Significance
Topoisomerases are enzymes that resolve DNA supercoiling during cell division and gene transcription. Inhibitors of these enzymes are used to treat multiple forms of cancer. Recently we found that topoisomerase inhibitors have profound effects on synaptic genes expressed in the brain. Here we examine the contribution of a clinically used topoisomerase inhibitor on the expression of synaptic proteins and synaptic transmission. We find that inhibition of topoisomerase 1 (TOP1) dampens excitatory and inhibitory synaptic transmission in cortical neurons. Additionally, these effects are fully reversible, because synaptic protein levels and synaptic transmission recover upon washout of the TOP1 inhibitor. These findings provide insights into how inhibition of TOP1 impacts synaptic function in neurons.
Keywords: synapse, topoisomerase, transcription
Abstract
Topotecan is a topoisomerase 1 (TOP1) inhibitor that is used to treat various forms of cancer. We recently found that topotecan reduces the expression of multiple long genes, including many neuronal genes linked to synapses and autism. However, whether topotecan alters synaptic protein levels and synapse function is currently unknown. Here we report that in primary cortical neurons, topotecan depleted synaptic proteins that are encoded by extremely long genes, including Neurexin-1, Neuroligin-1, Cntnap2, and GABAAβ3. Topotecan also suppressed spontaneous network activity without affecting resting membrane potential, action potential threshold, or neuron health. Topotecan strongly suppressed inhibitory neurotransmission via pre- and postsynaptic mechanisms and reduced excitatory neurotransmission. The effects on synaptic protein levels and inhibitory neurotransmission were fully reversible upon drug washout. Collectively, our findings suggest that TOP1 controls the levels of multiple synaptic proteins and is required for normal excitatory and inhibitory synaptic transmission.
Topoisomerase inhibitors such as topotecan (Hycamtin) are widely used to treat multiple forms of cancer, including brain metastases, ovarian cancer, and small cell lung cancer (1). Topoisomerases resolve DNA supercoiling during cell division and during gene transcription (2). Type I topoisomerases, encoded by Top1, Top3a, and Top3b in mammals, resolve supercoiling by cleaving a single strand of DNA, whereas type II topoisomerases, encoded by Top2a and Top2b, cleave both DNA strands (2). Recently, both types of topoisomerases were associated with neurodevelopment (3–5). For instance, topoisomerase 1 (TOP1) and topoisomerase 2 (TOP2) inhibitors transcriptionally up-regulate the paternal copy of Ubiquitin-protein ligase E3A (Ube3a) (5), a gene that affects synaptic activity and that is deleted or duplicated in distinct neurodevelopmental disorders (Angelman syndrome and autism, respectively) (6, 7). Moreover, a de novo mutation in Top1 and de novo mutations in genes that interact with Top1 and Top3b were identified recently in patients with autism (8, 9), whereas deletion of Top3b increases the risk for schizophrenia and intellectual disability (10, 11). Top2b is also required for axon outgrowth in different regions of the nervous system and for the survival of postmitotic neurons (12–15).
TOP1 is localized primarily in the nucleus of postmitotic neurons and is expressed throughout the developing and adult brain (16), suggesting a nuclear function. Indeed, we recently found that topotecan, a selective TOP1 inhibitor, reduced the expression of extremely long genes (>200 kb) in postmitotic neurons by impairing transcription elongation (17). Topotecan and related camptothecin analogs inhibit TOP1 by covalently trapping the enzyme on DNA (2). TOP1 inhibitors also reduce the expression of long genes in cancer cell lines (17–19), revealing a gene length-dependent component to transcription that is common to several mammalian cell types.
In neurons, many genes down-regulated by topotecan are associated with synapses, cell adhesion, and neurotransmission (17). Moreover, a number of these down-regulated long genes are associated with autism, including Neurexin-1 (Nrxn1; 1,059 kb), Neuroligin-1 (Nlgn1; 900 kb), and Contactin-associated protein 2 (Cntnap2; 2,241 kb), genes that are well known to regulate inhibitory and excitatory synaptic function (20–22). In contrast, topotecan had minimal effects on most other genes expressed in neurons (the average length of expressed genes in mouse neurons is 59.3 kb) (17).
Because the transcriptional effects following TOP1 inhibition were strongly biased toward long, synapse-associated genes, we hypothesized that TOP1 might be particularly important for maintaining normal synaptic functions (17). In this study we tested this hypothesis using biochemical and physiological approaches with cultured mouse cortical neurons. We found that multiple synaptic proteins, each encoded by extremely long genes, were depleted after neurons were treated with topotecan, a TOP1 inhibitor. TOP1 inhibition also profoundly suppressed excitatory and inhibitory synaptic activity without affecting overall neuron health. Our study shows that TOP1 regulates the levels of multiple synaptic proteins and is required for proper synaptic function. Moreover, our work provides insights into how TOP1 dysfunction could impact synaptic activity.
Results
Depletion of Synaptic Cell Adhesion Proteins That Originate from Long Genes.
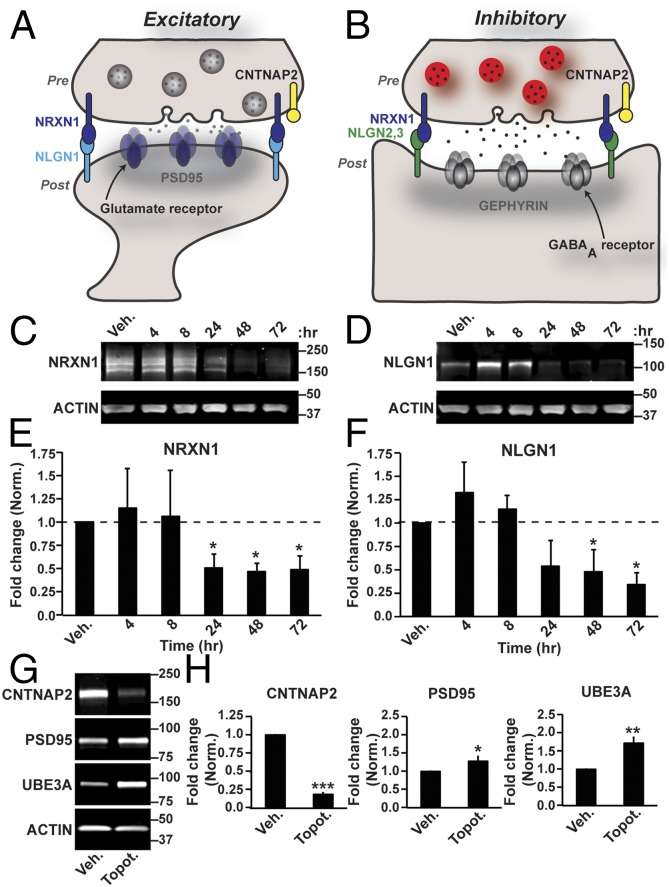
NRXN1 and NLGN1 are well-characterized synaptic adhesion molecules that interact with each other on opposing sides of the synaptic terminal (Fig. 1 A and B) (21, 23) and are encoded by extremely long genes (17). We previously found that topotecan, a selective TOP1 inhibitor, dose-dependently reduced the expression of Nrxn1 and Nlgn1, as well as many other long autism- and synapse-associated genes (17). To determine if topotecan reduces the protein levels of NRXN1 and NLGN1, we treated cultured cortical neurons at 7 d in vitro (DIV 7) with vehicle or 300 nM topotecan and then harvested cells at various time points for Western analyses. We detected multiple NRXN1 bands in vehicle-treated samples (Fig. 1C), consistent with there being multiple NRXN1 isoforms (21, 24). After topotecan was added, NRXN1 levels declined within 24 h and remained low when exposed to drug for 72 h (Fig. 1 C and E). Likewise, NLGN1 levels declined in topotecan-treated neurons and remained low when exposed to drug for 72 h (Fig. 1 D and F). The protein level of CNTNAP2, another synaptic adhesion molecule that is encoded by an extremely long gene (17), also was depleted in topotecan-treated neurons (Fig. 1 G and H).
Fig. 1.
TOP1 inhibition reduces the expression of synaptic adhesion molecules. (A and B) Schematics of an excitatory (A) and inhibitory (B) synapse. Pre, presynaptic; Post, postsynaptic. (C and D) Cortical neurons were treated with vehicle or with 300 nM topotecan on DIV 7 and then were harvested 4, 8, 24, 48, or 72 h later. Shown are representative immunoblots with antibodies to NRXN1 (C) or NLGN1 (D) and actin. (E and F) Quantification of fold change in NRXN1 (E) and NLGN1 (F) expression normalized to actin. Data are shown as means ± SEM; n = 3 cultures. *P < 0.05 (unpaired Student t test). (G and H) Cortical neurons were treated with vehicle or 300 nM topotecan from DIV 7–10 and then were harvested for Western blot analysis. (G) Shown are representative immunoblots of the indicated proteins. (H) Fold change of the indicated proteins, normalized to actin. Data are shown as means ± SEM; n = 4–6 cultures. *P < 0.05, **P < 0.005 (unpaired Student t test).
In contrast, postsynaptic density protein 95 (PSD95, DLG4; gene length, 26.9 kb), a scaffolding protein that interacts with NLGN1 (25), was slightly elevated at the 72-h time point (Fig. 1 G and H). We did not examine NLGN2 or NLGN3 because the expression of these short genes (14.6 kb and 26.1 kb, respectively) was not altered in topotecan-treated neurons (17). Consistent with previous work, topotecan increased protein levels of UBE3A (Fig. 1 G and H) by reducing the expression of an extremely long antisense transcript that overlaps Ube3a (5, 17).
Prolonged treatment (up to 7 d) with topotecan did not alter MAP2 staining or activate caspase-3 (Fig. S1), a marker of apoptotic cells; this result is consistent with our previous finding showing that 300 nM topotecan is not toxic to neurons (17). As positive controls, a high concentration of topotecan (10 μM) or glutamate (250 μM) resulted in disorganized MAP2 staining and activated caspase-3 (Fig. S1).
Taken together, these experiments show that three synaptic adhesion molecules, each originating from extremely long genes, are depleted with a drug that inhibits TOP1. This finding is noteworthy, given that mutations in any one of these long genes can impair synaptic function and neurodevelopment (21, 26, 27).
TOP1 Inhibition Suppresses Spontaneous Network Activity in Cortical Neurons.
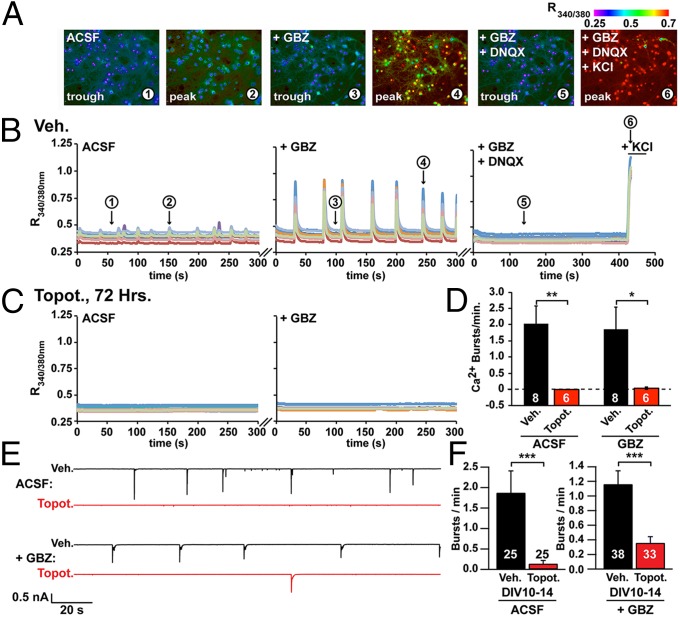
Because the expression of Nrxn1, Nlgn1, Cntnap2, and other genes linked to synaptic function was reduced in topotecan-treated neurons (17), we hypothesized that topotecan might impair synaptic activity in neuronal cultures. To test this hypothesis, we took advantage of the fact that after approximately 2 weeks in culture (on DIV 14–15) cortical neurons generate spontaneous, synchronous bursts of electrical activity that can be monitored with Ca2+-sensitive dyes or by whole-cell electrophysiology (28–32). Because bursting begins after about 2 weeks, we delayed the addition of topotecan from DIV 7–10 to DIV 11–14. Importantly, NRXN1 and NLGN1 protein levels were decreased to a similar extent regardless of whether cultures were treated with topotecan from DIV 7–10 (Fig. 1 C–F) or from DIV 11–14 (Fig. S2 A–D). Using the ratiometric Ca2+ indicator fura-2, we found that vehicle-treated cultures displayed synchronous, low-amplitude Ca2+ bursts (R340/380: 0.032 ± 0.001) with a frequency of 2.0 ± 0.6 bursts/min (Fig. 2 A, B, and D and Movie S1). The addition of gabazine (GBZ), a GABAA receptor antagonist, increased the amplitude of these Ca2+ bursts (R340/380: 0.328 ± 0.005) without changing burst frequency (1.9 ± 0.7 bursts/min) (Fig. 2 A, B, and D and Movie S1). Coapplication of GBZ and 6,7-dinitroquinoxaline-2,3-dione disodium salt (DNQX), an AMPA-type glutamate receptor antagonist, eliminated these bursts (Fig. 2B). In contrast, no Ca2+ bursts were detected in most cultures treated with topotecan for 72 h [six of six cultures in artificial cerebrospinal fluid (ACSF); five of six cultures in ACSF+GBZ] (Fig. 2 C and D and Movie S2). The nearly complete loss of activity was not caused by compromised neuronal health or overt impairment of voltage-gated calcium channels, because vehicle and topotecan-treated neurons displayed equivalent calcium responses following KCl-induced depolarization (Fig. 2B, Fig. S2E, and Movies S1 and S2).
Fig. 2.
TOP1 inhibition suppresses spontaneous calcium bursts and electrical activity in cortical neuron cultures. Fura2-calcium imaging of spontaneous network activity in neurons at DIV 14–15 after the addition of 5 μM GBZ, after the addition of GBZ plus 5 μM DNQX, and after all neurons were depolarized with 55 mM KCl. (A) Representative images from vehicle-treated neurons taken at time points 1–6 indicated in B. (B) Activity in neurons treated with vehicle (Veh.). (C) Activity in neurons treated with 300 nM topotecan (Topot.) at 72 h. (D) Ca2+ burst frequency. Data are shown as means ± SEM from three independent cultures; n = 6–8 coverslips per condition; n = 466 neurons for vehicle treatment, and n = 280 neurons for topotecan treatment. *P < 0.05, **P < 0.005 (unpaired Student t test). (E and F) Spontaneous burst activity monitored by whole-cell electrophysiology in cortical neuron cultures treated with vehicle (black trace) or 300 nM topotecan (red trace). Treatment started on DIV 7 and ended on the day of recording. (E) Representative voltage-clamp recordings in ACSF and after the addition of 5 μM GBZ. (F) Burst frequency integrated from DIV 10–14. ***P < 0.0005 (significantly different from vehicle by Mann–Whitney test).
We also monitored synchronous bursts of activity in vehicle- and topotecan-treated cultures by whole-cell recordings (all activity was eliminated when GBZ and tetrodotoxin were added) (Fig. S3 A–E). Consistent with our Ca2+ imaging results, topotecan (300 nM) significantly reduced burst activity in ACSF and GBZ conditions (Fig. 2 E and F and Fig. S3 F–H). Based on cumulative distribution analyses, no bursts were observed during the recording session in a large percentage of topotecan-treated cultures (Fig. S3H). In contrast, bursts were detected in the majority of vehicle-treated cultures. Taken together, these data indicate that topotecan suppresses spontaneous synaptic activity without affecting neuron health and suggests a role for TOP1 in maintaining neurotransmission at normal levels.
TOP1 Inhibition Impairs Excitatory Synapse Formation and Function.
TOP1 is well positioned to regulate gene expression in excitatory and inhibitory neurons, because TOP1 is present in 93% of all NeuN+ neurons in culture, including 99% of all MAP2+ neurons and 100% of all glutamic acid decarboxylase 67-positive (GAD67+) inhibitory neurons (Fig. S4). To provide insights into how TOP1 inhibition affects network activity, we examined intrinsic electrical properties and miniature excitatory postsynaptic currents (mEPSCs) in GAD67− pyramidal neurons and GAD67+ interneurons (see SI Materials and Methods for details of cell classification). We found that mEPSCs were undetectable until DIV 11 in cultured mouse cortical neurons, analogous to others’ observations in rat neurons (33), so we extended the topotecan treatment by an additional 1–4 days. Note that over this longer time course NRXN1 and NLGN1 protein levels remained low (Fig. S5 A–D) without affecting cell health (Fig. S1). We observed no significant differences in the resting membrane potential or action potential threshold in pyramidal neurons or inhibitory neurons (Fig. S5E). However, there was a significant increase in input resistance and a significant reduction in membrane capacitance and rheobase in pyramidal neurons but not in inhibitory neurons (Fig. S5E). These data suggest that topotecan-treated pyramidal neurons are more excitable. However, increased pyramidal neuron excitability would be predicted to increase network activity, not reduce network activity as we observed.
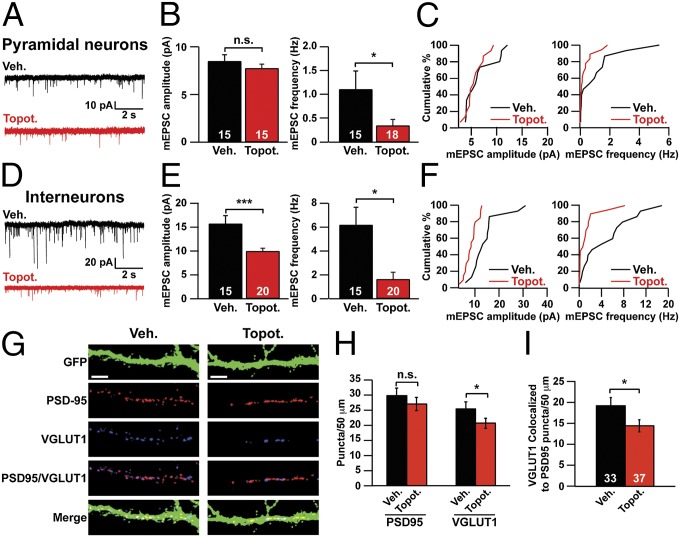
Thus we next explored the possibility that TOP1 inhibition reduced synaptic activity. To test this possibility, we treated neurons with topotecan on DIV 7 and then recorded mEPSCs on DIV 11–13. We found that treatment of cortical neurons with topotecan decreased mEPSC frequency but not mEPSC amplitude in pyramidal neurons (Fig. 3 A–C). In the interneuron population, topotecan strongly decreased both mEPSC amplitude and frequency (Fig. 3 D–F). Moreover, topotecan reduced the number of excitatory synapses, as evidenced by fewer vesicular glutamate transporter 1 (VGLUT1, a presynaptic marker) puncta and fewer VGLUT1/PSD95 colocalized puncta in topotecan-treated neurons (Fig. 3 G–I). There was no change in the number of PSD95 (a postsynaptic marker) puncta (Fig. 3H). We were unable to measure a change in the probability of neurotransmitter release at excitatory synapses because in vehicle-treated cultures electrical stimulation evoked a barrage of polysynaptic activity that obscured the monosynaptically evoked events. Taken together, these data suggest that TOP1 activity is required for the formation and function of excitatory synapses.
Fig. 3.
TOP1 inhibition impairs excitatory synaptic transmission and synapse formation. (A–C) Pyramidal neurons were treated with vehicle or 300 nM topotecan beginning on DIV 7 and ending on the day of recording (DIV 11–13). (A) Representative mEPSC traces. (B) mEPSC amplitude (Left) and frequency (Right). *P < 0.05 (Mann–Whitney test); n.s., not significantly different. (C) Cumulative distribution of mean mEPSC amplitudes (Left) and frequencies (Right). (D–F) Interneurons were treated with vehicle or 300 nM topotecan starting on DIV 7 and ending on the day of recording (DIV 11–13). (D) Representative mEPSC traces. (E) mEPSC amplitude (Left) and frequency (Right). ***P < 0.0005, *P < 0.05 (Mann–Whitney test). (F) Cumulative distribution of mean mEPSC amplitudes (Left) and frequencies (Right). (G–I) Cortical neuron cultures were infected with GFP lentivirus on DIV 3. Neurons then were treated with vehicle or 300 nM topotecan for 72 h, beginning on DIV 7. (G) On DIV 10, neurons were immunostained for VGLUT1 (an excitatory presynaptic marker), PSD95 (an excitatory postsynaptic marker), and GFP. (Scale bars, 10 μm.) (H) Number of PSD95 and VGLUT1 puncta per 50 μm of dendrite. n = 33 (Veh.), 37 (Topot.) neurons. *P < 0.05 (unpaired Student t test). (I) Number of colocalized VGLUT1/PSD95 puncta per 50 μm of dendrite. Data in B, E, H, and I are presented as means ± SEM. *P < 0.05 (unpaired Student t test).
TOP1 Inhibition Profoundly Impairs Inhibitory Synapse Function Without Affecting Inhibitory Synapse Number.
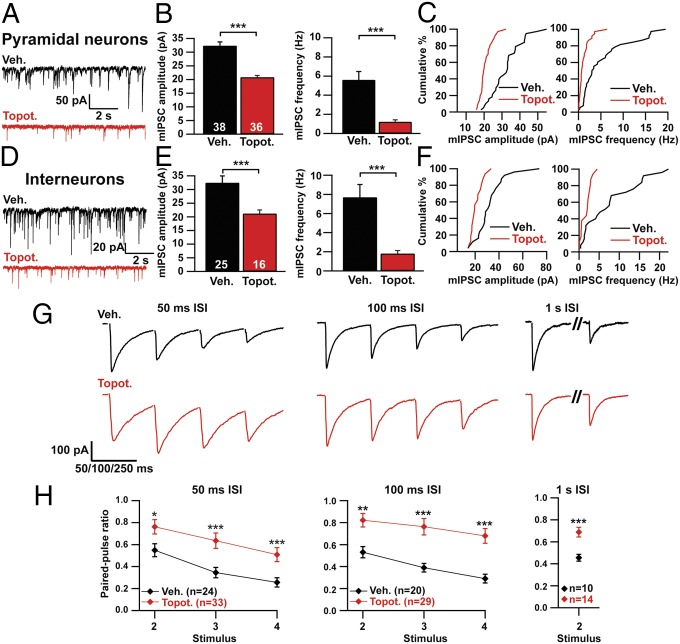
Next, we examined how TOP1 inhibition affects inhibitory synapses by measuring miniature inhibitory postsynaptic currents (mIPSCs) in GAD67– pyramidal neurons and GAD67+ interneurons. We found that mIPSC amplitude and frequency were profoundly reduced in pyramidal neurons and interneurons in topotecan-treated cultures (Fig. 4 A–F). Additionally, we examined release probability by testing the paired-pulse ratio of evoked IPSCs (Fig. 4 G and H). Topotecan-treated neurons had higher paired-pulse ratios at 50-ms, 100-ms, and 1-s interstimulus intervals (ISIs), suggesting a decrease in the probability of neurotransmitter release.
Fig. 4.
Topotecan suppresses inhibitory synaptic transmission. (A–C) Pyramidal neurons were treated with vehicle or 300 nM topotecan starting on DIV 7 and ending on the day of recording (DIV 10–14). (A) Representative mIPSC traces. (B) mIPSC amplitude (Left) and frequency (Right). ***P < 0.0005 (Mann–Whitney test). (C) Cumulative distribution of mean mIPSC amplitudes (Left) and mIPSC frequencies (Right). (D–F) Interneurons were treated with vehicle or 300 nM topotecan starting on DIV 7 and ending on the day of recording (DIV 10–14). (D) Representative mIPSC traces. (E) mIPSC amplitude (Left) and frequency (Right). ***P < 0.0005 (Mann–Whitney test). (F) Cumulative distribution of mean mIPSC amplitudes (Left) and mIPSC frequencies (Right). (G) Representative traces of evoked IPSCs at different ISIs from cultured cortical neurons treated with vehicle or 300 nM topotecan. Treatment started on DIV 7 and ended on the day of recording (DIV 10–14). Glutamatergic transmission was blocked with DNQX and d-2-amino-5-phosphonovaleric acid (DL-AP5). Stimulus artifacts have been erased for clarity. (H) Paired-pulse ratios for bursts elicited at 50-ms, 100-ms, and 1-s ISIs. *P < 0.05, **P < 0.005, ***P < 0.0005 (Mann–Whitney test). The number of neurons recorded is indicated in graphs. Data in B, E, and H are presented as means ± SEM.
To determine if these effects on inhibitory synapse activity were associated with a reduction in the number of inhibitory synapses, we immunostained for the inhibitory postsynaptic marker GEPHYRIN and the inhibitory presynaptic marker vesicular GABA transporter (VGAT) (Fig. S6A). There was no significant change in the number of GEPHYRIN, VGAT, or colocalized VGAT/GEPHYRIN puncta (Fig. S6 A–C), indicating that TOP1 inhibition did not alter the number of inhibitory synapses.
GABAA receptors mediate synaptic transmission at many inhibitory synapses (34). We previously found that the expression of the GABAAβ1 and GABAAβ3 receptor subunits was reduced in topotecan-treated cortical neurons (17). These GABAA receptor subunits are encoded by long genes (437.2 kb and 238.3 kb, respectively). Because spontaneous mIPSC amplitude was reduced in topotecan-treated neurons, we next explored the possibility that overall GABAA receptor levels were reduced in topotecan-treated neuronal cultures. Based on Western blot analysis from whole-cell lysates, we found that the GABAA receptor subunits GABAAβ3 and GABAAγ2 were reduced at the protein level following TOP1 inhibition (Fig. S6 D–G). GABAAβ1 was reduced at the transcript level (17) but not at the protein level, possibly reflecting a longer protein half-life (35).
GABAA receptors are pentameric receptors made up of multiple subunits (34). Disruption of individual GABAA receptor subunits can reduce the surface levels of interacting subunits (36, 37). To determine if TOP1 inhibition altered the surface levels of multiple GABAA receptor subunits, we labeled surface GABAA receptors with biotin and then quantified surface and total protein levels. We found that TOP1 inhibition reduced the surface expression of GABAAβ3, GABAAγ2, and GABAAα1 (Fig. S6 H and I), subunits that are expressed in cortical neurons at this developmental age (38). We were unable to detect surface expression of the GABAAβ1 subunit, possibly because it is not trafficked to the membrane or incorporated into GABAA receptors at this stage of development. Additionally, we did not observe a change in GABAAα5 subunit levels (Fig. S6 H and I).
Taken together, our data indicate that TOP1 inhibition decreased neurotransmission at inhibitory synapses without changing the number of inhibitory synapses. Moreover, our data suggest that deficits in neurotransmitter release probability and postsynaptic GABAA receptor density could account for reduced inhibitory synapse function.
Synaptic Effects of Topotecan Are Reversible.
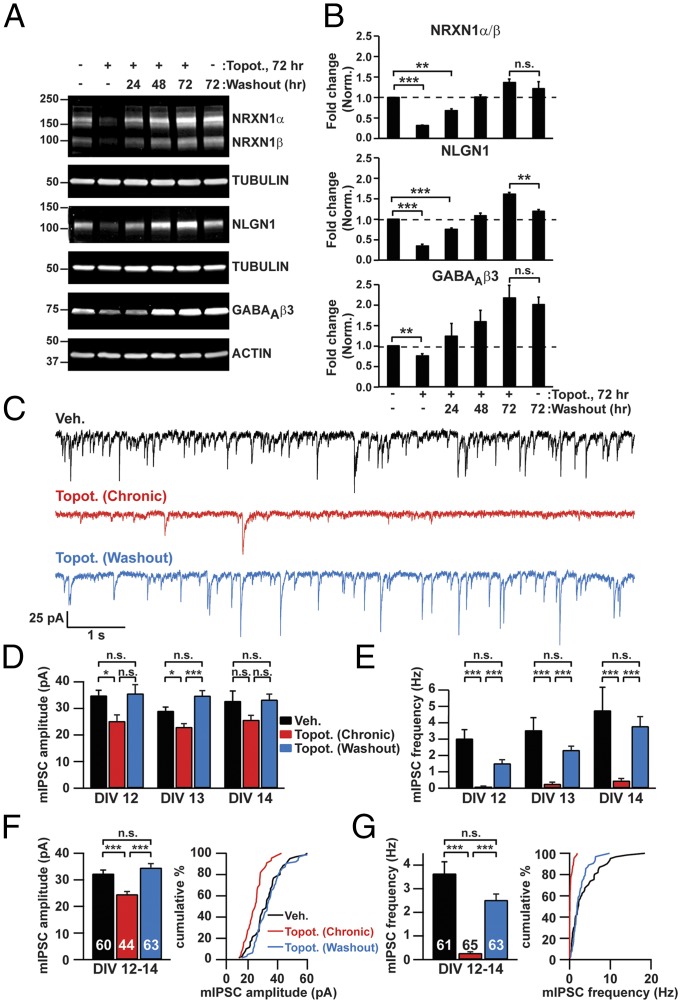
We previously found that the genomewide transcriptional effects of topotecan were fully reversible upon drug washout (17), suggesting that the synaptic phenotypes caused by TOP1 inhibition also might be reversible. To test this possibility, we treated neurons on DIV 7 with topotecan for 72 h, washed out the drug, and then assessed recovery 24–72 h later. As above, topotecan reduced the levels of NRXN1, NLGN1, and GABAAβ3 at the 72-h time point (Fig. 5 A and B). Remarkably, the levels of all these proteins were restored to pretreatment levels or higher upon drug washout. GABAAβ3 levels were higher at the 72-h time point in both vehicle- and topotecan-treated neurons, most likely because the expression of this subunit increases over time in culture (38). Additionally, the profound reduction in mIPSC amplitude and frequency in topotecan-treated pyramidal neurons was restored within 48 h of drug washout (Fig. 5 C–G), paralleling the biochemical recovery of NRXN1, NLGN1, and GABAAβ3 (Fig. 5 A and B). Taken together, our data indicate that the transcriptional, posttranscriptional, and synaptic deficits in topotecan-treated neurons are directly correlated and are all fully reversed upon drug washout, thus suggesting that TOP1 is required in these processes.
Fig. 5.
Effects of TOP1 inhibition on synaptic protein levels and synaptic transmission are reversible following topotecan washout. (A and B) Beginning on DIV 7, cortical neurons were treated for 72 h with vehicle or 300 nM topotecan. Topotecan was washed out, conditioned medium was added, and neurons were harvested 24 (DIV 11), 48 (DIV 12), or 72 (DIV 13) h later. (A) Representative immunoblots for the indicated proteins. (B) Quantification of fold change in protein levels normalized to tubulin or actin. n = 3–6 culture wells. **P < 0.005, ***P < 0.0005 (unpaired Student t test). (C–G) Neurons were treated with vehicle or 300 nM topotecan for 72 h. Topotecan was washed out 72 h later (on DIV 10) (blue trace and bars), and neurons were recorded 48 h (DIV 12), 72 h (DIV 13), and 4 d (DIV 14) later. Alternatively, neurons were kept in medium containing vehicle (black trace and bars) or topotecan (red trace and bars) until recording. (C) Representative mIPSC traces on DIV 13. (D and E) mIPSC amplitude (D) and frequency (E) from 48 h (DIV 12; n = 24, 22, 23), 72 h (DIV 13; n = 22, 22, 19), and 4 d (DIV 14; n = 15, 21, 21) of drug washout. (F and G) mIPSC amplitude (F, Left), frequency (G, Left), and cumulative distribution data (F and G, Right) averaged across DIV 12–14. *P < 0.05, **P < 0.005, ***P < 0.0005 (Mann–Whitney test). Data in B and D–G are presented as means ± SEM.
Discussion
We previously found that TOP1 inhibitors reduce the expression of a large number of long genes linked to synapses and to autism (17). However, whether TOP1 inhibitors affect synaptic protein levels or synaptic function was unknown. Here, we report that multiple synaptic proteins encoded by long genes, including cell adhesion molecules linked to autism and GABA receptor subunits, are depleted in topotecan-treated neurons. These findings suggest that TOP1 inhibitors could reduce the protein levels of many genes with long transcripts (17), especially if their protein half-lives are relatively short (35). Moreover, our data suggest that a basic biological feature—gene length—can be used to predict whether TOP1 inhibitors will reduce gene expression and protein levels. Thus, neurons appear to have an Achilles heel in the form of TOP1, because inhibition of this single transcriptional regulator depletes multiple proteins required for normal synaptic function.
Remarkably, spontaneous burst activity was eliminated almost completely in topotecan-treated cultures. Burst activity presumably was shaped by GABAA and glutamate receptor activation, because bursts were sensitive to antagonists of these receptors, consistent with previous studies (28–32). Inhibition of TOP1 also profoundly impaired inhibitory synaptic activity without affecting the number of inhibitory synapses or neuron health, as measured by the absence of caspase-3 activation and the lack of effect on depolarization-induced (voltage-gated) calcium influx. Given that the loss and restoration of transcripts, synaptic proteins, and synaptic function were coincident, our data suggest that TOP1 serves as a master transcriptional regulator for many synaptic proteins and for synaptic function.
Because TOP1 inhibition affects multiple synaptic proteins, it was not possible to predict a priori how deficits in synaptic transmission would manifest. Intriguingly, some of the phenotypes observed with topotecan bear similarities to knockdown or KO models for NRXN1, NLGN1, and CNTNAP2. For instance, in neocortex, Nrxn1α-KO mice exhibit synaptic deficits, such as a decrease in mIPSC frequency (39). Moreover, Nrxn1α/2α double-knockout mice have decreased neurotransmitter release, which is similar to our findings at inhibitory synapses (39). Although synaptic transmission and synapse number in NLGN1-KO mice are phenotypically normal (40–42), shRNA knockdown of NLGN1 decreases the number of excitatory and inhibitory synapses in hippocampal neuron culture (43). Additionally, sparse knockdown of NLGN1 in a subset of neurons in utero leads to decreases in mEPSC frequency (44). Moreover, in a culture system similar to ours, Südhof and colleagues found that excitatory and inhibitory neurotransmission were reduced after knockdown of Cntnap2 (32), a long gene whose protein levels are reduced upon TOP1 inhibition (Fig. 1 G and H). Thus, depending on the system, brain region, and the context of the reduction/deletion, NRXN1, NLGN1, and CNTNAP2 can have varying effects on synaptic transmission, and some of these effects phenocopy our findings in topotecan-treated cortical neurons.
We previously found that the transcriptional effects of topotecan on long synaptic genes were dose dependent (17). Given these graded transcriptional effects, synaptic function and brain development are likely to be impacted in proportion to how strongly TOP1 is inhibited by chemicals or mutations. Moreover, TOP1 activity increases from birth to maturity (16) and can be inhibited by the activation of glutamate or GABA receptors in mouse cerebellum (45). Thus, several mechanisms may regulate the transcriptional activities of TOP1 in neurons in a graded manner.
Because the expression of multiple long synaptic genes was reduced in topotecan-treated neurons, it seems unlikely that adding back one of these down-regulated genes (such as Nrxn1) would fully rescue synaptic deficits. Moreover, adding back one synaptic cell-adhesion molecule would not likely restore the protein levels of all affected synaptic cell-adhesion molecules as well as multiple GABAA receptor subunits. Note that we were unable to rescue phenotypes in topotecan-treated neurons genetically because TOP1, when overexpressed, was just as sensitive to topotecan as endogenous TOP1.
Our study was focused on using topotecan, a drug that poisons and depletes TOP1 protein (2, 5). Irinotecan (used to treat gastrointestinal malignancies) (1) is another TOP1 inhibitor derived from camptothecin, and ICRF-193 is a TOP2 inhibitor. Both drugs had transcriptional effects in neurons similar to those of topotecan (17). Thus, we speculate that other drugs that inhibit TOP1 or TOP2 enzymes could have similar effects on synapse function.
Our findings in mammalian neurons may have parallels to research with TOP1 in Drosophila. TOP1 deletion is lethal in flies and mice (46, 47). However, a mutation that reduced TOP1 expression in the fly brain was not lethal and suppressed seizure-like behavior (47). Feeding flies TOP1 inhibitors, including camptothecin (a derivative of topotecan) and apigenin (a flavone found in fruits and vegetables) also suppressed seizures (48). Mechanistically how TOP1 inhibition blocked seizures in flies was not resolved. Given that topotecan reduced spontaneous, synchronous (seizure-like) activity in mouse cortical cultures (Fig. 2), it will be interesting to determine if TOP1 inhibitors suppress seizures in flies via a synaptic mechanism similar the mechanism we identified in mouse neurons. TOP1 inhibition reduces nascent RNA at the 3′ end of long, but not short, genes in Drosophila S2 cells (49), suggesting that TOP1 regulates gene expression in a length-dependent manner in flies. Thus, TOP1 could regulate neurotransmission by facilitating long gene expression in vertebrates and invertebrates.
Intriguingly, a de novo mutation in TOP1 and mutations in genes that interact with TOP1 were identified in patients with autism (8, 9), and these genes cluster, based on annotation criteria, with other transcriptional regulators implicated in autism (50). Furthermore, TOP1 cleavage complexes and DNA damage accumulate in mouse models of the neurodegenerative syndromes ataxia telangiectasia and spinocerebellar ataxia with axonal neuropathy (51). Cancer patients (including children) treated with topotecan report memory impairment and confusion suggestive of deficits in synaptic function and neurotransmission (52–54). Thus, transient TOP1 inhibition has the potential to impair brain function reversibly, whereas a persistent change in TOP1 activity has the potential to disrupt neurodevelopment and promote neurodegeneration. In summary, our findings indicate that synapses are particularly vulnerable to TOP1 inhibitors and suggest that this synaptic vulnerability could have profound effects on the developing and adult brain.
Materials and Methods
All experimental animal procedures were carried out according to the National Institutes of Health Guide for the Care and Use of Laboratory Animals (55) and were approved by the Institutional Animal Care and Use Committee at the University of North Carolina at Chapel Hill. Mouse cortical neuron culture, biochemistry, immunocytochemistry, and physiology procedures are described in detail in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Tal Kafri and Ping Zhang at the University of North Carolina (UNC) Lentiviral Core for assistance with the preparation of lentiviral vectors and Vladimir Ghukasyan at the UNC Confocal and Multiphoton Imaging Core, Ian King, and Gabriela Salazar for technical assistance. This work was supported by a grant from the Angelman Syndrome Foundation, Simons Foundation Grant SFARI 10-3625, and National Institute of Mental Health Grant R01MH093372 (to B.D.P. and M.J.Z.) and by National Institutes of Health Pioneer Award DP1ES024088 (to M.J.Z.). A.M.M. was supported by a Joseph E. Wagstaff Postdoctoral Research Fellowship from the Angelman Syndrome Foundation. The Confocal and Multiphoton Imaging Core is funded by the National Institute of Neurological Disorders and Stroke Grant P30NS045892 and The Eunice Kennedy Shriver National Institute of Childhood Health and Development Grant P30HD03110.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1413204111/-/DCSupplemental.
References
- 1.Pommier Y. Drugging topoisomerases: Lessons and challenges. ACS Chem Biol. 2013;8(1):82–95. doi: 10.1021/cb300648v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pommier Y. DNA topoisomerase I inhibitors: Chemistry, biology, and interfacial inhibition. Chem Rev. 2009;109(7):2894–2902. doi: 10.1021/cr900097c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beaudet AL. Angelman syndrome: Drugs to awaken a paternal gene. Nature. 2012;481(7380):150–152. doi: 10.1038/nature10784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miller G. Neurodevelopmental disorders. New hope for a devastating neurological disorder. Science. 2011;334(6063):1615. doi: 10.1126/science.334.6063.1615. [DOI] [PubMed] [Google Scholar]
- 5.Huang HS, et al. Topoisomerase inhibitors unsilence the dormant allele of Ube3a in neurons. Nature. 2012;481(7380):185–189. doi: 10.1038/nature10726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mabb AM, Judson MC, Zylka MJ, Philpot BD. Angelman syndrome: Insights into genomic imprinting and neurodevelopmental phenotypes. Trends Neurosci. 2011;34(6):293–303. doi: 10.1016/j.tins.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cook EH, Jr, et al. Autism or atypical autism in maternally but not paternally derived proximal 15q duplication. Am J Hum Genet. 1997;60(4):928–934. [PMC free article] [PubMed] [Google Scholar]
- 8.Neale BM, et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012;485(7397):242–245. doi: 10.1038/nature11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iossifov I, et al. De novo gene disruptions in children on the autistic spectrum. Neuron. 2012;74(2):285–299. doi: 10.1016/j.neuron.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu D, et al. Top3β is an RNA topoisomerase that works with fragile X syndrome protein to promote synapse formation. Nat Neurosci. 2013;16(9):1238–1247. doi: 10.1038/nn.3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stoll G, et al. Deletion of TOP3β, a component of FMRP-containing mRNPs, contributes to neurodevelopmental disorders. Nat Neurosci. 2013;16(9):1228–1237. doi: 10.1038/nn.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Y, et al. Topoisomerase IIbeta is required for proper retinal development and survival of postmitotic cells. Biol Open. 2014;3(2):172–184. doi: 10.1242/bio.20146767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang X, Li W, Prescott ED, Burden SJ, Wang JC. DNA topoisomerase IIbeta and neural development. Science. 2000;287(5450):131–134. doi: 10.1126/science.287.5450.131. [DOI] [PubMed] [Google Scholar]
- 14.Heng X, et al. Nurr1 regulates Top IIβ and functions in axon genesis of mesencephalic dopaminergic neurons. Mol Neurodegener. 2012;7:4. doi: 10.1186/1750-1326-7-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tiwari VK, et al. Target genes of Topoisomerase IIβ regulate neuronal survival and are defined by their chromatin state. Proc Natl Acad Sci USA. 2012;109(16):E934–E943. doi: 10.1073/pnas.1119798109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Plaschkes I, Silverman FW, Priel E. DNA topoisomerase I in the mouse central nervous system: Age and sex dependence. J Comp Neurol. 2005;493(3):357–369. doi: 10.1002/cne.20793. [DOI] [PubMed] [Google Scholar]
- 17.King IF, et al. Topoisomerases facilitate transcription of long genes linked to autism. Nature. 2013;501(7465):58–62. doi: 10.1038/nature12504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Solier S, et al. Transcription poisoning by Topoisomerase I is controlled by gene length, splice sites, and miR-142-3p. Cancer Res. 2013;73(15):4830–4839. doi: 10.1158/0008-5472.CAN-12-3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Veloso A, et al. Genome-wide transcriptional effects of the anti-cancer agent camptothecin. PLoS ONE. 2013;8(10):e78190. doi: 10.1371/journal.pone.0078190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Betancur C, Sakurai T, Buxbaum JD. The emerging role of synaptic cell-adhesion pathways in the pathogenesis of autism spectrum disorders. Trends Neurosci. 2009;32(7):402–412. doi: 10.1016/j.tins.2009.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Südhof TC. Neuroligins and neurexins link synaptic function to cognitive disease. Nature. 2008;455(7215):903–911. doi: 10.1038/nature07456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peñagarikano O, Geschwind DH. What does CNTNAP2 reveal about autism spectrum disorder? Trends Mol Med. 2012;18(3):156–163. doi: 10.1016/j.molmed.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sheng M, Kim E. The postsynaptic organization of synapses. Cold Spring Harb Perspect Biol. 2011;3(12):1–20. doi: 10.1101/cshperspect.a005678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ullrich B, Ushkaryov YA, Südhof TC. Cartography of neurexins: More than 1000 isoforms generated by alternative splicing and expressed in distinct subsets of neurons. Neuron. 1995;14(3):497–507. doi: 10.1016/0896-6273(95)90306-2. [DOI] [PubMed] [Google Scholar]
- 25.Irie M, et al. Binding of neuroligins to PSD-95. Science. 1997;277(5331):1511–1515. doi: 10.1126/science.277.5331.1511. [DOI] [PubMed] [Google Scholar]
- 26.Zoghbi HY. Postnatal neurodevelopmental disorders: Meeting at the synapse? Science. 2003;302(5646):826–830. doi: 10.1126/science.1089071. [DOI] [PubMed] [Google Scholar]
- 27.Peñagarikano O, et al. Absence of CNTNAP2 leads to epilepsy, neuronal migration abnormalities, and core autism-related deficits. Cell. 2011;147(1):235–246. doi: 10.1016/j.cell.2011.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baltz T, de Lima AD, Voigt T. Contribution of GABAergic interneurons to the development of spontaneous activity patterns in cultured neocortical networks. Front Cell Neurosci. 2010;4:15. doi: 10.3389/fncel.2010.00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muramoto K, Ichikawa M, Kawahara M, Kobayashi K, Kuroda Y. Frequency of synchronous oscillations of neuronal activity increases during development and is correlated to the number of synapses in cultured cortical neuron networks. Neurosci Lett. 1993;163(2):163–165. doi: 10.1016/0304-3940(93)90372-r. [DOI] [PubMed] [Google Scholar]
- 30.Opitz T, De Lima AD, Voigt T. Spontaneous development of synchronous oscillatory activity during maturation of cortical networks in vitro. J Neurophysiol. 2002;88(5):2196–2206. doi: 10.1152/jn.00316.2002. [DOI] [PubMed] [Google Scholar]
- 31.Voigt T, Opitz T, de Lima AD. Synchronous oscillatory activity in immature cortical network is driven by GABAergic preplate neurons. J Neurosci. 2001;21(22):8895–8905. doi: 10.1523/JNEUROSCI.21-22-08895.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anderson GR, et al. Candidate autism gene screen identifies critical role for cell-adhesion molecule CASPR2 in dendritic arborization and spine development. Proc Natl Acad Sci USA. 2012;109(44):18120–18125. doi: 10.1073/pnas.1216398109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin YC, et al. Development of excitatory synapses in cultured neurons dissociated from the cortices of rat embryos and rat pups at birth. J Neurosci Res. 2002;67(4):484–493. doi: 10.1002/jnr.10077. [DOI] [PubMed] [Google Scholar]
- 34.Whiting PJ, et al. Molecular and functional diversity of the expanding GABA-A receptor gene family. Ann N Y Acad Sci. 1999;868:645–653. doi: 10.1111/j.1749-6632.1999.tb11341.x. [DOI] [PubMed] [Google Scholar]
- 35.Cohen LD, et al. Metabolic turnover of synaptic proteins: Kinetics, interdependencies and implications for synaptic maintenance. PLoS ONE. 2013;8(5):e63191. doi: 10.1371/journal.pone.0063191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ortinski PI, Lu C, Takagaki K, Fu Z, Vicini S. Expression of distinct alpha subunits of GABAA receptor regulates inhibitory synaptic strength. J Neurophysiol. 2004;92(3):1718–1727. doi: 10.1152/jn.00243.2004. [DOI] [PubMed] [Google Scholar]
- 37.Nusser Z, et al. Alterations in the expression of GABAA receptor subunits in cerebellar granule cells after the disruption of the alpha6 subunit gene. Eur J Neurosci. 1999;11(5):1685–1697. doi: 10.1046/j.1460-9568.1999.00581.x. [DOI] [PubMed] [Google Scholar]
- 38.Laurie DJ, Wisden W, Seeburg PH. The distribution of thirteen GABAA receptor subunit mRNAs in the rat brain. III. Embryonic and postnatal development. J Neurosci. 1992;12(11):4151–4172. doi: 10.1523/JNEUROSCI.12-11-04151.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Missler M, et al. Alpha-neurexins couple Ca2+ channels to synaptic vesicle exocytosis. Nature. 2003;423(6943):939–948. doi: 10.1038/nature01755. [DOI] [PubMed] [Google Scholar]
- 40.Varoqueaux F, et al. Neuroligins determine synapse maturation and function. Neuron. 2006;51(6):741–754. doi: 10.1016/j.neuron.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 41.Kim J, et al. Neuroligin-1 is required for normal expression of LTP and associative fear memory in the amygdala of adult animals. Proc Natl Acad Sci USA. 2008;105(26):9087–9092. doi: 10.1073/pnas.0803448105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blundell J, et al. Neuroligin-1 deletion results in impaired spatial memory and increased repetitive behavior. J Neurosci. 2010;30(6):2115–2129. doi: 10.1523/JNEUROSCI.4517-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chih B, Engelman H, Scheiffele P. Control of excitatory and inhibitory synapse formation by neuroligins. Science. 2005;307(5713):1324–1328. doi: 10.1126/science.1107470. [DOI] [PubMed] [Google Scholar]
- 44.Kwon HB, et al. Neuroligin-1-dependent competition regulates cortical synaptogenesis and synapse number. Nat Neurosci. 2012;15(12):1667–1674. doi: 10.1038/nn.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zehorai E, Eitan E, Hershfinkel M, Sekler I, Priel E. Glutamate regulates the activity of topoisomerase I in mouse cerebellum. Mol Neurobiol. 2008;38(3):242–252. doi: 10.1007/s12035-008-8044-x. [DOI] [PubMed] [Google Scholar]
- 46.Morham SG, Kluckman KD, Voulomanos N, Smithies O. Targeted disruption of the mouse topoisomerase I gene by camptothecin selection. Mol Cell Biol. 1996;16(12):6804–6809. doi: 10.1128/mcb.16.12.6804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Song J, Hu J, Tanouye M. Seizure suppression by top1 mutations in Drosophila. J Neurosci. 2007;27(11):2927–2937. doi: 10.1523/JNEUROSCI.3944-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Song J, Parker L, Hormozi L, Tanouye MA. DNA topoisomerase I inhibitors ameliorate seizure-like behaviors and paralysis in a Drosophila model of epilepsy. Neuroscience. 2008;156(3):722–728. doi: 10.1016/j.neuroscience.2008.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Teves SS, Henikoff S. Transcription-generated torsional stress destabilizes nucleosomes. Nat Struct Mol Biol. 2014;21(1):88–94. doi: 10.1038/nsmb.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Parikshak NN, et al. Integrative functional genomic analyses implicate specific molecular pathways and circuits in autism. Cell. 2013;155(5):1008–1021. doi: 10.1016/j.cell.2013.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Katyal S, et al. Aberrant topoisomerase-1 DNA lesions are pathogenic in neurodegenerative genome instability syndromes. Nat Neurosci. 2014;17(6):813–821. doi: 10.1038/nn.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bruce JN, et al. Regression of recurrent malignant gliomas with convection-enhanced delivery of topotecan. Neurosurgery. 2011;69(6):1272–1279, discussion 1279–1280. doi: 10.1227/NEU.0b013e3182233e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nagel BJ, et al. Early patterns of verbal memory impairment in children treated for medulloblastoma. Neuropsychology. 2006;20(1):105–112. doi: 10.1037/0894-4105.20.1.105. [DOI] [PubMed] [Google Scholar]
- 54.Potter SL, et al. Phase 2 clinical trial of intrathecal topotecan in children with refractory leptomeningeal leukemia: A Children’s Oncology Group trial (P9962) Pediatr Blood Cancer. 2012;58(3):362–365. doi: 10.1002/pbc.23317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Committee on Care and Use of Laboratory Animals (1996) Guide for the Care and Use of Laboratory Animals (Natl Inst Health, Bethesda), DHHS Publ No (NIH) 85-23.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.