Significance
Bacteria possess many regulatory systems to monitor their environment and adapt their physiology accordingly. Whereas most systems sense one specific signal, the general stress response (GSR) is activated by many signals and protects cells against a wide range of adverse conditions. In Alphaproteobacteria, the GSR is controlled by the response regulator PhyR, but little is known about the upstream pathways. Here, we establish the GSR as a complex regulatory network composed of a particular family of partially redundant sensor kinases and of additional response regulators that modulate PhyR activity in Sphingomonas melonis. Given the broad conservation of this kinase family, it is possible that it plays a general role in the GSR in Alphaproteobacteria.
Keywords: general stress response, two-component system, histidine kinase, response regulator, ECF sigma factor
Abstract
The general stress response (GSR) in Alphaproteobacteria was recently shown to be controlled by a partner-switching mechanism that is triggered by phosphorylation of the response regulator PhyR. Activation of PhyR ultimately results in release of the alternative extracytoplasmic function sigma factor σEcfG, which redirects transcription toward the GSR. Little is known about the signal transduction pathway(s) controlling PhyR phosphorylation. Here, we identified the single-domain response regulator (SDRR) SdrG and seven histidine kinases, PakA to PakG, belonging to the HWE/HisKA2 family as positive modulators of the GSR in Sphingomonas melonis Fr1. Phenotypic analyses, epistasis experiments, and in vitro phosphorylation assays indicate that Paks directly phosphorylate PhyR and SdrG, and that SdrG acts upstream of or in concert with PhyR, modulating its activity in a nonlinear pathway. Furthermore, we found that additional SDRRs negatively affect the GSR in a way that strictly requires PhyR and SdrG. Finally, analysis of GSR activation by thermal, osmotic, and oxidative stress indicates that Paks display different degrees of redundancy and that a specific kinase can sense multiple stresses, suggesting that the GSR senses a particular condition as a combination of, rather than individual, molecular cues. This study thus establishes the alphaproteobacterial GSR as a complex and interlinked network of two-component systems, in which multiple histidine kinases converge to PhyR, the phosphorylation of which is, in addition, subject to regulation by several SDRRs. Our finding that most HWE/HisKA2 kinases contribute to the GSR in S. melonis Fr1 opens the possibility that this notion might also be true for other Alphaproteobacteria.
In addition to specific stress responses, many bacteria use a so-called “general stress response” (GSR) that can be induced by a number of different, unrelated stresses and that confers multiple stress resistance. In Alphaproteobacteria, the GSR is controlled by an alternative extracytoplasmic function (ECF) sigma factor, usually called σEcfG (1) or ECF15 sigma factor (2), the activity of which is regulated by a conserved partner-switching mechanism (1, 3–14). In unstressed conditions, σEcfG is sequestered by its anti-sigma factor NepR. Upon stress, the anti-anti-sigma factor PhyR becomes activated and binds NepR, thereby releasing σEcfG and allowing it to bind RNA polymerase and redirect transcription toward stress genes. Because PhyR uses a degenerate N-terminal sigma factor-like output domain to compete with σEcfG for NepR binding, this partner switch was coined “sigma factor mimicry” (1). The importance of the alphaproteobacterial GSR in natural environments is underlined by several studies demonstrating its requirement for survival and competitiveness in the phyllosphere in Sphingomonas melonis (10) and Methylobacterium extorquens (8), the establishment of symbiotic interactions in Bradyrhizobium japonicum (9), and host–pathogen interactions in Brucella (6, 11) and Bartonella (3) species.
PhyR is a member of the response regulator family and harbors a C-terminal receiver (REC) domain in addition to its N-terminal output domain. As is the case for other response regulators, the activity of PhyR is controlled by phosphorylation of a conserved Asp residue in its REC domain. Despite a rather advanced understanding of the partner switch itself (15–17), little is known about the decisive step of GSR activation, that is, regulation of PhyR phosphorylation. As a response regulator, PhyR is expected to be part of a two-component system, or phosphorelay, working together with one or several histidine kinases that control PhyR phosphorylation upon sensing a particular stimulus. In agreement with this notion, histidine kinase-encoding genes are often found at the phyR locus [referred to as cis-encoded kinases in the following, according to Sauviac and Bruand (18)]. These kinases are usually annotated as HWE or HisKA2 kinases based on the Pfam dimerization and histidine phosphotransferase (DHp) domain classification (19), although this annotation does not always conform to the original definition of HWE kinases based on the HWE motif in the catalytic and ATPase (CA) domain (20). The cis-encoded kinases have only been studied in a few cases. In Caulobacter crescentus, the gene is called phyK and has been shown to be required for PhyR phosphorylation and GSR activation in vivo for the different stresses tested, with a phyK mutant virtually displaying the same phenotype and stress induction as a phyR mutant (12, 21). In contrast, a recent study in Sinorhizobium meliloti has demonstrated a role for the cis-encoded kinase RsiC in phosphorylation of the PhyR orthologs RsiB1/RsiB2 in response to starvation and heat shock (HS) in vivo, but the rsiC mutant only partially phenocopies an rsiB1 rsiB2 mutant, and the involvement of one additional kinase, SMc00322, was proposed (18). In other Alphaproteobacteria, no genes coding for histidine kinases are found at the phyR locus, but several HWE and/or HisKA2 kinases are encoded elsewhere in the genome (22). For example, M. extorquens PA1 encodes 21 kinases of the HWE and/or HisKA2 type, but there is no clear indication from sequence homology or genomic context for a prime candidate as PhyR kinase (23).
Our previous work characterized phyP, which is located at the phyR locus in S. melonis Fr1, as a PhyR phosphatase rather than a kinase. Among other evidence, this conclusion is based on the findings that phyP is essential unless in a ΔphyR or ΔecfG background (10), and that PhyP depletion leads to lethal overactivation of the GSR in a manner that requires the phosphorylatable Asp residue (Asp194) of PhyR (24). Assuming that PhyP acts solely as a phosphatase, this notion implies that alternative phosphodonors for PhyR must exist.
In the present study, we describe our efforts to identify and characterize additional regulators of the PhyR cascade in S. melonis Fr1. Our results suggest that most HWE/HisKA2 kinases (collectively referred to as HRXXN kinases in the following) in S. melonis Fr1 contribute to GSR activity by directly phosphorylating PhyR. In addition, we identified several single-domain response regulators (SDRRs) that positively or negatively affect the GSR in a PhyR-dependent manner. This study thus establishes the GSR as a complex and interwoven network of two-component systems in S. melonis Fr1 and suggests that HRXXN kinases might play a major role in the GSR of other Alphaproteobacteria as well.
Results
Identification of a SDRR, SdrG, Involved in the GSR.
To identify factors positively regulating the PhyR-NepR-σEcfG cascade, we conducted a genetic screen, performing transposon mutagenesis on a strain expressing the dominant streptomycin-sensitive rpsL1 allele (25) under control of the σEcfG-dependent ecfG2 promoter (10, 24). This construct would allow selection for mutants with an impaired PhyR-NepR-σEcfG cascade on medium containing streptomycin (details are provided in SI Materials and Methods). Because the GSR is activated in standard LB-Lennox medium (σEcfG-dependent promoter activity of the WT compared with phyR and ecfG deletion strains is shown in Fig. 1B), there was no need to apply particular stresses for GSR induction. Among the genes identified during the selection were both ecfG and phyR, confirming the general validity of the selection. No histidine kinase mutant was found, in line with the partial redundancy of kinases in PhyR activation (discussed below). The only other gene identified with an obvious link to signal transduction was Sphme2DRAFT_3354, which encodes a stand-alone receiver domain that will be referred to as sdrG in the following (for SDRR involved in the GSR).
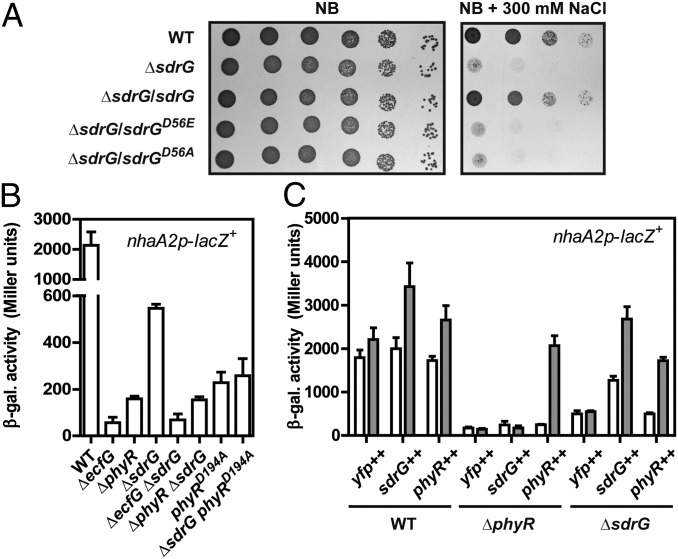
Fig. 1.
Salt sensitivity phenotype of the ΔsdrG strain (A) and epistatic interactions of sdrG with phyR and ecfG (B and C). (A) Salt sensitivity of sdrG mutant strains complemented with different sdrG alleles expressed from plasmid pAK126a as indicated. Control strains harbored the empty plasmid pAK126a. (B) β-Gal activity of the σEcfG-dependent nhaA2p-lacZ+ fusion in various mutant backgrounds. (C) Activity of the nhaA2p-lacZ+ fusion in various backgrounds (Bottom) upon overexpression of SYFP2 alone (yfp++, plasmid pQY), a SYFP2-PhyR fusion (phyR++, plasmid pQYD-PhyR), or a SYFP2-SdrG fusion (sdrG++, plasmid pQYD-SdrG) from the cumate-inducible promoter PQ5 (plasmid pQYD). White bars represent β-gal activity before and gray bars represent β-gal activity after 4 h of induction with 25 μM cumate. Values are given as mean ± SD of three independent experiments.
To verify its involvement in the GSR, we constructed an in-frame deletion of sdrG. The ΔsdrG strain was more sensitive to osmotic and oxidative stress and to HS than the WT (Fig. 1A and Fig. S1 A and B), similar to the ecfG or phyR mutant, and this defect could be complemented by expressing the WT sdrG allele in trans (Fig. 1A). In addition, monitoring GSR activity by use of a transcriptional fusion of the σEcfG-dependent promoter nhaA2p to lacZ (nhaA2p-lacZ+) showed markedly reduced activity in ΔphyR, phyRD194A, ΔecfG, and ΔsdrG strains compared with the WT strain (Fig. 1B), confirming that sdrG is involved in the GSR. To place sdrG in the PhyR-NepR-σEcfG cascade genetically, we next performed epistasis experiments. Deleting sdrG in ΔphyR, phyRD194A, or ΔecfG background had no further effect on nhaA2p-lacZ+ activity compared with the parent strains (Fig. 1B), suggesting that SdrG acts upstream or in concert with PhyR. Similar results were obtained using artificial induction of the cascade by phyR or sdrG overexpression. Although both sdrG and phyR overexpression led to increased nhaA2p-lacZ+ activity in the WT background, this effect was strictly dependent on phyR in the case of sdrG overexpression, whereas phyR overexpression could partially bypass the requirement for sdrG (Fig. 1C). We next asked whether phosphorylation of SdrG was essential for its function. We thus tested in salt stress phenotypic assays the sdrGD56A and sdrGD56E alleles encoding Ala and Glu substitutions of the putatively phosphorylatable Asp56 in SdrG, respectively, the latter of which may mimic the phosphorylated state of response regulators (e.g., ref. 26). Both alleles failed to complement the stress sensitivity to salt of ΔsdrG (Fig. 1A), suggesting that phosphorylated SdrG is likely the active form and the D56E substitution in SdrG does not mimic the active form and/or that the activity of SdrG involves phosphoryl group shuttling. Finally, because SdrG apparently acts in the same pathway as PhyR, we wondered whether the essential PhyR phosphatase PhyP would be dispensable in a ΔsdrG background, similar to ecfG and phyR mutant backgrounds (10). We could not obtain sdrG phyP double mutants, suggesting that PhyR phosphatase activity is still required in a ΔsdrG background. In summary, these results suggest that SdrG is involved in the GSR by directly or indirectly modulating PhyR activity, and this activity requires phosphorylation of SdrG.
Multiple Kinases and SDRRs Contribute to GSR Activity in Vivo.
Because the above results suggested that PhyR and SdrG need to be phosphorylated for proper function of the GSR and the genetic selection did not result in the identification of histidine kinases, a candidate approach was undertaken. As mentioned in the introduction, histidine kinases encoded at the phyR locus are classified by Pfam as either HWE or HisKA2 kinases (19), with both classes showing a typical HRXXN motif in DHp helix α1 (Fig. 2A), a region comprising major determinants for response regulator recognition/specificity in the HisKA histidine kinase family (27, 28). S. melonis Fr1 possesses eight HRXXN kinases (Fig. 2B) in addition to the PhyR phosphatase PhyP, and we renamed seven of the eight kinases PhyR-activating kinase A (PakA) through PakG for reasons outlined below. These HRXXN kinases are either orphans (PakA, PakE, and PakG) or encoded in a predicted operon with or in close proximity to a gene encoding an SDRR, referred to as “cognate” response regulators in the following (PakB, PakC, PakD, and PakF) (Fig. S2A). We named the genes encoding the cognate response regulators pkrB, pkrC, pkrD and pkrF, standing for pakB-, pakC-, pakD- and pakF-associated response regulator, respectively.
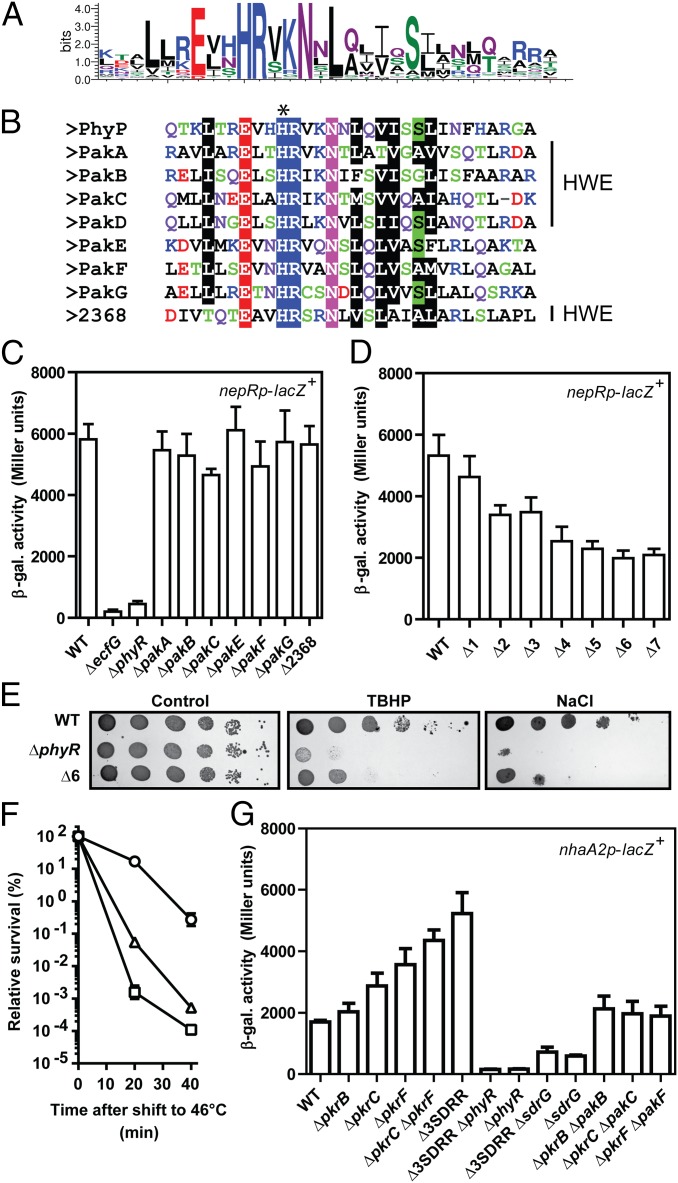
Fig. 2.
Multiple kinases, but not their cognate response regulators, positively regulate GSR activity in vivo. (A) Consensus sequence of DHp helix α1 of cis-encoded kinases derived from an alignment of 63 kinases generated with the WebLogo tool (51). (B) Alignment of DHp helix α1 of HRXXN kinases encoded in the S. melonis Fr1 genome. The predicted phosphorylatable His residue is highlighted by an asterisk, and the presence of the HWE motif in the CA domain is indicated (Right). (C) β-Gal activity of the nepRp-lacZ+ fusion in single pak mutants and ΔSphme2DRAFT_2368 (Δ2368) as indicated. (D) β-Gal activity of the nepRp-lacZ+ fusion in multiple mutant backgrounds: Δ1 (ΔpakC), Δ2 (ΔpakC ΔpakB), Δ3 (ΔpakC ΔpakB ΔpakA), Δ4 (ΔpakC ΔpakB ΔpakA ΔpakF), Δ5 (ΔpakC ΔpakB ΔpakA ΔpakF ΔpakE), Δ6 (ΔpakC ΔpakB ΔpakA ΔpakF ΔpakE ΔpakG), and Δ7 (ΔpakC ΔpakB ΔpakA ΔpakF ΔpakE ΔpakG ΔSphme2DRAFT_2368). (E) Oxidative and osmotic stress resistance phenotypes of WT, ΔphyR, and Δ6 strains. Tenfold dilution series were plated on TB (control) and TB containing 240 μM TBHP or 325 mM NaCl. (F) HS phenotypes of WT (○), ΔphyR (□), and Δ6 (△) strains. (G) β-Gal activity of the nhaA2p-lacZ+ fusion in single- and multiple-cognate response regulator mutants, alone or in combination with phyR, sdrG, or their cognate kinases. Δ3SDRR denotes the SDRR triple mutant (ΔpkrB ΔpkrC ΔpkrF). Values are given as mean ± SD of three independent experiments.
To test the involvement of kinases in the GSR, we generated single in-frame deletions. No clean deletion of pakD could be obtained despite several attempts. pakD is located on a megaplasmid, and the entire megaplasmid was apparently lost in most cases during the mutagenesis procedure when applying counterselection to select for resolution of the merodiploid. In the few cases where the second homologous recombination occurred and the megaplasmid was retained, only WT was recovered. A clean pakD mutation also could not be obtained when pakD was covered in trans or when we attempted to delete it together with its adjacent response regulator pkrD, for which also no clean deletion mutant could be recovered. It is not clear at the moment whether pakD is truly essential, because it would imply that its essentiality depended on some cis-acting factor(s) on the megaplasmid, or whether the failure of getting these mutants merely represents a technical issue.
The pak mutants were tested for a defect in GSR activity using the nepRp-lacZ+ reporter fusion, a reporter of GSR activity comparable to the nhaA2p-lacZ+ fusion. Strains were grown in LB-Lennox medium without applying a particular stress, because GSR activity was already high under these conditions (compare the WT with the phyR mutant in Fig. 2C, also Fig. 1B). None of the single pak mutants showed robust reduction in GSR activity (Fig. 2C). Because kinases might be redundant, we then constructed multiple kinase mutants by consecutively deleting pak genes. Here, a progressive reduction in GSR activity was observed as pak genes were successively deleted (Fig. 2D), suggesting that multiple kinases regulate the GSR. No decrease in β-gal activity was seen for a control transcriptional fusion (rpmBp-lacZ+) in which lacZ was under control of the σEcfG-independent rpmB promoter (Fig. S2C). When tested in phenotypic assays, the sextuple mutant showed increased sensitivity to osmotic and oxidative stress (Fig. 2E) and HS (Fig. 2F) compared with the WT, corroborating the idea that Paks are important for the GSR. Also in agreement with a role of Paks in the GSR, we were able to delete phyP in the sextuple or septuple kinase mutant backgrounds. Because the PhyR phosphatase PhyP is essential in a WT background preventing lethal hyperphosphorylation of PhyR (10, 24), the successful deletion of phyP suggested that a major source of PhyR phosphorylation is absent in these genetic backgrounds. However, because the GSR activity in the sextuple or septuple kinase mutants was still substantially higher than in a phyR mutant (Fig. 2 C and D) and the phyR mutant was more sensitive than the sextuple mutant in phenotypic stress assays (Fig. 2 E and F), other phosphodonors for PhyR in these backgrounds likely exist, with one obvious candidate being pakD.
Similarly, we constructed single and multiple mutants of cognate response regulators (pkrB, pkrC, and pkrF) and tested their involvement in the GSR using the nhaA2p-lacZ+ fusion. As for pakD itself, we were not able to obtain deletion of its cognate response regulator-encoding gene pkrD. For pkrC and pkrF mutants, in contrast to the sdrG mutant, GSR activity was not reduced, but rather increased, in the single mutants (Fig. 2G). The effect of pkrC and pkrF deletion on GSR activity was additive in the corresponding double mutant (Fig. 2G). For pkrB, no significant difference was observed in the single mutant or in the pkrB pkrC pkrF triple mutant compared with the ΔpkrC ΔpkrF background (Fig. 2G). The increased GSR activity in the ΔpkrC ΔpkrF ΔpkrB background was entirely dependent on PhyR and SdrG (Fig. 2G). Furthermore, the effect on GSR activity in pkrC and pkrF single mutants also depended on their cognate kinases, PakC and PakF, respectively (Fig. 2G). These results suggest that the cognate response regulators PkrC and PkrF are negative regulators of the GSR acting upstream of PhyR, SdrG, and their cognate kinases, perhaps by competing with PhyR and/or SdrG for phosphorylation (results are discussed below and in Discussion). In agreement, single- and multiple-deletion mutants showed the same or even slightly increased resistance to osmotic, oxidative, and thermal stress when tested in phenotypic assays compared with the WT (Fig. S1 C and D). In conclusion, our data suggest that several HRXXN kinases are positive regulators of the GSR, whereas at least two of the cognate response regulators are negative regulators.
HRXXN Kinases Activate the GSR in a PhyR-Dependent Fashion.
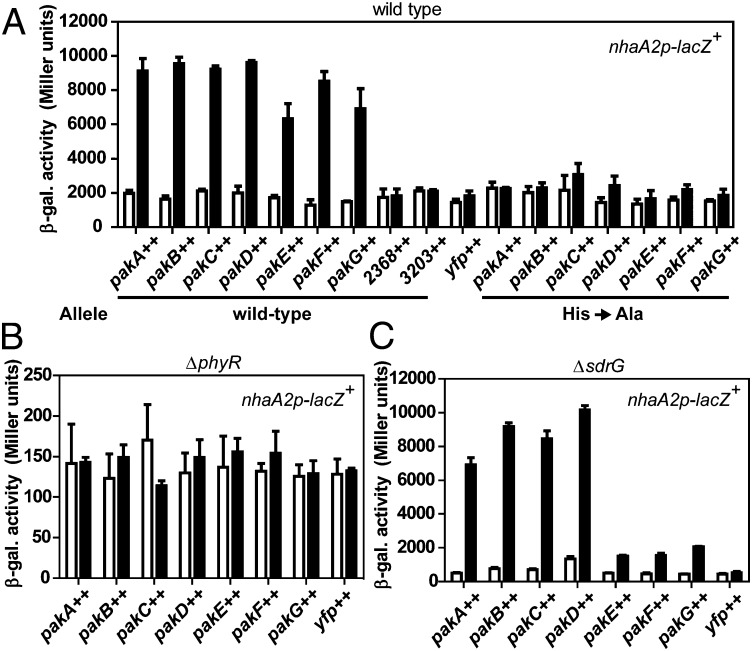
Although there was a clear difference in GSR activity between the WT and the septuple kinase mutant (Fig. 2D), it was not evident whether all kinases or only some were involved in the GSR. To address this question, we individually overexpressed kinases from the cumate-inducible promoter PQ5 (24) in the WT background carrying the nhaA2p-lacZ+ fusion; to follow protein levels by Western blotting, kinases were fused C-terminally to SYFP2, a GFP derivative. Overexpression of all pak genes (i.e., pakA to pakG) activated the GSR threefold to fivefold, whereas overproduction of the remaining HRXXN kinase (Sphme2DRAFT_2368), an unrelated NtrB-like kinase from S. melonis Fr1 (Sphme2DRAFT_3203), or SYFP2 alone failed to do so (Fig. 3A). As expected, no change in expression of the rpmBp-lacZ+ control fusion was observed when kinases were overexpressed (Fig. S3A). To exclude the possibility that GSR activation was simply a result of proteotoxic stress upon overexpression, we also tested whether activity of kinases was required for GSR activation by overexpressing histidine kinases that carried Ala substitutions of the predicted phosphorylatable His residue. None of these mutant kinases caused activation of the GSR (Fig. 3A), suggesting that phosphorylation of kinases and, as a consequence, their target(s) is required. We next examined whether GSR activation by kinase overexpression requires PhyR or SdrG. No GSR activation was seen in a phyR mutant (Fig. 3B), indicating that kinases do not act in parallel to, but upstream of, phyR. In the ΔsdrG background, pakA through pakD were able to activate the GSR to a similar extent as in the WT background, whereas GSR activation by pakE, pakF, and pakG was diminished, albeit still observable (Fig. 3C). In each case, the reduction/lack of GSR activation in the different backgrounds or with the pak mutant alleles was not due to reduced expression levels of kinases, as verified by Western blotting (Fig. S3 B and C). These data suggest that Sphme2DRAFT_2368 is the only HRXXN kinase that cannot activate the GSR when overproduced, although we cannot exclude the possibility that the SYFP2-kinase fusion is nonfunctional. In this respect, we note that when using bacterial two-hybrid assays, in contrast to the other kinases, we did not observe homodimer formation for Sphme2DRAFT_2368 (Fig. S4C).
Fig. 3.
Effect of HRXXN kinase overexpression on GSR activity. Kinases (WT or His-to-Ala mutant versions as described in the main text) were overexpressed in the WT (A), ΔphyR (B), or ΔsdrG (C) background. The nhaA2p-lacZ+ activity was measured before (white bars) and after 2 h of (black bars) induction of kinase overexpression (++) from promoter PQ5 (plasmid pQYD) with 25 μM cumate. The control expressed only SYFP2 from plasmid pQY. 3203++ designates overexpression of a cytoplasmic, NtrB-like histidine kinase that does not harbor the HRXXN motif (Sphme2DRAFT_3203). Values are given as mean ± SD of three independent experiments. Expression of kinases was verified by immunoblotting (Fig. S3 B and C).
Altogether, these results indicate that overexpression of seven of the eight HRXXN kinases in S. melonis Fr1 allows GSR activation in a strictly PhyR-dependent way. Our data further suggest that different kinases depend on SdrG to different extents.
HRXXN Kinases Phosphorylate PhyR and SdrG in Vitro.
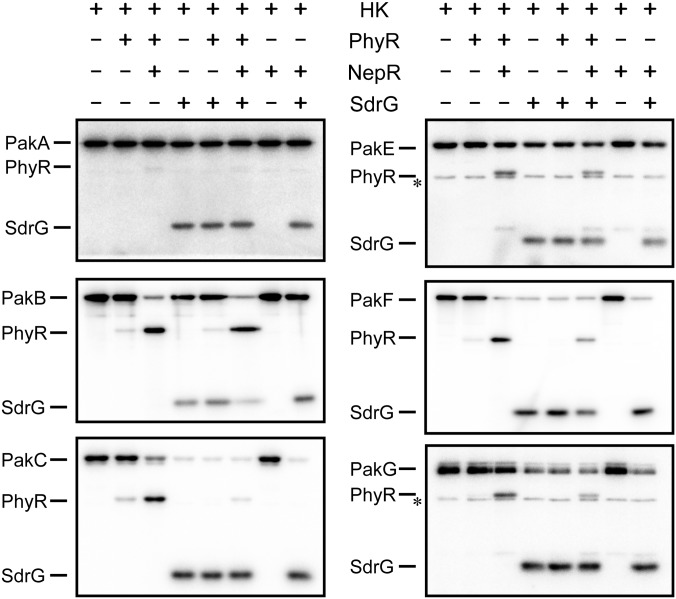
A simple explanation for the kinases’ different dependencies on SdrG in GSR activation (Fig. 3C) would be that some kinases (PakA–PakD) directly phosphorylated PhyR, whereas others (PakE–PakG) preferentially phosphorylated SdrG, possibly being the first step in a His-Asp phosphorelay resulting in PhyR phosphorylation. We thus sought to test phosphotransfer of Paks to PhyR and SdrG directly in vitro. We were able to obtain soluble versions active in autophosphorylation of all Paks, except PakD. When phosphotransfer to PhyR was tested, all kinases appeared to be inefficient phosphodonors to PhyR after 30 s of incubation (Fig. 4). Because there is circumstantial evidence that in the absence of NepR, PhyR is either inefficiently phosphorylated or PhyR∼P is rapidly hydrolyzed to its unphosphorylated form (Fig. S4 A and B), we also tested phosphotransfer to PhyR in the presence of NepR. When NepR was included in phosphotransfer reactions, the signal of the band representing phosphorylated PhyR was markedly enhanced (Fig. 4). This effect was not due to inhibition of PhyR∼P dephosphorylation by NepR but to increased phosphotransfer from the kinases to PhyR, as evidenced by the depletion of the radiolabel from the kinases in reactions containing NepR (e.g., compare lanes 2 and 3 for PakB in Fig. 4). Although the mechanistic basis of this phenomenon is unknown, these results suggest that NepR enhances PhyR phosphorylation in vitro.
Fig. 4.
In vitro phosphotransfer reactions (30 s) of HRXXN kinases autophosphorylated with [γ-32P]ATP (Left of each autoradiograph) to PhyR and/or SdrG containing or not containing NepR (Top). The asterisk next to some autoradiographs (PakE and PakG) indicates an impurity that was present in the kinase preparation.
In addition to phosphorylating PhyR in presence of NepR, all kinases were able to phosphorylate SdrG after 30 s, and addition of NepR to the reactions had no effect here (Fig. 4). To test whether the kinases preferred one substrate over the other, we performed competition experiments in which the reaction mixtures contained NepR and equimolar amounts of PhyR and SdrG. No clear preference could be observed except for PakA and PakC toward SdrG and PakB toward PhyR. Notably, there was no correlation between the in vitro p` of a kinase for SdrG or PhyR and the dependence of this same kinase on SdrG for GSR activation when overexpressed in vivo (Fig. 3C), suggesting that a simple model of specific PhyR and SdrG kinases was probably not true. Because these results were unexpected, we wondered whether some kinases might form heteromers, which could explain the differences between in vivo and in vitro data. Bacterial two-hybrid assays, however, revealed that kinases only formed homomers (Fig. S4C).
Thus, although the in vitro conditions do not recapitulate the intracellular environment and preferences might be different in vivo, our results demonstrate that all kinases phosphorylate both PhyR and SdrG on a time scale that is generally considered to represent true histidine kinase-response regulator pairs in vivo (29, 30). Importantly, these results, together with the in vivo overexpression experiments (Figs. 1C and 3C) and the essentiality of phyP in the ΔsdrG background, also suggest that PhyR and SdrG are unlikely to act in a strictly linear pathway, such as a phosphorelay, and that SdrG probably does not simply act as a phosphoshuttle for PhyR phosphorylation.
Multiple Kinases Sense Distinct Stresses.
Having identified several PhyR and SdrG kinases, we next wondered whether they are involved in sensing specific signals or stresses. Because it was not possible to activate the GSR further applying several different stresses in standard LB-Lennox medium, we tested different alternative media for decreased basal GSR activity. It was found that the WT strain grown in 1% tryptone broth (TB) displayed a fourfold reduction of nhaA2p-lacZ+ activity compared with the same strain grown in LB-Lennox medium, and that several stress conditions could induce the GSR twofold to threefold, including a shift to 37 °C (HS), ethanol (EtOH), salt (NaCl), and the organic peroxide t-butyl hydroperoxide (TBHP) (Fig. 5A and Fig. S5A). To test the involvement of Paks in response to these stresses, multiple pak mutants, rather than single pak mutants, were first tested because of the possibility of functional redundancy. Because the basal nhaA2p-lacZ+ activity of multiple pak mutants was already reduced compared with the WT (Fig. S5A), similar to what was observed in LB-Lennox medium (Fig. 2D), the difference in activation (“nhaA2p-lacZ+ activity with stress” − “nhaA2p-lacZ+ activity without stress”) relative to the WT [“relative stress activation” (RSA)] rather than the absolute nhaA2p-lacZ+ activity was compared. The same approach (measurement of RSA) has been used before in the Bacillus subtilis GSR to discriminate between the contribution of sensory proteins to the basal activity level and their roles in stress activation (31, 32). As shown in Fig. 5B, stress activation was completely abolished in a phyR mutant for all stresses tested and greatly reduced (for NaCl and HS) or completely eliminated (EtOH and TBHP) in an sdrG mutant, confirming their central roles in the GSR. Sharp declines in RSA of a particular multiple mutant compared with its parent strain allowed us to propose major contributions for several kinases in stress sensing. In brief, the results (Fig. 5B) suggested roles for PakC and PakF in HS sensing, for PakB in TBHP sensing, and for PakF in NaCl sensing, whereas no clearcut role for any individual kinase in EtOH sensing was evident. To delineate the individual contribution of the different Paks in stress activation and to test for redundancy, we next measured stress activation in single pakB, pakC, and pakF mutants and the three possible double mutants (Fig. 5C and Fig. S5B). Here, the strains showing the lowest RSA were ΔpakCF for HS, ΔpakF for salt stress, and ΔpakBC in case of TBHP stress, confirming the results obtained with the multiple mutants. However, it was also evident that besides the major kinases responsible for GSR activation in a given stress condition, other kinases contributed to the response in a minor way that was (partially) masked in the presence of the major kinases. For example, the ΔpakF strain showed no RSA reduction in response to TBHP, whereas when the pakF mutation was combined with a mutation in either one of the major kinases (strains ΔpakBF and ΔpakCF), RSA was reduced compared with the pakB and pakC single mutants. In addition, the results indicated different degrees of redundancy of kinases: For HS, the single pakC and pakF mutants already showed reduced RSA (50%) and this effect was additive (7% RSA), whereas for TBHP, the presence of either pakB or pakC was sufficient to activate the response almost fully and only the corresponding double mutant showed a drastic reduction in RSA (13%).
Fig. 5.
Induction of the GSR by various stresses in single- and multiple pak mutants. (A) β-Gal activity of the nhaA2p-lacZ+ fusion in the WT background upon application of various stresses. (B) RSA of the nhaA2p-lacZ+ fusion by various stresses (Top Left in each panel) in multiple pak mutants. Multiple mutants are labeled as follows: Δ1 (ΔpakC), Δ2 (ΔpakC ΔpakB), Δ3 (ΔpakC ΔpakB ΔpakA), Δ4 (ΔpakC ΔpakB ΔpakA ΔpakF), Δ5 (ΔpakC ΔpakB ΔpakA ΔpakF ΔpakE), and Δ6 (ΔpakC ΔpakB ΔpakA ΔpakF ΔpakE ΔpakG). For clarity, the kinase that is additionally deleted in a particular mutant that shows reduced RSA compared with its parent is indicated above the bars. (C) RSA of the nhaA2p-lacZ+ fusion in selected single and double mutants. Values are given as mean ± SD of three independent experiments, except for the results shown in A, which are mean ± SD of six independent experiments. (D) TBHP and NaCl sensitivity phenotypes of various mutant strains as indicated. Raw data (absolute β-gal activity) to calculate RSA in B and C are shown in Fig. S5 A and B, respectively.
We next tested the single and double mutants in phenotypic stress assays. Here, PakB and PakC, the two major kinases for activation by TBHP, were also required for resistance to this organic peroxide (Fig. 5D). For salt stress, all mutants were more sensitive than the WT, with the pakF mutant and the three double mutants exhibiting the highest sensitivity. These strains were, however, not as sensitive as the Δ6 strain was, indicating a contribution of the other kinases to resistance to salt. For HS, no decrease in viability was observed when strains were incubated up to 2 h at 37 °C (Fig. S5C), the conditions used to measure β-gal activity. We thus tested our standard HS condition (46 °C). Note that it was not possible to check activation of the response at 46 °C, because β-gal was apparently inactivated at this temperature. When shifted to 46 °C, all single and double mutants showed decreased viability, with the ΔpakBC strain showing the strongest phenotype, although the strain was still more resistant than the phyR mutant (Fig. S5D). Given that the ΔpakBC strain shows the strongest phenotypes, one possibility is that the same signals are produced during severe HS (46 °C) and exposure to TBHP, which, in turn, are different from the signals generated at 37 °C. In summary, depending on the stressor considered, resistance is conferred by the kinases that are activated by this stress (TBHP) or by the full complement of kinases (salt), with contribution of the basal level provided by the other “nonsensing” kinases. Because noninducing conditions are currently not known, if they exist at all, basal vs. inducible contributions of kinases to stress resistance could not be assessed.
In conclusion, these experiments clearly demonstrate that kinases are not fully redundant but, instead, are able to respond to varying degrees to different stresses. Hence, the GSR monitors a range of different signals through different kinases and integrates this information through PhyR to adjust pathway output.
Discussion
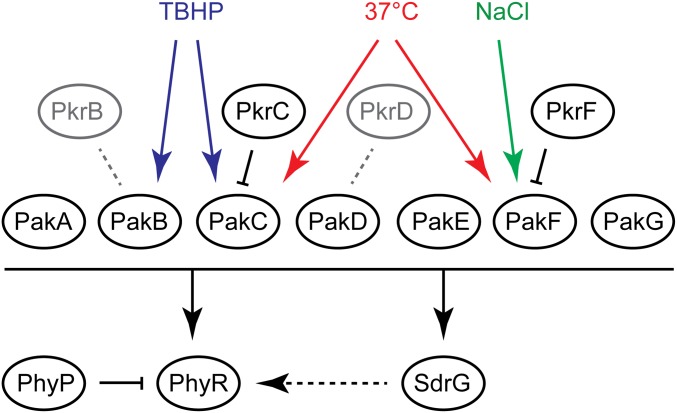
In the present study, we identified a complex signal transduction network regulating the GSR in S. melonis Fr1. In the current model (Fig. 6), PhyR phosphorylation is controlled by seven cytoplasmic histidine kinases (Paks) and the previously identified transmembrane phosphatase PhyP. Paks also phosphorylate the SDRR SdrG, which plays a central role in the GSR as a positive regulator upstream of PhyR. Finally, additional SDRRs negatively affect the GSR in a way that requires their cognate kinases.
Fig. 6.
Summary of the current model of GSR regulation in S. melonis Fr1 (details are provided in the main text). Colored arrows indicate activating stress conditions for kinases. Black arrows and bars represent activating and inhibitory relationships, respectively. Gray dotted lines denote possible but unknown interactions between cognate kinase-response regulator pairs.
Stress Sensing by Paks.
For three kinases, activating conditions could be identified, namely, thermal, oxidative, and/or osmotic stress. Given the fact that kinases responding to the same stress possess different sensor domains (Fig. S2B), and that one kinase can respond to at least two different stresses, we speculate that stresses are sensed indirectly, through stress-induced molecular cues. Hence, it is conceivable that a particular stress leads to multiple molecular cues that can each be sensed by one kinase and, in turn, different stresses can produce a common molecular cue. On the other hand, some kinases possess multiple sensor domains, suggesting that they might also be able to sense several unrelated signals, or that multiple signals are necessary for activation. Indeed, PakD, PakB, and PakA all possess multiple PAS and/or GAF domains. Because of low sequence identity among these domain classes, it is generally not possible to predict the actual signal sensed by PAS or GAF domains (33–35), and the presence of multiple domains complicates the picture. Another kinase, PakG, does not harbor any domains other than the DHp and CA domains of the catalytic core, suggesting that it might be the protein level of PakG that is regulated in response to stress or that its activity might be regulated by an accessory protein. A future challenge will be to identify activating conditions for the other Paks (and PhyP) and the nature of signals specifically recognized by each sensor.
Role of SDRRs in the GSR.
Regarding the role of SdrG in the signal transduction pathway leading to GSR activation, our data indicate that SdrG acts upstream or at the level of PhyR. This finding would be in agreement with a model in which SdrG is part of a phosphorelay eventually resulting in PhyR phosphorylation. However, the observation that all kinases tested efficiently phosphorylated both SdrG and PhyR in vitro and that the requirement of SdrG for GSR induction in vivo can be bypassed by either PhyR or Pak overexpression is not in agreement with this model. In addition, whereas mutations in phyP, which encodes the essential PhyR phosphatase PhyP, are readily obtained in a ΔphyR background (10), we were unable to obtain phyP mutants in the ΔsdrG background. This result indicates that a substantial amount of PhyR is phosphorylated in the sdrG mutant and argues against a model in which PhyR phosphorylation is controlled by SdrG through a strictly linear phosphorelay.
An alternative model for SdrG function is the modulation of histidine kinase activity, as was proposed for DivK (36) of C. crescentus (37). In such a model, upon sensing a specific stimulus, a particular kinase would phosphorylate both SdrG and PhyR, and phosphorylated SdrG would further stimulate the autophosphorylation activity of the same and/or other kinases, which would result in even more PhyR∼P and SdrG∼P. Similarly (or alternatively), SdrG∼P could switch off or reduce the phosphatase activity of PhyP toward PhyR∼P, also resulting in elevated PhyR∼P levels. In this positive feedback loop, SdrG would play the role of an amplifier of the response. Another possible role of SdrG is at the level of the partner switch itself, where SdrG might be required during PhyR phosphorylation and/or NepR binding to PhyR∼P in vivo. This possible function might also relate to the observation that NepR stimulates PhyR phosphorylation in vitro (Fig. 4), which is perhaps due to a cooperative effect of PhyR phosphorylation and NepR binding. It is thus conceivable that SdrG facilitates or stabilizes PhyR phosphorylation upon stress activation during the time that NepR is still bound to σEcfG. If this were the case, however, SdrG would likely do so indirectly, because no stimulatory effect of SdrG on PhyR phosphorylation could be observed in vitro (Fig. 4). Regardless of the exact mechanism of its function, SdrG appears to have a similarly central role to the GSR as PhyR under physiological conditions, because a ΔsdrG strain is impaired in stress resistance (Fig. 1A and Fig. S1) and sensing of various stresses (Fig. 5). Putative SdrG orthologs are found in other Alphaproteobacteria, and, interestingly, a previous genetic study in M. extorquens PA1 showed that its SdrG ortholog, encoded by Mext_0407, is similarly involved in the GSR (23). These observations suggest that SdrG might be central to the GSR in several Alphaproteobacteria.
Based on kinase in vivo overexpression data (Fig. 3C), dependencies on SdrG seem different for kinases but do not correlate with the in vitro preference of kinases for SdrG and PhyR in terms of phosphorylation. Currently, we do not have a simple explanation reconciling these results. We initially speculated that, in vivo, kinases might form heteromers that would show different preferences for SdrG and PhyR compared with the homomers used in in vitro phosphotransfer reactions. Because bacterial two-hybrid experiments showed only homomer formation among kinases, we did not pursue this possibility, although we cannot exclude that heterodimerization might happen in vivo. The actual preferences in vivo might be different from what was observed in vitro due to the involvement of additional proteins or factors that, for example, modulate the kinases’ preferences for SdrG and PhyR or spatially restrict interactions between kinases and substrates.
Some of the cognate SDRRs were shown to affect the GSR negatively (Fig. 2G). One obvious explanation is that they compete with SdrG and PhyR for phosphorylation by their cognate kinase. A possible role for these response regulators could be as a dedicated phosphate sink. Alternatively, they might act in a parallel, PhyR-independent pathway, allowing coregulation of the GSR and another response. In C. crescentus, based on genetic experiments, the SDRR LovR has been proposed to regulate PhyR activity negatively by functioning as a phosphate sink, together with its cognate kinase LovK (21). In addition, LovK controls cell adhesion by an unknown mechanism (38), but independent of the σEcfG ortholog, SigT (39). Although the physiological relevance was not explored, these data suggest that both functions might coexist. Such an organization to coordinate different physiological responses is poorly documented and distinct from the paradigm of insulated two-component systems (40, 41) but might have regulatory advantages in the case of the GSR. Clearly, more work is needed to clarify the mechanisms by which SdrG and the cognate response regulators influence the GSR and to understand their physiological implications.
Conservation of the Role of HRXXN Kinases in the GSR of Alphaproteobacteria.
All histidine kinases encoded at the phyR locus carry the HRXXN motif in their DHp domain. Like S. melonis Fr1, most Alphaproteobacteria also possess additional HRXXN kinases encoded elsewhere in their genomes, and the question arises as to whether these kinases also play a role in the GSR in other Alphaproteobacteria. Although this possibility was not explored systematically in another organism, recent data indeed suggest links to the PhyR cascade for some of these kinases. In C. crescentus, in addition to PhyK, which is responsible for sensing several activating cues (12), another kinase, LovK, was implicated as a negative regulator of the PhyR cascade (21). In S. meliloti, a recent genetic study (18) has found both positive and negative effects of the cis-encoded kinases RsiC on GSR activity in different conditions, leading the authors of that study to propose a bifunctional role of RsiC as kinase/phosphatase of PhyR. The deletion of another HRXXN kinase, SMc00322, shows a synthetic phenotype with the rsiC mutation in GSR induction by thermal stress. This observation is similar to our results for GSR induction in response to TBHP stress, for which only the pakBC double mutant, but not the pakB and pakC single mutants, showed a strongly reduced response. Interestingly, in S. meliloti, Sauviac and Bruand (18) also observed that although the rsiC SMc00322 double mutant fails to induce the GSR in response to thermal stress, it still responds to GSR induction in the stationary phase, another GSR-inducing condition in this organism. These observations are in agreement with the existence of other regulators of the GSR in S. meliloti, with one or more of the six remaining HRXXN kinases possibly being responsible for this phenotype. For Erythrobacter litoralis, a recent in vitro study using phosphotransfer profiling identified PhyR and another SDRR (the ortholog of LovR and PkrB) as substrates of the HRXXN kinases EL368 and EL346 (42). However, the involvement of these two kinases, or the PhyR-NepR-σEcfG core cascade itself, in the GSR has not yet been studied in E. litoralis. Finally, the phylogenetic distribution of PhyR and HRXXN kinases also supports a general link between these regulators. The PhyR cascade is exclusively present in Alphaproteobacteria, and members of the HWE and HisKA2 kinase families, for both of which the HRXXN motif in the DHp domain is diagnostic, are primarily associated with Alphaproteobacteria (20, 22), with their presence in other bacterial groups likely being the result of horizontal gene transfer. Notably, the few Alphaproteobacteria that have lost PhyR, NepR, and σEcfG also lack HRXXN kinases (Table S1). One example is Magnetococcus marinus MC-1, which has a genome size of 4.7 Mbp and encodes 172 genes with predicted functions in two-component system signal transduction (43). This observation suggests that there is a correlation between the presence of PhyR and HRXXN kinases that is not simply due to genome reduction or a small repertoire of two-component systems. Taken together, it thus seems reasonable to assume that, besides S. melonis Fr1, other Alphaproteobacteria also possess several HRXXN kinases/phosphatases that converge to PhyR to control the GSR.
In our study, however, we found no induction of the GSR upon overexpression of the HRXXN kinase Sphme2DRAFT_2368 in vivo, and in another study in E. litoralis, the HRXXN kinase EL_362 did not phosphorylate PhyR in vitro (42), suggesting that not all HRXXN kinases are PhyR kinases. In yet other studies, the role of HRXXN kinases in cellular processes other than the GSR has been demonstrated (44–46), although, at the same time, their involvement in the GSR has not been assessed, and thus cannot be excluded at this time. For example, in Rhizobium leguminosarum, the orphan HRXXN kinase R-LOV-HK (pLR110320) regulates light-dependent cell attachment, exopolysaccharide production, and nodulation, but the response regulator presumably mediating this response was not identified and the involvement of PhyR in these phenotypes has not been studied (44). In another example, the HRXXN kinase SMa0113 of S. meliloti was shown to regulate both succinate-mediated catabolite repression (SMCR) and polyhydroxybutyrate (PHB) production, but its cognate SDRR, SMa0114, seems dispensable for SMCR regulation and plays a noncanonical role in PHB production. It has been proposed that SMa0113 might mediate these responses through a yet-unidentified response regulator and that SMa0114 would modulate (possibly inhibit) SMa0113 activity (45). This model is somehow reminiscent of the negative roles of the SDRRs on their cognate kinases in activation of the GSR in S. melonis Fr1 (Fig. 2G). Finally, as discussed above, LovK of C. crescentus is proposed to serve a dual role in GSR regulation and cell adhesion (21, 38, 39). Thus, it might be possible that other HRXXN kinases function with PhyR and a second response regulator in branched pathways. More experimental data are needed on HRXXN kinases to identify additional characteristics besides the HRXXN motif that would dictate substrate specificity and qualify them as PhyR kinases/phosphatases.
Conclusion
The signal transduction system leading to PhyR phosphorylation in S. melonis Fr1, and possible other Alphaproteobacteria, represents one of the few examples of complex two-component system networks. Compared with prototypical two-component systems, it seems reasonable that the GSR has evolved to respond to many different signals (through kinases) while maintaining a common output (through PhyR). Thus, duplication of kinases and diversification of the sensory part without changes in response regulator substrate specificity would allow a particular organism to expand the range of signals sensed and enable niche adaptation. Indeed, it has been noted before, based on phylogenetic analyses, that HWE kinases are rapidly evolving with respect to their sensor domains (20). In such a scenario, the driving forces shaping evolution of two-component signaling in the GSR would be clearly different from the driving forces thought to dictate the evolution of paralogous histidine kinase-response regulator pairs, which results in pathway insulation (47, 48). It will be interesting to see if and how the repertoire of HRXXN kinases and their sensor domains relates to the lifestyles of different Alphaproteobacteria.
Materials and Methods
Strains and Plasmids.
Details on mutant strain and plasmid construction are given in SI Materials and Methods. Strains are listed in Table S2, and primers are listed in Table S3. A high-quality permanent draft of the S. melonis Fr1 genome is available from Integrated Microbial Genomes on the Department of Energy Joint Genome Institute website (img.jgi.doe.gov/cgi-bin/w/main.cgi), and locus tags of described genes within this study are listed in Table S4. Escherichia coli TOP10 (Invitrogen) was used for cloning, and E. coli BL21(DE3) (Invitrogen) or Rosetta2(DE3) (Novagen) was used for protein expression. All S. melonis Fr1 strains are derivatives of the WT strain JVZ857 (49) and were routinely grown at 28 °C in baffled flasks with shaking at 160 rpm in LB-Lennox medium, nutrient broth without NaCl (NB) (Fluka), or tryptone broth (TB) (Oxoid). When appropriate, antibiotics were used at the following concentrations: chloramphenicol (34 μg/mL), tetracycline (10 μg/mL), kanamycin (50 μg/mL), streptomycin (100 μg/mL), and carbenicillin (50 μg/mL).
β-Gal Assays and Stress Phenotypes.
If not otherwise stated, strains were grown from a single colony overnight in LB-Lennox medium to exponential phase and β-gal activity was measured according to the method of Miller (50). For overexpression experiments, β-gal activity was measured before and 2 h or 4 h after induction with 25 μM cumate as detailed in the figure legends. For the stress induction experiments shown in Fig. 5, strains were grown in TB; precultures were grown during the day and used to inoculate 100 mL of main overnight culture at an OD600 of 0.0004. The next day, cultures (OD600 of 0.15–0.25) were split into five 15-mL samples and left untreated (control); stressed by the addition of NaCl (50 mM), TBHP (1.6 mM), or ethanol (1%); or shifted to 37 °C (mild HS), and β-gal activity was measured after 1 h of incubation. RSA was calculated by subtracting the β-gal activity of the control from the β-gal activity of the stressed cultures, relative to the value of a WT strain, which was included in all experiments as a reference. All measurements represent the mean ± SD of at least three biological replicates. SDs for RSA were obtained by error propagation. Stress phenotypes presented in Fig. 1A and salt and HS phenotypes shown in Fig. S1 A and B were performed as described previously (10). Other stress phenotypes were performed in essentially the same way, but in TB medium instead of NB medium. Briefly, for TBHP and salt sensitivity assays, strains were grown in TB to exponential phase (OD600 of 0.15–0.25), 10-fold serially diluted, and spotted on TB containing 240 μM TBHP or 325 mM NaCl. For severe HS phenotypes, exponentially growing cultures (OD600 of 0.15–0.25) at 28 °C were transferred to a water bath at 46 °C and incubated with shaking, and aliquots were taken after 20 and 40 min, 10-fold serially diluted, and spotted on TB plates. Similarly, for mild HS phenotypes, cultures were transferred to 37 °C and incubated for 2 h, after which cells were serially diluted and spotted on TB plates. Aliquots taken before temperature upshift served as controls. Plates were incubated at 28 °C for 3–5 d.
Phosphotransfer Reactions.
Protein expression and purification are described in SI Materials and Methods. Two hundred microliters of purified kinase (5 μM) in kinase buffer [10 mM Hepes-KOH (pH 8.0), 50 mM KCl, 10% (vol/vol) glycerol, 0.1 mM EDTA, 1 mM DTT] were equilibrated to room temperature for 30 min, and autophosphorylation was initiated by addition of MgCl2 (5 mM final concentration) and 10 μCi [γ-32P]ATP (5,000 Ci/mmol; Hartmann Analytic GmbH). After 5–10 min (for PakC and PakF) or 1 h (for the other kinases) of autophosphorylation, 15 μL of kinase solution was mixed with an equal volume of response regulator(s) (5 μM each) and/or NepR (7.5 μM) in kinase buffer containing 5 mM MgCl2, and the phosphotransfer reaction was allowed to proceed for 30 s before it was quenched by addition of 3× Lämmli buffer. For the control, the kinase was mixed with kinase buffer containing 5 mM MgCl2. Samples were then subjected to SDS/PAGE [15% (wt/vol) acrylamide], the gel was dried, the dye front and unincorporated ATP were removed, the gel was exposed to a phosphorimager screen, and the screen was scanned using an FX Imager Pro Plus (Biorad).
Supplementary Material
Acknowledgments
We thank Urs Jenal, Wladimir Malaga, Herbert P. Schweizer, Martin Thanbichler, and David S. Waughn for plasmids. This work was supported by Swiss National Science Foundation (Grant 310030B-152835).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1410095111/-/DCSupplemental.
References
- 1.Francez-Charlot A, et al. Sigma factor mimicry involved in regulation of general stress response. Proc Natl Acad Sci USA. 2009;106(9):3467–3472. doi: 10.1073/pnas.0810291106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Staroń A, et al. The third pillar of bacterial signal transduction: Classification of the extracytoplasmic function (ECF) sigma factor protein family. Mol Microbiol. 2009;74(3):557–581. doi: 10.1111/j.1365-2958.2009.06870.x. [DOI] [PubMed] [Google Scholar]
- 3.Abromaitis S, Koehler JE. The Bartonella quintana extracytoplasmic function sigma factor RpoE has a role in bacterial adaptation to the arthropod vector environment. J Bacteriol. 2013;195(11):2662–2674. doi: 10.1128/JB.01972-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alvarez-Martinez CE, Lourenço RF, Baldini RL, Laub MT, Gomes SL. The ECF sigma factor sigma(T) is involved in osmotic and oxidative stress responses in Caulobacter crescentus. Mol Microbiol. 2007;66(5):1240–1255. doi: 10.1111/j.1365-2958.2007.06005.x. [DOI] [PubMed] [Google Scholar]
- 5.Bastiat B, Sauviac L, Bruand C. Dual control of Sinorhizobium meliloti RpoE2 sigma factor activity by two PhyR-type two-component response regulators. J Bacteriol. 2010;192(8):2255–2265. doi: 10.1128/JB.01666-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delory M, Hallez R, Letesson JJ, De Bolle X. An RpoH-like heat shock sigma factor is involved in stress response and virulence in Brucella melitensis 16M. J Bacteriol. 2006;188(21):7707–7710. doi: 10.1128/JB.00644-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gourion B, Francez-Charlot A, Vorholt JA. PhyR is involved in the general stress response of Methylobacterium extorquens AM1. J Bacteriol. 2008;190(3):1027–1035. doi: 10.1128/JB.01483-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gourion B, Rossignol M, Vorholt JA. A proteomic study of Methylobacterium extorquens reveals a response regulator essential for epiphytic growth. Proc Natl Acad Sci USA. 2006;103(35):13186–13191. doi: 10.1073/pnas.0603530103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gourion B, et al. The PhyR-σ(EcfG) signalling cascade is involved in stress response and symbiotic efficiency in Bradyrhizobium japonicum. Mol Microbiol. 2009;73(2):291–305. doi: 10.1111/j.1365-2958.2009.06769.x. [DOI] [PubMed] [Google Scholar]
- 10.Kaczmarczyk A, et al. Role of Sphingomonas sp. strain Fr1 PhyR-NepR-σEcfG cascade in general stress response and identification of a negative regulator of PhyR. J Bacteriol. 2011;193(23):6629–6638. doi: 10.1128/JB.06006-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim HS, Caswell CC, Foreman R, Roop RM, 2nd, Crosson S. The Brucella abortus general stress response system regulates chronic mammalian infection and is controlled by phosphorylation and proteolysis. J Biol Chem. 2013;288(19):13906–13916. doi: 10.1074/jbc.M113.459305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lourenço RF, Kohler C, Gomes SL. A two-component system, an anti-sigma factor and two paralogous ECF sigma factors are involved in the control of general stress response in Caulobacter crescentus. Mol Microbiol. 2011;80(6):1598–1612. doi: 10.1111/j.1365-2958.2011.07668.x. [DOI] [PubMed] [Google Scholar]
- 13.Martínez-Salazar JM, Salazar E, Encarnación S, Ramírez-Romero MA, Rivera J. Role of the extracytoplasmic function sigma factor RpoE4 in oxidative and osmotic stress responses in Rhizobium etli. J Bacteriol. 2009;191(13):4122–4132. doi: 10.1128/JB.01626-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sauviac L, Philippe H, Phok K, Bruand C. An extracytoplasmic function sigma factor acts as a general stress response regulator in Sinorhizobium meliloti. J Bacteriol. 2007;189(11):4204–4216. doi: 10.1128/JB.00175-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Campagne S, et al. Structural basis for sigma factor mimicry in the general stress response of Alphaproteobacteria. Proc Natl Acad Sci USA. 2012;109(21):E1405–E1414. doi: 10.1073/pnas.1117003109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herrou J, Foreman R, Fiebig A, Crosson S. A structural model of anti-anti-σ inhibition by a two-component receiver domain: The PhyR stress response regulator. Mol Microbiol. 2010;78(2):290–304. doi: 10.1111/j.1365-2958.2010.07323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herrou J, Rotskoff G, Luo Y, Roux B, Crosson S. Structural basis of a protein partner switch that regulates the general stress response of α-proteobacteria. Proc Natl Acad Sci USA. 2012;109(21):E1415–E1423. doi: 10.1073/pnas.1116887109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sauviac L, Bruand C. A putative bifunctional histidine kinase/phosphatase of the HWE family exerts positive and negative control on the Sinorhizobium meliloti general stress response. J Bacteriol. 2014;196(14):2526–2535. doi: 10.1128/JB.01623-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Finn RD, et al. Pfam: Tthe protein families database. Nucleic Acids Res. 2014;42(Database issue):D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karniol B, Vierstra RD. The HWE histidine kinases, a new family of bacterial two-component sensor kinases with potentially diverse roles in environmental signaling. J Bacteriol. 2004;186(2):445–453. doi: 10.1128/JB.186.2.445-453.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Foreman R, Fiebig A, Crosson S. The LovK-LovR two-component system is a regulator of the general stress pathway in Caulobacter crescentus. J Bacteriol. 2012;194(12):3038–3049. doi: 10.1128/JB.00182-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Staroń A, Mascher T. General stress response in α-proteobacteria: PhyR and beyond. Mol Microbiol. 2010;78(2):271–277. doi: 10.1111/j.1365-2958.2010.07336.x. [DOI] [PubMed] [Google Scholar]
- 23.Metzger LC, Francez-Charlot A, Vorholt JA. Single-domain response regulator involved in the general stress response of Methylobacterium extorquens. Microbiology. 2013;159(Pt 6):1067–1076. doi: 10.1099/mic.0.066068-0. [DOI] [PubMed] [Google Scholar]
- 24.Kaczmarczyk A, Vorholt JA, Francez-Charlot A. Cumate-inducible gene expression system for sphingomonads and other Alphaproteobacteria. Appl Environ Microbiol. 2013;79(21):6795–6802. doi: 10.1128/AEM.02296-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaczmarczyk A, Vorholt JA, Francez-Charlot A. Markerless gene deletion system for sphingomonads. Appl Environ Microbiol. 2012;78(10):3774–3777. doi: 10.1128/AEM.07347-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klose KE, Weiss DS, Kustu S. Glutamate at the site of phosphorylation of nitrogen-regulatory protein NTRC mimics aspartyl-phosphate and activates the protein. J Mol Biol. 1993;232(1):67–78. doi: 10.1006/jmbi.1993.1370. [DOI] [PubMed] [Google Scholar]
- 27.Casino P, Rubio V, Marina A. Structural insight into partner specificity and phosphoryl transfer in two-component signal transduction. Cell. 2009;139(2):325–336. doi: 10.1016/j.cell.2009.08.032. [DOI] [PubMed] [Google Scholar]
- 28.Skerker JM, et al. Rewiring the specificity of two-component signal transduction systems. Cell. 2008;133(6):1043–1054. doi: 10.1016/j.cell.2008.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laub MT, Biondi EG, Skerker JM. Phosphotransfer profiling: Systematic mapping of two-component signal transduction pathways and phosphorelays. Methods Enzymol. 2007;423:531–548. doi: 10.1016/S0076-6879(07)23026-5. [DOI] [PubMed] [Google Scholar]
- 30.Skerker JM, Prasol MS, Perchuk BS, Biondi EG, Laub MT. Two-component signal transduction pathways regulating growth and cell cycle progression in a bacterium: A system-level analysis. PLoS Biol. 2005;3(10):e334. doi: 10.1371/journal.pbio.0030334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gaidenko TA, Bie X, Baldwin EP, Price CW. Substitutions in the presumed sensing domain of the Bacillus subtilis stressosome affect its basal output but not response to environmental signals. J Bacteriol. 2011;193(14):3588–3597. doi: 10.1128/JB.00060-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gaidenko TA, Bie X, Baldwin EP, Price CW. Two surfaces of a conserved interdomain linker differentially affect output from the RST sensing module of the Bacillus subtilis stressosome. J Bacteriol. 2012;194(15):3913–3921. doi: 10.1128/JB.00583-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Crosson S, Rajagopal S, Moffat K. The LOV domain family: Photoresponsive signaling modules coupled to diverse output domains. Biochemistry. 2003;42(1):2–10. doi: 10.1021/bi026978l. [DOI] [PubMed] [Google Scholar]
- 34.Henry JT, Crosson S. Ligand-binding PAS domains in a genomic, cellular, and structural context. Annu Rev Microbiol. 2011;65:261–286. doi: 10.1146/annurev-micro-121809-151631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hurley JH. GAF domains: Cyclic nucleotides come full circle. Sci STKE. 2003;2003(164):PE1. doi: 10.1126/stke.2003.164.pe1. [DOI] [PubMed] [Google Scholar]
- 36.Sommer JM, Newton A. Pseudoreversion analysis indicates a direct role of cell division genes in polar morphogenesis and differentiation in Caulobacter crescentus. Genetics. 1991;129(3):623–630. doi: 10.1093/genetics/129.3.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paul R, et al. Allosteric regulation of histidine kinases by their cognate response regulator determines cell fate. Cell. 2008;133(3):452–461. doi: 10.1016/j.cell.2008.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Purcell EB, Siegal-Gaskins D, Rawling DC, Fiebig A, Crosson S. A photosensory two-component system regulates bacterial cell attachment. Proc Natl Acad Sci USA. 2007;104(46):18241–18246. doi: 10.1073/pnas.0705887104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fiebig A, et al. A cell cycle and nutritional checkpoint controlling bacterial surface adhesion. PLoS Genet. 2014;10(1):e1004101. doi: 10.1371/journal.pgen.1004101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Laub MT, Goulian M. Specificity in two-component signal transduction pathways. Annu Rev Genet. 2007;41:121–145. doi: 10.1146/annurev.genet.41.042007.170548. [DOI] [PubMed] [Google Scholar]
- 41.Podgornaia AI, Laub MT. Determinants of specificity in two-component signal transduction. Curr Opin Microbiol. 2013;16(2):156–162. doi: 10.1016/j.mib.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 42.Correa F, Ko WH, Ocasio V, Bogomolni RA, Gardner KH. Blue light regulated two-component systems: Enzymatic and functional analyses of light-oxygen-voltage (LOV)-histidine kinases and downstream response regulators. Biochemistry. 2013;52(27):4656–4666. doi: 10.1021/bi400617y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ulrich LE, Zhulin IB. The MiST2 database: A comprehensive genomics resource on microbial signal transduction. Nucleic Acids Res. 2010;38(Database issue):D401–D407. doi: 10.1093/nar/gkp940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bonomi HR, et al. Light regulates attachment, exopolysaccharide production, and nodulation in Rhizobium leguminosarum through a LOV-histidine kinase photoreceptor. Proc Natl Acad Sci USA. 2012;109(30):12135–12140. doi: 10.1073/pnas.1121292109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Garcia PP, Bringhurst RM, Arango Pinedo C, Gage DJ. Characterization of a two-component regulatory system that regulates succinate-mediated catabolite repression in Sinorhizobium meliloti. J Bacteriol. 2010;192(21):5725–5735. doi: 10.1128/JB.00629-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Swartz TE, et al. Blue-light-activated histidine kinases: Two-component sensors in bacteria. Science. 2007;317(5841):1090–1093. doi: 10.1126/science.1144306. [DOI] [PubMed] [Google Scholar]
- 47.Capra EJ, Laub MT. Evolution of two-component signal transduction systems. Annu Rev Microbiol. 2012;66:325–347. doi: 10.1146/annurev-micro-092611-150039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Capra EJ, Perchuk BS, Skerker JM, Laub MT. Adaptive mutations that prevent crosstalk enable the expansion of paralogous signaling protein families. Cell. 2012;150(1):222–232. doi: 10.1016/j.cell.2012.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Innerebner G, Knief C, Vorholt JA. Protection of Arabidopsis thaliana against leaf-pathogenic Pseudomonas syringae by Sphingomonas strains in a controlled model system. Appl Environ Microbiol. 2011;77(10):3202–3210. doi: 10.1128/AEM.00133-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miller JH, editor. Experiments in Molecular Genetics. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1972. pp. 352–355. [Google Scholar]
- 51.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: A sequence logo generator. Genome Res. 2004;14(6):1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.