Significance
Small noncoding RNAs optimize bacterial gene expression under stress and increase the virulence of many bacterial pathogens. The RNA-binding protein Hfq (host factor Q-beta phage) promotes base pairing between small RNAs and target mRNAs, but it is not known how Hfq brings the two RNAs together in the proper orientation. We used chemical footprinting, small-angle X-ray scattering, and molecular dynamics simulations to model the structure of Hfq bound to an mRNA in solution. The surprising result is that the mRNA wraps entirely around the Hfq protein, specifically contacting both surfaces. This destabilizes the mRNA structure around the small RNA target site, poising it to base pair with a complementary small RNA also bound to Hfq.
Keywords: small noncoding RNA, RNA–protein interactions, SAXS, Lsm protein, bacterial posttranscriptional control
Abstract
The Sm-like protein Hfq (host factor Q-beta phage) facilitates regulation by bacterial small noncoding RNAs (sRNAs) in response to stress and other environmental signals. Here, we present a low-resolution model of Escherichia coli Hfq bound to the rpoS mRNA, a bacterial stress response gene that is targeted by three different sRNAs. Selective 2′-hydroxyl acylation and primer extension, small-angle X-ray scattering, and Monte Carlo molecular dynamics simulations show that the distal face and lateral rim of Hfq interact with three sites in the rpoS leader, folding the RNA into a compact tertiary structure. These interactions are needed for sRNA regulation of rpoS translation and position the sRNA target adjacent to an sRNA binding region on the proximal face of Hfq. Our results show how Hfq specifically distorts the structure of the rpoS mRNA to enable sRNA base pairing and translational control.
The bacterium Escherichia coli encodes 80 small noncoding RNAs (sRNAs) that fine-tune gene expression for different growth environments, increasing survival under various stress conditions (1, 2). Base pairing between an sRNA and an mRNA can inhibit gene expression by masking the ribosome binding site or by increasing mRNA turnover (3). Alternatively, sRNAs increase translation by changing the mRNA structure and exposing the ribosome binding site. In E. coli, sRNA regulation depends on Hfq (host factor Q-beta phage), a protein that stabilizes and accelerates base pairing between many known sRNAs and their mRNA targets (3, 4).
Hfq belongs to the Sm/Lsm protein family (5) and recognizes diverse RNA targets by three distinct RNA binding surfaces (6). The distal face of the Sm ring binds AAN triplets (7, 8) present in many mRNA targets of sRNA regulation (9). The inner surface of the proximal face binds U-rich single strands (10, 11), which are a common feature of bacterial sRNA terminators and important for Hfq action (12, 13). Finally, a patch of conserved basic residues (R16, R17, R19, and K47) on the rim interacts with internal U-rich sequences in sRNAs, increasing the accessibility of the seed region (14) and catalyzing base pairing between complementary strands (15).
Although Hfq is known to bind specific sequences in sRNAs and mRNAs, how it restructures its targets for translational control is not understood. We address this question using E. coli rpoS, a well-studied target of posttranscriptional regulation by sRNAs and Hfq. rpoS encodes σS, a major stress-response regulator that is up-regulated by DsrA, RprA, and ArcZ sRNAs in E. coli (16). Genetic experiments showed that an inhibitory stem loop in the rpoS mRNA blocks ribosome binding; sRNAs open this inhibitory stem by base pairing to its upstream strand (17, 18). Hfq must be recruited to an (AAN)4 motif in an upstream domain of the rpoS mRNA to facilitate sRNA base pairing and regulation (19, 20). Biochemical experiments showed that Hfq interacts weakly with the rpoS inhibitory stem loop, cycling off the sRNA–mRNA antisense duplex as it is formed (21). These experiments left unanswered why Hfq must interact with two domains of the rpoS mRNA, how it remodels the rpoS mRNA to seed base pairing by a complementary sRNA (22), and why sRNA binding displaces Hfq from the inhibitory stem loop.
Here, we show that Hfq enables sRNA regulation by folding the rpoS mRNA leader into a specific tertiary structure that partially unwinds the inhibitory stem and poises Hfq to bring both RNAs together. Small-angle X-ray scattering (SAXS), functional assays, and SHAPE (selective 2′-hydroxyl acylation and primer extension) footprinting revealed that Hfq contacts three distinct sites in the rpoS mRNA, folding the 5′ leader of the rpoS mRNA into a compact structure. Three-dimensional models of the rpoS•Hfq complex refined against the SAXS data show that the two domains of the rpoS mRNA wrap around the Hfq hexamer, placing the inhibitory stem over the arginine patch and adjacent to the sRNA binding sites on the rim and proximal face. These results demonstrate that multiple RNA binding surfaces on Hfq enable the protein to distort the structure of the rpoS mRNA, poising the complex for sRNA entry and translation.
Results
Hfq Binds A-Rich and U-Rich Motifs in rpoS mRNA.
We used SHAPE footprinting to identify Hfq interaction sites in the rpoS leader RNA. Previous experiments showed that the (AAN)4 motif upstream of the sRNA target site binds the distal face of the Hfq and recruits Hfq to the rpoS mRNA (20, 22, 23). Hfq has the potential to also interact with a “U5” loop motif (5′ UUAUUU) downstream of the sRNA target site (21, 24).
For footprinting experiments, we used rpoS301, a 284-nt variant of the 576-nt rpoS leader that lacks a nonessential upstream domain but retains the Hfq binding domain and inhibitory stem needed for translational control and Hfq and sRNA binding (24). The rpoS301 RNA folds homogeneously in vitro and retains the native secondary structure (Fig. S1 A and B) based on its similar SHAPE modification as the full-length rpoS leader (24). We compared the SHAPE modification levels of free rpoS301 RNA with rpoS301 RNA bound to DsrA sRNA or to Hfq (Fig. 1A and Fig. S1C). We then categorized the decrease or increase in relative SHAPE reactivity based on a histogram of the entire dataset (Fig. S1D), which reflects a change in the accessibility of the ribose 2′OH or the flexibility of the RNA backbone (25).
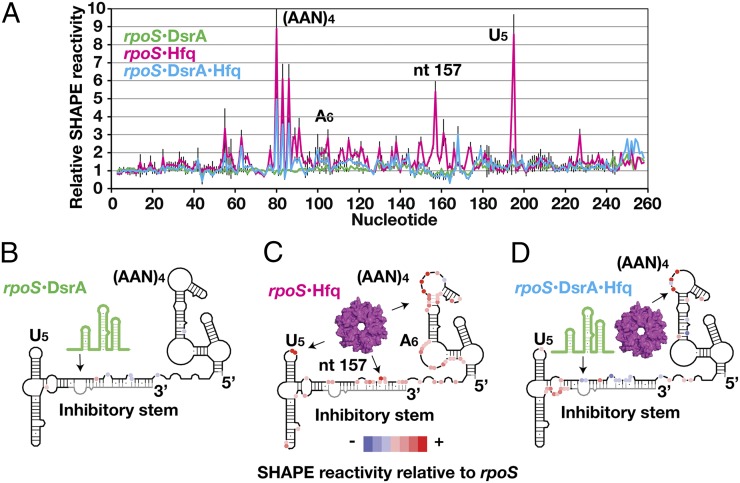
Fig. 1.
Conformational changes in rpoS mRNA from binding of DsrA and Hfq. (A) SHAPE reactivity of 50 nM rpoS301 RNA in complex with 200 nM DsrA (green trace), 333 nM Hfq (magenta), or DsrA and Hfq (blue) relative to rpoS RNA alone (Fig. S1). (AAN)4 motif, nucleotides 77–88; inhibitory stem, nucleotides 149–184 and 249–284; U5 motif, nucleotides 192–197. NMIA modification was carried out at 37 °C (Materials and Methods) and the extent of modification was measured by primer extension (Fig. S1C). Error bars represent ±SD for at least three independent experiments. (B–D) Schematic of SHAPE reactivity relative to free rpoS RNA for each complex, from a histogram of the entire dataset (Fig. S1D). Red circles, nucleotides with enhanced SHAPE reactivity; blue circles, nucleotides with reduced SHAPE reactivity; black line, regions with unchanged SHAPE reactivity; gray line, regions with no SHAPE data. Arrows indicate DsrA and Hfq binding regions in rpoS RNA.
As expected, base pairing between rpoS mRNA and DsrA sRNA protected the DsrA binding site in the inhibitory stem from modification, reducing the SHAPE reactivity by ∼30–40% (Fig. 1B). The SHAPE reactivity of the upstream and downstream domains did not change appreciably, however, suggesting they are unaffected by DsrA (green trace in Fig. 1 A and B).
By contrast, Hfq remodeled the rpoS mRNA structure extensively (magenta trace in Fig. 1 A and C). First, the reactivity of the inhibitory stem and the helix connecting the (AAN)4 and A6 motifs increased two- to threefold over that of the free RNA. These residues were uniformly and moderately modified in the rpoS•Hfq complex, suggesting that Hfq partially opens the mRNA secondary structure. An Hfq-induced structural change in the inhibitory stem was also reported based on RNase footprinting experiments (26).
Second, Hfq binding resulted in unusually strong modification of three regions that we deduced make specific contacts with Hfq: the (AAN)4 motif previously known to bind Hfq, the U5 motif in the downstream domain, and A157 in the inhibitory stem near the 5′ end of the sRNA target site. The first A of every AAN triplet was four to nine times more modified in the Hfq complex than in the RNA control (A80, A83, and A85 in Fig. 1A) (also Fig. S1C). This hyperreactivity was explained by a structure showing that the A-specific pocket on the distal face of E. coli Hfq (7) locks the ribose into a highly reactive C2′-endo conformation (27). The third and fourth residues in the U5 motif were also eight times more modified in the rpoS•Hfq complex (A194 and U195 in Fig. 1A) (Fig. S1C, Bottom Left), indicating that Hfq also interacts with this U-rich loop as suggested by previous RNase footprinting experiments (21). Finally, A157 in the inhibitory stem was six times more reactive in the rpoS•Hfq complex (Fig. 1A), pointing to a previously unsuspected interaction between Hfq and the start of the inhibitory stem.
When both DsrA and Hfq were added, the modification pattern of the DsrA•rpoS• Hfq ternary complex showed that Hfq releases the inhibitory stem and U5 motif but remains bound to the upstream (AAN)4 motif (Fig. 1D). This is consistent with in vitro annealing experiments showing that Hfq cycles off the sRNA–mRNA duplex after the RNAs have base paired (21). The DsrA target site was ∼50–80% less modified in the ternary complex than in the DsrA•rpoS complex (Fig. 1D), consistent with tighter DsrA•rpoS binding in the presence of Hfq (19). Meanwhile, nucleotides upstream of the Shine–Dalgarno sequence became two- to threefold more accessible in the ternary complex.
A U5 Motif Binds the Lateral Rim of Hfq.
To test which surfaces of Hfq contact rpoS mRNA, we repeated the SHAPE experiments with Hfq mutants Y25D, R16A, and K56A that disrupt RNA binding to the distal face, the lateral rim, and the proximal face, respectively (8, 14, 28). As expected, the Y25D mutation selectively disturbed the hypermodification of the (AAN)4 motif (Fig. 2A), consistent with its binding to Hfq’s distal face (7, 22). Binding was only partially impaired by the Y25D mutation, as it eliminated the hyperreactivity of only the third (AAN) triplet and shifted the modification pattern 1 nt upstream (Fig. 2A, purple trace). The SHAPE reactivities of the inhibitory stem and the downstream domain were the same as in the WT Hfq complex, indicating that those regions interact with a different surface of Hfq.
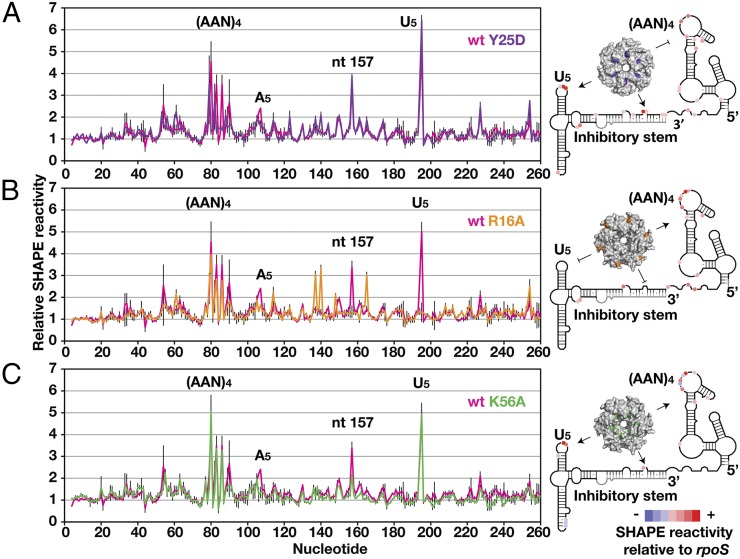
Fig. 2.
Mapping rpoS interaction sites on Hfq. (Left) SHAPE modification of rpoS RNA in complex with wt Hfq (pink trace) or an RNA binding surface mutation. Error bars are as in Fig. 1. (A) Distal face Y25D mutation (purple) disrupts Hfq binding to (AAN)4 motif. (B) Rim R16A mutation (orange) disrupts Hfq binding to inhibitory stem and U5 motif. (C) Proximal face K56A mutation (green) retained the three direct binding sites, but did not appreciably change the RNA secondary structure.
Strikingly, the R16A rim mutation abolished interactions with the inhibitory stem and the U5 motif while leaving intact interactions with the (AAN)4 motif (Fig. 2B, orange trace). The lost hypermodification of the U5 motif (A194 and U195) suggested that this loop directly contacts the lateral rim of Hfq. Modification of A157 returned to the average level, and modification of C137, C140, and C165 increased approximately threefold (Fig. 2B, orange and gray traces), indicating that the perturbed interaction with the rim also changed the conformation of the inhibitory stem. Finally, the K56A mutant did not appreciably change the modification pattern (Fig. 2C), confirming that the proximal face of Hfq does not bind rpoS mRNA directly.
U5 Motif Binding at Hfq Rim Facilitates DsrA Annealing.
The SHAPE results showed that the lateral rim of Hfq contacts the downstream U5 motif in the rpoS mRNA leader, whereas the distal face remains bound to the upstream (AAN)4 motif. To investigate whether the U5 motif is required for regulation of rpoS translation by Hfq and sRNAs, we replaced the UUAUUU loop with UCGC (Fig. 3A, ΔU5), shortened the stem by 3 bp (Fig. 3A, U5SS), or enlarged the loop by 9 nt (Fig. 3A, U5UL).
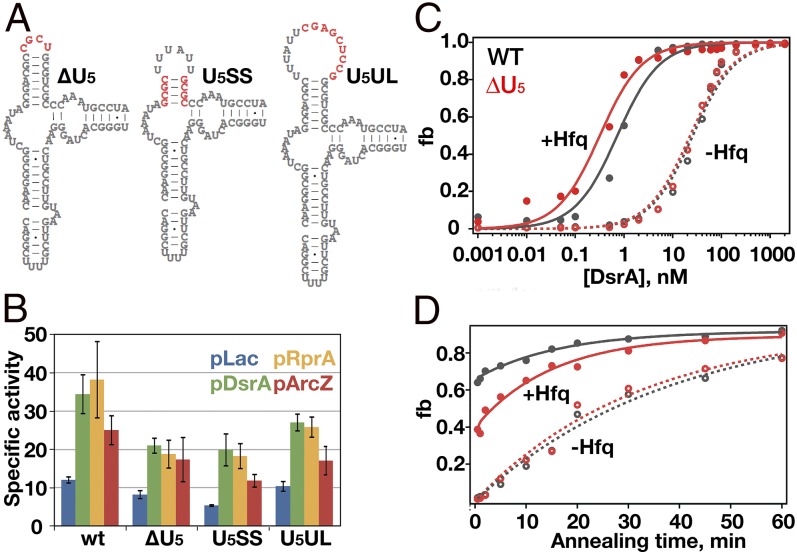
Fig. 3.
Function of U5 motif in sRNA binding and regulation. (A) Mutations in the U5 motif (red) delete the U-rich sequence in the loop (ΔU5), shorten the helix (U5SS), or insert a GC-rich sequence in the loop (U5UL). (B) β-Galactosidase activity assays measure translation of rpoS::lacZ in E. coli when sRNAs are overexpressed from IPTG-inducible plasmids. Empty pLac vector (blue), pDsrA (green), pRprA (orange), and pArcZ (red) are shown. (C and D) DsrA binding to WT rpoS RNA (gray) and ΔU5 rpoS RNA (red) without Hfq (open circles and dashed lines) or with Hfq (solid circles and solid lines). Binding was measured by native gel mobility shift (Fig. S2 C and D). (C) Equilibrium binding. A fraction of rpoS•DsrA (RD) or rpoS•DsrA•Hfq (RDH) vs. [DsrA] was fitted to a single-site binding isotherm. (D) rpoS-DsrA annealing kinetics. Data were fitted to single- (no Hfq) or double- (+Hfq) exponential equations (20). Error bars represent ±SD for at least three independent experiments.
All three mutations in the U5 stem loop diminished the ability of DsrA and RprA sRNAs to up-regulate expression of full-length rpoS::lacZ fusions in the E. coli chromosome by 20–40% (Fig. 3B, green and gold bars). The magnitude of this effect was similar to that of mutating the upstream (AAN)4 and A6 motifs (Fig. S2A, Δ2), although the U5 mutations had a smaller effect on up-regulation by ArcZ sRNA (Fig. 3B, red bars). When the (AAN)4, A6, and U5 motifs were all mutated, expression of rpoS::lacZ fusions was reduced a further 50% (compare Δ2 and Δ3 in Fig. S2 A and B), showing that the (AAN)4, A6, and U5 motifs not only interact with different surfaces of Hfq, but also make distinct contributions to the regulation of rpoS translation by sRNAs and Hfq.
To investigate whether the U5 motif is important for DsrA annealing in vitro, we next measured the stability of the DsrA•rpoS complex, using native gel mobility shift assays (Fig. S2 C and D) (19). We titrated 32P-labeled rpoS mRNA with DsrA sRNA (0–2 μM) and quantified the total fraction of rpoS•DsrA and rpoS•DsrA•Hfq complexes as a function of DsrA concentration (Fig. 3C). Without Hfq, the ΔU5 mutation did not change the strength of the DsrA–rpoS RNA interaction, suggesting that this mutation does not alter the structure of free rpoS mRNA (Table S1). With Hfq present, however, DsrA bound the ΔU5 complex about twofold better than the WT rpoS•DsrA•Hfq complex, perhaps owing to better release of the downstream domain (Table S1).
We next measured the ability of Hfq to increase the rate of DsrA annealing with rpoS mRNA (Fig. 3D). Without Hfq, both WT and ΔU5 rpoS mRNA base paired with DsrA at the same rate (0.03 min−1). In the presence of Hfq, however, a lower proportion of ΔU5 than WT rpoS mRNA annealed with DsrA during the first 30 s (Table S1). Thus, these results suggest that interactions between Hfq and the U5 motif distort the rpoS mRNA conformation for efficient DsrA entry.
Further SHAPE footprinting results on the ΔU5 mRNA confirmed that this defect in DsrA annealing was due to impaired Hfq binding at the U5 motif, based on the loss of hyperreactivity at this position (Fig. S3A). The ΔU5 mutation also lowered modification of the upstream (AAN)4 motif by ∼80%, consistent with an overall reduction in Hfq affinity (24). Surprisingly, we still observed strong modification of A157 in the inhibitory stem, suggesting this contact depends on recruitment of Hfq by the (AAN)4 motif rather than U5 motif. The SHAPE reactivity of the inhibitory stem was no longer enhanced, however, consistent with our previous conclusion that the U5 motif is needed for Hfq to open the inhibitory stem. The U5SS and U5UL mutations also disrupted Hfq binding at the U5 motif (Fig. S3 B and C) and reduced in vivo expression of rpoS::lacZ.
Hfq Folds rpoS mRNA.
If the distal face of Hfq binds the upstream (AAN)4 motif while the lateral rim interacts with the downstream U5 motif, Hfq binding likely alters the tertiary conformation of the rpoS mRNA. To test that hypothesis, we used SAXS to compare the global shape of free rpoS RNA and the rpoS•Hfq complex in solution, at molar ratios from 1:0.5 to 1:3 RNA:Hfq6 (Fig. 4). The scattering profile of free Hfq protein (Fig. S4 A and B) was consistent with its known structure as previously reported (29, 30). The scattering profile of the free rpoS mRNA (284 nt) revealed an extended structure with radius of gyration (Rg) = 68.1 ± 0.6 Å (Fig. S4 C and D). A dimensionless Kratky plot (31) of the scattering intensity exhibited the plateau at higher-momentum transfer (q), indicating an extended or flexible conformation (Fig. 4A, black symbols).
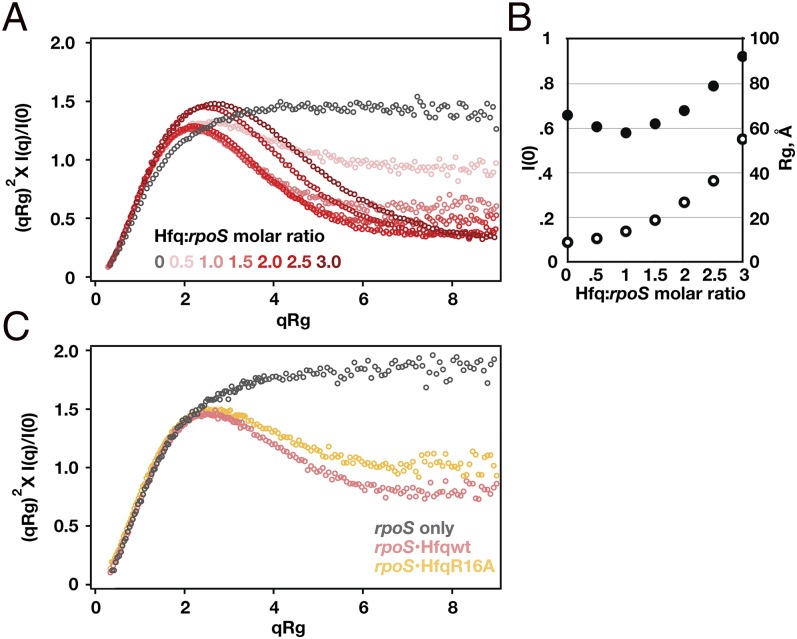
Fig. 4.
SAXS of rpoS•Hfq complexes reveals a compact structure. (A) Dimensionless Kratky plot (31) of SAXS profiles for rpoS RNA alone (black) and rpoS•Hfq complexes at increasing protein:RNA ratios (pink to red). Bell-shaped curves indicate compact structures. See Fig. S4 and Table S2 for further data. (B) Hfq binding increased I(0) (open circles) and decreased Rg (solid circles) compared with free rpoS RNA. At 1:1 mol ratio, ∼95% of RNA is bound to Hfq. Correction for scattering from the free RNA and protein reduces the experimental Rg of the complex by ∼1 Å. (C) Kratky plots of rpoS RNA alone (black), with 1:1 WT Hfq (pink) and with 1:1 Hfq:R16A (gold).
The shape of the Kratky scattering curves changed dramatically when Hfq was added, forming the symmetric maximum characteristic of globular particles (Fig. 4A, red symbols). This change in shape corresponded with a drop in Rg (Fig. 4B, solid circles) despite the greater mass of the rpoS•Hfq complex (Fig. 4B, open circles). The smallest Rg value of 58 ± 1 Å was reached at 1:1 rpoS:Hfq6 (Fig. S4 E and F), at which concentration 95% of the RNA is expected to be bound with Hfq (24). The change in the scattering profile cannot be explained by scattering from the protein alone, as Hfq has a much smaller X-ray scattering contrast than the RNA (Fig. S4E). Instead, we inferred that the flexible rpoS leader must adopt a more compact tertiary structure when bound to Hfq. This compact structure is stabilized by interactions between rpoS mRNA and the rim of Hfq, because the Hfq:R16A mutant formed a more extended complex with rpoS mRNA than did WT Hfq (Fig. 4C, gold symbols).
Structure Models of rpoS and Hfq.
We next used the SAXS and SHAPE footprinting results to model the 3D structures of the free rpoS mRNA and the rpoS•Hfq complex. Molecular envelopes calculated ab initio from the SAXS data revealed an elongated “L” for the free rpoS RNA (Fig. S5A), which curled inward when Hfq was present (Fig. S5B). Nevertheless, many structural features were lost when these envelopes were averaged, presumably because the RNA is flexible and poorly constrained by the scattering curves. In addition, Hfq was nearly invisible in the molecular envelopes, owing to its lower scattering contrast relative to the RNA. Therefore, we used rigid-body methods to build atomistic models of the rpoS•Hfq complex, using the information available from SAXS, crystal structures, and biochemical footprinting. Although these data cannot specify the conformation of individual residues, we obtained low-resolution models that were consistent with all of the available data and that suggested how Hfq enables sRNA regulation of rpoS translation.
We first built an all-atom model of the full-length E. coli Hfq hexamer by appending disordered N- and C-terminal residues to a crystallographic model of the stable Sm core [Protein Data Bank (PDB) ID: 4HT8] (32). We used the program SASSIE (33) to simulate conformations of the N and C termini that fitted the experimental SAXS data (Fig. S6A). In the best structures, the N termini (amino acids 1–5) projected from the center of the proximal face (Fig. 5A, purple), whereas the C termini were mostly oriented toward the distal face (Fig. 5A, pink). This distal orientation differs from the radial projection of the C termini in previous ab initio models (30) (Fig. S5C).
Fig. 5.
Model of the rpoS RNA•Hfq regulatory complex. Shown are all-atom models of (A and B) full-length Hfq, (C and D) rpoS RNA, and (E and F) rpoS•Hfq complex. The U5-rim contact was constrained in the closed model (SASREF) (35); the open model is from the SASSIE (33) trajectory. Hfq is rendered as a surface; Sm core [residues (res) 6–65], wheat color; N termini (res 1–5), slate color; C termini (res 66–102), pink. rpoS RNA ribbon, gray; (AAN)4 motif, purple; U5 motif, orange; sRNA binding site, green; Shine–Dalgarno site, violet. See Fig. S7 for details of the RNA model. (B, D, and F) Scattering curves predicted by models (red or blue) compared with experimental scattering (gray) for (B) full-length Hfq, (D) rpoS RNA, and (F) rpoS•Hfq complex in closed or open conformation. (G) Open structures from SASSIE. The best-fitting 917 models from the trajectory (H) were clustered (UCSF Chimera), and each cluster is represented by a semitransparent surface to illustrate the wedge of conformations that describe the scattering curve.
To model the tertiary structure of the free rpoS mRNA, we divided the rpoS301 sequence into six fragments, using our SHAPE-determined secondary structure as a guide (Fig. S7A). We generated structures for each fragment with MC-Sym (34) and arranged the fragments in space by rigid-body modeling (SASREF) (35) against the experimental SAXS data (Fig. S7B and SI Materials and Methods). In the resulting model, the upstream and inhibitory domains again form an L connected by a flexible hinge at nucleotides 128–129 (Fig. 5C). Because these domains likely sample different orientations in solution, we used this hinge as a pivot point in a SASSIE Monte Carlo simulation, which generated an ensemble of 27,427 structures spanning the experimental Rg (Fig. S6B). The best-fit structures from this ensemble resembled the initial L-shaped model.
Structural Models of the rpoS•Hfq Complex.
We repeated this modeling procedure to visualize the structure of the rpoS•Hfq complex, using the scattering data from the 1:1 rpoS:Hfq6 sample as an experimental constraint. We used a crystallographic structure of the Hfq core bound to rA7 (32) to model the interaction between the (AAN)4 motif and the distal face of Hfq. In addition, as our SHAPE data showed that the rpoS U5 motif and A157 in the inhibitory stem both interact with the rim of Hfq, we constrained those residues to be within 7 Å of R16 in any Hfq monomer.
The resulting model (Fig. 5 E and F) showed the rpoS mRNA wrapped around the Hfq hexamer, with the U5 motif on the proximal side of the rim opposite the second AAN triplet and A157 at the rim on the other side of the ring. Strikingly, this orientation projected the inhibitory stem across the proximal face of Hfq, with the sRNA complementary strand toward Hfq and the ribosome binding site away from Hfq. This wrapped structure necessitates a slight unwinding of the inhibitory stem, consistent with the moderate increase in SHAPE modification of this region when Hfq binds. Hfq may induce additional RNA conformational changes that are not captured by our rigid-body modeling procedure. Overall, the model explained how Hfq folds the rpoS mRNA into a more compact structure and why interactions with both the AAN motif and the U5 motif are needed for efficient sRNA entry.
To determine whether other conformations also fit the SAXS data, we used SASSIE to vary the orientation of the downstream RNA domain about the flexible hinge (nucleotides 128–129). Structures of the rpoS•Hfq complex that best represent the data (χ2 < 1.5; 917 structures) were symmetrically distributed about Rg = 55 Å (Fig. 5H) and collectively sampled a restricted wedge of space that could reflect an oscillatory path of the inhibitory stem in which the U5 motif detaches and rebinds the Hfq lateral rim (Fig. 5G and Fig. S6D). This ensemble of “open” structures described the scattering data nearly as well as the initial “closed” structure (Fig. 5F). In all of these structures, nucleotide A157 remained close to the Hfq rim, consistent with our SHAPE data showing that hypermodification of this residue in the inhibitory stem depends on the (AAN)4 motif binding rather than the U5 motif. By contrast, the sRNA annealing site, the ribosome binding site, and the U5 motif moved away from Hfq in the more open structures.
Discussion
Our SHAPE footprinting results, SAXS data, and all-atom models collectively show that Hfq folds the rpoS mRNA leader into a compact tertiary structure. This folded structure positions the inhibitory stem of the rpoS leader over the proximal face of Hfq where sRNAs are known to bind. This unexpected result explains many features of rpoS regulation by sRNAs and Hfq, such as how Hfq brings together the complementary regions of the mRNA and sRNA near the arginine patches along the rim and why sequences upstream and downstream of the sRNA target site are important. Moreover, our SHAPE results show that Hfq partially opens the secondary structure of the inhibitory stem to enhance sRNA annealing and ribosome binding (22, 26). Remodeling of the rpoS mRNA requires interactions with both (AAN)4 and U5 motifs.
As the SAXS data do not provide information about local structure, our model cannot capture the details of the RNA–Hfq interactions. Moreover, the model does not account for local perturbations to the RNA structure. Nevertheless, the overall arrangement of the rpoS mRNA leader with respect to Hfq in our model is well supported by experimental data. First, the dramatic change in the scattering function provides direct physical evidence for compaction of the RNA by Hfq. Second, the marked change in RNA backbone modification (SHAPE) in response to the Hfq and ΔU5 mutations is consistent with specific Hfq interactions, rather than nonspecific effects of the protein on the RNA structure. Unusually strong ribose modification may serve as a diagnostic for direct Hfq–RNA interactions. Third, mutational studies showed that the position and orientation of the (AAN)4 and U5 sequences are important for Hfq-mediated sRNA regulation, suggesting they bind Hfq simultaneously (Fig. S3) (24). Finally, an unbiased search of structural models indicated that only a subset of RNA conformations recapitulates the SAXS data (Fig. 5 G and H and Fig. S6D).
Our data show that Hfq folds the rpoS leader into a compact, closed conformation by simultaneously recognizing an upstream (AAN)4 motif and downstream U5 motif flanking the sRNA target site. In this closed mode, the inhibitory stem is partially melted, and the 5′ end of the target site interacts with the Hfq rim where we propose the arginine patch promotes base pairing with a complementary sRNA. The SHAPE data show that Hfq disengages from the downstream U5 motif after a sRNA base pairs with the inhibitory stem, while remaining bound to the (AAN)4 motif. The potential to form more open structures explains how the rpoS leader can flex to allow Hfq to cycle off the DsrA–rpoS duplex, exposing the ribosome binding site.
The potential for opening and closing the rpoS•Hfq complex is clearly captured in our structural models. The closed rpoS•Hfq model obtained by constraining the U5 motif to interact with the Hfq rim was reasonably consistent with the SAXS data. However, the Monte Carlo simulations showed that more open structures fitted the scattering data equally well, even assuming a small fraction of free RNA. The rpoS•Hfq complex may fluctuate between open and closed conformations in solution. As the scattering curves for 2:1 Hfq:rpoS also indicate a folded structure, our data do not exclude models in which the open rpoS leader binds a second Hfq hexamer.
Although AAN sequences are known to recruit Hfq via its distal face (7, 8, 20), here we find that the U5 motif in rpoS also contributes to sRNA annealing by interacting with the Hfq rim. This distorts the mRNA structure, making it more accessible to sRNAs (22). Multilateral Hfq interactions may be widespread among bacterial sRNA–mRNA pairs and important for regulation. The fhlA mRNA leader was proposed to contact both distal and proximal faces of Hfq based on competitive binding experiments (36). Hfq inhibits translation of cirA by binding to an upstream (AAN) motif and two U-rich patches close to the Shine–Dalgarno sequence (37), raising the possibility that Hfq also folds the cirA mRNA for translational control. Our results show that Hfq forms a specific, folded rpoS mRNP that spring loads the regulatory helix for sRNA entry.
Materials and Methods
SHAPE Footprinting.
Complexes of 50 nM rpoS301 RNA, 333 nM E. coli Hfq hexamer, and 200 nM DsrA sRNA were prepared as previously described (24) in 10 µL annealing buffer [50 mM Tris⋅HCl, pH 7.5, 50 mM NaCl, 50 mM KCl, 50 mM NH4Cl, 2% (vol/vol) glycerol] at 25 °C for 2 h. Complexes were modified with N-methylisatoic anhydride (Molecular Probes) and analyzed by reverse transcription as described in SI Materials and Methods. Reported values of relative SHAPE reactivities are the average of at least three independent experiments.
Hfq Binding and Translational Activation.
E. coli strains and β-galactosidase assays of rpoS::lacZ expression were performed as previously described (19, 24). Gel mobility shift binding assays with ∼70 nM 32P-labeled rpoS301 RNA and DsrA or Hfq were performed in annealing buffer for 2 h at 25 °C as previously described (20, 24) before native 6% polyacrylamide gel electrophoresis in 66 mM Hepes, 34 mM Tris, 0.1 mM EDTA, and 2 mM MgCl2.
SAXS.
rpoS301 RNA and Hfq protein were prepared under native conditions as described in SI Materials and Methods. Small-angle X-ray scattering data were collected at room temperature at the Advanced Photon Source 12-ID-B, over the range 0.005 < q < 1.007 Å−1 after background subtraction. Data collected at three different sample concentrations showed the expected increase in I(0) and constant Rg and ratios of scattering intensity, indicating a lack of interparticle interactions (Fig. S4). Parameters of the fits and estimates of the particle mass are listed in Table S2.
Structural Models.
Three-dimensional models of rpoS mRNA secondary structure fragments (Fig. S7) were generated using MC-Sym web server (34) and oriented in three dimensions with SASREF (35), using the RNA chain connectivity and the SAXS experimental data as constraints. CORAL was used to model the full rpoS•Hfq complex against the SAXS data for the 1:1 RNA:Hfq sample (35). In the complex, rpoS P 195 (U5 motif) and P 157 (inhibitory stem) were constrained to ≤12 or 15 Å, respectively, from R16 Cα in any Hfq monomer. Monte Carlo simulations were performed using the program SASSIE (33) to identify conformations of free Hfq, free rpoS mRNA, and the rpoS•Hfq complex consistent with the scattering data for each sample. The coordinates of the Hfq core were fixed during the simulations, whereas the N and C termini (amino acids 1–5 and amino acids 66–102) were allowed to move. The RNA was allowed to pivot between nucleotides 128 and 129. Whereas the residuals between the best 917 models and the experimental data for the 1:1 rpoS•Hfq complex showed some positive serial correlation (Durbin–Watson <2), the magnitudes of the residuals were on the order of the statistical error of the data (Fig. S5E). See SI Materials and Methods for details of the modeling.
Certain commercial equipment, instruments, materials, suppliers, or software are identified in this paper to foster understanding. Such identification does not imply recommendation or endorsement by the National Institute of Standards and Technology, nor does it imply that the materials or equipment identified are necessarily the best available for the purpose.
Supplementary Material
Acknowledgments
The authors thank X. Zuo and the APS 12-ID-B staff, S. Krueger, S. Gottesman, G. Storz, and Y.-X. Wang for helpful discussion and D. Kilburn, S. Panja, N. Majdalani, N. Kim, and T. Schlick for their assistance. This work was supported by the National Institute of General Medicine (Grant R01 GM46686 to S.A.W.) and the National Cancer Institute Center for Cancer Research (Y.-X. Wang). This work benefited from CCP-SAS software developed through a joint Engineering and Physical Sciences Research Council (EP/K039121/1) and National Science Foundation (CHE-1265821) grant. Use of the Advanced Photon Source was supported by the US Department of Energy under Contract DE-AC02-06CH11357.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1410114111/-/DCSupplemental.
References
- 1.Gottesman S, et al. Small RNA regulators and the bacterial response to stress. Cold Spring Harb Symp Quant Biol. 2006;71:1–11. doi: 10.1101/sqb.2006.71.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Michaux C, Verneuil N, Hartke A, Giard JC. Physiological roles of small RNA molecules. Microbiology. 2014;160(Pt 6):1007–1019. doi: 10.1099/mic.0.076208-0. [DOI] [PubMed] [Google Scholar]
- 3.Storz G, Opdyke JA, Zhang A. Controlling mRNA stability and translation with small, noncoding RNAs. Curr Opin Microbiol. 2004;7(2):140–144. doi: 10.1016/j.mib.2004.02.015. [DOI] [PubMed] [Google Scholar]
- 4.Vogel J, Luisi BF. Hfq and its constellation of RNA. Nat Rev Microbiol. 2011;9(8):578–589. doi: 10.1038/nrmicro2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mura C, Randolph PS, Patterson J, Cozen AE. Archaeal and eukaryotic homologs of Hfq: A structural and evolutionary perspective on Sm function. RNA Biol. 2013;10(4):636–651. doi: 10.4161/rna.24538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sauer E. Structure and RNA-binding properties of the bacterial LSm protein Hfq. RNA Biol. 2013;10(4):610–618. doi: 10.4161/rna.24201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Link TM, Valentin-Hansen P, Brennan RG. Structure of Escherichia coli Hfq bound to polyriboadenylate RNA. Proc Natl Acad Sci USA. 2009;106(46):19292–19297. doi: 10.1073/pnas.0908744106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mikulecky PJ, et al. Escherichia coli Hfq has distinct interaction surfaces for DsrA, rpoS and poly(A) RNAs. Nat Struct Mol Biol. 2004;11(12):1206–1214. doi: 10.1038/nsmb858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lorenz C, et al. Genomic SELEX for Hfq-binding RNAs identifies genomic aptamers predominantly in antisense transcripts. Nucleic Acids Res. 2010;38(11):3794–3808. doi: 10.1093/nar/gkq032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sauer E, Weichenrieder O. Structural basis for RNA 3′-end recognition by Hfq. Proc Natl Acad Sci USA. 2011;108(32):13065–13070. doi: 10.1073/pnas.1103420108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schumacher MA, Pearson RF, Møller T, Valentin-Hansen P, Brennan RG. Structures of the pleiotropic translational regulator Hfq and an Hfq-RNA complex: A bacterial Sm-like protein. EMBO J. 2002;21(13):3546–3556. doi: 10.1093/emboj/cdf322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ishikawa H, Otaka H, Maki K, Morita T, Aiba H. The functional Hfq-binding module of bacterial sRNAs consists of a double or single hairpin preceded by a U-rich sequence and followed by a 3′ poly(U) tail. RNA. 2012;18(5):1062–1074. doi: 10.1261/rna.031575.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Otaka H, Ishikawa H, Morita T, Aiba H. PolyU tail of rho-independent terminator of bacterial small RNAs is essential for Hfq action. Proc Natl Acad Sci USA. 2011;108(32):13059–13064. doi: 10.1073/pnas.1107050108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sauer E, Schmidt S, Weichenrieder O. Small RNA binding to the lateral surface of Hfq hexamers and structural rearrangements upon mRNA target recognition. Proc Natl Acad Sci USA. 2012;109(24):9396–9401. doi: 10.1073/pnas.1202521109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Panja S, Schu DJ, Woodson SA. Conserved arginines on the rim of Hfq catalyze base pair formation and exchange. Nucleic Acids Res. 2013;41(15):7536–7546. doi: 10.1093/nar/gkt521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Battesti A, Majdalani N, Gottesman S. The RpoS-mediated general stress response in Escherichia coli. Annu Rev Microbiol. 2011;65:189–213. doi: 10.1146/annurev-micro-090110-102946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lease RA, Cusick ME, Belfort M. Riboregulation in Escherichia coli: DsrA RNA acts by RNA:RNA interactions at multiple loci. Proc Natl Acad Sci USA. 1998;95(21):12456–12461. doi: 10.1073/pnas.95.21.12456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Majdalani N, Cunning C, Sledjeski D, Elliott T, Gottesman S. DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proc Natl Acad Sci USA. 1998;95(21):12462–12467. doi: 10.1073/pnas.95.21.12462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soper T, Mandin P, Majdalani N, Gottesman S, Woodson SA. Positive regulation by small RNAs and the role of Hfq. Proc Natl Acad Sci USA. 2010;107(21):9602–9607. doi: 10.1073/pnas.1004435107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Soper TJ, Woodson SA. The rpoS mRNA leader recruits Hfq to facilitate annealing with DsrA sRNA. RNA. 2008;14(9):1907–1917. doi: 10.1261/rna.1110608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lease RA, Woodson SA. Cycling of the Sm-like protein Hfq on the DsrA small regulatory RNA. J Mol Biol. 2004;344(5):1211–1223. doi: 10.1016/j.jmb.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 22.Soper TJ, Doxzen K, Woodson SA. Major role for mRNA binding and restructuring in sRNA recruitment by Hfq. RNA. 2011;17(8):1544–1550. doi: 10.1261/rna.2767211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Updegrove T, Wilf N, Sun X, Wartell RM. Effect of Hfq on RprA-rpoS mRNA pairing: Hfq-RNA binding and the influence of the 5′ rpoS mRNA leader region. Biochemistry. 2008;47(43):11184–11195. doi: 10.1021/bi800479p. [DOI] [PubMed] [Google Scholar]
- 24.Peng Y, Soper TJ, Woodson SA. Positional effects of AAN motifs in rpoS regulation by sRNAs and Hfq. J Mol Biol. 2014;426(2):275–285. doi: 10.1016/j.jmb.2013.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chamberlin SI, Weeks KM. Mapping local nucleotide flexibility by selective acylation of 2′-amine substituted RNA. J Am Chem Soc. 2000;122(2):216–224. [Google Scholar]
- 26.Hämmerle H, Večerek B, Resch A, Bläsi U. Duplex formation between the sRNA DsrA and rpoS mRNA is not sufficient for efficient RpoS synthesis at low temperature. RNA Biol. 2013;10(12):1834–1841. doi: 10.4161/rna.27100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Steen KA, Rice GM, Weeks KM. Fingerprinting noncanonical and tertiary RNA structures by differential SHAPE reactivity. J Am Chem Soc. 2012;134(32):13160–13163. doi: 10.1021/ja304027m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Updegrove TB, Wartell RM. The influence of Escherichia coli Hfq mutations on RNA binding and sRNA•mRNA duplex formation in rpoS riboregulation. Biochim Biophys Acta. 2011;1809(10):532–540. doi: 10.1016/j.bbagrm.2011.08.006. [DOI] [PubMed] [Google Scholar]
- 29.Beich-Frandsen M, et al. Structural insights into the dynamics and function of the C-terminus of the E. coli RNA chaperone Hfq. Nucleic Acids Res. 2011;39(11):4900–4915. doi: 10.1093/nar/gkq1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Henderson CA, et al. Hfq binding changes the structure of Escherichia coli small noncoding RNAs OxyS and RprA, which are involved in the riboregulation of rpoS. RNA. 2013;19(8):1089–1104. doi: 10.1261/rna.034595.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Durand D, et al. NADPH oxidase activator p67(phox) behaves in solution as a multidomain protein with semi-flexible linkers. J Struct Biol. 2010;169(1):45–53. doi: 10.1016/j.jsb.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 32.Wang W, et al. Cooperation of Escherichia coli Hfq hexamers in DsrA binding. Genes Dev. 2011;25(19):2106–2117. doi: 10.1101/gad.16746011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Curtis JE, Raghunandan S, Nanda H, Krueger S. SASSIE: A program to study intrinsically disordered biological molecules and macromolecular ensembles using experimental scattering restraints. Comput Phys Commun. 2012;183(2):382–389. [Google Scholar]
- 34.Parisien M, Major F. The MC-Fold and MC-Sym pipeline infers RNA structure from sequence data. Nature. 2008;452(7183):51–55. doi: 10.1038/nature06684. [DOI] [PubMed] [Google Scholar]
- 35.Petoukhov MV, Svergun DI. Global rigid body modeling of macromolecular complexes against small-angle scattering data. Biophys J. 2005;89(2):1237–1250. doi: 10.1529/biophysj.105.064154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Salim NN, Feig AL. An upstream Hfq binding site in the fhlA mRNA leader region facilitates the OxyS-fhlA interaction. PLoS ONE. 2010;5(9):e13028. doi: 10.1371/journal.pone.0013028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Salvail H, Caron MP, Bélanger J, Massé E. Antagonistic functions between the RNA chaperone Hfq and an sRNA regulate sensitivity to the antibiotic colicin. EMBO J. 2013;32(20):2764–2778. doi: 10.1038/emboj.2013.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.