Significance
To our knowledge, as the first genetic-based approach for the simultaneous isolation and intracellular genetic analysis of live circulating tumor cells, NanoFlares provide opportunities for invasive cancer study, diagnosis, prognosis, and personalized therapy.
Keywords: cancer metastasis, nanotechnology, diagnostic, mRNA, NanoFlares
Abstract
Metastasis portends a poor prognosis for cancer patients. Primary tumor cells disseminate through the bloodstream before the appearance of detectable metastatic lesions. The analysis of cancer cells in blood—so-called circulating tumor cells (CTCs)—may provide unprecedented opportunities for metastatic risk assessment and investigation. NanoFlares are nanoconstructs that enable live-cell detection of intracellular mRNA. NanoFlares, when coupled with flow cytometry, can be used to fluorescently detect genetic markers of CTCs in the context of whole blood. They allow one to detect as few as 100 live cancer cells per mL of blood and subsequently culture those cells. This technique can also be used to detect CTCs in a murine model of metastatic breast cancer. As such, NanoFlares provide, to our knowledge, the first genetic-based approach for detecting, isolating, and characterizing live cancer cells from blood and may provide new opportunities for cancer diagnosis, prognosis, and personalized therapy.
Metastatic tumor formation is instigated by dissemination of cancer cells from the primary tumor through the bloodstream to new organ sites (1, 2). Current methods to definitively identify metastatic cancer, including clinical presentation and radiographic evaluation, often rely on the existence of overt distal tumors (3, 4). Clinical treatments administered after metastatic tumor diagnosis are often ineffective, and survival rates are poor (3, 5, 6). The grave impact of metastasis on mortality necessitates the development of novel methodologies that enable early and accurate assessment of metastatic risk before the establishment of secondary tumors so that available therapies can be administered and patient prognoses can be improved.
Tumor metastasis is believed to progress through a series of steps by which cancer cells originating at a primary tumor intravasate into the blood stream, travel through the blood stream, extravasate, and form secondary tumors at distal sites (2, 7). Recent studies have indicated that metastatic tumor cells frequently hijack the developmental program of epithelial–mesenchymal transition (EMT) for dissemination to distant organs (7, 8). At the molecular level, EMT involves the cellular loss of epithelial markers (e.g., E-cadherin, a transmembrane cell adhesion protein) and increased expression of mesenchymal markers (e.g., Vimentin, a type II intermediate filament protein; Fibronectin, an extracellular matrix binding glycoprotein) (7–10). The loss of intercellular contacts and apical–basal polarity during EMT results in a more invasive and migratory phenotype of tumor cells. Ultimately, it is believed that cancer cells that have entered the bloodstream must also reverse the process of EMT to colonize distal organs (11).
Circulating tumor cells (CTCs) (3–5, 12–14) offer the opportunity to detect cancer metastasis before distal tumor formation. Recent studies indicate that the presence of CTCs before treatment indicates poor survival in patients with metastatic cancer, and the presence of CTCs after treatment is associated with tumor relapse (3–5, 12–14). CTC molecular expression profiles, however, are largely heterogeneous within a cancer and a single patient, and evidence suggests that certain subpopulations are prone to seed metastases (15). Current methods for isolating CTCs from patient peripheral blood rely on immunomagnetic separation via cell-surface antigen recognition (16). Sole reliance on the signal from one or a few cell-surface proteins can exclude CTC subpopulations that do not express significant levels of the appropriate proteins necessary for isolation [e.g., epithelial cell adhesion molecule (EpCAM)] or detection (e.g., cytokeratins 8 and 12) (16) and does not offer genetic information at the single-cell level (14, 16). New genetic-based technologies that isolate live CTC peripheral blood populations with high recovery and fidelity and facilitate downstream single-cell molecular analysis will provide a method for clinicians to assess metastatic risk and response to treatment.
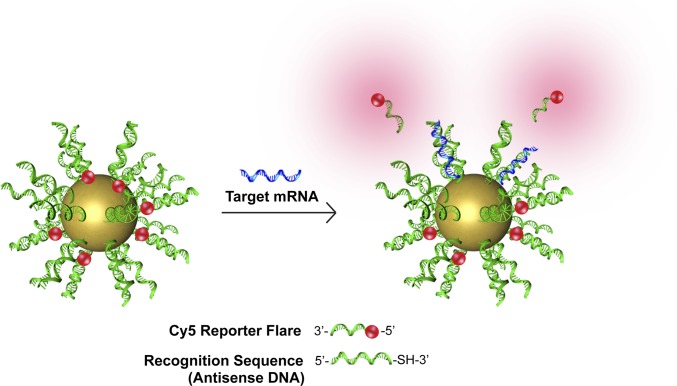
Recently, we introduced the NanoFlare (17–19), a spherical nucleic acid (SNA) (20) nanoconstruct capable of detecting intracellular mRNA levels in live cells at the single-cell level (Fig. 1). The NanoFlare consists of a spherical gold nanoparticle densely functionalized with a monolayer of single-stranded DNA (ssDNA) containing a 3′ thiol that is complementary to mRNA for a target gene. The ssDNA “recognition sequence” is prehybridized to a shorter DNA complement containing a fluorescent reporter (the “reporter flare”) whose fluorescence is quenched based on its proximity to the gold particle. When target mRNA binds the recognition sequence, the reporter flare strand is displaced, providing a fluorescent readout. These constructs have been shown to enter over 60 cell lines and primary cells tested to date, and they are believed to do so by engaging scavenger receptors that facilitate caveolin-mediated endocytosis (17). They are stable to nuclease degradation (21), lack cytotoxicity (22), and are effective for single-gene and multigene detection from a single nanoconstruct (18). These properties make the NanoFlare suited for simultaneously labeling, isolating, and genetically characterizing live CTCs at the single-cell level.
Fig. 1.
Schematic of NanoFlare structure and function. The NanoFlare contains a monolayer of antisense DNA (recognition sequence) adsorbed to the surface of a 13-nm spherical gold nanoparticle. A reporter flare sequence is hybridized to the recognition sequence, which contains a fluorophore (red). The dye is quenched in close proximity to the gold surface. The reporter flare is displaced when complementary mRNA (blue) binds the recognition sequence, providing a fluorescent signal.
We report the application of NanoFlares for the capture of live circulating breast cancer cells. These NanoFlares are able to detect target mRNA from model metastatic breast cancer cell lines in human blood with high recovery and up to 99% fidelity (≤1% incidence of false-negative results). We also report the retrieval of live recurrent breast cancer cells spiked into whole human blood using NanoFlares and subsequent culture as mammospheres. NanoFlare technology allows for the simultaneous isolation and intracellular characterization of live cancer cells from whole blood. NanoFlares may enable the genetic analysis of CTC subpopulations on an individual patient basis, providing information about metastatic proclivity and therapeutic efficacy throughout treatment regimens.
Results
Design and Synthesis of NanoFlares for EMT Marker Genes.
To evaluate the potential of NanoFlares as agents for identifying and isolating CTCs based upon unique genetic signatures, we designed and synthesized NanoFlares targeting genes involved in EMT, including the mesenchymal marker genes, Twist, Vimentin, and Fibronectin, and the epithelial marker E-cadherin. (Table 1). NanoFlare function was initially determined by performing a buffer test in which the fluorescence increase of each NanoFlare was measured in the presence of excess complementary or noncomplementary target DNA (Fig. S1). NanoFlares that showed a robust fluorescence increase in the presence of complementary, but not noncomplementary, DNA were used for subsequent CTC enumeration and isolation experiments.
Table 1.
NanoFlare recognition and reporter flare DNA sequences
| Sequence name | Sequence |
| E-cadherin recognition | 5′-AGA GCC CAA TAA TTT CTG CAA AAA AA-SH-3′ |
| E-cadherin flare | 5′-Cy5-GCA GAA ATT ATT GGG-3′ |
| Twist recognition | 5′-TCT GAA TCT TGC TCA GCT TAA AAA AA-SH-3′ |
| Twist flare | 5′-Cy5-AAG CTG AGC AAG-3′ |
| Vimentin recognition | 5′-TAC CAA GAC CTG CTC AAT GTT AAA AAA A-SH-3′ |
| Vimentin flare | 5′-Cy5-AA CAT TGA GCA GC-3′ |
| Fibronectin recognition | 5′-ACA ATG GTG TGA ACT ACA AGA AAA AAA A-SH-3′ |
| Fibronectin flare | 5′-Cy5-TC TTG TAG TTC ACA C-3′ |
| Scrambled recognition | 5′-ACC ATT AAC CGA TCA TCC AAG AAA AAA A-SH-3′ |
| Scrambled flare | 5′-Cy5-CT TGG ATG ATC GGT T-3′ |
Validation of EMT NanoFlares in Cell-Culture Models.
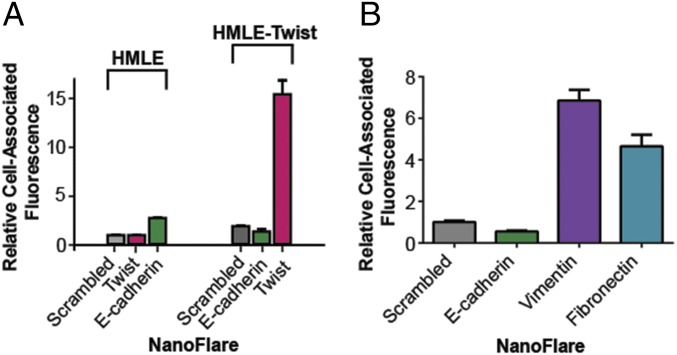
To determine whether NanoFlares could distinguish differences in mRNA levels for a target gene in two different cell types, we evaluated E-cadherin and Twist levels in two mammary cell lines designed to mimic EMT: immortalized human mammary epithelial cells (HMLE) cells and mesenchymal HMLE-Twist cells. In the latter cell type, ectopic expression of the transcription repressor Twist results in a mesenchymal phenotype (23, 24). In a typical experiment, Twist, E-cadherin, or Scrambled Control NanoFlares were incubated with HMLE or HMLE-Twist cells overnight, followed by analysis of NanoFlare fluorescence by flow cytometry. A significant 15-fold enrichment in Twist-NanoFlare signal was observed in HMLE-Twist cells compared with HMLE cells (Fig. 2A). In contrast, a threefold enhancement of E-cadherin NanoFlare signal was observed in HMLE cells compared with HMLE-Twist cells. Importantly, these results accurately reflect the mRNA levels of Twist and E-cadherin in each of these cell lines as measured by quantitative RT-PCR (Fig. S2 A and B).
Fig. 2.
NanoFlares detect mesenchymal markers in EMT model cell lines. (A) NanoFlares detect E-cadherin and Twist mRNA in model mammary cell lines, HMLE, and HMLE-Twist. Results represent flow-cytometric analysis of cells treated with NanoFlares (100 pM) for 16 h. (B) NanoFlares detect EMT markers in metastatic breast cancer cell line MDA-MB-231. NanoFlares were designed for mesenchymal markers (Vimentin and Fibronectin), an epithelial marker (E-cadherin), and a nontargeting Scrambled Control NanoFlare. Results represent flow-cytometric analysis of cells treated with NanoFlares (100 pM) for 8 h.
To further explore the ability of NanoFlares for cancer cell labeling and analysis, a human triple-negative metastatic breast cancer cell line, MDA-MB-231, was studied. MDA-MB-231 cells were treated overnight with Vimentin, Fibronectin, E-cadherin, or Scrambled Control NanoFlares. MDA-MB-231 cells express high levels of Vimentin and Fibronectin and relatively low levels of the epithelial marker E-cadherin, which was used as a low-expressing genetic control (Fig. 2B and Fig. S2C). After incubation with NanoFlares, cells were collected, and fluorescence was measured using flow cytometry. The Vimentin and Fibronectin NanoFlare showed an approximate six- and fourfold increase in fluorescence, respectively, compared with a nontargeting Scrambled Control NanoFlare (Fig. 2B). By contrast, the E-cadherin NanoFlare did not show an enriched signal compared with the control. Significantly, independent analysis of Vimentin, Fibronectin, and E-cadherin mRNA levels by RT-PCR correlated with observed fluorescent differences detected by NanoFlares (Fig. S2C). These results indicate that NanoFlares can be used to label and detect genetically relevant targets in breast cancer cell lines.
Isolation and Characterization of Breast cancer Cells from Human Whole Blood.
Vimentin and Fibronectin NanoFlares were evaluated for their ability to identify breast cancer cells seeded in human whole blood as a proxy for the detection of CTCs from patient blood samples. mCherry cDNA was expressed in MDA-MB-231 cells and used for tracking MDA-MB-231 cells in human blood samples. A known number of mCherry MDA-MB-231 cells were spiked into human whole blood from a healthy male volunteer. Vimentin, Fibronectin, or Scrambled Control NanoFlares were added, and samples were incubated at 37 °C for 8 h. After incubation, the samples were processed to remove red blood cells and CD45+ cells. To determine the reliability of NanoFlares to isolate CTCs, final samples were analyzed for mCherry (Cy3 channel) and NanoFlare (Cy5 channel) fluorescence by flow cytometry (Fig. 3).
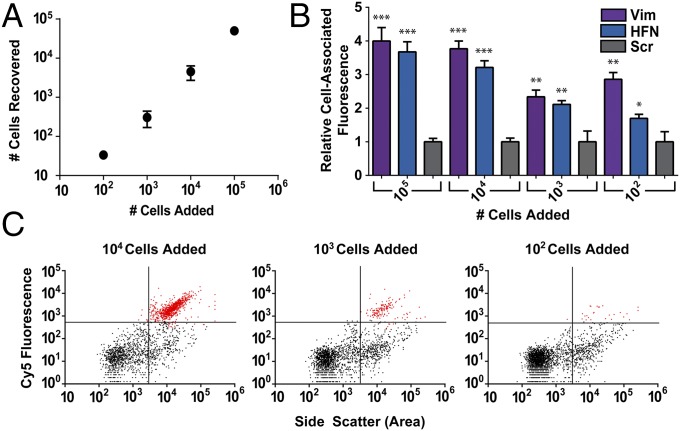
Fig. 3.
NanoFlares isolate metastatic breast cancer cells (MDA-MB-231) spiked into human whole blood. (A) Recovery of mCherry MDA-MB-231 cells from whole blood upon isolation with NanoFlares. (B) Relative cell-associated fluorescence of Vimentin (Vim), Fibronectin (HFN), or nontargeted Scrambled (Scr) Control NanoFlares in 100–100,000 MDA-MB-231 added to whole blood. (C) Scatter plots showing ability of Vimentin NanoFlare to detect small numbers of cells added to whole blood, with a range of 10,000, 1000, and 100 cells. ***P < 0.0005, **P < 0.005, *P < 0.05.
Volume counting beads were added to each sample and used to calculate the total recovery of spiked mCherry MDA-MB-231 cells (Fig. 3A). The average recovery from these experiments was ∼68% ± 14%, which is consistent with other methods of CTC isolation that use a densitometric CTC enrichment step such as a Ficoll gradient (25).
In samples treated with the Vimentin or Fibronectin NanoFlare, ∼99% of the cells that expressed high mCherry fluorescence in the Cy3 channel also expressed a robust NanoFlare fluorescence signal in the Cy5 channel (Fig. S3). NanoFlares were able to detect as few as 100 mCherry MDA-MB-231 cells from whole-blood samples (Fig. 3C). In addition, the Vimentin and Fibronectin NanoFlare showed a four- and threefold fluorescent increase over the nontargeting Scrambled Control NanoFlare after an 8-h incubation (Fig. 3B). These experiments demonstrate the use of NanoFlares as tools for isolating live cancer cells from whole blood based upon intracellular markers.
Isolation and Characterization of Circulating Tumor Cells from a Mouse Xenograft Model of Human Metastatic Breast Cancer.
To determine whether NanoFlares can detect CTCs in a murine model, we used an orthotopic model of triple-negative breast cancer (TNBC) with widespread metastases to several organs (26). Blood was obtained 6.5 wk after tumor-cell inoculation and was treated with Vimentin, Fibronectin, or nontargeting Scrambled Control NanoFlares, or PBS of an equivalent volume for 6 h. Samples were processed to remove red blood cells and CD45+ and ter119+ cells and then analyzed for mCherry (Cy3 channel) and NanoFlare (Cy5 channel) fluorescence using flow cytometry (Fig. 4).
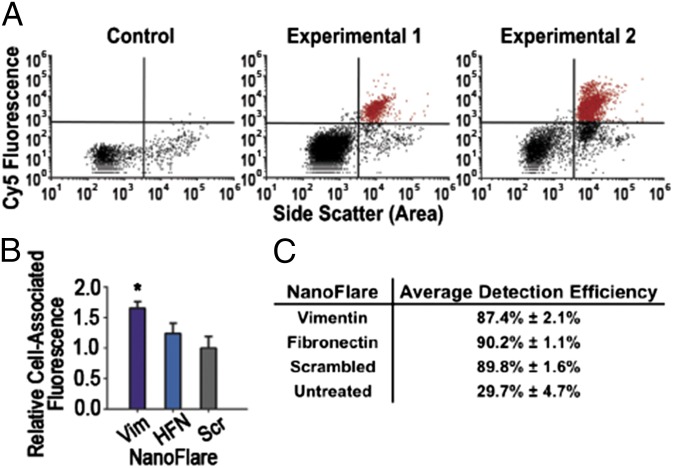
Fig. 4.
NanoFlares detect circulating breast cancer cells from whole blood in a murine model of metastatic breast cancer. (A) Representative scatter plots (n = 1 per scatter plot) show mCherry MDA-MB-231 cells recovered from whole blood by Vimentin NanoFlares from two example tumor-bearing mice (Experimental 1 and 2) (n = 12 per group) or non–tumor-bearing (Control) mice (n = 12 per group). (B) Relative cell associated fluorescence of isolated blood samples treated with Vimentin (Vim), Fibronectin (HFN), or Scrambled (Scr) Control NanoFlares added 6 h before flow-cytometric analysis. (C) Efficiency of mCherry-MDA-MB-231 cell detection by NanoFlares; values were calculated by correlating mCherry fluorescence to NanoFlare fluorescence. Untreated population detection indicates channel bleed-through of mCherry to NanoFlare channel. Samples were collected after 6.5-wk tumor inoculation. Cy5 represents NanoFlare signal. *P < 0.05.
Representative scatter plots (n = 1 per scatter plot) from blood treated with Vimentin NanoFlares demonstrate a positive Cy5 cell population in tumor cell-injected mice (Experimental 1 and Experimental 2) compared with the noninjected (Control) mice (Fig. 4A). The Vimentin NanoFlare and Fibronectin NanoFlare showed a 1.5- and 1.25-fold increase in fluorescence, respectively, over the nontargeting Scrambled Control NanoFlare (Fig. 4B). Although the Fibronectin NanoFlare fluorescence response was not statistically significant with respect to the Scrambled Control NanoFlare, the Vimentin NanoFlare was sufficient for recovering over 87% of CTCs based upon comparison to our internal mCherry control (Fig. 4C). Most likely, the cause of the lower fluorescent response in this experiment was the shorter incubation time of the murine blood with the NanoFlares.
Isolation and Characterization of Seeded GFP Recurrent Cells from Human Whole Blood.
In contrast to many of the antibody-based CTC isolation systems (22), exposing cells to NanoFlares does not result in cell death, providing an opportunity to characterize tumor cells after NanoFlare-based CTC isolation. Toward this end, a GFP-expressing recurrent cell line isolated from recurrent tumors of a human epidermal growth factor receptor (HER2) mouse model of breast cancer, previously established as capable of forming mammospheres, was used to test the feasibility of using NanoFlares to retrieve live cells for further characterization and subsequent analysis. One hundred thousand GFP-expressing recurrent cells were spiked into 200 μL of human blood, followed by incubation with Vimentin NanoFlares for 8 h at 37 °C. The samples were then processed to remove red blood cells and CD45+ cells and sorted via FACS. Cells were gated for GFP and Cy5 fluorescence. Upon FACS analysis, samples treated with Vimentin NanoFlares exhibited a 5.4-fold increase in Cy5 fluorescence compared with Scrambled Control NanoFlare, indicating uptake of NanoFlares and mRNA-triggered increase in fluorescence (Fig. 5 A and B). For sorting, only cells above the background level of Cy5 fluorescence (background is demonstrated in Fig. 5A) were retained and cultured (cells in the red gate in Fig. 5B). To demonstrate that the sample processing step had no appreciable effect on subsequent mammosphere culture, GFP-expressing recurrent cells were isolated using GFP as a marker from a blood sample that was not treated with NanoFlares.
Fig. 5.
Cell isolation and mammosphere formation post NanoFlare treatment and flow cytometry analysis. Representative scatter plots show Cy5 fluorescence (NanoFlare) of GFP recurrent cells spiked into (A) untreated human whole blood or (B) Vimentin NanoFlare-treated blood. Upon treatment with NanoFlares, Cy5 fluorescence of GFP-positive cells increases 5.4-fold. Cells in the red gate in the Vimentin sample were sorted for mammosphere culture. Cells retrieved from blood form mammospheres (C) untreated or (D) Vimentin NanoFlare-treated.
Three replicates from each isolation were pooled and cultured for mammosphere analysis. One thousand GFP-positive cells were seeded into wells of a 96-well plate and monitored for 9 d, after which time mammospheres, spherical clusters enriched in cancer stem cells (CSCs), were observed. Mammospheres successfully resulted from cells retrieved from untreated (Fig. 5C) or Vimentin NanoFlare-treated (Fig. 5D) samples, demonstrating that neither the sample-processing steps nor NanoFlare treatment had any appreciable effect on cell viability. Moreover, the mammospheres expressed GFP, confirming their identity as the GFP recurrent cells originally spiked into the blood sample.
Discussion
NanoFlares, when combined with flow cytometry, provide a method for detecting, isolating, and genetically characterizing circulating cancer cells from whole blood. The modular and rapidly adaptable structure of the NanoFlare enables the detection of a wide variety of cancer-relevant genetic targets simply by changing the identities of the recognition sequence and complementary reporter flare. We have demonstrated this method in the design, synthesis, and evaluation of NanoFlares for four genetic targets relevant to EMT and breast cancer metastasis. NanoFlares containing an antisense recognition motif for E-Cadherin, Twist, Vimentin, and Fibronectin show robust fluorescence responses in the presence of target DNA in buffer tests, as well as target mRNA in breast cancer cell culture model systems. For each cell line studied, the mRNA levels for target genes determined by NanoFlares mirror those quantified by cell-averaging RT-PCR. In addition, NanoFlares for E-cadherin and Twist are able to accurately distinguish epithelial and mesenchymal cells, suggesting that NanoFlares can be used to identify mesenchymal-like cells by assaying for the presence or absence of key markers and have the potential to act as a powerful new tool for isolating and characterizing circulating cancer cells.
Importantly, the NanoFlare-based flow cytometry assay can be used to enumerate, isolate, and survey circulating cancer cells in whole blood, which has direct implications for CTC analysis in cancer patients. NanoFlares for Vimentin and Fibronectin can be used to assess metastatic breast cancer cells spiked into human whole blood. NanoFlares are able to identify mCherry-MDA-MB-231 cells in whole blood with ∼99% fidelity and provide a robust (four- and threefold above a nontargeting Scrambled Control NanoFlare, respectively) fluorescent readout for targets associated with a mesenchymal-like phenotype. Furthermore, circulating cancer cells can successfully be isolated from the peripheral blood of a murine xenograft model of triple-negative metastatic breast cancer using NanoFlares. Although the Vimentin and Fibronectin NanoFlare fluorescent responses are not as robust in this system, the Vimentin NanoFlare can still be used to detect and enumerate mCherry MDA-MB-231 cells from the peripheral circulation with over 87% CTC recovery. Vimentin NanoFlares can also be used to identify and collect murine recurrent mammary tumor cells from whole blood. Further, NanoFlare treatment and subsequent sample processing do not affect the ability of these cells to be further cultured and to form mammospheres suggesting it may be possible to isolate, culture, and further study live CTCs from cancer patients.
NanoFlares change the paradigm of looking at protein markers on CTCs to tracking CTCs based upon unique intracellular genetic signatures. NanoFlares are readily taken up by many epithelial cell types, and, in contrast to conventional genetic analyses such as RT-PCR, allow for live-cell genetic analysis at the single-cell level. The modular structure of the NanoFlare allows for the detection of a wide variety of intracellular genetic targets. Further, with no obvious toxicity, NanoFlares enable the collection of CTCs for subsequent culture and study. In addition, NanoFlares provide the ability to combine intracellular markers with cell-surface markers (e.g., EpCAM and HER2) for the dual identification of putative CTCs. This combined approach may increase the ability to more specifically identify CTCs, and subpopulations of CTCs, that are clearly tumor-derived and that correlate with patient disease status, treatment response, and prognosis. The ability to detect and quantify live patient-derived CTCs will provide vital information both in the study of cancer metastasis and the application of personalized medicine.
Materials and Methods
Oligonucleotide Synthesis.
Oligonucleotides were synthesized with an Expedite 8909 Nucleotide Synthesis System (ABI) using standard solid-phase phosphoramidite chemistry. Oligonucleotide reagents were purchased from Glen Research. Oligonucleotides were purified by reverse-phase high-performance liquid chromatography (HPLC) on a Varian Microsorb C18 column (10 μm, 300 × 10 mm).
Spherical Nucleic Acid Synthesis.
A 3′-alkyl-thiol–modified oligonucleotide recognition sequence (1 aliquot, final concentration 6 μM) was added to a 10-nM solution of citrate-stabilized 13 ± 1-nm spherical gold nanoparticles. After 1 h, SDS and sodium phosphate buffer (pH 7.4) were added to a final concentrations of 0.01% and 20 mM, respectively, and were incubated overnight at room temperature with shaking. The following morning, sodium chloride was added three times at 4-h intervals to a final concentration of 0.1 M, 0.2 M, and 0.3 M. The particles were incubated overnight at room temperature with shaking. The particles were purified by three successive rounds of centrifugation (16,000 × g, 20 min). After each centrifugation, the supernatant was removed, and the functionalized particles were resuspended in PBS (137 mM NaCl, 10 mM phosphate, 2.7 mM KCl, pH 7.4). The concentration of the purified nanoconstructs was determined by ultraviolet-visible (UV-Vis) spectroscopy (λmax = 524 nm; ε = 2.7 × 108 M-1⋅cm−1).
Quantification of Oligonucleotide Loading.
Purified nanoparticles were treated with 0.1 M aqueous KCN to oxidatively dissolve the gold nanoparticles and liberate the surface-bound oligonucleotides. Oligonucleotide concentration was determined using the QuantIT OliGreen oligonucleotide quantification assay (Life Technologies) following the manufacturer’s protocol. From a standard curve of fluorescence versus free oligonucleotide of the same sequence at known concentrations, the average number of oligonucleotides per sample was obtained. The average number of oligonucleotides per nanoparticle was then calculated by dividing the concentration of oligonucleotides by the concentration of nanoparticles in each sample. Average oligonucleotide recognition sequence loading per gold nanoparticle was ∼62 ± 9 strands per particle.
Reporter-Strand Hybridization.
Purified ssDNA nanoparticles (100 nM) were incubated with 10-fold excess complementary reporter “flare” oligonucleotides for 60 min at 65 °C in the dark. The nanoparticles were then immediately refrigerated at 4 °C overnight. The final NanoFlares were isolated from unreacted flare strands by three successive rounds of centrifugation (16,000 × g, 20 min) and resuspension in PBS (137 mM NaCl, 10 mM phosphate, 2.7 mM KCl, pH 7.4); the purified product contained ∼10 hybridized reporter oligonucleotides per gold nanoparticle (AuNP).
Fluorescence Measurements (Buffer Test).
Fluorescence measurements were obtained using a Biotek Synergy Mx Monochromator-Based Multimode Microplate Reader (Biotek). To determine the response of NanoFlares to complementary nucleotides, 1 nM NanoFlares were exposed to cDNA (10 μM, final concentration). The plate was then incubated at 37 °C in the dark for 10 min; after incubation, fluorescence was measured on the multiplate reader using λexc = 630 nm and λem = 670 nm.
Cell Culture.
Human MDA-MB-231 triple-negative breast cancer (TNBC) cells labeled with mCherry fluorescent protein have been described previously (27), were cultured in DMEM-F12 media with 5% (vol/vol) FBS and blasticidin (1 µg/mL), and were supplemented with pen/strep, l-glutamate, nonessential amino acids, and sodium pyruvate. HMLE and HMLE-Twist cells were cultured in 1:1 mammary epithelial cell growth medium (MEGM)/DMEM-F12 supplemented with EGF (5 ng/mL), insulin (2.5 μg/mL), and hydrocortisone (0.25 μg/mL), as described (24). Recurrent breast tumor cells isolated from recurrent tumors of a HER2-mouse model of breast cancer (28) were lentivirally labeled with GFP and cultured in DMEM with 10% calf serum (CS), EGF (10 ng/mL), and insulin (5 μg/mL).
RT-PCR.
RNA was isolated using the RNeasy Kit (QIAGEN) or the E.Z.N.A. Total RNA Kit (Omega Bio-Tek). cDNA was generated by reverse transcription using GoScript RT (Promega). Quantitative RT-PCR (qRT-PCR) was performed using GoTaq qPCR Master Mix (Promega), and mRNA was normalized to TATA-binding protein (TBP).
Flow Cytometry of Cultured Cells.
Cells were plated at 20,000 cells per well in 24-well plates 24 h before NanoFlare treatment. Cells were treated with NanoFlares (100 pM, final concentration) for 8 h or 16 h at 37 °C. Cells collected for flow cytometry were washed twice with PBS, trypsinized, and resuspended in media. A BD LSR Fortessa Analyzer was used for cytometric analysis. The fluorescence associated with each cell was recorded (Cy5 channel) relative to the background fluorescence of untreated cells. The gain settings remained the same throughout all experiments.
Flow Cytometry of mCherry-MDA-MB-231 Cells Spiked into Human Blood.
Northwestern University Institutional Review Board approval was obtained to procure blood samples from healthy volunteers. A known number of mCherry-MDA-MB-231 cells were added to 200 μL of whole human blood from a healthy male volunteer donor. NanoFlares were added to spiked blood samples to a final concentration of 6 nM. Whole-blood samples were incubated at 37 °C with agitation for 8 h. Blood samples were then diluted with 200 μL of cell culture media and treated with 20 μL of RosetteSep CD45+ Cell Depletion Mixture (cat. no. 15122; Stem Cell Technologies) for 20 min. Samples were pipetted over 300 μL of Histopaque 1077 (cat. no. 10771; Sigma-Aldrich) and spun for 20 min at 400 × g. The plasma and lymphocyte layers were removed, and 4 mL of red blood cell lysis buffer was added (cat. no. 11814389001; Roche Applied Science). Samples were exposed to lysis buffer for 10 min at room temperature with shaking and then centrifuged at room temperature for 5 min at 400 × g. The supernatants were aspirated, and pellets were resuspended in 350 μL of cell-culture media. Ten thousand (10 μm) volume counting beads were added. Samples were analyzed by flow cytometry (BD LSR Fortessa Analyzer) using the same method described in the previous subsection for flow cytometry of cultured cells.
Flow Cytometry of Murine Blood Samples from mCherry MDA-MB-231 Xenograft Model.
All animal experiments were approved by the Institutional Animal Care and Use Committee at the University of Wisconsin-Madison. mCherry-labeled MDA-MB-231 cells (2.5 × 105) were resuspended in 100% Matrigel (BD Bioscience) and injected intraductally into the fourth mammary glands of 4- to 5-wk-old female immunodeficient NOD scid IL2 receptor γ chain knockout (NSG) mice (n = 12; The Jackson Laboratory) as described previously (26). Control mice were not injected with tumor cells (n = 12). Mammary xenograft tumors were allowed to grow for 6.5 wk. At 6.5 wk, 0.8–1.0 mL of blood was drawn from anesthetized mice by cardiac puncture before euthanasia. NanoFlares were added to 100 μL of blood samples to a final concentration of 6 nM. Blood samples were then allowed to incubate at 37 °C with agitation for 6 h. After incubation, blood samples were diluted with 200 μL of cell culture media and treated with 100 μL/mL EasySep Custom Murine CD45 and ter119 Depletion Mixture (designed for these experiments by Stem Cell Technologies) for 15 min. Each sample was then placed into an EasySep Magnet (cat. no. 18000; Stem Cell Technologies) for 5 min. The supernatant was collected and added to 4 mL of red blood cell lysis buffer (cat. no. 11814389001; Roche Applied Science); samples were incubated with agitation for 10 min at room temperature before centrifugation (5 min, 400 × g, room temperature). After centrifugation, the supernatant was aspirated, and the pellet was resuspended in 350 μL of cell-culture media. Samples were analyzed by flow-cytometric analysis using the same method listed for cultured cells; the gain settings remained the same throughout all experiments.
Fluorescence-Activated Cell Sorting of GFP Recurrent Cells Spiked into Human Blood.
Northwestern University Institutional Review Board approval was obtained to procure blood samples from healthy volunteers. Cultured GFP recurrent cells were trypsinized, diluted with cell-culture media, and counted using a Countess automated cell counter (Invitrogen). One hundred thousand cells were spiked into 200 μL of whole human blood. Vimentin NanoFlares were added to spiked blood samples to a final concentration of 6 nM. Samples were incubated at 37 °C with agitation for 8 h. After incubation, blood samples were diluted with 200 μL of cell-culture media and treated with 20 μL of RosetteSep CD45+ Cell Depletion Mixture for 20 min. Samples were then pipetted over 300 μL of Histopaque 1077 and centrifuged for 20 min at 0.4 × g. The plasma and lymphocyte layers were removed, and samples were added to 4 mL of red blood cell lysis buffer. Samples were incubated in lysis buffer for 10 min at room temperature with shaking and then centrifuged at room temperature for 5 min at 400 × g. The supernatant was aspirated, and the pellet was resuspended in 350 μL of cell-culture media. Cells were sorted into fresh media on a BD FACSAria cell sorter. GFP-positive cells were identified and gated, and then side scatter versus Cy5 fluorescence was plotted for the GFP-positive cells. From the Vimentin NanoFlare-treated group, only GFP-positive cells with Cy5 fluorescence above background were collected. Cells exhibiting a positive NanoFlare fluorescence signal were cultured. GFP-expressing cells were collected from the untreated group as a positive control to demonstrate that the blood sample-processing steps had no adverse effect on the ability to culture mammospheres. Note that these experiments were performed using murine cells and using a FACS instrument so total fluorescent values are slightly lower than observed in experiments with the human MDA-MB-231 cell line.
Culture of Mammospheres from GFP-Expressing Recurrent Tumor Cell Spiked Human Blood Samples.
Isolated GFP-expressing recurrent tumor cells were centrifuged at 1,200 × g for 5 min and resuspended in MEBM supplemented with 0.5% methylcellulose, 4 μg/mL heparin, 0.02 μg/mL EGF, 0.02 μg/mL basic fibroblast growth factor (bFGF), and B27 supplement (1:50 dilution) at a concentration of 10 GFP positive cells per μL, and 100-μL aliquots were distributed into a 96-well tissue culture plate. Cells were supplemented with additional media every 4–5 d and imaged on day 9 using a Zeiss Axiovert 200 microscope.
Supplementary Material
Acknowledgments
C.S.T., C.A.M., C.C., and V.L.C. thank the National Cancer Institute for financial support for this project through the Center for Cancer Nanotechnology Excellence initiative of the NIH under Award U54CA151880. C.S.T. acknowledges generous support from the Howard Hughes Medical Institute for a Physician Scientist Early Career Award, Air Force Office of Scientific Research Grant FA95501310192, and National Cancer Institute Grant R01CA167041. C.C. acknowledges generous support from American Cancer Society Grant RSG-09-252-01-RMC and NIH Grant R01 CA182467. K.M.M. thanks the Ryan family and the Malkin family for generous financial support for this research project. A.B.C. was supported by the Department of Defense through the National Defense Science and Engineering Graduate fellowship.
Footnotes
Conflict of interest statement: C.A.M. and C.S.T. are cofounders of AuraSense, LLC, a start-up biotechnology company that licensed the NanoFlare technology from Northwestern University.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1418637111/-/DCSupplemental.
References
- 1.Liotta LA, Kohn EC. The microenvironment of the tumour-host interface. Nature. 2001;411(6835):375–379. doi: 10.1038/35077241. [DOI] [PubMed] [Google Scholar]
- 2.Chiang AC, Massagué J. Molecular basis of metastasis. N Engl J Med. 2008;359(26):2814–2823. doi: 10.1056/NEJMra0805239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cristofanilli M, et al. Circulating tumor cells: A novel prognostic factor for newly diagnosed metastatic breast cancer. J Clin Oncol. 2005;23(7):1420–1430. doi: 10.1200/JCO.2005.08.140. [DOI] [PubMed] [Google Scholar]
- 4.Budd GT, et al. Circulating tumor cells versus imaging: Predicting overall survival in metastatic breast cancer. Clin Cancer Res. 2006;12(21):6403–6409. doi: 10.1158/1078-0432.CCR-05-1769. [DOI] [PubMed] [Google Scholar]
- 5.Hayes DF, et al. Circulating tumor cells at each follow-up time point during therapy of metastatic breast cancer patients predict progression-free and overall survival. Clin Cancer Res. 2006;12(14 Pt 1):4218–4224. doi: 10.1158/1078-0432.CCR-05-2821. [DOI] [PubMed] [Google Scholar]
- 6.Pantel K, Brakenhoff RH, Brandt B. Detection, clinical relevance and specific biological properties of disseminating tumour cells. Nat Rev Cancer. 2008;8(5):329–340. doi: 10.1038/nrc2375. [DOI] [PubMed] [Google Scholar]
- 7.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119(6):1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sarrió D, et al. Epithelial-mesenchymal transition in breast cancer relates to the basal-like phenotype. Cancer Res. 2008;68(4):989–997. doi: 10.1158/0008-5472.CAN-07-2017. [DOI] [PubMed] [Google Scholar]
- 9.Zajchowski DA, et al. Identification of gene expression profiles that predict the aggressive behavior of breast cancer cells. Cancer Res. 2001;61(13):5168–5178. [PubMed] [Google Scholar]
- 10.May CD, et al. Epithelial-mesenchymal transition and cancer stem cells: A dangerously dynamic duo in breast cancer progression. Breast Cancer Res. 2011;13(1):202. doi: 10.1186/bcr2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gunasinghe NPAD, Wells A, Thompson EW, Hugo HJ. Mesenchymal-epithelial transition (MET) as a mechanism for metastatic colonisation in breast cancer. Cancer Metastasis Rev. 2012;31(3-4):469–478. doi: 10.1007/s10555-012-9377-5. [DOI] [PubMed] [Google Scholar]
- 12.Allard WJ, et al. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin Cancer Res. 2004;10(20):6897–6904. doi: 10.1158/1078-0432.CCR-04-0378. [DOI] [PubMed] [Google Scholar]
- 13.Liu MC, et al. Circulating tumor cells: A useful predictor of treatment efficacy in metastatic breast cancer. J Clin Oncol. 2009;27(31):5153–5159. doi: 10.1200/JCO.2008.20.6664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Riethdorf S, et al. Detection of circulating tumor cells in peripheral blood of patients with metastatic breast cancer: A validation study of the CellSearch system. Clin Cancer Res. 2007;13(3):920–928. doi: 10.1158/1078-0432.CCR-06-1695. [DOI] [PubMed] [Google Scholar]
- 15.Gupta GP, et al. Identifying site-specific metastasis genes and functions. Cold Spring Harb Symp Quant Biol. 2005;70:149–158. doi: 10.1101/sqb.2005.70.018. [DOI] [PubMed] [Google Scholar]
- 16.Yu M, Stott S, Toner M, Maheswaran S, Haber DA. Circulating tumor cells: Approaches to isolation and characterization. J Cell Biol. 2011;192(3):373–382. doi: 10.1083/jcb.201010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choi CHJ, Hao L, Narayan SP, Auyeung E, Mirkin CA. Mechanism for the endocytosis of spherical nucleic acid nanoparticle conjugates. Proc Natl Acad Sci USA. 2013;110(19):7625–7630. doi: 10.1073/pnas.1305804110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prigodich AE, et al. Multiplexed nanoflares: mRNA detection in live cells. Anal Chem. 2012;84(4):2062–2066. doi: 10.1021/ac202648w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seferos DS, Giljohann DA, Hill HD, Prigodich AE, Mirkin CA. Nano-flares: Probes for transfection and mRNA detection in living cells. J Am Chem Soc. 2007;129(50):15477–15479. doi: 10.1021/ja0776529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giljohann DA, et al. Gold nanoparticles for biology and medicine. Angew Chem Int Ed Engl. 2010;49(19):3280–3294. doi: 10.1002/anie.200904359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prigodich AE, Alhasan AH, Mirkin CA. Selective enhancement of nucleases by polyvalent DNA-functionalized gold nanoparticles. J Am Chem Soc. 2011;133(7):2120–2123. doi: 10.1021/ja110833r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Massich MD, Giljohann DA, Schmucker AL, Patel PC, Mirkin CA. Cellular response of polyvalent oligonucleotide-gold nanoparticle conjugates. ACS Nano. 2010;4(10):5641–5646. doi: 10.1021/nn102228s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang J, et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117(7):927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 24.Brown RL, et al. CD44 splice isoform switching in human and mouse epithelium is essential for epithelial-mesenchymal transition and breast cancer progression. J Clin Invest. 2011;121(3):1064–1074. doi: 10.1172/JCI44540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Noble PBC, Cutts JH. Separation of blood leukocytes by Ficoll gradient. Can Vet J. 1967;8(5):110–111. [PMC free article] [PubMed] [Google Scholar]
- 26.Malin D, et al. αB-crystallin: A novel regulator of breast cancer metastasis to the brain. Clin Cancer Res. 2014;20(1):56–67. doi: 10.1158/1078-0432.CCR-13-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Malin D, Chen F, Schiller C, Koblinski J, Cryns VL. Enhanced metastasis suppression by targeting TRAIL receptor 2 in a murine model of triple-negative breast cancer. Clin Cancer Res. 2011;17(15):5005–5015. doi: 10.1158/1078-0432.CCR-11-0099. [DOI] [PubMed] [Google Scholar]
- 28.Moody SE, et al. The transcriptional repressor Snail promotes mammary tumor recurrence. Cancer Cell. 2005;8(3):197–209. doi: 10.1016/j.ccr.2005.07.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.