Significance
Expanding the genetic code is an important aim of synthetic biology, but some organisms developed naturally expanded genetic codes over the course of evolution. To understand the selective advantage of genetically encoding more than 20 amino acids, we investigated the proteome-wide response to reducing the genetic code of Methanosarcina acetivorans from 21 to 20 amino acids. The data show how a natural proteome adapts to genetic code reduction and indicate that the selective value of an expanded genetic code is related to carbon source range and metabolic efficiency.
Keywords: evolution, genetic code expansion, methanogenesis, pyrrolysine, tRNAPyl
Abstract
Expanding the genetic code is an important aim of synthetic biology, but some organisms developed naturally expanded genetic codes long ago over the course of evolution. Less than 1% of all sequenced genomes encode an operon that reassigns the stop codon UAG to pyrrolysine (Pyl), a genetic code variant that results from the biosynthesis of Pyl-tRNAPyl. To understand the selective advantage of genetically encoding more than 20 amino acids, we constructed a markerless tRNAPyl deletion strain of Methanosarcina acetivorans (ΔpylT) that cannot decode UAG as Pyl or grow on trimethylamine. Phenotypic defects in the ΔpylT strain were evident in minimal medium containing methanol. Proteomic analyses of wild type (WT) M. acetivorans and ΔpylT cells identified 841 proteins from >7,000 significant peptides detected by MS/MS. Protein production from UAG-containing mRNAs was verified for 19 proteins. Translation of UAG codons was verified by MS/MS for eight proteins, including identification of a Pyl residue in PylB, which catalyzes the first step of Pyl biosynthesis. Deletion of tRNAPyl globally altered the proteome, leading to >300 differentially abundant proteins. Reduction of the genetic code from 21 to 20 amino acids led to significant down-regulation in translation initiation factors, amino acid metabolism, and methanogenesis from methanol, which was offset by a compensatory (100-fold) up-regulation in dimethyl sulfide metabolic enzymes. The data show how a natural proteome adapts to genetic code reduction and indicate that the selective value of an expanded genetic code is related to carbon source range and metabolic efficiency.
Synthesizing whole genomes (1) and eliminating codons (2) are novel methods for rewriting the genetic code that may dramatically alter the repertoire of genetically encoded amino acids. Expansion of the genetic code has led to exciting technologies, including site-directed protein labeling and production of proteins with hardwired posttranslational modifications (3). The current approaches to cotranslationally insert noncanonical amino acids (ncAAs) into proteins rely on the reassigning of one of three stop codons (4).
Although these approaches were highly successful in incorporating over 100 ncAAs into proteins (3), they limit the expansion of the code to no more than 2 additional amino acids at a time and significantly challenge the cellular production host by unnaturally extending proteins and reducing growth rate (5). Alternate methods focus on quadruplet codons (6, 7) and recoding (8) or reassigning sense codons (9–13). Attempts to reassign a sense codon in Mycoplasma capricolum were defied by tRNA misacylation by endogenous aminoacyl-tRNA synthetases (9). This result indicates that, although extensively rewriting the genetic code may be possible, it comes with unexpected challenges related to cellular fitness and translation fidelity. These considerations will impact efforts to engineer cells to synthesize proteins with multiple ncAAs or create biologically contained strains that require an expanded code for survival (14).
Opening codons by reducing the genetic code is highly promising, but it is unknown how removing 1 amino acid from the genetic code might impact the proteome or cellular viability. Many genetic code variations are found in nature (15), including stop or sense codon reassignments, codon recoding, and natural code expansion (16). Pyrrolysine (Pyl) is a rare example of natural genetic code expansion. Evidence for genetically encoded Pyl is found in <1% of all sequenced genomes (17). In these organisms, Pyl is encoded by the UAG codon, which requires tRNAPyl, pyrrolysyl-tRNA synthetase (PylRS), and the products of three genes (pylBCD) that synthesize Pyl from two molecules of lysine (18). The PylRS enzyme was engineered to genetically encode >100 ncAAs (19). The Pyl encoding system has already been used to expand the genetic codes of Escherichia coli (20–22), mammalian cells, and animals (23).
Despite the use of Pyl in synthetic biology, little is known about the role of Pyl in its native environment or the evolutionary pressures that sustain expanded genetic codes in nature. The Pyl-decoding trait is found in methanogenic archaea of the orders Methanosarcinales and Methanomassiliicoccales (24) and certain anaerobic bacteria (17). In addition to producing 74% of global methane emissions, methanogens are remarkable for their ability to survive with only the most basic carbon and energy sources (25). Methanosarcina shows the greatest substrate range among methanogens and survives on acetate, carbon monoxide, methylamines, methanol, or dimethyl sulfide (DMS). Their broad substrate range depends, in part, on the presence of Pyl in the active site of several methylamine methyltransferases (26). Hundreds of Methanosracina genes contain in-frame TAG codons (27), but natural Pyl incorporation was only shown in methylamine methyltransferases (17, 28) and tRNAHis guanylyltransferase (Thg1) (29).
Methanosarcina acetivorans provides an ideal model system to identify Pyl-containing proteins and study the impact of genetic code reduction on the proteome and physiology of the cell. We constructed a markerless tRNAPyl deletion (ΔpylT) strain of M. acetivorans C2A and used three independent mass spectrometry (MS) approaches to characterize soluble proteomes from M. acetivorans grown on minimal medium containing trimethylamine (TMA) or methanol and ΔpylT cells grown on methanol. The data reveal previously unidentified biochemical roles for Pyl and Pyl-containing proteins and indicate that the expanded genetic code of Methanosarcina is intricately linked with cellular metabolism and the composition of the proteome.
Results
M. acetivorans with a Reduced Genetic Code.
There are 267 ORFs in the M. acetivorans genome with one or multiple in-frame UAG codon(s) (Figs. S1 and S2 and Table S1). Except for Thg1 and the methylamine methyltransferase (mtxB), it is unknown if these ORFs are expressed or the resulting protein contains Pyl. To uncover more Pyl-containing proteins and investigate the role of Pyl in the M. acetivorans proteome, we constructed and characterized a tRNAPyl deletion strain of M. acetivorans C2A (Fig. 1). We monitored the growth rate of three independently obtained markerless tRNAPyl deletion mutants and compared these cells with wild type (WT) cells grown on minimal medium containing TMA or methanol (Fig. 1 and Table 1). As expected (26), the tRNAPyl deletion strain cannot use TMA as a growth substrate. In rich medium containing yeast extract, previous studies indicated that tRNAPyl deletion did not affect growth on methanol (26). In minimal medium containing methanol, ΔpylT shows significant increases in lag time (10%) and doubling time (30%) (Fig. 1). The data show that the expanded genetic code of M. acetivorans confers a selective advantage.
Fig. 1.
Characterization of M. acetivorans variants. (A) Structure of the Pyl operon showing StyI sites flanking the tRNAPyl gene. (B) The Southern blot shows the genotype of WT M. acetivorans (lane 1) and three independently obtained tRNAPyl markerless deletion mutants (lanes 2–4). In the deletion strains, only 73 bases of the tRNAPyl gene were deleted. As anticipated, the digoxigenin-labeled molecular weight marker (M) migrates slower than the digested genomic DNA. (C) Growth curve of WT and ΔpylT strains in minimal media containing methanol (MeOH) or TMA as the sole carbon source. Data are based on triplicate measurements; ΔpylT data are based on triplicate measurements of three independently obtained deletion strains. Error bars show 1 SD.
Table 1.
Growth statistics for M. acetivorans strains
| Strain | Carbon source | Doubling time (h) | Maximum A578 | Lag time (h) |
| WT | MeOH | 5.9 ± 0.7 | 1.02 ± 0.01 | 39.9 ± 1.6 |
| ΔpylT | MeOH | 7.9 ± 0.5 | 0.99 ± 0.02 | 45.2 ± 1.5 |
| WT | TMA | 7.5 ± 0.3 | 1.06 ± 0.40 | 52.4 ± 0.9 |
From 21 to 20—Proteome Adaptation to Genetic Code Reduction.
To better understand the nature of the selective value of Pyl, we characterized the soluble proteomes of WT and ΔpylT strains. Of 4,721 potential protein coding genes in M. acetivorans, 841 proteins were identified, including ∼300 proteins identified by gel-based methods; the liquid chromatography (LC) -MS/MS approach identified 583 proteins. Proteins were considered identified if two or more significant peptides (peptide score > 35) were detected and verified by MS/MS spectra. All peptides identified by LC-MS/MS are listed in Dataset S1.
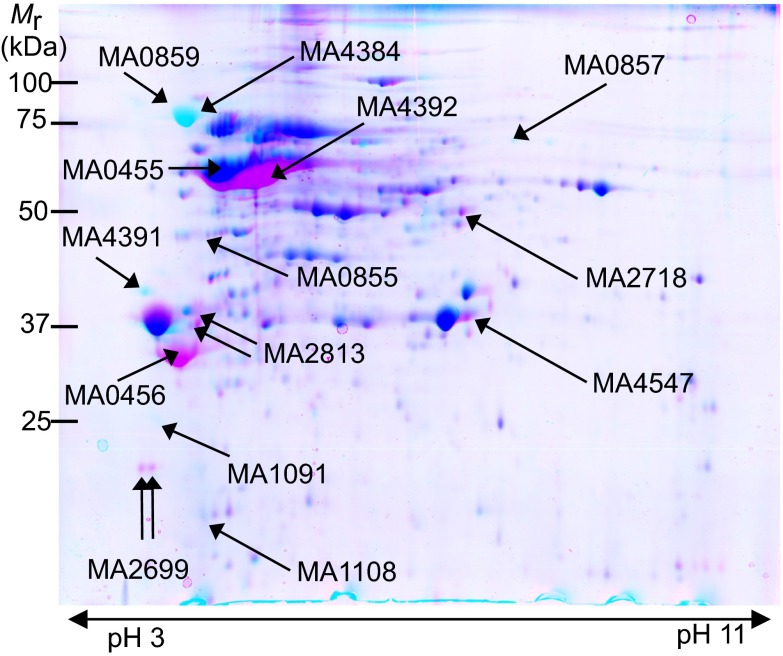
The ΔpylT strain has a globally altered proteome (Fig. 2). We identified 347 differentially regulated proteins showing more than twofold change (Tables S2 and S3), most of which are proteins that do not contain Pyl. The most affected pathways include stress response, methanogenesis, methylsulfide metabolism, translation, and amino acid metabolism (Table 2, Figs. S3 and S4, and Tables S2 and S3). In ΔpylT, proteins involved in heat shock (Hsp60 and GroEL/GroES) and oxidative stress response pathways were significantly up-regulated (approximately fivefold). We observed enhanced expression of eight distinct methanosarcina disulfide reductases (Table S3), which were suggested to play a major role in oxidative defense (30). Prematurely truncated or mistranslated proteins may activate this stress response. Protein mistranslation and misfolding elicit similar stress responses in other organisms, such as E. coli (31).
Fig. 2.
Deletion of tRNAPyl leads to a globally altered proteome. Overlapped 2D gel image showing soluble proteomes (pH range 3–11) of the WT (magenta) and ΔpylT (cyan) M. acetivorans strains. Protein spots that overlap exactly are shown in blue. Proteins that showed high levels of differential expression are marked. Mr, molecular mass.
Table 2.
Most affected proteins in ΔpylT compared with WT M. acetivorans
| Locus | Gene name | Function | x-Fold change |
| MA3790 | ilvC | Ile biosynthesis | 0.03 |
| MA2273 | S-layer domain | ABC transporter | 0.1 |
| MA0821 | pgi | Glycolysis | 0.1 |
| MA1778 | Exosome | RNA metabolism | 0.1 |
| MA0182 | eif2b | Protein synthesis | 0.1 |
| MA3052 | valS | Protein synthesis | 0.1 |
| MA0076 | eif2B1 | Protein synthesis | 0.2 |
| MA4391 | mtaC2 | Methanogenesis | 0.3 |
| MA0855 | mtaA | Methanogenesis | 1.8 |
| MA1567 | pdxS | Cofactor synthesis | 4.1 |
| MA4043 | Hypothetical | ? | 4.5 |
| MA4127 | Recombinase | Mobile element | 4.5 |
| MA1317 | gatD | Protein synthesis | 4.9 |
| MA1275 | ahcY | Cys biosynthesis | 5.0 |
| MA3564 | argJ | Arg biosynthesis | 5.8 |
| MA4386 | hsp60-4 | Stress response | 6.8 |
| MA0857 | hsp60-5 | Stress response | 6.9 |
| MA1091 | rpl18p | Protein synthesis | 10.3 |
| MA1108 | rps13p | Protein synthesis | 37.7 |
| MA4384 | mtsF | Methanogenesis | 35.5 |
| MA0859 | mtsD | Methanogenesis | 112.2 |
| MA4558 | mtsH | Methanogenesis | Distinct in ΔpylT* |
Distinct peptides observed in the LC-MS/MS experiment refer to proteins identified with more than two significant peptides in which the peptides observed in one strain (methanol-grown WT or ΔpylT) are completely absent in the other strain. Fig. S5 shows typical spectra for distinct peptides. x-Fold change indicates the differential abundance ΔpylT/WT.
Ribosomal proteins S13 and L18 are >10-fold more abundant in ΔpylT. S13 is 40-fold up-regulated in ΔpylT and one of the most perturbed proteins identified. S13 interacts with the tRNA binding site and central protuberance of the ribosome, and it is also in contact with L5 (32). L18 is essential in E. coli, and its association with L5 and the 5S rRNA is required for proper ribosomal assembly (33). Because deletion of tRNAPyl creates a cell with up to 267 new stop codons, overexpression of S13 and L18 may help reassemble or stabilize stalled ribosomes.
Metabolic Adaptations in Methanogensis Without Pyl.
The proteomic analysis indicates the tRNAPyl deletion strain is metabolically less efficient than WT. In methanogenesis from methanol, the MtaB protein abstracts a methyl group from the methanol substrate and transfers it to a cognate corrinoid protein (MtaC). In the next step, the MtaA protein transfers the methyl group from MtaC to convert coenzyme M (CoM) into methyl-CoM, which is a substrate for methanogenesis. Proteins responsible for methanogenesis from methanol are two- to fourfold down-regulated in ΔpylT (MtaB1, MtaC, MtaB2, MtaA, MtaC2), including a newly identified Pyl-containing corrinoid:CoM methyltransferase (MtaA; MA0855) (Table 2). The full-length Pyl-containing MtaA was observed in a gel that contained only the WT proteome. Because of its low abundance, the observed value of 1.8 ± 0.4-fold up-regulated in ΔpylT may be inaccurate, simply reflecting the fact that the spot intensity is close to background. The molecular mass of the protein in the gel indicates that Pyl is present (Table 3 and Table S4).
Table 3.
Newly identified Pyl proteins in M. acetivorans
| Group/locus | Name | Carbon source | UAG translation | Mr stop | Mr Pyl | Mr observed |
| i | Essential | |||||
| MA0528 | MttB | TMA | Mr | 36.1 | 53.8 | 50 |
| MA0855 | MtaA | MeOH | Mr | 9.7 | 38.4 | 38 |
| MA0932 | MttB | TMA | Mr | 36.2 | 54.1 | 52 |
| ii | Read through | |||||
| MA0864 | His kinase | TMA | 1 peptide | 25.2 | 111.9 | 110 |
| MA3625 | Endonuclease | MeOH | 6 peptides | 9.3 | 51.9 | 50 |
| iii | Extension | |||||
| MA1887 | Hypo | TMA | 1 peptide | 35.2 | 36.2 | 36 |
| MA2509 | Hypo | TMA | Mr | 18.6 | 19.8 | 20 |
| iv | Unassigned | |||||
| MA0154 | PylB | MeOH | Pyl peptide | 40.3 | 42.9 | 42 |
Molecular mass (Mr) is in kilodaltons. The UAG translation column indicates the data source that confirmed read through of UAGs: Mr indicates UAG translation confirmed by the mass of the protein observed in gel; number of peptides indicates the number of significant peptides identified downstream of the Pyl locus; Pyl peptide indicates MS/MS identification of the Pyl-containing peptide. Hypo, hypothetical protein.
Proteins catalyzing methanogensis from methanol are less abundant in ΔpylT compared with WT. Global changes in the proteome reduce the fraction of soluble protein devoted to methanol methanogenesis in ΔpylT cells (Table S5). Methanol methanogenesis proteins account for 33% of the soluble protein in the WT cell, but this fraction is reduced to 21% in ΔpylT. The most striking examples are the MtaB2 (MA1616) and MtaC2 (MA4391) proteins, which each make up 6% of the soluble proteome in WT cells and ≤0.4% of the proteome in ΔpylT cells (Table S5).
Reduced abundance of methanol methanogensis enzymes in ΔpylT may be responsible for the observed growth defects, but the largest protein abundance changes in the ΔpylT proteome involve a related metabolism—methanogensis from methylsulfides. Methylsulfide methanogenesis proceeds similarly to the methanol route, including a methylsulfide methyltransferase (MtsX), a cognate corrinoid protein, and transfer of the methyl group to CoM with subsequent entry into the generic methanogenesis pathway. M. acetivorans was known to grow on DMS (34). Characterization of the methyltransferases was completed recently (35). MtsX enzymes are distinct from their relatives involved in methanol and methylamine methanogenesis, because both methyltransferase and corrinoid protein exist in a single polypeptide. M. acetivorans encodes three MtsX genes (mtsD, mtsF, and mtsH). All three proteins were identified by MS. According to LC-MS/MS, MtsH is distinct in ΔpylT cells. MtsF (30-fold) and MtsD (112-fold) were the most up-regulated proteins in ΔpylT. MtsF increased to 1.2% of the soluble proteome in ΔpylT from 0.03% in WT, and MtsD increased to 5.0% in ΔpylT compared with 0.04% in WT. The data indicate a compensatory switch away from methanol and toward methyl sulfide metabolism in M. acetivorans cells that are only able to encode 20 amino acids.
There are ArsR-type transcriptional regulators in the genomic context of each of the three mtsX genes (35, 36). We confirmed expression of one of these putative regulators (msrC; MA4383), which is twofold more abundant in WT (Table S2). We detected one significant peptide each for MsrB (MA0460) and MsrD (MA4397) in ΔpylT. An M. acetivorans msrC deletion mutant showed no phenotype during TMA growth, but methanol-grown cells showed twofold increases in both generation and lag time. Expression of methanol methyltransferase genes was also inhibited (36). This observation gave rise to the name methanol-specific regulators (msrs).
The msrC gene is located adjacent to mtsF, and MtsF protein expression was abolished in the msrC deletion mutant, suggesting a regulatory role for MsrC in DMS metabolism (37). The fact that we observe reduced abundance of MsrC in ΔpylT along with increased MtsX expression indicates that the regulatory system associated with DMS metabolism is complex and may involve multiple regulatory components. Indeed, MA4561 [methyl sulfide methyltransferase-associated sensor (MsmS)] was implicated as another component in MtsF regulation. An msmS deletion mutant constitutively expresses MtsF, whereas in WT cells, MtsF is only expressed in the presence of DMS (38). We hypothesize that, in the pylT mutant, MsrC is one component of a regulatory system that senses reduced metabolic efficiency from methanol methanogenesis and stimulates production of MtsF and MtsD. Expression profiling of the MtsX genes indicated that their abundance is low in methanol-grown cells, but their mRNA abundance increases between 2- and 50-fold when DMS is added to the media (37). The MtsX enzymes are essential for DMS catabolism when DMS is the sole carbon source, but the same enzymes are involved in DMS formation in cells grown on carbon monoxide (35, 39). These reports suggest an alternative possibility that ΔpylT produces increased levels of DMS.
Proteomes of Methanol- and TMA-Grown M. acetivorans.
To gain insight into the metabolic shift in WT cells as they adapt from methanol to TMA substrates, we compared proteomes from WT grown in minimal medium containing either methanol or TMA. We found that enzymes involved in methanol methanogenesis MtaB1, MtaC1, and MtaC2 all show 10-fold up-regulation in methanol-containing media (Table S6). In a proteomic analysis comparing M. acetivorans grown on methanol vs. acetate, methanol methyltransferase components MtaB1 (500-fold), MtaC1 (500-fold), and MtaC2 (90-fold) were also significantly up-regulated in methanol-grown cells (40). A central component of the methanogensis pathway, formyl-methanofuran dehydrogenase (Fmd) subunits A–F, was 10- to 40-fold up-regulated in methanol compared with acetate media (40). We found that FmdA is another 20-fold up-regulated in TMA-metabolizing cells. A flavodoxin (MA2699) was 10-fold up-regulated in methanol vs. TMA. The MA2699 transcript was 29-fold up-regulated on methanol compared with acetate (40), suggesting a functional role in electron transport specific to methanol metabolism. MtsD showed 2.5-fold higher abundance in methanol-grown cells, suggesting that DMS methanogenesis is somewhat up-regulated normally during growth on methanol compared with TMA.
Proteins up-regulated in TMA-grown cells included those expressed from loci MA0146 (fivefold) and MA0527 encoding enzymes responsible for methanogensis from dimethylamine (Table S6). The most differentially regulated gene is MA1362 (O-linked GlcNAc transferase). The enzyme is involved in protein glycosylation and induced over 20-fold in TMA- vs. methanol-grown cells, which could represent an uncharacterized mechanism to modulate protein function in TMA metabolism. Enzymes involved in lysine (lysA; 4-fold) and arginine (argJ; 17-fold) biosynthesis were also significantly more abundant in TMA-grown cells. An archaeal peptide chain release factor 1 paralog (SI Text) and three hypothetical proteins (MA0864, MA1887, and MA3997) are 3- to 10-fold up-regulated in TMA-grown cells. Two of these genes (MA0864 and MA1887) contain in-frame TAGs. We confirmed translation of the UAG codon in the histidine kinase MA0864 (Table 3 and Table S4). Histidine kinases are one of a small number of protein families enriched with Pyl proteins (Fig. S2). Identifying Pyl in MA0864 may indicate that the larger family of histidine kinases (of which there are eight in M. acetivorans) also encodes Pyl and possibly requires the residue for activity.
Expanding the Pyl Proteome.
Our proteomic and mRNA expression data (Fig. S1) expanded the known Pyl proteome from few previously experimentally verified Pyl proteins (28, 29) to the now confirmed expression of 19 UAG-containing ORFs (Table 3, Fig. S1, and Tables S1 and S4). In three cases (histidine kinases MA2732 and MA3962, recombinase MA4127, and Thg1 MA0816), the truncated protein was detected and identified by molecular mass that resulted from decoding a UAG as stop in ΔpylT. The data show that M. acetivorans is capable of terminating peptide chain extension at UAG and indicate that UAG was converted to a stop codon by deletion of pylT. We detected no peptides suggesting that UAG is read as a stop codon in WT. Several putative Pyl-containing proteins (Table 3, Fig. S2, and Table S4) were identified as expressed by detection of peptides upstream of the Pyl locus. We were unable to identify peptides including or downstream of the Pyl locus in all cases, and therefore, it remains unclear if these UAGs are translated. For many of these proteins, Pyl is located at or near the C terminus; therefore, it is not possible to confirm Pyl insertion based on molecular mass of the protein, and there are few or no downstream peptides to confirm UAG read through.
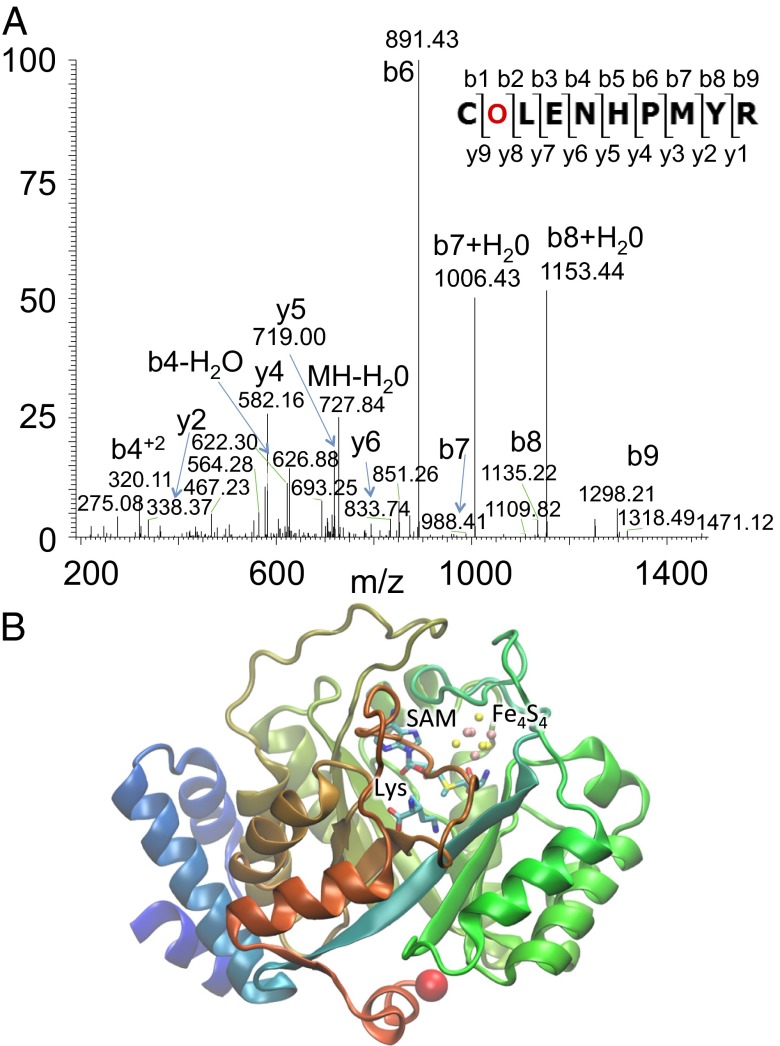
For three proteins (MA0864, MA1887, and MA3625), MS/MS analysis identified peptides beyond the UAG codon that showed UAG read through and indicated the presence of Pyl in the corresponding protein. Determination of the relative molecular masses confirmed UAG translation for four additional proteins (MA0528, MA0855, MA0932, and MA2509). Among these confirmed full-length proteins are the methanogenesis protein MtaA, two MttB homologs, and three hypothetical proteins (Table 3 and Table S4). We identified a Pyl-containing peptide by MS/MS for the PylB protein, which catalyzes the first step of Pyl biosynthesis (41) (Fig. 3).
Fig. 3.
Pyl incorporation in the Pyl biosynthesis enzyme methylornithine synthase (pylB). (A) MS/MS spectra identified Pyl (O) in the relevant tryptic peptide from WT M. acetivorans cells grown on methanol. In the peptide, Cys (C) is carbamidomethylated, and Met (M) is oxidized. (B) A view of the crystal structure of PylB (42) is shown with its substrate lysine (Lys) and cofactors S-adenosyl methionine (SAM) and an iron-sulfur (Fe4S4) cluster in the active site. The protein is colored to highlight the N (blue) and C (red) termini. The structure does not include the Pyl peptide. When pylB is expressed in WT M. acetivorans cells, the protein contains an additional 21 amino acids, including one Pyl residue, which would extend the C terminus beyond the red sphere.
Interestingly, we were able to detect the C-terminal fragment of the tRNA editing enzyme Thg1 in ΔpylT. In WT cells, Thg1 is expressed as a full-length Pyl-containing protein (29). Thg1 had initially been annotated as two separate proteins in M. acetivorans, and it seems that, in the absence of Pyl, the C-terminal fragment of Thg1 is expressed independently. We previously showed that the two recombinant purified Thg1 halves could function in trans in in vitro activity assays (29). Our proteomic data showed that the second peptide is expressed independently, and therefore, the protein may reassemble in the cell to form functional Thg1 without Pyl.
Discussion
Roles of Pyl in M. acetivorans.
Methanogenesis is the major source of biogenic methane production on the planet. Methane is, at the same time, a potent greenhouse gas and a promising biofuel. These facts are primary reasons why M. acetivorans and its relatives are not only of biological interest but also a likely source of biotechnological applications. It was assumed that the biological role of Pyl was restricted to its catalytic role in the active site of methylamine methyltransferases (28) until we showed that its cotranslational incorporation is essential for formation of full-length Thg1 in M. acetivorans (29). Taking these prior observations into account, Pyl proteins can be classified into three categories (Table 3 and Table S4): (i) Pyl is required for enzymatic activity, (ii) Pyl is noncatalytic but required for translation of full-length protein, and (iii) Pyl is inserted at intended stop codons, resulting in a short peptide extension. Group iv includes PylB, where the role of the extension is unclear. Most potential and experimentally verified Pyl proteins that we found fall into class iii.
Pyl in the Active Site.
We confirmed that both MttB homologs (MA0582 and MA0932) contain Pyl, which is in line with the essential catalytic role of Pyl in these enzymes. Excitingly, we also identified a Pyl-containing methylcobamide:CoM methyltransferase (mtaA). Although it is unclear whether Pyl contributes to the catalytic activity of MtaA, Pyl is located within the characteristic triosephosphate isomerase (TIM)-barrel fold, like the catalytically active Pyl residues in MttB, suggesting a catalytic role for Pyl in MtaA. Other Pyl proteins may use the unusual amino acids in their active sites, but because many are uncharacterized, it is not possible to determine the role of Pyl at this time. We observed that Pyl proteins (potential and experimentally verified) are, indeed, overrepresented in methanogenesis-related enzymes. Histidine kinases, recombinases, transposases, and radical S-adenosylmethionine enzymes are other biochemical activities that are enriched with Pyl proteins (Fig. S2), indicating the potential of a catalytic Pyl in these enzymes.
Pyl for UAG Read Through.
Pyl is only required for read through of an in-frame UAG in Thg1 but not its activity (29). Pyl incorporation in two more proteins, MA0864 and MA3625, extends the proteins from 215 to 975 amino acids and from 79 to 446 amino acids, respectively. Pyl incorporation in both MA0864 and MA3625 links an N-terminal hypothetical protein to a C-terminal protein with known homologs. The C-terminal part of MA3625 encodes a hypothetical tRNA splicing endonuclease, and the C terminus of MA0864 encodes a histidine kinase. Although our data clearly show that a full-length protein is formed, the function and potential effect of joining these proteins or the role of Pyl (if any) remains to be characterized.
Pyl at the End.
A third group of proteins includes candidates that cannot be easily assigned to one of the other groups (i.e., PylB, the hypothetical protein MA3459, and the putative histidine kinase MA3962). For these proteins, several amino acids residues are added as a result of UAG read through, with the potential of a regulatory function. Although MA3459 and MA3962 are clearly expressed in cells, we do not have experimental evidence that their protein products include Pyl. For PylB, however, we were able to detect the Pyl-containing peptide (Fig. 3A). PylB is the first enzyme in the Pyl biosynthesis pathway catalyzing the isomerization of lysine to 3-methylornithine (18). PylB is a TIM barrel (Fig. 3B), with a central cavity containing the catalytic (4Fe-4S) cluster and S-adenosylmethionine (42). The recombinant PylB is catalytically active without the extension of the 21-amino acid residue Pyl peptide (18). Nevertheless the Pyl-containing peptide could modulate PylB activity or contribute to a regulatory mechanism in which the level of available Pyl is sensed, reminiscent of the Trp operon (43).
UAG Is Reassigned to Pyl.
It has not been conclusively shown whether UAG has a dual meaning (i.e., stop and Pyl) in M. acetivorans or if all UAG codons are read as Pyl. Earlier work with the Pyl-containing methylamine methyl transferases (26) suggested the existence of an RNA recoding element (Pyl insertion sequence, PYLIS) downstream of the Pyl codon, similar to the selenocysteine (Sec) insertion sequence (SECIS) found in selenoprotein mRNAs (44). In the case of Sec, SECIS designates particular UGA codons as Sec, whereas other UGA codons are stop signals. The putative PYLIS element was not subsequently identified in other Pyl-containing proteins, such as Thg1, or those listed in Table 3. Pyl can be incorporated into normally non-Pyl proteins, such as a recombinant uidA gene product in M. acetivorans (26) or β-galactosidase in E. coli expressing a recombinant Pyl system (22). We found no evidence that UAG is read as stop in WT M. acetivorans, but we did identify peptides from UAG stop codons in ΔpylT. Taken together, this evidence indicates that the UAG codon is reassigned to Pyl and not selectively recoded.
UAG Codon Evolution.
In contrast to M. acetivorans, the Pyl-decoding bacterium Acetohalobium arabaticum dynamically expands and reduces its genetic code depending on the carbon source, encoding Pyl only when TMA is present (17). Some bacteria have the genes required to reassign UAG to Pyl but do not express Pyl-containing proteins. Under the conditions tested and even in the presence of TMA, Desulfitobacterium dehalogenans and Desulfitobacterium hafniense did not produce Pyl-tRNAPyl or detectable Pyl-containing protein in vivo (17). In contrast, all archaea examined so far [Methanosarcina barkeri (28), M. acetivorans (26), Methanosarcina mazei (45), and Methanococcoides burtonii (46)] that genetically encode Pyl seem to express Pyl-containing proteins in cells grown on methylamines or methanol (29). Underlying these different interpretations of the UAG codon are clear differences in UAG codon use. M. acetivorans and other Pyl-decoding archaea have significantly fewer UAG codons (∼5%) than both non–Pyl-decoding archaea and Pyl-decoding bacteria (∼20%) (17). The data suggest that, in Pyl-decoding archaea, selective pressure eliminated UAG codons at positions unfavorable to Pyl insertion and protein extension. Conversely, Pyl-decoding bacteria control the meaning of the UAG codon by either silencing the Pyl operon or only synthesizing Pyl-tRNAPyl when TMA is present.
Impact of Expanding the Genetic Code on Cellular Physiology.
Microbiome (47) and environmental microbial sequencing (48) work is revealing organisms with new genetic code variations. At the same time, advances in synthetic biology are leading to new engineered genetic codes (3). Despite these achievements, little is known about the impact of an expanded genetic code on the proteome, physiology, or fitness of the cell. Initial studies indicate that there is a fitness cost for engineering genetic code expansion. Expanding the genetic code of E. coli by reassigning the UAG codon to phosphoserine leads to slowly growing cells, which is caused by unnatural extension of proteins beyond their normal UAG stop signal and incorporation of phosphoserine throughout the proteome (5, 49).
We observed that M. acetivorans derives a selective advantage from maintaining an expanded genetic code with Pyl. Methanosarcina are able to survive using more chemically diverse carbon sources [acetate, methanol, methylamines, or DMS (25)] than non–Pyl-decoding methanogens. Furthermore, Pyl confers a fitness advantage in methanol growth. The proteomic comparison of WT and ΔpylT indicates repression of methanol methanogenesis enzymes (one of which contains Pyl) with a significant metabolic shift toward DMS metabolism. It is possible that, by inducing DMS metabolism, the cells are compensating for the reduced metabolic efficiency of methanol methanogenesis without Pyl. Deletion of the MtsX genes in addition to tRNAPyl should result in more drastic phenotypic defects. Such experiments would help to establish a mechanism for the metabolic compensation suggested by the proteomic data.
M. acetivorans is, in a way, prepared to survive without Pyl but at a phenotypic cost that reduces carbon source range, growth rate, and metabolic efficiency. Although M. acetivorans can survive with a reduced genetic code, we found that Pyl is essential for optimal growth on methanol and that removing Pyl has a tremendous impact on the proteome. The data show that reassignment of a rarely used codon can dramatically alter cellular metabolism. These effects will have to be considered in approaches to reduce the genetic code of organisms to open new codons for genetic code expansion.
Materials and Methods
We conducted three independent proteomic investigations. In two gel-based methods, we separated three soluble proteomes: WT cells grown on TMA, WT cells grown on methanol, and ΔpylT cells grown on methanol. Proteomes were separated on independent gels (2D gels) and overlaid to quantitate differential protein abundance or simultaneously, by difference gel electrophoresis. For both gel-based methods, spots showing differential regulation >1.8-fold were characterized by MS/MS. We separated proteomes from methanol-grown cells (ΔpylT or WT) by LC, and fractions were analyzed by MS/MS. Proteins identified with two or more significant peptides in only one of the samples (e.g., WT or ΔpylT) but not both were labeled as distinct. Full details of the markerless deletion of tRNAPyl, growth conditions, and proteomic analysis are in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dieter Jahn and Martina Jahn for providing laboratory space to perform initial proteomic experiments. We also thank Terrence Wu (Keck MS and Proteomics Resource, Yale University), Manfred Nimtz (Helmholtz Centre for Infection Research), and Thorsten Johl (Helmholtz Centre for Infection Research) for support with MS analysis and Yuchen Liu, Hans Aerni, and Ava Artaiz for critical discussions of the manuscript. We thank Bill Metcalf, Gary Olsen, Claudia Reich, and Carl Woese for reagents, training, discussions, and their enthusiasm for the Archaea. This work was supported by Natural Sciences and Engineering Research Council of Canada Grants RGPIN 04282-2014 (to P.O.) and RGPIN 04776-2014 (to I.U.H.), National Institutes of Health Grant GM22854 (to D.S.), and Defense Advanced Research Projects Agency Contract N66001-12-C-4211 (to D.S.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1420193111/-/DCSupplemental.
References
- 1.Gibson DG, et al. Complete chemical synthesis, assembly, and cloning of a Mycoplasma genitalium genome. Science. 2008;319(5867):1215–1220. doi: 10.1126/science.1151721. [DOI] [PubMed] [Google Scholar]
- 2.Lajoie MJ, et al. Genomically recoded organisms expand biological functions. Science. 2013;342(6156):357–360. doi: 10.1126/science.1241459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O’Donoghue P, Ling J, Wang YS, Söll D. Upgrading protein synthesis for synthetic biology. Nat Chem Biol. 2013;9(10):594–598. doi: 10.1038/nchembio.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu CC, Schultz PG. Adding new chemistries to the genetic code. Annu Rev Biochem. 2010;79:413–444. doi: 10.1146/annurev.biochem.052308.105824. [DOI] [PubMed] [Google Scholar]
- 5.Heinemann IU, et al. Enhanced phosphoserine insertion during Escherichia coli protein synthesis via partial UAG codon reassignment and release factor 1 deletion. FEBS Lett. 2012;586(20):3716–3722. doi: 10.1016/j.febslet.2012.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moore B, Persson BC, Nelson CC, Gesteland RF, Atkins JF. Quadruplet codons: Implications for code expansion and the specification of translation step size. J Mol Biol. 2000;298(2):195–209. doi: 10.1006/jmbi.2000.3658. [DOI] [PubMed] [Google Scholar]
- 7.Wang K, et al. Optimized orthogonal translation of unnatural amino acids enables spontaneous protein double-labelling and FRET. Nat Chem. 2014;6(5):393–403. doi: 10.1038/nchem.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bröcker MJ, Ho JM, Church GM, Söll D, O’Donoghue P. Recoding the genetic code with selenocysteine. Angew Chem Int Ed Engl. 2014;53(1):319–323. doi: 10.1002/anie.201308584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krishnakumar R, et al. Transfer RNA misidentification scrambles sense codon recoding. ChemBioChem. 2013;14(15):1967–1972. doi: 10.1002/cbic.201300444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kwon I, Kirshenbaum K, Tirrell DA. Breaking the degeneracy of the genetic code. J Am Chem Soc. 2003;125(25):7512–7513. doi: 10.1021/ja0350076. [DOI] [PubMed] [Google Scholar]
- 11.Zeng Y, Wang W, Liu WR. Towards reassigning the rare AGG codon in Escherichia coli. ChemBioChem. 2014;15(12):1750–1754. doi: 10.1002/cbic.201400075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lajoie MJ, et al. Probing the limits of genetic recoding in essential genes. Science. 2013;342(6156):361–363. doi: 10.1126/science.1241460. [DOI] [PubMed] [Google Scholar]
- 13.Krishnakumar R, Ling J. Experimental challenges of sense codon reassignment: An innovative approach to genetic code expansion. FEBS Lett. 2014;588(3):383–388. doi: 10.1016/j.febslet.2013.11.039. [DOI] [PubMed] [Google Scholar]
- 14.Hammerling MJ, et al. Bacteriophages use an expanded genetic code on evolutionary paths to higher fitness. Nat Chem Biol. 2014;10(3):178–180. doi: 10.1038/nchembio.1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Knight RD, Freeland SJ, Landweber LF. Rewiring the keyboard: Evolvability of the genetic code. Nat Rev Genet. 2001;2(1):49–58. doi: 10.1038/35047500. [DOI] [PubMed] [Google Scholar]
- 16.Ambrogelly A, Palioura S, Söll D. Natural expansion of the genetic code. Nat Chem Biol. 2007;3(1):29–35. doi: 10.1038/nchembio847. [DOI] [PubMed] [Google Scholar]
- 17.Prat L, et al. Carbon source-dependent expansion of the genetic code in bacteria. Proc Natl Acad Sci USA. 2012;109(51):21070–21075. doi: 10.1073/pnas.1218613110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gaston MA, Zhang L, Green-Church KB, Krzycki JA. The complete biosynthesis of the genetically encoded amino acid pyrrolysine from lysine. Nature. 2011;471(7340):647–650. doi: 10.1038/nature09918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wan W, Tharp JM, Liu WR. Pyrrolysyl-tRNA synthetase: An ordinary enzyme but an outstanding genetic code expansion tool. Biochim Biophys Acta. 2014;1844(6):1059–1070. doi: 10.1016/j.bbapap.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Longstaff DG, et al. A natural genetic code expansion cassette enables transmissible biosynthesis and genetic encoding of pyrrolysine. Proc Natl Acad Sci USA. 2007;104(3):1021–1026. doi: 10.1073/pnas.0610294104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nozawa K, et al. Pyrrolysyl-tRNA synthetase-tRNA(Pyl) structure reveals the molecular basis of orthogonality. Nature. 2009;457(7233):1163–1167. doi: 10.1038/nature07611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ambrogelly A, et al. Pyrrolysine is not hardwired for cotranslational insertion at UAG codons. Proc Natl Acad Sci USA. 2007;104(9):3141–3146. doi: 10.1073/pnas.0611634104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chin JW. Expanding and reprogramming the genetic code of cells and animals. Annu Rev Biochem. 2014;83:379–408. doi: 10.1146/annurev-biochem-060713-035737. [DOI] [PubMed] [Google Scholar]
- 24.Borrel G, et al. Unique characteristics of the pyrrolysine system in the 7th order of methanogens: Implications for the evolution of a genetic code expansion cassette. Archaea. 2014;2014:374146. doi: 10.1155/2014/374146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu Y, Whitman WB. Metabolic, phylogenetic, and ecological diversity of the methanogenic archaea. Ann N Y Acad Sci. 2008;1125:171–189. doi: 10.1196/annals.1419.019. [DOI] [PubMed] [Google Scholar]
- 26.Mahapatra A, et al. Characterization of a Methanosarcina acetivorans mutant unable to translate UAG as pyrrolysine. Mol Microbiol. 2006;59(1):56–66. doi: 10.1111/j.1365-2958.2005.04927.x. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Y, Gladyshev VN. High content of proteins containing 21st and 22nd amino acids, selenocysteine and pyrrolysine, in a symbiotic deltaproteobacterium of gutless worm Olavius algarvensis. Nucleic Acids Res. 2007;35(15):4952–4963. doi: 10.1093/nar/gkm514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hao B, et al. A new UAG-encoded residue in the structure of a methanogen methyltransferase. Science. 2002;296(5572):1462–1466. doi: 10.1126/science.1069556. [DOI] [PubMed] [Google Scholar]
- 29.Heinemann IU, et al. The appearance of pyrrolysine in tRNAHis guanylyltransferase by neutral evolution. Proc Natl Acad Sci USA. 2009;106(50):21103–21108. doi: 10.1073/pnas.0912072106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lessner DJ, Ferry JG. The archaeon Methanosarcina acetivorans contains a protein disulfide reductase with an iron-sulfur cluster. J Bacteriol. 2007;189(20):7475–7484. doi: 10.1128/JB.00891-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ling J, et al. Protein aggregation caused by aminoglycoside action is prevented by a hydrogen peroxide scavenger. Mol Cell. 2012;48(5):713–722. doi: 10.1016/j.molcel.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cukras AR, Green R. Multiple effects of S13 in modulating the strength of intersubunit interactions in the ribosome during translation. J Mol Biol. 2005;349(1):47–59. doi: 10.1016/j.jmb.2005.03.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spierer P, Bogdanov AA, Zimmermann RA. Parameters for the interaction of ribosomal proteins L5, L18, and L25 with 5S RNA from Escherichia coli. Biochemistry. 1978;17(25):5394–5398. doi: 10.1021/bi00618a012. [DOI] [PubMed] [Google Scholar]
- 34.Ni S, Woese CR, Aldrich HC, Boone DR. Transfer of Methanolobus siciliae to the genus Methanosarcina, naming it Methanosarcina siciliae, and emendation of the genus Methanosarcina. Int J Syst Bacteriol. 1994;44(2):357–359. doi: 10.1099/00207713-44-2-357. [DOI] [PubMed] [Google Scholar]
- 35.Oelgeschläger E, Rother M. In vivo role of three fused corrinoid/methyl transfer proteins in Methanosarcina acetivorans. Mol Microbiol. 2009;72(5):1260–1272. doi: 10.1111/j.1365-2958.2009.06723.x. [DOI] [PubMed] [Google Scholar]
- 36.Bose A, Metcalf WW. Distinct regulators control the expression of methanol methyltransferase isozymes in Methanosarcina acetivorans C2A. Mol Microbiol. 2008;67(3):649–661. doi: 10.1111/j.1365-2958.2007.06075.x. [DOI] [PubMed] [Google Scholar]
- 37.Bose A, Kulkarni G, Metcalf WW. Regulation of putative methyl-sulphide methyltransferases in Methanosarcina acetivorans C2A. Mol Microbiol. 2009;74(1):227–238. doi: 10.1111/j.1365-2958.2009.06864.x. [DOI] [PubMed] [Google Scholar]
- 38.Molitor B, et al. A heme-based redox sensor in the methanogenic archaeon Methanosarcina acetivorans. J Biol Chem. 2013;288(25):18458–18472. doi: 10.1074/jbc.M113.476267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moran JJ, House CH, Vrentas JM, Freeman KH. Methyl sulfide production by a novel carbon monoxide metabolism in Methanosarcina acetivorans. Appl Environ Microbiol. 2008;74(2):540–542. doi: 10.1128/AEM.01750-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li L, et al. Quantitative proteomic and microarray analysis of the archaeon Methanosarcina acetivorans grown with acetate versus methanol. J Proteome Res. 2007;6(2):759–771. doi: 10.1021/pr060383l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krzycki JA. The path of lysine to pyrrolysine. Curr Opin Chem Biol. 2013;17(4):619–625. doi: 10.1016/j.cbpa.2013.06.023. [DOI] [PubMed] [Google Scholar]
- 42.Quitterer F, List A, Eisenreich W, Bacher A, Groll M. Crystal structure of methylornithine synthase (PylB): Insights into the pyrrolysine biosynthesis. Angew Chem Int Ed Engl. 2012;51(6):1339–1342. doi: 10.1002/anie.201106765. [DOI] [PubMed] [Google Scholar]
- 43.Babitzke P, Gollnick P, Yanofsky C. The mtrAB operon of Bacillus subtilis encodes GTP cyclohydrolase I (MtrA), an enzyme involved in folic acid biosynthesis, and MtrB, a regulator of tryptophan biosynthesis. J Bacteriol. 1992;174(7):2059–2064. doi: 10.1128/jb.174.7.2059-2064.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yoshizawa S, Böck A. The many levels of control on bacterial selenoprotein synthesis. Biochim Biophys Acta. 2009;1790(11):1404–1414. doi: 10.1016/j.bbagen.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 45.Veit K, Ehlers C, Schmitz RA. Effects of nitrogen and carbon sources on transcription of soluble methyltransferases in Methanosarcina mazei strain Go1. J Bacteriol. 2005;187(17):6147–6154. doi: 10.1128/JB.187.17.6147-6154.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goodchild A, et al. A proteomic determination of cold adaptation in the Antarctic archaeon, Methanococcoides burtonii. Mol Microbiol. 2004;53(1):309–321. doi: 10.1111/j.1365-2958.2004.04130.x. [DOI] [PubMed] [Google Scholar]
- 47.Campbell JH, et al. UGA is an additional glycine codon in uncultured SR1 bacteria from the human microbiota. Proc Natl Acad Sci USA. 2013;110(14):5540–5545. doi: 10.1073/pnas.1303090110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ivanova NN, et al. Stop codon reassignments in the wild. Science. 2014;344(6186):909–913. doi: 10.1126/science.1250691. [DOI] [PubMed] [Google Scholar]
- 49.Aerni HR, Shifman MA, Rogulina S, O’Donoghue P, Rinehart J. Revealing the amino acid composition of proteins within an expanded genetic code. Nucleic Acids Res. 2014 doi: 10.1093/nar/gku1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.