Significance
Breaks in DNA are repaired by homologous recombination. Because the structure of DNA ends at a break site can be variable, the repair machinery must be designed to act on a variety of heterogeneous DNA break sites. Bacterial RecQ helicase and RecJ nuclease initiate the repair of double-stranded DNA breaks; however, neither protein alone can deal with the broad range of physiological ends. Human Bloom syndrome helicase (BLM) is the homolog of RecQ, and it functions in DNA resection, contributing to genomic stability in humans. We establish that RecQ and RecJ complement one another by acting on DNA ends and intermediates that the other cannot. Thus, by leveraging complementary substrate preferences, recombination initiation from all types of DNA ends, in many organisms, is ensured.
Keywords: DNA repair, homologous recombination, DNA break, helicase, nuclease
Abstract
Recombinational DNA repair by the RecF pathway of Escherichia coli requires the coordinated activities of RecA, RecFOR, RecQ, RecJ, and single-strand DNA binding (SSB) proteins. These proteins facilitate formation of homologously paired joint molecules between linear double-stranded (dsDNA) and supercoiled DNA. Repair starts with resection of the broken dsDNA by RecQ, a 3′→5′ helicase, RecJ, a 5′→3′ exonuclease, and SSB protein. The ends of a dsDNA break can be blunt-ended, or they may possess either 5′- or 3′-single-stranded DNA (ssDNA) overhangs of undefined length. Here we show that RecJ nuclease alone can initiate nucleolytic resection of DNA with 5′-ssDNA overhangs, and that RecQ helicase can initiate resection of DNA with blunt-ends or 3′-ssDNA overhangs by DNA unwinding. We establish that in addition to its well-known ssDNA exonuclease activity, RecJ can display dsDNA exonuclease activity, degrading 100–200 nucleotides of the strand terminating with a 5′-ssDNA overhang. The dsDNA product, with a 3′-ssDNA overhang, is an optimal substrate for RecQ, which unwinds this intermediate to reveal the complementary DNA strand with a 5′-end that is degraded iteratively by RecJ. On the other hand, RecJ cannot resect duplex DNA that is either blunt-ended or terminated with 3′-ssDNA; however, such DNA is unwound by RecQ to create ssDNA for RecJ exonuclease. RecJ requires interaction with SSB for exonucleolytic degradation of ssDNA but not dsDNA. Thus, complementary action by RecJ and RecQ permits initiation of recombinational repair from all dsDNA ends: 5′-overhangs, blunt, or 3′-overhangs. Such helicase–nuclease coordination is a common mechanism underlying resection in all organisms.
Homologous recombination is a relatively error-free mechanism to repair double-stranded DNA (dsDNA) breaks (DSBs) and single-stranded DNA (ssDNA) gaps, which are produced by UV light, γ-irradiation, and chemical mutagens (1). In wild-type Escherichia coli, the labor of recombinational repair is divided between the RecBCD and RecF pathways of recombination, which are responsible for the repair of DSBs and ssDNA gaps, respectively (2–5). However, the proteins of the RecF pathway are capable of DSB repair, as well as ssDNA gap repair: in recBC mutant cells containing the suppressor mutations, sbcB and sbcC (suppressors of recBC), the proteins of the RecF pathways provide the needed recombinational DNA repair functions (2, 6).
The RecF pathway in E. coli involves the functions of RecA, RecF, RecG, RecJ, RecN, RecO, RecQ, RecR, RuvA, RuvB, RuvC, and single-strand DNA binding (SSB) proteins (1, 7). The RecF pathway of recombination is evolutionarily conserved across Bacteria, with most of components present in all bacteria (8). In addition, orthologs of RecF pathway proteins are found in Eukarya. RecA promotes DNA strand invasion and exchange (9–11), as does eukaryotic Rad51 (12, 13). RecO can both anneal SSB–ssDNA complexes (14, 15) and, in conjunction with RecR (and RecF), mediate loading of RecA onto SSB–ssDNA complexes (16–18). Saccharomyces cerevisiae Rad52 is a functional homolog of RecO in that it also displays both DNA-annealing and Rad51-loading activities (19–22). The RecFOR complex promotes the loading of RecA onto SSB-coated gapped DNA at ssDNA–dsDNA junctions (17, 18) and, when mutated, is suppressed by hyperactive alleles of recA (23), a property that is shared with the yeast Rad55/57 proteins (24). Furthermore, human BRCA2 protein and a fungal analog, Brh2, are partial functional analogs of the RecFOR proteins (25–27).
RecQ helicase plays several roles in both early and late steps of recombination (28, 29), as do the RecQ-family helicases in Eukarya [e.g., Sgs1 and Bloom Syndrome helicase (BLM)] (30–32). In addition, eukaryotic Exonuclease 1 (Exo1) and Dna2 helicase/nuclease function somewhat analogously, although not identically, to RecJ nuclease (33–36). The in vitro reconstitution of DSB repair in E. coli, yeast, and human have shown that resection involves specific pairs of a helicase and nuclease for DNA end resection: RecQ/RecJ, Sgs1/Dna2, BLM/DNA2, and BLM/EXO1 (28, 37–39).
A comparison of DSB repair by the RecBCD and RecF pathways shows that repair starts with the processing a DSB into resected dsDNA with a 3′-ssDNA overhang (7). RecJ has a 5′ to 3′ exonuclease activity on ssDNA and the action of RecJ is facilitated by RecQ, which has a 3′ to 5′ helicase activity (40, 41). The resulting processed DNA has a 3′-ssDNA overhang. The RecFOR complex binds to the 5′-end at the junction between ssDNA and dsDNA, and loads RecA protein onto the adjacent ssDNA (17, 18). Finally, the RecA nucleoprotein filament promotes pairing with homologous dsDNA (9). These steps have been reconstituted in vitro in a coordinated reaction using RecAFORQJ and SSB proteins (28).
Despite progress, most studies have used DNA substrates with simple blunt-ends. However, in vivo, there are many potential structures at the end of a DSB. When the DSB is created by a replication fork encountering nicked DNA, the break can be blunt-ended (5). However, related mechanisms can produce dsDNA with either 5′- or 3′-ssDNA overhangs. Similarly, the actual intermediates of DNA processing may result in dsDNA with either 5′- or 3′-ssDNA ends. Clearly, a DNA repair pathway must be capable of dealing with such a variety of DNA end structures. In this study, we investigated the processing of DSBs by RecJ and RecQ, both individually and together. We found that a DNA with a 5′-ssDNA overhang end was degraded by RecJ nuclease and converted into an intermediate with a 3′-ssDNA overhang. Although this intermediate was no longer a substrate for RecJ, RecQ could bind to this intermediate and initiate unwinding, thereby supplying 5′-tailed ssDNA for further resection by RecJ. In addition, we established that RecQ allows RecJ to initiate nucleolytic resection on otherwise poor substrates (e.g., blunt-end DNA or DNA with 3′-ssDNA overhangs). Thus, RecQ and RecJ cooperate biochemically to create DNA intermediates for one another that enable resection of all types of broken DNA molecules.
Results
The Structure of DNA Ends Affects the Efficiency of Joint Molecule Formation by RecAFORQJ and SSB Proteins.
As previously reported (28), the RecA, RecF, RecO, RecR, RecQ, RecJ, and SSB proteins promote joint molecule formation between linear dsDNA and homologous supercoiled DNA, as illustrated in Fig. 1A. In the previous study (28), we used EcoRI to make the linear dsDNA, which produces 4-nt 5′-ssDNA overhangs at both ends. To investigate the relationship between the structure of the DNA end and the efficiency of joint molecule formation by these proteins, we examined linear dsDNA with different end-structures. Initially, linear dsDNA with 5′-ssDNA overhangs, blunt-ends, and 3′-ssDNA overhangs was created by digesting pUC19 circular DNA with EcoRI, SmaI, and PstI, respectively, and then tested for DNA pairing with supercoiled DNA in reactions mediated by RecAFORQJ and SSB proteins at the previously used standard temperature of 30 °C (Fig. 1B). The dsDNA with 5′-ssDNA overhangs was a good substrate for joint molecule formation, but DNA with blunt-ends or 3′-ssDNA overhangs was not (Fig. 1B). The structure at the DNA end clearly affected the efficiency of dsDNA processing by RecQ and RecJ as evident by the heterogeneous smear below the linear dsDNA band for DNA with 5′-ssDNA overhangs (Fig. 1B, lanes 3 and 4), which was not apparent for blunt-ended DNA or DNA with 3′-ssDNA overhangs (Fig. 1B, lanes 7, 8, 11, and 12). Thus, the nature of the DNA end affects the efficiency of dsDNA resection, which is the first step of joint molecule formation.
Fig. 1.
RecAFORQJ-mediated joint molecule formation is sensitive to the end structure of the linear dsDNA to be resected. (A) Schematic of RecAFORQJ-mediated joint molecule formation between linear dsDNA and supercoiled dsDNA (28). The reaction comprises two concerted steps. In the first step, RecJ—with the help of RecQ and SSB—processes the linear dsDNA. In the second step, the RecFOR proteins load RecA onto the processed linear DNA, which is followed by RecA-promoted joint molecule formation. (B) Joint molecule formation using linear dsDNA with 4-nt 5′-overhangs (lanes 1–4), blunt-ends (lanes 5–8), or 4-nt 3′-overhangs (lanes 9–12). EcoRI, SmaI, and PstI were used to generate the linear dsDNA. Samples were analyzed by agarose gel electrophoresis followed by ethidium bromide staining. (C) Effect of linear DNA-end structure on joint molecule formation was investigated using EcoRI-cut pUC19 DNA that was filled-in to produce the overhangs indicated. Joint molecule assays, such as those shown in B, were quantified to determine the percentage of the limiting linear dsDNA that was converted into joint molecules. Each experiment was performed twice; the error bars show standard error (SE), unless smaller than the symbol.
To determine the effect of DNA-end structure in more detail, we constructed substrates with 4-, 3-, 2-, and 1-nt overhangs and blunt-ends by treating the EcoRI- or HindIII-cut DNA with Klenow fragment and various combinations of deoxynucleoside triphosphates (dNTPs) and dideoxynucleoside triphosphates (ddNTPs). For the substrates generated from EcoRI-cleaved DNA (Fig. 1C), the 4-nt overhang showed the highest efficiency of joint molecule formation (24% at 60 min), whereas the 3-nt overhang showed a lower efficiency (7% at 60 min); and the other DNA substrates (0-, 1-, and 2-nt overhangs) showed less than 3% joint molecule formation after 60 min (Fig. 1C). Similar results were obtained using substrates generated from HindIII-cut DNA (Fig. S1A): DNA with 4- and 3-nt 5′-overhangs produced 10% and 13% joint molecules at 60 min, respectively, whereas DNA with 0-, 1-, or 2-nt overhangs produced less than 2% at 60 min. The difference in the efficiencies of joint molecule formation also corresponded to the difference in efficiencies of processing (Fig. S1 B and C). These results demonstrate that DNA with the longer 5′-ssDNA overhang was the better substrate for resection by RecQ and RecJ.
RecJ Can Process DNA with a 5′-ssDNA Overhang to Degrade That Strand into the DNA Duplex Region to Produce Resected DNA with a 3′-ssDNA Overhang.
The structure of the dsDNA end affected not only the accumulation of joint molecule products, but also resection of the linear dsDNA (Fig. 1B and Fig. S1 B and C). Because RecJ nuclease is responsible for resection in this reconstituted system (28), this finding suggested that RecJ’s nucleolytic activity is sensitive to DNA end structure. Therefore, we investigated the processing of linear dsDNA with different end structures by RecJ in the absence and presence of SSB and RecQ. In this study, we use two different methods to detect the DNA resection. One procedure uses linear plasmid dsDNA (2.7 kb) and agarose gel electrophoresis, as this assay was also used to detect joint molecule formation; this procedure needs a change of a few percent or more in length of DNA for detection, requiring resection of ∼200 nt or more to be scored in this assay. The other procedure uses shorter (∼250 bp) linear dsDNA and denaturing PAGE; this procedure can detect changes of a few nucleotides in length. We used the former method to detect extensive, long-range (>200 nt) resection of DNA and the latter method to detect more limited, short-range (<200 nt) resection.
Fig. 2 shows resection of the short (∼250 bp) dsDNA by RecJ detected by denaturing PAGE. The DNA substrates are depicted at the top of Fig. 2; in each case, the bottom strand was labeled with 32P at 3′-end using DNA polymerase so that the left end of each dsDNA is blunt. The right end of each dsDNA is the product of restriction enzyme cleavage, so that the structure is a 5′-ssDNA overhang, blunt-end, or 3′-ssDNA overhang. Because degradation by RecJ occurs in 5′ to 3′ direction (40), the assay monitors resection of the bottom DNA strand from its right side. Fig. 2A shows the products produced by RecJ in the presence of SSB. Surprisingly, even in the absence of RecQ, RecJ resected the dsDNA with a 4-nt 5′-overhang well into the region of DNA duplex (Fig. 2A, lanes 1–6). Although most of the products represented degradation of ∼50 nt into the dsDNA, resection of 100–200 nt of duplex DNA is detectable as shown by the appearance of products near the 64-nt marker (Fig. 2A, lane 2). These results are consistent with our previous results, where the RecJ nuclease—but not RecQ helicase—was shown to be necessary for RecAFOR-dependent joint molecule formation (28). Interestingly, although substrates of three different lengths were used, the patterns of product formation are similar (Fig. 2A, lanes 2, 4, and 6), suggesting the existence of common sites for termination (see below). Degradation of DNA with a 2-nt, 5′-overhang produced a similar pattern, but with a lower efficiency of processing: 94% of the substrate remained unprocessed after 60 min of incubation (Fig. 2A, lane 8). On the other hand, DNA with a blunt-end or a 3′-ssDNA overhang was not significantly resected (Fig. 2A, lanes 9–16).
Fig. 2.
The RecJ nuclease processes linear dsDNA together with RecQ helicase and SSB protein. (A) Processing by RecJ and SSB, but in the absence of RecQ. EcoRI (lanes 1–2), SalI (lanes 3 and 4), HindIII (lanes 5 and 6), AccI (lanes 7 and 8), SmaI (lanes 9 and 10), HincII (lanes 11 and 12), KpnI (lanes 13 and 14), and PstI (lanes 15 and 16) were used to prepare the substrates as shown at the top of panels. Analysis was at 0 and 60 min by the denaturing PAGE assay. (B) Processing by RecJ, RecQ, and SSB; same DNA and analysis as in A. (C) The processing in the presence of the protein indicated, using HindIII-cut DNA. Analysis was at 0 and 60 min by the denaturing PAGE assay.
The same resection reactions mediated by RecJ were also performed in the presence of RecQ and SSB (Fig. 2B). Each of the three different DNA substrates with a 4-nt 5′-overhang was processed within 60 min (Fig. 2B, lanes 1–6). However, in this case, most of the DNA was completely degraded, resulting in nearly complete disappearance of substrate bands at 60 min, and only a few percent remaining as products that were shorter by a few bases. In contrast, processing of DNA with a 2-nt 5′-overhang, a blunt-end, and a 4-nt 3′-overhang under the same condition was not detected (Fig. 2B, lanes 7–16). These observations are consistent with the results obtained in Fig. 1. The failure of RecQ to process the blunt-end and 3′-ssDNA overhanging DNA (Fig. 2B, lanes 9–16) is not unexpected because the buffer contained 10 mM Mg2+ and, at this concentration of Mg2+, RecQ does not efficiently unwind dsDNA (42). Note that a low amount of resection of the 2-nt 5′-overhang was expected in the presence of RecQ, but the extent was too low to be detected in this assay (see below). Furthermore, the failure to detect processing of the blunt-ended DNA verifies that RecQ cannot initiate unwinding from either blunt-ends (Fig. 2B, lanes 10 and 12) or 4-nt 3′-overhangs (Fig. 2B, lanes 14 and 16) at these conditions (but see below). However, even though RecQ cannot initiate unwinding, it is clear from the resection of DNA with a 5′-ssDNA overhang (Fig. 2B, lanes 1–6) that, even though RecJ initiates resection, RecQ facilitates more extensive resection.
To determine whether SSB is required for resection, DNA with a 4-nt 5′-overhang was treated with RecJ in the absence of SSB (Fig. 2C, lanes 7 and 8). Resection occurred, but most of processing terminated within 50 bases (compare Fig. 2C, lanes 4 and 8), indicating that SSB stimulated nucleolytic degradation by RecJ. RecQ also stimulated the processing by RecJ nuclease in the absence of SSB (Fig. 2C, compare lanes 6 and 8). In the presence of both SSB and RecQ, the processing by RecJ was almost complete as described above (Fig. 2C, lanes 1 and 2). Fig. 2C also shows that the RecJ is necessary for resection (lanes 9 and 10).
Degradation by RecJ Nuclease Stops Predominantly Upstream of GC-Rich Sequences.
To determine the positions of termination during RecJ-mediated degradation, the electrophoretic pattern of resection products was compared with a Maxam–Gilbert sequencing ladder (Fig. S2). Both in the absence and presence of SSB, RecJ degradation stopped at the same positions, but with different frequencies (Fig. S2B); however, in the absence of SSB, RecJ terminated resection earlier than in its presence. Five major termination sites observed both in the absence and presence of SSB are indicated with black arrows. These sites are positioned to the 5′-side of GC-rich sequences (Fig. S2B, underlined sequences), suggesting that the increased stability of GC-rich duplex DNA is inhibitory to the RecJ-mediated resection in the absence of RecQ. There are also some termination sites that are unique to either the absence or presence of SSB (Fig. S2), but the nature or cause of termination at these sites is unknown.
DNA Resection by RecJ Nuclease Is Processive.
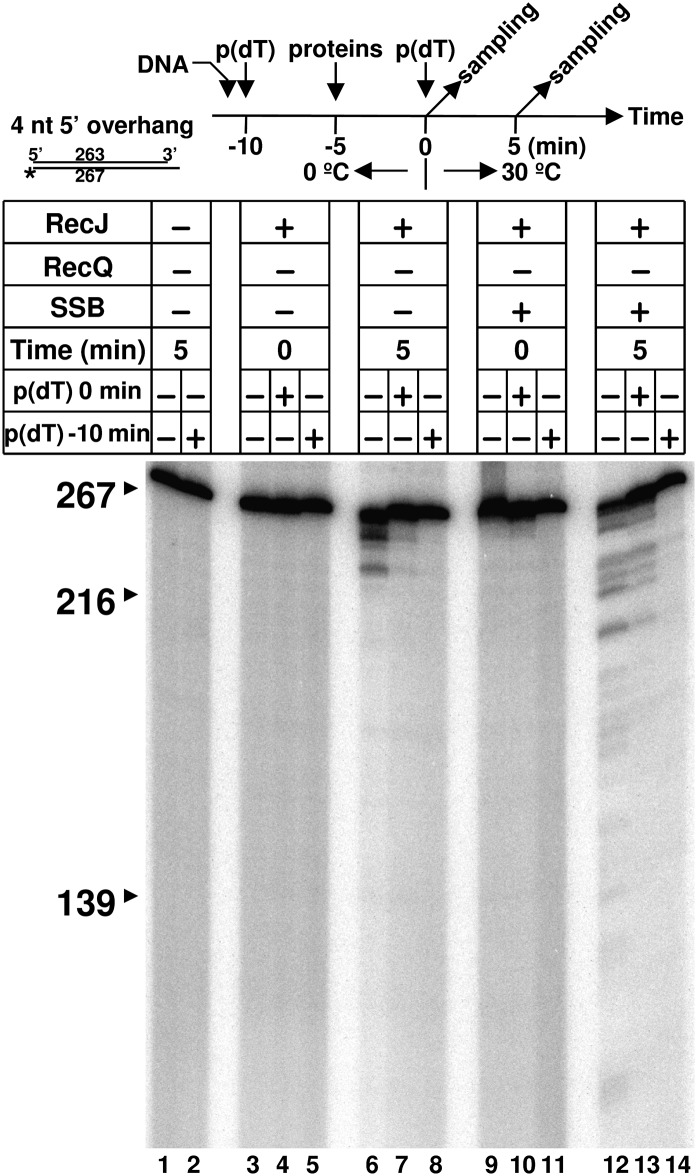
The results shown in Fig. 2 suggest that resection of DNA with a 5′-ssDNA overhang by RecJ is processive: if the reaction had occurred in a stepwise distributive manner, then blunt-ended DNA would have been generated as an intermediate and the resection would have stopped completely. To confirm this conclusion, we performed RecJ-mediated resection reactions in the presence of a DNA scavenger added after the formation of a RecJ–DNA complex (Fig. 3). RecJ nuclease was first mixed with the DNA on ice. After a 5-min incubation, resection was initiated by adding the scavenger DNA and then shifting the temperature to 30 °C. At time 0 (Fig. 3, lanes 3–5 and 9–11) and after a 5-min incubation (Fig. 3, lanes 6–8 and 12–14), a sample was taken and analyzed by PAGE. In the absence of scavenger DNA, resection occurred as shown previously, with a different pattern of product in absence and presence of SSB (Fig. 3, lanes 6 and 12, respectively). This resection did not occur before the temperature shift from 0 °C to 30 °C (Fig. 3, lanes 3 and 9). Similar patterns of products were also observed in the presence of scavenger DNA (Fig. 3, compare lanes 6 and 7 and lanes 12 and 13), but the band intensities are lower, implying that RecJ degrades a defined processive distance before dissociating, which is shorter than the substrate and, in the absence of scavenger, multiple rounds occur but RecJ cannot act on the 3′-ssDNA–tailed intermediates. As a control, resection did not occur when scavenger DNA was added before addition of RecJ to the sample (Fig. 3, lanes 8 and 14). These results demonstrate that resection by RecJ of dsDNA with a 5′-ssDNA overhang is processive either in the presence or absence of SSB. The products of this reaction have a 3′-ssDNA overhang, which is not used for further resection by RecJ nuclease in the absence of RecQ helicase (Fig. S3). Thus, resection of DNA with a 5′-ssDNA overhang—in the absence of RecQ helicase—is processive and effectively a single-turnover reaction of self-limited extent because the 3′-ssDNA–tailed product is not a suitable substrate for RecJ.
Fig. 3.
Resection of dsDNA by RecJ nuclease is processive. Resection performed in the presence of scavenger DNA, poly(dT) (40-μM nucleotides). 32P-labeled HindIII-cut dsDNA was used as described in Materials and Methods. After incubation for the time indicated (0 or 5 min), samples were analyzed by PAGE. Reactions without scavenger DNA are lanes 1, 3, 6, 9, and 12. In the reactions of lanes 4, 7, 10, and 13, scavenger DNA were added at 0 min. To show that the scavenger is effective, poly(dT) was added 5 min before mixing dsDNA with RecJ [shown as “p(dT) -10 min”: lanes 2, 5, 8, 11, and 14]. The reaction scheme is summarized at the top.
The Efficiency of Resection Is Sensitive to Duplex DNA Stability.
In the absence of RecQ, RecJ could resect dsDNA with 5′-ssDNA overhangs only for a short distance; even though resection lengths of 100–200 nt could be detected, the majority of products were resected <100 nt (Fig. 2A). However, in the presence of RecQ, the resection lengths were much longer (>200 nt) (Figs. 1B and 2B). This RecQJ-coupled resection was performed in the presence of 10 mM Mg2+, a reaction condition where the RecQ cannot unwind dsDNA efficiently even in the presence of SSB (42). However, the DNA substrates used for RecQ-mediated reactions in the previous studies and in this current study differ significantly. Typically, linearized plasmid DNA that had either blunt-ends or either 5′- or 3′-overhangs shorter than 4 nt was used in the helicase assays, but in this study the RecJ-resected linear DNA would have contained a stretch of 3′-ssDNA substantially longer than 4 nt before the duplex region. Given that RecQ is a 3′ to 5′ helicase, we compared unwinding of three different synthetic DNA substrates: 80-bp blunt-ended dsDNA, 40-bp blunt-ended dsDNA, and 40-bp dsDNA with a 40-nt 3′-ssDNA overhang, in the presence of SSB (Fig. S4A). RecQ could not efficiently unwind 80-bp and 40-bp blunt-ended DNA at the higher Mg2+ concentrations (Fig. S4A, lanes 2 and 4, and Fig. S4B), as expected. On the other hand, DNA with the 3′-ssDNA overhang was unwound efficiently (Fig. S4A, lane 6, and Fig. S4B).
It is also known that RecQ can efficiently unwind dsDNA when the free Mg2+ concentration is lowered (42). As expected, when the concentration of Mg2+ was decreased progressively, more of the 80-bp and 40-bp blunt-ended dsDNA was unwound (Fig. S4B, red and blue symbols). In contrast, the DNA with the 3′-ssDNA overhang was only slightly sensitive to Mg2+ concentration over this range (Fig. S4B, black plots). These results show that the RecQ helicase is active on DNA with 40-nt 3′-ssDNA overhangs even in the presence of 10 mM Mg2+, and they are consistent with the findings in Fig. 2 B and C.
To determine the effect of Mg2+ concentration on RecQJ-coupled resection, we next monitored resection using agarose gel electrophoresis, which detects only extensive, long-range resection but not short-range resection. Fig. 4A shows resection by RecQ, RecJ, and SSB proteins in the presence of 1, 5, and 10 mM Mg2+. In 10 mM Mg2+, only DNA with a 5′-ssDNA overhang was efficiently processed (Fig. 4A, lanes 21–32), as demonstrated earlier (Fig. 1B). In 5 mM Mg2+, processing of dsDNA with either a 3′-ssDNA or a 5′-ssDNA overhang was detected (Fig. 4A, lanes 9–16), but the blunt-end DNA remained refractory to processing (Fig. 4A, lanes 17–20). When the Mg2+ concentration was reduced further to 1 mM, different degradation patterns were observed (Fig. 4A, lanes 1–8). The dsDNA (either 5′-ssDNA overhang or blunt-end) was first converted to ssDNA by RecQ unwinding (Fig. 4A, lanes 2 and 6), and then it was slowly degraded by RecJ nuclease (Fig. 4A, lanes 3–4 and 7–8). Under these conditions, DNA unwinding by RecQ occurs readily, not only from DNA ends but also from sites internal to the DNA (42). On the other hand, RecJ showed less nucleolytic degradation of ssDNA in 1 mM Mg2+ than in 10 mM Mg2+ (Fig. S4C). Quantification of data such as those in Fig. 4A is shown in Fig. 4B, where the amount of dsDNA substrate processed at 60 min is plotted as a function of a more complete Mg2+ titration. Under all conditions, dsDNA with a 5′-ssDNA overhang is resected efficiently (Fig. 4B, black circles) [note that at 1 mM Mg2+ more than 90% of dsDNA was converted to ssDNA by RecQ helicase but was not completely degraded by RecJ nuclease (Fig. 4A, lanes 1–8)]. The processing of DNA with a 3′-ssDNA overhang was efficient at low Mg2+ concentrations (<6 mM) but not at higher Mg2+ concentration (Fig. 4B, red squares). For blunt-end DNA, the threshold for efficient processing was less than 2 mM Mg2+ for these experiments, which were performed at 30 °C (Fig. 4B, blue triangles); when the temperature was increased to 37 °C, however, the threshold shifted to ∼4 mM, and more than 80% of DNA was unwound and degraded at 2 mM Mg2+ (Fig. S4 D and E). Thus, the efficiency of resection depends on the concentration of Mg2+, temperature, and the structure of the DNA end. When either RecJ or RecQ was omitted, this extensive resection was not experimentally detectable by the agarose gel assay (Fig. S4F); nonetheless, limited resection of shorter lengths occurred in the absence of RecQ, as detected by the PAGE assay (Fig. 2 A and C).
Fig. 4.
Both the structure of the DNA end and the concentration of magnesium ion affect the efficiency of dsDNA resection by RecQJ proteins. (A) Extensive long-range resection of dsDNA was followed by agarose gel electrophoresis. DNA with 5′-ssDNA overhangs, blunt-ends, and 3′-overhangs was prepared by EcoRI, SmaI, and PstI digestion of pUC19 DNA. Reactions contained RecQ, RecJ, and SSB, and indicated concentration of magnesium acetate. (B) Quantification and plot of dsDNA processed at 60 min as a function of Mg2+ concentration. The experiments were performed with RecQ, RecJ, and SSB. The experiments were performed twice and error bars show SEs, unless smaller than the symbol. “dsDNA processed” represents the loss of full-length dsDNA substrate at 60 min. Note that at 1 mM Mg2+, this value represents mainly unwinding by RecQ helicase. (C) Joint molecule formation as a function of Mg2+ concentrations. The experiments were performed with RecQ, RecJ, SSB, and RecAFOR proteins as done in Fig. 1, but in the presence of the indicated concentration of magnesium acetate, using DNA with 5′-ssDNA overhangs (EcoRI-cut), blunt-ends (SmaI-cut), or 3′-ssDNA overhangs (PstI-cut), and assayed at 60 min. (D) Quantification of joint molecule formation after 60 min in the presence of both RecQ and RecJ, RecQ, or RecJ with EcoRI-cut pUC19 (5′-overhang), PstI-cut pUC19 (3′-overhang), or SmaI-cut pUC19 (blunt-end) as the linear dsDNA substrate. RecA, RecFOR, and SSB proteins were always present. The magnesium acetate concentration was 5 mM. The reactions were performed at 30 °C for DNA with 5′- and 3′-overhang DNA, and at 37 °C for blunt-end DNA. The experiments were performed two to four times and error bars show SEs.
Fig. 4B shows that DNA with 3′-overhangs or blunt-ends was poorly processed by RecQJ when the concentration of Mg2+ was increased to 10 mM. This low rate of resection could explain the paucity of joint molecules observed at 10 mM Mg2+, when DNA with 3′-overhangs or blunt-ends was used (Fig. 1). To confirm this possibility, we measured joint molecule formation at increasing concentrations of Mg2+ (Fig. 4C). For DNA with 5′-overhangs, joint molecule formation increased steadily up to 10 mM Mg2+ (Fig. 4C, black circles). Because the extent of resection within this range of Mg2+ concentration was relatively unchanged (Fig. 4B), this finding implied that either RecFOR-mediated loading of RecA or RecA-mediated heteroduplex formation was the rate-limiting step. In fact, the Mg2+-dependence of this reaction is similar to that of RecA-mediated joint molecule formation between ssDNA and homologous dsDNA (43, 44), suggesting that RecA-mediated heteroduplex formation was rate-limiting. On the other hand, joint molecule formation involving DNA with 3′-overhangs showed a peak at 4–5 mM Mg2+ and then decreased above this concentration (Fig. 4C, red squares). Because the efficiency of RecQJ-mediated resection of DNA with 3′-overhangs decreased from 6 to 10 mM Mg2+ (Fig. 4B), the efficiency of joint molecule formation should decrease even though the efficiency of DNA heteroduplex formation by RecA protein increased. Similarly, joint molecule formation with blunt-end DNA also showed a peak at 5 mM Mg2+ for the same reasons (Fig. S4G), but because processing of blunt-ended DNA was the poorest (Fig. 4B), the overall yields of pairing products were the lowest at all conditions tested (Fig. 4C and Fig. S4G).
To see the requirement of RecQ helicase in resection, we used DNA possessing 3′-overhangs at 5 mM Mg2+, where the reaction could not be initiated by RecJ nuclease but only by RecQ helicase; joint molecule formation was measured as a function of RecQ concentration (Fig. S4H). At 0-nM RecQ helicase, the reaction efficiency was lower than 2%. In the presence of 5- to 20-nM RecQ helicase, joint molecules increased to 4–5% then decreased as the concentration of RecQ increased, as observed previously (28). This result is clearly different from the same experiment using DNA with 5′-overhangs: 5- to 10-nM RecQ helicase only slightly increased the joint molecule formation (28) (Fig. 4D). Joint molecule formation with blunt-end DNA also required RecQ helicase (Fig. 4D). RecJ nuclease was required for joint molecule formation with all three different linear DNA molecules (Fig. 4D). These observations demonstrate that in addition to RecJ nuclease, RecQ helicase has an important role in the initiation of homologous recombination, especially when the DNA is blunt-ended or has 3′-overhangs.
RecJ Requires the C Terminus of SSB Protein for Resection of ssDNA but Not for Resection of dsDNA.
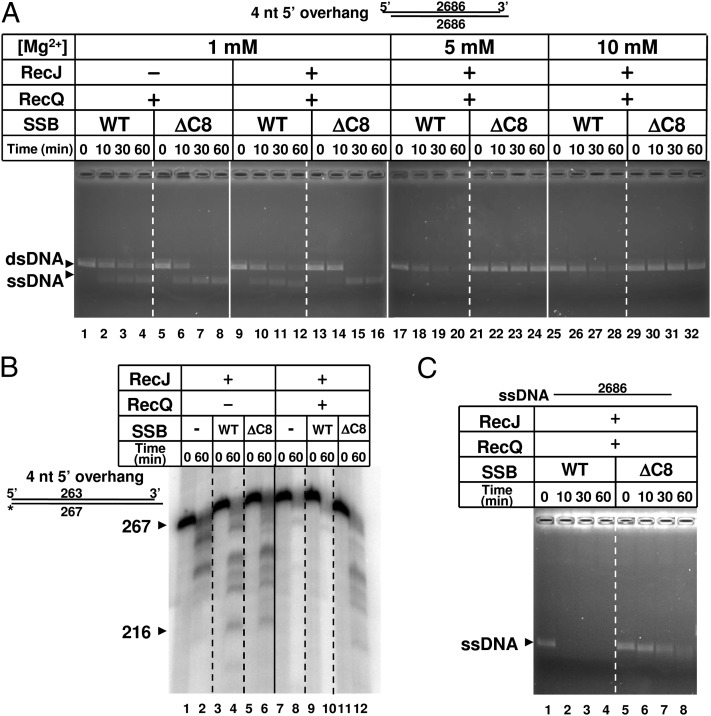
SSB physically interacts with both RecQ and RecJ (45, 46). The interaction between SSB and RecQ is mediated via interactions with the C terminus of SSB (47). To determine whether this interaction is required for RecQJ-mediated resection, we used a truncated SSB that lacks eight amino acids from the C terminus, and is known to be defective for these protein–protein interactions (Fig. 5). Fig. 5A shows the effect of this C-terminally truncated SSB on RecQJ-mediated resection as measured by agarose gel electrophoresis. At 1 mM Mg2+, both wild-type and truncated SSB supported unwinding by RecQ (Fig. 5A, lanes 1–8), showing that this interaction domain was not essential for RecQ helicase function. When RecJ was also present, the ssDNA was slightly but clearly degraded with wild-type SSB present, as seen by the small change in electrophoretic mobility (Fig. 5A, lanes 9–12; see also Fig. 4A, lanes 1–4), but not with truncated SSB (Fig. 5A, lanes 13–16). The difference in resection was more significant when the Mg2+ concentration was increased to 5 mM (Fig. 5A, lanes 17–24) or 10 mM (Fig. 5A, lanes 25–32). Extensive resection by RecQ and RecJ was not observed in the presence of truncated SSB (Fig. 5A, lanes 21–24 and 29–32), although it occurred efficiently in the presence of wild-type SSB (Fig. 5A, lanes 17–20 and 25–28). These results demonstrate that the C terminus of SSB is involved in the extensive resection of dsDNA by RecQJ proteins, likely because of interaction with RecJ.
Fig. 5.
The C terminus of SSB protein is required for resection of ssDNA by RecJ nuclease, but for neither resection of dsDNA by RecJ nuclease nor unwinding of dsDNA by RecQ helicase. (A) Extensive long-range resection of dsDNA in the presence of wild-type (WT) or truncated (ΔC8) SSB was followed by agarose gel electrophoresis. The experiments were in the presence of the proteins indicated at the indicated concentration of magnesium acetate. The substrate was EcoRI-cut pUC19 DNA. (B) Short-range resection of dsDNA in the absence or presence of wild-type (WT) or truncated (ΔC8) SSB protein as followed by denaturing PAGE. The experiments were done in the presence of the proteins indicated at 10 mM magnesium acetate. The substrate was 32P-labeled, HindIII-cut pUC19 DNA as used in Fig. 2. (C) Degradation of ssDNA in the presence of wild-type (WT) or truncated (ΔC8) SSB was followed by agarose gel electrophoresis. The EcoRI-cut pUC19 DNA was converted to ssDNA by RecQ at 1 mM magnesium acetate before adding RecJ and 9 mM magnesium acetate.
To determine whether the SSB truncation affected the more limited, short-range resection of dsDNA by RecJ, reactions in 10 mM Mg2+ were analyzed by PAGE (Fig. 5B). In the absence of RecQ, truncated SSB supported resection by RecJ (Fig. 5B, lane 6) as efficiently as the wild-type SSB (Fig. 5B, lane 4). Most of this resection is because of exonucleolytic degradation of dsDNA, not ssDNA, showing that interaction with SSB has little role in resection of dsDNA by RecJ. Addition of the RecQ to the reaction without SSB or with wild-type SSB resulted in disappearance of resection intermediates (Fig. 5B, lanes 8 and 10), signifying extensive resection; however, in the presence of the truncated SSB, the same gel pattern was observed regardless of the presence of RecQ (Fig. 5B, lane 6 vs. 12), indicating that the intermediates created by RecJ could not be further processed by RecQJ-mediated resection. Extensive resection by RecQ and RecJ requires exonucleolytic degradation of dsDNA by RecJ, which is unaffected by the SSB truncation, as shown above. This step is then followed by RecQ unwinding, which also is unaffected by SSB truncation, and finally, this is followed by exonucleolytic resection of the SSB-coated ssDNA, which evidently must be affected by SSB truncation.
To confirm this inference regarding the role of this SSB interaction on the capacity of RecJ to degrade ssDNA, dsDNA was first unwound by RecQ at 1 mM Mg2+ in the presence of either wild-type SSB (Fig. 5C, lane 1) or truncated SSB (Fig. 5C, lane 5) but in the absence of RecJ. Subsequently, the concentration of Mg2+ was increased to 10 mM and resection of the SSB–ssDNA complex was initiated by adding RecJ (Fig. 5C, lanes 2–4 and 6–8). In the presence of wild-type SSB, the ssDNA was almost completely degraded by RecJ nuclease within 10 min (Fig. 5C, lane 2); in contrast, in the presence of the truncated SSB, degradation was slower and resection intermediates remained at 60 min (Fig. 5C, lane 6). These results indicate that the C-terminal region of SSB is important for the efficient processing of SSB-coated ssDNA by RecJ nuclease. Thus, the C terminus of SSB is important for extensive processing by RecQ and RecJ proteins, because the interaction with RecJ is needed for exonucleolytic degradation of SSB-ssDNA but not for exonucleolytic degradation of dsDNA.
Discussion
In this study, we investigated the processing of DNA by RecJ nuclease and RecQ helicase with regards to their functions in the initiation of homologous recombination. We discovered that resection of a DSB is the consequence of coordination between RecQ and RecJ proteins, and that it depends on the nature of DNA end. We established that RecQ prefers DNA with 3′-ssDNA ends over blunt-ends, and that it acts poorly on DNA with 5′-ssDNA ends. In contrast, RecJ provides the complement to this bias: we discovered that RecJ degrades the strand of dsDNA that is terminated with just a few nucleotides of ssDNA at its 5′-end; this nucleolytic resection is processive, continuing for up to 100–200 nt. In this regard, although initiating on ssDNA, RecJ is mostly acting as a dsDNA exonuclease. The resulting DNA product has a 3′-ssDNA overhang structure, which provides RecQ with a binding site to initiate unwinding of the dsDNA region using its 3′ to 5′ helicase activity. This unwinding, in turn, now provides a fresh 5′-ssDNA tail for subsequent exonucleolytic degradation by RecJ; in this latter capacity, RecJ acts as an ssDNA exonuclease. Thus, in a single round of resection by RecQ and RecJ, up to several hundred nucleotides can be resected, but the repeated unwinding by RecQ and iterative degradation by RecJ can resect thousands of nucleotides. We also found that SSB is involved in this choreographed reaction. Interaction with the C-terminal region of SSB promotes extensive resection by RecQ and RecJ, and we established that this interaction is required for exonucleolytic degradation of ssDNA but not for degradation of dsDNA. RecJ is known to interact with SSB (45), which we infer from the results here is via its C-terminal acidic tail; however, this interaction is dispensable for unwinding by RecQ.
Even though RecJ is known and considered to be a potent ssDNA exonuclease, we show here that RecJ can degrade one strand of dsDNA from the 5′-end for several hundred nucleotides, provided there are just 3–4 nt of ssDNA at the 5′-end. Such an activity had been already observed (28, 45), but only limited resection could be detected in the previous assays (<10 nt) (45). Thus, the resection of dsDNA by RecJ was not considered to be potentially significant with regard to productive resection of DSBs in recombinational repair. However, we now established that the resection of dsDNA by RecJ is processive (Fig. 3) and that it persists for distances of at 100–200 nt (Fig. 2A). In vitro, RecA requires only ∼15 nt of ssDNA to promote the homology search (48), and in vivo the minimal efficient processing segment for recombination by the RecF pathway is 44–90 bp (49). Consequently, the capability of RecJ to resect dsDNA in the presence of SSB is sufficient to provide ssDNA for homologous pairing by RecA (Fig. 2 and Fig. S2). These observations demonstrate that RecJ nuclease has an intrinsic ability to process a DSB in the absence of a helicase to create ssDNA sufficient for RecA nucleoprotein filament formation and DNA strand exchange. However, RecQ helicase can act on these intermediates and unwind them to provide 5′-tailed ssDNA for subsequent and repeated degradation by RecJ nuclease (Figs. 2 and 4). RecQ does not unwind linear plasmid-length dsDNA efficiently in the presence of high concentrations of free Mg2+ (42), but here we showed that RecQ efficiently unwinds DNA with a 3′-ssDNA overhang for at least 40 bp in 10 mM Mg2+ (Fig. S4A) to produce a substrate for RecJ-mediated resection (Figs. 2 and 4). These observations agree with the previous finding that RecQ helicase only slightly stimulated joint molecule formation that required resection of linear DNA (28).
Genetic analyses have established that RecJ plays a more important role in recombination than RecQ, possibly because of redundant helicases that can assume unwinding responsibilities in a recQ cell (50, 51). This in vivo finding is consistent with our in vitro analysis. The recombination of bacteriophage-λ carrying mutation in its own recombination system, Red, requires the recA and recJ genes, but not recQ, in cells which are recBC− and carry sbcBC suppressor mutations (51). Recombination by bacteriophage-λ initiates at its DNA ends, which have a 12-nt 5′-ssDNA overhang (52). As shown in Fig. 2, such DNA is a good substrate for RecJ-mediated resection without the need for RecQ. RecJ could convert the 12-nt 5′-ssDNA overhang to a 3′-ssDNA overhang of more than ∼150 nt, where RecA could be loaded. For bacteriophage-λ, the Orf protein mediates this loading; when the orf gene is mutated, the RecFOR proteins can substitute (51, 53, 54). These genetic observations are readily explained by our finding that processive degradation of DNA with 5′-ssDNA overhangs, as found on the bacteriophage-λ genome, is mediated by RecJ nuclease without the need for RecQ.
The free Mg2+ concentration in E. coli cells is 1–2 mM (55), which corresponds to ∼2–3 mM of total Mg2+ as used in this study, because the ATP (1 mM) binds approximately an equivalent of Mg2+. At this physiological concentration of Mg2+, blunt-end DNA and DNA with 5′-or 3′-ssDNA overhangs are good substrates for combined RecQJ-mediated resection (Fig. 4B and Fig. S4E). For joint molecule formation, a slightly higher Mg2+ concentration is optimal (Fig. 4C); however, this particular optimum may reflect the absence of molecular crowding, which affects intracellular interactions between macromolecules. It has been established that volume-excluding agents, such as polyvinyl alcohol or polyethylene glycol, stimulate DNA strand exchange by RecA at conditions that are otherwise suboptimal or nonpermissive, such as low concentrations of Mg2+ (56). Consequently, we conclude that both RecQ and RecJ cooperate to resect duplex DNA—with any type of end structure—over a range of conditions that are approximately physiological.
Fig. 6 provides an illustration of how RecQ and RecJ cooperate to process various dsDNA breaks. The end of a DSB can have either a 3′-ssDNA overhang (Fig. 6A), blunt-end (Fig. 6B), or a 5′-ssDNA overhang (Fig. 6C). Because RecQ is a 3′→5′ helicase, dsDNA with a 3′-ssDNA overhang is the best substrate for RecQ-mediated unwinding; on the other hand, because RecJ is a 5′→3′ exonuclease, DNA with a 5′-ssDNA overhang is the best substrate for degradation. Therefore, the processing of DSBs with a 3′-ssDNA overhang DNA is primarily initiated by RecQ helicase capacity (Fig. 6A). In contrast, the processing of DSBs with a 5′-ssDNA overhang is initiated by the ssDNA exonuclease activity of RecJ; however, this degradative capacity can progress several hundred nucleotides into the duplex region by virtue of the dsDNA exonuclease capability of RecJ, without the need for RecQ (Fig. 6C). The blunt-end DSB requires unwinding by RecQ, followed by exonucleolytic degradation of the 5′-terminated ssDNA by RecJ (Fig. 6B). For all of the different DSBs (Fig. 6 A–C), the first intermediate is dsDNA with a 3′-ssDNA overhang. This intermediate is a good substrate for further resection by the combined activities of RecQ and RecJ, with RecQ binding to the 3′-ssDNA tail and unwinding additional dsDNA, and with RecJ subsequently degrading the ssDNA, this process can continue iteratively and indefinitely (Fig. 6D). Finally, the RecFOR complex recognizes the ssDNA–dsDNA junction and loads RecA onto the ssDNA in a 5′→3′ direction (Fig. 6E) (18).
Fig. 6.
The model for resection of dsDNA breaks by RecQ helicase and RecJ nuclease. The DNA end at a DSB is either a 3′-ssDNA overhang, blunt-end or 5′-ssDNA overhang. RecQ, a 3′ to 5′ helicase, preferentially unwinds DNA with a 3′-ssDNA overhang structure. Conversely, RecJ, a 5′ to 3′ exonuclease, preferentially degrades DNA with a 5′-ssDNA overhang. (A) For DNA with a 3′-ssDNA overhang, RecQ initiates resection by unwinding the DNA. RecJ then exonucleolytically degrades the 5′-terminated DNA strand. (B) For blunt-ended DNA, RecQ also initiates resection by unwinding the DNA, although the initiation efficiency is lower than with a 3′-overhang DNA. RecJ then exonucleolytically degrades the 5′-terminated DNA strand. (C) For DNA with a 5′-ssDNA overhang, RecJ initiates resection by first degrading the ssDNA tail, using its dsDNA exonuclease activity, the duplex region is resected. RecQ is not required. (D) The intermediate generated in A–C is DNA containing a 3′-overhang of more than 100 nt of ssDNA. RecQ repeatedly unwinds this intermediate, and RecJ exonucleolytically degrades the 5′-terminated DNA strand. (E) RecFOR proteins recognize the ssDNA–dsDNA junction and load RecA onto the ssDNA.
The crystal structure of full length Thermus thermophilus RecJ revealed a hole that accommodates a ssDNA strand, but not dsDNA, located on one side of an oligonucleotide-binding (OB) domain, offering a structural explanation and mechanism for ssDNA strand specificity of RecJ exonuclease (57). Such a mechanism can explain why RecJ-mediated resection of dsDNA frequently stopped upstream of GC-rich regions (Fig. S2). For the resection of dsDNA, RecJ would need to locally melt dsDNA and bind to the resultant 5′-ssDNA for exonucleolytic degradation. Degradation of dsDNA did not always stop precisely at the GC-rich regions but also ∼2–3 nt before the GC-rich region (Fig. S2), suggesting that the OB-fold of RecJ binds 2 or 3 nt of ssDNA just upstream of the nucleolytic center.
Truncation of eight amino acids from C terminus of SSB inhibited degradation of ssDNA by RecJ (Fig. 5C) but not degradation of dsDNA by RecJ in the absence of RecQ (Fig. 5B). The C-terminal tail is highly acidic, and its removal would diminish the interaction between RecJ and SSB (45) and increase the affinity of the SSB to ssDNA (58); both of these alterations could contribute to an inability of RecJ to degrade ssDNA complexed with the truncated SSB. Interestingly, both intact and truncated SSB stimulated RecJ-mediated resection of duplex DNA (i.e., DNA with a 4-nt 5′-ssDNA overhang) (Fig. 5B). We suggest that SSB stimulates this reaction by binding to the 3′-ended DNA strand produced during the resection, which cannot be degraded by RecJ, preventing the nonproductive binding of RecJ to this competing ssDNA, in a manner analogous to the stimulation of eukaryotic Exo1 by the eukaryotic ssDNA binding protein, replication protein-A (RPA) (59).
Recent studies of homologous recombination in Eukarya showed that the RecQ-family helicases, BLM from human, and Sgs1 from budding yeast are involved in the resection of DSBs (37, 39, 60–62). As expected from their structural homology to RecQ, BLM and Sgs1 are also 3′→5′ helicases (32, 63). Also involved is the eukaryotic Exo1 protein (38, 64). Eukaryotic Exo1 is unrelated to prokaryotic exonuclease I, and is an exonuclease that degrades the 5′-terminated strand of dsDNA from an end (65) but it also exonucleolytically degrades ssDNA from its 5′ end (59). We showed here that RecJ is also both a dsDNA and ssDNA exonuclease, as is Exo1, although with opposite substrate preferences. This functional similarity between the prokaryotic and eukaryotic exonucleases involved in DSB resection is striking despite the absence of structural similarity. In addition to the similarity between exonucleases, there is a BLM/Sgs1-mediated pathway for dsDNA resection that involves another exonuclease DNA2/Dna2 (61, 66). The BLM/DNA2-mediated resection of dsDNA requires ATP hydrolysis (37–39); these and other findings highlight the similarities of the eukaryotic Sgs1/Dna2 and BLM/DNA2 to the bacterial RecBCD/AddAB-family of recombination resection enzymes. Thus, the mechanism of DSB resection by the cooperative action of a 3′→5′ helicase and 5′→3′ nuclease is a mechanism common to all organisms.
In addition, given that duplex DNA with a 3′-ssDNA overhang is the common intermediate of resection (Fig. 6D), such resected DNA is expected to be a good substrate for BLM/Sgs1-mediated 3′→5′ unwinding, and once initiated, resection is expected to be driven by continued helicase activity. However, in all of the characterized resection pathways, a nuclease is required in vivo to cleave the 5′-ssDNA generated by the RecQ ortholog: either RecJ in Bacteria or DNA2/Dna2 in Eukarya. In the presence of RPA, both yeast and human Dna2 degrade ssDNA in the 5′→3′ direction (37–39). Although different in mechanistic detail, the nuclease and helicase activities of the eukaryotic proteins function analogously to those of RecQ and RecJ. The other pathway for long-rang resection in Eukarya requires Exonuclease 1. Although eukaryotic Exo1 is a dsDNA-specific exonuclease, its behavior is similar to that of RecJ in that both yeast and human enzymes display a DNA end-preference. Being a dsDNA-preferring, rather than ssDNA-preferring nuclease, Exo1 displays the opposite substrate bias, preferring blunt dsDNA or dsDNA with a 3′-ssDNA rather than 5′-ssDNA overhang (59). Interestingly, for human EXO1, the binding of BLM enhances the affinity of EXO1 for DNA ends, thereby promoting resection at ends; although a noncatalytic function, in this regard, the evidence for cooperation between nuclease and helicase extends from bacteria to humans. Finally, in both long-range resection pathways of both yeast and humans, the Mre11-Rad50-Xrs2 complex (or MRE11-RAD50-NBS1 in humans) promotes resection by Sgs1/Dna2 (BLM/DNA2) and Exo1 (EXO1) (37, 39, 61, 67) by recruiting these proteins to DNA ends. In E. coli, ironically the blunt-ended break is the poorest substrate for RecQJ-dependent resection. RecQ helicase or RecJ nuclease can initiate resection from DNA with 3′- or 5′-overhangs, but poorly from blunt-end DNA (Fig. 4). Interestingly, RecN is a member of the SMC (structural maintenance of chromosomes) family DNA binding proteins, as is the Rad50 protein, and it is therefore a candidate for stimulating DSB resection from blunt-end DNA by perhaps recruiting these resection proteins to DNA ends in a manner analogous to the Mre11-Rad50-Xrs2 complex.
Materials and Methods
Proteins.
RecQ (68), SSB (69), RecA (70), RecF (17), and RecO and RecR (15) were purified as described previously. RecJ and truncated SSB lacking C-terminal eight amino acids were purified as described in SI Materials and Methods.
Joint Molecule Formation Assays.
Joint molecule formation was performed as previously described (28), with minor modifications. The final concentrations of components were 5 µM RecA, 15 nM RecF, 100 nM RecO, 1 µM RecR, 3 µM SSB, 0.05 U/µL RecJ (3.3 nM), 10 nM RecQ, 10 µM (nt) linear pUC19, and 20 µM (nt) supercoiled pUC19 in buffer containing 20 mM Tris-acetate (pH 7.5), 10 mM magnesium acetate, 1 mM ATP, 1 mM phosphoenolpyruvate (PEP), 20 U/mL pyruvate kinase (PK), 0.1 mg/mL bovine serum albumin (BSA), 1 mM dithiothreitol (DTT), and 5% (vol/vol) glycerol. After reaction at 30 °C, products were analyzed by agarose gel (0.8%) electrophoresis in 1× TAE buffer [40 mM Tris-acetate (pH 8.0), 1 mM EDTA], followed by ethidium bromide staining. Joint molecule formation was determined as the percentage of the limiting linear dsDNA that was incorporated into joint molecules. It was quantified from the band intensity of joint molecule products at the indicated time, and band intensities of linear dsDNA and supercoiled dsDNA at 0 time.
Short-Range DNA Resection Assay.
32P-labeled linear dsDNA cut by several restriction endonucleases were prepared as described in SI Materials and Methods. Briefly, pUC19 dsDNA was cleaved by NdeI, labeled by Klenow fragment with [α-32P]dATP and cold dTTP, and cleaved by the indicated restriction endonuclease.
The reaction buffer for resection by RecJ nuclease was 20 mM Tris acetate (pH 7.5), 10 mM magnesium acetate, 0.1 µg/mL BSA, 5% (vol/vol) glycerol, and 1 mM DTT. When RecQ was used, 1 mM ATP, 1 mM PEP, and 20 U/mL pyruvate kinase were added; these components were also added to the reaction without RecQ when the results with and without RecQ helicase were being compared. Standard reactions contained 10 µM (nt) 32P-dsDNA, 0.125 U/µL RecJ (8.1 nM), 0.1 µM RecQ, and 3 µM SSB. After the incubation for 0 or 60 min at 30 °C, a 5-µL sample was taken, mixed with 5 µL of stop solution (40 mM EDTA, 0.8% SDS, and 0.2% bromophenol blue), and chilled on ice. The samples (total 10 µL) were treated with Proteinase K (Boehringer Mannheim; 1 µL of 2 mg/mL Proteinase K solution) followed by incubation at 37 °C for 20 min. Before loading on a gel, 10 µL of formamide mix [90% (vol/vol) formamide, 10 mM EDTA, 0.08% xylene cyanol, and 0.08% bromophenol blue] were added, and the sample was incubated at 90 °C for 10 min. Electrophoresis was performed using a DNA sequencing gel (13 cm × 38 cm × 0.3 mm) composed of 6% polyacrylamide (29:1), 8 M urea, and 1× TBE buffer [90 mM Tris-borate (pH 8.0) and 2 mM EDTA], at 1,500–2,000 V (voltage was adjusted to set the gel temperature to 50 °C) until the xylene cyanol migrated 25 cm. After the electrophoresis, the gel was dried and analyzed by autoradiography using a Storm 820 PhosphorImager with ImageQuaNT software. The size markers shown to the left side of each gel images indicate the electrophoretic mobility of the HindIII-, EcoRI-, and BglI-digested substrates (267, 216, and 64 nt) and the major RecJ-resection products (139 and 96 nt, determined from the Maxam–Gilbert sequencing ladder), respectively.
Long-Range Resection Assay.
Extensive, long-range dsDNA resection was assayed as follows. The sample contained 5 µM (nt) linear pUC19 dsDNA (that had been cleaved using the restriction enzymes indicated in the figure legends), 0.05 U/µL RecJ (3.3 nM), 10 nM RecQ, and 1 µM SSB in 90-µL solution. The reaction buffer was 20 mM Tris-acetate (pH 7.5), 1–10 mM magnesium acetate, 1 mM ATP, 1 mM PEP, 20 U/mL pyruvate kinase, 0.1 µg/mL BSA, 5% (vol/vol) glycerol, and 1 mM DTT. The reaction was started by adding RecJ. After the indicated time at 30 °C, an aliquot (20 µL) was taken, mixed with stop solution (5.4 µL) containing 4.7% SDS, 235 mM EDTA, and 0.047% bromophenol blue, and then stored on ice. After finishing all reactions, the samples were analyzed by agarose gel (0.8%) electrophoresis in 1× TAE, followed by staining with ethidium bromide. The amounts of dsDNA processed were quantified as the decrease of band intensities.
Resection of ssDNA was performed as done for the dsDNA resection assay above, but with the following modifications. The EcoRI-cut pUC19 DNA was first unwound by 50 nM RecQ in the presence of SSB but in the absence of RecJ at 30 °C for 30 min. After the unwinding reaction, the resection reaction was started by adding 9 mM magnesium acetate and RecJ.
Supplementary Material
Acknowledgments
We thank Ichiro Amitani, Máté Gyimesi, Naofumi Handa, Hsu-Yang Lee, and Satona Ono for their comments on this manuscript, and all members of the S.C.K. laboratory for helpful discussions and insights on this work. This work was supported, in whole or in part, by National Institutes of Health Grant GM62653 (to S.C.K.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1420009111/-/DCSupplemental.
References
- 1.Persky NS, Lovett ST. Mechanisms of recombination: Lessons from E. coli. Crit Rev Biochem Mol Biol. 2008;43(6):347–370. doi: 10.1080/10409230802485358. [DOI] [PubMed] [Google Scholar]
- 2.Horii Z, Clark AJ. Genetic analysis of the recF pathway to genetic recombination in Escherichia coli K12: Isolation and characterization of mutants. J Mol Biol. 1973;80(2):327–344. doi: 10.1016/0022-2836(73)90176-9. [DOI] [PubMed] [Google Scholar]
- 3.Wang TV, Smith KC. recF-dependent and recF recB-independent DNA gap-filling repair processes transfer dimer-containing parental strands to daughter strands in Escherichia coli K-12 uvrB. J Bacteriol. 1984;158(2):727–729. doi: 10.1128/jb.158.2.727-729.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tseng YC, Hung JL, Wang TC. Involvement of RecF pathway recombination genes in postreplication repair in UV-irradiated Escherichia coli cells. Mutat Res. 1994;315(1):1–9. doi: 10.1016/0921-8777(94)90021-3. [DOI] [PubMed] [Google Scholar]
- 5.Kowalczykowski SC. Initiation of genetic recombination and recombination-dependent replication. Trends Biochem Sci. 2000;25(4):156–165. doi: 10.1016/s0968-0004(00)01569-3. [DOI] [PubMed] [Google Scholar]
- 6.Lloyd RG, Buckman C. Identification and genetic analysis of sbcC mutations in commonly used recBC sbcB strains of Escherichia coli K-12. J Bacteriol. 1985;164(2):836–844. doi: 10.1128/jb.164.2.836-844.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kowalczykowski SC, Dixon DA, Eggleston AK, Lauder SD, Rehrauer WM. Biochemistry of homologous recombination in Escherichia coli. Microbiol Rev. 1994;58(3):401–465. doi: 10.1128/mr.58.3.401-465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rocha EP, Cornet E, Michel B. Comparative and evolutionary analysis of the bacterial homologous recombination systems. PLoS Genet. 2005;1(2):e15. doi: 10.1371/journal.pgen.0010015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shibata T, DasGupta C, Cunningham RP, Radding CM. Purified Escherichia coli recA protein catalyzes homologous pairing of superhelical DNA and single-stranded fragments. Proc Natl Acad Sci USA. 1979;76(4):1638–1642. doi: 10.1073/pnas.76.4.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DasGupta C, Shibata T, Cunningham RP, Radding CM. The topology of homologous pairing promoted by RecA protein. Cell. 1980;22(2 Pt 2):437–446. doi: 10.1016/0092-8674(80)90354-2. [DOI] [PubMed] [Google Scholar]
- 11.Cox MM, Lehman IR. recA protein-promoted DNA strand exchange. Stable complexes of recA protein and single-stranded DNA formed in the presence of ATP and single-stranded DNA binding protein. J Biol Chem. 1982;257(14):8523–8532. [PubMed] [Google Scholar]
- 12.Shinohara A, Ogawa H, Ogawa T. Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell. 1992;69(3):457–470. doi: 10.1016/0092-8674(92)90447-k. [DOI] [PubMed] [Google Scholar]
- 13.Sung P. Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast RAD51 protein. Science. 1994;265(5176):1241–1243. doi: 10.1126/science.8066464. [DOI] [PubMed] [Google Scholar]
- 14.Luisi-DeLuca C, Kolodner R. Purification and characterization of the Escherichia coli RecO protein. Renaturation of complementary single-stranded DNA molecules catalyzed by the RecO protein. J Mol Biol. 1994;236(1):124–138. doi: 10.1006/jmbi.1994.1123. [DOI] [PubMed] [Google Scholar]
- 15.Kantake N, Madiraju MV, Sugiyama T, Kowalczykowski SC. Escherichia coli RecO protein anneals ssDNA complexed with its cognate ssDNA-binding protein: A common step in genetic recombination. Proc Natl Acad Sci USA. 2002;99(24):15327–15332. doi: 10.1073/pnas.252633399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Umezu K, Chi NW, Kolodner RD. Biochemical interaction of the Escherichia coli RecF, RecO, and RecR proteins with RecA protein and single-stranded DNA binding protein. Proc Natl Acad Sci USA. 1993;90(9):3875–3879. doi: 10.1073/pnas.90.9.3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morimatsu K, Kowalczykowski SC. RecFOR proteins load RecA protein onto gapped DNA to accelerate DNA strand exchange: A universal step of recombinational repair. Mol Cell. 2003;11(5):1337–1347. doi: 10.1016/s1097-2765(03)00188-6. [DOI] [PubMed] [Google Scholar]
- 18.Morimatsu K, Wu Y, Kowalczykowski SC. RecFOR proteins target RecA protein to a DNA gap with either DNA or RNA at the 5′ terminus: Implication for repair of stalled replication forks. J Biol Chem. 2012;287(42):35621–35630. doi: 10.1074/jbc.M112.397034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shinohara A, Ogawa T. Stimulation by Rad52 of yeast Rad51-mediated recombination. Nature. 1998;391(6665):404–407. doi: 10.1038/34943. [DOI] [PubMed] [Google Scholar]
- 20.New JH, Sugiyama T, Zaitseva E, Kowalczykowski SC. Rad52 protein stimulates DNA strand exchange by Rad51 and replication protein A. Nature. 1998;391(6665):407–410. doi: 10.1038/34950. [DOI] [PubMed] [Google Scholar]
- 21.Sugiyama T, New JH, Kowalczykowski SC. DNA annealing by RAD52 protein is stimulated by specific interaction with the complex of replication protein A and single-stranded DNA. Proc Natl Acad Sci USA. 1998;95(11):6049–6054. doi: 10.1073/pnas.95.11.6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sugiyama T, Kowalczykowski SC. Rad52 protein associates with replication protein A (RPA)-single-stranded DNA to accelerate Rad51-mediated displacement of RPA and presynaptic complex formation. J Biol Chem. 2002;277(35):31663–31672. doi: 10.1074/jbc.M203494200. [DOI] [PubMed] [Google Scholar]
- 23.Volkert MR, Hartke MA. Suppression of Escherichia coli recF mutations by recA-linked srfA mutations. J Bacteriol. 1984;157(2):498–506. doi: 10.1128/jb.157.2.498-506.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fortin GS, Symington LS. Mutations in yeast Rad51 that partially bypass the requirement for Rad55 and Rad57 in DNA repair by increasing the stability of Rad51-DNA complexes. EMBO J. 2002;21(12):3160–3170. doi: 10.1093/emboj/cdf293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang H, Li Q, Fan J, Holloman WK, Pavletich NP. The BRCA2 homologue Brh2 nucleates RAD51 filament formation at a dsDNA-ssDNA junction. Nature. 2005;433(7026):653–657. doi: 10.1038/nature03234. [DOI] [PubMed] [Google Scholar]
- 26.Jensen RB, Carreira A, Kowalczykowski SC. Purified human BRCA2 stimulates RAD51-mediated recombination. Nature. 2010;467(7316):678–683. doi: 10.1038/nature09399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu J, Doty T, Gibson B, Heyer WD. Human BRCA2 protein promotes RAD51 filament formation on RPA-covered single-stranded DNA. Nat Struct Mol Biol. 2010;17(10):1260–1262. doi: 10.1038/nsmb.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Handa N, Morimatsu K, Lovett ST, Kowalczykowski SC. Reconstitution of initial steps of dsDNA break repair by the RecF pathway of E. coli. Genes Dev. 2009;23(10):1234–1245. doi: 10.1101/gad.1780709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harmon FG, Brockman JP, Kowalczykowski SC. RecQ helicase stimulates both DNA catenation and changes in DNA topology by topoisomerase III. J Biol Chem. 2003;278(43):42668–42678. doi: 10.1074/jbc.M302994200. [DOI] [PubMed] [Google Scholar]
- 30.Watt PM, Louis EJ, Borts RH, Hickson ID. Sgs1: A eukaryotic homolog of E. coli RecQ that interacts with topoisomerase II in vivo and is required for faithful chromosome segregation. Cell. 1995;81(2):253–260. doi: 10.1016/0092-8674(95)90335-6. [DOI] [PubMed] [Google Scholar]
- 31.Ellis NA, et al. The Bloom’s syndrome gene product is homologous to RecQ helicases. Cell. 1995;83(4):655–666. doi: 10.1016/0092-8674(95)90105-1. [DOI] [PubMed] [Google Scholar]
- 32.Cejka P, Kowalczykowski SC. The full-length Saccharomyces cerevisiae Sgs1 protein is a vigorous DNA helicase that preferentially unwinds holliday junctions. J Biol Chem. 2010;285(11):8290–8301. doi: 10.1074/jbc.M109.083196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fiorentini P, Huang KN, Tishkoff DX, Kolodner RD, Symington LS. Exonuclease I of Saccharomyces cerevisiae functions in mitotic recombination in vivo and in vitro. Mol Cell Biol. 1997;17(5):2764–2773. doi: 10.1128/mcb.17.5.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsubouchi H, Ogawa H. Exo1 roles for repair of DNA double-strand breaks and meiotic crossing over in Saccharomyces cerevisiae. Mol Biol Cell. 2000;11(7):2221–2233. doi: 10.1091/mbc.11.7.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nimonkar AV, Ozsoy AZ, Genschel J, Modrich P, Kowalczykowski SC. Human exonuclease 1 and BLM helicase interact to resect DNA and initiate DNA repair. Proc Natl Acad Sci USA. 2008;105(44):16906–16911. doi: 10.1073/pnas.0809380105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Budd ME, Choe WC, Campbell JL. DNA2 encodes a DNA helicase essential for replication of eukaryotic chromosomes. J Biol Chem. 1995;270(45):26766–26769. doi: 10.1074/jbc.270.45.26766. [DOI] [PubMed] [Google Scholar]
- 37.Cejka P, et al. DNA end resection by Dna2-Sgs1-RPA and its stimulation by Top3-Rmi1 and Mre11-Rad50-Xrs2. Nature. 2010;467(7311):112–116. doi: 10.1038/nature09355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nimonkar AV, et al. BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev. 2011;25(4):350–362. doi: 10.1101/gad.2003811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Niu H, et al. Mechanism of the ATP-dependent DNA end-resection machinery from Saccharomyces cerevisiae. Nature. 2010;467(7311):108–111. doi: 10.1038/nature09318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lovett ST, Kolodner RD. Identification and purification of a single-stranded-DNA-specific exonuclease encoded by the recJ gene of Escherichia coli. Proc Natl Acad Sci USA. 1989;86(8):2627–2631. doi: 10.1073/pnas.86.8.2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Umezu K, Nakayama K, Nakayama H. Escherichia coli RecQ protein is a DNA helicase. Proc Natl Acad Sci USA. 1990;87(14):5363–5367. doi: 10.1073/pnas.87.14.5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harmon FG, Kowalczykowski SC. Biochemical characterization of the DNA helicase activity of the Escherichia coli RecQ helicase. J Biol Chem. 2001;276(1):232–243. doi: 10.1074/jbc.M006555200. [DOI] [PubMed] [Google Scholar]
- 43.Honigberg SM, Gonda DK, Flory J, Radding CM. The pairing activity of stable nucleoprotein filaments made from recA protein, single-stranded DNA, and adenosine 5′-(γ-thio)triphosphate. J Biol Chem. 1985;260(21):11845–11851. [PubMed] [Google Scholar]
- 44.Roman LJ, Kowalczykowski SC. Relationship of the physical and enzymatic properties of Escherichia coli recA protein to its strand exchange activity. Biochemistry. 1986;25(23):7375–7385. doi: 10.1021/bi00371a020. [DOI] [PubMed] [Google Scholar]
- 45.Han ES, et al. RecJ exonuclease: Substrates, products and interaction with SSB. Nucleic Acids Res. 2006;34(4):1084–1091. doi: 10.1093/nar/gkj503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shereda RD, Reiter NJ, Butcher SE, Keck JL. Identification of the SSB binding site on E. coli RecQ reveals a conserved surface for binding SSB’s C terminus. J Mol Biol. 2009;386(3):612–625. doi: 10.1016/j.jmb.2008.12.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shereda RD, Bernstein DA, Keck JL. A central role for SSB in Escherichia coli RecQ DNA helicase function. J Biol Chem. 2007;282(26):19247–19258. doi: 10.1074/jbc.M608011200. [DOI] [PubMed] [Google Scholar]
- 48.Hsieh P, Camerini-Otero CS, Camerini-Otero RD. The synapsis event in the homologous pairing of DNAs: RecA recognizes and pairs less than one helical repeat of DNA. Proc Natl Acad Sci USA. 1992;89(14):6492–6496. doi: 10.1073/pnas.89.14.6492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shen P, Huang HV. Homologous recombination in Escherichia coli: dependence on substrate length and homology. Genetics. 1986;112(3):441–457. doi: 10.1093/genetics/112.3.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Luisi-DeLuca C, Lovett ST, Kolodner RD. Genetic and physical analysis of plasmid recombination in recB recC sbcB and recB recC sbcA Escherichia coli K-12 mutants. Genetics. 1989;122(2):269–278. doi: 10.1093/genetics/122.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sawitzke JA, Stahl FW. Phage lambda has an analog of Escherichia coli recO, recR and recF genes. Genetics. 1992;130(1):7–16. doi: 10.1093/genetics/130.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stahl FW, Kobayashi I, Stahl MM. In phage lambda, cos is a recombinator in the red pathway. J Mol Biol. 1985;181(2):199–209. doi: 10.1016/0022-2836(85)90085-3. [DOI] [PubMed] [Google Scholar]
- 53.Sawitzke JA, Stahl FW. Roles for lambda Orf and Escherichia coli RecO, RecR and RecF in lambda recombination. Genetics. 1997;147(2):357–369. doi: 10.1093/genetics/147.2.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sawitzke JA, Stahl FW. The phage lambda orf gene encodes a trans-acting factor that suppresses Escherichia coli recO, recR, and recF mutations for recombination of lambda but not of E. coli. J Bacteriol. 1994;176(21):6730–6737. doi: 10.1128/jb.176.21.6730-6737.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Alatossava T, Jütte H, Kuhn A, Kellenberger E. Manipulation of intracellular magnesium content in polymyxin B nonapeptide-sensitized Escherichia coli by ionophore A23187. J Bacteriol. 1985;162(1):413–419. doi: 10.1128/jb.162.1.413-419.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lavery PE, Kowalczykowski SC. Enhancement of recA protein-promoted DNA strand exchange activity by volume-occupying agents. J Biol Chem. 1992;267(13):9307–9314. [PubMed] [Google Scholar]
- 57.Wakamatsu T, et al. Structure of RecJ exonuclease defines its specificity for single-stranded DNA. J Biol Chem. 2010;285(13):9762–9769. doi: 10.1074/jbc.M109.096487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Williams KR, Spicer EK, LoPresti MB, Guggenheimer RA, Chase JW. Limited proteolysis studies on the Escherichia coli single-stranded DNA binding protein. Evidence for a functionally homologous domain in both the Escherichia coli and T4 DNA binding proteins. J Biol Chem. 1983;258(5):3346–3355. [PubMed] [Google Scholar]
- 59.Cannavo E, Cejka P, Kowalczykowski SC. Relationship of DNA degradation by Saccharomyces cerevisiae exonuclease 1 and its stimulation by RPA and Mre11-Rad50-Xrs2 to DNA end resection. Proc Natl Acad Sci USA. 2013;110(18):E1661–E1668. doi: 10.1073/pnas.1305166110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nimonkar AV, Sica RA, Kowalczykowski SC. Rad52 promotes second-end DNA capture in double-stranded break repair to form complement-stabilized joint molecules. Proc Natl Acad Sci USA. 2009;106(9):3077–3082. doi: 10.1073/pnas.0813247106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008;134(6):981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gravel S, Chapman JR, Magill C, Jackson SP. DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev. 2008;22(20):2767–2772. doi: 10.1101/gad.503108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Karow JK, Chakraverty RK, Hickson ID. The Bloom’s syndrome gene product is a 3′-5′ DNA helicase. J Biol Chem. 1997;272(49):30611–30614. doi: 10.1074/jbc.272.49.30611. [DOI] [PubMed] [Google Scholar]
- 64.Nicolette ML, et al. Mre11-Rad50-Xrs2 and Sae2 promote 5′ strand resection of DNA double-strand breaks. Nat Struct Mol Biol. 2010;17(12):1478–1485. doi: 10.1038/nsmb.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Szankasi P, Smith GR. A DNA exonuclease induced during meiosis of Schizosaccharomyces pombe. J Biol Chem. 1992;267(5):3014–3023. [PubMed] [Google Scholar]
- 66.Imamura O, Campbell JL. The human Bloom syndrome gene suppresses the DNA replication and repair defects of yeast dna2 mutants. Proc Natl Acad Sci USA. 2003;100(14):8193–8198. doi: 10.1073/pnas.1431624100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mimitou EP, Symington LS. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455(7214):770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Harmon FG, Kowalczykowski SC. RecQ helicase, in concert with RecA and SSB proteins, initiates and disrupts DNA recombination. Genes Dev. 1998;12(8):1134–1144. doi: 10.1101/gad.12.8.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lohman TM, Green JM, Beyer RS. Large-scale overproduction and rapid purification of the Escherichia coli ssb gene product. Expression of the ssb gene under lambda PL control. Biochemistry. 1986;25(1):21–25. doi: 10.1021/bi00349a004. [DOI] [PubMed] [Google Scholar]
- 70.Morimatsu K, Horii T, Takahashi M. Interaction of Tyr103 and Tyr264 of the RecA protein with DNA and nucleotide cofactors. Fluorescence study of engineered proteins. Eur J Biochem. 1995;228(3):779–785. doi: 10.1111/j.1432-1033.1995.tb20323.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.