Significance
Serogroup B meningococcus (MenB) causes severe sepsis and invasive meningococcal disease, particularly affecting young children and adolescents. The genome-derived vaccine 4CMenB that targets MenB, has now been approved in over 30 countries worldwide. Here we report the crystal structure of the trimeric autotransporter Neisserial adhesin A (NadA), one of the three protein antigens included in 4CMenB, and the epitope mapping of a bactericidal mAb monoclonal antibody that targets the functional head domain of NadA. These results provide important insights into the structure and vaccine-induced immune response of this meningococcal antigen and may inform the engineering of improved immunogens by structure-based design.
Keywords: meningitis, coiled coil, thermostability, hydrogen–deuterium exchange, trimeric autotransporter adhesin
Abstract
Serogroup B Neisseria meningitidis (MenB) is a major cause of severe sepsis and invasive meningococcal disease, which is associated with 5–15% mortality and devastating long-term sequelae. Neisserial adhesin A (NadA), a trimeric autotransporter adhesin (TAA) that acts in adhesion to and invasion of host epithelial cells, is one of the three antigens discovered by genome mining that are part of the MenB vaccine that recently was approved by the European Medicines Agency. Here we present the crystal structure of NadA variant 5 at 2 Å resolution and transmission electron microscopy data for NadA variant 3 that is present in the vaccine. The two variants show similar overall topology with a novel TAA fold predominantly composed of trimeric coiled-coils with three protruding wing-like structures that create an unusual N-terminal head domain. Detailed mapping of the binding site of a bactericidal antibody by hydrogen/deuterium exchange MS shows that a protective conformational epitope is located in the head of NadA. These results provide information that is important for elucidating the biological function and vaccine efficacy of NadA.
The Gram-negative encapsulated bacterium Neisseria meningitidis causes severe sepsis and meningococcal meningitis. Invasive meningococcal disease (IMD) is associated with 5–15% mortality; furthermore, devastating long-term sequelae such as amputations, hearing loss, and neurodevelopmental disabilities are observed in 11–19% of IMD survivors (1). Meningococcal serogroups are distinguished by the composition of their capsular polysaccharides. The five serogroups most commonly associated with invasive disease are A, B, C, W, and Y. (2). Effective mono- or polyvalent-conjugated polysaccharide vaccines against N. meningitidis serogroups A, C, W, and Y have been available since the early 1990s (3). However, serogroup B meningococcus (MenB) is responsible for the majority of endemic and epidemic meningococcal disease in developed countries (4–6). The development of an efficient capsular polysaccharide-based vaccine against MenB has been hampered by potential autoimmunity issues, namely, the structural similarity between the MenB capsular polysaccharide and the neuraminic acid present on the surface of human fetal neural tissues (7).
In early 2013 the European Medicines Agency approved 4CMenB, to our knowledge the first broadly protective vaccine against MenB, for the prevention of IMD in all age groups. 4CMenB is a multicomponent vaccine formulation composed of three surface-exposed meningococcal proteins originally identified by the reverse vaccinology approach (8) plus outer membrane vesicles from the New Zealand epidemic clone. The three antigenic proteins are factor H-binding protein (fHbp), neisserial heparin-binding antigen (NHBA), and neisserial adhesin A (NadA) (9, 10).
The gene encoding NadA is present in ∼30% of pathogenic meningococcal isolates and is associated mostly with strains that belong to three of the four hypervirulent serogroup B lineages (11–14). NadA expression levels can vary among isolates by more than 100-fold, and its expression is up-regulated in vivo by niche-specific signals (15). NadA induces high levels of bactericidal antibodies in humans (16–18) and is recognized by serum antibodies of children convalescent after IMD (19), suggesting that it is expressed and is immunogenic during IMD. Two main genetically distinct groups of NadA have been identified that share overall amino acid sequence identities of 45–50%. Group I includes the three most common variants (NadA1, NadA2, and NadA3, the latter being the vaccine variant), which share ∼95% sequence identity and are immunologically cross-reactive (11). Group II includes three rarer variants: NadA4, primarily associated with carriage strains (11); NadA5, found mainly in strains of clonal complex 213 (20, 21); and NadA6 (Fig. S1A); these three share ∼90% sequence identity (Fig. S1B) (22).
Functionally, NadA3 expressed on the surface of Escherichia coli promotes adhesion to and invasion of Chang epithelial cells (23). This adhesive activity has been mapped, at least partially, to an N-terminal region extending to residue T132 (23, 24). Recently, interactions of NadA3 with β-1 integrin (25) and with the heat shock protein Hsp90 (26) have been reported.
Structurally, NadA belongs to the class of trimeric autotransporter adhesins (TAAs) (27, 28), which are known to mediate adhesion through interaction with extracellular matrix proteins and are involved in invasion of target cells (29). TAAs are obligate homotrimers, and accordingly the recombinant NadA3 vaccine antigen, lacking the C-terminal membrane anchor region, forms soluble, stable trimers (23, 30). TAAs generally are made of a conserved C-terminal integral membrane β-barrel, which anchors the proteins to the outer membrane, and an N-terminal “passenger” domain responsible for adhesion (31). The TAA passenger domain typically is made of a central α-helical domain (stalk) that forms coiled-coil structures and a distinct N-terminal domain (head) that is mainly responsible for binding to host cellular receptors.
Here we present the X-ray structure of a large ectodomain fragment of NadA5 and a structural analysis by transmission electron microscopy (TEM) of the vaccine variant NadA3. In addition, epitope mapping shows that the head of NadA3 contains immunogenic regions responsible for the generation of a protective bactericidal response.
Results
NadA Coiled-Coil Composition Greatly Influences Protein Thermostability.
Extensive attempts to crystallize NadA3 using the vaccine construct that includes the predicted head and entire stalk domains (30) were unsuccessful. We hypothesized that a long, flexible stalk might have hindered crystallization. Therefore, multiple NadA3 constructs were generated, each with a progressively shorter stalk (Fig. S2), and these constructs were screened for increased thermostability using differential scanning calorimetry (DSC), because higher melting-point (Tm) values may increase the probability of crystallization (32). Further, based on sequence alignments, corresponding NadA4 and NadA5 constructs were designed and tested. The DSC screens did not reveal Tm gains in the NadA3 constructs (Fig. S3A) but showed remarkably higher thermostability in NadA4 and NadA5 (ΔTm ∼14 °C) (Fig. 1). Five additional NadA4 constructs with progressive C-terminal truncations did not yield further Tm increases, and a NadA4 “head-only” construct was not detected by DSC, suggesting that the head domain is unable to fold when isolated from the stalk (Fig. S3B). Ultimately, reproducible diffraction-quality crystals were obtained only from the construct NadA5A24–R220.
Fig. 1.
NadA4 and NadA5 show high thermostability. DSC profiles show a ΔTm of ∼14 °C between the full-length construct of NadA3 (blue) and constructs of NadA4 and NadA5 (green and red).
High-Resolution Structure of NadA.
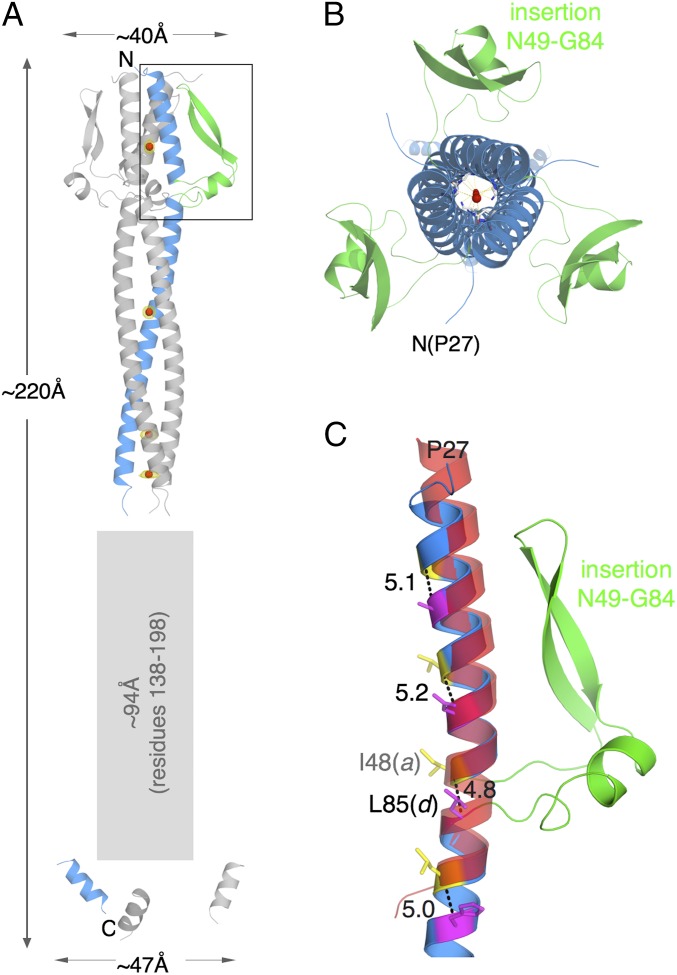
The crystal structure of NadA5 was solved at 2 Å resolution by single anomalous dispersion (SAD), using a dataset collected at 1.54 Å wavelength on a crystal soaked in a solution containing 1 M NaI (Table S1). The crystals of NadA5 belonged to space group R 3 2 (H), had a solvent content of 60%, and contained one monomer of NadA5 in the asymmetric unit positioned on the crystallographic threefold axis along c (Fig. S4). The NadA5 biological homotrimer was visualized using the two threefold crystallographic symmetry operators along the c axis of the crystal (Fig. 2A).
Fig. 2.
The crystal structure of NadA5. (A) Cartoon diagram of trimeric NadA5. Monomeric NadA5 that occupies the asymmetric unit is colored in blue for the coiled-coil region and in green for the wing-like insertion of the head; two symmetry-related molecules are shown in gray. Red spheres show the positions of iodide ions buried in the coiled-coil, and yellow meshes around these spheres show anomalous difference electron densities. The large rectangular gray-shaded outline shows the region of low-σ electron density. (B) Top view of NadA5, looking down the c axis, after rotation of 90° around x. (C) A zoomed-in view of the region of the insertion in the head of NadA5 (boxed region in A). The ribbon of NadA is colored as in A, and a superimposed α-helix, chosen from the trimeric transcription factor GCN4 (PDB ID code 4DME) to show the continuity of the coiled-coil of NadA5 around the region of the interruption, is shown as a semitransparent red ribbon. Yellow and magenta sticks depict NadA heptad residues a and d, respectively, and the distance in angstroms between their Cα carbons is shown.
Overall, the structure of NadA5 (residues A24–R220) is made of an elongated coiled-coil, or stalk, that runs from the N terminus to the C terminus and extends in length for roughly 220 Å with an average width of 15 Å in its central stalk region. A broader N-terminal region (width ∼40 Å) forming the head domain is readily apparent (Fig. 2). High-quality electron density was observed allowing residues P27–A137 to be fitted with confidence (Fig. S5). However, after residue A137 only low σ-level and discontinuous electron densities were observed, preventing confident fitting of the polypeptide chain in this region. Further observations of clear electron density toward the C terminus revealed another continuous stretch spanning T199–A210, although discontinuous, low σ-level electron densities in the region between A137 and T199 retained a few notable peaks with threefold symmetry features (Fig. S5). This observation suggests that the three helices of the NadA5 stalk are less stable in this region, likely resulting in partial unwinding of the coil and therefore in some flexibility and disorder. These conclusions are also supported by the observation that the fragments of the last visible residues, T199–A210, are arranged as short uncoiled helices, with a relatively large diameter of ∼47 Å (Fig. 2A).
The N Terminus of NadA5 Presents a Previously Unidentified Head Domain Organization.
In contrast to other TAAs, NadA lacks a truly independent globular head domain. Instead, NadA possesses an almost exclusively coiled-coil architecture that can be divided in two fragments (made of residues A34–I48 and L85–A137, respectively) separated by a sequence insertion (residues N49–G84) (Fig. 2 and Fig. S6). Quite remarkably, this interruption does not result in a discontinuity or structural perturbation of the coiled-coil, and it forms wing-like structures (three per trimer) that protrude from the stalk and pack against the N-terminal helices. Although this arrangement differentiates NadA from other TAAs, the overall larger dimensions of the N-terminal region are reminiscent of the typical TAA architecture, and the peculiar structure of the head domain might reflect particular host-receptor specificity.
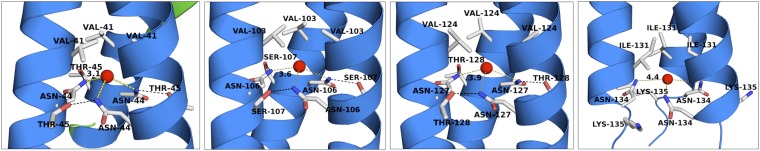
N at Position d (N@d) Layers in the NadA Coiled-Coil.
The coiled-coil of NadA5 begins in the N terminus with the first heptad repeat formed by residues A34 (position a) and A37 (position d) (Fig. S6). A second heptad is made of V41 at position a and N44 at position d, with N44 being the first of four total Asn residues observed in position d of the heptads (Fig. 3). As previously observed for other TAAs (33), the N at position d layers (termed “N@d” layers) are found to coordinate ions in the buried core of the hydrophobic coiled-coil. For NadA, because of the high concentration of NaI in the crystal soaking experiments, we observed the presence of buried iodide ions. Residues I48 (a) and L85 (d) form the third heptad repeat, which, as described above, is characterized by a remarkably long insertion (36 residues) or interruption that replaces the expected bc residue pair of the heptad with residues 49–84 (Fig. 2C and Fig. S6A). Finally, a continuous series of seven additional canonical heptads can be observed for the remaining visible stalk, spanning residues L85–N134.
Fig. 3.
The arrangement of the four N@d layers of NadA5 and their environment are shown. The coiled-coil is depicted in blue, iodides are shown as red spheres, and residues of the heptad repeat in position a, e, and d for each layer are shown as sticks. Black dashed lines show the interactions between the side chains of Asn and residues in position e of a neighboring helix of the coiled-coil. Yellow dashed lines show the distance in angstroms between iodide ions and Asn side chains.
NadA Has No Close Structural Homologs.
A database search with the DALI server (34) identified several coiled-coil protein structures, such as tropomyosin, myosin, and vimentin, but the only TAA structure identified as similar to NadA, although with high rmsd values (>15 Å), was SadA from Salmonella enterica. This result suggests that, although the helical fold is a predominant quality that biases the DALI selections, NadA has no overall close structural homologs (Fig. S7).
Electron Microscopy of NadA3 Shows a Conserved Overall Topology.
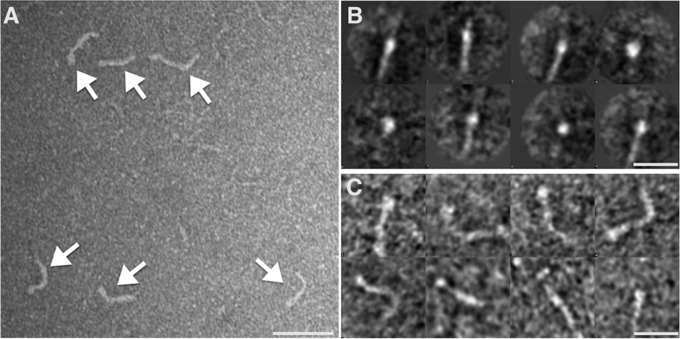
NadA3 was studied by negative stain TEM to determine if it possessed the same overall topology as NadA5. NadA3 was readily identified in TEM images (Fig. 4A), and single boxed NadA3 particles first were band-pass filtered to increase the signal-to-noise ratio, then were rotationally and translationally aligned, and then centered before undergoing multivariate statistical analysis for classification (35). Reference-free 2D class averages showed that NadA3 has a thin elongated structure that is broader at one end, likely corresponding to the N-terminal head domain (Fig. 4B). The NadA3 molecules displayed a maximum length of ∼300 Å and were observed both in linear conformations (similar to those in the NadA5 crystals) and in curved conformations where the deviation from linearity was 30–90° (hairpins or multiple inflections were rarely observed) (Fig. 4C). As expected from the high sequence conservation (51% identity), TEM suggests that the NadA3 and NadA5 variants share the same topology.
Fig. 4.
Negative stain TEM of NadA3. (A) Elongated structures corresponding to negatively stained NadA3 are indicated by white arrows. (Scale bar, 50 nm.) (B) A set of representative class averages (∼10 images per class) of NadA3 is shown. Some of the class averages, presenting side views, show an elongated structure with an enlarged terminus. (C) Single particles bandpass-filtered showing different degrees of curvature are shown. (Scale bars, 20 nm in B and C.)
Insights into Full-Length NadA3 and NadA5 from Homology Modeling.
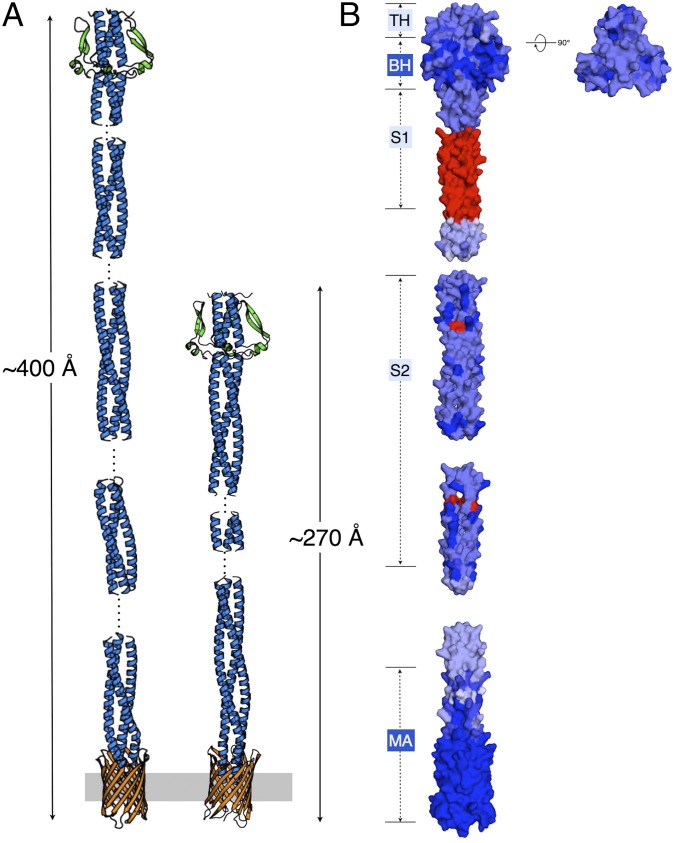
Full-length in silico models of NadA3 and NadA5 were generated using structure-based sequence alignments of the five NadA variants, along with predictions and manual analyses of coiled-coil periodicities, and omitting regions where sequence analyses showed loss or low probability of coiled-coil formation. Full-length NadA3 and NadA5 have different overall lengths, with NadA3 extending for ∼400 Å and NadA5 for ∼270 Å, predominantly because of the 81-residue difference in the stalks (Fig. 5A and Fig. S1B). A map of sequence conservation between NadA variants reveals a patch of highly conserved residues in the N-terminal head domain in addition to the wholly conserved transmembrane β-barrel. Specifically, although the apical region of the head domain is less conserved, the grooves formed between the wing-like protrusions and the coiled-coil are mostly conserved, suggesting that this basal head region is of particular interest for future functional studies (Fig. 5B).
Fig. 5.
Full-length models of NadA3 and NadA5 displaying sequence conservation. (A) Models of NadA3 and NadA5 are shown as cartoons; the stalk is shown in blue, the wing-like insertions of the head domain are shown in green, and the transmembrane anchor is shown in orange. The outer membrane where NadA is anchored is shown as a gray bar. Dashed lines indicate either regions missing in the crystal structure of NadA5 or regions with low sequence homology or unknown secondary structure that were not included in the models. (B) The sequence conservation between NadA3 and NadA5 is mapped onto the surface of the model of NadA3 using a gradient of blue from low (light blue) to high (dark blue). Red surface patches indicate sequence insertions in NadA3 that are missing in NadA5 and thus were omitted from the sequence-conservation analyses. Light- and dark-blue boxes show regions of medium and high conservation, respectively, as described in the main text. The representation in B was generated using ProtSkin (40). BH, basal head; MA, membrane anchor; S1, stalk1; S2, stalk2; TH, top head.
Bactericidal mAb 33E8 Is Specific for NadA Group I Variants.
Despite sharing a similar 3D fold, NadA group I variants are not cross-reactive with NadA group II variants (11). To investigate the molecular basis for this lack of cross-reactivity, epitope-mapping experiments were performed using the bactericidal mAb 33E8 specific for group I variants. mAb 33E8 originally was obtained from mice immunized with NadA1 and displayed binding to purified recombinant NadA1, 2, and 3 but not to NadA4 and 5 by surface plasmon resonance (SPR) experiments (Fig. S8). Similarly, FACS analyses showed that mAb 33E8 bound to meningococcal strains expressing NadA variants 1, 2, and 3 but not to a strain that expresses NadA5. Finally, in a serum bactericidal assay (SBA) experiment, mAb 33E8 elicited positive bactericidal titers (>2,048) only against meningococcal strains DE11445, 5/99, and NMB, which express NadA variants 1, 2, and 3, respectively.
Bactericidal mAb 33E8 Binds to the NadA3 Head Domain.
SPR experiments performed on different NadA3 fragments showed that both full-length NadA3 and the constructs containing the head+stalk bound to mAb 33E8 with high affinity. In contrast, all other NadA3 constructs made of the coiled-coil only, as well as those of NadA4 and NadA5 made of head+stalk, were not able to bind to mAb 33E8 (Fig. S8A). Interestingly, a monomeric construct containing only the head domain (NadA3A24–V89) also bound to mAb 33E8 but with significantly decreased affinity. The binding affinities calculated by SPR for the NadA3 full-length, head+stalk, and head-only constructs revealed interactions with equilibrium binding dissociation constants (Kd) of 0.6 nM, 3.4 nM, and 1.5 μM, respectively (Fig. S8B). DSC experiments suggested that the head-only construct is unfolded (Fig. S8C), perhaps explaining the ∼1,000-fold lower affinity observed in SPR. Overall, these data indicate that the head or the head+stalk region of NadA3 contains the protective epitope recognized by mAb 33E8 and that this epitope is either mutated or absent in NadA4 and Nad5.
Size-exclusion chromatography and nondenaturing nanoelectrospray MS then were used to study the stoichiometry of the 33E8/NadA3 interaction. We used Fab 33E8 in a high-affinity complex with the NadA3 construct A24–A170. The native mass spectrum of the complex showed that the major species was observed in the 6,300–7,750 m/z range, with a molecular mass ranging from 193.5 to 197.2 kDa, thus in agreement with a single trimer of NadA3A24–A170 (calculated molecular mass, 50.8 kDa) simultaneously bound to three Fab 33E8 fragments (corresponding to a molecular mass in the range 47.4–49.7 kDa) (Fig. S8D).
Hydrogen–Deuterium Exchange MS Identified the Protective Epitope on NadA.
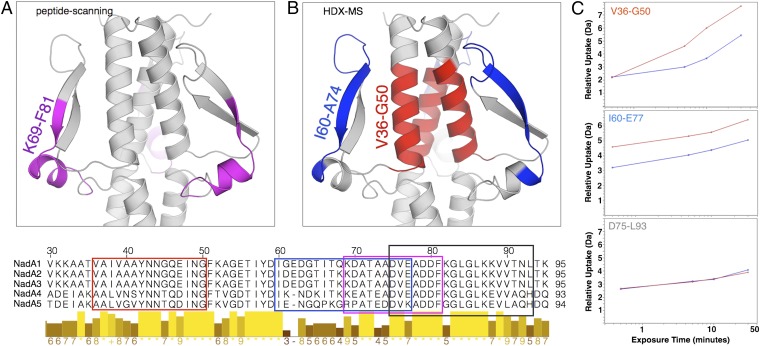
To locate accurately the epitope targeted by mAb 33E8, we first performed peptide-scanning experiments using overlapping 13mer fragments covering the entire NadA3 vaccine antigen. This approach revealed only one linear peptide recognized by mAb 33E8, made of residues K69–F81 localized on the most solvent-accessible wing region of the homology model of the NadA3 head domain (Fig. 6A). Subsequently we performed epitope mapping by hydrogen–deuterium exchange (HDX)-MS using the construct NadA3A24–A170. The binding of Fab 33E8 induced a significant reduction of deuterium uptake for two nonconsecutive peptides, allowing mapping of an epitope involving segments V36–G50 and I60–A74 and taking into account a third peptide (D75–L93) not affected by deuterium protection (Fig. 6 B and C) as mapped on the NadA3 homology model.
Fig. 6.
Epitope mapping of monoclonal antibody 33E8. (A and B, Upper) Zoomed-in views of the head of the NadA3 homology model showing the epitope recognized by mAb 33E8 as detected by peptide scanning (magenta in A) and by HDX-MS (red and blue in B). (Lower) Residues of the 33E8 epitope also are shown in a sequence alignment of five NadA variants, marked with boxes colored according to the scheme above, plus a black rectangle that contains peptide 75-93, which is not affected by the binding. Similarity scores as calculated with Clustal are shown graphically below the alignment. (C) Deuterium uptake measured by HDX-MS. Blue curves show peptides of the NadA3A24–A170–Fab 33E8 complex, and red curves show the NadA3A24–A170 peptides alone.
In summary, epitope mapping by peptide scanning and HDX-MS revealed the location of the binding region of Fab 33E8 on the head domain of NadA, in agreement with SPR measurements. In addition, HDX-MS considerably expands the total epitope previously detected by peptide scanning to include residues V36–G50, which reside on the coiled-coil in the center of the head. This finding strongly suggests a conformational nature of the epitope of 33E8. In addition, these results could explain the residual binding of construct NadA3A24–V89 to 33E8, which, despite the lack of a proper trimeric folding, still may display poorly structured portions of the epitope in a conformation partially competent for binding.
Discussion
The structure of NadA5 reveals a novel trimeric autotransporter adhesin that has no close homologs among other TAAs or among other proteins present in the Protein Data Bank (PDB) but with an overall architecture reminiscent of several other surface molecules of known pathogens (Fig. S7) (36). A single coiled-coil includes both the apical N-terminal region and the main stalk of NadA5, and a peculiar sequence insertion of 36 residues between the a and d position of the third N-terminal heptad repeat gives rise to wing-like structures without altering the coiled-coil geometry. These lateral protrusions are the most solvent-exposed regions of the head, and mapping of the epitope of the bactericidal mAb 33E8 on the NadA3 homology model shows how residues located in this region contribute to form an epitope that is likely to be conformational and that is protective, at least in the mouse model used herein.
TEM analysis of the vaccine variant 3 shows how NadA variants share similar structures despite a lack of immunological cross-reactivity. This structural similarity presumably reflects the moderately conserved sequence and the conserved function among NadA variants, in particular the previously demonstrated ability of both NadA3 and NadA4 to bind to Chang cells (11). TEM imaging also reveals how the coiled-coil stalk of NadA undergoes bending, which presumably confers functional advantages and also may explain the difficulties in crystallizing a longer NadA variant.
Although our analyses confirm the same overall structure for different NadA variants, the lack of immunological cross-reactivity between the two groups of variants (11) suggests structural and/or surface-localized differences. First, the full-length models of NadA3 and NadA5 allow us to discern how the stalks of different variants possess different lengths, slightly different or shifted periodicity in the coiled-coils, and interruptions made of sequences of currently unknown structure. Analyses of sequence conservation also reveal how the apical portion of the head of NadA, although structurally conserved, possesses a low degree of sequence conservation as compared with the rest of the molecule. Therefore it is tempting to speculate that both the top of the head of NadA and the variable stalk regions underlie the lack of cross-reactivity between the two variant groups. Also, sequence conservation of the basal head (BH in Fig. 5B) of NadA, which contains the protective epitope, shows that this region, although conserved overall, possesses moderate local sequence variability that might help explain the differences in antibody binding (Fig. 6).
In this study we used a rational, biophysically driven approach to solve the first (to our knowledge) 3D structure of the vaccine antigen NadA, which previously had been recalcitrant to crystallization. Overall, our findings shed light on the architecture of this meningococcal antigen and may help elucidate the molecular mechanism of its biological function. The structure revealed here provides working models that can aid in the interpretation of previous studies and guide further investigation to elucidate fully the function of NadA as both an adhesin and a vaccine antigen. For example, these insights now raise the possibility for a structural vaccinology approach for the design of a more broadly cross-protective antigen, as described previously for the highly variable meningococcal fHbp (37), and/or represent a starting point for an epitope-focused strategy (38). Furthermore, additional structural and epitope-mapping studies to elucidate at high resolution the interaction between antibodies generated by human immunization with NadA are ongoing and promise to deliver a deeper understanding of the role of NadA in the human immune response to the first (to our knowledge) licensed and broadly protective vaccine against MenB.
Materials and Methods
Cloning, Expression, and Purification.
All NadA constructs were PCR-amplified from MenB chromosomal DNA template strains 2996 (NadA3), NGE28 (NadA4), and M01-240320 (NadA5). Full details can be found in SI Materials and Methods.
DSC.
Thermal stability experiments of NadA proteins at 10-μM concentration were performed using a MicroCal VP-Capillary DSC instrument (GE Healthcare), with a temperature scan range from 10–110 °C, a thermal ramp rate of 180 °C/h, and a 4-s filter period. Data were analyzed with Origin 7 software.
Crystallization, Data Collection, and Structure Determination.
Crystallization experiments were performed using a Gryphon crystallization robot (Art Robbins Instruments). X-ray diffraction data were collected at the Swiss Light Source (Paul Scherrer Institute, Villigen, Switzerland) beamline X06DA on a Pilatus 2M detector. The NadA5 structure was solved by the SAD method. Data collection and refinement statistics are reported in Table S1. Atomic coordinates have been deposited in the PDB (ID code 4CJD). Figures were generated using PyMOL (www.pymol.org). Full details can be found in SI Materials and Methods.
Negative Stain TEM.
NadA3Δ351–405 was loaded onto glow-discharged 200-square mesh copper/nickel grids and observed using a TEM Tecnai G2 spirit transmission electron microscope (FEI). Images were collected with a CCD camera, Olympus SIS Morad data collect 2K*4K. Full details can be found in SI Materials and Methods.
In silico Molecular Modeling of Full-Length NadA3 and NadA5.
Amino acid sequences of the translocation domains of NadA3 and NadA5, residues 86–345 and 85–264, respectively, were threaded onto the crystallographic coordinates of the translocation domain of the trimeric autotransporter Hia (PDB ID code 3EMO). Full details can be found in SI Materials and Methods.
Antibody Generation and Fab Purification.
The murine IgG2b isotype mAb 33E8 was produced and purified by Areta International. The antigen-binding fragment (Fab 33E8) was prepared by papain digestion of mAb 33E8 followed by purification using a Protein A column (Thermo Scientific). Full details can be found in SI Materials and Methods.
SPR.
All SPR experiments were performed using a Biacore T200 instrument (GE Healthcare) at 25 °C. Full details can be found in SI Materials and Methods.
Complement-Mediated Bactericidal Activity.
Bactericidal activity of mAb 33E8 was evaluated against strains DE11445, 5/99, and NMB, which express NadA variants 1, 2, and 3 respectively. The bactericidal activity was defined as the mAb concentration that resulted in a 50% decrease in colony-forming units per milliliter after 1-h incubation in the reaction mixture compared with the colony-forming units per milliliter in negative control wells at time 0. Full details can be found in SI Materials and Methods.
Flow Cytometry-FACS.
The ability of mAb 33E8 to bind surface-exposed variants of NadA on live bacteria was determined by FACS analysis. Full details can be found in SI Materials and Methods.
Nondenaturing Nanoelectrospray MS.
NadA samples at 100-μM concentration were analyzed on a SynaptG2 HDMS mass spectrometer (Waters) equipped with a nano-electrospray ionization (ESI) source. Full details can be found in SI Materials and Methods.
Epitope Mapping by Synthetic Peptide Scanning.
Screening of a cellulose-bound peptide library of NadA3 fragments for binding to mAb 33E8 was performed as described previously (39). Full details can be found in SI Materials and Methods.
Epitope Mapping by HDX-MS.
The averaged deuterium exchange behaviors of 19 peptides covering 100% of the NadA3A24–A170 sequence were measured at different time points (from 30 s to 30 min) in the absence or presence of Fab 33E8. Binding experiments were facilitated by working at low temperature (on ice, 0 °C). Full details can be found in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank S. Strelkov (Katholieke Universiteit Leuven) for providing the executable file of the program TWISTER; V. Nardi-Dei for technical assistance with size-exclusion HPLC; J. Diez and S. Russo (Expose GmbH) for assistance with diffraction data collection; B. Aricó, S. Bambini, D. Serruto, M. Merola, A. Carfi, C. Ciferri, and P. Costantino for discussions and critical reading of the draft; Catherine Mallia for editorial assistance; and M. Nissum and D. Maione for their support. Strain M01-240320 was kindly provided by R. Borrow (Health Protection Agency); strains 2996, NMB, and NGE28 by E. R. Moxon (University of Oxford); strains DE11445 and DE11458 by U. Vogel (University of Würzburg); strain 5/99 by D. A. Caugant (Norwegian Institute of Public Health); and strain IB4846 by D. W. Kim (Hanyang University). M.B. held a Novartis Academy PhD fellowship at the University of Siena and also was supported by W. F. Anderson (Center for Structural Genomics of Infectious Disease).
Footnotes
Conflict of interest statement: All authors except M.B., I.V., and G.S. are employees of Novartis Vaccines.
Data deposition: The atomic coordinates reported in this paper have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4CJD).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1419686111/-/DCSupplemental.
References
- 1.Pace D, Pollard AJ. Meningococcal disease: Clinical presentation and sequelae. Vaccine. 2012;30(Suppl 2):B3–B9. doi: 10.1016/j.vaccine.2011.12.062. [DOI] [PubMed] [Google Scholar]
- 2.Harrison LH, Trotter CL, Ramsay ME. Global epidemiology of meningococcal disease. Vaccine. 2009;27(Suppl 2):B51–B63. doi: 10.1016/j.vaccine.2009.04.063. [DOI] [PubMed] [Google Scholar]
- 3.Pace D, Pollard AJ, Messonier NE. Quadrivalent meningococcal conjugate vaccines. Vaccine. 2009;27(Suppl 2):B30–B41. doi: 10.1016/j.vaccine.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 4.Rosenstein NE, Perkins BA, Stephens DS, Popovic T, Hughes JM. Meningococcal disease. N Engl J Med. 2001;344(18):1378–1388. doi: 10.1056/NEJM200105033441807. [DOI] [PubMed] [Google Scholar]
- 5.Black S, Pizza M, Nissum M, Rappuoli R. Toward a meningitis-free world. Sci Transl Med. 2012;4(123):123ps125. doi: 10.1126/scitranslmed.3003859. [DOI] [PubMed] [Google Scholar]
- 6.Viner RM, et al. Outcomes of invasive meningococcal serogroup B disease in children and adolescents (MOSAIC): A case-control study. Lancet Neurol. 2012;11(9):774–783. doi: 10.1016/S1474-4422(12)70180-1. [DOI] [PubMed] [Google Scholar]
- 7.Finne J, Bitter-Suermann D, Goridis C, Finne U. An IgG monoclonal antibody to group B meningococci cross-reacts with developmentally regulated polysialic acid units of glycoproteins in neural and extraneural tissues. J Immunol. 1987;138(12):4402–4407. [PubMed] [Google Scholar]
- 8.Pizza M, et al. Identification of vaccine candidates against serogroup B meningococcus by whole-genome sequencing. Science. 2000;287(5459):1816–1820. doi: 10.1126/science.287.5459.1816. [DOI] [PubMed] [Google Scholar]
- 9.Giuliani MM, et al. A universal vaccine for serogroup B meningococcus. Proc Natl Acad Sci USA. 2006;103(29):10834–10839. doi: 10.1073/pnas.0603940103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Su EL, Snape MD. A combination recombinant protein and outer membrane vesicle vaccine against serogroup B meningococcal disease. Expert Rev Vaccines. 2011;10(5):575–588. doi: 10.1586/erv.11.32. [DOI] [PubMed] [Google Scholar]
- 11.Comanducci M, et al. NadA diversity and carriage in Neisseria meningitidis. Infect Immun. 2004;72(7):4217–4223. doi: 10.1128/IAI.72.7.4217-4223.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vogel U, et al. Predicted strain coverage of a meningococcal multicomponent vaccine (4CMenB) in Europe: A qualitative and quantitative assessment. Lancet Infect Dis. 2013;13(5):416–425. doi: 10.1016/S1473-3099(13)70006-9. [DOI] [PubMed] [Google Scholar]
- 13.Wang X, et al. Prevalence and genetic diversity of candidate vaccine antigens among invasive Neisseria meningitidis isolates in the United States. Vaccine. 2011;29(29-30):4739–4744. doi: 10.1016/j.vaccine.2011.04.092. [DOI] [PubMed] [Google Scholar]
- 14.de Filippis I, et al. Molecular epidemiology of Neisseria meningitidis serogroup B in Brazil. PLoS ONE. 2012;7(3):e33016. doi: 10.1371/journal.pone.0033016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fagnocchi L, et al. Transcriptional regulation of the nadA gene in Neisseria meningitidis impacts the prediction of coverage of a multicomponent meningococcal serogroup B vaccine. Infect Immun. 2013;81(2):560–569. doi: 10.1128/IAI.01085-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bowe F, et al. Mucosal vaccination against serogroup B meningococci: Induction of bactericidal antibodies and cellular immunity following intranasal immunization with NadA of Neisseria meningitidis and mutants of Escherichia coli heat-labile enterotoxin. Infect Immun. 2004;72(7):4052–4060. doi: 10.1128/IAI.72.7.4052-4060.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ciabattini A, et al. Intranasal immunization of mice with recombinant Streptococcus gordonii expressing NadA of Neisseria meningitidis induces systemic bactericidal antibodies and local IgA. Vaccine. 2008;26(33):4244–4250. doi: 10.1016/j.vaccine.2008.05.049. [DOI] [PubMed] [Google Scholar]
- 18.Findlow J, et al. Multicenter, open-label, randomized phase II controlled trial of an investigational recombinant Meningococcal serogroup B vaccine with and without outer membrane vesicles, administered in infancy. Clin Infect Dis. 2010;51(10):1127–1137. doi: 10.1086/656741. [DOI] [PubMed] [Google Scholar]
- 19.Litt DJ, et al. Putative vaccine antigens from Neisseria meningitidis recognized by serum antibodies of young children convalescing after meningococcal disease. J Infect Dis. 2004;190(8):1488–1497. doi: 10.1086/424464. [DOI] [PubMed] [Google Scholar]
- 20.Bambini S, et al. Distribution and genetic variability of three vaccine components in a panel of strains representative of the diversity of serogroup B meningococcus. Vaccine. 2009;27(21):2794–2803. doi: 10.1016/j.vaccine.2009.02.098. [DOI] [PubMed] [Google Scholar]
- 21.Lucidarme J, et al. Characterization of fHbp, nhba (gna2132), nadA, porA, sequence type (ST), and genomic presence of IS1301 in group B meningococcal ST269 clonal complex isolates from England and Wales. J Clin Microbiol. 2009;47(11):3577–3585. doi: 10.1128/JCM.00936-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bambini S, et al. Neisseria adhesin A variation and revised nomenclature scheme. Clin Vaccine Immunol. 2014;21(7):966–971. doi: 10.1128/CVI.00825-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Capecchi B, et al. Neisseria meningitidis NadA is a new invasin which promotes bacterial adhesion to and penetration into human epithelial cells. Mol Microbiol. 2005;55(3):687–698. doi: 10.1111/j.1365-2958.2004.04423.x. [DOI] [PubMed] [Google Scholar]
- 24.Tavano R, et al. Mapping of the Neisseria meningitidis NadA cell-binding site: Relevance of predicted alpha-helices in the NH2-terminal and dimeric coiled-coil regions. J Bacteriol. 2011;193(1):107–115. doi: 10.1128/JB.00430-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nägele V, et al. Neisseria meningitidis adhesin NadA targets beta1 integrins: Functional similarity to Yersinia invasin. J Biol Chem. 2011;286(23):20536–20546. doi: 10.1074/jbc.M110.188326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Montanari P, et al. Human heat shock protein (Hsp) 90 interferes with Neisseria meningitidis adhesin A (NadA)-mediated adhesion and invasion. Cell Microbiol. 2012;14(3):368–385. doi: 10.1111/j.1462-5822.2011.01722.x. [DOI] [PubMed] [Google Scholar]
- 27.Cotter SE, Surana NK, St Geme JW., III Trimeric autotransporters: A distinct subfamily of autotransporter proteins. Trends Microbiol. 2005;13(5):199–205. doi: 10.1016/j.tim.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 28.Linke D, Riess T, Autenrieth IB, Lupas A, Kempf VAJ. Trimeric autotransporter adhesins: Variable structure, common function. Trends Microbiol. 2006;14(6):264–270. doi: 10.1016/j.tim.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 29.Łyskowski A, Leo JC, Goldman A. Structure and biology of trimeric autotransporter adhesins. Adv Exp Med Biol. 2011;715:143–158. doi: 10.1007/978-94-007-0940-9_9. [DOI] [PubMed] [Google Scholar]
- 30.Magagnoli C, et al. Structural organization of NadADelta(351-405), a recombinant MenB vaccine component, by its physico-chemical characterization at drug substance level. Vaccine. 2009;27(15):2156–2170. doi: 10.1016/j.vaccine.2009.01.099. [DOI] [PubMed] [Google Scholar]
- 31.Dautin N, Bernstein HD. Protein secretion in gram-negative bacteria via the autotransporter pathway. Annu Rev Microbiol. 2007;61:89–112. doi: 10.1146/annurev.micro.61.080706.093233. [DOI] [PubMed] [Google Scholar]
- 32.Dupeux F, Röwer M, Seroul G, Blot D, Márquez JA. A thermal stability assay can help to estimate the crystallization likelihood of biological samples. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 11):915–919. doi: 10.1107/S0907444911036225. [DOI] [PubMed] [Google Scholar]
- 33.Hartmann MD, et al. A coiled-coil motif that sequesters ions to the hydrophobic core. Proc Natl Acad Sci USA. 2009;106(40):16950–16955. doi: 10.1073/pnas.0907256106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holm L, Rosenström P. Dali server: Conservation mapping in 3D. Nucleic Acids Res. 2010;38(Web Server issue):W545–549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.White HE, Saibil HR, Ignatiou A, Orlova EV. Recognition and separation of single particles with size variation by statistical analysis of their images. J Mol Biol. 2004;336(2):453–460. doi: 10.1016/j.jmb.2003.12.015. [DOI] [PubMed] [Google Scholar]
- 36.Barocchi MA, Masignani V, Rappuoli R. Opinion: Cell entry machines: A common theme in nature? Nat Rev Microbiol. 2005;3(4):349–358. doi: 10.1038/nrmicro1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scarselli M, et al. Rational design of a meningococcal antigen inducing broad protective immunity. Sci Transl Med. 2011;3(91):91ra62. doi: 10.1126/scitranslmed.3002234. [DOI] [PubMed] [Google Scholar]
- 38.Correia BE, et al. Proof of principle for epitope-focused vaccine design. Nature. 2014;507(7491):201–206. doi: 10.1038/nature12966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Giuliani MM, et al. The region comprising amino acids 100 to 255 of Neisseria meningitidis lipoprotein GNA 1870 elicits bactericidal antibodies. Infect Immun. 2005;73(2):1151–1160. doi: 10.1128/IAI.73.2.1151-1160.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deprez C, et al. Solution structure of the E.coli TolA C-terminal domain reveals conformational changes upon binding to the phage g3p N-terminal domain. J Mol Biol. 2005;346(4):1047–1057. doi: 10.1016/j.jmb.2004.12.028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.