Significance
Apurinic/apyrimidinic endonuclease 1 (APE1) has been shown to be a critical endonuclease required for class switch recombination (CSR). Here we show that APE1’s endonuclease activity, but not its redox regulation or transcriptional regulation activity, is important for CSR. Conversely, APE1 is dispensable for activation-induced cytidine deaminase (AID)-induced somatic hypermutation (SHM) as well as IgH/c-myc translocation. Moreover, during CSR, APE1 is not required for AID-induced S-region break formation, but is critical for the processing of cleaved ends because both repair protein Ku80 recruitment and end synapse formation are blocked by APE1 deficiency. APE1 deficiency only partially impairs CSR, and we found that two other end-processing enzymes, meiotic recombination 11 homolog (MRE11) and carboxy-terminal binding protein (CtBP)-interacting protein (CtIP), are responsible for the remaining CSR activity in the absence of APE1.
Keywords: class switch recombination, somatic hypermutation, DNA cleavage, DNA synapse formation, end processing
Abstract
Activation-induced cytidine deaminase (AID) is essential for antibody diversification, namely somatic hypermutation (SHM) and class switch recombination (CSR). The deficiency of apurinic/apyrimidinic endonuclease 1 (Ape1) in CH12F3-2A B cells reduces CSR to ∼20% of wild-type cells, whereas the effect of APE1 loss on SHM has not been examined. Here we show that, although APE1’s endonuclease activity is important for CSR, it is dispensable for SHM as well as IgH/c-myc translocation. Importantly, APE1 deficiency did not show any defect in AID-induced S-region break formation, but blocked both the recruitment of repair protein Ku80 to the S region and the synapse formation between Sμ and Sα. Knockdown of end-processing factors such as meiotic recombination 11 homolog (MRE11) and carboxy-terminal binding protein (CtBP)-interacting protein (CtIP) further reduced the remaining CSR in Ape1-null CH12F3-2A cells. Together, our results show that APE1 is dispensable for SHM and AID-induced DNA breaks and may function as a DNA end-processing enzyme to facilitate the joining of broken ends during CSR.
Upon encountering antigens in the periphery, mature B cells undergo two types of genetic alterations, somatic hypermutation (SHM) and class switch recombination (CSR), in the Ig gene during the G1 phase (1, 2). Although mechanistically different, both events are initiated by the activation-induced cytidine deaminase (AID) (3–5), which introduces single-strand DNA break (SSB) in the target DNA, namely the V region for SHM and the S region for CSR (6–9). Most SHMs are introduced during the repair of AID-induced SSBs through error-prone DNA synthesis by translesion polymerases, such as Polη and Polζ (10, 11).
By contrast, efficient CSR requires a series of biological steps including conversion of AID-induced SSBs to double-strand breaks (DSBs), the synapsis formation of S-region broken ends, and their recombination (8, 12). Among them, processing of SSBs to DSBs without replication involves many enzymes including exonucleases, endonucleases, and/or helicases. Furthermore, when the cleaved ends are blocked by unusual modified bases, they should be removed by end-processing enzymes such as carboxy-terminal binding protein (CtBP)-interacting protein (CtIP) and the MRN complex (13). Subsequently, the DSBs at the donor and acceptor S regions are joined by either the nonhomologous end joining (NHEJ) or alternative end joining (A-EJ) pathway (14–17). During the NHEJ pathway that requires DSBs with blunt end or short overhang, additional end processing could occur at several steps before the recruitment of repair protein Ku80, or after synapse formation (13). The A-EJ pathway is used for the joining of DSB with long overhangs and also is involved in aberrant chromosomal translocations, which depend on uracil DNA glycosylase (UNG) (18).
Both SHM and CSR take place during the G1 phase and are transcription-dependent (19). Because AID is known to introduce DNA cleavage in a limited number of non-Ig genes such as oncogene c-myc, AID targets are restricted (20). There are only a small number of enzymes that can cause DNA damage (genotoxic activity) in a transcription-dependent manner. Topoisomerase 1 (Top1) was recently shown to be genotoxic in neural cells that do not replicate (21). During transcription, Top1 normally regulates the DNA helix by transient SSBs, covalent association with DNA, DNA rotation around the helix, and relegation. However, Top1 occasionally may be trapped as covalently bound to DNA in the repetitive sequences that can form non-B structures (22).
We proposed that AID suppresses Top1 protein synthesis by deaminating cytidine (C) in miRNAs (23, 24). The reduction in the Top1 protein facilitates the formation of non-B DNA structures in the V and S regions of the Ig locus when they are actively transcribed. Such unusual DNA structure causes irreversible DNA cleavage by Top1 because this structure suppresses the ordinary rotation around the helix of DNA (20, 22–24). Conversely, AID was also proposed to directly deaminate DNA, generating G:U mismatches that are detected and cleaved either by the base excision repair (BER) pathway including UNG and apurinic/apyrimidinic endonuclease 1 (APE1) or by the mismatch repair (MMR) enzymes including MutS homolog 2/MutS homolog 6 (Msh2/Msh6) (9, 25, 26). It is presumed that APE1 nicks DNA at abasic sites after uracil (U) removal by UNG, creating SSBs that, after processing, give rise to SHM, CSR, or aberrant recombinations/translocations (9).
APE1 is a multifunctional protein that orchestrates multiple activities in the cell: (i) endonuclease activity for DNA damage repair (27), (ii) the redox activity for the maintenance of transcription factors (28), and (iii) binding to negative Ca2+-response elements for acetylation-dependent regulation of gene expression (29). The endonuclease activity of APE1 not only nicks at abasic sites, but also resects peptide-tyrosyl nucleotides at the 3′ end of DNA (30, 31). Because of the embryonic lethality of APE1 ablation in mice (32), the role of APE1 in CSR and SHM has not been conclusively examined, and several studies gave rise to contradictory conclusions (33, 34).
Recently, however, Masani et al. generated CH12F3-2A B cells completely lacking APE1 and found that the genetic ablation of Ape1 reduces the CSR efficiency in CH12F3-2A cells to 20% of the wild-type (WT) cells, whereas a deletion of APE2 has no effect on the CSR of CH12F3-2A cells (35). The results clearly demonstrated the involvement of APE1 in CSR, but at the same time raised several critical questions as to the role of APE1 in CSR. First, it is of particular importance to determine with which enzymatic activity and by what mechanism APE1 is involved in CSR. It is also important to assess whether APE1 is also required for AID-induced SHM. Furthermore, it is interesting to know which enzymes could account for the remaining CSR activity in APE1-deficient CH12F3-2A cells, although Masani et al. proposed that a latent endonuclease activity of the MMR factor MLH1/PMS2 complex may be responsible (35).
In the present study, we examined APE1’s role in CSR and SHM using APE1-deficient CH12F3-2A cells (35) and found that, although APE1’s endonuclease activity is required for CSR, it is dispensable for SHM and IgH/c-myc translocation. Surprisingly, the endonuclease activity of APE1 is dispensable for AID-induced S-region cleavage, but necessary for Ku80 recruitment and synapse formation of the broken ends. Our results suggest that APE1 functions as a DNA end resection enzyme and plays a critical role in processing AID-induced SSBs for efficient joining and recombination during CSR.
Results
The Endonuclease Activity of APE1 Is Required for CSR.
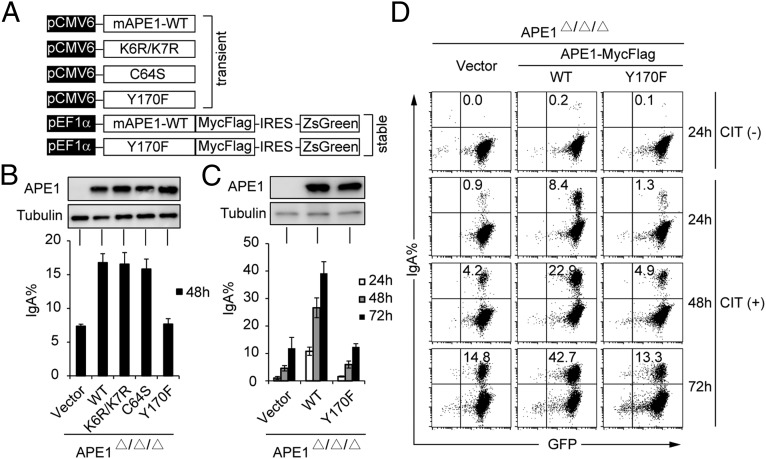
To elucidate which function of APE1 is important for CSR, we expressed APE1 WT and its loss-of-function mutant proteins in Ape1-null CH12F3-2A cells and tested the ability of their transfectants for restoration of IgA switching. Three mutant proteins include (i) APE1K6R/K7R that is defective in the acetylation-mediated gene regulation function (29); (ii) APE1C64S mutant that lacks the redox activity (36); and (iii) APE1Y170F mutant that is devoid of the endonuclease activity (37) (Fig. 1A). Western blotting analysis showed that WT and all APE1 mutant proteins were almost equally expressed in transiently transfected Ape1-null CH12F3-2A cells (Fig. 1B, Upper). The transfectants with WT, K6R/K7R, and C64S mutants switched to IgA at similar levels after stimulation with CD40L, IL-4, and TGF-β (CIT), whereas Y170F transfectant could not enhance switching compared with the vector control (Fig. 1B, Lower).
Fig. 1.
APE1's endonuclease activity is essential for CSR. (A) Schematic representation of the various APE1 mutants used in the CSR complementation experiments. (B) Representative IgA population of Ape1-null CH12F3-2A cells after transient introduction of various APE1 constructs. Data are represented as mean ± SD. (C) IgA switching efficiency of Ape1-null CH12F3-2A cells stably expressing the empty vector, WT, or Y170F mutant of APE1. APE1 protein expression was determined by Western blot (Upper), and CSR efficiency was measured by FACS as the percentage of cell surface IgA expression at indicated time after CIT stimulation (Lower). Data are represented as mean ± SD. (D) Representative flow cytometry (FACS) profile of the IgA switching population under CIT (−) and (+) conditions.
To further confirm the importance of the endonuclease activity of APE1 for CSR, we generated two transfectant lines from Ape1-null CH12F3-2A cells stably expressing WT APE1 (WT transfectant) or Y170F mutant (Y170F transfectant), as well as a control line stably transfected with the empty vector (vector transfectant). To minimize the potential variation among single clones, bulk GFP-positive cells were sorted after G418 selection and used for further analysis. Although Y170F and WT transfectants expressed similar levels of the APE1 protein, Y170F transfectant failed to rescue IgA switching at the three time points tested compared with vector and WT transfectants (Fig. 1 C and D). Together, these data indicate that the endonuclease activity of APE1 is required for efficient CSR.
APE1 Is Dispensable for AID-Induced S-Region Mutations.
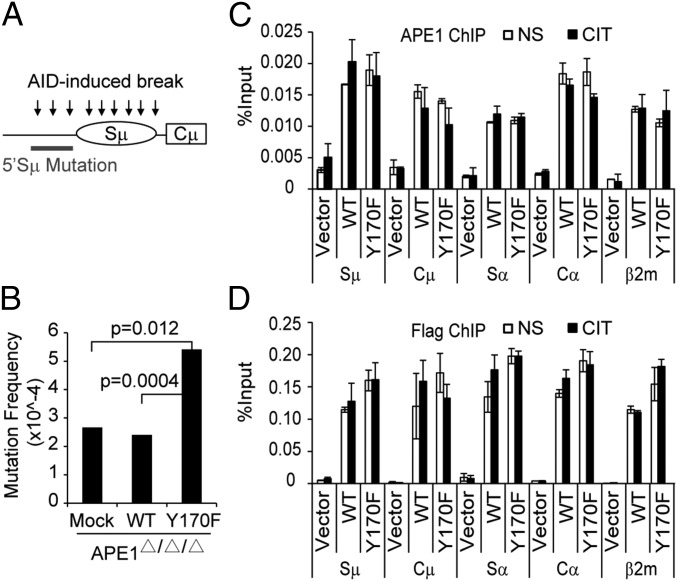
We then asked whether the endonuclease activity of APE1 is also required for SHM. Because the 5′ Sμ region is the efficient target for SHM in CH12F3-2A cells, we isolated and sequenced the 5′ Sμ region from the three transfectants (vector, WT, and Y170F) of Ape1-null CH12F3-2A cells 72 h after CIT stimulation (Fig. 2A). The mutation frequencies were almost equal between vector and WT transfectants (Fig. 2B and Table S1), and the mutation base profile remained unchanged (Table S2), indicating that APE1 is not required for 5′ Sμ mutation.
Fig. 2.
APE1 is dispensable for AID-induced 5′ Sμ mutation. Ape1-null cells stably expressing the vector, WT, or Y170F mutant of APE1 were stimulated with CIT for 72 h, followed by genomic DNA extraction for mutation analysis. (A) The diagram depicts the position of the 5′ Sμ region selected for the mutation analysis. (B) Result of the mutation analysis for the 5′ Sμ region. P values (Fisher’s exact test) for significant difference are shown in the graph. The detailed results are shown in Tables S1 and S2. (C and D) ChIP and quantitative PCR analysis for APE1 (C) and Flag (D) in cells stimulated (or not) with CIT for 24 h. Data are represented as mean ± SD.
Interestingly, Y170F transfectant had significantly more mutations, which could be due to the dominant-negative binding of Y170F to DNA that may prevent error-free repair of the break sites of the BER system. ChIP with APE1 or Flag antibody showed that the Y170F mutant bound to not only S-region DNA, but also nonimmunoglobulin DNA such as the β2m locus, as efficiently as WT APE1 (Fig. 2 C and D). Binding of APE1 to S-region DNA was independent of AID activation (Fig. 2 C and D), which is consistent with the idea that APE1 has a surveillance mechanism that allows it to slide along DNA for scanning damaged sites (38). Because mutations in the 5′ Sμ region are considered “footprints” of the AID-dependent DNA breaks (6, 7), our results indicate that AID-induced S-region cleavage takes place normally without APE1.
APE1 Is Dispensable for AID-Induced IgH/c-myc Translocation.
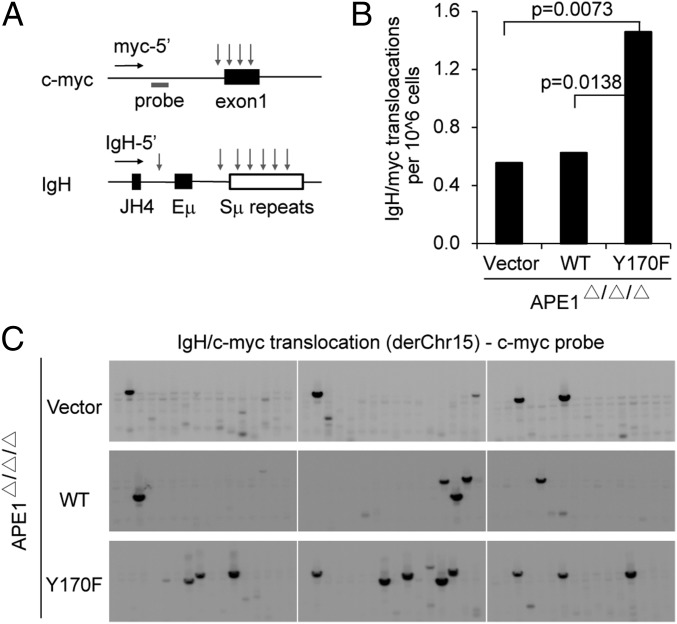
To confirm that AID induces normal levels of DNA cleavage in APE1-deficient cells, we examined the frequency of aberrant translocations between IgH and c-myc in the absence or presence of APE1. AID-induced IgH/c-myc translocation was measured by a pair of PCR primers (one in c-myc and the other in Ig JH4) as shown in Fig. 3A. The AID-induced translocation frequency was similar between vector and WT transfectants, whereas Y170F transfectant showed significantly more frequent translocations than vector and WT transfectants (Fig. 3 B and C). Because IgH/c-myc translocation depends on AID-induced DSB intermediates (18), equal levels of the translocation frequency with or without APE1 indicate that AID-induced cleavage in the S region as well as c-myc locus does not depend on APE1. Enhanced translocation by Y170F suggests again the dominant-negative effect of the catalytically inactive APE1 mutant on DNA repair enzymes, which could reduce the chromosomal translocation.
Fig. 3.
APE1 is dispensable for AID-induced IgH/c-myc translocation. Ape1-null cells stably expressing the vector, WT, or Y170F mutant of APE1 were stimulated with CIT for 72 h, followed by genomic DNA extraction for translocation analysis. (A) A schematic representation of the PCR assay used for IgH/c-myc translocations. Primers used to detect derivative chromosome 15 (derChr15) translocations are shown as horizontal arrows. Vertical arrows represent AID-induced DNA break sites. The gray bar shows the position of the c-myc probe used for Southern detection. (B) Result of the IgH/c-myc translocation derChr15 analysis (96 lanes total from two independent experiments). P values (Fisher’s exact test) for significant difference are shown in the graph. (C) Representative Southern blots with c-myc probe for IgH/c-myc translocation are shown (105 cells per lane).
APE1 Is Dispensable for the Formation of AID-Induced S-Region Breaks.
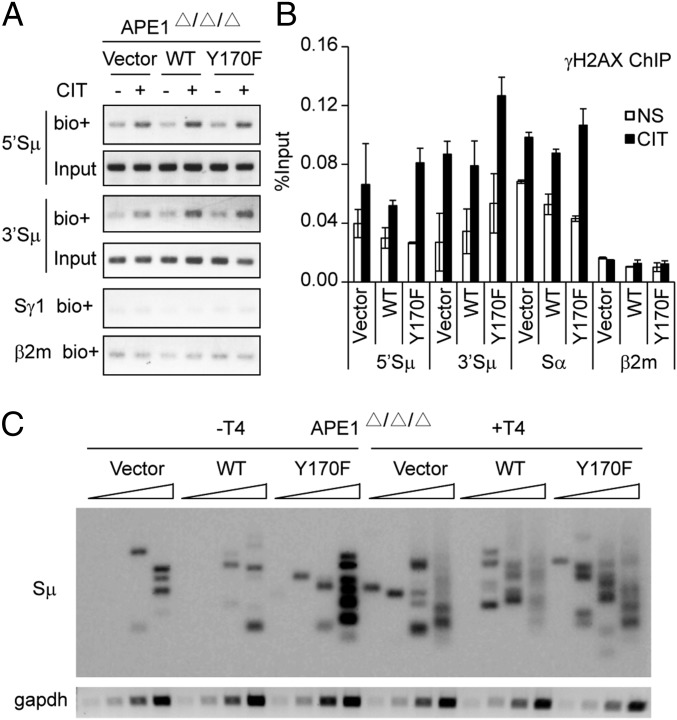
To examine directly the dispensability of APE1 in S-region cleavage, we assayed the level of DSBs in the three cell lines by the break-end labeling method, using biotin-labeled nucleotides and T4 polymerase (39). CIT stimulation induced similar levels of DNA breaks at both the 5′ and 3′ sequences to the core Sμ region in the presence or absence of APE1 (Fig. 4A). The histone γH2AX ChIP assay further confirmed that AID-induced DSBs occurred normally at both the Sμ and Sα regions in the absence of APE1 (Fig. 4B).
Fig. 4.
AID induces a similar level of DNA break at the Sμ region in the absence of functional APE1. (A) Cells were stimulated (or not) with CIT for 24 h and then harvested for biotin-labeling DNA break assay as described in Materials and Methods. Pulled-down DNA labeled with biotin–dUTP at the cleaved ends was subjected to region-specific PCR. (B) ChIP and quantitative PCR analysis for γH2AX in cells stimulated (or not) with CIT for 24 h. Data are represented as mean ± SD. (C) LM-PCR analysis of DNA isolated from cells stimulated with CIT for 24 h. DNA was left untreated (−T4) or treated (+T4) with T4 DNA polymerase and assessed with Sμ-specific primer or by amplification of gapdh (internal control for template loading). Wedges indicate a threefold increase in DNA.
We then carried out the linker ligation-mediated PCR (LM-PCR) assay for DSB formation at the Sμ region in Ape1-null cells and their transfectants in the presence or absence of T4 polymerase. We did not detect any change of DSBs between vector and WT transfectants with or without T4 polymerase (Fig. 4C). Interestingly, Y170F transfectant showed slightly enhanced DNA cleavage activity compared with vector or WT transfectants, suggesting that the Y170F mutant inhibits DNA repair. The recruitment of MMR protein Msh2 was not affected with or without APE1 (Fig. S1), making it less likely that elevated MMR activity is responsible for the cleavage detected in the absence of APE1. We conclude that APE1 is dispensable for AID-induced DNA break.
APE1 Is Required for Efficient End Joining of Cleaved S Regions During CSR.
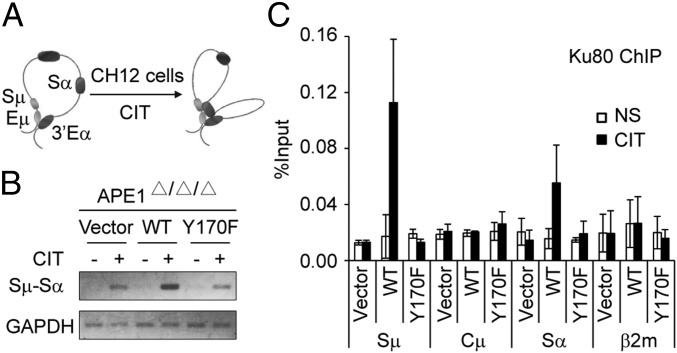
Because the DNA cleavage in the Sµ region remains unaltered in the absence of APE1 during CSR, we examined whether the next step after DNA cleavage, namely end joining, is affected by APE1 deficiency. We confirmed the previous report that the distribution of junction microhomology lengths of Sμ–Sα recombination is not drastically different between Ape1-null and WT CH12F3-2A cells (35), using the three transfectant lines derived from Ape1-null CH12F3-2A cells (Fig. S2). The results suggest that both the NHEJ and A-EJ pathways are defective in APE1 deficiency. Consistently, the chromosome confirmation capture (3C) assay showed that the Sμ–Sα synapse formation induced by CIT stimulation was suppressed in the vector and Y170F transfectants compared with the WT transfectant (Fig. 5 A and B). Furthermore, the accumulation of Ku80, a protein critical for NHEJ, was very much reduced at S regions of vector- and Y170F-transfectant cells compared with WT transfectant (Fig. 5C), indicating that the reduced CSR in vector and Y170F transfectants might be due to the less efficient generation of DSBs with blunt ends.
Fig. 5.
APE1 is required for efficient Sμ–Sα synapse formation during CSR. (A) Scheme of long-range interactions between Sμ–Sα elements in the IgH locus before and after AID activation. (B) Representative gel picture of the 3C assay detecting Sμ–Sα interaction in the three cell lines stimulated (or not) with CIT for 24 h. GAPDH was amplified as loading control. (C) ChIP and quantitative PCR analysis for Ku80 in cells stimulated (or not) with CIT for 24 h. Data are represented as mean ± SD.
APE1 May Function as Cleaved-End Processing Enzyme for Ig Diversification.
Although AID-induced S-region cleavage takes place normally in APE1 deficiency, both Ku80 accumulation and the synapse formation of the broken ends are severely affected by the absence of the endonuclease activity of APE1. We speculated that APE1 is involved in 3′ end processing of SSBs during CSR, because it is well established that the APE1 is involved in the 3′ end processing of SSBs (30, 40, 41).
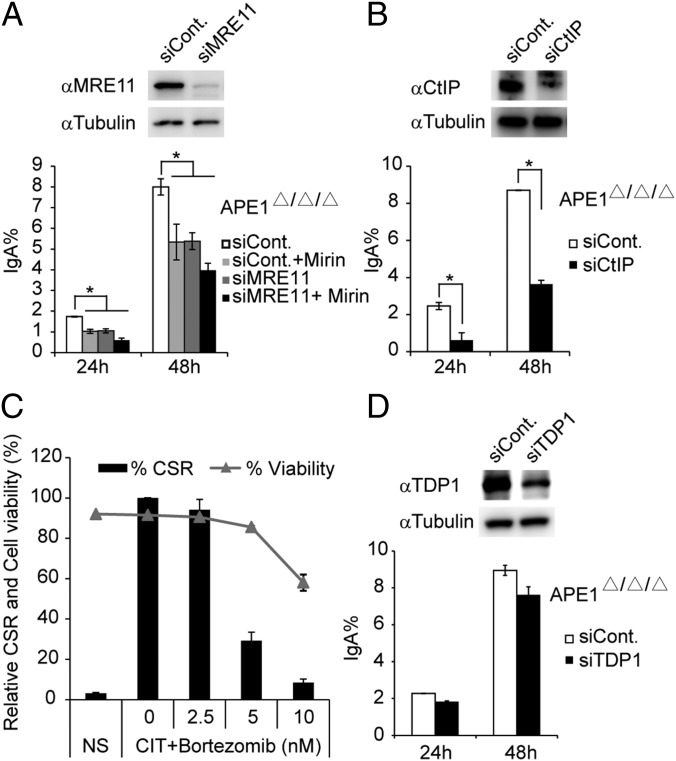
To test this possibility, we investigated whether other broken end-processing enzymes are responsible for the residual CSR activity in APE1-deficient CH12F3-2A cells. siRNA knockdown or drug inhibition of meiotic recombination 11 homolog (MRE11) could significantly reduce the remaining CSR activity in the absence of APE1 (Fig. 6A). Such reduction was more robust in case of CtIP knockdown (Fig. 6B).
Fig. 6.
The involvement of end-processing enzymes in the residual switching in Ape1-null CH12F3-2A cells. (A, B, and D) Protein expression (Upper) and IgA switching efficiency (Lower) of Ape1-null CH12F3-2A cells transfected with the indicated siRNA oligos and stimulated with CIT for 24 or 48 h. (C) Relative IgA switching efficiency (bar graph) and cell viability (dot plot) of CH12F3-2A–Bcl2 cells treated with various concentrations of the proteasome inhibitor Bortezomib. In all datasets, data are represented as mean ± SD.
Because both MRE11 and CtIP are known to be involved in the processing of the 3′ DNA end containing phospho-tyrosyl peptides to DNA–3′OH, which are generated by Top1 cleavage (42–44), these results suggest that the residual switching activity observed in Ape1-null CH12F3-2A cells may depend on the redundant Top1 cleavage complex (Top1-cc) processing enzymes. If so, it is expected that the inhibition of DNA-bound Top1 degradation with proteasome inhibitors and blocking the generation of DNA–3′-tyrosyl peptides could inhibit CSR in CH12F3-2A cells. Indeed, we found that CSR was severely blocked by this treatment, with modest cell death at low concentrations of Bortezomib (Fig. 6C). However, knockdown of tyrosyl–DNA phosphodiesterase 1 (TDP1), an enzyme that removes the 3′-tyrosyl residues of Top1 on DNA ends leaving a 3′-phosphate (45), did not further reduce IgA switching in Ape1-null CH12F3-2A cells (Fig. 6D). This finding is consistent with the report that TDP1-deficient patients do not show any immune defects (46).
Discussion
Requirement of APE1’s Endonuclease Activity for CSR but Not for DNA Cleavage.
Genetic ablation of APE1 in CH12F3-2A cells showed that APE1 is involved in AID-induced CSR (35). The present study has shown that the endonuclease activity among three APE1’s activities is required for efficient CSR. However, the endonuclease activity appears to function after the formation of AID-induced DNA strand cleavage, as evidenced by three different DNA break assays: (i) biotin–dUTP end labeling assay; (ii) γH2AX foci formation ChIP assay; and (iii) LM-PCR assay in the presence or absence of T4 polymerase.
In agreement with the normal level of AID-induced DNA cleavage in the absence of APE1, neither SHM nor IgH/c-myc translocation was reduced by APE1 deficiency. Because DNA break formation governs the frequency of recurrent translocation (47) and c-myc breaks are rate-limiting for IgH/c-myc translocation (48), the same frequency of IgH/c-myc translocation in the presence or absence of APE1 strongly suggests that AID-induced cleavage does not depend on APE1 in either the S-region or c-myc locus. In other words, APE1 is dispensable for AID-induced break formation during CSR.
Differential Roles of APE1 in SHM, IgH/c-myc Translocation, and CSR.
UNG suppresses SHM by recruiting faithful BER pathway components at cleaved DNA loci, with competition against error-prone polymerases (49). In contrast, APE1 is not involved in the correct repair after cleavage during SHM because SHM occurs similarly between vector and WT transfectants of Ape1-null CH12F3-2A cells. This finding indicates that during CSR and SHM, APE1 does not function as a canonical BER factor that cleaves abasic sites and activates the BER pathway.
Although both CSR and IgH/c-myc translocation are initiated by AID-induced breaks, B cells employ divergent repair pathways to join their break ends (18, 50). The differential requirement of APE1 in CSR and IgH/c-myc translocation may reflect the fact that the factors required for resolution of DNA breaks differ between the two reactions. In fact, IgH/c-myc translocation does not require either histone H2AX or Ku80 protein, both of which are essential to CSR (18). APE1 deficiency, which does not affect either accumulation of γH2AX at cleaved S regions or IgH/c-myc translocation, almost completely abolishes the recruitment of Ku80 and impairs CSR. Because the average lengths and distribution of microhomology in the residual CSR products did not change drastically, both NHEJ and A-EJ are reduced proportionally in APE1 deficiency. This finding is consistent with the severe reduction of the synapse formation between two broken ends. The reason why IgH/c-myc translocation is not reduced in APE1 deficiency may be due to the fact that this translocation is orders-of-magnitude inefficient compared with CSR. Under such circumstances, DNA cleavage rather than end-joining process may be rate limiting (47, 48), and the residual end-joining activity (∼20%) detected in the absence of APE1 could be more than enough to mediate such translocation.
It is interesting to compare the role of APE1 with Brd4, because we recently found that the acetylated histone reader Brd4 is involved in CSR and IgH/c-myc translocation, but not SHM (51). Although APE1 binds to S-region DNA constitutively, Brd4 is recruited to the S region during CSR in an AID-dependent manner. We examined the effect of APE1 deficiency on Brd4 recruitment by ChIP assay and found that Brd4 was recruited to the S region to the same level in either the presence or absence of APE1 (Fig. S3). The results are consistent with the fact that γH2AX formation is intact in the absence of APE1. Because both APE1 and Brd4 are required for CSR, it is likely that APE1 and Brd4 function cooperatively to initiate DNA repair at the DNA and histone levels, respectively, but independently of each other.
End Processing by APE1’s Endonuclease Activity.
Although APE1 deficiency drastically reduces the synapse formation (3C assay) and Ku80 recruitment, it does not affect DNA cleavage. The results suggest that APE1 is involved in a step between cleavage and cleaved-end joining. Because the endonuclease activity of APE1 is essential to CSR, APE1’s endonuclease activity is likely to be required in a step during SSB end processing to DSB, which is required for Ku80 recruitment and synapse formation.
Evidence showed that Top1 is the cutting enzyme that introduces cleavage immediately after AID activation (23, 24). Top1 covalently bound to the non–B-structure DNA in the S region or the Top1-cc must be degraded by proteasome, which leaves 3′-phosphotyrosyl peptide on DNA (22, 52). In agreement with Top1-mediated cleavage during CSR, blocking the degradation of Top1-cc with proteasome inhibitors strongly reduces CSR in CH12F3-2A cells. APE1’s ability that removes 3′-tyrosyl residue with its endonuclease activity generating a 3′-hydroxyl end (30, 31) makes it a good candidate for processing AID-induced SSB ends during CSR. Importantly, APE1 was identified as the major activity in human cell extracts for the removal of 3′-phosphoglycolate on DNA break ends (41).
APE1 deficiency impairs, but does not abolish, CSR in CH12F3-2A cells, suggesting that there are some other enzymes involved in similar SSB end processing. We found that the remaining CSR activity in APE1-deficient cells can be further reduced by knockdown of MRE11 or CtIP, two enzymes capable of removing covalently bound Top1 from the 3′ end of DNA (42–44). A recent study suggests that CtIP and MRE11 may use their nuclease activity to process 3′ end Top1-cc in the context of the DNA secondary structure that forms due to common fragile sites or repetitive sequence (53). In addition, the 3′-end processing activity of CtIP is clearly dissociated with its well-recognized 5′-end resection activity for homologous recombination, which does not depend on its nuclease activity (53, 54). Thus, AID-induced Top1-cc in the S region containing highly repetitive sequence could be removed by proteasome followed by either APE1 directly at the broken end or CtIP/MRE11 at inside positions when the broken end forms a secondary structure.
Top1-cc can also be processed by TDP1, which, instead of creating a 3′-hydorxyl end as do APE1, MRE11, and CtIP, leaves a phosphate on the 3′ end of DNA that needs to be further processed by polynucleotide kinase phosphatase (PNKP) to make a 3′-hydroxyl end (22). The different mechanism of processing Top1-cc by TDP1 may partially explain why its knockdown had no effect on CSR. Interestingly, both TDP1 and PNKP are involved in the repair of transcription-coupled Top1-induced SSB in neural cells, and their defect results in abnormal neurodevelopment and neurodegeneration (46, 55). Similarly, dysfunction of ATM results in ataxia telangiectasia with accumulation of pathogenic Top1-cc in neural tissue (21). Thus, Top1-cc processing is critical for maintaining genome stability and preventing neurodegeneration in neural cells on one hand, and is important for mediating proper CSR in B cells on the other hand.
In summary, we have shown that APE1, although essential for CSR with its endonuclease activity, is dispensable for SHM or Igh/c-myc translocation in CH12F3-2A cells. More importantly, APE1’s endonuclease activity is not required for the AID-induced DNA cleavage, in parallel with the findings that UNG is dispensable for the DNA cleavage step during SHM (56, 57) and CSR (39, 58). Recently, we reported that UNG enhances AID-dependent S–S synapse formation by recruiting p53-binding protein 1 and DNA-dependent protein kinase, catalytic subunit (49). We provide the evidence indicating that APE1 processes the broken ends of S-region DNA for the binding of Ku80 and mediates efficient S–S synapse formation during CSR. APE1’s endonuclease activity is likely to remove 3′-tyrosyl residues from the DNA end after Top1 cleavage and Top1-cc degradation, which is essential to efficient recombination of broken S regions.
Materials and Methods
Biotin-Labeling DNA Break Assay.
Cells were stimulated with CIT for 24 h, and switching efficiency was assayed by surface IgA staining. The biotin-labeling DNA break assay was performed as described (39). Primers for amplifying Sμ, Sγ1, and β2m loci are listed in Table S3.
LM-PCR.
Genomic DNA was prepared, and LM-PCR was performed as described (33) with minor modifications. Briefly, after 24 h of CIT stimulation, living cells were purified by gradient centrifugation in percoll, and DNA was extracted within low-melting agarose plugs. A total of 20 μL of DNA was treated with or without T4 polymerase (TAKARA) before being used for linker ligation in a 100-μL reaction. Ligation was stopped by adding 200 μL of H2O and heating at 70 °C for 10 min. Ligated DNA was assayed for gapdh DNA by PCR to adjust DNA input before LM-PCR. Threefold dilutions of input DNA were amplified by KOD-FX-Neo polymerase (TOYOBO) by using a touchdown PCR program. PCR products were electrophoresed on 1% agarose gels and validated by Southern blot using 5′ Sμ probe. Primers and probe sequences are shown in Table S3.
Antibodies.
Antibodies used for Western blot and ChIP analysis are listed in Table S4.
Other materials and methods are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Prof. Kefei Yu (Michigan State University) for providing Ape1-null CH12F3-2A cells; Nasim A. Begum and Andre Stanlie for their optimization of translocation and 3C assay protocols in CH12F3-2A cells; and members of the T.H. laboratory for sharing regents and useful technical information and for discussion and critical reading of the manuscript. This work was supported by Ministry of Education, Culture, Sports, Science, and Technology of Japan Grant-in-Aid for Specially Promoted Research 17002015 (to T.H.) and Grant-in-Aid for Scientific Research (C) 25440007 (to M.K.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1420221111/-/DCSupplemental.
References
- 1.Faili A, et al. AID-dependent somatic hypermutation occurs as a DNA single-strand event in the BL2 cell line. Nat Immunol. 2002;3(9):815–821. doi: 10.1038/ni826. [DOI] [PubMed] [Google Scholar]
- 2.Schrader CE, Guikema JE, Linehan EK, Selsing E, Stavnezer J. Activation-induced cytidine deaminase-dependent DNA breaks in class switch recombination occur during G1 phase of the cell cycle and depend upon mismatch repair. J Immunol. 2007;179(9):6064–6071. doi: 10.4049/jimmunol.179.9.6064. [DOI] [PubMed] [Google Scholar]
- 3.Muramatsu M, et al. Specific expression of activation-induced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family in germinal center B cells. J Biol Chem. 1999;274(26):18470–18476. doi: 10.1074/jbc.274.26.18470. [DOI] [PubMed] [Google Scholar]
- 4.Muramatsu M, et al. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102(5):553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 5.Revy P, et al. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2) Cell. 2000;102(5):565–575. doi: 10.1016/s0092-8674(00)00079-9. [DOI] [PubMed] [Google Scholar]
- 6.Petersen S, et al. AID is required to initiate Nbs1/gamma-H2AX focus formation and mutations at sites of class switching. Nature. 2001;414(6864):660–665. doi: 10.1038/414660a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nagaoka H, Muramatsu M, Yamamura N, Kinoshita K, Honjo T. Activation-induced deaminase (AID)-directed hypermutation in the immunoglobulin Smu region: Implication of AID involvement in a common step of class switch recombination and somatic hypermutation. J Exp Med. 2002;195(4):529–534. doi: 10.1084/jem.20012144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Honjo T, Kinoshita K, Muramatsu M. Molecular mechanism of class switch recombination: Linkage with somatic hypermutation. Annu Rev Immunol. 2002;20:165–196. doi: 10.1146/annurev.immunol.20.090501.112049. [DOI] [PubMed] [Google Scholar]
- 9.Stavnezer J, Guikema JE, Schrader CE. Mechanism and regulation of class switch recombination. Annu Rev Immunol. 2008;26:261–292. doi: 10.1146/annurev.immunol.26.021607.090248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saribasak H, et al. DNA polymerase ζ generates tandem mutations in immunoglobulin variable regions. J Exp Med. 2012;209(6):1075–1081. doi: 10.1084/jem.20112234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daly J, et al. Altered Ig hypermutation pattern and frequency in complementary mouse models of DNA polymerase ζ activity. J Immunol. 2012;188(11):5528–5537. doi: 10.4049/jimmunol.1102629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chaudhuri J, et al. Evolution of the immunoglobulin heavy chain class switch recombination mechanism. Adv Immunol. 2007;94:157–214. doi: 10.1016/S0065-2776(06)94006-1. [DOI] [PubMed] [Google Scholar]
- 13.Povirk LF. Processing of damaged DNA ends for double-strand break repair in mammalian cells. ISRN Mol Biol. 2012 doi: 10.5402/2012/345805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Casellas R, et al. Ku80 is required for immunoglobulin isotype switching. EMBO J. 1998;17(8):2404–2411. doi: 10.1093/emboj/17.8.2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pan-Hammarström Q, et al. Impact of DNA ligase IV on nonhomologous end joining pathways during class switch recombination in human cells. J Exp Med. 2005;201(2):189–194. doi: 10.1084/jem.20040772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chaudhuri J, Alt FW. Class-switch recombination: Interplay of transcription, DNA deamination and DNA repair. Nat Rev Immunol. 2004;4(7):541–552. doi: 10.1038/nri1395. [DOI] [PubMed] [Google Scholar]
- 17.Yan CT, et al. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature. 2007;449(7161):478–482. doi: 10.1038/nature06020. [DOI] [PubMed] [Google Scholar]
- 18.Ramiro AR, et al. Role of genomic instability and p53 in AID-induced c-myc-Igh translocations. Nature. 2006;440(7080):105–109. doi: 10.1038/nature04495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muramatsu M, Nagaoka H, Shinkura R, Begum NA, Honjo T. Discovery of activation-induced cytidine deaminase, the engraver of antibody memory. Adv Immunol. 2007;94:1–36. doi: 10.1016/S0065-2776(06)94001-2. [DOI] [PubMed] [Google Scholar]
- 20.Kato L, et al. An evolutionary view of the mechanism for immune and genome diversity. J Immunol. 2012;188(8):3559–3566. doi: 10.4049/jimmunol.1102397. [DOI] [PubMed] [Google Scholar]
- 21.Katyal S, et al. Aberrant topoisomerase-1 DNA lesions are pathogenic in neurodegenerative genome instability syndromes. Nat Neurosci. 2014;17(6):813–821. doi: 10.1038/nn.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pommier Y, et al. Repair of and checkpoint response to topoisomerase I-mediated DNA damage. Mutat Res. 2003;532(1-2):173–203. doi: 10.1016/j.mrfmmm.2003.08.016. [DOI] [PubMed] [Google Scholar]
- 23.Kobayashi M, et al. AID-induced decrease in topoisomerase 1 induces DNA structural alteration and DNA cleavage for class switch recombination. Proc Natl Acad Sci USA. 2009;106(52):22375–22380. doi: 10.1073/pnas.0911879106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kobayashi M, et al. Decrease in topoisomerase I is responsible for activation-induced cytidine deaminase (AID)-dependent somatic hypermutation. Proc Natl Acad Sci USA. 2011;108(48):19305–19310. doi: 10.1073/pnas.1114522108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petersen-Mahrt SK, Harris RS, Neuberger MS. AID mutates E. coli suggesting a DNA deamination mechanism for antibody diversification. Nature. 2002;418(6893):99–103. doi: 10.1038/nature00862. [DOI] [PubMed] [Google Scholar]
- 26.Di Noia JM, Neuberger MS. Molecular mechanisms of antibody somatic hypermutation. Annu Rev Biochem. 2007;76:1–22. doi: 10.1146/annurev.biochem.76.061705.090740. [DOI] [PubMed] [Google Scholar]
- 27.Wilson DM, 3rd, Barsky D. The major human abasic endonuclease: Formation, consequences and repair of abasic lesions in DNA. Mutat Res. 2001;485(4):283–307. doi: 10.1016/s0921-8777(01)00063-5. [DOI] [PubMed] [Google Scholar]
- 28.Kelley MR, Georgiadis MM, Fishel ML. APE1/Ref-1 role in redox signaling: Translational applications of targeting the redox function of the DNA repair/redox protein APE1/Ref-1. Curr Mol Pharmacol. 2012;5(1):36–53. doi: 10.2174/1874467211205010036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bhakat KK, Izumi T, Yang SH, Hazra TK, Mitra S. Role of acetylated human AP-endonuclease (APE1/Ref-1) in regulation of the parathyroid hormone gene. EMBO J. 2003;22(23):6299–6309. doi: 10.1093/emboj/cdg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilson DM., 3rd Properties of and substrate determinants for the exonuclease activity of human apurinic endonuclease Ape1. J Mol Biol. 2003;330(5):1027–1037. doi: 10.1016/s0022-2836(03)00712-5. [DOI] [PubMed] [Google Scholar]
- 31.Harrigan JA, et al. WRN exonuclease activity is blocked by DNA termini harboring 3′ obstructive groups. Mech Ageing Dev. 2007;128(3):259–266. doi: 10.1016/j.mad.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xanthoudakis S, Smeyne RJ, Wallace JD, Curran T. The redox/DNA repair protein, Ref-1, is essential for early embryonic development in mice. Proc Natl Acad Sci USA. 1996;93(17):8919–8923. doi: 10.1073/pnas.93.17.8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guikema JE, et al. APE1- and APE2-dependent DNA breaks in immunoglobulin class switch recombination. J Exp Med. 2007;204(12):3017–3026. doi: 10.1084/jem.20071289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sabouri Z, et al. Apex2 is required for efficient somatic hypermutation but not for class switch recombination of immunoglobulin genes. Int Immunol. 2009;21(8):947–955. doi: 10.1093/intimm/dxp061. [DOI] [PubMed] [Google Scholar]
- 35.Masani S, Han L, Yu K. Apurinic/apyrimidinic endonuclease 1 is the essential nuclease during immunoglobulin class switch recombination. Mol Cell Biol. 2013;33(7):1468–1473. doi: 10.1128/MCB.00026-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walker LJ, Robson CN, Black E, Gillespie D, Hickson ID. Identification of residues in the human DNA repair enzyme HAP1 (Ref-1) that are essential for redox regulation of Jun DNA binding. Mol Cell Biol. 1993;13(9):5370–5376. doi: 10.1128/mcb.13.9.5370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim WC, et al. Characterization of the endoribonuclease active site of human apurinic/apyrimidinic endonuclease 1. J Mol Biol. 2011;411(5):960–971. doi: 10.1016/j.jmb.2011.06.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beloglazova NG, et al. Thermodynamic, kinetic and structural basis for recognition and repair of abasic sites in DNA by apurinic/apyrimidinic endonuclease from human placenta. Nucleic Acids Res. 2004;32(17):5134–5146. doi: 10.1093/nar/gkh846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Begum NA, et al. Further evidence for involvement of a noncanonical function of uracil DNA glycosylase in class switch recombination. Proc Natl Acad Sci USA. 2009;106(8):2752–2757. doi: 10.1073/pnas.0813252106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Izumi T, et al. Requirement for human AP endonuclease 1 for repair of 3′-blocking damage at DNA single-strand breaks induced by reactive oxygen species. Carcinogenesis. 2000;21(7):1329–1334. [PubMed] [Google Scholar]
- 41.Parsons JL, Dianova II, Dianov GL. APE1 is the major 3′-phosphoglycolate activity in human cell extracts. Nucleic Acids Res. 2004;32(12):3531–3536. doi: 10.1093/nar/gkh676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sartori AA, et al. Human CtIP promotes DNA end resection. Nature. 2007;450(7169):509–514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hartsuiker E, Neale MJ, Carr AM. Distinct requirements for the Rad32(Mre11) nuclease and Ctp1(CtIP) in the removal of covalently bound topoisomerase I and II from DNA. Mol Cell. 2009;33(1):117–123. doi: 10.1016/j.molcel.2008.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murai J, et al. Tyrosyl-DNA phosphodiesterase 1 (TDP1) repairs DNA damage induced by topoisomerases I and II and base alkylation in vertebrate cells. J Biol Chem. 2012;287(16):12848–12857. doi: 10.1074/jbc.M111.333963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang SW, et al. A eukaryotic enzyme that can disjoin dead-end covalent complexes between DNA and type I topoisomerases. Proc Natl Acad Sci USA. 1996;93(21):11534–11539. doi: 10.1073/pnas.93.21.11534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takashima H, et al. Mutation of TDP1, encoding a topoisomerase I-dependent DNA damage repair enzyme, in spinocerebellar ataxia with axonal neuropathy. Nat Genet. 2002;32(2):267–272. doi: 10.1038/ng987. [DOI] [PubMed] [Google Scholar]
- 47.Hakim O, et al. DNA damage defines sites of recurrent chromosomal translocations in B lymphocytes. Nature. 2012;484(7392):69–74. doi: 10.1038/nature10909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang JH, et al. Mechanisms promoting translocations in editing and switching peripheral B cells. Nature. 2009;460(7252):231–236. doi: 10.1038/nature08159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yousif AS, Stanlie A, Mondal S, Honjo T, Begum NA. Differential regulation of S-region hypermutation and class-switch recombination by noncanonical functions of uracil DNA glycosylase. Proc Natl Acad Sci USA. 2014;111(11):E1016–E1024. doi: 10.1073/pnas.1402391111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Robbiani DF, et al. AID is required for the chromosomal breaks in c-myc that lead to c-myc/IgH translocations. Cell. 2008;135(6):1028–1038. doi: 10.1016/j.cell.2008.09.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stanlie A, Yousif AS, Akiyama H, Honjo T, Begum NA. Chromatin reader Brd4 functions in Ig class switching as a repair complex adaptor of nonhomologous end-joining. Mol Cell. 2014;55(1):97–110. doi: 10.1016/j.molcel.2014.05.018. [DOI] [PubMed] [Google Scholar]
- 52.Lin CP, Ban Y, Lyu YL, Liu LF. Proteasome-dependent processing of topoisomerase I-DNA adducts into DNA double strand breaks at arrested replication forks. J Biol Chem. 2009;284(41):28084–28092. doi: 10.1074/jbc.M109.030601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang H, et al. CtIP maintains stability at common fragile sites and inverted repeats by end resection-independent endonuclease activity. Mol Cell. 2014;54(6):1012–1021. doi: 10.1016/j.molcel.2014.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Makharashvili N, et al. Catalytic and noncatalytic roles of the CtIP endonuclease in double-strand break end resection. Mol Cell. 2014;54(6):1022–1033. doi: 10.1016/j.molcel.2014.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shen J, et al. Mutations in PNKP cause microcephaly, seizures and defects in DNA repair. Nat Genet. 2010;42(3):245–249. doi: 10.1038/ng.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nagaoka H, Ito S, Muramatsu M, Nakata M, Honjo T. DNA cleavage in immunoglobulin somatic hypermutation depends on de novo protein synthesis but not on uracil DNA glycosylase. Proc Natl Acad Sci USA. 2005;102(6):2022–2027. doi: 10.1073/pnas.0409491102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zahn A, et al. Separation of function between isotype switching and affinity maturation in vivo during acute immune responses and circulating autoantibodies in UNG-deficient mice. J Immunol. 2013;190(12):5949–5960. doi: 10.4049/jimmunol.1202711. [DOI] [PubMed] [Google Scholar]
- 58.Begum NA, et al. Uracil DNA glycosylase activity is dispensable for immunoglobulin class switch. Science. 2004;305(5687):1160–1163. doi: 10.1126/science.1098444. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.