Abstract
Determining the geographic scale at which to apply ecosystem-based management (EBM) has proven to be an obstacle for many marine conservation programs. Generalizations based on geographic proximity, taxonomy, or life history characteristics provide little predictive power in determining overall patterns of connectivity, and therefore offer little in terms of delineating boundaries for marine spatial management areas. Here, we provide a case study of 27 taxonomically and ecologically diverse species (including reef fishes, marine mammals, gastropods, echinoderms, cnidarians, crustaceans, and an elasmobranch) that reveal four concordant barriers to dispersal within the Hawaiian Archipelago which are not detected in single-species exemplar studies. We contend that this multispecies approach to determine concordant patterns of connectivity is an objective and logical way in which to define the minimum number of management units and that EBM in the Hawaiian Archipelago requires at least five spatially managed regions.
1. Introduction
Global catches of commercially fished species have declined by up to 90% under classic single-species fisheries models [1–3]. The high-profile failures of fisheries managed for maximum sustainable yield has led to widespread interest in a shift toward ecosystem-based management (EBM) of marine resources (reviewed by [4]). EBM can be broadly defined as an integrated approach that considers the entire ecosystem, including linkages and the cumulative impacts of all human activities within and as part of the system. As such, EBM is explicitly place-based and adaptive in nature, and therefore particularly attractive for management. In recognition of the need for explicit boundaries in ecosystem-based management, Spalding et al. [5] divided the oceans into 232 ecoregions. However, marine ecosystems are highly complex, with many linkages and feedbacks that occur across multiple scales of space and time in ways that have proven difficult to predict [4]. Existing approaches to EBM in marine systems include spatial control of human activities through the use of marine protected areas (MPAs) and/or ocean zoning, changes in governance, monitoring and evaluation via ecosystem indicators derived from multiple disciplines (e.g., oceanography, ecology, economics, political science, and sociology), risk assessment, and precautionary adaptive management [6]. Successful spatial management requires a complex system of zones, each of which seeks to match resource exploitation with biological productivity, local population levels, and socioeconomic payoffs [7]. Delineation of the appropriate spatial scales for management zones within a specific management network requires a detailed understanding of dispersal pathways and population connectivity (reviewed by [8–10]). Despite the central role of dispersal and connectivity in sustaining marine populations, our understanding of these processes is still largely underdeveloped, and “a strong commitment to understanding patterns of connectivity in marine populations will clearly be necessary to guide the practical design of networks of marine reserves” [10, p.113]. In effect, managers cannot practice EBM if they do not know the boundaries of the corresponding ecosystems.
Understanding connectivity in the sea is complicated by the fact that most marine organisms have a biphasic life cycle with benthic or sedentary adults and dispersing eggs and/or larvae, which may be pelagic for as little as a few minutes to more than a year. Following the pelagic phase, larvae settle onto a patch of suitable habitat, where they may remain throughout their lives, and in cases of sessile organisms such as corals, the act of settlement includes permanent attachment to a single site. Thus, long-distance dispersal is accomplished almost exclusively during the pelagic larval phase, which can potentially span large expanses of open ocean [11–15]. On the other hand, species which lack a pelagic larval phase, such as marine mammals and elasmobranchs, have the potential to range widely throughout the oceans and face few obvious barriers to dispersal. Despite the potential for long-distance movement in most marine species, the geographic limits of such dispersal remain uncertain, because it is virtually impossible to track microscopic juveniles during the pelagic phase (reviewed by [16]), making indirect methods of quantifying larval dispersal particularly attractive (reviewed by [8, 17– 19]). Intuitive expectations that larval dispersal is a function of pelagic larval duration (PLD) are not supported by recent meta-analyses ([20–25]). Despite considerable research, the scale of larval dispersal and the boundaries for EBM remain nebulous due to the complex interaction of larval biology, oceanographic regimes, habitat quality and distribution, and the variability of each through time [26].
Delineating management units is further complicated by the fact that single-species studies of genetic connectivity are often contradictory. Analyses of connectivity frequently focus on single-species exemplars which are then extrapolated to the level of the community, but the utility of exemplars in such cases is limited; even among closely related species with similar ecology, life histories, and geographic ranges, the corresponding patterns of connectivity can be very different [27, 28]. In other cases, animals with highly divergent biology can have surprisingly similar patterns of connectivity [26]. Such variability among species appears to be the rule rather than the exception, and has led to a call for multispecies comparisons of connectivity across trophic levels to broadly define the boundaries for management, and to determine shared avenues of exchange among ecosystems. Due to logistical difficulties in completing such comparisons in marine habitat, few such studies exist (e.g., [29–31]). The linear nature of the Hawaiian Archipelago (Figure 1) provides an excellent forum for resolving shared barriers to gene flow across species and trophic levels, with the goal of developing a geographic framework for EBM.
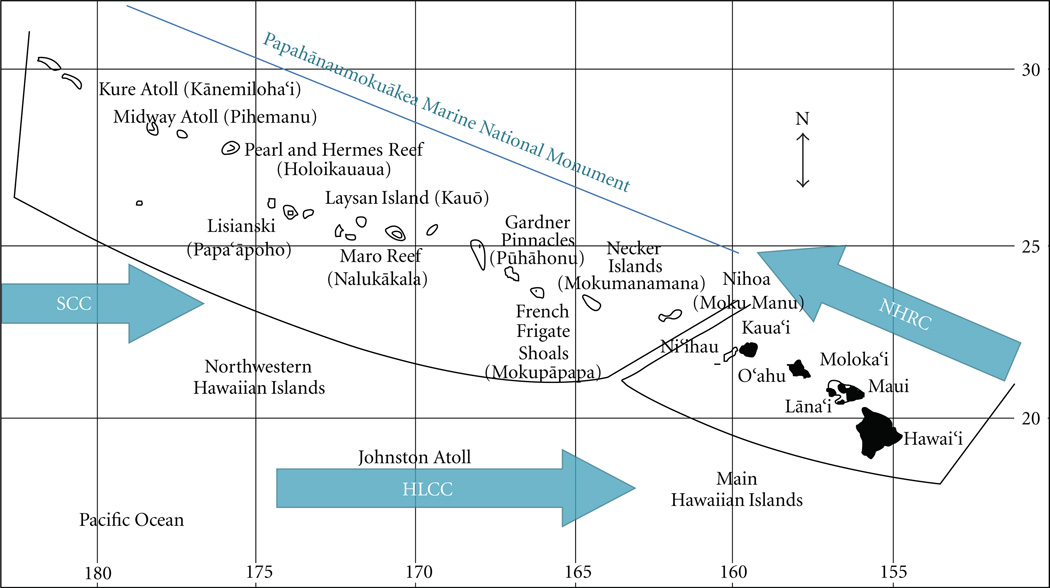
Figure 1.
Map of the Hawaiian Archipelago with major currents denoted: the North Hawaiian Ridge Current (NHRC), the Hawaiian Lee Countercurrent (HLCC), and the Subtropical Countercurrent (SCC). The lines around the two regions of the archipelago highlight the islands, atolls, and banks protected within the Papahānaumokuākea Marine National Monument in the Northwestern Hawaiian Islands (NWHI) and the inhabited high islands of Main Hawaiian Islands (MHI) with each of the 15 primary target areas for collection labeled. For purposes of this analysis, the islands of Lāna’i, Maui & Moloka’i are treated as a single site within the Maui Nui complex of the MHI. Listed from northwest to southeast, these are: Kure Atoll (Kānemiloha’i), Midway Atoll (Pihemanu), Pearl and Hermes Reef (Holoikauaua), Lisianski (Papa’āpoho), Laysan Island (Kauō), Maro Reef (Nalukākala), Gardner Pinnacles (Pūhāhonu), French Frigate Shoals (Mokupāpapa), Necker Island (Mokumanamana), Nihoa (Moku Manu), Ni’ihau, Kaua’i, O’ahu, Maui Nui, and Hawai’i.
The Hawaiian archipelago stretches more than 2600 km in length and consists of two regions: the Main Hawaiian Islands (MHI) which are high volcanic islands with a heavy human presence and the Northwestern Hawaiian Islands (NWHI) which are a string of tiny islands, atolls, shoals, and banks that are essentially uninhabited. Due to their isolation, the roughly 4,500 square miles of coral reefs in the NWHI are among the healthiest and most extensive remaining in the world [32] with abundant large apex predators, a high proportion of endemic species [33, 34], and few human impacts compared to the MHI [18, 35]. In contrast, coral reefs in the MHI are under considerable anthropogenic pressure from the 1.29 million residents (with over 900,000 of those living on the island of O’ahu) and the more than 7 million tourists that visit the state annually. Coral reefs in many of the urban areas and popular tourist destinations have sustained significant impacts, and many show ongoing declines [35– 37]. The primary impacts to coral reefs in the MHI are local and anthropogenic, including coastal development, land-based sources of pollution, overfishing, recreational overuse, and alien species. In contrast, the primary stressors in the NWHI are global in nature, including climate change, ocean acidification, and marine debris [18, 35–37].
On June 15, 2006, the President of the United States signed a proclamation creating the Papahānaumokuākea Marine National Monument (PMNM), encompassing the entire NWHI, at the time the world’s largest marine protected area (MPA). The monument designation affords the NWHI the greatest possible marine environmental protection under United States law. The PMNM spans nearly 140,000 square miles and is home to more than 7,000 currently described species including fishes, invertebrates, algae, marine mammals, and birds although many biologists believe that this is a gross underestimate of the true biodiversity in the region [38]. While the full extent of PMNM biodiversity is unknown, about 25% of the known species are found nowhere else on Earth [39–42]. In 2010, the PMNM was inscribed to the UNESCO World Heritage List, the first U.S. site to be designated in over 15 years. The remote PMNM and surrounding waters became the first primarily marine site to be named in the United States, and the first primarily marine location in the world to be designated as a mixed site for both its outstanding natural and cultural value.
Our research group has embarked on a genetic survey of approximately 60 species of reef-associated fishes, gastropods, crustaceans, echinoderms, cnidarians, and marine mammals, designed to address the issue of population connectivity across the Northwestern and Main Hawaiian Islands and linkages of the Hawaiian Archipelago to other locations throughout the Central Pacific. This effort seeks to inform ecosystem-based management of the PMNM and to evaluate the potential for spillover from the protected area of the NWHI to the heavily populated and exploited MHI. Here we take a molecular genetic approach to infer patterns and magnitude of connectivity in a suite of taxonomically diverse reef-associated species and present preliminary results from 27 species, a subset of the 60 or so target species being collected to understand connectivity across the Hawaiian Archipelago and aid in defining the spatial scale over which EBM should be considered. Although EBM is explicitly place-based, and superficially the definition of an ecosystem seems straightforward, the resolution of geographic boundaries is confounded by obscure biological and oceanographic processes in most marine locations that complicates direct application to management (reviewed by [43]). In managing reefs in the Hawaiian Archipelago, what exactly constitutes a coral reef ecosystem? Is it a reef complex, an island or atoll, an arbitrary geographic distance, a series of adjacent islands and atolls, or the entire Archipelago that is the appropriate geographic scale for management? This work seeks to resolve and quantify the direction and magnitude of exchange among reef habitats across a broad taxonomic spectrum, and to use this information to define objective boundaries, as a necessary prerequisite for the implementation of EBM.
2. Methods
2.1. Sample Collection, DNA Extraction, and Amplification
Tissue samples for DNA analyses were collected from approximately 60 species at as many of the 16 primary islands and atolls as possible in the Hawaiian Archipelago, including the remote and tightly regulated NWHI (Figure 1). It is important to note that sampling remote areas of the Pacific is difficult and expensive and requires extensive permitting and voyage planning compared to collections on the mainland; permitted collections are limited to a maximum of 50 individuals per species at each site, and there are only 1 or 2 days per location, during which the researchers are at the mercy of the weather as to whether or not they can even launch dive boats. Thus, we do not have complete coverage for all species, but in addition to the two species available in the published literature (e.g., [44]), we have currently analyzed 25 additional species (total 27, Tables 1 and 2) from many of the islands and atolls across the Hawaiian Archipelago (Figure 1). Details for the sampling protocols, tissue preservation, DNA extraction, and amplification can be found in Iacchei and Toonen [45] and Skillings and Toonen [46]. Briefly, tissue biopsy samples were taken in the field and stored in either 20% dimethyl sulfoxide salt-saturated buffer [47] or >70% ethanol. DNA was extracted using either a commercially available extraction kit (e.g., Qiagen DNeasy), the chloroform extraction protocol described in Concepcion et al. [48] or a modified salting-out protocol [49]. Following extraction, DNA was stored at −20°C. Most studies were conducted with direct sequencing of a mtDNA fragment using the polymerase chain reaction (PCR) as outlined in references from Table 1. In general, a segment of approximately 600–800 base pairs of the mtDNA cytochrome b (Cytb) was amplified from most of the fishes, and cytochrome oxidase subunit I (COI) was amplified from the majority of invertebrate species, but some used other mitochondrial or nuclear sequence regions or microsatellite markers (see Table 1 for details). PCR recipes and cycling conditions for individual species are provided in the publications cited in Table 1 and upon request from the authors. PCR products to be sequenced were treated with 1.5 units of Exonuclease I and 1.0 units of Fast Alkaline Phosphatase (ExoFAP, Fermentas) per 15 µL PCR products at 37°C for 60 minutes, followed by deactivation at 80°C for 10 minutes. DNA sequencing was performed with fluorescently–labeled dideoxy terminators on an ABI 3130XL Genetic Analyzer (Applied Biosystems) at the Hawai’i Institute of Marine Biology EPSCoR Sequencing Facility. All specimens were initially sequenced in one direction and unique genotypes were confirmed by sequencing in the opposite direction. For analysis of microsatellite loci, amplification products were visualized on an ABI 3130XL Genetic Analyzer using GS500LZ size standards, and analyzed using Genemapper 4.0 (Applied Biosystems).
Table 1.
Species of marine organisms, total sample size, total number of sites, genetic marker(s) used, and study citation for each of the organisms surveyed for population genetic structure across the Hawaiian Archipelago to date. Not all samples were included in subsequent analysis, therefore, the actual sample sizes by site for each species in this analysis are provided in Table 2 . Abbreviations for genetic markers used are: SSR = microsatellites; NIS = nuclear intron sequence data; Cytb = cytochrome b; COI = cytochrome oxidase subunit I; COII = cytochrome oxidase subunit II; CR = control region.
| Species name | Sample size | Number of sites | Marker | Reference |
|---|---|---|---|---|
| Fishes: | ||||
| (1) Epinephelus (=Hyporthodus) quernus | 301 | 10 | SSR, CR | Rivera et al. (see [50, 51]) |
| (2) Stegastes fasciolatus | 219 | 7 | CR | Ramon et al. [44]. |
| (3) Dascylus albisella | 102 | 7 | CR | Ramon et al. [44]. |
| (4) Ctenochaetus strigosus | 499 | 15 | Cytb | Eble et al. [52]. |
| (5) Zebrasoma flavescens | 528 | 15 | Cytb | Eble et al. [52, 53]. |
| (6) Acanthurus nigrofuscus | 305 | 11 | Cytb | Eble et al. [52]. |
| (7) Lutjanus kasmira | 385 | 9 | Cytb, NIS | Gaither et al. [54]. |
| (8) Squalus mitsukurii | 112 | 6 | CR | Daly-Engel et al. [55]. |
| Gastropods: | ||||
| (9) Cellana exarata | 150 | 7 | COI | Bird et al. [28]. |
| (10) Cellana sandwicensis | 109 | 6 | COI | Bird et al. [28]. |
| (11) Cellana talcosa | 105 | 5 | COI | Bird et al. [28]. |
| (12) Dendropoma gregaria | 176 | 15 | COI | Faucci et al. (unpubl. data) |
| (13) Dendropoma platypus | 143 | 15 | COI | Faucci et al. (unpubl. data) |
| (14) Dendropoma rhyssoconcha | 94 | 11 | COI | Faucci et al. (unpubl. data) |
| (15) Serpulorbis variabilis | 73 | 13 | COI | Faucci et al. (unpubl. data) |
| Crustaceans: | ||||
| (16) Calcinus haigae | 146 | 5 | COI | Baums et al. (unpubl. data) |
| (17) Calcinus hazletti | 179 | 12 | COI | Baums et al. (unpubl. data) |
| (18) Calcinus seurati | 161 | 4 | COI | Baums et al. (unpubl. data) |
| (19) Panulirus marginatus | 449 | 14 | COII | Iacchei et al. (unpubl. data) |
| (20) Panulirus penicillatus | 227 | 9 | COI | Iacchei et al. (unpubl. data) |
| Echinoderms: | ||||
| (21) Holothuria atra | 399 | 15 | COI | Skillings et al. [56] |
| (22) Holothuria whitmaei | 427 | 10 | COI | Skillings et al. (unpubl. data) |
| (23) Acanthaster planci | 338 | 11 | CR | Timmers et al. [57] |
| Scleractinian: | ||||
| (24) Montipora capitata | 551 | 13 | SSR | Concepcion et al. (unpubl. data) |
| (25) Porites lobata | 443 | 11 | SSR | Polato et al. [58] |
| Marine Mammals: | ||||
| (26) Monachus schauinslandi | 2409 | 8 | SSR | Schultz et al. [59, 60] |
| (27) Stenella longirostris | 386 | 8 | SSR, CR | Andrews et al. [61]. |
Table 2.
Sample size per location (refer to Figure 1 for Hawaiian names of sites) for each of the 27 species included in the combined multispecies analysis of population genetic structure across the Hawaiian Archipelago to date. Locations with fewer than 5 individuals were excluded from all analyses and are not included here (see text).
| Species Name | Hawai’i (=Big Island) |
Maui Nui |
O’ahu | Kaua’i | Ni’ihau | Nihoa | Necker Island |
French Frigate Shoals |
Gardner Pinnacles |
Maro Reef |
Laysan Island |
Lisianski | Pearl & Hermes Reef |
Midway Atoll’ |
Kure Atoll |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Hyporthodus (=Epinephelus) quernus | 36 | 30 | 9 | 30 | 44 | 30 | 30 | 46 | 29 | 27 | |||||
| Stegastes fasciohtus | 27 | 42 | 49 | 24 | 15 | 20 | 42 | ||||||||
| Dascylus albisella | 10 | 8 | 25 | 28 | 7 | 14 | |||||||||
| Ctenochaetus strigosus | 102 | 100 | 40 | 28 | 29 | 37 | 29 | 31 | 33 | 32 | 38 | ||||
| *Zebrasoma flavescens | 146 | 122 | 35 | 42 | 20 | 40 | 26 | 29 | 33 | 35 | |||||
| Acanthurus nigrofuscus | 33 | 65 | 39 | 26 | 39 | 33 | 28 | 28 | 6 | 9 | |||||
| Lutjanus kasmira | 101 | 39 | 50 | 36 | 49 | 40 | 21 | 40 | 9 | ||||||
| *Cellana exarata | 41 | 18 | 21 | 11 | 23 | 36 | |||||||||
| *Cellana sandwicensis | 42 | 21 | 20 | 18 | 8 | ||||||||||
| *Cellana talcosa | 43 | 24 | 38 | ||||||||||||
| Dendropoma gregaria | 53 | 25 | 39 | 20 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | ||||
| Dendropoma platypus | 16 | 20 | 40 | 21 | 5 | 5 | 5 | 5 | 5 | 6 | 5 | 5 | 5 | ||
| Dendropoma rhyssoconcha | 29 | 24 | 25 | 6 | 5 | ||||||||||
| Serpulorbis variabilis | 13 | 25 | 15 | 8 | |||||||||||
| Calcinus haigae | 51 | 21 | |||||||||||||
| Calcinus hazletti | 21 | 31 | 12 | 7 | 34 | 33 | 22 | 11 | |||||||
| Calcinus seurati | 9 | 30 | 78 | 44 | |||||||||||
| Panulirus marginatus | 19 | 48 | 56 | 56 | 53 | 53 | 36 | 47 | 29 | 46 | |||||
| Panulirus penicillatus | 46 | 43 | 47 | 5 | 33 | 5 | 18 | 30 | |||||||
| Holothuria atra | 30 | 24 | 30 | 5 | 28 | 12 | 37 | 35 | 23 | ||||||
| Holothuria whitmaei | 26 | 33 | 26 | 22 | 57 | 24 | 59 | ||||||||
| Acanthaster planci | 106 | 81 | 25 | 24 | 30 | 13 | 27 | ||||||||
| *Montipora capitata | 47 | 47 | 51 | 25 | 40 | 47 | 48 | 50 | 44 | 43 | 51 | ||||
| *Porites lobata | 23 | 16 | 18 | 21 | 23 | 34 | 33 | 44 | 21 | 22 | |||||
| Squalus mitsukurii | 7 | 91 | 8 | ||||||||||||
| Monachus schauinslandi | ________________54________________ | 7 | 766 | 656 | 310 | 260 | 134 | 222 | |||||||
| *Stenella longirostris | 79 | 59 | 40 | 32 | 45 | 33 | 47 | 119 | 51 | ||||||
Species with * show a significant isolation-by-distance signature across the archipelago.
2.2. Genetic Analyses
For each species, details of the analyses are provided in the studies cited in Table 1, or upon request from the authors. In brief, overall genetic variation was partitioned among sites as pairwise ΦST using the best fit model of sequence evolution, as determined by Modeltest 3.7 [62], that could be implemented by Arlequin 3.11 [63]. For most of the studies, FST was standardized for within population levels of heterozygosity [64, 65], and calculations of Dest [66] were done manually using formula macros in Microsoft Excel ([67] in review). The number and location of shared genetic breaks among species across the Archipelago are unchanged whether corrected or uncorrected F- statistics or Dest was used because the relative differences between these values are all highly correlated with our data set (data not shown). Because any set level of divergence selected is ultimately arbitrary, we use a significant pairwise FST among populations sampled on either side of the channel of interest as our metric of divergence. Significance of pairwise values was determined by permutation testing in Arlequin, with False Discovery Rate (FDR) correction for multiple tests [68] unless otherwise specified in the original publication (Table 1). Significant pairwise differences among adjacent islands, were overlaid visually on a map of the Hawaiian Archipelago (Figure 1) species-by-species. The number of significant pairwise differences among locations was summed across all 27 species and those that exceed random expectations (see below) are depicted in Figure 2.
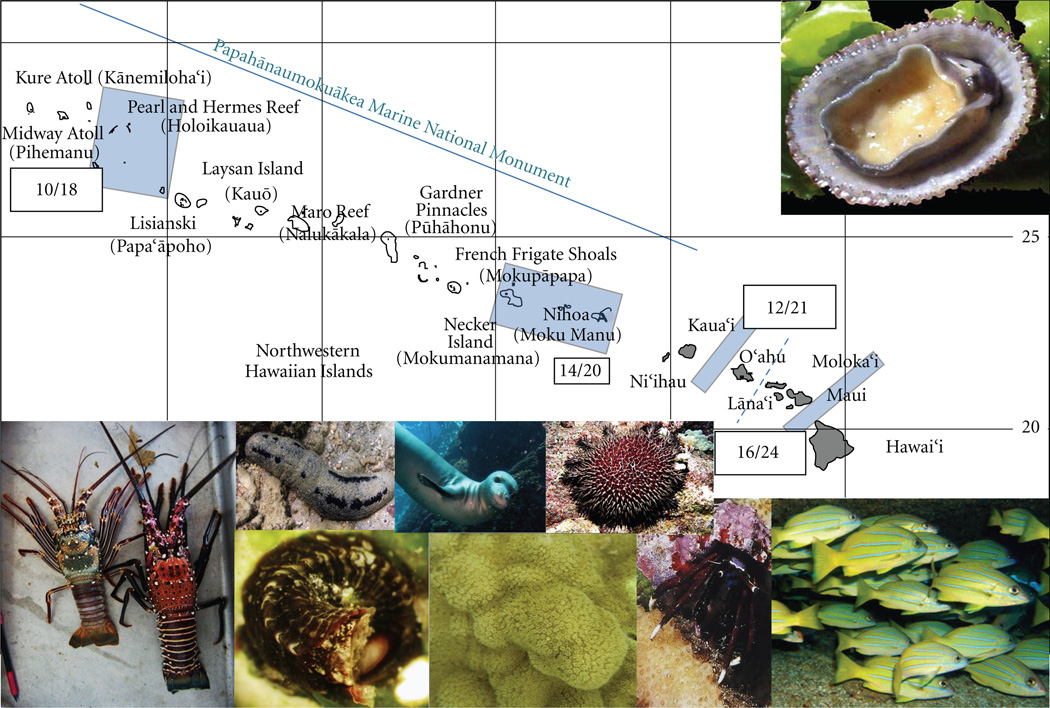
Figure 2.
Map of the Hawaiian Archipelago with significant consensus genetic breaks among the 27 taxa listed in Table 1 overlaid as blue bars between islands. In each bar, the number of species that show evidence for restricted gene flow across the barrier is listed in the numerator, and the total number of species for which we have data across that geographic area is listed in the denominator. The total number of sites included for each species is variable because not all species have been collected or analyzed at each site. The dotted line between Maui Nui and O’ahu highlights the location of the barrier that is shared by 8 of the surveyed species but is not significantly different than random expectations. The images include some of the species included in these analyses (left to right): Panulirus penicillatus, Panulirus marginatus, Holothuria atra, Dendropoma rhyssoconcha, Monachus schauinslandi, Porites lobata, Acanthaster planci, Calcinus hazletti, Lutjanus kasmira, and Cellana sandwicensis (photo credits to Derek Smith, Joe O’Malley, and the authors).
2.3. Statistical Testing of Shared Genetic Barriers
Because not all species are collected in all locations, we looked only at the channels between adjacent islands where samples of that species were available on both sides so that a test for pairwise population differentiation was possible at that site. We initially excluded any sites for which there were fewer than 20 individuals from each location on adjacent sides of the channel being tested, but found that the presence and location of barriers was unaffected in these analyses with any sample size greater than 5 individuals per site (data not shown). Thus, in the interest of including as much data as possible in this comparison, we include all sites for which the sample size was 5 or more (Table 2). We observed a total of 73 significant pairwise differences among the 178 possible pairwise tests for these species (Tables 1 and 2). The distribution of these pairwise differences was tested using a χ2 test with 13 degrees of freedom (14 between island channels); we calculate the expected number of the 73 pairwise differences that would occur, weighted by sample size between each island, at random within each channel. The validity of a shared genetic break at any given location was also tested using a χ2 to determine if the number of observed significant pairwise differences across the species sampled at that location differed significantly from the null expectation that all detected breaks were distributed equally among the 14 interisland channels.
3. Results
Although each species differs in the particular pattern of population structure and the inferred magnitude of larval exchange among sites, some consistent genetic breaks are apparent among these divergent species (Figure 2). In particular, the data indicate four strong barriers to gene flow in the channels between: (1) the Big Island of Hawai’i and Maui, (2) the islands of O’ahu and Kaua’i, (3) the MHI and NWHI, and (4) the far NW end and the rest of the NWHI chain around Pearl and Hermes Reef (Figure 2). The presence or absence and the strength of a given barrier vary among species (see references in Table 1). Likewise, there are some significant barriers that appear for only one or a few species, but do not appear in the majority of study organisms (e.g., Laysan Island for the sea cucumber, Holothuria atra, see Skillings et al. [56], or Gardner Pinnacles for the endemic grouper Epinephelus (=Hyporthodus) quernus, see Rivera et al. [51]).
Despite the vast differences in natural history among taxa, more than 50% of the species surveyed to date share the same four concordant barriers to gene flow across the Hawaiian Archipelago (Figure 2). Notably 8 of 19 species also show a break between O’ahu and Maui Nui, but this partition is not significantly different from random expectations (χ2 = 0.17, df = 1, p > 0.05). Essentially, roughly 50% of the sampled species must share a genetic discontinuity in order to deviate from the random expectation of 5.2 significant differences in each channel (χ2 = 4.4, df = 1, p < 0.05). Other than the four significant breaks depicted in Figure 2, and the nonsignificant split between O’ahu and Maui Nui, no other inter-island channel constitutes a barrier for more than 4 of the sampled species. Thus, with the caveat that additional sampling may yet demonstrate a fifth significant barrier between O’ahu and Maui Nui, there are currently four significant shared barriers to gene flow that divide the Hawaiian Archipelago into a minimum of five distinct ecoregions with limited exchange. In stark contrast to those locations, other inter-island channels have significantly fewer barriers than expected by chance (e.g., the region between French Frigate Shoals and Pearl & Hermes Atoll in the NWHI, χ2 = 3.85, df = 1, p = 0.05). This overall pattern of high connectivity among some locations and shared genetic barriers in others across the archipelago is significantly nonrandom (χ2 = 56.18, df = 13, p < 0.0001).
Distance alone is a poor predictor of the locations of these barriers to dispersal. The distance between areas that are isolated can be quite small (such as the ‘Alenuihāhā Channel between Hawai’i and Maui, ~45 km) whereas much larger distances between atolls in the NWHI generally show no consistent barriers to dispersal (for example Gardner Pinnacles is ~180km northwest of French Frigate Shoals). Likewise, more of the significant barriers to dispersal are found in the geographically smaller (600 km) MHI with the significant absence of barriers occurring in the geographically larger (2000 km) NWHI. Because adjacent sites can be highly differentiated whereas more distant sites are not, relatively few species (7/27) show a significant signal of isolation-by-distance across the Hawaiian archipelago (see Table 2 for highlighted exceptions).
4. Discussion
These data are striking in that more than half of the species surveyed show significant concordant barriers to gene flow concentrated in the four highlighted regions of the Archipelago (Figure 2). Given the broad differences in taxonomy, life history, and ecology of the species surveyed, including limpets, sea cucumbers, vermetid tube snails, reef fishes, monk seals, and spinner dolphins (Table 1), there is no a priori reason to expect that patterns of connectivity would be shared among the majority of the species. However, the four shared barriers to dispersal highlighted here indicate that these species are responding to common factors that limit dispersal and delineate independent units in terms of connectivity over management-relevant time scales. We hypothesize that the dominant factors are likely abiotic as opposed to biotic, given the diversity of species with radically divergent life histories that share the pattern of isolation.
4.1. Discordance between Genetic and Oceanographic Predictions
The most obvious candidates for such physical barriers to gene flow are geographic distance and oceanic currents. For most species there are enigmatic restrictions to dispersal that appear to have little to do with geographic distance. Many of the studies listed in Table 1 provide cases of divergence among proximate sites in the face of lower divergence among more distant sites elsewhere in the archipelago. Regardless, the overall dataset indicates that much of the NWHI is well connected despite greater average distances among the sites whereas the MHI show greater structure on average despite geographic proximity. Although some species do show isolation-by-distance, there appears to be a substantial taxonomic effect because three of the seven cases are sister species of Cellana limpets, and two of the remaining four are scleractinian corals (Table 2). While we cannot rule out the role of distance in limiting dispersal within the Hawaiian Archipelago, the impact of distance on the probability of dispersal does not appear to be a simple linear effect for the majority of species surveyed to date. This discord is not particularly surprising given the complexity of oceanographic current patterns. Recent analyses of larval dispersal in the Southern California Bight showed that probability of exchange among sites was uncorrelated with geographic distance, but strongly correlated with a derived “oceanographic distance” including realistic annual water movement patterns across many years [26, 69].
In Hawai’i, however, the patterns of genetic differentiation do not generally match predictions for larval dispersal based on water movement information from either a two-dimensional Eulerian advection-diffusion model [70, 71] or a Lagrangian particle-tracking model [72, 73]. One of the primary predictions of both simulation models is that the average distance of larval dispersal is short, roughly on the order of 50–150 km, and that the Main Hawaiian Islands (MHI) ought to be consistently connected and well mixed whereas the NWHI ought to show a number of isolated populations [70, 71]. For a PLD of less than about 45 days, the larval dispersal simulations predict a majority of local recruitment of larvae to their natal island/atoll or the adjacent ones (see [51]). In stark contrast to the primary prediction of the available larval dispersal simulation models (a well-mixed MHI and comparatively patchy NWHI), the consensus finding across 27 taxa to date is the opposite: the MHI show far more population structure than any equivalent geographical scale within the NWHI, and the primary dispersal barrier predicted by Eulerian simulation models is located in the region of the NWHI in which there is a significant paucity of population structure among surveyed species (Figure 2). Possible reasons for oceanographic simulations failing to predict the structure observed in the empirical genetic data are many (reviewed by [22, 74, 75]), but given the number and diversity of taxa across which the pattern holds, the genetic inference of isolation between the four regions highlighted in Figure 2 is robust.
4.2. Multispecies Approaches to Measuring Connectivity
All connectivity studies face practical limitations in terms of the number of specimens, sample sites, and taxonomic scope of study, which is why the vast majority of studies to date have focused on one or a few exemplar species to draw generalizations. Exemplar species are an attractive compromise to guide conservation and management efforts given the imposing logistic and resources challenges of conducting connectivity studies on all species of management relevance. However, the utility of exemplar species depends on whether they represent the community as a whole. Unfortunately, in most cases where this assumption has been tested explicitly, the patterns of dispersal and genetic structure differ significantly and unpredictably even among closely related species with similar life histories (e.g., [28, 52, 54, 76–78]). Despite the perceived potential for long-distance dispersal and broad mixing in the ocean, many taxa show unique archipelagic diversity (e.g., [79]) and even finer scale population structure than expected (e.g., [31, 80]). Regardless of whether we compare within taxonomic groups or between them, some of the species we have surveyed (e.g., Myripristis berndti, Centropyge loricula, Lutjanus kasmira, Acanthaster planci, and Calcinus spp.) appear to live up to their expected potential for dispersal and show no significant population structure across the Central Pacific (see [54, 57, 81–84]). In contrast, other species that appear capable of extensive dispersal (Epinephelus quernus, Ctenochaetus strigosus, Stenella longirostris, and Zebrasoma flavescens) show significant population differentiation within the Hawaiian Archipelago [50, 52, 53, 61, 85] and island-by-island or in some cases even site-by-site differences in population structure (e.g., [28, 44] Faucci et al. unpubl. data). Despite the potential for wide dispersal, Christie et al. [85] use individual parentage analyses to document self-recruitment in the Yellow Tang (Zebrasoma flavescens) and illustrate that at least some larvae recruit to the same region of the Kona coastline from which they were originally spawned.
Such variability among species greatly complicates efforts to generalize management implications from single-species studies and severely restricts the utility of exemplar species for decision making in conservation and management. While there is a consistent push to move beyond single-species management plans and implement EBM at a national and international level (e.g., [86, 87]), the exact geographic scale at which EBM should be applied is seldom obvious, and the accumulating data indicate that studies of marine connectivity cannot be generalized easily for this purpose. It is clearly impractical to study every species individually, and even if we could, how would the connectivity matrix from all those species be combined into a single coherent data set to guide EBM? For example, the multispecies conservation plan for U.S. federal lands states: “conservation objectives will not be achieved with a single reserve or a single population. Rather, local populations widely scattered across the landscape, but connected by movement, will be necessary. Few of these populations will be large enough to avoid problems faced by small populations, such as extirpation due to stochastic factors and inbreeding depression. Connectivity maintenance is therefore one of the most critical aspects of multispecies conservation. Connectivity, however, is notoriously difficult to directly measure” [88, p.64]. A variety of landscape genetic approaches to identifying cryptic barriers to connectivity have been proposed (e.g., [69, 89, 90]), but with few exceptions (e.g., [26]) such work has also been conducted on single-species. The push to implement EBM highlights an explicit need for multi-species comparisons of connectivity across all trophic levels to define the boundaries for management and resolve shared avenues of exchange among ecosystems.
Due to resource constraints as well as the logistical difficulties in completing such multispecies comparisons, only a few such studies exist. The few explicit multispecies connectivity studies that have been conducted to date (e.g., [29–31]) all face the limitation that there is no generally accepted method by which to analyze the aggregate connectivity data. Thus, like the study presented here, the primary method of analyzing shared genetic breaks is by counting the number or proportion of species that share a genetic discontinuity among locations. For example, a survey of 50 coastal marine species along the west coast of North America concluded that a greater proportion of species show significant genetic differentiation between the central (40% of species between Monterey, CA and Cape Blanco, OR) and northern sites (33% of species between Cape Blanco and Sitka, AK) than between the southern sites (15% of species between Monterey and Santa Barbara, CA; [30]). Likewise, a survey of 9 species of fish and 10 species of invertebrates in Indonesia defines partitions where more than two or three species share a phylogenetic break [31]. We have employed a similar approach with counting up shared genetic discontinuities in the data set, but elected to test whether these shared breaks deviate significantly from random. In our study, a surprisingly high number of species need to share a break to deviate significantly from random: even where 8 of the 19 species show differentiation between O’ahu and Maui Nui, that result was non-significant. The overall pattern of genetic divergence among sites within the Hawaiian Archipelago is highly non-random, with the central region of the NWHI having significantly fewer genetic breaks, and four individual channels emerge as having significantly more species sharing a break than expected at random (Figure 2).
There are substantial caveats to comparing FST and ΦST values directly among studies and marker classes (reviewed by [91–93]). Further, several recent publications have pointed out that the maximum attainable FST is inversely proportional to the mean within-population heterozygosity [64, 65], and therefore does not accurately measure population differentiation [66]. Thus, for highly polymorphic genetic markers, such as microsatellite loci, the maximum attainable FST is reduced far below one [64]. Contrary to the intuition that more polymorphic loci will reveal finer population structure, FST values are actually constrained to be lower as allelic diversity gets higher [67]. This limitation has led some to advocate the use of “true genetic differentiation” (Dest) as the primary or only means of comparison (e.g., [66]). While an attractive alternative in theory, there is as yet no means of significance testing for Dest, and researchers have to pick an arbitrary value at which to determine a genetic break before comparisons can be made; however, in the absence of statistics any cutoff value selected can be arbitrary and problematic [94]. For example, Kelly and Palumbi [30] chose ΦST = 0.10 as the delineation between strong (ΦST = 0.11–0.60) and moderate (ΦST = 0.02–0.10) population structure. While there is nothing wrong with this delineation, one could have also chosen ΦST = 0.05 or ΦST = 0.15 with equal justification, and there is no consistent and defensible level of population structure that determines the cut-off at which management decisions ought to be made [60]. Most published estimates of population structure remain uncorrected for marker variation and heterozygosity; thus, a value of 0.10 in one species may be on a completely different scale than in the next species if they have different levels of mean within population heterozygosity [64, 66, 67]. For this reason we use statistical significance as our cutoff and draw no inferences regarding the magnitude of the barriers beyond the number of species that share them. A method by which the boundaries of an ecosystem can be defined with multi-species data sets, and linkages between ecosystems can be quantified, is a logical prerequisite for successful implementation of EBM in the sea.
4.3. Connectivity in the Hawaiian Archipelago: Not 1 But at Least 5 Distinct Regions
The primary finding of this work is that the Hawaiian Archipelago is not a single, well-mixed community, but rather there are at least four significant multi-species barriers to dispersal along the length of the island chain. Additional sampling or more sophisticated statistical analyses may reveal additional barriers, but we report four strong concordant breaks here. As outlined above, some species cross these barriers, others do not, and the patterns of connectivity can, and do, vary dramatically among individual species (see refs. in Table 1). Regardless, a strong and consistent pattern emerges from the multispecies comparison in which the majority of 27 taxonomically diverse species share four significant concordant genetic breaks across the archipelago. It is noteworthy that the variability among individual species studies of connectivity published to date certainly does not lend itself to an expectation of such strong concordant patterns. Despite the suite of taxonomic, ecological, and biological differences that might lead us to expect highly divergent patterns among these diverse taxa, some unknown barriers appear to consistently limit dispersal in a majority of the 27 species surveyed to date. These results illustrate that while a single species is rarely representative of the average connectivity, concordant patterns can emerge when many species are examined simultaneously. Insofar as this is a general result, it would mandate that a broad suite of reef species across multiple taxonomic groups and ecological niches ought to be surveyed to resolve general trends and to provide connectivity information pertinent to management of any large marine management area such as the PMNM.
The two primary caveats to this finding are that: (1) the basis for these shared genetic restrictions is poorly understood and discovering the location of these barriers is only the first step, and (2) it is an overly simplistic statistical model to show significant deviations from random pairwise differences across species as a measure of the strength of dispersal barriers. Nonetheless, such summing is the primary means of comparison available at this time, and this is the only multispecies study that employs even this simplistic statistical approach. In terms of the first caveat, it will be valuable to determine the ecological and oceanographic factors driving regional, island, or site specific genetic structure; this will likely be important for ecosystem-based management of both the Main and Northwestern Hawaiian Islands, and may provide general characteristics to predict ecosystem-level partitions among coral reefs elsewhere. Discovering the existence and location of these barriers leads to questions about the underlying cause for so many species sharing these concordant patterns, and what maintains those barriers to dispersal among taxa as diverse as limpets and dolphins. In terms of the second caveat, as outlined above, we need to develop a quantitative method for multispecies studies of connectivity among many locations. Ultimately, it would be ideal to bring the multispecies data sets together in a single analysis to determine both the relative strength and statistical confidence in each of the detected barriers, but no such method exists currently.
4.4. Conclusions and Management Implications
This multi-species approach to understanding population connectivity across the Hawaiian Archipelago reveals four previously unrecognized barriers to dispersal that delineate five relatively isolated regions of the Hawaiian Archipelago. In contrast to predictions based on either geographic distance between islands (isolation by distance) or on larval dispersal model predictions using pelagic larval duration, there are more barriers to dispersal within the Main Hawaiian Islands (MHI, ~600 km) than the Northwestern Hawaiian Islands (NWHI, ~2000 km). The underlying mechanism of this isolation remains unknown, but the concordance across 52% to 70% (depending on the barrier) of the 27 taxonomically and ecologically divergent species sampled here demonstrates that the pattern is robust and likely to derive from physical rather than biologically intrinsic factors.
These data provide information pertinent to current management issues facing the broader Pacific and efforts to implement ecosystem-based management (EBM) in Hawai’i. In particular, these data directly address the controversy about whether the NWHI is a series of isolated (and therefore relatively fragile) island ecosystems, and whether the Papaphānaumokuākea Marine National Monument provides spillover benefits to the highly exploited waters of the MHI [35]. We find that the NWHI are far more connected on average (and therefore comparatively robust) than the MHI, but that connectivity between the MHI and NWHI is limited. The results highlight that the Main Hawaiian Islands are isolated in terms of resource management and will not receive substantial subsidy from the Papahānaumokuākea Marine National Monument; the MHI must stand alone in management of marine resources. Furthermore, even the comparatively small MHI are not a single panmictic unit, and future management plans should incorporate knowledge of the substantial isolation among multiple regions within the MHI. For example, Bird et al. [28] argue that each island should be considered a separate management unit for the culturally important Hawaiian limpets (‘opihi, genus Cellana). Likewise, the impact of invasive species is felt globally and with 343 alien marine species documented in Hawai’i thus far [95], there is considerable concern regarding the vulnerability of Hawaiian reefs to invasion and the likely spread of aliens that are already introduced. Our findings predict barriers through which invasive species should have difficulty advancing, and indeed recent studies of several species of invasive fishes and invertebrates appear to corroborate those predictions (e.g., [96, 97].
This study is one of the few multispecies surveys of marine connectivity to date and confirms that this approach can illuminate general patterns pertinent to management that do not emerge from single-species exemplar studies. The manner in which policy makers delineate the boundaries for ecosystem-based management remains a subject of considerable debate, but we argue this multispecies approach offers a possible solution. Here, we resolve concordant patterns of connectivity in an objective and quantitative manner to define a minimum of five marine spatial management units in the Hawaiian Archipelago.
Acknowledgments
The authors thank the Papahānaumokuākea Marine National Monument, US Fish and Wildlife Services, and Hawai’i Division of Aquatic Resources (DAR) for coordinating research activities and permitting, and the U.S. National Oceanic and Atmospheric Administration (NOAA) research vessel Hi’ialakai and her crew for years of outstanding service and support. Special thanks go to J. Leong, S. Karl, S. Godwin, R. Kosaki and the members of the ToBo Lab. We could not have completed this work without the assistance of the UH Dive Safety Program, U.S. National Marine Fisheries Service, the Pacific Island Fisheries Science Center, National Marine Sanctuaries Program, and Coral Reef Ecosystem Division, especially: A. Tom, A.Wilhelm, H. Johnson, M. Pai, D. Carter, C. Kane, C. Meyer, D. Smith, C. Kelley, D. Minton, P. Reath, J. Zardus, D. Croswell, B. Holland, M. Stat, X. Pochon, M. Rivera, E. Brown, M. Ramsay, J. Maragos, L. Eldredge, H. Bollick, S. Coles, W. Walsh, B. Carmen, I. Williams, A. Friedlander, J. Randall, S. Cotton, A. Montgomery, S. Pooley, M. Seki, J. Zamzow, E. DeMartini, J. Polovina, R. Humphreys, D. Kobayashi, F. Parrish, R. Moffitt, G. DiNardo, J. O’Malley, R. Brainard, J. Kenyon, K. Schultz, M. Duarte, H. Kawelo, E. Fielding, L. Sorenson, L. Basch, A. Alexander, K. Selkoe, M. Craig, L. Rocha, Z. Forsman, Z. Szabo, C. Musberger, D. White, K. Tenggardjaja, Y. Papastamatiou, K. Gorospe, B. Wainwright, S. Daley, M. Crepeau, A. Eggers, and the HIMB EPSCoR Core Genetics Facility a sincere thanks to you all. We also appreciate the feedback of the anonymous reviewers and B. Halpern whose helpful comments greatly improved and clarified the text. This work was funded in part by grants from the National Science Foundation (DEB no. 99–75287, OCE no. 04–54873, OCE no. 05–50294, OCE no. 06–23678, OCE no. 09–29031), National Marine Sanctuaries NWHICRER-HIMB partnership (MOA-2005-008/6882), University of Hawai’i Sea Grant College Program, National Park Service PICRP, National Marine Fisheries Service, Western Pacific Regional Fishery Management Council, NOAA’s Coral Reef Conservation Program, the Hawai’i Coral Reef Initiative, NSF EPSCoR, EPA STAR Fellowship, the Watson T. Yoshimoto Foundation, the Jessie D. Kay Memorial Fellowship, UH Graduate Student Organization Grants Program, PADI Foundation Research Grant, Charles and Margaret Edmondson Research Fund, American Malacological Society Student Award, Conchologists of America Research Grant, Sigma Xi Grants-in-Aid, Society for Integrative and Comparative Biology Student Award, Western Society of Malacologists Student Award, and the Ecology, Evolution, and Conservation Biology (EECB) NSF GK-12 fellowships. This is HIMB contribution no. 1422 and SOEST no. 8051.
References
- 1.Pauly D, Christensen V, Dalsgaard J, Froese R, Torres F. Fishing down marine food webs. Science. 1998;279(5352):860–863. doi: 10.1126/science.279.5352.860. [DOI] [PubMed] [Google Scholar]
- 2.Pauly D, Watson R. Counting the last fish. Scientific American. 2003;289(1):42–47. doi: 10.1038/scientificamerican0703-42. [DOI] [PubMed] [Google Scholar]
- 3.Worm B, Barbier EB, Beaumont N, et al. Impacts of biodiversity loss on ocean ecosystem services. Science. 2006;314(5800):787–790. doi: 10.1126/science.1132294. [DOI] [PubMed] [Google Scholar]
- 4.McLeod KL, Leslie HM. Why ecosystem-based management? In: McLeod KL, Leslie HM, editors. Ecosystem-Based Management for the Oceans. Washington, DC, USA: Island Press; 2009. [Google Scholar]
- 5.Spalding MD, Fox HE, Allen GR, et al. Marine ecoregions of the world: a bioregionalization of coastal and shelf areas. BioScience. 2007;57(7):573–583. [Google Scholar]
- 6.Kappel CV. Threats to marine biodiversity and considerations for its conservation: from species to seascapes, Ph.D. dissertation. Stanford University; 2006. [Google Scholar]
- 7.Sanchirico JN, Wilen JE. Optimal spatial management of renewable resources: matching policy scope to ecosystem scale. Journal of Environmental Economics and Management. 2005;50(1):23–46. [Google Scholar]
- 8.Hedgecock D, Barber PH, Edmands S. Genetic approaches to measuring connectivity. Oceanography. 2007;20:70–79. [Google Scholar]
- 9.Thorrold SR, Zacherl DC, Levin LA. Population connectivity and larval dispersal: using geochemical signatures in calcified structures. Oceanography. 2007;20:80–89. [Google Scholar]
- 10.Fogarty MJ, Botsford L. Population connectivity and spatial management of marine fisheries. Oceanography. 2007;20:112–123. [Google Scholar]
- 11.Thorson G. Reproductive and larval ecology of marine bottom invertebrates. Biological Review. 1950;25:1–45. doi: 10.1111/j.1469-185x.1950.tb00585.x. [DOI] [PubMed] [Google Scholar]
- 12.Strathmann RR. Hypotheses on the origins of marine larvae. Annual Review of Ecology and Systematics. 1993;24:89–117. [Google Scholar]
- 13.Kinlan BP, Gaines SD, Lester SE. Propagule dispersal and the scales of marine community process. Diversity and Distributions. 2005;11(2):139–148. [Google Scholar]
- 14.Lessios HA, Robertson DR. Crossing the impassable: genetic connections in 20 reef fishes across the eastern Pacific barrier. Proceedings of the Royal Society B. 2006;273(1598):2201–2208. doi: 10.1098/rspb.2006.3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dawson MN, Hamner WM. A biophysical perspective on dispersal and the geography of evolution in marine and terrestrial systems. Journal of the Royal Society Interface. 2008;5(19):135–150. doi: 10.1098/rsif.2007.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levin LA. Recent progress in understanding larval dispersal: new directions and digressions. Integrative and Comparative Biology. 2006;46(3):282–297. doi: 10.1093/icb/icj024. [DOI] [PubMed] [Google Scholar]
- 17.Grosberg RK, Cunningham CW. Genetic structure in the sea: from populations to communities. In: Bertness MD, Gaines SD, Hay ME, editors. Marine Community Ecology. Sunderland, Mass, USA: Sinauer; 2001. pp. 61–84. [Google Scholar]
- 18.Selkoe KA, Halpern BS, Ebert CM, et al. A map of cumulative impacts to a “pristine” coral reef ecosystem, the Papahānaumokuākea. Coral Reefs. 2009;28(3):635–650. [Google Scholar]
- 19.Hellberg ME. Gene flow and isolation among populations of marine animals. Annual Review of Ecology, Evolution, and Systematics. 2009;40:291–310. [Google Scholar]
- 20.Bradbury IR, Bentzen P. Non-linear genetic isolation by distance, life history, and dispersal estimation in aquatic organisms. Marine Ecology Progress Series. 2007;340:245–257. [Google Scholar]
- 21.Bradbury IR, Laurel B, Snelgrove PVR, Bentzen P, Campana SE. Global patterns in marine dispersal estimates: the influence of geography, taxonomic category and life history. Proceedings of the Royal Society B. 2008;275(1644):1803–1810. doi: 10.1098/rspb.2008.0216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weersing K, Toonen RJ. Population genetics, larval dispersal, and connectivity in marine systems. Marine Ecology Progress Series. 2009;393:1–12. [Google Scholar]
- 23.Shanks AL. Pelagic larval duration and dispersal distance revisited. Biological Bulletin. 2009;216(3):373–385. doi: 10.1086/BBLv216n3p373. [DOI] [PubMed] [Google Scholar]
- 24.Ross PM, Hogg ID, Pilditch CA, Lundquist CJ. Phylogeography of New Zealand’s coastal benthos. New Zealand Journal of Marine & Freshwater Research. 2009;43:1009–1027. [Google Scholar]
- 25.Riginos C, Douglas KE, Jin Y, Shanahan DF, Treml EA. Effects of geography and life history traits on genetic differentiation in benthic marine fishes. Ecography. In press. [Google Scholar]
- 26.Selkoe KA, Watson JR, White C, et al. Taking the chaos out of genetic patchiness: seascape genetics reveals ecological and oceanographic drivers of genetic patterns in three temperate reef species. Molecular Ecology. 2010;19(17):3708–3726. doi: 10.1111/j.1365-294X.2010.04658.x. [DOI] [PubMed] [Google Scholar]
- 27.Rocha LA, Bass AL, Robertson DR, Bowen BW. Adult habitat preferences, larval dispersal, and the comparative phylogeography of three Atlantic surgeonfishes (Teleostei: Acanthuridae) Molecular Ecology. 2002;11(2):243–252. doi: 10.1046/j.0962-1083.2001.01431.x. [DOI] [PubMed] [Google Scholar]
- 28.Bird CE, Holland BS, Bowen BW, Toonen RJ. Contrasting phylogeography in three endemic Hawaiian limpets (Cellana spp.) with similar life histories. Molecular Ecology. 2007;16(15):3173–3186. doi: 10.1111/j.1365-294X.2007.03385.x. [DOI] [PubMed] [Google Scholar]
- 29.Avise JC. Molecular population structure and the biogeographic history of a regional fauna: a case history with lessons for conservation biology. Oikos. 1992;63:62–76. [Google Scholar]
- 30.Kelly RP, Palumbi SR. Genetic structure among 50 species of the northeastern Pacific rocky intertidal community. PLoS One. 2010;5(1) doi: 10.1371/journal.pone.0008594. Article ID e8594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carpenter KE, Barber PH, Crandall E, et al. Comparative phylogeography of the coral triangle and implications for marine management. Journal of Marine Biology. 2011:14. Article ID 396982 2011. [Google Scholar]
- 32.Pandolfi JM, Bradbury RH, Sala E, et al. Global trajectories of the long-term decline of coral reef ecosystems. Science. 2003;301(5635):955–958. doi: 10.1126/science.1085706. [DOI] [PubMed] [Google Scholar]
- 33.DeMartini EE, Friedlander AM. Spatial patterns of endemism in shallow-water reef fish populations of the Northwestern Hawaiian Islands. Marine Ecology Progress Series. 2004;271:281–296. [Google Scholar]
- 34.Friedlander AM, DeMartini EE. Contrasts in density, size, and biomass of reef fishes between the Northwestern and the Main Hawaiian islands: the effects of fishing down apex predators. Marine Ecology Progress Series. 2002;230:253–264. [Google Scholar]
- 35.Williams ID, Walsh WJ, Schroeder RE, Friedlander AM, Richards BL, Stamoulis KA. Assessing the importance of fishing impacts on Hawaiian coral reef fish assemblages along regional-scale human population gradients. Environmental Conservation. 2008;35(3):261–272. [Google Scholar]
- 36.Friedlander AM, Aeby GS, Brainard RB, et al. The state of coral reef ecosystems of the Northwestern Hawaiian Islands. In: Waddell JE, editor. The State of Coral Reef Ecosystems of the United States and Pacific Freely Associated States. NOAA/NCCOS Center for Coastal Monitoring and Assessment’s Biogeography Team; 2005. pp. 270–311. NOAA Technical Memorandum NOS NCCOS 11. [Google Scholar]
- 37.Rodgers KS, Jokiel PL, Bird CE, Brown EK. Quantifying the condition of Hawaiian coral reefs. Aquatic Conservation: Marine and Freshwater Ecosystems. 2010;20(1):93–105. [Google Scholar]
- 38.Fautin D, Dalton P, Incze LS, et al. An overview of marine biodiversity in United States waters. PLoS One. 2010;5(8) doi: 10.1371/journal.pone.0011914. Article ID e11914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abbott IA. Marine Red Algae of the Hawaiian Islands. Honolulu, Hawaii, USA: Bishop Museum Press; 1999. [Google Scholar]
- 40.Ziegler AC. Hawaiian Natural History, Ecology & Evolution. Honolulu, Hawaii, USA: University of Hawaii Press; 2002. [Google Scholar]
- 41.Eldredge LG, Evenhuis NL. Hawaii’s biodiversity: a detailed assessment of the numbers of species in the Hawaiian Islands. Bishop Museum Occasional Papers 76. 2003:28. http://hbs.bishopmuseum.org/pdf/op76.pdf.
- 42.Randall JE. Reef and Shore Fishes of the Hawaiian Islands. Honolulu, Hawaii, USA: University of Hawaii Sea Grant Program; 2007. [Google Scholar]
- 43.Selkoe KA, Henzler CM, Gaines SD. Seascape genetics and the spatial ecology of marine populations. Fish and Fisheries. 2008;9(4):363–377. [Google Scholar]
- 44.Ramon M, Nelson P, DeMartini E, Walsh W, Bernardi G. Phylogeography, historical demography, and the role of post-settlement ecology in two Hawaiian damselfish species. Marine Biology. 2008;153(6):1207–1217. [Google Scholar]
- 45.Iacchei M, Toonen RJ. Caverns, compressed air, and crustacean connectivity: insights into Hawaiian spiny lobster populations. In: Pollock NW, editor. Diving for Science 2010. Proceedings of the 29th American Academy of Underwater Sciences Symposium. Dauphin Island, Ala, USA: American Academy of Underwater Sciences; 2010. [Google Scholar]
- 46.Skillings DJ, Toonen RJ. It’s just a flesh wound: non-lethal sampling for conservation genetics studies. In: Pollock NW, editor. Diving for Science. Proceedings of the 29th American Academy of Underwater Sciences Symposium; Proceedings of the American Academy of Underwater Sciences; Dauphin Island, Ala, USA: AAUS; 2010. [Google Scholar]
- 47.Seutin G, White BN, Boag PT. Preservation of avian blood and tissue samples for DNA analyses. Canadian Journal of Zoology. 1991;69:82–90. [Google Scholar]
- 48.Concepcion GT, Medina M, Toonen RJ. Noncoding mitochondrial loci for corals. Molecular Ecology Notes. 2006;6(4):1208–1211. [Google Scholar]
- 49.Sunnucks P, Hales DF. Numerous transposed sequences of mitochondrial cytochrome oxidase I-II in aphids of the genus Sitobion (Hemiptera: Aphididae) Molecular Biology and Evolution. 1996;13(3):510–524. doi: 10.1093/oxfordjournals.molbev.a025612. [DOI] [PubMed] [Google Scholar]
- 50.Rivera MAJ, Kelley CD, Roderick GK. Subtle population genetic structure in the Hawaiian grouper, Epinephelus quernus (Serranidae) as revealed by mitochondrial DNA analyses. Biological Journal of the Linnean Society. 2004;81(3):449–468. [Google Scholar]
- 51.Rivera M, Andrews KR, Kobayashi DR, et al. Genetic analyses and simulations of larval dispersal reveal distinct populations and directional connectivity across the range of the Hawaiian Grouper (Epinephelus quernus) Journal of Marine Biology. 2011;11:11. Article ID 765353. [Google Scholar]
- 52.Eble JA, Toonen RJ, Bowen BW. Endemism and dispersal: comparative phylogeography of three surgeonfishes across the Hawaiian Archipelago. Marine Biology. 2009;156(4):689–698. [Google Scholar]
- 53.Eble JA, Sorenson LS, Papastamatiou YP, Basch L, Toonen RJ, Bowen BW. Escaping paradise: Larval export from Hawaii in an Indo-Pacific reef fish, the Yellow Tang. Marine Ecology Progress Series. doi: 10.3354/meps09083. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gaither MR, Bowen BW, Toonen RJ, et al. Genetic consequences of introducing allopatric lineages of Bluestriped Snapper (Lutjanus kasmira) to Hawaii. Molecular Ecology. 2010;19(6):1107–1121. doi: 10.1111/j.1365-294X.2010.04535.x. [DOI] [PubMed] [Google Scholar]
- 55.Daly-Engel TS, Grubbs RD, Feldheim KA, Bowen BW, Toonen RJ. Is multiple mating beneficial or unavoidable? Low multiple paternity and genetic diversity in the shortspine spurdog Squalus mitsukurii. Marine Ecology Progress Series. 2010;403:255–267. [Google Scholar]
- 56.Skillings DJ, Bird CE, Toonen RJ. Gateways to Hawai’i: genetic population structure of the tropical sea cucumber Holothuria atra. Journal of Marine Biology. 2011:16. Article ID 783030. [Google Scholar]
- 57.Timmers MA, Andrews KR, Bird CE, de Maintenon MJ, Brainard RE, Toonen RJ. Widespread dispersal of the crown-of-thorns sea star, Acanthaster planci, across the Hawaiian Archipelago and Johnston Atoll. Journal of Marine Biology. 2011:10. Article ID 934269. [Google Scholar]
- 58.Polato NR, Concepcion GT, Toonen RJ, Baums IB. Isolation by distance across the Hawaiian Archipelago in the reef-building coral Porites lobata. Molecular Ecology. 2010;19(21):4661–4677. doi: 10.1111/j.1365-294X.2010.04836.x. [DOI] [PubMed] [Google Scholar]
- 59.Schultz JK, Baker JD, Toonen RJ, Bowen BW. Extremely low genetic diversity in the endangered Hawaiian monk seal (Monachus schauinslandi) Journal of Heredity. 2009;100(1):25–33. doi: 10.1093/jhered/esn077. [DOI] [PubMed] [Google Scholar]
- 60.Schultz JK, Baker JD, Toonen RJ, Harting AL, Bowen BW. Range-wide genetic connectivity of the Hawaiian Monk Seal and implications for translocation. Conservation Biology. 2010 doi: 10.1111/j.1523-1739.2010.01615.x. In press. [DOI] [PubMed] [Google Scholar]
- 61.Andrews KR, Karczmarski L, Au WWL, et al. Rolling stones and stable homes: social structure, habitat diversity and population genetics of the Hawaiian spinner dolphin (Stenella longirostris) Molecular Ecology. 2010;19(4):732–748. doi: 10.1111/j.1365-294X.2010.04521.x. [DOI] [PubMed] [Google Scholar]
- 62.Posada D, Crandall KA. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14(9):817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 63.Excoffier L, Laval LG, Schneider S. ARLEQUIN ver.3.0: an integrated software package for population genetics data analysis. Evolutionary Bioinformatics. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- 64.Hedrick PW. A standardized genetic differentiation measure. Evolution. 2005;59(8):1633–1638. [PubMed] [Google Scholar]
- 65.Meirmans PG. Using the AMOVA framework to estimate a standardized genetic differentiation measure. Evolution. 2006;60(11):2399–2402. [PubMed] [Google Scholar]
- 66.Jost L. GST and its relatives do not measure differentiation. Molecular Ecology. 2008;17(18):4015–4026. doi: 10.1111/j.1365-294x.2008.03887.x. [DOI] [PubMed] [Google Scholar]
- 67.Bird CE, Karl SA, Toonen RJ. Koenemann S, Held C, Schubart C, editors. Detecting and measuring genetic differentiation. Crustacean Issues: Phylogeography and Population Genetics in Crustacea. in review. [Google Scholar]
- 68.Benjamini Y, Yekutieli D. The control of the false discovery rate in multiple testing under dependency. Annals of Statistics. 2001;29(4):1165–1188. [Google Scholar]
- 69.White C, Selkoe KA, Watson J, Siegel DA, Zacherl DC, Toonen RJ. Ocean currents help explain population genetic structure. Proceedings of the Royal Society B. 2010;277(1688):1685–1694. doi: 10.1098/rspb.2009.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Treml EA, Halpin PN, Urban DL, Pratson LF. Modeling population connectivity by ocean currents, a graph-theoretic approach for marine conservation. Landscape Ecology. 2008;23(1):19–36. [Google Scholar]
- 71.Friedlander AM, Kobayashi D, Bowen BW, et al. Papahānaumokuākea Monument Biogeographic Assessment Report. NOAA Technical Memorandum; 2009. Chapter 9: connectivity and integrated ecosystem studies; pp. 291–330. [Google Scholar]
- 72.Kobayashi DR. Colonization of the Hawaiian Archipelago via Johnston Atoll: a characterization of oceanographic transport corridors for pelagic larvae using computer simulation. Coral Reefs. 2006;25(3):407–417. [Google Scholar]
- 73.Kobayashi DR, Polovina JJ. Simulated seasonal and interannual variability in larval transport and oceanography in the Northwestern Hawaiian Islands using satellite remotely sensed data and computer modeling. Atoll Research Bulletin. 2006;543:365–390. [Google Scholar]
- 74.Woodson CB, McManus MA. Foraging behavior can influence dispersal of marine organisms. Limnology and Oceanography. 2007;52(6):2701–2709. [Google Scholar]
- 75.Paris CB, Chérubin LM, Cowen RK. Surfing, spinning, or diving from reef to reef: effects on population connectivity. Marine Ecology Progress Series. 2007;347:285–300. [Google Scholar]
- 76.Marko PB. ‘What’s larvae got to do with it?’ Disparate patterns of post-glacial population structure in two benthic marine gastropods with identical dispersal potential. Molecular Ecology. 2004;13(3):597–611. doi: 10.1046/j.1365-294x.2004.02096.x. [DOI] [PubMed] [Google Scholar]
- 77.Crandall ED, Frey MA, Grosberg RK, Barber PH. Contrasting demographic history and phylogeographical patterns in two Indo-Pacific gastropods. Molecular Ecology. 2008;17(2):611–626. doi: 10.1111/j.1365-294X.2007.03600.x. [DOI] [PubMed] [Google Scholar]
- 78.Rocha LA, Craig MT, Bowen BW. Phylogeography and the conservation of coral reef fishes. Coral Reefs. 2007;26(3):501–512. [Google Scholar]
- 79.Meyer CP, Geller JB, Paulay G. Fine scale endemism on coral reefs: archipelagic differentiation in turbinid gastropods. Evolution. 2005;59(1):113–125. [PubMed] [Google Scholar]
- 80.Paulay G, Meyer C. Dispersal and divergence across the greatest ocean region: do larvae matter? Integrative and Comparative Biology. 2006;46(3):269–281. doi: 10.1093/icb/icj027. [DOI] [PubMed] [Google Scholar]
- 81.Craig MT, Eble JA, Bowen BW, Robertson DR. High genetic connectivity across the Indian and Pacific Oceans in the reef fish Myripristis berndti (Holocentridae) Marine Ecology Progress Series. 2007;334:245–254. [Google Scholar]
- 82.Schultz JK, Pyle RL, DeMartini E, Bowen BW. Genetic connectivity among color morphs and Pacific archipelagos for the flame angelfish, Centropyge loriculus. Marine Biology. 2007;151(1):167–175. [Google Scholar]
- 83.Craig MT, Eble JA, Bowen BW. Origins, ages and population histories: comparative phylogeography of endemic Hawaiian butterflyfishes (genus Chaetodon) Journal of Biogeography. 2010;37(11):2125–2136. [Google Scholar]
- 84.Eble JA, Rocha LA, Craig MT, Bowen BW. Not all larvae stay close to home: insights into marine population connectivity with a focus on the Brown Surgeonfish. Journal of Marine Biology. 2010 doi: 10.1155/2011/518516. Article ID 518516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Christie MR, Tissot BN, Albins MA, et al. Larval connectivity in an effective network of marine protected areas. PLoS One. 2010;5(12) doi: 10.1371/journal.pone.0015715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Slocombe DS. Implementing ecosystem-based management. BioScience. 1993;43(9):612–622. [Google Scholar]
- 87.Lubchenco J, Palumbi SR, Gaines SD, Andelman S. Plugging a hole in the ocean: the emerging science of marine reserves. Ecological Applications. 2003;13(1):S3–S7. [Google Scholar]
- 88.Noon BR, McKelvey KS, Dickson BG. Multispecies conservation planning on U.S. federal lands. In: Millspaugh JJ, Thompson FR III, editors. Models for Planning Wildlife Conservation in Large Landscapes. New York, NY, USA: Elsevier; 2009. pp. 51–83. [Google Scholar]
- 89.Manel S, Schwartz MK, Luikart G, Taberlet P. Landscape genetics: combining landscape ecology and population genetics. Trends in Ecology and Evolution. 2003;18(4):189–197. [Google Scholar]
- 90.Storfer A, Murphy MA, Evans JS, et al. Putting the ‘landscape’ in landscape genetics. Heredity. 2007;98(3):128–142. doi: 10.1038/sj.hdy.6800917. [DOI] [PubMed] [Google Scholar]
- 91.Whitlock MC, Mccauley DE. Indirect measures of gene flow and migration: FST doesn’t equal 1/(4Nm + 1) Heredity. 1999;82(2):117–125. doi: 10.1038/sj.hdy.6884960. [DOI] [PubMed] [Google Scholar]
- 92.Neigel JE. A comparison of alternative strategies for estimating gene flow from genetic markers. Annual Review of Ecology and Systematics. 1997;28:105–128. [Google Scholar]
- 93.Neigel JE. Is FST obsolete? Conservation Genetics. 2002;3(2):167–173. [Google Scholar]
- 94.Gerlach G, Jueterbock A, Kraemer P, Deppermann J, Harmand P. Calculations of population differentiation based on GST and D: forget GST but not all of statistics. Molecular Ecology. 2010;19(18):3845–3852. doi: 10.1111/j.1365-294X.2010.04784.x. [DOI] [PubMed] [Google Scholar]
- 95.Eldredge LG, Smith CM, editors. “A guidebook of introduced marine species in Hawai’i,” Bishop Museum Technical Report. 2001;21:65. [Google Scholar]
- 96.Concepcion GT, Kahng SE, Crepeau MW, Franklin EC, Coles SL, Toonen RJ. Resolving natural ranges and marine invasions in a globally distributed octocoral (genus Carijoa) Marine Ecology Progress Series. 2010;401:113–127. [Google Scholar]
- 97.Gaither MR, Toonen RJ, Robertson DR, Planes S, Bowen BW. Genetic evaluation of marine biogeographical barriers: perspectives from two widespread Indo-Pacific snappers (Lutjanus kasmira and Lutjanus fulvus) Journal of Biogeography. 2010;37(1):133–147. [Google Scholar]