–994. doi: 10.1016/j.biochi.2012.12.006. [DOI] [PubMed] [Google Scholar]
- Antony AC. Vegetarianism and vitamin B-12 (cobalamin) deficiency. Am J Clin Nutr. 2003;78:3–6. doi: 10.1093/ajcn/78.1.3. [DOI] [PubMed] [Google Scholar]
- Banerjee R, Chowdhury S. Methylmalonyl-CoA mutase. In: Banerjee R, editor. Chemistry and Biochemistry of B12. New York: John Wiley & Sons, Inc.; 1999. pp. 707–729. [Google Scholar]
- Bito T, Matsunaga Y, Yabuta Y, Kawano T, Watanabe F. Vitamin B12 deficiency in Caenorhabditis elegans results in loss of fertility, extended life cycle, and reduced lifespan. FEBS Open Bio. 2013;3:112–117. doi: 10.1016/j.fob.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouslimani A, Sanchez LM, Garg N, Dorrestein PC. Mass spectrometry of natural products: current, emerging and future technologies. Natural product reports. 2014;31:718–729. doi: 10.1039/c4np00044g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbeer C, Kenley JS, Di Masi DR, Leighton M. Transport of vitamin B12 in Escherichia coli. Corrinoid specificities of the periplasmic B12-binding protein and of energy-dependent B12 transport. J Biol Chem. 1978;253:1347–1352. [PubMed] [Google Scholar]
- Brandt LJ, Bernstein LH, Wagle A. Production of vitamin B 12 analogues in patients with small-bowel bacterial overgrowth. Ann Intern Med. 1977;87:546–551. doi: 10.7326/0003-4819-87-5-546. [DOI] [PubMed] [Google Scholar]
- Brigidi P, Vitali B, Swennen E, Altomare L, Rossi M, Matteuzzi D. Specific detection of bifidobacterium strains in a pharmaceutical probiotic product and in human feces by polymerase chain reaction. Syst Appl Microbiol. 2000;23:391–399. doi: 10.1016/S0723-2020(00)80070-3. [DOI] [PubMed] [Google Scholar]
- Campbell GR, Taga ME, Mistry K, Lloret J, Anderson PJ, Roth JR, Walker GC. Sinorhizobium meliloti bluB is necessary for production of 5,6-dimethylbenzimidazole, the lower ligand of B12. Proc Natl Acad Sci USA. 2006;103:4634–4639. doi: 10.1073/pnas.0509384103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CH, Escalante-Semerena JC. ArsAB, a novel enzyme from Sporomusa ovata activates phenolic bases for adenosylcobamide biosynthesis. Mol Microbiol. 2011;81:952–967. doi: 10.1111/j.1365-2958.2011.07741.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlop-Powers Z, Milshteyn A, Brady SF. Metagenomic small molecule discovery methods. Curr Opin Microbiol. 2014;19C:70–75. doi: 10.1016/j.mib.2014.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z, Li K, Hammad LA, Karty JA, Bauer CE. Vitamin B12 regulates photosystem gene expression via the CrtJ antirepressor AerR in Rhodobacter capsulatus. Mol Microbiol. 2014;91:649–664. doi: 10.1111/mmi.12491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimermancic P, Medema MH, Claesen J, Kurita K, Wieland Brown LC, Mavrommatis K, Pati A, Godfrey PA, Koehrsen M, Clardy J, et al. Insights into secondary metabolism from a global analysis of prokaryotic biosynthetic gene clusters. Cell. 2014;158:412–421. doi: 10.1016/j.cell.2014.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft MT, Lawrence AD, Raux-Deery E, Warren MJ, Smith AG. Algae acquire vitamin B12 through a symbiotic relationship with bacteria. Nature. 2005;438:90–93. doi: 10.1038/nature04056. [DOI] [PubMed] [Google Scholar]
- Dadd RH. Insect nutrition: current developments and metabolic implications. Annual Review of Entomology. 1973;18:381–420. doi: 10.1146/annurev.en.18.010173.002121. [DOI] [PubMed] [Google Scholar]
- Dantas G, Sommer MO, Degnan PH, Goodman AL. Experimental approaches for defining functional roles of microbes in the human gut. Annu Rev Microbiol. 2013;67:459–475. doi: 10.1146/annurev-micro-092412-155642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degnan PH, Barry NA, Mok KC, Taga ME, Goodman AL. Human gut microbes use multiple transporters to distinguish vitamin B12 analogs and compete in the gut. Cell Host Microbe. 2014;15:47–57. doi: 10.1016/j.chom.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz Heijtz R, Wang S, Anuar F, Qian Y, Bjorkholm B, Samuelsson A, Hibberd ML, Forssberg H, Pettersson S. Normal gut microbiota modulates brain development and behavior. Proc Natl Acad Sci U S A. 2011;108:3047–3052. doi: 10.1073/pnas.1010529108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimroth P. Sodium ion transport decarboxylases and other aspects of sodium ion cycling in bacteria. Microbiol. Rev. 1987;51:320–340. doi: 10.1128/mr.51.3.320-340.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorrestein PC, Mazmanian SK, Knight R. Finding the Missing Links among Metabolites, Microbes, and the Host. Immunity. 2014;40:824–832. doi: 10.1016/j.immuni.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escalante-Semerena JC. Conversion of cobinamide into adenosylcobamide in bacteria and archaea. J Bacteriol. 2007;189:4555–4560. doi: 10.1128/JB.00503-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faith JJ, Rey FE, O'Donnell D, Karlsson M, McNulty NP, Kallstrom G, Goodman AL, Gordon JI. Creating and characterizing communities of human gut microbes in gnotobiotic mice. Isme J. 2010;4:1094–1098. doi: 10.1038/ismej.2010.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix MA, Duveau F. Population dynamics and habitat sharing of natural populations of Caenorhabditis elegans and C. briggsae. BMC biology. 2012;10:59. doi: 10.1186/1741-7007-10-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischbach MA, Sonnenburg JL. Eating for two: how metabolism establishes interspecies interactions in the gut. Cell Host Microbe. 2011;10:336–347. doi: 10.1016/j.chom.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenkel EP, Kitchens RL. Intracellular localization of hepatic propionyl-CoA carboxylase and methylmalonyl-CoA mutase in humans and normal and vitamin B12 deficient rats. Br J Haematol. 1975;31:501–513. doi: 10.1111/j.1365-2141.1975.tb00885.x. [DOI] [PubMed] [Google Scholar]
- Garsin DA. Ethanolamine utilization in bacterial pathogens: roles and regulation. Nat Rev Microbiol. 2010;8:290–295. doi: 10.1038/nrmicro2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gherasim C, Lofgren M, Banerjee R. Navigating the B(12) road: assimilation, delivery, and disorders of cobalamin. J Biol Chem. 2013;288:13186–13193. doi: 10.1074/jbc.R113.458810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannella RA, Broitman SA, Zamcheck N. Vitamin B12 uptake by intestinal microorganisms: mechanism and relevance to syndromes of intestinal bacterial overgrowth. J Clin Invest. 1971;50:1100–1107. doi: 10.1172/JCI106581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannella RA, Broitman SA, Zamcheck N. Competition between bacteria and intrinsic factor for vitamin B 12 : implications for vitamin B 12 malabsorption in intestinal bacterial overgrowth. Gastroenterology. 1972;62:255–260. [PubMed] [Google Scholar]
- Girard CL, Santschi DE, Stabler SP, Allen RH. Apparent ruminal synthesis and intestinal disappearance of vitamin B12 and its analogs in dairy cows. J Dairy Sci. 2009;92:4524–4529. doi: 10.3168/jds.2009-2049. [DOI] [PubMed] [Google Scholar]
- Goodman AL, Gordon JI. Our unindicted coconspirators: human metabolism from a microbial perspective. Cell Metab. 2010;12:111–116. doi: 10.1016/j.cmet.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman AL, McNulty NP, Zhao Y, Leip D, Mitra RD, Lozupone CA, Knight R, Gordon JI. Identifying genetic determinants needed to establish a human gut symbiont in its habitat. Cell Host Microbe. 2009;6:279–289. doi: 10.1016/j.chom.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman AL, Wu M, Gordon JI. Identifying microbial fitness determinants by insertion sequencing using genome-wide transposon mutant libraries. Nat Protoc. 2011;6:1969–1980. doi: 10.1038/nprot.2011.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopinath K, Venclovas C, Ioerger TR, Sacchettini JC, McKinney JD, Mizrahi V, Warner DF. A vitamin B12 transporter in Mycobacterium tuberculosis. Open Biology. 2013;3:120175. doi: 10.1098/rsob.120175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon HA, Pesti L. The gnotobiotic animal as a tool in the study of host microbial relationships. Bacteriological reviews. 1971;35:390–429. doi: 10.1128/br.35.4.390-429.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray MJ, Escalante-Semerena JC. The cobinamide amidohydrolase (cobyric acid-forming) CbiZ enzyme: a critical activity of the cobamide remodelling system of Rhodobacter sphaeroides. Mol Microbiol. 2009;74:1198–1210. doi: 10.1111/j.1365-2958.2009.06928.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen EE, Lozupone CA, Rey FE, Wu M, Guruge JL, Narra A, Goodfellow J, Zaneveld JR, McDonald DT, Goodrich JA, et al. Pan-genome of the dominant human gut-associated archaeon, Methanobrevibacter smithii, studied in twins. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4599–4606. doi: 10.1073/pnas.1000071108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell KE, Wheeler GL, Leptos KC, Goldstein RE, Smith AG. Insights into the evolution of vitamin B12 auxotrophy from sequenced algal genomes. Molecular Biology and Evolution. 2011;28:2921–2933. doi: 10.1093/molbev/msr124. [DOI] [PubMed] [Google Scholar]
- Ikeda M, Hosotani T, Kurimoto K, Mori T, Ueda T, Kotake Y, Sakakibara B. The differences of the metabolism related to vitamin B6-dependent enzymes among vitamin B6-deficient germ-free and conventional rats. J Nutr Sci Vitaminol (Tokyo) 1979;25:131–139. doi: 10.3177/jnsv.25.131. [DOI] [PubMed] [Google Scholar]
- Johnson DR, Nemir A, Andersen GL, Zinder SH, Alvarez-Cohen L. Transcriptomic microarray analysis of corrinoid responsive genes in Dehalococcoides ethenogenes strain 195. FEMS Microbiol Lett. 2009;294:198–206. doi: 10.1111/j.1574-6968.2009.01569.x. [DOI] [PubMed] [Google Scholar]
- Jones KM, Kobayashi H, Davies BW, Taga ME, Walker GC. How rhizobial symbionts invade plants: the Sinorhizobium-Medicago model. Nat Rev Microbiol. 2007;5:619–633. doi: 10.1038/nrmicro1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller S, Ruetz M, Kunze C, Krautler B, Diekert G, Schubert T. Exogenous 5,6-dimethylbenzimidazole caused production of a non-functional tetrachloroethene reductive dehalogenase in Sulfurospirillum multivorans. Environ Microbiol. 2013 doi: 10.1111/1462-2920.12268. [DOI] [PubMed] [Google Scholar]
- Klug G. Beyond catalysis: vitamin B12 as a cofactor in gene regulation. Mol Microbiol. 2014;91:635–640. doi: 10.1111/mmi.12490. [DOI] [PubMed] [Google Scholar]
- Kräutler B. Vitamin B12: chemistry and biochemistry. Biochem Soc Trans. 2005;33:806–810. doi: 10.1042/BST0330806. [DOI] [PubMed] [Google Scholar]
- Lau KS, Gottlieb C, Wasserman LR, Herbert V. Measurement of Serum Vitamin B12 Level Using Radioisotope Dilution and Coated Charcoal. Blood. 1965;26:202–214. [PubMed] [Google Scholar]
- Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489:220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur DG, Balasubramanian S, Frankish A, Huang N, Morris J, Walter K, Jostins L, Habegger L, Pickrell JK, Montgomery SB, et al. A systematic survey of loss-of-function variants in human protein-coding genes. Science. 2012;335:823–828. doi: 10.1126/science.1215040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacNeil LT, Watson E, Arda HE, Zhu LJ, Walhout AJ. Diet-induced developmental acceleration independent of TOR and insulin in C. elegans. Cell. 2013;153:240–252. doi: 10.1016/j.cell.2013.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macy JM, Ljungdahl LG, Gottschalk G. Pathway of succinate and propionate formation in Bacteroides fragilis. J Bacteriol. 1978;134:84–91. doi: 10.1128/jb.134.1.84-91.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews RG. Cobalamin-dependent methionine synthase. In: Banerjee R, editor. Chemistry and Biochemistry of B12. New York: John Wiley & Sons, Inc.; 1999. pp. 681–706. [Google Scholar]
- McCutcheon JP, McDonald BR, Moran NA. Convergent evolution of metabolic roles in bacterial co-symbionts of insects. Proc Natl Acad Sci USA. 2009;106:15394–15399. doi: 10.1073/pnas.0906424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNulty NP, Yatsunenko T, Hsiao A, Faith JJ, Muegge BD, Goodman AL, Henrissat B, Oozeer R, Cools-Portier S, Gobert G, et al. The impact of a consortium of fermented milk strains on the gut microbiome of gnotobiotic mice and monozygotic twins. Science Translational Medicine. 2011;3:106ra106. doi: 10.1126/scitranslmed.3002701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Men Y, Seth EC, Yi S, Allen RH, Taga ME, Alvarez-Cohen L. Sustainable growth of Dehalococcoides mccartyi 195 by corrinoid salvaging and remodeling in defined lactate-fermenting consortia. Applied and environmental microbiology. 2014a;80:2133–2141. doi: 10.1128/AEM.03477-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Men Y, Seth EC, Yi S, Crofts TS, Allen RH, Taga ME, Alvarez-Cohen L. Identification of specific corrinoids reveals corrinoid modification in dechlorinating microbial communities. Environ Microbiol. 2014b doi: 10.1111/1462-2920.12500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mickelsen O. Intestinal synthesis of vitamins in the nonruminant. Vitamins and Hormones. 1956;14:1–95. doi: 10.1016/s0083-6729(08)60298-8. [DOI] [PubMed] [Google Scholar]
- Mok KC, Taga ME. Growth Inhibition of Sporomusa ovata by Incorporation of Benzimidazole Bases into Cobamides. J Bacteriol. 2013;195:1902–1911. doi: 10.1128/JB.01282-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran NA, McCutcheon JP, Nakabachi A. Genomics and evolution of heritable bacterial symbionts. Annu Rev Genet. 2008;42:165–190. doi: 10.1146/annurev.genet.41.110306.130119. [DOI] [PubMed] [Google Scholar]
- Murphy MF, Sourial NA, Burman JF, Doyle DV, Tabaqchali S, Mollin DL. Megaloblastic anaemia due to vitamin B12 deficiency caused by small intestinal bacterial overgrowth: possible role of vitamin B12 analogues. Br J Haematol. 1986;62:7–12. doi: 10.1111/j.1365-2141.1986.tb02894.x. [DOI] [PubMed] [Google Scholar]
- Nahvi A, Sudarsan N, Ebert MS, Zou X, Brown KL, Breaker RR. Genetic control by a metabolite binding mRNA. Chem Biol. 2002;9:1043. doi: 10.1016/s1074-5521(02)00224-7. [DOI] [PubMed] [Google Scholar]
- Nielsen MJ, Rasmussen MR, Andersen CB, Nexo E, Moestrup SK. Vitamin B12 transport from food to the body's cells--a sophisticated, multistep pathway. Nature reviews. Gastroenterology & hepatology. 2012;9:345–354. doi: 10.1038/nrgastro.2012.76. [DOI] [PubMed] [Google Scholar]
- Ortiz-Guerrero JM, Polanco MC, Murillo FJ, Padmanabhan S, Elias-Arnanz M. Light-dependent gene regulation by a coenzyme B12-based photoreceptor. Proc Natl Acad Sci U S A. 2011;108:7565–7570. doi: 10.1073/pnas.1018972108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pol A, van der Drift C, Vogels GD. Corrinoids from Methanosarcina barkeri: structure of the alpha-ligand. Biochem Biophys Res Commun. 1982;108:731–737. doi: 10.1016/0006-291x(82)90890-7. [DOI] [PubMed] [Google Scholar]
- Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raven JL, Robson MB, Morgan JO, Hoffbrand AV. Comparison of three methods for measuring vitamin B12 in serum: Radioisotopic, Euglena gracilis and Lactobacillus leichmannii. Br J Haematol. 1972;22:21–31. doi: 10.1111/j.1365-2141.1972.tb08783.x. [DOI] [PubMed] [Google Scholar]
- Reinhold A, Westermann M, Seifert J, von Bergen M, Schubert T, Diekert G. Impact of vitamin B12 on formation of the tetrachloroethene reductive dehalogenase in Desulfitobacterium hafniense strain Y51. Applied and environmental microbiology. 2012;78:8025–8032. doi: 10.1128/AEM.02173-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renz P. Biosynthesis of the 5,6-dimethylbenzimidazole moiety of cobalamin and of the other bases found in natural corrinoids. In: Banerjee R, editor. Chemistry and Biochemistry of B12. New York: John Wiley & Sons, Inc.; 1999. pp. 557–575. [Google Scholar]
- Rodionov DA, Vitreschak AG, Mironov AA, Gelfand MS. Comparative genomics of the vitamin B12 metabolism and regulation in prokaryotes. J Biol Chem. 2003;278:41148–41159. doi: 10.1074/jbc.M305837200. [DOI] [PubMed] [Google Scholar]
- Roeselers G, Mittge EK, Stephens WZ, Parichy DM, Cavanaugh CM, Guillemin K, Rawls JF. Evidence for a core gut microbiota in the zebrafish. Isme J. 2011;5:1595–1608. doi: 10.1038/ismej.2011.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenblatt DS, Fenton WA. Inborn errors of cobalamin metabolism. In: Banerjee R, editor. Chemistry and biochemistry of B12. New York: Wiley; 1999. pp. 367–384. [Google Scholar]
- Roth JR, Lawrence JG, Bobik TA. Cobalamin (coenzyme B12): synthesis and biological significance. Annu Rev Microbiol. 1996;50:137–181. doi: 10.1146/annurev.micro.50.1.137. [DOI] [PubMed] [Google Scholar]
- Ryzhkova EP, Brukhanov AL. Effect of a corrinoid on Methanosarcina barkeri DNA synthesis. Microbiology. 2009;78:1–6. [PubMed] [Google Scholar]
- Santos F, Vera JL, Lamosa P, de Valdez GF, de Vos WM, Santos H, Sesma F, Hugenholtz J. Pseudovitamin B(12) is the corrinoid produced by Lactobacillus reuteri CRL1098 under anaerobic conditions. FEBS Lett. 2007;581:4865–4870. doi: 10.1016/j.febslet.2007.09.012. [DOI] [PubMed] [Google Scholar]
- Sañudo-Wilhelmy SA, Cutter LS, Durazo R, Smail EA, Gómez-Consarnau L, Webb EA, Prokopenko MG, Berelson WM, Karl DM. Multiple B-vitamin depletion in large areas of the coastal ocean. Proc Natl Acad Sci U S A. 2012;109:14041–14045. doi: 10.1073/pnas.1208755109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seekatz AM, Aas J, Gessert CE, Rubin TA, Saman DM, Bakken JS, Young VB. Recovery of the gut microbiome following fecal microbiota transplantation. mBio. 2014;5:e00893–e00814. doi: 10.1128/mBio.00893-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seetharam B, Alpers DH. Absorption and transport of cobalamin (vitamin B12) Annu Rev Nutr. 1982;2:343–369. doi: 10.1146/annurev.nu.02.070182.002015. [DOI] [PubMed] [Google Scholar]
- Seth EC, Taga ME. Nutrient cross-feeding in the microbial world. Frontiers in Microbiology. 2014;5:350. doi: 10.3389/fmicb.2014.00350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegl A, Kamke J, Hochmuth T, Piel J, Richter M, Liang C, Dandekar T, Hentschel U. Single-cell genomics reveals the lifestyle of Poribacteria, a candidate phylum symbiotically associated with marine sponges. Isme J. 2011;5:61–70. doi: 10.1038/ismej.2010.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K, McCoy KD, Macpherson AJ. Use of axenic animals in studying the adaptation of mammals to their commensal intestinal microbiota. Seminars in Immunology. 2007;19:59–69. doi: 10.1016/j.smim.2006.10.002. [DOI] [PubMed] [Google Scholar]
- Stabler SP, Allen RH. Vitamin B12 deficiency as a worldwide problem. Annu Rev Nutr. 2004;24:299–326. doi: 10.1146/annurev.nutr.24.012003.132440. [DOI] [PubMed] [Google Scholar]
- Sugita H, Miyajima C, Deguchi Y. The vitamin B12-producing ability of the intestinal microflora of freshwater fish. Aquaculture. 1991;92:267–276. [Google Scholar]
- Sumi Y, Miyakawa M, Kanzaki M, Kotake Y. Vitamin B-6 deficiency in germfree rats. J Nutr. 1977;107:1707–1714. doi: 10.1093/jn/107.9.1707. [DOI] [PubMed] [Google Scholar]
- Taga ME, Walker GC. Sinorhizobium meliloti requires a cobalamin-dependent ribonucleotide reductase for symbiosis with its plant host. Molecular Plant-Microbe Interactions. 2010;23:1643–1654. doi: 10.1094/MPMI-07-10-0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenaka S, Sugiyama S, Ebara S, Miyamoto E, Abe K, Tamura Y, Watanabe F, Tsuyama S, Nakano Y. Feeding dried purple laver (nori) to vitamin B12-deficient rats significantly improves vitamin B12 status. Br J Nutr. 2001;85:699–703. doi: 10.1079/bjn2001352. [DOI] [PubMed] [Google Scholar]
- Tanioka Y, Miyamoto E, Yabuta Y, Ohnishi K, Fujita T, Yamaji R, Misono H, Shigeoka S, Nakano Y, Inui H, et al. Methyladeninylcobamide functions as the cofactor of methionine synthase in a Cyanobacterium, Spirulina platensis NIES-39. FEBS letters. 2010;584:3223–3226. doi: 10.1016/j.febslet.2010.06.013. [DOI] [PubMed] [Google Scholar]
- Thomas T, Rusch D, DeMaere MZ, Yung PY, Lewis M, Halpern A, Heidelberg KB, Egan S, Steinberg PD, Kjelleberg S. Functional genomic signatures of sponge bacteria reveal unique and shared features of symbiosis. Isme J. 2010;4:1557–1567. doi: 10.1038/ismej.2010.74. [DOI] [PubMed] [Google Scholar]
- Torrents E, Trevisiol C, Rotte C, Hellman U, Martin W, Reichard P. Euglena gracilis ribonucleotide reductase: the eukaryote class II enzyme and the possible antiquity of eukaryote B12 dependence. J Biol Chem. 2006;281:5604–5611. doi: 10.1074/jbc.M512962200. [DOI] [PubMed] [Google Scholar]
- Tucker BJ, Breaker RR. Riboswitches as versatile gene control elements. Curr Opin Struct Biol. 2005;15:342–348. doi: 10.1016/j.sbi.2005.05.003. [DOI] [PubMed] [Google Scholar]
- Vitreschak AG, Rodionov DA, Mironov AA, Gelfand MS. Regulation of the vitamin B12 metabolism and transport in bacteria by a conserved RNA structural element. RNA. 2003;9:1084–1097. doi: 10.1261/rna.5710303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe F, Yabuta Y, Bito T, Teng F. Vitamin B12-containing plant food sources for vegetarians. Nutrients. 2014;6:1861–1873. doi: 10.3390/nu6051861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters LS, Storz G. Regulatory RNAs in bacteria. Cell. 2009;136:615–628. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson E, MacNeil LT, Ritter AD, Yilmaz LS, Rosebrock AP, Caudy AA, Walhout AJ. Interspecies systems biology uncovers metabolites affecting C. elegans gene expression and life history traits. Cell. 2014;156:759–770. doi: 10.1016/j.cell.2014.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wostmann BS. The germfree animal in nutritional studies. Annu Rev Nutr. 1981;1:257–279. doi: 10.1146/annurev.nu.01.070181.001353. [DOI] [PubMed] [Google Scholar]
- Wostmann BS, Knight PL. Antagonism between vitamins A and K in the germfree rat. J Nutr. 1965;87:155–160. doi: 10.1093/jn/87.2.155. [DOI] [PubMed] [Google Scholar]
- Yi S, Seth EC, Men YJ, Stabler SP, Allen RH, Alvarez-Cohen L, Taga ME. Versatility in corrinoid salvaging and remodeling pathways supports corrinoid-dependent metabolism in Dehalococcoides mccartyi. Appl. Environ. Microbiol. 2012;78:7745–7752. doi: 10.1128/AEM.02150-12. [DOI] [PMC free article] [PubMed] [Google Scholar]