Significance
We determine the number of broadly neutralizing antibodies required to inhibit influenza virus membrane fusion by simultaneously observing individual viral particles undergoing fusion and counting the number of antibodies bound to them. The viral membrane fusion process is mediated by fusion proteins whose activity is blocked through the binding of these antibodies to evolutionarily conserved epitopes. Surprisingly, the number of antibodies required for inhibition is markedly lower than the number of fusion proteins present, indicating virus neutralization does not require saturation of epitope occupancy. Overall, our results support a model of membrane fusion requiring several fusion proteins working together in a coordinated, stochastic fashion, and the inhibition of this process through disruption of fusion protein coordination.
Keywords: influenza, neutralizing antibody, hemagglutinin, neutralization stoichiometry, membrane fusion
Abstract
The ability of antibodies binding the influenza hemagglutinin (HA) protein to neutralize viral infectivity is of key importance in the design of next-generation vaccines and for prophylactic and therapeutic use. The two antibodies CR6261 and CR8020 have recently been shown to efficiently neutralize influenza A infection by binding to and inhibiting the influenza A HA protein that is responsible for membrane fusion in the early steps of viral infection. Here, we use single-particle fluorescence microscopy to correlate the number of antibodies or antibody fragments (Fab) bound to an individual virion with the capacity of the same virus particle to undergo membrane fusion. To this end, individual, infectious virus particles bound by fluorescently labeled antibodies/Fab are visualized as they fuse to a planar, supported lipid bilayer. The fluorescence intensity arising from the virus-bound antibodies/Fab is used to determine the number of molecules attached to viral HA while a fluorescent marker in the viral membrane is used to simultaneously obtain kinetic information on the fusion process. We experimentally determine that the stoichiometry required for fusion inhibition by both antibody and Fab leaves large numbers of unbound HA epitopes on the viral surface. Kinetic measurements of the fusion process reveal that those few particles capable of fusion at high antibody/Fab coverage display significantly slower hemifusion kinetics. Overall, our results support a membrane fusion mechanism requiring the stochastic, coordinated action of multiple HA trimers and a model of fusion inhibition by stem-binding antibodies through disruption of this coordinated action.
Recently described antibodies capable of neutralizing a broad range of influenza viruses through recognition of highly conserved epitopes on the hemagglutinin protein (HA) have potential use as antiinfluenza therapeutics and for rational design of vaccines, HA-binding proteins, and small molecules (1–5). Such diverse applications require a detailed understanding of the infection mechanisms that are disrupted by antibody binding. An important parameter for in vitro antibody characterization is the binding stoichiometry, which relates the number of antibodies that must bind a virus in order to achieve a functional output, such as viral neutralization (6, 7). Here, we use fluorescence microscopy to visualize individual virus particles and measure the stoichiometry of broadly neutralizing antibodies as they disrupt the fusogenic activity of the HA protein.
The homotrimeric HA transmembrane protein consists of two disulfide-linked domains, HA1 and HA2 (8, 9), and exhibits substantial antigenic drift, having two phylogenetically distinct groups—groups 1 and 2—with 18 subtypes (10). The HA1 head domain, distal from the viral envelope, contains a binding site for sialic acid moieties that binds virions to a target cell and facilitates their entry into a cellular endosome via clathrin-mediated endocytosis. The HA2 stem domain comprises mostly the envelope-proximal ectodomain and the transmembrane domain. The low pH of late endosomes triggers the stem to unfold and embed its hydrophobic N-terminal region into the endosomal membrane. Refolding of the protein brings the viral and endosomal membranes close together and catalyzes their fusion (11, 12). Several biophysical studies indicate that multiple HA trimers must work together by coordinating their conformational changes for membrane fusion to occur (13–16).
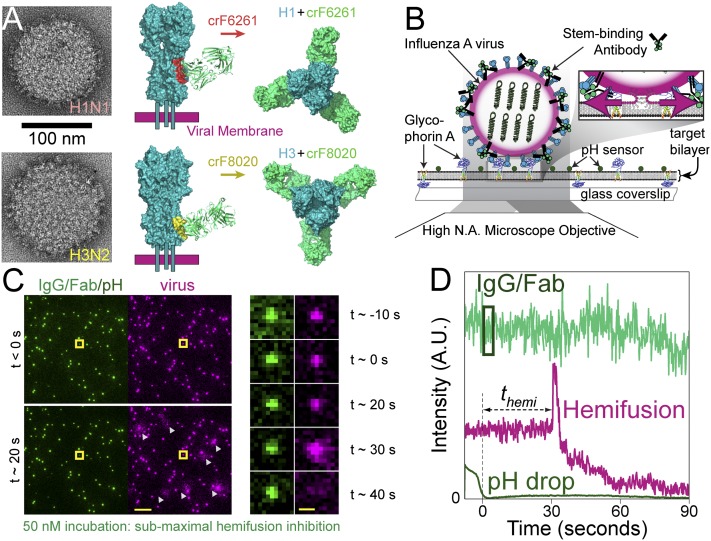
Head-binding antibodies typically recognize variable loop regions surrounding the receptor site and show serotype-specific neutralization (1, 17, 18), although some can neutralize a limited set of viral serotypes (19–21). In contrast, stem-binding antibodies recognize an epitope region that is highly conserved between influenza strains and possess a broad neutralization capacity across many viral subtypes (19, 22–26) or even across groups (19, 27, 28). We recently demonstrated that binding of HA by the broadly neutralizing, stem-binding antibodies CR6261 (group 1-specific) and CR8020 (group 2-specific) (22–24) results in inhibition of HA-mediated viral membrane fusion (29). The ability of antibodies to block fusion confirms the accessibility of their membrane-proximal epitope on intact viruses despite the dense packing of surface proteins (Fig. 1A), an aspect also recently shown by cryoelectron tomography (30).
Fig. 1.
Experimental design and readouts. (A, Left) Transmission EM images of the two influenza A viruses, H1N1 (Top) and H3N2 (Bottom), depicting the high density of spike proteins present on the viral surface. (A, Middle and Left) Side and top views, respectively, showing a space-filling model of crF6261 bound to H1 [Top, Protein Data Bank (PDB) ID code 3GBN with epitope in red] and crF8020 bound to H3 (Bottom, PDB ID code 3SDY with epitope in yellow). These crystal structures highlight differences in proximity of the two epitopes to the viral membrane and of the HA–Fab angle upon binding. (B) Schematic depiction of experimental design. Alexa-488–labeled IgG (or Fab) are bound to R18-labeled influenza A viruses (magenta-edged sphere) immobilized on a glass-supported planar bilayer through interaction with glycophorin A; pH-sensitive fluorescein (pKa 6.4) is also bound to the bilayer surface. Fluorescence is excited and detected via objective TIRF microscopy. Zoom: Acidification of the virus particles causes membrane fusion, resulting in escape of the R18 dye from the viral membrane into the target bilayer and producing a dequenching signal. (C, Left) False-color still frames from a fusion movie at time points before (Top) and 20 s after (Bottom) the pH drop. (Scale bar: 10 μm.) IgG/Fab (green spots, 50 nM incubation) and fluorescein (diffuse background) are visualized on the left, whereas the R18-labeled viruses (magenta) and their low pH-induced dequenching (white triangles) are simultaneously visualized on the right. (C, Right) Image montage of the virus highlighted by the yellow square, which is covered with a subinhibitory number of IgG molecules (green). Its fusion to the bilayer is seen as a flash of R18 intensity (magenta) followed by outward R18 diffusion. (Scale bar: 1 μm.) (D) Fluorescence time trajectory for the highlighted virus in C. Time t = 0 is set to loss of the fluorescein signal (dark green) upon arrival of the fusion-inducing pH 5.0 buffer. The time to hemifusion, themi, occurs at t ≈ 30 s for this virus particle and is observed as the abrupt increase in R18 fluorescence (magenta). The virus-bound IgG/Fab fluorescence (light green) used for stoichiometry measurements is indicated by the box: 1 s after the pH drop and enclosing 3 s of fluorescence information.
Radioisotope labeling of neutralizing IgG antibodies previously indicated that neutralization stoichiometries required large numbers of epitopes to be bound (18, 31). These results were in line with multihit and coating models of virus neutralization, as opposed to single-hit models involving a critical site vulnerable to neutralization (6, 32, 33). However, such experiments are complicated by a convolution of cell-binding inhibition, inhibition of membrane fusion, aggregation of virus particles, and the abrogation of infection postentry (18, 33).
Here, we use a novel fluorescence imaging approach to simultaneously quantify the number of CR6261 or CR8020 antibodies (IgG) or their Fab-fragments (Fab) bound to an individual, infectious virus while directly visualizing its capacity for membrane fusion. This measurement strategy allows us to directly correlate binding stoichiometry to its functional effect of inhibiting HA-mediated membrane fusion. We find that a virus can be rendered nonfusogenic while still having large numbers of unbound HA epitopes. Furthermore, binding of the neutralizing IgG/Fab molecules causes a delay in the kinetics of membrane fusion. Our methodology provides insight into the mechanism of HA-mediated fusion that is difficult to obtain via conventional approaches (34).
Results
Fluorescently Labeled IgG and Fab Inhibit Hemifusion.
We observed individual influenza virus particles by labeling their envelope with the lipophilic dye R18 at concentrations causing partial self-quenching of the dye’s fluorescence. We have previously confirmed that the viruses maintain their infectivity following R18 labeling (29). R18-labeled viruses (A/Puerto Rico/8/34 H1N1 or A/Aichi/68-X31 H3N2) were incubated with the broadly neutralizing, stem-binding antibodies CR6261 or CR8020 (collectively IgG), or their Fab fragments (crF6261 or crF8020, respectively), at increasing concentrations before immobilization. We immobilized the viruses on a planar, lipid bilayer that is formed on top of a glass microscope coverslip placed on an inverted fluorescence microscope (Fig. 1B). Incubation conditions ensured that IgG/Fab binding had achieved equilibrium before immobilization, and dissociation over the course of the experiment was found to be negligible (Fig. S1). One hundred or more virus particles per ∼140 × 70-μm2 field of view were capable of binding to the glass-supported bilayers even at IgG/Fab concentrations as high as 1 μM (Fig. S2). After lowering the pH, we visualize influenza HA-mediated hemifusion of the viral envelope with the bilayer through R18 dequenching at the site of each virus particle (Fig. 1 C and D and refs. 15, 16, and 29). We quantify the hemifusion efficiency as the ratio of virions that underwent fusion to the total number of viral particles visualized. Data in Fig. 2A demonstrates that binding of Alexa Fluor 488-labeled IgG (5.4 ± 0.8 dyes/CR6261 IgG and 5.0 ± 0.5 dyes/CR8020 IgG; Fig. S3 and Table S1) or Fab (2.4 ± 0.3 dyes/crF6261 Fab and 2.3 ± 0.1 dyes/crF8020 Fab) to viral HA causes a group-specific, dose-dependent reduction in the hemifusion efficiency of the H1N1 and H3N2 viral strains, similar to our previous reports (29). This observation indicates that both monovalent and bivalent binding (valency referring to the number of paratopes bound to epitopes) can lead to hemifusion inhibition directly through epitope recognition.
Fig. 2.
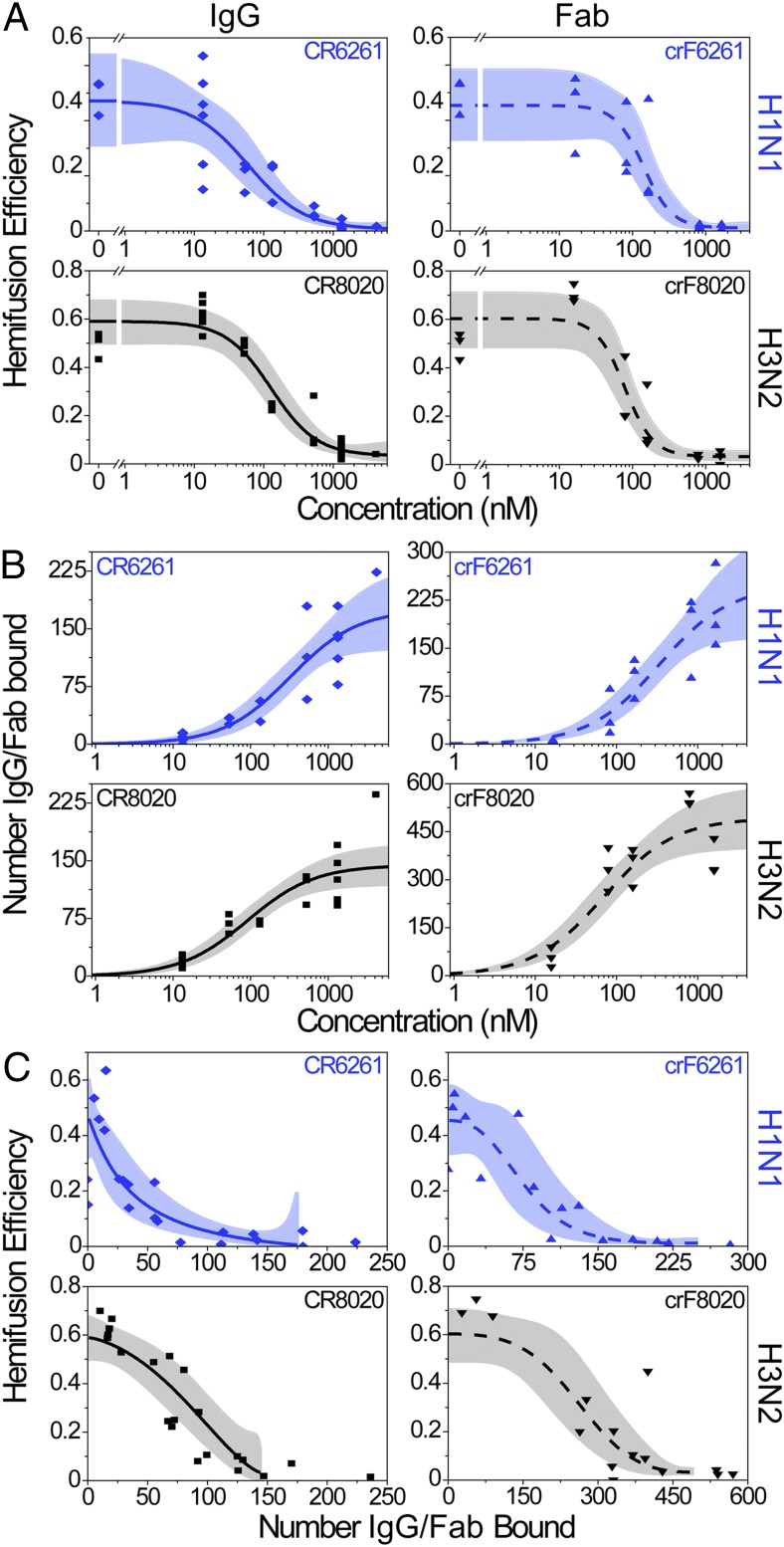
Hemifusion inhibition and antibody stoichiometry. In A–C, IgG data are on the left-hand graphs (solid fit lines), Fab data are on the right (dashed fit lines); top rows are the H1N1 strain (blue) and bottom rows are H3N2 (black). Each data point represents a single experimental run (CR6261 IgG shown as diamonds, crF6261 Fab as upward triangles, CR8020 IgG as squares, and crF8020 as downward triangles); the best-fit lines are in blue or black and their 95% confidence bands are in light blue or gray. (A) Hemifusion efficiency decreases as the concentration of neutralizing IgG or Fab is increased. (B) The median number of neutralizing IgG or Fab bound to virions increases as the concentration used for incubation with virus increases. (C) Plot of hemifusion data (A) versus the number of IgG or Fab bound to the viral surface (B), allowing for the estimation of the number of IgG/Fab required for a given reduction in hemifusion efficiency. Fit lines used are the logistic function (A), hyperbolic function (B), and a combination of the two (C) (SI Materials and Methods). Fit lines in C are truncated at high coverage and correspond to the plateau values obtained in A and B at high IgG/Fab concentrations.
Using bilayers supported by a PEG or dextran cushion (15) (rather than by glass), we were able to observe viral content release and found that the requisite formation of a fusion pore was inhibited to a similar extent as hemifusion, indicating that hemifusion is, indeed, a functional readout for full fusion (Fig. S4). In a few instances at low IgG concentration, antibody-labeled hemagglutinin proteins could be seen to diffuse into the target bilayer after successful content release (Movies S1–S3). This observation demonstrates a complete merger of the viral membrane with the target-supported bilayer.
Maximal Hemifusion Inhibition by IgG Binding Occurs Below Full Epitope Occupancy.
Binding stoichiometry for each virus particle is determined as a quotient of two integrated fluorescence intensity measurements: intensity from IgG/Fab bound to a virus (Fig. 1), Ibound, and intensity from individual IgG/Fab molecules, Iindividual. The latter is measured by visualizing individual, labeled IgG/Fab molecules under identical illumination conditions in the absence of virus and target bilayer (Fig. S5). The number of molecules bound to an individual virion is calculated as Nbound = Ibound/Iindividual. IgG/Fab detection efficiency was >99% for IgG and ∼90% for Fab (Table S1), and reported numbers of bound IgG/Fab were corrected for the small amounts of unlabeled molecules.
The median number of IgG/Fab bound to individual virions for each experiment performed is presented in Fig. 2B. Upon increasing IgG/Fab concentrations, the number of IgG/Fab molecules bound to a virus (fusing and nonfusing) increases sigmoidally, with the upper plateau corresponding to the region of Fig. 2A where the hemifusion efficiency is zero. Hence, this plateau value provides an estimate for the number of IgG/Fab molecules that must bind a virus to achieve maximum inhibition of hemifusion. This upper limit (asymptote) does not correspond to saturation of epitope binding, which we were not able to ascertain experimentally due to technical limitations of achievable labeled antibody concentrations and of required virus concentrations (SI Materials and Methods). Moreover, virus saturation by antibody binding occurs at higher concentrations than those required for neutralization in viral neutralization assays (VNAs) (18). Rather, the upper limit more relevantly reports the functional outcome of epitope binding, namely hemifusion inhibition, and the number of IgG/Fab needed to cause it. These and other stoichiometric values are summarized in Table 1, with calculated fit parameters reported in Table S2, including the 95% confidence intervals.
Table 1.
Summary of stoichiometric and kinetic results
| Virus | Neutralizing molecule | IgG/Fab bound at maximum hemifusion inhibition | IgG/Fab bound at half-maximum hemifusion inhibition | Fractional occupancy at half-maximum inhibition* | Fold increase of hemifusion time |
| H1N1 | CR6261 IgG | 175 (122–229) | 27 (6–65) | 0.16 (0.03–0.39) | 2.2 (1.6–3.0) |
| crF6261 Fab | 248 (158–338) | 74 (39–119) | 0.30 (0.12–0.55) | 2.7 (1.6–4.5) | |
| H3N2 | CR8020 IgG | 144 (119–170) | 84 (62–110) | 0.59 (0.41–0.79) | 2.1 (1.5–2.9) |
| crF8020 Fab | 493 (394–592) | 261 (192–340) | 0.53 (0.36–0.73) | 2.6 (2.2–3.0) |
Values in parenthesis report 95% confidence intervals.
Quotient of IgG/Fab bound at half-maximum and maximum hemifusion inhibition.
Under conditions of maximum hemifusion inhibition, we find that both viral strains are bound by comparable numbers of IgG molecules (175 for H1N1 and 144 for H3N2), whereas there is a difference in the number of Fab bound (248 for H1N1 and 493 for H3N2). Rationales for this difference are considered in Discussion. At the highest IgG concentrations we could achieve, ∼4,200 nM, we observed full hemifusion inhibition and measured 224 CR6261 IgG and 236 CR8020 IgG bound to the H1N1 and H3N2 viruses, respectively. Thereby, we show that more stem-binding IgG can sterically fit on the surface of hemifusion-inhibited virions than what is required for maximal inhibition.
To gain insight into the mechanism of hemifusion inhibition through IgG/Fab binding, we first compared the number bound at maximum hemifusion inhibition to estimates of the number of HA trimers on the viral surface. Previous studies have estimated 300–500 HA trimers per 120-nm diameter spherical virion (35–37), a size comparable to our viral strains (125 nm diameter, Fig. S6). Concordantly, recent cryoelectron tomography reconstruction of viruses produced identically to those that we have used (30, 38) indicate there are ∼375 HA trimers on each virus. Using this estimate and the data for the maximum fusion inhibition (Table 1) we calculate an IgG:HA stoichiometry of 0.47 and 0.38 for CR6261 and CR8020 IgG, respectively. For the Fab we estimate a Fab:HA stoichiometry of 0.69 and 1.31 for crF6261 and crF8020, respectively. These observations are consistent with predictions that the larger, bivalent IgG molecules would have lower stoichiometries relative to the smaller, monovalent Fab (30). For the IgG specifically, stoichiometry values less than one indicate that not every HA trimer needs an IgG bound for hemifusion to be inhibited and suggests that bivalent binding may play a role in hemifusion inhibition.
Secondly, we investigated steric interactions on the viral surface and binding valency in silico through a coarse-grained 2D Monte Carlo simulation (SI Materials and Methods) of protein packing. Thousands of random, viral surfaces with dense protein packing were generated using the crystallographic dimensions of the envelope proteins (HA, neuraminidase, and M2 proton channel) at appropriate relative concentrations. Surface density was set to mimic a viral surface having 375 HA trimers, then increasing numbers of IgG/Fab were bound to HA in accord with their cocrystal structure (23, 24). The number of IgG/Fab bound when overlap between neighboring proteins could no longer be prevented was taken as the sterically allowed maximum.
Simulations indicated that a maximum of 270–300 IgG sterically fit on a viral surface (Fig. S7). These values are in agreement with 3D cryo-EM results that steric interactions alone permit 75% of the 375 HA to complex with IgG (30). Our simulations revealed that bivalent IgG binding would prevail until ∼175 IgG molecules had bound, at which point monovalent binding would become dominant (Fig. S7). This result suggests that the 175 CR6261 or 144 CR8020 IgG bound at maximum hemifusion inhibition (Table 1) would bind bivalently to abrogate the fusogenicity of up to 350 or 288 HA trimers, respectively. These numbers are lower than estimates for the total number of HA trimers present on a virus and substantially lower than the number of epitopes present on a virus. Our simulations predict that ∼500 Fab molecules could fit on a virus (Fig. S7), although no previous predictions have been made for the number of Fab that could bind a virus. This steric maximum agrees with the experimentally determined number of crF8020, however, only about half as many crF6261 bind at maximum hemifusion inhibition. Overall, we conclude that not all sterically available epitopes need to be bound by IgG/Fab for the membrane fusion capacity of a virus particle to be blocked.
Low Occupancies Can Significantly Reduce the Extent of Hemifusion.
Combining measurements of hemifusion efficiency with the number of virus-bound IgG/Fab molecules (Fig. 2C; see SI Materials and Methods for data fitting) allows us to calculate the number of IgG/Fab molecules needed to reduce hemifusion efficiency by half. Such a value is akin to an EC50, but is a direct measurement of the number of virus-bound IgG/Fab causing the reduction in hemifusion efficiency rather than reporting a concentration. For example, we calculate that 27 CR6261 IgG need to bind the H1N1 virus for the hemifusion efficiency to be reduced 50% from 0.47 to 0.24 (Fig. 2C). Results of these calculations are listed in Table 1.
We next define the fractional occupancy as the ratio of the number of IgG/Fab bound at half-maximum hemifusion inhibition to the number of IgG/Fab bound at maximum hemifusion inhibition. For H1N1 at half-maximum hemifusion inhibition, the fractional occupancy is 27/175 = 0.16 (Table 1). This definition focuses on the functional consequence of IgG/Fab binding by defining it relative to the number of occupied binding sites required for maximum hemifusion inhibition rather than the number of sterically available sites. Fig. 2C and Table 1 show that fractional occupancy significantly lower than unity leads to half-maximum hemifusion inhibition. Comparing the fractional occupancies, we find that they are similar for the IgG and Fab of both 6261 and 8020, respectively. We also detect a difference in the fractional occupancy between the two viral strains being bound by the two IgG/Fab used in our experiments.
IgG/Fab Binding Delays the Time to Hemifusion.
The time to hemifusion is measured as the time between disappearance of the fluorescein signal (pH drop) and the onset of dequenching caused by lipophilic dye escape from the site of viral fusion (Fig. 1C). Concurrent with decreasing hemifusion efficiency and increasing numbers of bound IgG/Fab, Fig. 3 shows that the time required for the remaining fusion-competent particles to undergo hemifusion becomes longer as the concentration of IgG/Fab increases.
Fig. 3.
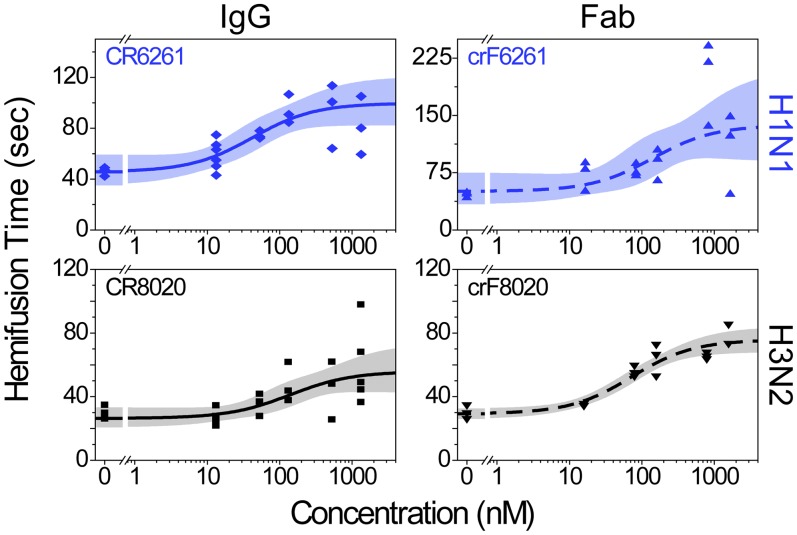
Hemifusion is delayed at higher IgG/Fab concentrations. Data are displayed as in Fig. 2 and are fit with a hyperbolic function having a constant offset (SI Materials and Methods); each data point is the geometric mean hemifusion time from a single experimental run. Fold increases in hemifusion times between zero and the highest IgG/Fab concentrations are listed in Table 1.
Hemifusion times increase in a sigmoidal fashion from baseline values of 46 and 30 s at zero IgG/Fab for the H1N1 and H3N2 strains, respectively (Table S2), to two- to threefold larger plateau values at high IgG/Fab concentrations. The existence of this upper plateau is surprising; a decreasing number of available HA trimers would be expected to result in a continuous increase of hemifusion times. From the existence of the upper plateau, it would appear that HA trimers have a temporal window of opportunity following acidification to induce fusion.
To gain further insight into the fusion mechanism, we obtained hemifusion kinetics for a large number of virus particles and analyzed the shape of the hemifusion-time distributions by fitting them to a gamma distribution. In this manner, we obtain information about the speed of the rate-limiting step along the fusion pathway and the number of rate-limiting steps. This latter value has been shown to represent the number of HA required for fusion (15). This kinetic analysis requires at least 50 events to have the statistical power to determine the number of HA trimers involved (39) and because increasing IgG/Fab concentrations results in fewer fusing virions, not all concentrations could be analyzed (SI Materials and Methods). The rates extracted by this gamma distribution analysis (Fig. S8) report slower kinetics at high IgG/Fab concentrations and faster overall kinetics of H3N2, similar to data in Fig. 3. Such kinetic differences could arise from sequence variations between H1N1 and H3N2 HA, giving rise to slight differences in protein structure and conformational energetics. Analysis of the number of rate-limiting steps suggest that both virus strains require two to three HA trimers at all IgG/Fab concentrations analyzed (Fig. S8). From this observation, we conclude that IgG/Fab binding does not alter the molecular mechanism of fusion. Rather, when inhibition is not complete and hemifusion still occurs, it does so with the same requirement of two to three HA trimers regardless of the presence of IgG/Fab bound to HA on the viral surface.
Discussion
We describe an experimental method that relates the stoichiometry of antibody or Fab binding to the efficiency and kinetics of viral membrane fusion at the single virus particle level. Increased binding of broadly neutralizing, stem-binding antibodies or Fab concomitantly causes a decrease in hemifusion efficiency and a maximally two- to threefold increase in the time to hemifusion. We find hemifusion efficiency is reduced by half with ∼30–80 stem-binding IgG bound to virus particles, and that hemifusion is fully inhibited by ∼140–180 stem-binding IgG. Thus, a significant effect is elicited with quantities of IgG that are substantially lower than the ∼270–300 IgG we and others (30) estimate could sterically fit on the viral surface. Relating the number of IgG/Fab required to achieve half-maximum hemifusion inhibition to the number needed to fully inhibit hemifusion, we conclude that even the rather low fractional occupancies of ∼0.16–0.6 have a potent effect. Further, Fab also inhibit membrane fusion, albeit with higher stoichiometry.
Using radioisotope-labeled IgG, Taylor et al. found that ∼50 HC2 or HC10 monoclonal, head-binding IgG per virion could reduce infectivity in VNAs by half (31), drastically lower than their estimates of 1,000 HA trimers per H7N1 virion. In contrast, Knossow et al. (18) used comparable IgG and methodologies with the H3N2 virus to find that complete viral neutralization occurred with IgG:HA ratios of 0.2:0.33, interestingly in a similar range as the 0.38:0.47 ratios we have determined. Whereas Knossow et al. attributed neutralization solely to inhibition of cellular binding, we recently demonstrated that head-binding antibodies also prevent the release of viral progeny (29). Our single-particle fusion assay avoids the convolution of multiple outcomes from IgG binding and focuses solely on membrane fusion. Overall, our results indicate that many epitopes on HA, and likely many HA trimers, remain unbound on the surface of fusion-inhibited virions.
Our data reveal a need for two to three HA trimers for fusion and strongly support a kinetic model of HA-mediated fusion requiring simultaneous action of multiple, neighboring trimers (15, 16). This model requires only the stochastic behavior of the HA trimers in combination with a high HA surface density. It also implies the existence of a dynamic network of potential folding partners between HA on the viral surface that is established by their relative geometry and spacing, and not necessarily by physical interactions between them. Network connections become activated to mediate fusion when HA trimers stochastically unfold at low pH. If a sufficient number of HA trimers are close enough in space and are refolding within a sufficiently short window of time, then they are able to coordinate their work and mediate the onset of lipid hemifusion. Prolonged exposure to low pH induces nonproductive HA refolding (40) and serves to disrupt the HA coordination network. Timing of the fusion process, thus, represents the interplay between (i) low pH-induced, stochastic firing of individual HA trimers; (ii) stochastic coordination between refolding HA trimers to mediate membrane fusion; and (iii) low pH-induced inactivation of the unfolded HA (Fig. 4).
Fig. 4.
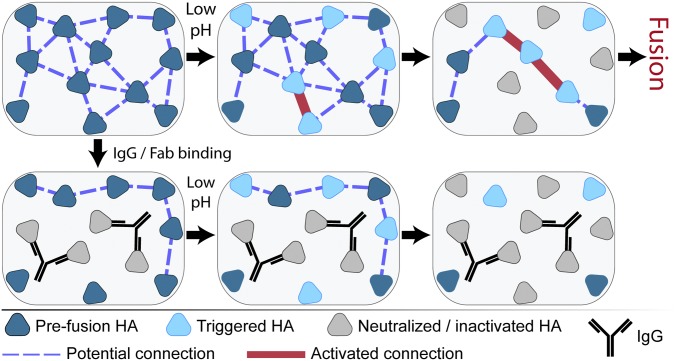
Cartoon illustrating inter-HA network disruption by IgG binding leading to fusion inhibition. Fusogenic HA at neutral pH in the prefusion conformation (dark blue triangles) initially have a network of connections (blue dashed lines) dictated by their spatial geometry relative to one another. Binding of IgG (black Y) neutralizes the HA (light gray triangles) by preventing their low pH-induced conformational changes and disrupts coordination with neighboring HA. Exposure to low pH conditions triggers HA to unfold (light blue triangles) and activates the inter-HA network (thick red lines) between neighboring, triggered HA. Continued low pH exposure causes HA inactivation by nonproductive refolding (light gray triangles), also removing inter-HA connections. Although productive fusion could arise at locations with a sufficiently high density of activated inter-HA connections, both IgG/Fab binding and low pH inactivation can inhibit accumulation of this density even in the presence of fusogenic HA. NA and M2 proteins are not depicted for clarity.
To account for the observed kinetic changes and low stoichiometries, we propose that IgG/Fab binding to block HA low-pH unfolding (23, 24, 29) inactivates network connections and lowers the probability of having a critical number of active HA trimers that are spaced closely enough to mediate fusion (Fig. 4). The elimination of neighbor–neighbor connections would then prevent the amassing of triggered HA into a density sufficient for membrane fusion to occur, even though some HA were still unbound and, likely, responsive to acidification. When a virus bound by IgG/Fab does fuse, the removal of active HA could lead to a longer elapsed time before sufficient neighboring HA trimers are able to coordinate their conformational changes. Such an interpretation is consistent with our finding that IgG/Fab binding causes slower hemifusion kinetics and with the results of a recent kinetic simulation of HA-mediated fusion (16).
In comparing the two viral strains, we find that maximum hemifusion inhibition of the H3N2 strain requires ∼3.8-fold more Fab than IgG whereas the H1N1 strain requires only ∼1.6-fold more Fab. Additionally, the H3N2 strain displays a larger fractional occupancy than does H1N1. Kinetically, the two viral strains exhibit differences in their baseline fusion rates (Fig. 3), indicative of inherent differences between them. As such, stoichiometry differences could be related to these interstrain differences, or, among other possible variations, to differences in the affinity and/or accessibility of the different epitopes on the viral surface, variations in surface protein density, or variation in virus strain composition (7, 37, 41). Additionally, it is plausible that the proximity of the CR(F)8020 epitope to the viral membrane, compared with the CR(F)6261 epitope (Fig. 1A), could potentially influence the binding angle of a Fab differently than an IgG. Further, large variations in the number of Fab compared with IgG may be common. Using the aforementioned HC2 and HC10, Schofield et al. (42) reported a requirement of 86- and >1,900-fold more Fab than IgG, respectively, to achieve similar levels of viral neutralization. Together, these results emphasize the enhancement in stoichiometric effectiveness that can be imparted to IgG through bivalent binding. Moreover, we find similar outcomes for both viral serotypes bound with similar numbers of IgG in terms of membrane fusion inhibition and kinetic delay.
In conclusion, we report the use of a single virus particle assay to measure the binding stoichiometry of broadly neutralizing, stem-binding antibodies and relate it solely to their functional effect of inhibiting HA-mediated membrane fusion. The data we report here sheds light both on the mechanism of inhibition by IgG/Fab and the function of HA itself during the membrane fusion process.
Materials and Methods
Proteoliposomes containing the sialoglycoprotein glycophorin A (GYPA) (full-length with GST tag; Abnova) were formed as previously described (29). Multilamellar proteoliposomes were incubated in glass-bottomed microfluidic channels to form contiguous, planar bilayers.
Influenza A virus strains A/Puerto Rico/8/34 (H1N1) and A/Aichi/68/X:31 (H3N2) were purchased from Charles River (2 mg/mL total viral protein) and used without further purification. Labeling involved virus diluted at 1:3 into Hepes buffer and mixed with octadecyl rhodamine B lipophilic dye (R18; Invitrogen, Inc.) in DMSO with a final concentration of 1 μM dye and <0.5% DMSO for 3 h. PD-10 desalting columns (GE Healthcare) removed unincorporated dye; three fractions of 200 μL with the highest particle count were pooled and used for fusion experiments.
Fusion experiments were conducted as previously described (29): 1-h virus incubation with labeled IgG/Fab, bilayer immobilization, and fluorescein-labeled streptavidin (Invitrogen) addition. An Olympus IX-71 inverted microscope was used with a 60×, TIRF (Total Internal Reflection Fluorescence) objective and 488- and 561-nm lasers (Sapphire; Coherent, Inc.). Green and red emissions were visualized on either half of an electron-multiplying CCD camera (Hamamatsu Photonics) using dual-view systems (either home-built or from Photometrics). Fusion was initiated by adding pH 5.0 citric acid buffer and fusion events recorded with 200-ms exposure at pH 5.0. Fluorescence intensity from individual IgG/Fab molecules adsorbed to clean glass in pH 5.0 buffer was acquired under identical illumination conditions.
Data analysis was performed using custom MATLAB (Mathworks, Inc.) scripts, similar to those previously described (15, 29). Briefly, viruses were identified in the red channel and paired with separately identified locations in the green channel. After correction for illumination profile and background, fluorescence trajectories were extracted and analyzed manually to select particles showing dequenching behavior. Experiments with fewer than 25 virions in a field of view were rejected. For stoichiometry calculations, green-channel trajectories were averaged for 15 frames (3 s) beginning 1 s after the pH drop (Fig. 1D). The number of virus-bound IgG/Fab molecules was quantified as a ratio of their integrated intensity when virus-bound to the integrated intensity of IgG/Fab individually (SI Materials and Methods). Time to hemifusion is the elapsed time from pH drop to the onset of hemifusion dequenching.
Supplementary Material
Acknowledgments
J.J.O. thanks Dr. Marc Stuart for assistance with electron microscopy, Drs. Fabrizia Fusetti and Alicja Filipowicz-Szymanska for assistance with mass spectrometry, and the NIH and the National Institute of General Medical Sciences for a Molecular Biophysics Training Grant.
Footnotes
Conflict of interest statement: B.B., M.H.K., J.J., C.T., T.K., J.G., R.V., and R.H.E.F. are current employees of the Crucell Vaccine Institute.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1411755111/-/DCSupplemental.
References
- 1.Han T, Marasco WA. Structural basis of influenza virus neutralization. Ann N Y Acad Sci. 2011;1217:178–190. doi: 10.1111/j.1749-6632.2010.05829.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fleishman SJ, et al. Computational design of proteins targeting the conserved stem region of influenza hemagglutinin. Science. 2011;332(6031):816–821. doi: 10.1126/science.1202617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burton DR, Poignard P, Stanfield RL, Wilson IA. Broadly neutralizing antibodies present new prospects to counter highly antigenically diverse viruses. Science. 2012;337(6091):183–186. doi: 10.1126/science.1225416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Steel J, et al. Influenza virus vaccine based on the conserved hemagglutinin stalk domain. MBio. 2010;1(1) doi: 10.1128/mBio.00018-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vanderlinden E, et al. Novel inhibitors of influenza virus fusion: Structure-activity relationship and interaction with the viral hemagglutinin. J Virol. 2010;84(9):4277–4288. doi: 10.1128/JVI.02325-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klasse PJ, Sattentau QJ. Occupancy and mechanism in antibody-mediated neutralization of animal viruses. J Gen Virol. 2002;83(Pt 9):2091–2108. doi: 10.1099/0022-1317-83-9-2091. [DOI] [PubMed] [Google Scholar]
- 7.Klasse PJ. In: Desk Encyclopedia of General Virology. Mahy BWJ, van Regenmortel MHV, editors. Academic; Oxford: 2008. pp. 404–410. [Google Scholar]
- 8.Wilson IA, Skehel JJ, Wiley DC. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature. 1981;289(5796):366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- 9.Waterfield M, Scrace G, Skehel J. Disulphide bonds of haemagglutinin of Asian influenza virus. Nature. 1981;289(5796):422–424. doi: 10.1038/289422a0. [DOI] [PubMed] [Google Scholar]
- 10.Tong S, et al. New world bats harbor diverse influenza A viruses. PLoS Pathog. 2013;9(10):e1003657. doi: 10.1371/journal.ppat.1003657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bullough PA, Hughson FM, Skehel JJ, Wiley DC. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature. 1994;371(6492):37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 12.Harrison SC. Viral membrane fusion. Nat Struct Mol Biol. 2008;15(7):690–698. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mittal A, Bentz J. Comprehensive kinetic analysis of influenza hemagglutinin-mediated membrane fusion: Role of sialate binding. Biophys J. 2001;81(3):1521–1535. doi: 10.1016/S0006-3495(01)75806-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Danieli T, Pelletier SL, Henis YI, White JM. Membrane fusion mediated by the influenza virus hemagglutinin requires the concerted action of at least three hemagglutinin trimers. J Cell Biol. 1996;133(3):559–569. doi: 10.1083/jcb.133.3.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Floyd DL, Ragains JR, Skehel JJ, Harrison SC, van Oijen AM. Single-particle kinetics of influenza virus membrane fusion. Proc Natl Acad Sci USA. 2008;105(40):15382–15387. doi: 10.1073/pnas.0807771105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ivanovic T, Choi JL, Whelan SP, van Oijen AM, Harrison SC. Influenza-virus membrane fusion by cooperative fold-back of stochastically induced hemagglutinin intermediates. ELife Sciences. 2013;2:e00333. doi: 10.7554/eLife.00333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bizebard T, et al. Structure of influenza virus haemagglutinin complexed with a neutralizing antibody. Nature. 1995;376(6535):92–94. doi: 10.1038/376092a0. [DOI] [PubMed] [Google Scholar]
- 18.Knossow M, et al. Mechanism of neutralization of influenza virus infectivity by antibodies. Virology. 2002;302(2):294–298. doi: 10.1006/viro.2002.1625. [DOI] [PubMed] [Google Scholar]
- 19.Dreyfus C, et al. Highly conserved protective epitopes on influenza B viruses. Science. 2012;337(6100):1343–1348. doi: 10.1126/science.1222908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ekiert DC, et al. Cross-neutralization of influenza A viruses mediated by a single antibody loop. Nature. 2012;489(7417):526–532. doi: 10.1038/nature11414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee PS, et al. Heterosubtypic antibody recognition of the influenza virus hemagglutinin receptor binding site enhanced by avidity. Proc Natl Acad Sci USA. 2012;109(42):17040–17045. doi: 10.1073/pnas.1212371109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Throsby M, et al. Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory B cells. PLoS ONE. 2008;3(12):e3942. doi: 10.1371/journal.pone.0003942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ekiert DC, et al. Antibody recognition of a highly conserved influenza virus epitope. Science. 2009;324(5924):246–251. doi: 10.1126/science.1171491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ekiert DC, et al. A highly conserved neutralizing epitope on group 2 influenza A viruses. Science. 2011;333(6044):843–850. doi: 10.1126/science.1204839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sui J, et al. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat Struct Mol Biol. 2009;16(3):265–273. doi: 10.1038/nsmb.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Okuno Y, Isegawa Y, Sasao F, Ueda S. A common neutralizing epitope conserved between the hemagglutinins of influenza A virus H1 and H2 strains. J Virol. 1993;67(5):2552–2558. doi: 10.1128/jvi.67.5.2552-2558.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Corti D, et al. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science. 2011;333(6044):850–856. doi: 10.1126/science.1205669. [DOI] [PubMed] [Google Scholar]
- 28.Nakamura G, et al. An in vivo human-plasmablast enrichment technique allows rapid identification of therapeutic influenza A antibodies. Cell Host Microbe. 2013;14(1):93–103. doi: 10.1016/j.chom.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 29.Brandenburg B, et al. Mechanisms of hemagglutinin targeted influenza virus neutralization. PLoS ONE. 2013;8(12):e80034. doi: 10.1371/journal.pone.0080034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harris AK, et al. Structure and accessibility of HA trimers on intact 2009 H1N1 pandemic influenza virus to stem region-specific neutralizing antibodies. Proc Natl Acad Sci USA. 2013;110(12):4592–4597. doi: 10.1073/pnas.1214913110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taylor HP, Armstrong SJ, Dimmock NJ. Quantitative relationships between an influenza virus and neutralizing antibody. Virology. 1987;159(2):288–298. doi: 10.1016/0042-6822(87)90466-1. [DOI] [PubMed] [Google Scholar]
- 32.Parren PW, Burton DR. The antiviral activity of antibodies in vitro and in vivo. Adv Immunol. 2001;77:195–262. doi: 10.1016/S0065-2776(01)77018-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reading SA, Dimmock NJ. Neutralization of animal virus infectivity by antibody. Arch Virol. 2007;152(6):1047–1059. doi: 10.1007/s00705-006-0923-8. [DOI] [PubMed] [Google Scholar]
- 34.Otterstrom J, van Oijen AM. Visualization of membrane fusion, one particle at a time. Biochemistry. 2013;52(10):1654–1668. doi: 10.1021/bi301573w. [DOI] [PubMed] [Google Scholar]
- 35.Tiffany JM, Blough HA. Models of structure of the envelope of influenza virus. Proc Natl Acad Sci USA. 1970;65(4):1105–1112. doi: 10.1073/pnas.65.4.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Inglis SC, Carroll AR, Lamb RA, Mahy BW. Polypeptides specified by the influenza virus genome I. Evidence for eight distinct gene products specified by fowl plague virus. Virology. 1976;74(2):489–503. doi: 10.1016/0042-6822(76)90355-x. [DOI] [PubMed] [Google Scholar]
- 37.Moulès V, et al. Importance of viral genomic composition in modulating glycoprotein content on the surface of influenza virus particles. Virology. 2011;414(1):51–62. doi: 10.1016/j.virol.2011.03.011. [DOI] [PubMed] [Google Scholar]
- 38.Harris A, et al. Influenza virus pleiomorphy characterized by cryoelectron tomography. Proc Natl Acad Sci USA. 2006;103(50):19123–19127. doi: 10.1073/pnas.0607614103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Floyd DL, Harrison SC, van Oijen AM. Analysis of kinetic intermediates in single-particle dwell-time distributions. Biophys J. 2010;99(2):360–366. doi: 10.1016/j.bpj.2010.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weber T, et al. Evidence for H(+)-induced insertion of influenza hemagglutinin HA2 N-terminal segment into viral membrane. J Biol Chem. 1994;269(28):18353–18358. [PubMed] [Google Scholar]
- 41.Shaw ML, Stone KL, Colangelo CM, Gulcicek EE, Palese P. Cellular proteins in influenza virus particles. PLoS Pathog. 2008;4(6):e1000085. doi: 10.1371/journal.ppat.1000085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schofield DJ, Stephenson JR, Dimmock NJ. Variations in the neutralizing and haemagglutination-inhibiting activities of five influenza A virus-specific IgGs and their antibody fragments. J Gen Virol. 1997;78(Pt 10):2431–2439. doi: 10.1099/0022-1317-78-10-2431. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.