Ice Age burial and Paleoindian mortuary ritual

Researchers excavate a double infant burial at the Upward Sun River site in Alaska.
In 2010, researchers recovered the remains of a cremated 3-year-old child dating to roughly 11,500 years ago that were lying within the hearth of a residential feature at the Upward Sun River archaeological site in Alaska. While excavating below the residence in 2013, the team found a second burial directly below the cremation hearth—a circular pit that contained the skeletons of two unburned infants. Ben Potter et al. (pp. 17060–17065) report the context and contributions of this double infant burial to the relatively scant record of mortuary and ritual behaviors at the time of the last Ice Age in North America. In addition to organic grave goods, the authors report that the infants were interred with the earliest known examples of North American hafted bifaces, which are dart or spear points that support theories about the form and function of Paleoindian weaponry. The authors also provide skeletal and dental analyses that reveal that the first infant died shortly after birth and that the second was a late-term fetus. The find represents a rare opportunity to examine how Pleistocene North Americans dealt with the death of the youngest members of their populations, according to the authors. — T.J.
Early diagnosis of metastatic cancers
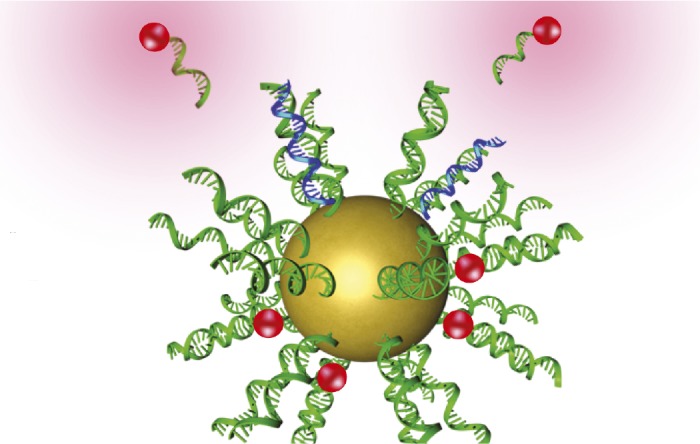
Nanostructure containing recognition sequences (green) that bind cancer cell target sequences (blue) and release a fluorescent signal (red).
Metastatic cancers are frequently undiagnosed until secondary tumors form, lowering therapeutic effectiveness and patient survival. Tiffany Halo et al. (pp. 17104–17109) engineered nanostructures to identify cancerous cells circulating in the bloodstream before they form secondary tumors. The structures, called NanoFlares, consist of a spherical gold nanoparticle to which a layer of single-stranded DNA (ssDNA) is attached. Hybridized to the ssDNA strands are short, fluorophore-containing ssDNA sequences called flare sequences. During circulation of the NanoFlares in the bloodstream, the ssDNA strands recognize the mRNA of a target gene inside live cancerous cells. Upon binding with the target mRNA, the flare sequences release from the ssDNA strands and provide a detectable cue inside the cancerous cells. The authors tested the construct in vitro and in a mouse model of metastatic breast cancer, finding that the nanostructures detected target cells in human blood in vitro with up to 99% accuracy and higher than 87% accuracy in the mouse model. Once labeled, the breast cancer cells could also be isolated from the blood and propagated in culture. According to the authors, the technique may facilitate early diagnosis and characterization of metastatic cancers, potentially improving patient survival. — J.P.J.
Mapping the vulnerability of Ebola virus
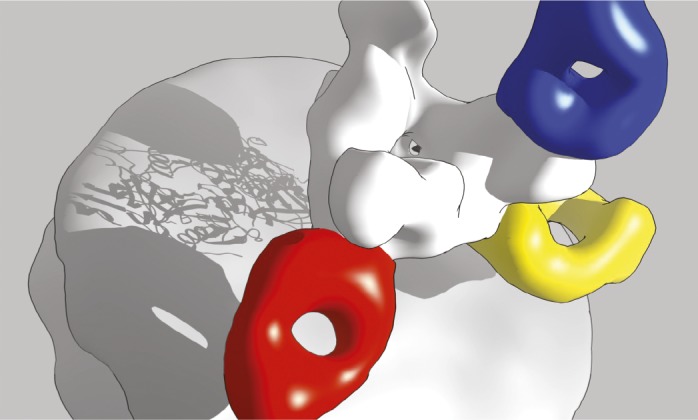
ZMapp in complex with Ebola virus glycoprotein GP (white). Antibody c13C6 (blue) binds the GP glycan cap. Antibodies c2G4 (red) and c4G7 (yellow) bind overlapping epitopes at the base.
The experimental antibody cocktail called ZMapp, now under development, was compassionately administered to some patients during the ongoing 2014 Ebola virus outbreak. The cocktail’s monoclonal antibodies were raised in rodents and modified for human use, but the antibodies’ mechanism of action remains unclear. Charles Murin et al. (pp. 17182–17187) used single-particle electron microscopy to reconstruct the structure of every monoclonal antibody in ZMapp, as well as related antibodies, in complex with an Ebola virus surface glycoprotein produced in cultured cells. The authors report that two of the antibodies, which appear to block the entry of the virus into human cells, bind to overlapping epitopes at the base of the viral glycoprotein, whereas the third antibody, which might flag viral particles and infected cells for immune destruction, binds to a glycan cap at the top of the glycoprotein. Together with those two binding sites, a distal-most region of the glycoprotein constitutes three distinct, immunologically vulnerable spots on the surface of the virus that might represent attractive targets for potential preventive or therapeutic agents. According to the authors, the findings suggest a likely mode of action of therapeutic antibody cocktails against Ebola virus and provide a plausible structural basis for the efficient design of future treatments against Ebola and related viruses. — P.N.
Genetic underpinnings of cat domestication

Abyssinian cat.
Humans have coexisted with cats for at least 9,000 years, and cat breeds emerged around 150 years ago, but little is known about the genetic changes that took place during domestication. Michael Montague et al. (pp. 17230–17235) sequenced the genome of a female Abyssinian cat, a domestic breed, and compared it with genome assemblies of six other domestic breeds, two wild cat species, and four other mammals to identify genomic differences that may underlie cat biology and domestication. The authors found that, compared with wild cat genomes, domestic cat genomes displayed evidence of natural selection in genes linked to memory, fear-conditioning behavior, and stimulus-reward learning, suggesting that the genetic changes may underlie the evolution of tameness. Domestic cats also displayed genetic variations that may explain certain aspects of feline biology including genes involved in lipid metabolism that support a hypercarnivorous diet, increased auditory and visual acuity, and large functional repertoires of vomeronasal receptor genes that enhance feline ability to sense pheromones in comparison with dogs. According to the authors, the genetic signatures may illuminate selective pressures that led to cat domestication. — J.P.J.
Virus implicated in sea-star wasting disease

Deflated sunflower sea star displays symptoms of sea-star wasting disease.
Since 2013, at least 20 sea star species have suffered heavy losses along the Pacific coast between Alaska and Mexico due to an unknown cause. Symptoms of sea-star wasting disease (SSWD) include tissue ulcers, inflammation, and necrosis leading to lethargy, limb curling and deflation, limb loss, and death. Ian Hewson et al. (pp. 17278–17283) conducted field surveys of healthy and infected sea stars and laboratory infection studies to identify a likely causative agent. Inoculation of healthy sea stars with material from diseased sea stars that included only virus-sized particles resulted in onset of disease. A metagenomic analysis revealed that sea star-associated densovirus (SSaDV) was the most prevalent contagious element contained in the material. Laboratory sea stars with SSWD displayed increasing levels of SSaDV as disease symptoms progressed, and infected sea stars from natural marine environments displayed higher levels of SSaDV than healthy sea stars, suggesting that the virus may be responsible for disease symptoms. Sampling of museum sea star specimens and the marine environment also revealed that the virus has been present in sea stars for at least 72 years and persists in plankton and marine sediments. The findings suggest an environmental reservoir for the virus that may explain past outbreaks, according to the authors. — J.P.J.
Brain response to a lost first language
During early exposure to a language, the brain forms representations of sound stimuli, but it is unknown whether the brain maintains or degrades sound representations if exposure to the language is discontinued. Lara Pierce et al. (pp. 17314–17319) conducted functional MRI studies on 48 female children with different levels of French and Chinese language exposure to determine whether the brain retains memory of an early, but discontinued, language. The authors played vocal recordings containing tonal sounds characteristic of Chinese, but not French, to three groups of children between 9 and 17 years of age. In one group, the children were born into a French-speaking family and had remained monolingual. In a second group, the children were adopted from a Chinese family into a French-speaking family before age 3, had stopped speaking Chinese, and had become French monolingual. In a third group, children had heard Chinese from birth, had started learning French by age 3, and had maintained fluency in both languages. Children in the two groups that had been exposed to Chinese early in life demonstrated similar neural activity in a brain region that was not activated in the children with French-only exposure. According to the authors, language representations created by the brain early in childhood can persist despite a loss of ability to speak the language. — J.P.J.


