Abstract
Intrinsic β-lactam resistance in Stenotrophomonas maltophilia is caused by blaL1 and/or blaL2, a kind of metallo-β-lactamase with a broad substrate spectrum including carbapenems. A rapid and sensitive molecular method for the detection of blaL1 in clinical samples is needed to guide therapeutic treatment. In present study, we first described a loop-mediated isothermal amplification (LAMP) method for the rapid detection of blaL1 in clinical samples by using two methods including a chromogenic method using calcein/Mn2+ complex and the real-time turbidity monitoring to assess the reaction. Then dissemination of L1-producing S. maltophilia was investigated from ICU patients in three top hospital in Beijing, China. The results showed that both methods detected the target DNA within 60 min under isothermal conditions (65°C). The detection limit of LAMP was 3.79 pg/μl DNA, and its sensitivity 100-fold greater than that of conventional PCR. All 21 test strains except for S. maltophilia were negative for blaL1, indicative of the high-specificity of the primers for the blaL1. A total of 22 L1-positive isolates were identified for LAMP-based surveillance of blaL1 from 105 ICU patients with clinically suspected multi-resistant infections. The sequences of these blaL1 genes were conservative with only a few sites mutated, and the strains had highly resistant to β-lactam antibiotics. The MLST recovered that 22 strains belonged to seven different S. maltophilia sequence types (STs). Furthermore, co-occurrence of blaL1 and blaL2 genes were detected in all of isolates. Strikingly, S. maltophilia DCPS-01 was recovered to contain blaL1, blaL2, and blaNDM-1 genes, possessing an ability to hydrolyse all β-lactams antibiotics. Our data showed the diversity types of S. maltophilia carrying blaL1 and co-occurrence of many resistant genes in the clinical strains signal an ongoing and fast evolution of S. maltophilia resulting from their wide spread in the respiratory infections, and therefore will be difficult to control.
Keywords: L1 metallo-β-lactamase, S. maltophilia, LAMP, rapid diagnosis, prevalence
INTRODUCTION
Stenotrophomonas maltophilia, commonly associated with respiratory infections in children and adults, is an emerging Gram-negative MDRO (multi-drug-resistant organism) of global significance (Brooke, 2012). Currently, the incidence of S. maltophilia nosocomial infection is increasing, particularly for the immunocompromised (cancer, cystic fibrosis, drug addicts, and AIDS patients), dialysis patients, recipients of organ transplants, in addition to the reported cases of community-acquired S. maltophilia. The low outer membrane permeability of S. maltophilia renders it inherently resistant to most antibiotics, whilst the production of group 2e β-lactamase (L2) and group 3c β-lactamase (L1) confers resistance to β-lactam containing antibiotics (Alonso and Martínez, 1997). The blaL1 existed on a 200-kb plasmid, encoding a broad-spectrum metallo-β-lactamase which hydrolyses carbapenems (Walsh et al., 1994; Avison et al., 2001), is usually produced at higher levels (induced) that hydrolyzes almost all known penicillins, cephalosporins, and carbapenems during β-lactam challenge. The ease of acquisition and spread of this antibiotic resistant gene of blaL1 in S. maltophilia emphasizes the need for antibiotic susceptibility testing of clinical isolates. At present, blaL1 has been identified in some clinical isolates of S. maltophilia with important drug resistance against carbapenems (Avison et al., 2001), so detection of blaL1 plays an important role to indicate the infection of S. maltophilia in the clinical work (Alonso and Martínez, 2000; AI-Jasser, 2006; Gould et al., 2006).
Recently, a number of molecular biology techniques have been used to detect different strains of S. maltophilia (Nakamura et al., 2010). PCR amplification of the 16S rRNA gene was used to detect S. maltophilia in blood samples of patients who are undergoing chemotherapy for acute leukemia or myelodysplastic syndrome (Nakamura et al., 2010). However, PCR requires specialized high-cost instruments and consumables. In addition, Taq DNA polymerase in PCR assays can be inactivated by inhibitors present in crude biological samples but the large fragment of Bst DNA polymerase (the large fragment of Bst DNA polymerase is part of Bacillus stearothermophilus DNA polymerase, it has 5′>–3′> DNA polymerase activity) in loop-mediated isothermal amplification (LAMP) assays is more resistant to inhibitors present in crude biological samples (de Franchis et al., 1988; Kaneko et al., 2007). Thus, another rapid, simple and cost effective assay is needed to complement current PCR methods. The LAMP method which was developed in 2000 relies on auto-cycling strand displacement DNA synthesis which proceeds under isothermal conditions, typically within 60 min, and in the presence of Bst DNA polymerase (Notomi et al., 2000; Song et al., 2005). In this study, we develop this new method to detect the blaL1 of S. maltophilia. The LAMP method has been shown to amplify target DNA with high-specificity, and it is used widely in the clinical detection of bacteria (Hara-Kudo et al., 2005; Ohtsuka et al., 2005), viruses (Okafuji et al., 2005), parasites (Chen et al., 2011; Kong et al., 2012), and for fetal sex identification (Hirayama et al., 2006).
Data on the prevalence of blaL1 in S. maltophilia from ICU of Chinese hospitals are lacking. The objective of the current study is to develop a rapid, simple assay for S. maltophilia and to further investigate the infection status and the species distribution of blaL1 in clinic. At first, we designed five primer sets which each set targets six or eight sequences on the blaL1. The specificity and sensitivity of the primers for blaL1 was confirmed, and the LAMP method used for the detection of blaL1 in clinical samples. Then, basing on this LAMP assays, dissemination and molecular characterization of L1-producing S. maltophilia isolates was investigated at ICU patients in three top hospitals (the hospitals that have large scale and many patients) in Beijing, China.
MATERIALS AND METHODS
BACTERIAL ISOLATES, IDENTIFICATION, MLST TYPING, AND ANTIMICROBIAL SUSCEPTIBILITY TESTING
A total of 37 bacterial strains were used in this study to develop the LAMP assays, and their sources are listed in Table 1. S. maltophilia K279a carrying blaL1 and blaL2 with the typical antimicrobial resistance properties was used as the positive control. The other species including common clinical infectious species and homologous species with S. maltophilia stored at our laboratory were used for estimating the sensitivity and specificity of the LAMP assay. 105 clinical nasopharyngeal swabs and sputum samples were collected from ICU hospitalized patients with clinically suspected multi-resistant infections in the 307 hospital, 302 hospital, and 301 hospital in China, and species identification was carried out using an automated system (Phoenix and BD systems) and matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS). 16S rDNA and blaL1 were validated by PCR-based sequencing, and their sequence showed 100% (for 16S rDNA) and 98–100% (for blaL1) identity with the sequences of previously reported genes, respectively. The allele number for each gene was assigned on the basis of the information in the MLST database 1. A combination of the allelic sequences of the seven genes yielded the allelic profile. Antimicrobial susceptibility testing was performed by microbroth dilution according to the Clinical and Laboratory Standards Institute (CLSI, Clinical and Laboratory Standards Institute Performance standards for antimicrobial susceptibility testing; Twentieth informational supplement CLSI Document M100-S20, Wayne, PA, USA 2010.), and Etest strips (bioMérieux) for carbapenems. The carbapenemase activity of isolates was assessed by Etest MBLs.
Table 1.
Bacterial strains used in the current study.
| Species | Source |
|---|---|
| Stenotrophomonas maltophilia-2 | Clinical isolate |
| S. maltophilia-17 | Clinical isolate |
| S. maltophilia-24 | Clinical isolate |
| S. maltophilia-25 | Clinical isolate |
| S. maltophilia-36 | Clinical isolate |
| S. maltophilia-41 | Clinical isolate |
| S. maltophilia-51 | Clinical isolate |
| S. maltophilia-58 | Clinical isolate |
| S. maltophilia-63 | Clinical isolate |
| S. maltophilia-65 | Clinical isolate |
| S. maltophilia-66 | Clinical isolate |
| S. maltophilia-67 | Clinical isolate |
| S. maltophilia -3859 | Clinical isolate |
| S. maltophilia-4621 | Clinical isolate |
| S. maltophilia-WJ2 | Clinical isolate |
| S. maltophilia-K279a | Our microorganism center |
| Acinetobacter baumannii B260 | Our microorganism center |
| A. baumannii H18 | Our microorganism center |
| Bacillus megatherium 4623 | Our microorganism center |
| Beta hemolytic Streptococcus group A CMCC32213 | Our microorganism center |
| Bordetella pertussis ATCC 18530 | Our microorganism center |
| Brucella suis 3572 | Clinical isolate |
| Corynebacterium diphtheriae CMCC38001 | Our microorganism center |
| Enterotoxigenic Escherichia coli 44824 | Our microorganism center |
| Mycobacterium tuberculosis 8362 | Our microorganism center |
| Neisseria meningitidis group B CMCC29022 | Our microorganism center |
| Salmonella aberdeen 9264 | Our microorganism center |
| Salmonella enteritidis 50326-1 | Our microorganism center |
| Salmonella paratyphi 86423 | Our microorganism center |
| Shigella flexneri 4536 | Our microorganism center |
| Shigella sonnei 2531 | Our microorganism center |
| Staphylococcus aureus 2740 | Our microorganism center |
| Vibrio carchariae 5732 | Our microorganism center |
| V. cholera 3802 | Our microorganism center |
| V. parahaemolyticus 5474 | Our microorganism center |
| Yersinia enterocolitica 1836 | Our microorganism center |
| Y. pestis 2638 | Our microorganism center |
The strains were screened for the presence of known MBL genes (blaV IM, blaIMP, blaSPM-1, blaGIM-1, blaSIM-1, blaAIM-1, and blaNDM-1) by PCR with primers as reported previously (Patzer et al., 2009). The strains were also screened for the presence of other β-lactamase genes (blaCTX, blaCMY, etc.; Poirel et al., 2007).
ISOLATION OF GENOMIC DNA
The 37 bacterial strains and the 105 clinical samples were cultured in brain heart infusion (BHI) broth at 37°C according to a standard protocol. Chelex® 100 was used to extract total genomic DNA (including plasmid DNA) from 5 ml overnight bacterial cultures. Briefly, 500 μl bacterial suspension was centrifuged at 10,000 × g for 2 min and the supernatant discarded. The pellet was resuspended in 500 μl distilled water and 500 μl Chelex DNA extraction buffer (25 mM NaOH, 10 mM Tris-HCl, 1% Triton X-100, 1% NP-40, 0.1 mM EDTA, 2% Chelex-100) added. The cell suspension was heated in boiling water for 10 min, held on ice for 5 min, and centrifuged at 14,000 × g for 2 min. The extracted DNA was used as template in the LAMP and PCR reactions.
As for isolation of DNA from clinical sputum samples, DNA was extracted directly from 200 μl clinical sputum samples with the TIANamp Genomic DNA Kit (TIANGEN Biotech Co., Ltd., Beijing, China). The DNA was purified with the SV GEL and PCR Clean-Up System (Promega Co., USA). The DNA concentration was detected using the Spectrophotometer ND-1000 (Thermo Fisher Scientific, Inc., USA).
PRIMER DESIGN
A total of 20 blaL1 in the NCBI GenBank database (GenBank: HQ822273.1; EF126060.1; EF126061.1; AM743169.1; AB294542.1; AJ251814.1; JF705927.1; JF705926.1; EF126051.1; EF126054.1; EF126053.1; EF601224.1; AB294547.1; AB294545.1; AJ291672.1; AF010282.1; AB194306.1; AJ289085.1; AJ289086.1; AB194305.1) were compared, then the sequences of conserved regions were chosen to design the primer sets. Primer Explorer V4 software 2 was used to design the outer forward primer (F3), outer backward primer (B3), forward inner primer (FIP), backward inner primer (BIP) and backward loop primer (loop B), used to accelerate the amplification reaction. Mergers of bases are used to circumvent the mutational site (Table 2). The FIP and BIP primers were linked by a four thymidine spacer (TTTT). Conventional PCR was performed using primers labeled L1-23-F3 and L1-23-B3. The primers were synthesized by Sangon Biotech Co., Ltd. (Shanghai, China).
Table 2.
Primers used for the amplification of blaL1.
| Primer | Type | Sequence (5’–3’) |
|---|---|---|
| L1-23F3 | Forward outer | CGGCATGCCACAGATGG |
| L1-23B3 | Backward outer | GCAGCACCGCCGTTTCT |
| L1-23FIP | Forward inner | TCAATCGCAGGTCCTGCGGTTTTCGGTCACCTGCTGGACAAC |
| L1-23BIP | Backward inner | CTY(C/T)AGCCATGCGCAY(T/C)GCS(C/G)GATTTTGCATTGGCCGCCACATG |
| L1-23LB | Loop backward | TCGCCGAGCTCAAGCGT |
LAMP REACTION
A 25 μl reaction volume was used for all LAMP reactions and contained the following components (final concentration): 20 mM Tris-HCl (pH 8.8), 10 mM KCl, 10 mM (NH4)2SO4, 0.1% Tween 20, 0.8 M betaine, 8 mM MgSO4, 1.4 mM each dNTP, and 8 U Bst DNA polymerase. The amount of primer per reaction was 40 pmol FIP and BIP, 20 pmol LB, 5 pmol F3 and B3. The appropriate amount of DNA template was included in the reaction volume. The LAMP assay proceeded in a reaction tube (Eiken Chemical Co., Ltd., Tochigi, Japan) for 60 min at 65°C.
DETECTION OF LAMP PRODUCTS
Two independent methods, based on either sample turbidity or fluorescence were used to detect LAMP products. Real-time changes in turbidity were monitored by measuring the optical density (λ650 nm) at 6 s intervals, for each LAMP reaction in a Loopamp real-time turbidimeter (LA-320c; Eiken Chemical Co., Ltd.). The changes in turbidity arose from the presence of the amplification by-product Mg2P2O7 (a white precipitate).
The second method used direct visual inspection to assess color changes in the presence of the fluorescent metal ion indicator calcein/Mn2+ complex. One microliter of calcein/Mn2+ complex (Eiken Chemical Co., Ltd.) was added to 25μl LAMP reaction volume prior to the commencement of the LAMP assay. On completion of the reaction a change in color from orange to green indicated a positive reaction, whilst no color change indicated a negative reaction. The color change was observed by the naked eye under natural light or under UV light at 365 nm.
PCR DETECTION
A 25 μl reaction volume was used for all PCR reactions and contained the following components: 12.5 μl PCR Taq MasterMix (Tiangen Biotech Co., Ltd.), 9.5 μl double distilled water, 1 μM L1-23F3 and L1-23B3 primers, and DNA template. The oligonucleotide primers used for cloning blaL1 are F: 5′>-atgcgttctaccctgctcgccttcgcc-3′> and R: 5′>-tcagcgggccccggccgtttccttggccag-3′>. The PCR was carried out as follows: initial PCR activation step, 94°C for 2 min; amplification, 35 cycles of 94°C for 30 s, 59°C for 30 s, and 72°C for 30 s; final extension step, 72°C for 10 min. The amplicons were purified using a PCR Purification Kit (TIANGEN Biotech Co., Ltd., Beijing, China) and sequenced by Beijing AuGCT DNASYN Biotechnology Co., Ltd. The sequences were compared with sequences in the GenBank database.
RESULTS
THE OPTIMAL PRIMER SETS FOR LAMP ASSAY
Five primer sets were detected in the same reaction condition using real-time turbidimeter and their turbidity curves were draw at 650 nm according to the amplified results. The optimal primer sets amplified the target sequence with the shortest time among them was chosen for further investigation (see Table 2).
SENSITIVITY OF THE LAMP METHOD FOR blaL1 DETECTION
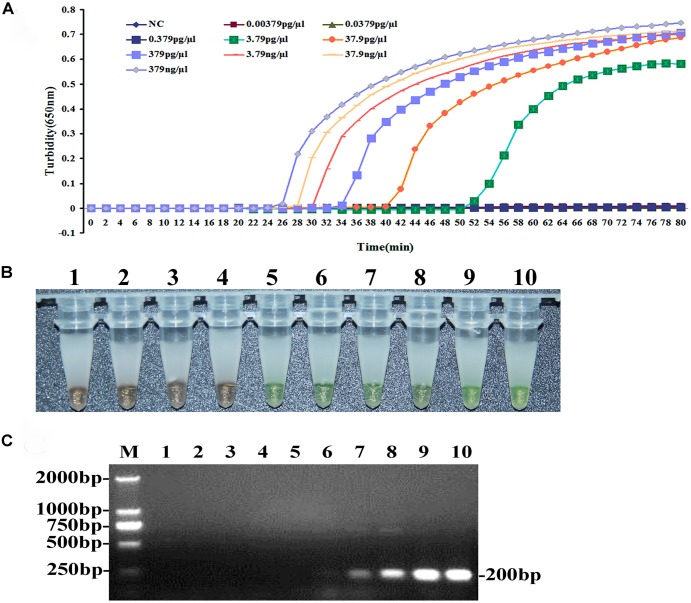
The sensitivity of the LAMP method for detecting blaL1 was evaluated using genomic DNA extracted from S. maltophilia K279a (Wizard Genomic DNA purification Kit), serially diluted 10-fold from 379 ng/μl to 0.00379 pg/μl. As shown in Figure 2A, the detection limit of the LAMP assay for blaL1 was 3.790 pg/μl. Visual inspection of the color change, post-LAMP assay, and in the presence of calcein/Mn2+ complex confirmed reactions positive (green) and negative (orange) for blaL1(Figure 2B). The results from the two detection methods were in agreement with sensitivity for blaL1. PCR reactions on the serially diluted DNA using primers L1-23F3 and L1-23B3 were also conducted, and the detection limit for blaL1 was established as 379 pg/μl (Figure 2C).
FIGURE 2.
Comparison of the sensitivities for blaL1 gene detection by LAMP and conventional PCR methods. Pure genomic DNA extracted from S. maltophilia-K279a was diluted tenfold (379.0 ng/μl to 0.00379 pg/μl) and the DNA assayed by LAMP (A,B) and PCR (C). (A) Turbidity was monitored using the Loopamp real-time turbidimeter and the OD recorded at 650 nm, at 6 s intervals. (B) Visual inspection of the color change, post-LAMP assay, and in the presence of calcein/Mn2+ complex. (C) PCR products were analyzed by 2% agarose gel electrophoresis and stained with ethidium bromide. The DNA marker is D2000 DNA Marker (Tiangen Biotech Co., Ltd.) The size is about 179 bp.
SPECIFICITY OF THE LAMP METHOD FOR blaL1 DETECTION
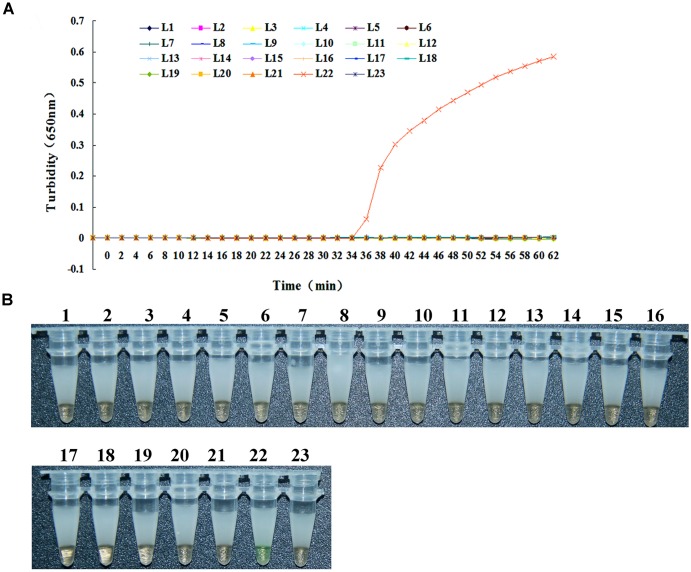
The specificity of the LAMP method for detecting blaL1 was evaluated using S. maltophilia K279a with blaL1 as the positive control, distilled water as the negative control, and 21 strains without carrying blaL1 including common clinical infectious species and homologous species with S. maltophilia as test subjects. As shown in Figure 1A, turbidity increased only when S. maltophilia K279a with blaL1 was used as template DNA in the LAMP assay. When distilled water and the 21 remaining bacterial species were used as template, no changes in turbidity were recorded. These results suggest that the primers had good specificity for blaL1. In addition, these results are consistent with those obtained using the fluorescent indicator calcein/Mn2+ complex. Whereby, only the LAMP assay with S. maltophilia K279a with blaL1 recorded a color change from orange to green, indicative of a positive reaction (Figure 1B). Whilst, all test samples negative for blaL1 and the negative control remained orange, indicative of a negative reaction.
FIGURE 1.
Specificity of the LAMP method for blaL1 gene detection. It has two parts, (A) is the graphic and (B) is the photography of microtubes. The reaction proceeded at 65°C for 65 min. Turbidity was monitored in the Loopamp real-time turbidimeter and the OD(λ650nm) recorded at 6 s intervals. L1, Brucella suis 3572; L2, Bacillus megatherium 4623; L3, Vibrio carchariae 5732; L4, Acinetobacter baumannii B260; L5, Corynebacterium diphtheriae CMCC38001; L6, Acinetobacter baumannii H18; L7, Mycobacterium tuberculosis 8362; L8, Shigella sonnei 2531; L9, Shigella flexneri 4536; L10, Salmonella enteritidis 50326-1; L11, Yersinia enterocolitica 1836; L12, Vibrio parahaemolyticus 5474; L13, Salmonella paratyphi 86423; L14, Neisseria meningitidis group B CMCC29022; L15, Enterotoxigenic E. coli 44824; L16, Beta hemolytic Streptococcus group A CMCC32213; L17, Yersinia pestis 2638; L18, Salmonella aberdeen 9264; L19, Vibrio cholera 3802; L20, Staphylococcus aureus 2740; L21, Bordetella pertussis ATCC 18530; L22, positive control (S. maltophilia -K279a); L23, negative control (distilled water).
DISSEMINATION OF L1-PRODUCING S. maltophilia IN CLINICAL
A total of 105 clinical sputum samples and nasopharyngeal swabs were collected for LAMP-based surveillance of blaL1 from 105 ICU patients with clinically suspected multi-resistant infections from the department of Respiratory Diseases in three top hospitals. Ten pairs of sputum samples and nasopharyngeal swabs from healthy people were collected as controls.
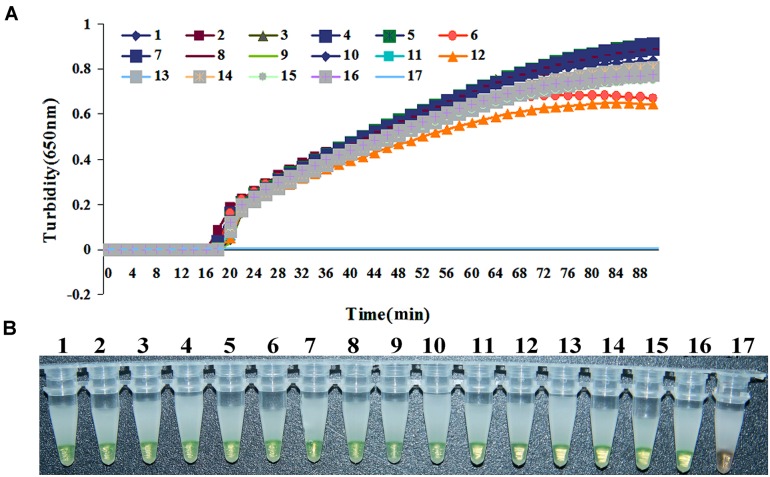
All clinical samples were analyzed by LAMP and PCR simultaneously. Of the 105 patients samples, 22 were confirmed to be infected with S. maltophilia with blaL1 and 83 negative samples by the LAMP assay (Figure 3), whilst the PCR assay detected 13 positive samples, and 92 negative samples. Then 22 strains of S. maltophilia were isolated and identified from all of clinical sputum samples and swabs samples, which the positive samples were in accordence with those in LAMP assay. S. maltophilia with blaL1 was positively identified 100% by LAMP and 86.7% by PCR, respectively. None of the samples from healthy people was tested as positive for blaL1. Thus, the results showed the LAMP assays is more sensitive and the specific than PCR for diagnosis of S. maltophilia in clinical practice.
FIGURE 3.
Loop-mediated isothermal amplification results for 15 S. maltophilia strains positive for blaL1 isolated from 15 clinical samples. (A) Turbidity was monitored using Loopamp, and the OD measured at 650 nm every 6 s. (B) Visual inspection of calcein/Mn2+ complex associated color changes post-LAMP assay. 1, S. maltophilia-2; 2, S. maltophilia-17; 3, S. maltophilia-24; 4, S. maltophilia-25; 5, S. maltophilia-36; 6, S. maltophilia-41; 7, S. maltophilia-51; 8, S. maltophilia-58; 9, S. maltophilia-63; 10, S. maltophilia-65; 11, S. maltophilia-66; 12, S. maltophilia-67; 13, S. maltophilia-3859; 14, S. maltophilia-4621; 15, S. maltophilia -WJ2; 16, positive control (S. maltophilia-K279a); 17, negative control (distilled water).
The sequence analysis of the blaL1 genes from S. maltophilia isolates confirmed conservation with the nucleotide sequences of reported genes or with only a few sites mutated. In the MLST analysis of S. maltophilia, the results of seven housekeeper genes recovered that 22 strains belonged to different sequence type (ST) including ST4, ST8, ST 25, ST 28, ST29, or ST31, respectively. To further characterize the 22 strains carrying blaL1 genes, the susceptibility pattern was detected and clearly showed that all isolates had highly resistant to β-lactam antibiotics. The isolates also tested positive for MBLs in both the imipenem-EDTA double-disk synergy test (DDST) and modified Hodge test (MHT).
Furthermore, PCR screening of the isolates were performed for the known MBL genes including blaNDM-1, blaV IM, blaIMP, blaSPM-1, blaGIM-1, blaSIM-1, blaAIM-1, and blaL2 (Patzer et al., 2009). PCR yielded products of 22 isolates with expected sizes for blaL1, and sequencing of these genes showed 100% identities with previously reported genes. It’s interesting to note that the isolate named as S. maltophilia DCPS-01 contained L1 and L2 β-lactamase genes with a novel blaNDM-1 which has attracted wide attention because of its superior resistance to all β-lactam antibiotics, which presented increased carbapenemase activity to all β-lactams (MIC >128 μg/mL for imipenem and meropenem), aminoglycosides and quinolones, and was only susceptible to tigecycline and colistin.
Therefore, our data showed the diversity genotypic features of S. maltophilia carrying blaL1 indicated wide spread in the respiratory infections. Importantly, the emergence of these powerful co-occurring resistance mechanisms described here provides warning that future therapeutic options may be seriously limited.
DISCUSSION
Stenotrophomonas maltophilia is a widespread environmental bacterium that has become a nosocomial pathogen of increasing importance. It is currently the third most common nosocomial non-fermenting bacteria, behind Pseudomonas aeruginosa and Acinetobacter baumannii, and is associxated with crude mortality rates ranging from 14 to 69% in patients with bacteraemias (Jang et al., 1992; Victor et al., 1994). Of the 1661 antibiotic resistant strains of S. maltophilia recovered from 14 hospitals across several regions of China during 2010, the majority (97.5%) were isolated from patients. Approximately 68.3% of strains were isolated from patients ≥60 years of age, whilst only 4.8% were from patients <18 years of age. Most (83.0%) isolates were recovered from sputum and respiratory tract secretions (The data and information from CHINET: CHINET 2010 surveillance of antibiotic resistance in S. maltophilia in China). Within hospitals, the bacterium is most often found in water sources, and can be a contaminant of hospital equipment such as nebulizers and intravenous catheters. From these sources, the organism can infect patients, resulting in a wide spectrum of symptoms dependent upon the site of infection, though, most commonly, S. maltophilia causes bacteraemias or respiratory tract infections (Denton and Kerr, 1998). β-Lactam resistance is due to the expression of the β-lactamases L1, which together hydrolyze the full range of β-lactam drugs, with the exception of monobactams (Walsh et al., 1994). It is therefore necessary to detect and monitor antibiotic resistance, persistence and spread of S. maltophilia within the community and in health care settings.
Loop-mediated isothermal amplification assays are generally less time and labor intensive compared with traditional methods of pathogen detection, in part because the amplification of the target gene is performed at a constant temperature, and the reaction times are usually less than an hour. To date, a method for detecting S. maltophilia based on LAMP assays has not been reported. In the current study, we designed primers specific for the metallo-β-lactamase blaL1 for use in a LAMP assay to detect S. maltophilia in clinical samples. Results from the specificity and sensitivity analyses demonstrated that the LAMP method detected genomic DNA at 3.79 pg/μl, and was specific for the β-lactamase blaL1. In the specificity and sensitivity detection, we only use the blaL1 of S. maltophilia K279a as the target gene, although many other S. maltophilia strains do not have the same sequence of blaL1, the most conserve regions of blaL1 were chosed to design the decisive primers. In LAMP reaction, it is not true that all the primers should combine with the target sequence. The decisive primers are the FIP and BIP, if the FIP and BIP can combine with the target sequence, the reaction is certain to occur, well, of course, more novel experiments should be made to ensure this conclusion. Although the LAMP method has complex amplification principle, the assay is rapid, easy to operate, highly sensitive and specific, and proceeds under isothermal conditions. We believe this assay would be suitable for use in inspection and quarantine departments and in health care units to test for S. maltophilia, and we anticipate its routine use in hospital testing regimes, particularly for rapid clinical testing.
A drawback of the LAMP method is the relatively high false-positive rates; a consequence of the assay’s high sensitivity (Notomi et al., 2000). Strict spatial separation of reagent preparation from the testing area is necessary to avoid contamination. In the current study, a sealing agent was applied to the reaction tube once the reaction mixture had been prepared, and its presence is useful in preventing contamination.
In conclusion, we designed a detection method based on LAMP for the specific, sensitive, rapid, and effective detection of the metallo-β-lactamase blaL1 of S. maltophilia. We believe this technique would greatly benefit hospitals and health units, and we anticipate that LAMP will become the gold standard for the rapid detection of pathogens in clinical samples. At the same time, this report provides new insights into the mechanisms of drug resistance and warning that future therapeutic options may be seriously limited.
AUTHOR CONTRIBUTIONS
Jing Yuan helped conceive project and designed experiments. Zhan Yang, Wei Liu, and Qian Cui performed and wrote the manuscript. Huan Li, Xiangna Zhao, Xiao Wei, Xuesong Wang, Wenkai Niu, Changqing Bai, Yan Li, and Liuyu Huang designed and executed experiments. Simo Huang, Derong Dong, and Sijing Lu helped to edit the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are indebted to the Department of Respiratory Diseases, Affiliated Hospital of Academy of Military Medical Science for kindly providing S. maltophilia and helpful information. We are grateful to LI Fengjing and XIAO Shengli from Beijing Lanpu Bio-tech Co., Ltd. for providing technical assistance and for participating in helpful discussions. This work was supported by Mega-projects of Science and Technology Research of China Grant 2011ZX10004-001 and 2013ZX10004-203, National Natural Science Foundation of China Grant 31370093 and 81201320, and National High Technology Research and Development Program of China 863 Program Grant SS2014AA022210.
Footnotes
REFERENCES
- AI-Jasser A. M. (2006). Stenotrophomonas maltophilia resistant to trimethoprim sulfamethoxazole: an increasing problem. Ann. Clin. Microbiol. Antimicrob. 5 23 10.1186/1476-0711-5-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso A., Martínez J. L. (1997). Multiple antibiotic resistance in Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 41 1140–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso A., Martínez J. L. (2000). Cloning and characterization of SmeDEF, a novel multidrug effux pump from Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 44 3079–3086 10.1128/AAC.44.11.3079-3086.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avison M. B., Higgins C. S., von Heldreich C. J., Bennett P. M., Walsh T. R. (2001). Plasmid location and molecular heterogeneity of the L1 and L2 β-lactamase genes of Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 45 413–419 10.1128/AAC.45.2.413-419.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooke J. S. (2012). Stenotrophomonas maltophilia: an emerging global opportunistic pathogen. Clin. Microbiol. Rev. 25 2–41 10.1128/CMR.00019-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R., Tong Q., Zhang Y., Lou D., Kong Q., Lv S., et al. (2011). Loop-mediated isothermal amplification: rapid detection of Angiostrongylus cantonensis infection in Pomacea canaliculata. Parasit. Vectors 4 204 10.1186/1756-3305-4-204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Franchis R., Cross N. C., Foulkes N. S., Cox T. M. (1988). A potent inhibitor of Taq polymerase copurifies with human genomic DNA. Nucleic Acids Res. 16 10355 10.1093/nar/16.21.10355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denton M., Kerr K. G. (1998). Microbiological and clinical aspects of infection associated with Stenotrophomonas maltophilia. Clin. Microbiol. Rev. 11 57–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould V. C., Okazaki A., Avison M. B. (2006). Beta-lactam resistance and beta-lactamase expression in clinical Stenotrophomonas maltophilia isolates having defined phylogenetic relationships. J. Antimicrob. Chemother. 57 199–203 10.1093/jac/dki453 [DOI] [PubMed] [Google Scholar]
- Hara-Kudo Y., Yoshino M., Kojima T., Ikedo M. (2005). Loop-mediated isothermal amplification for the rapid detection of Salmonella. FEMS Microbiol. Lett. 253 155–161 10.1016/j.femsle.2005.09.032 [DOI] [PubMed] [Google Scholar]
- Hirayama H., Kageyama S., Takahashi Y., Moriyasu S., Sawai K., Onoe S., et al. (2006). Rapid sexing of water buffalo (Bubalus bubalis) embryos using loop-mediated isothermal amplification. Theriogenology 66 1249–1256 10.1016/j.theriogenology.2006.03.036 [DOI] [PubMed] [Google Scholar]
- Jang T. N., Wang F. D., Wang L. S., Liu C. Y., Liu I. M. (1992). Xanthomonas maltophilia bacteremia: an analysis of 32 cases. J. Formos. Med. Assoc. 91 1170–1176 10.1093/jac/dkn194 [DOI] [PubMed] [Google Scholar]
- Kaneko H., Kawana T., Fukushima E., Suzutani T. (2007). Tolerance of loop-mediated isothermal amplification to a culture medium and biological substances. J. Biochem. Biophys. Methods 70 499–501 10.1016/j.jbbm.2006.08.008 [DOI] [PubMed] [Google Scholar]
- Kong Q. M., Lu S. H., Tong Q. B., Lou D., Chen R., Zheng B., et al. (2012). Loop-mediated isothermal amplification (LAMP): early detection of Toxoplasma gondii infection in mice. Parasit. Vectors 5 2 10.1186/1756-3305-5-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura A., Sugimoto Y., Ohishi K., Sugawara Y., Fujieda A., Monma F., et al. (2010). Diagnostic value of PCR analysis of bacteria and fungi from blood in empiric-therapy-resistant febrile neutropenia. J. Clin. Microbiol. 48 2030–2036 10.1128/JCM.01700-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N., et al. (2000). Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 28:E63 10.1093/nar/28.12.e63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuka K., Yanagawa K., Takatori K., Hara-Kudo Y. (2005). Detection of Salmonella enterica naturally contaminated liquid eggs by loop-mediated isothermal amplification, and characterization of Salmonella isolates. Appl. Environ. Microbiol. 71 6730–6735 10.1128/AEM.71.11.6730-6735.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okafuji T., Yoshida N., Fujino M., Motegi Y., Ihara T., Ota Y., et al. (2005). Rapid diagnostic method for detection of mumps virus genome by loop-mediated isothermal amplification. J. Clin. Microbiol. 43 1625–1631 10.1128/JCM.43.4.1625-1631.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patzer J. A., Walsh T. R., Weeks J., Dzierzanowska D., Toleman M. A. (2009). Emergence and persistence of integron structures harbouring VIM genes in the Children’s Memorial Health Institute, Warsaw, Poland, 1998–2006. J. Antimicrob. Chemother. 63 269–273 10.1093/jac/dkn512 [DOI] [PubMed] [Google Scholar]
- Poirel L., Pitout J. D., Nordmann P. (2007). Carbapenemases: molecular diversity and clinical consequences. Future Microbiol. 2 501–512 10.2217/17460913.2.5.501 [DOI] [PubMed] [Google Scholar]
- Song T., Toma C., Nakasone N., Iwanaga M. (2005). Sensitive and rapid detection of Shigella and enteroinvasive Escherichia coli by a loop-mediated isothermal amplification method. FEMS Microbiol. Lett. 243 259–263 10.1016/j.femsle.2004.12.014 [DOI] [PubMed] [Google Scholar]
- Victor M. A., Arpi M., Bruun B., Jønsson V., Hansen M. M. (1994). Xanthomonas maltophilia bacteremia in immunocompromised hematological patients. Scand. J. Infect. Dis. 26 163–170 10.3109/00365549409011780 [DOI] [PubMed] [Google Scholar]
- Walsh T. R., Hall L., Assinder S. J., Nichols W. W., Cartwright S. J., MacGowan A. P., et al. (1994). Sequence analysis of the L1 metallo-β-lactamase from Xanthomonas maltophilia. Biochim. Biophys. Acta 1218 199–201 10.1016/0167-4781(94)90011-6 [DOI] [PubMed] [Google Scholar]