Significance
One of the central questions in biopharmaceutical development is how to extend circulation residence times of protein-based drugs to improve efficacy and patient comfort. Immunoglobulin G and serum albumin are rescued from lysosomal degradation by the pH-dependent binding to the neonatal Fc receptor and consequently have naturally long circulation half-lives. Here we describe the development and characterization of small affinity proteins (affibody molecules) that can hitchhike on the neonatal Fc receptor rescue system. We find that addition of the affibody molecules to an already half-life–extended recombinant protein leads to a nearly threefold longer circulation residence time in mice. Such affibody molecules may have a general use as fusion partners with biopharmaceuticals to extend their circulation residence times.
Keywords: affibody molecule, neonatal Fc receptor, ABD, albumin-binding domain, pharmacokinetics
Abstract
Proteins endocytosed from serum are degraded in the lysosomes. However, serum albumin (SA) and IgG, through its Fc part, bind to the neonatal Fc receptor (FcRn) at low pH in the endosome after endocytosis, and are transported back to the cellular surface, where they are released into the bloodstream, resulting in an extended serum circulation time. Association with Fc or SA has been used to prolong the in vivo half-life of biopharmaceuticals, using the interaction with FcRn to improve treatment regimens. This has been achieved either directly, by fusion or conjugation to Fc or SA, or indirectly, using SA-binding proteins. The present work takes this principle one step further, presenting small affinity proteins that bind directly to FcRn, mediating extension of the serum half-life of fused biomolecules. Phage display technology was used to select affibody molecules that can bind to FcRn in the pH-dependent manner required for rescue by FcRn. The biophysical and binding properties were characterized in vitro, and the affibody molecules were found to bind to FcRn more strongly at low pH than at neutral pH. Attachment of the affibody molecules to a recombinant protein, already engineered for increased half-life, resulted in a nearly threefold longer half-life in mice. These tags should have general use as fusion partners to biopharmaceuticals to extend their half-lives in vivo.
The use of many proteins and peptides as drugs is hampered by their relatively short plasma half-life (1), and several methods of achieving half-life extension have been investigated (2). For peptides and smaller proteins, a common approach is to attach polymers, such as polyethylene glycol (PEG), which increase hydrodynamic radius and extend half-life by decreasing kidney filtration rate (3–5). Another approach is to directly or indirectly use IgG or serum albumin (SA) for half-life extension. Both of these abundant blood proteins have an unusually long biological half-life, because they are rescued from lysosomal degradation by the neonatal Fc receptor (FcRn) (6–11).
FcRn is a heterodimeric protein residing primarily in the endosomal compartments of cells of the vascular endothelia and cells of hematopoietic origin (7). One of its functions is to rescue endocytosed IgG and SA from lysosomal degradation by binding in the endosomes, where the pH is below 6.5 (12). Subsequently, the ligand–receptor complex is transported back to the plasma membrane via recycling endosomes, followed by release of IgG and SA as the pH increases to above 7.
Expression of biopharmaceutical proteins as a genetic fusion with the Fc part of IgG molecules, SA, or fragments of SA provides a direct approach for utilization of the FcRn- mediated rescue mechanism (13), whereas fusion with a SA-binding protein provides an indirect approach (14). Several Fc-fused biopharmaceuticals have been approved by the Food and Drug Administration (15), and numerous SA-fused and SA-binding protein-fused biopharmaceuticals are in clinical development.
Both Fc-fusion proteins and SA-fusion proteins are large and complex and generally require an eukaryotic host cell for production. Additionally, wild-type Fc also has natural effector functions, including triggering of ADCC and binding to other Fc receptors, which might be undesired in some situations. Thus, it would be desirable to have smaller and simpler fusion partners that preferably can be produced in less complex hosts and with only the desired half-life extension functionality. Several SA-binding proteins, including the albumin-binding protein domain GA148-GA3 from streptococcal protein G (an ABD) (16), are small and simple and may be produced in bacterial hosts, such as Escherichia coli. However, one limitation of using SA or SA-binding proteins is that they rely on the SA–FcRn interaction, which is relatively weak. A peptide that binds directly to FcRn with independent folding and that does not require an eukaryotic host for production could be superior.
The aim of the present study was to investigate whether it is possible to generate small affinity proteins, so-called affibody molecules, to interact directly with FcRn in a pH-dependent fashion, similar to the interactions between FcRn and IgG or SA. Such affinity proteins could serve as fusion tags for extending the in vivo half-life of biopharmaceuticals, and might have complementary or even additive effects on the pharmacokinetic profile compared with currently used strategies.
Affibody molecules are a class of small affinity proteins, consisting of only 58 amino acids, with no cysteine residues in their framework (17). They can be efficiently produced recombinantly in bacteria and, as demonstrated in previous studies, their small size and independent folding allow straightforward fusion with other proteins or peptides with minimal effects on the inherent properties of the fusion partner (18). Moreover, their generally high thermostability and stability to proteolysis might be advantageous in vivo compared with unstructured peptides. In addition, clinical data support the functionality of affibody molecules in humans (19). Taken together, these characteristics suggest that affibody molecules might be suitable fusion tags for biopharmaceuticals.
In the present work, several affibody molecules with desired pH-dependent binding to FcRn were generated by selection from a combinatorial library displayed on phage. Three candidates were evaluated in vivo, showing significant half-life extension of a recombinant protein already engineered for increased half-life by indirect binding to FcRn via SA.
Results
Selection of Affibody Molecules to FcRn.
Affibody molecules interacting with the ectodomain of human FcRn (hFcRnECD) (20) were selected by biopanning from a library displayed on phage. The library was constructed essentially as described previously (21, 22), with equal representation of all amino acids, except that cysteines and prolines were omitted in the randomized positions. Selection was performed in four parallel tracks of four rounds each, with increased washing after binding and decreased amounts of target protein in later rounds (Table S1). For all tracks, the library was allowed to interact with hFcRnECD at pH 5.5. The tracks differed in elution conditions (pH 2.2 or 8.0) and in the presence or absence of human SA (HSA), which was added to prevent the selection of variants interacting with the HSA-binding site on hFcRn. The rationale was that there likely will be much competition for this site in vivo owing to the very high serum level of HSA.
After selection, an initial screen of enriched clones revealed that a majority could interact with hFcRnECD at pH ≤ 6.0; however, several of those were found to interact with the β2m part of the receptor. These clones were excluded from further work, because this interaction might lead to off-target binding to MHC class I, another heterodimeric receptor containing β2m. The variants were clustered using a nearest-neighbor algorithm. Analysis of the results from the initial screen together with the clustering results identified four related variants that did not interact with β2m and had the desired pH-dependent binding profile that would be required for FcRn-mediated rescue. These variants appeared only in selection tracks in which HSA was included. Thus, another 930 clones from these two tracks were analyzed, resulting in 12 additional related but unique variants.
ELISA-Based Screening of Selected Variants.
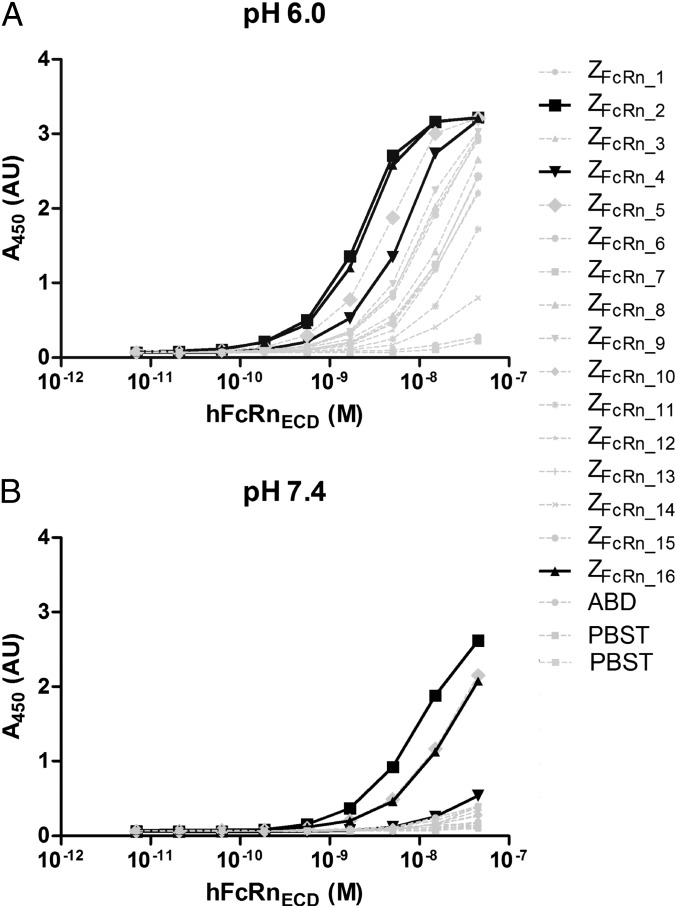
The 16 variants were expressed as fusions to an ABD, followed by an ELISA analysis demonstrating that all could interact with hFcRnECD at pH 6.0 (Fig. 1A). All variants showed drastically reduced binding when the assay was performed at pH 7.4 (Fig. 1B). The data presented in Fig. 1 were used to rank the affibody molecules’ affinities for FcRn at pH 6.0 and 7.4. The affibody molecules with the highest affinities at pH 6.0 were ZFcRn_2, ZFcRn_16, ZFcRn_5, and ZFcRn_4, with ZFcRn_2 and ZFcRn_16 exhibiting similar profiles. At pH 7.4, ZFcRn_2, ZFcRn_5, and ZFcRn_16 were the strongest binders, with ZFcRn_5 and ZFcRn_16 showing similar binding profiles. All variants had a substantially lower affinity for hFcRnECD at pH 7.4 than at pH 6.0.
Fig. 1.
ELISA analysis of the interaction between the selected affibody molecules and hFcRnECD. An anti-ABD antibody was immobilized in the bottom of the wells and used to capture the affibody-ABD fusion proteins. A dilution series of biotinylated hFcRnECD was added to the wells at pH 6.0 (A) or 7.4 (B), followed by development of the ELISA by the addition of HRP-conjugated streptavidin and 3,3′,5,5′-tetramethylbenzidine (TMB) substrate. The x-axis corresponds to the amount of added hFcRnECD, and the y-axis corresponds to the A450 measured after development.
Based on the foregoing results, ZFcRn_2 and ZFcRn_16 were chosen for further investigation, because they had the highest affinity at pH 6.0. ZFcRn_4 was also included in further studies, because the difference in affinity between pH 6.0 and pH 7.4 appeared to be large, which could prove advantageous for its ability to be recycled by FcRn; that is, it would be efficiently captured in the endosomes and efficiently released after recycling when encountering pH 7.4.
Further ELISA analysis of the 16 variants revealed no measurable affinity for human β2m or for a control protein (human polyclonal IgG) at pH 6.0 or 7.4 (Fig. S1). Therefore, it is likely that they interact solely with the α-chain of hFcRn or with both chains in a manner requiring the presence of the α-chain. The absence of binding to polyclonal human IgG indicates that these binders do not nonspecifically interact with proteins in general.
Binding of Selected Affibody Variants to FcRn.
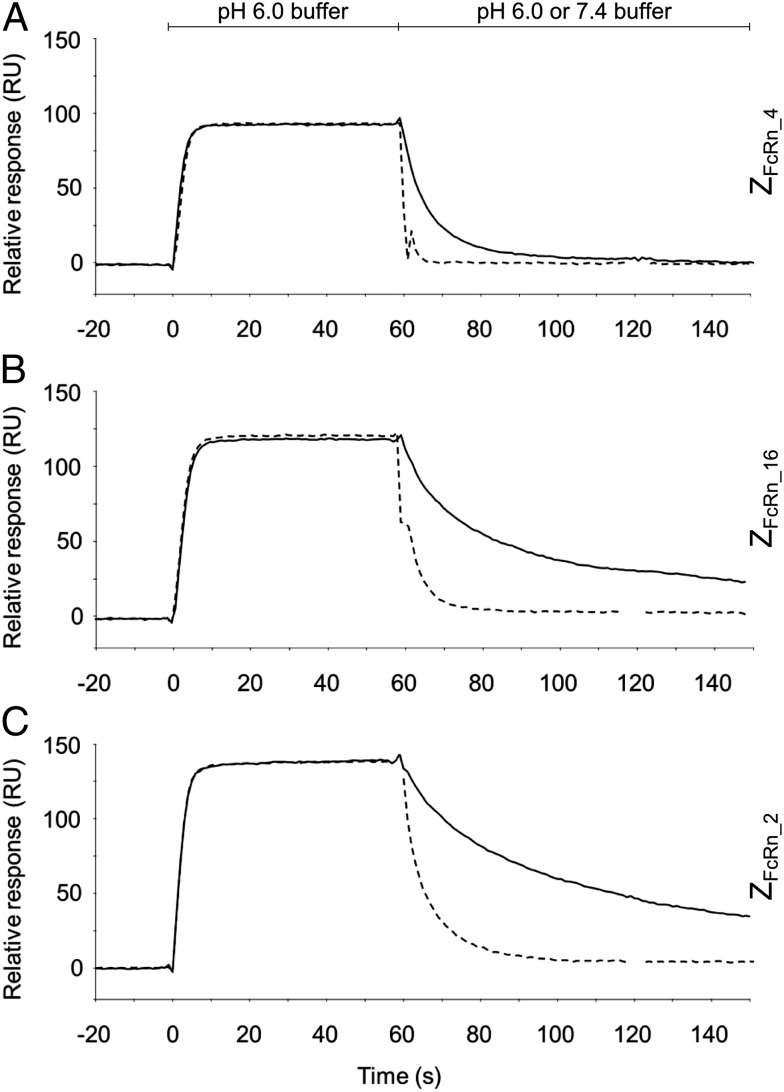
ZFcRn_4, ZFcRn_16, and ZFcRn_2 were subcloned and produced with an N-terminal His6 tag. The affinity between the affibody molecules and human and murine FcRnECD were determined by biosensor analysis (Table 1). The affinities for all three variants against both receptors were considerably higher at pH 6.0 than at pH 7.4. In addition, the off-rates at pH 6.0 and pH 7.4 were compared by biosensor analysis. The affibody molecules were injected at pH 6.0 over a flow cell with immobilized hFcRnECD, followed by injection of buffer at pH 6.0 or 7.4. All three variants displayed considerably faster off-rates at pH 7.4 than at pH 6.0 (Fig. 2).
Table 1.
Characterization of affibody molecules
| Affibody molecule | Affinity for hFcRn, nM | Affinity for mFcRn, nM | Melting temperature, °C | ||
| pH 6.0 | pH 7.4 | pH 6.0 | pH 7.4 | ||
| ZFcRn_4 | 50 | >1,000 | 350 | >5,000 | 58 |
| ZFcRn_16 | 14 | >500 | 95 | >500 | 59 |
| ZFcRn_2 | 12 | >500 | 90 | >500 | 49 |
Fig. 2.
Off-rate analysis of the interaction between affibody molecules and hFcRnECD by surface plasmon resonance. Affibody molecules were injected over a flow cell with immobilized hFcRnECD at pH 6.0, followed by immediate injection of buffer with pH 6.0 (solid line) or 7.4 (dashed line). (A) ZFcRn_4. (B) ZFcRn_16. (C) ZFcRn_2.
Analysis of the Selected Affibody Molecules by Circular Dichroism Spectroscopy.
The melting temperature of the affibody molecules as measured by circular dichroism spectroscopy, ranged from 49 to 59 °C (Table 1). Spectra from 250 to 195 nm were also collected before and after heating to 90 °C (Fig. S2). These were highly similar, suggesting reversible protein folding after heat denaturation.
Binding to the Human and Mouse Orthologs of Full-Length FcRn on Cells.
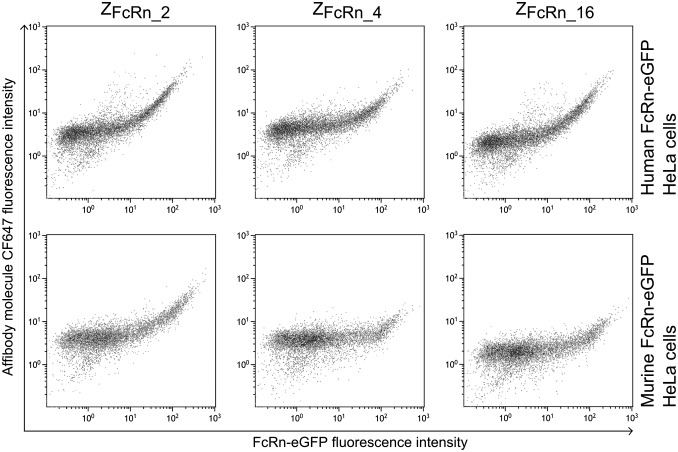
HeLa cells expressing the human or murine orthologs of FcRn as a fusion to eGFP with varying eGFP/FcRn expression (Fig. S3) level were constructed previously (20). The cells were incubated with CF647-labeled ZFcRn_4, ZFcRn_16, or ZFcRn_2 at pH 6.0, followed by flow cytometry analysis (Fig. 3). The intensity of the affibody signal increased with increasing amounts of FcRn on the target cell; that is, cells with a higher eGFP/FcRn signal had a higher CF647/affibody signal. All three affibody molecules were found to bind to both the human and the murine orthologs of FcRn.
Fig. 3.
Flow cytometry analysis of affibody molecules binding the human and murine orthologs of FcRn. HeLa cells expressing hFcRn-eGFP or mFcRn-eGFP was permeabilized and stained with CF647-labeled affibody molecules. The cells were analyzed by flow cytometry, and CF647 fluorescence was plotted on the y-axis as a function of eGFP fluorescence on the x-axis.
Construction of Fusion Proteins and Biochemical Characterization.
In preparation for in vivo analysis, ZFcRn_4, ZFcRn_16, and ZFcRn_2 were subcloned and expressed in the format ZFcRn-ABD-ZTaq, where ZFcRn is the FcRn-binding affibody molecule, ZTaq is a model protein (an affibody molecule binding to DNA polymerase from Thermus aquaticus), and ABD is a deimmunized high-affinity ABD (23, 24). A control fusion protein, ZTaq-ABD, was constructed as well. Biosensor analysis of the affinity between the fusion proteins and hFcRn showed an approximately fourfold weaker interaction compared with the affinity between the free ZFcRn domains and hFcRn.
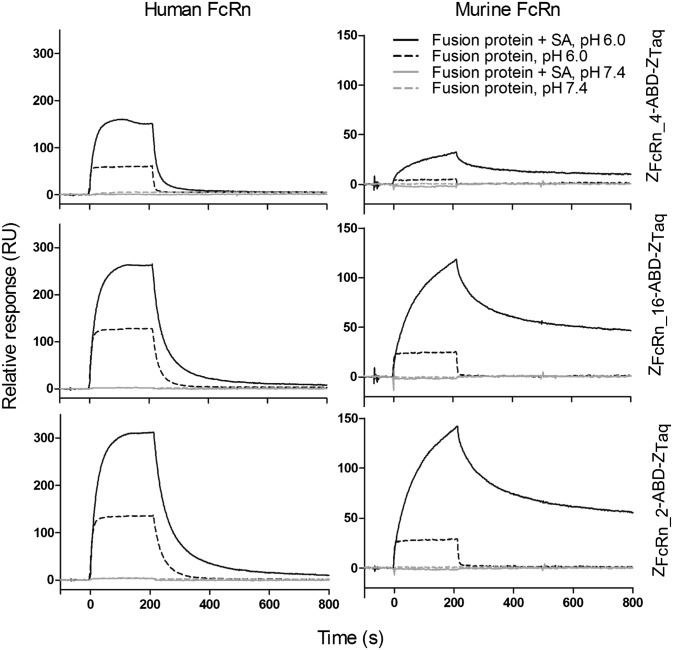
For investigating possible cooperativity between ZFcRn and ABD/SA when binding to FcRn, the fusion proteins were injected over surfaces with immobilized human or murine FcRn in the absence or presence of SA (Fig. 4). At pH 6.0, the fusion proteins can bind to hFcRn and mFcRn in both the presence and the absence of SA. The difference in off-rate for the interactions between the fusion proteins and murine FcRn was two orders of magnitude lower in the presence of mouse SA (MSA), indicating cooperativity between ZFcRn and ABD/SA in the interaction (Table 2). The difference in off-rate for the interactions between the fusion proteins and human FcRn was only marginally slower in the presence of HSA, indicating cooperativity in this case as well, but to a much lesser extent.
Fig. 4.
Surface plasmon resonance analysis of the interaction between the fusion proteins and FcRnECD. Fusion proteins were injected with or without incubation for 4 h with SA at pH 6.0 or 7.4. HSA or MSA was used, depending on the origin of FcRnECD.
Table 2.
Analysis of the apparent off-rate and in vivo half-lives of the fusion proteins
| Fusion protein | koff hFcRn, s−1* | koff mFcRn, s−1† | In vivo half-life, h | ||
| −HSA | +HSA | −MSA | +MSA | ||
| ZFcRn_4-ABD-ZTaq | 0.2 | 0.05 | ---‡ | 0.006 | 73 |
| ZFcRn_16-ABD-ZTaq | 0.04 | 0.02 | 0.3 | 0.006 | 91 |
| ZFcRn_2-ABD-ZTaq | 0.03 | 0.02 | 0.2 | 0.006 | 61 |
| ZTaq-ABD | NB | NB | NB | NB | 33 |
NB, not binding.
The analysis was performed in the presence or absence of HSA.
The analysis was performed in the presence or absence of MSA.
The off-rate was too fast to be measured with the current setup.
For both receptors, the equilibrium responses were clearly higher in the presence of SA, indicating that it is the complex of the fusion protein and SA that is interacting with FcRn, not just the fusion protein by itself. In addition, free SA did not interact with the receptor at the concentration used (200 nM), further supporting that it is the complex and not free SA that is interacting (Fig. S4) (20). At pH 7.4, no interaction between the fusion proteins and hFcRn or mFcRn was observed at the concentration used (Fig. 4). As expected, the control, ZTaq-ABD, did not interact with human or murine FcRn at the concentrations used (Fig. S4).
Serum Circulation Half-Life Extension by the Affibody Molecules.
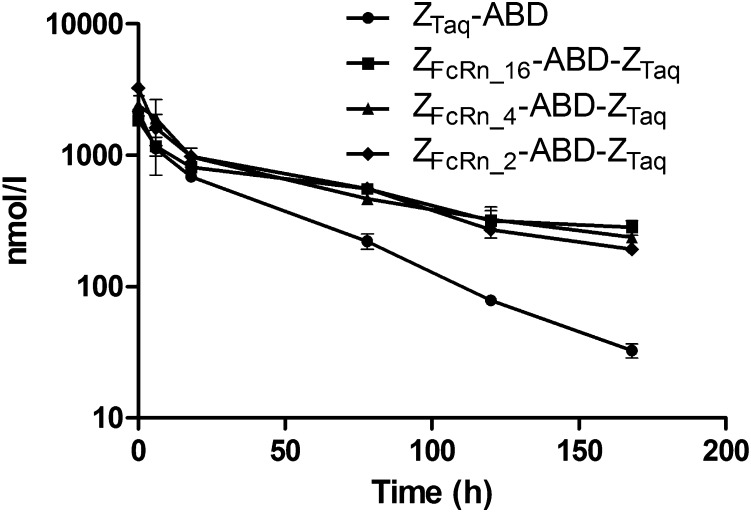
Even though the affibody molecules were selected to bind to the human ortholog of FcRn, the reactivity to the murine ortholog allowed evaluation of half-life extension in mice. The three ZFcRn-ABD-ZTaq fusion proteins and the control were injected into mice, and serum concentrations were measured at different time points. ABD fusion proteins have been shown to have a long circulation half-life caused by ABD binding to SA, followed by FcRn-mediated rescue from lysosomal degradation (25, 26). In agreement with the previous results, the terminal half-life of ZTaq-ABD was ∼33 h. This half-life is also comparable to the half-life of MSA (35 h). The terminal half-lives of the constructs containing ZFcRn_4, ZFcRn_16, or ZFcRn_2 were 1.8- to 2.7-fold longer than the half-life of the control protein (Fig. 5). The calculated terminal half-lives were 91 h for ZFcRn_16, 73 h for ZFcRn_4, and 61 h for ZFcRn_2, demonstrating that the FcRn-binding affibody molecules contribute to extension of the half-life, and that the combination with ABD yields a significantly longer half-life compared with the half-life extension achieved by ABD alone.
Fig. 5.
Measurement of the half-life of the fusion proteins in vivo. The constructs were i.v. injected into mice at time 0, and groups of three mice were euthanized at different time points. The concentration of fusion protein in serum samples was determined by ELISA and plotted on the y-axis as a function of time after injection on the x-axis. Error bars correspond to 1 SD.
Discussion
Taking advantage of FcRn-mediated recycling is an efficient way to prolong the half-lives of biological molecules, and has been a topic of intense study in recent years. Successful fusion partners for extending the half-life include the Fc part of IgG, SA, and SA-binding proteins. Recently, short peptides interacting directly with FcRn were described (27). These peptides were found to allow FcRn-based recycling of an attached protein by FcRn-overexpressing cells in vitro. It is presently not known whether these peptides are also able to extend the serum half-life of a fused protein in vivo.
The most widely studied class of proteins using FcRn-mediated recycling is IgG. It has been found that, on average, a small fraction of IgG fails to be recycled by FcRn in each endocytosis event and is subsequently degraded (28). This realization led to the development of therapeutic mAbs with enhanced affinity for FcRn at pH 6, with significantly extended half-life compared with natural IgG (29–31). These mAbs compete favorably with endogenous IgG for FcRn rescue and thus have a lower probability of becoming degraded. Conflicting findings have been published, however, in which an increase in affinity at pH 6 was not accompanied by an extended half-life (32). It has been suggested that half-life extension is instead dependent on the combination of several parameters, including favorable competition at lower pH (after endocytosis) and very low affinity at pH 7.4 (33). The results of the present study tend to support this suggestion. We found that the fusion protein containing ZFcRn_4 had a longer terminal half-life than the protein containing ZFcRn_2 even though ZFcRn_2 had a higher affinity for mFcRnECD at pH 6.0 (Table 1). At pH 7.4, ZFcRn_4 had a considerably lower affinity for mFcRnECD than ZFcRn_2, which could explain why the fusion protein containing ZFcRn_4 had a longer serum half-life than the fusion protein containing ZFcRn_2.
A conserved sequence motif was seen in ZFcRn_2, ZFcRn_4, and ZFcRn_16 (Table S2), which included two histidines in positions 14 and 32. Histidine is the only amino acid that normally changes its protonation state in the relevant pH interval, and thus a possible explanation for the large difference in affinity at pH 6.0 and 7.4 could be that one or both histidines are involved in the interaction and change their protonation state with changes in pH. Also, histidines are known to be important for the pH-dependent interaction between IgG or SA and FcRn (6, 34, 35).
Three FcRn-binding affibody variants were evaluated in vivo. This was done in the context of a fusion protein consisting of an FcRn-specific affibody variant genetically fused to a high-affinity ABD and the model protein ZTaq. This setup allowed for a direct comparison with previous data with the ABD (26). In vivo, ABD will associate with SA to make the fusion protein complex sufficiently large to avoid renal clearance and will increase the half-life by FcRn-mediated recycling. This led to a half-life extension from less than 1 h for the model protein to up to 33 h. The addition of FcRn-binding affibody molecules to this already half-life–extended fusion protein led to a nearly threefold further increase in serum circulation time. How much of the increased half-life is a consequence of the individual domains (FcRn-binding affibody molecule or ABD) and how much is a consequence of a potential cooperativity when interacting with FcRn is not known; however, surface plasmon resonance analysis revealed cooperativity between ZFcRn and ABD (through SA), which might be present in vivo as well (Fig. 4). The cooperativity is dependent on the concentration of FcRn, and because the local concentration of FcRn in the endosomes is not known, the contribution of the cooperativity in vivo cannot be quantified. Nonetheless, the half-life disparity among the fusion proteins demonstrates that the FcRn-binding affibody molecules contribute significantly to the half-life extension. There likely is room for further improvement in half-life of the ZFcRn-ABD fusion tag, at least to that of Fc-engineered IgGs, which might not be the upper limit.
In conclusion, we have used phage display technology with functional pH elution (binding at pH 5.5 and elution at pH 2.2 or 8.0) to isolate affibody variants with specific pH-dependent binding to FcRn. The variants were found to bind to the full-length versions of both the human and murine orthologs. The best-performing variant could increase the serum circulation half-life of a model protein, already engineered for an extended half-life, by close to threefold. We envision that affibody molecules, such as those described in this work, as well as ABDs described previously, can be coupled in a modular fashion to endow biopharmaceuticals with desired FcRn-mediated functions, such as an extended half-life.
Materials and Methods
Selection of Affibody Molecules.
The ectodomain of human FcRn (hFcRnECD) (20), produced in SKOV-3 cells, was biotinylated by EZ-Link Sulfo-NHS-LC-Biotin (Thermo Scientific) and used as a target for the selection of affibody molecules. The selections were performed by four rounds of biopanning, allowing phages to bind in selection buffer (100 mM sodium phosphate pH 5.5, 150 mM sodium chloride, 0.1% Tween-20, and 0.1% gelatin) in the presence or absence of 1.5 µM HSA. Elution was performed at pH 8.0 (100 mM sodium phosphate, 150 mM sodium chloride) or at pH 2.2 (50 mM glycin-HCl). The DNA sequences of enriched clones were determined, and the corresponding amino acid sequences were clustered based on sequence similarity using an average-link hierarchical clustering method (36).
Protein Production from the Phagemid Vector and ELISA.
The clones were directly expressed from the phagemid vector as an N-terminal fusion to ABD. The clones were expressed in 1 mL of cultures in Escherichia coli at 37 °C and the protein accumulated in the periplasmic space after induction with 1 mM isopropyl-β-d-1-thiogalactopyranoside (IPTG). The periplasmic fractions of the cultures were released by freeze-thawing and the interactions with hFcRnECD, β2m, and hIgG were investigated by ELISA using standard techniques. In brief, a goat anti-ABD antibody (in-house produced) at a concentration of 2 µg/mL was immobilized in the bottom of a multiwell plate, followed by capture of the expressed affibody variant from the periplasmic extracts. For affinity ranking, a dilution series (1:3 dilution from 2 µg/mL to 0.3 ng/mL) of biotinylated hFcRnECD was added to the wells. For specificity analysis, biotinylated hβ2m (5 µg/mL) or hIgG (5 µg/mL) was added to the wells. The ELISAs were developed by HRP-conjugated streptavidin (1:30,000; Thermo Scientific) and substrate (Thermo Scientific), followed by measurement of A450.
Subcloning and Expression of Affibody Molecules.
Sixteen affibody variants were subcloned and produced under control of the T7-promoter as single domains with an N-terminal His6-tag in E. coli BL21(DE3) at 37 °C. The affibody molecules were subsequently purified by immobilized metal-ion affinity chromatography using His GraviTrap columns (GE Healthcare) according to the manufacturer’s protocol. ZFcRn_4, ZFcRn_16, and ZFcRn_2 were also subcloned as fusions to a high-affinity ABD (23) and ZTaq (37) with (G4S)4 linkers connecting the domains, resulting in fusion proteins with the format ZFcRn-ABD-ZTaq. During the subcloning, the two N-terminal amino acids in the FcRn-binding affibody molecules were changed from VD to AE. The control was constructed similarly and consisted of ZTaq and ABD connected by a (G4S)4 linker. The fusion proteins were expressed in E. coli BL21(DE3) at 37 °C after induction by 1 mM IPTG and were purified by affinity chromatography. After purification, potential residual endotoxins were removed by passing the proteins through 1-mL EndoTrap red columns (Hyglos), according to the manufacturer’s protocol.
In Vitro Characterization of Proteins.
The molecular masses of all proteins were analyzed with an Agilent 1100 LC-MSD, and data agreed with the expected molecular masses. Circular dichroism spectra of ZFcRn_4, ZFcRn_16, and ZFcRn_2 were collected at a concentration of 0.5 mg/mL in PBS on a Jasco J-810 spectropolarimeter. Each sample was analyzed from 195 to 250 nm at 20 °C, before and after variable temperature measurement (VTM). The VTM was performed by measuring the ellipticity at 221 nm, during heating from 20 to 90 °C with a temperature gradient of 5 °C/min.
The affinity of the interactions between the human and murine FcRnECD and the affibody molecules, expressed as monomers with a His6 tag or as fusion to ABD and ZTaq, were measured by biosensor analysis on a Biacore 3000 instrument (GE Healthcare). Human and murine FcRnECD was immobilized on separate flow cells on a CM5 chip in acetate buffer at pH 4.65. The immobilization level was ∼500 RU for experiments with the fusion proteins and ∼2,000 RU for experiments with the affibody monomers. Reference flow cells were created by activation and deactivation. McIlvaine’s phosphate-citrate buffer (pH 6.0 or 7.4), supplemented with 0.005% Tween-20, was used as running buffer and for dilution of the analytes. All analyses were performed at 25 °C with a flow rate of 50 μL/min. The affinity constants were determined by injecting a dilution series from 1,024 nM to 0.5 nM (pH 6.0) or from 10,240 nM to 5 nM (pH 7.4). The affinities were derived using GraphPad Prism 5 software, using a one-site binding saturation model.
For off-rate analysis of monomeric constructs, the proteins were injected in running buffer (pH 6.0), followed by injection of buffer at pH 6.0 (running buffer) or buffer at pH 7.4 (McIlvaine’s phosphate-citrate buffer, pH 7.4, supplemented with 0.005% Tween-20) using the coinject procedure.
For cooperativity analysis (Fig. 4 and Fig. S4), 100 nM fusion protein was injected with or without previous incubation (4 h) with 200 nM HSA or MSA over the flow cells with immobilized human or murine FcRnECD, respectively. The off-rates were calculated using GraphPad Prism 5 software.
Binding of the affibody molecules to full-length human and murine FcRn, expressed as fusion to eGFP in HeLa cells (20), was investigated by flow cytometry. Direct labeling of the affibody molecules with CF647 (Sigma-Aldrich) was performed according to the manufacturer’s protocol. Cells were detached by trypsination and were fixed with 50 µL of formaldehyde (2%; Sigma-Aldrich) in PBS for 10 min at room temperature, followed by staining of ∼100,000 cells/sample with the labeled variants in PBS containing 0.1% saponin at pH 6.0. A total of 10,000 cells/sample were analyzed in a Gallios flow cytometer (Beckman Coulter), and the data were analyzed using Kaluza software (Beckman Coulter).
In Vivo Analysis of Fusion Proteins.
The study was reviewed and approved by the regional animal experimental ethics committee in Stockholm (North) and performed by Adlego AB. The affibody variants as fusions to ABD and ZTaq were administered i.v. to male NMRI mice (Charles River) at a dose of 92 nmol/kg body weight. Sera from groups of three mice per time point were obtained at 0.08, 6, 18, 78, 120, and 168 h. The concentration of each affibody variant was determined by ELISA. Half-area 96-well ELISA plates (Corning) were coated with a goat anti-affibody antibody, followed by the addition of sera samples diluted 1:100–1:100 000. Bound affibody variants were detected with rabbit anti-ABD IgG (4 µg/mL; Affibody AB), followed by an HRP-conjugated donkey anti-rabbit antibody (1:20,000; Jackson Laboratories). The wells were developed by ImmunoPure TMB substrate (Pierce) according to the manufacturer’s protocol, and A450 was determined. The concentration of the fusion proteins was determined by comparing the A450 of the samples with the A450 obtained from standard curves using the pure proteins of known concentration. The concentrations in the serum samples were then plotted as their natural logarithms against time. The resulting curves followed a two-compartment model, and the terminal half-life was calculated from the slopes.
Supplementary Material
Acknowledgments
We thank Finn Dunås, Per Jonasson, and Gunilla Fant for technical assistance, and Eva Allerbring and Adnane Achour for their kind gift of the β2m. Funding of the project was provided by Knut och Alice Wallenbergs Stiftelse.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1417717111/-/DCSupplemental.
References
- 1.Beck A, Reichert JM. Therapeutic Fc-fusion proteins and peptides as successful alternatives to antibodies. MAbs. 2011;3(5):415–416. doi: 10.4161/mabs.3.5.17334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kontermann RE. Strategies for extended serum half-life of protein therapeutics. Curr Opin Biotechnol. 2011;22(6):868–876. doi: 10.1016/j.copbio.2011.06.012. [DOI] [PubMed] [Google Scholar]
- 3.Yamaoka T, Tabata Y, Ikada Y. Distribution and tissue uptake of poly(ethylene glycol) with different molecular weights after intravenous administration to mice. J Pharm Sci. 1994;83(4):601–606. doi: 10.1002/jps.2600830432. [DOI] [PubMed] [Google Scholar]
- 4.Caliceti P, Veronese FM. Pharmacokinetic and biodistribution properties of poly(ethylene glycol)-protein conjugates. Adv Drug Deliv Rev. 2003;55(10):1261–1277. doi: 10.1016/s0169-409x(03)00108-x. [DOI] [PubMed] [Google Scholar]
- 5.Alconcel SNS, Baas AS, Maynard HD. FDA-approved poly(ethylene glycol)–protein conjugate drugs. Polym Chem. 2011;2:1442–1448. [Google Scholar]
- 6.Andersen JT, et al. Structure-based mutagenesis reveals the albumin-binding site of the neonatal Fc receptor. Nat Commun. 2012;3:610. doi: 10.1038/ncomms1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roopenian DC, Akilesh S. FcRn: The neonatal Fc receptor comes of age. Nat Rev Immunol. 2007;7(9):715–725. doi: 10.1038/nri2155. [DOI] [PubMed] [Google Scholar]
- 8.Brambell FW. The transmission of immunity from mother to young and the catabolism of immunoglobulins. Lancet. 1966;2(7473):1087–1093. doi: 10.1016/s0140-6736(66)92190-8. [DOI] [PubMed] [Google Scholar]
- 9.Brambell FW, Hemmings WA, Morris IG. A theoretical model of gamma-globulin catabolism. Nature. 1964;203:1352–1354. doi: 10.1038/2031352a0. [DOI] [PubMed] [Google Scholar]
- 10.Junghans RP, Anderson CL. The protection receptor for IgG catabolism is the beta2-microglobulin–containing neonatal intestinal transport receptor. Proc Natl Acad Sci USA. 1996;93(11):5512–5516. doi: 10.1073/pnas.93.11.5512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andersen JT, Sandlie I. The versatile MHC class I-related FcRn protects IgG and albumin from degradation: Implications for development of new diagnostics and therapeutics. Drug Metab Pharmacokinet. 2009;24(4):318–332. doi: 10.2133/dmpk.24.318. [DOI] [PubMed] [Google Scholar]
- 12.Raghavan M, Gastinel LN, Bjorkman PJ. The class I major histocompatibility complex related Fc receptor shows pH-dependent stability differences correlating with immunoglobulin binding and release. Biochemistry. 1993;32(33):8654–8660. doi: 10.1021/bi00084a037. [DOI] [PubMed] [Google Scholar]
- 13.Chuang VTG, Kragh-Hansen U, Otagiri M. Pharmaceutical strategies utilizing recombinant human serum albumin. Pharm Res. 2002;19(5):569–577. doi: 10.1023/a:1015396825274. [DOI] [PubMed] [Google Scholar]
- 14.Stork R, Campigna E, Robert B, Müller D, Kontermann RE. Biodistribution of a bispecific single-chain diabody and its half-life extended derivatives. J Biol Chem. 2009;284(38):25612–25619. doi: 10.1074/jbc.M109.027078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Czajkowsky DM, Hu J, Shao Z, Pleass RJ. Fc-fusion proteins: New developments and future perspectives. EMBO Mol Med. 2012;4(10):1015–1028. doi: 10.1002/emmm.201201379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Makrides SC, et al. Extended in vivo half-life of human soluble complement receptor type 1 fused to a serum albumin-binding receptor. J Pharmacol Exp Ther. 1996;277(1):534–542. [PubMed] [Google Scholar]
- 17.Nygren P-A. Alternative binding proteins: Affibody binding proteins developed from a small three-helix bundle scaffold. FEBS J. 2008;275(11):2668–2676. doi: 10.1111/j.1742-4658.2008.06438.x. [DOI] [PubMed] [Google Scholar]
- 18.Löfblom J, et al. Affibody molecules: Engineered proteins for therapeutic, diagnostic and biotechnological applications. FEBS Lett. 2010;584(12):2670–2680. doi: 10.1016/j.febslet.2010.04.014. [DOI] [PubMed] [Google Scholar]
- 19.Baum RP, et al. Molecular imaging of HER2-expressing malignant tumors in breast cancer patients using synthetic 111In- or 68Ga-labeled affibody molecules. J Nucl Med. 2010;51(6):892–897. doi: 10.2967/jnumed.109.073239. [DOI] [PubMed] [Google Scholar]
- 20.Seijsing J, Lindborg M, Löfblom J, Uhlén M, Gräslund T. Robust expression of the human neonatal Fc receptor in a truncated soluble form and as a full-length membrane-bound protein in fusion with eGFP. PLoS ONE. 2013;8(11):e81350. doi: 10.1371/journal.pone.0081350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nord K, et al. Binding proteins selected from combinatorial libraries of an alpha-helical bacterial receptor domain. Nat Biotechnol. 1997;15(8):772–777. doi: 10.1038/nbt0897-772. [DOI] [PubMed] [Google Scholar]
- 22.Grönwall C, et al. Selection and characterization of Affibody ligands binding to Alzheimer amyloid beta peptides. J Biotechnol. 2007;128(1):162–183. doi: 10.1016/j.jbiotec.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 23.Jonsson A, Dogan J, Herne N, Abrahmsén L, Nygren P-A. Engineering of a femtomolar affinity binding protein to human serum albumin. Protein Eng Des Sel. 2008;21(8):515–527. doi: 10.1093/protein/gzn028. [DOI] [PubMed] [Google Scholar]
- 24.Frejd F. 2012. in Therapeutic Proteins: Strategies to Modulate Their Plasma Half-Lives, ed Kontermann R (Wiley-Blackwell, Hoboken, NJ), pp 269–283.
- 25.Hopp J, et al. The effects of affinity and valency of an albumin-binding domain (ABD) on the half-life of a single-chain diabody-ABD fusion protein. Protein Eng Des Sel. 2010;23(11):827–834. doi: 10.1093/protein/gzq058. [DOI] [PubMed] [Google Scholar]
- 26.Orlova A, et al. Site-specific radiometal labeling and improved biodistribution using ABY-027, a novel HER2-targeting affibody molecule albumin-binding domain fusion protein. J Nucl Med. 2013;54(6):961–968. doi: 10.2967/jnumed.112.110700. [DOI] [PubMed] [Google Scholar]
- 27.Mezo AR, et al. Reduction of IgG in nonhuman primates by a peptide antagonist of the neonatal Fc receptor FcRn. Proc Natl Acad Sci USA. 2008;105(7):2337–2342. doi: 10.1073/pnas.0708960105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim J, et al. Albumin turnover: FcRn-mediated recycling saves as much albumin from degradation as the liver produces. Am J Physiol Gastrointest Liver Physiol. 2006;290(2):G352–G360. doi: 10.1152/ajpgi.00286.2005. [DOI] [PubMed] [Google Scholar]
- 29.Hinton PR, et al. Engineered human IgG antibodies with longer serum half-lives in primates. J Biol Chem. 2004;279(8):6213–6216. doi: 10.1074/jbc.C300470200. [DOI] [PubMed] [Google Scholar]
- 30.Petkova SB, et al. Enhanced half-life of genetically engineered human IgG1 antibodies in a humanized FcRn mouse model: Potential application in humorally mediated autoimmune disease. Int Immunol. 2006;18(12):1759–1769. doi: 10.1093/intimm/dxl110. [DOI] [PubMed] [Google Scholar]
- 31.Zalevsky J, et al. Enhanced antibody half-life improves in vivo activity. Nat Biotechnol. 2010;28(2):157–159. doi: 10.1038/nbt.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gurbaxani B, Dela Cruz LL, Chintalacharuvu K, Morrison SL. Analysis of a family of antibodies with different half-lives in mice fails to find a correlation between affinity for FcRn and serum half-life. Mol Immunol. 2006;43(9):1462–1473. doi: 10.1016/j.molimm.2005.07.032. [DOI] [PubMed] [Google Scholar]
- 33.Datta-Mannan A, et al. FcRn affinity-pharmacokinetic relationship of five human IgG4 antibodies engineered for improved in vitro FcRn binding properties in cynomolgus monkeys. Drug Metab Dispos. 2012;40(8):1545–1555. doi: 10.1124/dmd.112.045864. [DOI] [PubMed] [Google Scholar]
- 34.Andersen JT, Dee Qian J, Sandlie I. The conserved histidine 166 residue of the human neonatal Fc receptor heavy chain is critical for the pH-dependent binding to albumin. Eur J Immunol. 2006;36(11):3044–3051. doi: 10.1002/eji.200636556. [DOI] [PubMed] [Google Scholar]
- 35.Medesan C, Matesoi D, Radu C, Ghetie V, Ward ES. Delineation of the amino acid residues involved in transcytosis and catabolism of mouse IgG1. J Immunol. 1997;158(5):2211–2217. [PubMed] [Google Scholar]
- 36.Orlova A, et al. Tumor imaging using a picomolar affinity HER2 binding affibody molecule. Cancer Res. 2006;66(8):4339–4348. doi: 10.1158/0008-5472.CAN-05-3521. [DOI] [PubMed] [Google Scholar]
- 37.Gunneriusson E, Nord K, Uhlén M, Nygren P. Affinity maturation of a Taq DNA polymerase-specific affibody by helix shuffling. Protein Eng. 1999;12(10):873–878. doi: 10.1093/protein/12.10.873. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.