Abstract
The establishment, maintenance and modulation of cell-type specific neural architectures are critically important to the formation of functional neural networks. At the neuroanatomical level, differential patterns of dendritic arborization directly impact neural function and connectivity, however the molecular mechanisms underlying the specification of distinct dendrite morphologies remain incompletely understood. To address this question, we analyzed global gene expression from purified populations of wild-type class I and class IV Drosophila melanogaster dendritic arborization (da) sensory neurons compared to wild-type whole larval RNA using oligo DNA microarray expression profiling. Herein we present detailed experimental methods and bioinformatic analyses to correspond with our data reported in the Gene Expression Omnibus under accession number GSE46154. We further provide R code to facilitate data accession, perform quality controls, and conduct bioinformatic analyses relevant to this dataset. Our cell-type specific gene expression datasets provide a valuable resource for guiding further investigations designed to explore the molecular mechanisms underlying differential patterns of neuronal patterning.
Keywords: Genomics, Microarrays, Gene expression, Neurons, Dendrites
| Specifications | |
|---|---|
| Organism/cell line/tissue | Drosophila melanogaster |
| Sequencer or array type | Agilent-018972 D. melanogaster oligo microarray 4x44K |
| Data format | Raw and processed |
| Experimental factors | Cell type |
| Experimental features | Gene expression profiling of purified class I and class IV Drosophila dendritic arborization (da) neurons was performed at the third instar larval stage of development and compared against age matched whole larval RNA to identify differentially enriched genes that potentially contribute to class-specific dendrite morphogenesis. |
| Consent | n/a |
Direct link to deposited data
Deposited data can be found here: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE46154
Experimental design, materials and methods
Drosophila strains and genetics
Drosophila melanogaster strains were raised on standard cornmeal–molasses–agar media at 25 °C. The GAL4ppk.1.9,UASmCD8::GFP [1], [2] strain was used for specific labeling of class IV da neurons, whereas ppk-GAL80;GAL4[221],UAS-mCD8::GFP [3], [4], [5] was used for specific labeling of class I da neurons. UAS-cut was independently used in qRT-PCR quality control experiments for the purity of the cell isolations [5]. Oregon-R was used as the wild-type strain for these studies.
Cell isolation, purification, and qRT-PCR
The isolation and purification of class I and class IV da neurons were performed as previously described [6]. Briefly, 40–50 age-matched third instar larvae expressing mCD8::GFP under the control of the either the class IV-specific GAL4ppk.1.9, or class I-specific GAL80ppk.1.9; GAL4[221] drivers were collected and washed several times in ddH20. The larvae were then rinsed in RNAse away, ddH20 and finally dissected. The tissue was then dissociated using a combination of enzymatic and mechanical perturbations to yield single cell suspensions which were filtered using a 30 μm membrane. The filtrate is then incubated with superparamagnetic beads (Dynabeads MyOne Streptavidin T1, Invitrogen) coupled with biotinylated mouse anti-CD8a antibody (eBioscience) for 60 min. Finally the da neurons attached to the magnetic beads were then separated using a powerful magnetic field. The isolated neurons were washed at least five times with 1× PBS to remove any potential non-specific cells and the quality and purity of isolated neurons was assessed under a stereo-fluorescent microscope equipped with phase contrast for examining the number of fluorescent (GFP-positive) vs. non-fluorescent (GFP-negative) cells. Only if the isolated cells were free of cellular debris and non-specific (i.e. non-fluorescing) contaminants were they retained for subsequent RNA extraction.
The purified class I and class IV neuron populations were then lysed in SuperAmp™ (Miltenyi Biotec) RNA lysis buffer followed by storage at − 80 °C. For whole larvae controls, 10 age-matched third instar larvae were homogenized in PicoPure RNA isolation buffer (Invitrogen), following which RNA was eluted as per manufacturer protocols. In order to process the whole larvae RNA identically to the da neuron samples, 10 μl of whole-larvae RNA, diluted to the same levels as da neuron samples, was added to SuperAmp™ (Miltenyi Biotec) buffer and stored at − 80 °C. As a quality control for the specificity of the cell-type specific isolations, we conducted qRT-PCR analyses [5]. Briefly, RNA was extracted from isolated class I or class IV da neurons (in the presence or absence of a UAS-cut transgene) as an independent measure of cell purity. These analyses were performed as previously described [7], [8] using the following pre-validated Qiagen QuantiTect Primer Assays (Qiagen, Germantown, MD, USA): cut (QT00501389) and expression data was normalized using primers for GAPDH2 (QT00922957) and RpL32 (QT00985677).
Microarray analysis
mRNA isolation, amplification, labeling, hybridization, and microarray analysis were conducted by Miltenyi Biotec. mRNA was isolated from independent pools of da neurons (1000–1500 neurons/pool) via magnetic bead technology. Sample quality was verified on an Agilent Bioanalyzer 2100 prior to amplification. The whole larval RNA lysate which was used as the control sample, had the same quantity as the isolated neuron subtype mRNA. SuperAmp RNA amplification was performed on all the samples according to Miltenyi Biotec's global PCR protocol using mRNA-derived cDNA. The amplified cDNA samples were quantified using the ND-1000 Spectrophotometer (NanoDrop Technologies) and the sample integrity verified via Agilent 2100 Bioanalyzer analysis (Agilent Technologies). 250 ng of each of the cDNAs was used as template for Cy3 labeling followed by hybridization to Agilent whole D. melanogaster genome oligo microarrays (4x44K). The microarray data, including metadata, raw data, and normalized datasets, has been deposited into the Gene Expression Omnibus (GEO) under the accession number GSE46154.
Agilent Feature Extraction Software (AFE) was used to read out and process the replicate microarray image files into text files, which contain annotation for the probe IDs associated to particular location of the microarray chip, as well as raw and background corrected intensities. There are a total of (7) arrays comprised of (2) technical replicates of class 1 da neurons, (2) technical replicates of class 4 da neurons and (3) technical replicates of whole third instar larval RNA. The analysis of the microarray data was performed in R [9] using the Bioconductor [10] suite of packages. We provide R code to facilitate data accession, perform quality controls, and conduct bioinformatic analyses relevant to this dataset (Supplementary data). The primary R file, FuncCall.r, calls the function AgilentProcess (which is contained in the analyzeAgilentMicroarray.R file) which internally calls the other required functions. The FuncCall.r file and the analyzeAgilentMicroarray.R. For pre-processing of the microarray data, the Agi4X44PreProcess [11] package was used for pre-processing the microarray data. The data was read in using a modified version of the read.AgilentFE (readAgilentFE) function to read in data which were processed by a newer version of AFE (version later than 9.5.3.1). The data was then background corrected by using the normexp method and normalized by the quantile normalization method, in the function BgandNorm.
For the next step in pre-processing, we used a modified version of the function filter.probes (the new function is probe_filters) (Supplementary data). This modified version of the filter.probes function enables the analyses of data processed by versions of the AFE later than 9.5.3.1. The probe_filters function removes saturated signals, as well as signals below the background and removes negative controls. This function requires an annotation database for annotating the probe IDs. This database (dros.db) was created using the AnnotationForge [12] package of the Bioconductor suite of tools derived from Agilent's annotation file for 4x44K oligonucleotide microarrays (https://earray.chem.agilent.com/earray/).
Quality controls and visualization
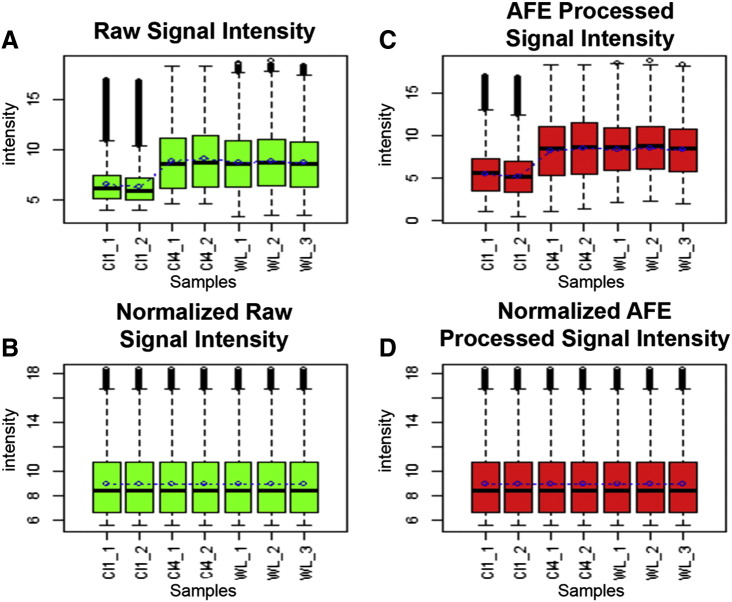
Box-plots (Fig. 1) were constructed showing a comparison study between raw intensity and background corrected processed signals. The comparison study shows box-plots of AFE extracted raw signal intensity or mean signal intensity (colored in green) and background corrected intensity (colored in red), before and after background correction and normalization. The intensity values (before normalization) and expression values (after normalization) are depicted on the y-axis and the x-axis contains the sample names. The intensity values (before normalization) are log normalized to base 2 for construction of the box-plot. It is evident from the box-plot that after the usage of the Normalization and the Background Correction functions the data is properly normalized having a reduction in the number of outliers and median for both the raw and AFE background corrected signals, being the same.
Fig. 1.
Box-plots showing the mean raw (A) and normalized signal (B) intensity expression data. The mean raw signals and mean processed signals represent the raw mean signal intensity of the spot on the microarrays and the normalized and background corrected mean signal intensities, respectively. The AFE processed signal intensity is the Agilent Feature Extracted background corrected signal (C), whereas the normalized AFE processed signal intensity (D) represents the normalized processed signal. Raw signal intensity is color-coded in green, whereas AFE processed signal intensity is color-coded in red. Class I samples are (Cl1_1; Cl1_2); class IV samples are (Cl4_1; Cl4_2); whole larvae samples are (WL_1; WL_2; WL_3).
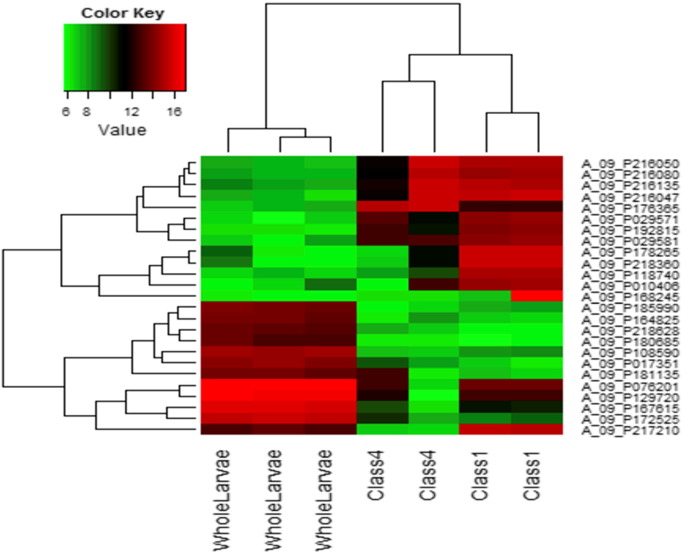
A heat map (Fig. 2) was also constructed which depicts the variation of gene expression values across the arrays for the top 25 most variant genes, from the background corrected and quantile normalized expression values. The heat map was constructed using a modified heat map function of Agi4x44PreProcess [11] (Heatmap_mod), to print the classes in the x-axis instead of the file names. The color ranges from green (least expressed) to red (most expressed). This analyses reveals clear clustering of class I vs. class IV vs. whole larvae and demonstrates that the variation is more observed across classes (i.e. between class I, class IV and whole larvae), compared to those between the arrays (technical replicates), confirming the fact that the gene expression remains similar within a cell type (class I vs. class IV) or tissue type (whole larvae). Within a class, these data indicate a higher degree of variance (at least among the top 25 most variant genes) in the class IV da neuron samples, relative to class I or whole larvae.
Fig. 2.
Heat map shows the expression values of the top 25 high variance genes across class I, class 4 and whole larvae arrays. The green color represents the lowest expression values, whereas red represents the highest expression values. The expression values are background corrected, quantile normalized and filtered (removal of saturated, below background intensities).
Expression analysis
The expression analysis was performed using the Limma [13] package of the Bioconductor suite [10]. Benjamini Hochberg multiple testing (False Discovery Rate test) correction was used with a q-value (False Discovery Rate adjusted p-value) threshold of 0.01. The analysis was conducted between Class 1 vs Whole Larvae and Class 4 vs Whole Larvae, with the t-value (t test value) determining whether a gene is up or downregulated (e.g. in Class 1 vs Whole Larvae, Class 1 genes having a t-value greater than 0 and a q-value less than 0.01, are upregulated compared to Whole Larvae and ones having t-value less than 0 and q value less than 0.01 are downregulated).
Bioinformatic analysis
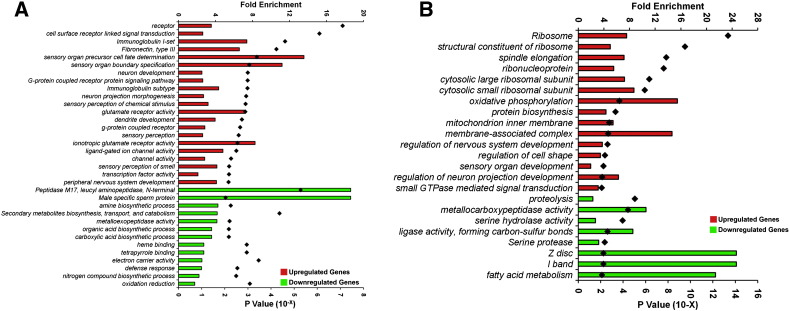
Functional enrichment analysis was performed using DAVID [14], [15] to identify statistically over-represented functional gene classes. The up/downregulated gene list of class 1 and class 4 da neurons from the microarray analysis was used as input and all genes represented in the microarray were used as background. DAVID computes the enrichment value, which is deduced by doing a modified Fisher exact test (also known as EASE test). This is used to determine whether the proportion of genes falling into each category differ by group [14]. The test provides an enrichment score (whereby the higher the score, the greater the chance of the gene being enriched) and a p-value (whereby the lower the p-value, the greater the chance of the gene being enriched). A p-value threshold of 0.01 was used as a cut-off for enrichment in the annotation categories. Fig. 3 illustrates DAVID-based gene ontology (GO) functional characterization of differentially expressed gene-sets in class I vs. class IV neurons. These analyses shed molecular insight into biological processes that are uniquely up or downregulated in class I vs. class IV da neurons which may contribute to differential patterns of dendritic arborization.
Fig. 3.
Functional characterization of differentially expressed gene-sets in class I and class IV da neurons. Analyses of gene ontology (GO) categories for genes that are enriched/depleted in class I (A) and class IV (B) da neurons. The histogram represents GO categories that are significantly over-represented (p < 0.01) in the population of differentially expressed genes that are uniquely regulated in class I (A) or class IV (B) neurons, when compared to whole larval controls. Bars indicate the fold enrichment (top x-axis) of the genes belonging to a given GO term in the population of regulated genes in comparison to the total population of genes in the Agilent 4x44K array. Black diamonds indicate the modified Fisher 's exact p-value (EASE score, bottle X axis) for each category.
Figure adapted from [5].
Acknowledgments
The authors gratefully acknowledge the National Institutes of Health (MH086928-02) for support of this research. The funders had no role in the study design, data collection and analyses, decision to publish, or preparation of the manuscript.
Footnotes
To run the R code package analyses, download the microarray data files extracted by the Agilent Feature Extractor (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE46154) and place the files in the Supplementary folder. Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.gdata.2014.10.011.
Appendix A. Supplementary data
-
•readAgilentFE —It is a modified version of read.AgilentFE function of Agi4X44PreProcess package to read AFE extracted data after version 9.5.3.1. The modification commented out the check for sequence and chr_coord which were column headers for AFE extracted files for version 9.5.3.1.
-
•probe_filter —It is a modified version of filter.probes function of Agi4X44PreProcess function to filter AFE normalized data after version 9.5.3.1. The modified part is write.filter.out, write.filt.out and write.control.out commented out and countFLAG, filter.nas, filterFLAGall, filterFLAG and filterWellAboveSIGNALv2 overriden with Agi4x44PreProcess package.
-
•make_flydb —Constructs the Drosophila annotation database dros.db.
-
•Heatmap_mod —It is a modification of Heatmap function in Agi4X44PreProcess package. The modification allows one to print the classes in the x-axis instead of the file names.
Annotation file for Agilent 4x44 Oligonucleotide array downloaded from https://earray.chem.agilent.com/earray/. Used by make_flydb function to produce fly annotation database dros.db.
The design file. It maps the microarray dataset file to the corresponding classes.
File containing commands to call the AgilentProcess function which processes the microarray dataset.
References
- 1.Grueber W.B., Ye B., Yang C.-H., Younger S., Borden K. Projections of Drosophila multidendritic neurons in the central nervous system: links with peripheral dendrite morphology. Development. 2007;134:55–64. doi: 10.1242/dev.02666. [DOI] [PubMed] [Google Scholar]
- 2.Ainsley J.A., Pettus J.M., Bosenko D., Gerstein C.E., Zinkevich N. Enhanced locomotion caused by loss of the Drosophila DEG/ENaC protein Pickpocket1. Curr. Biol. 2003;13:1557–1563. doi: 10.1016/s0960-9822(03)00596-7. [DOI] [PubMed] [Google Scholar]
- 3.Yang C.-H., Rumpf S., Xiang Y., Gordon M.D., Song W. Control of the postmating behavioral switch in Drosophila females by internal sensory neurons. Neuron. 2009;61:519–526. doi: 10.1016/j.neuron.2008.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grueber W.B., Jan L.Y., Jan Y.N. Different levels of the homeodomain protein Cut regulate distinct dendrite branching patterns of Drosophila multidendritic neurons. Cell. 2003;112:805–818. doi: 10.1016/s0092-8674(03)00160-0. [DOI] [PubMed] [Google Scholar]
- 5.Iyer E.P.R., Iyer S.C., Sullivan L., Wang D., Meduri R., Graybeal L.L., Cox D.N. Functional genomic analyses of two morphologically distinct classes of Drosophila sensory neurons: post-mitotic roles of transcription factors in dendritic patterning. PLoS ONE. 2013;8(8):e72434. doi: 10.1371/journal.pone.0072434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iyer E.P.R., Iyer S.C., Sulkowski M.J., Cox D.N. Isolation and purification of Drosophila peripheral neurons by magnetic bead sorting. J. Vis. Exp. 2009 doi: 10.3791/1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iyer S.C., Wang D., Iyer E.P.R., Trunnell S.A., Meduri R. The RhoGEF trio functions in sculpting class specific dendrite morphogenesis in Drosophila sensory neurons. PLoS ONE. 2012;7:e33634. doi: 10.1371/journal.pone.0033634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sulkowski M.J., Iyer S.C., Kurosawa M.S., Iyer E.P.R., Cox D.N. Turtle functions downstream of Cut in differentially regulating class specific dendrite morphogenesis in Drosophila. PLoS ONE. 2011;6:e22611. doi: 10.1371/journal.pone.0022611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.R Core Team . R Foundation for Statistical Computing; Vienna, Austria: 2013. R: a language and environment for statistical computing. (URL http://www.R-project.org/) [Google Scholar]
- 10.Gentleman V.J., Carey D.M., Bates B., Bolstad M., Dettling S., Dudoit B., Ellis L., Gautier Y. Bioconductor: open software development for computational biology and bioinformatics R. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lopez-Romero P. Agi4x44PreProcess: preprocessing of Agilent 4x44 array data. R package version 1.22.0. http://www.bioconductor.org/packages//2.11/bioc/html/Agi4x44PreProcess.html
- 12.Carlson M., Pages H. AnnotationForge: code for building annotation database packages. R package version 1.4.4. http://www.bioconductor.org/packages/release/bioc/html/AnnotationForge.html
- 13.Smyth G.K. Limma: linear models for microarray data. In: Gentleman R., Carey V., Dudoit S., Irizarry R., Huber W., editors. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. Springer; New York: 2005. pp. 397–420. [Google Scholar]
- 14.Dennis G., Sherman B.T., Hosack D.A., Yang J., Gao W. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:R60. [PubMed] [Google Scholar]
- 15.Huang D.W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
-
•readAgilentFE —It is a modified version of read.AgilentFE function of Agi4X44PreProcess package to read AFE extracted data after version 9.5.3.1. The modification commented out the check for sequence and chr_coord which were column headers for AFE extracted files for version 9.5.3.1.
-
•probe_filter —It is a modified version of filter.probes function of Agi4X44PreProcess function to filter AFE normalized data after version 9.5.3.1. The modified part is write.filter.out, write.filt.out and write.control.out commented out and countFLAG, filter.nas, filterFLAGall, filterFLAG and filterWellAboveSIGNALv2 overriden with Agi4x44PreProcess package.
-
•make_flydb —Constructs the Drosophila annotation database dros.db.
-
•Heatmap_mod —It is a modification of Heatmap function in Agi4X44PreProcess package. The modification allows one to print the classes in the x-axis instead of the file names.
Annotation file for Agilent 4x44 Oligonucleotide array downloaded from https://earray.chem.agilent.com/earray/. Used by make_flydb function to produce fly annotation database dros.db.
The design file. It maps the microarray dataset file to the corresponding classes.
File containing commands to call the AgilentProcess function which processes the microarray dataset.