Significance
Mechanical properties of the extracellular environment provide important cues that regulate cell behavior. Understanding this mechanical signaling has become important in disease treatment as well as tissue engineering. To efficiently study cellular responses to rigidity signals, we have created a model system of micropatterned composite material based on the “cell-on-a-chip” concept. We demonstrate that a migrating fibroblast uses filopodia to probe substrate rigidity, such that it “feels” its way based on the deformability of a material before occupying an area. Myosin II plays a key role in rigidity sensing and responses. This mechanism allows cells to migrate efficiently by avoiding mechanically unfavorable areas without backtracking.
Keywords: rigidity sensing, durotaxis, focal adhesions, myosin II, polyacrylamide
Abstract
Rigidity sensing and durotaxis are thought to be important elements in wound healing, tissue formation, and cancer treatment. It has been challenging, however, to study the underlying mechanism due to difficulties in capturing cells during the transient response to a rigidity interface. We have addressed this problem by developing a model experimental system that confines cells to a micropatterned area with a rigidity border. The system consists of a rigid domain of one large adhesive island, adjacent to a soft domain of small adhesive islands grafted on a nonadhesive soft gel. This configuration allowed us to test rigidity sensing away from the cell body during probing and spreading. NIH 3T3 cells responded to the micropatterned rigidity border similarly to cells at a conventional rigidity border, by showing a strong preference for staying on the rigid side. Furthermore, cells used filopodia extensions to probe substrate rigidity at a distance in front of the leading edge and regulated their responses based on the strain of the intervening substrate. Soft substrates inhibited focal adhesion maturation and promoted cell retraction, whereas rigid substrates allowed stable adhesions and cell spreading. Myosin II was required for not only the generation of probing forces but also the retraction in response to soft substrates. We suggest that a myosin II-driven, filopodia-based probing mechanism ahead of the leading edge allows cells to migrate efficiently, by sensing physical characteristics before moving over a substrate to avoid backtracking.
Substrate rigidity has been shown to influence cell growth, differentiation, and migration in vitro (1–3). Different tissues show different characteristic stiffness, and changes in tissue stiffness represent a common marker of diseases including cancer, liver fibrosis, and arteriosclerosis. Consideration of substrate rigidity has become an essential aspect in not only disease treatment, such as the prevention of cancer metastasis (4–7), but also tissue engineering (8, 9).
There has been intense interest in understanding rigidity sensing at the cellular and molecular level. Of particular interest are cellular responses to spatial or temporal changes in rigidity, such as the ability of fibroblasts to migrate preferentially toward stiffer substrates at a rigidity interface known as durotaxis (10). However, limitations in the experimental system have hampered the understanding of how cells detect and respond to a spatial or temporal transition in substrate rigidity. Most studies on cellular response to substrate rigidity have been performed with cells on a surface of homogeneous rigidity (11–13). Whereas some studies have examined cellular responses to changing rigidity, few have focused on the dynamic behavior of how cells probe and respond to temporal or spatial changes in substrate rigidity. One such study, which involved pushing and pulling a deformable substrate by micromanipulation to simulate changes in substrate rigidity, showed that cells extended protrusions and moved toward the direction of increased tension and recoiled from the direction of reduced tension (10). A more recent study using a similar approach further indicated that durotaxis involved dynamic tugging of adhesions (14). However, this micromanipulation approach was time and labor intensive.
Alternatively, rigidity responses may be studied by placing migrating cells on engineered substrates with juxtaposed elastic and rigid domains (15–17). One study used micropost arrays of different dimensions to create a rigidity border (18). However, as in the original study of durotaxis, this system still relied on spontaneous, time-consuming migration of cells to the rigidity border, where the cell stays for only a limited period in the actual area of interest. In addition, it was often difficult to create sufficiently soft substrates with microposts. Studies using this, and other approaches, have suggested that the cytoskeleton, specifically actomyosin contractility, is crucial for rigidity sensing during durotaxis (18, 19), and one recent study with patterned microposts suggested that a traction force imbalance at a rigidity border contributes to durotaxis (20).
Using an approach that traps cells at a micropatterned rigidity border, we report here a rigidity sensing mechanism based on filopodia extensions that allows cells to probe substrate rigidity ahead of their leading edge. We have previously described a method for micropatterning arrays of micrometer-sized adhesive photoresist islands on the surface of nonadhesive polyacrylamide hydrogels (21). The study indicated that long range strain, dictated by the rigidity of underlying hydrogels rather than the local rigidity of photoresist islands in direct contact with the cell, determines cellular rigidity responses. Thus, soft substrates may be created using an array of small rigid adhesive islands grafted onto a soft nonadhesive hydrogel, whereas a rigidity border may be generated by juxtaposing such an island array with a large island, each covering half of the spreading area of a cell (Fig. 1A). In combination, this pattern provides a tool to assess rigidity sensing by trapping cells at a rigidity border, thereby avoiding prolonged recording waiting for the transient localization of cells to the transition region. Another advantage of this system is that the nonadhesive hydrogel that separates the adhesive islands allows the investigation of whether, and how, cells probe material rigidity across a distance beyond their border.
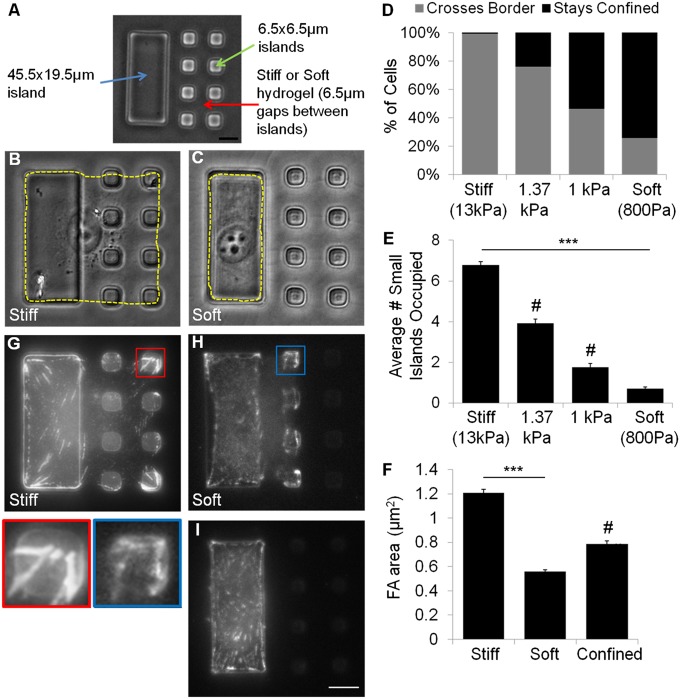
Fig. 1.
A micropatterned composite substrate that confines cells to a rigidity border allows highly efficient detection of durotaxis-like behavior. The testing substrate consists of a soft hydrogel (A, red arrow), grafted with one large 45.5 × 19.5 μm rectangular island to create the rigid domain (A, blue arrow), and two rows of four 6.5 × 6.5 μm islands to create the soft domain (A, green arrow). Control substrates are prepared by grafting the same pattern of islands onto a rigid hydrogel. NIH 3T3 cells spread over both large and small islands 16 h after plating on control substrates (B), but remain on the large island (rigid domain) when plated on testing substrates (C). Cell outlines are shown by yellow dashed lines. Bar graph shows almost all of the cells are able to cross the border between large and small islands on control substrates (D, left bar, n = 132), whereas most cells are confined to the large island on testing substrates (D, right bar, n = 233). Hydrogels of higher rigidity cause progressively more cells to spread beyond the large island (C and D, middle bars, n = 208 and 181). After 16 h of plating, cells on control substrates are able to spread over many more small islands (E, left bar) than cells on testing substrates (E, right bar). Immunofluorescence images of paxillin show large focal adhesions on both large and small islands for cells on control substrates (G), but only small focal adhesions in cells that spread onto some small islands on testing substrates (H). Cells confined to the large island also form small focal adhesions (I). Morphometry indicates that focal adhesions on control substrates (F, Left; n = 815 focal adhesions from 30 cells) are about twice the size of focal adhesions in cells that spread across soft gels on testing substrates (F, Middle; n = 696 focal adhesions from 19 cells). Cells that are confined to the large island show focal adhesions of intermediate size, regardless of the rigidity of the underlying gel (F, Right; 412 focal adhesions from 18 cells). (Scale bars, 10 μm.) Error bars represent SEM ***P < 0.001. #, statistically significant from both stiff and soft conditions P < 0.001.
Results
NIH 3T3 Cells Respond to a Micropatterned Rigidity Border on a Composite Substrate.
We have developed a substrate with a rigidity border across a small square region the area of a spread fibroblast, using a previously developed method to create micropatterned composite materials (21, 22). The micropattern, as shown in Fig. 1A, consists of one large rectangular adhesive island of rigid photoresist, 45.5 × 19.5 μm2 in area, flanked by two rows of four small square islands of 6.5 × 6.5 μm2, grafted on top of a nonadhesive polyacrylamide hydrogel. The pattern was designed so that the overall area was ∼2000 μm2, which is within the normal range of spreading for NIH 3T3 cells on rigid adhesive substrates. Therefore, under normal conditions, we expect the cell to cover the overall square area without experiencing a serious constraint. Our previous study using this composite system indicated that the deformation of soft hydrogels between islands should lead cells to interpret the region occupied by small islands as being soft, whereas the large island provides an adjacent rigid domain of a similar area. Therefore, this combination creates a “testing substrate” that traps cells at or near a rigidity border. The same pattern of islands grafted on stiff hydrogels creates a “control substrate” of a similar chemical composition without a rigidity transition. In addition to testing the response to a rigidity border, the distance of 6.5 μm between the islands allowed us to ask if and how cells sense substrate rigidity across a distance. We found no difference in the amount of fibronectin or gelatin adsorbing to the surface of SU-8 islands on control or testing substrates (Fig. S1 A and B). Moreover, the islands were of a similar height regardless of the composition of the underlying polyacrylamide, suggesting an identical Young’s modulus for the cured SU-8 on control and testing substrates (assuming that the photochemical curing reaction of SU-8 was unaffected by the underlying dried layer of polyacrylamide (SI Methods).
NIH 3T3 cells were first allowed to adhere to the composite substrate for 16 h before imaging. Cells on control substrates readily crossed from the large island to occupy some or all adjacent small islands; <1% of cells were confined entirely to the rigid large island after 16 h (Fig. 1 B and D). In contrast, 74% of cells plated on testing substrates remained completely confined to the large island (Fig. 1 C and D). The response was further examined based on the size of focal adhesions, which is known to be a sensitive indicator of substrate rigidity and cell shape (2, 11). Focal adhesions, as shown by paxillin immunofluorescence, decreased from an average area of 0.79 μm2 for cells confined to the large island (Fig. 1 F and I), to 0.56 μm2 for cells that spread across the soft gel to adhere to one or more small islands (Fig. 1 F and H). In contrast, cells that spread across the rigid gel of control substrates showed an increase in the size of focal adhesions (1.21 μm2, Fig. 1 F and G), which reflected the dependence of focal adhesion size on cell spreading (23, 24). These results confirmed that cells were indeed able to sense the difference in rigidity in the present model system, as they do on conventional rigid or soft substrates (11), and that an increase in the size of focal adhesions took place only when cells adhered and spread across the rigid control substrate. By varying the rigidity of the polyacrylamide layer, we found that the transition took place around 1000 Pa, such that substrates with gels softer than 1000 Pa inhibited cell spreading and focal adhesion growth (Fig. 1 D and E).
NIH 3T3 Cells Are Able to Detect Substrate Rigidity Outside Their Border.
The above observations raise the possibility that cells were able to detect substrate rigidity outside their border before deciding if they should occupy the area. To understand how cells behave at the micropatterned rigidity border, we performed time-lapse recording starting 30 min after plating to determine how cells spread from the large to small islands. The process appeared to involve fine probing structures close to the detection limit of phase contrast optics (Fig. 2 A and B, yellow arrowheads).
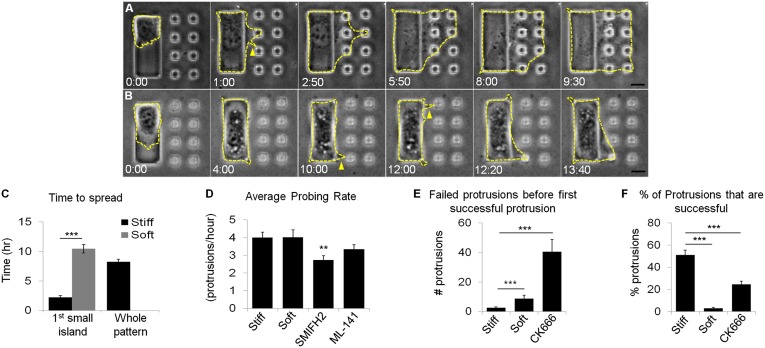
Fig. 2.
NIH 3T3 cells are able to detect substrate rigidity beyond the cell border. NIH 3T3 cells are observed with time-lapse phase contrast microscopy and outlines are indicated by yellow dashed lines. On control substrates, cells that initially attach to the large island are able to detect small islands across the nonadhesive hydrogel and migrate over all of the islands in less than 10 h (A). On testing substrates, most cells that initially attach to the large island remain confined, although transient protrusions can be seen extending toward small islands (B, yellow arrows). A fraction of cells are able to occupy a small island after an extended period of incubation (B, Lower Right). Numbers at the bottom of each image indicate the lapse time in hours and minutes after initial cell attachment to a large island, which takes place ∼30 min after plating. The average time required to occupy the first small island is much longer on testing substrates (C, gray bar, n = 18; of the 78 cells recorded, only 18 occupied at least one small island) than on control substrates (C, black bar on left, n = 116). It takes on average 8 h for cells on control substrates to occupy all of the small islands (C, black bar on right, n = 49). Cells on testing substrates were never seen to occupy all of the small islands. Cells on both stiff and soft substrates extend protrusions and probe the small islands at the same rate (D, n = 36 and 73). Formin inhibition (SMIFH2) significantly reduces the probing rate, whereas Cdc42 inhibition (ML-141) causes a slight decrease in probing rate (D, n = 66 and 64). Cells on soft substrates extend more failed protrusions before successfully occupying one small island (E, n = 35 and 15). The percentage of successful protrusions is much higher on stiff substrates than soft substrates (F, n = 36 and 68). Arp 2/3 inhibition (CK666) increases the number of failed protrusions and reduces the percentage of successful protrusions on control substrates (E and F, n = 20 and 22). (Scale bars, 10 μm.) Error bars represent SEM ***P < 0.001 **P < 0.01.
Cells extended these thin probing extensions at a similar rate on both control and testing substrates (Fig. 2D); however, a much higher percentage of cells showed subsequent spreading onto small islands on control substrates than on testing substrates (Fig. 2F). A spreading response involved the adhesion of a probing extension to a small island followed by the formation of lamellipodia to occupy the island area. Once adhered to the first small island on control substrates, most cells continued to spread onto the rest of the islands to occupy the full patterned area (Fig. 2A, Movie S1, and Fig. S2A). It took on average 2 h after initial plating for a cell to occupy the first island and by 8 h, the majority of cells had spread over the remaining seven islands (Fig. 2C, black bars).
On testing substrates, NIH 3T3 cells formed thin extensions similar to those seen on control substrates. Minute deflections of the small islands were occasionally visible, indicative of probing forces (Movie S2). However, most of the thin extensions failed to lead to spreading onto adjacent small islands (Fig. 2 E and F and Movie S3). It took on average 10 h for a fraction of cells on testing substrates to occupy one small island (Fig. 2C, gray bar) and no cell was found to occupy all of the small islands 16 h after plating (Fig. S2A). These observations suggest that cells use thin extensions to probe substrate rigidity outside their border. The mechanical signal then determines the stability of the extension and the rate and efficiency of subsequent cell spreading. Soft substrates cause a low probability of spreading and a high probability of retraction.
To ensure that the composite materials used in our substrates did not significantly affect cell spreading behavior, NIH 3T3 cells were plated on uniformly rigid polyacrylamide gels coated with adhesion proteins in the same micropattern as for the composite substrate. Cell spreading on these substrates followed a similar time course and reached a similar extent as for cells on control composite substrates (Fig. S3 and Movie S4). Thus, although the 3D topography of the SU-8 islands may affect some properties of the cell, it did not affect the extent or rate of cells spreading, which is the focus of this study.
To determine the role of actin-related protein 2/3 (Arp 2/3)-mediated actin polymerization and lamellipodial protrusion in the response to rigid substrates (25), we treated cells with Arp 2/3 inhibitor, CK666 (26). CK666 treatment decreased the percentage of successful spreading onto small islands (Fig. 2F and Fig. S2A) and increased the percentage of failed attempts (Fig. 2E), but increased the probing rate (Fig. S2B). Inhibition of the small GTPase Rac1 with inhibitor NSC23766, which also interferes with lamellipodia formation (27), had a similar effect (Fig. S2A). These results indicate that cell spreading across rigid substrates requires Arp 2/3- and Rac1-mediated actin polymerization and lamellipodia protrusion.
Filopodia Are Involved in Probing Substrate Rigidity.
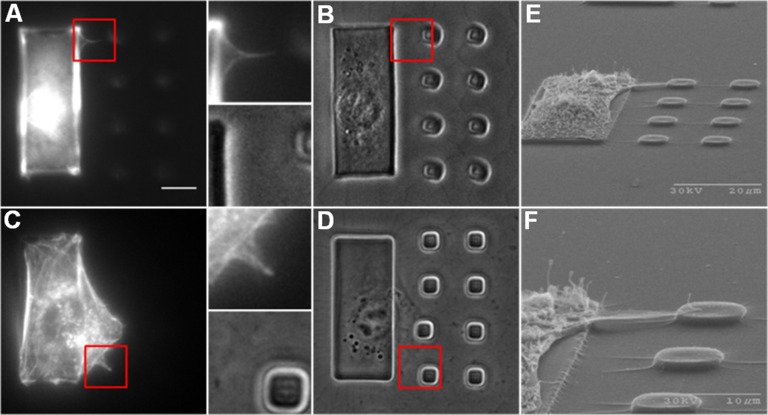
The above results suggest that thin extensions close to the detection limit of phase optics were involved in probing substrate rigidity in front of the cell. To capture these structures, cells on testing substrates were fixed and stained with fluorescent phalloidin 7 h after plating, when cells were expected to probe actively across the rigidity border. Thin actin-containing filopodia protrusions were found to connect the cell body with adjacent small islands (Fig. 3 A and B). Similar structures were observed in cells that managed to occupy one small island and started to probe additional small island (Fig. 3 C and D). Scanning electron microscopy of cells on control substrates also revealed fine protrusions extending from the cell body on the large island to adjacent small islands (Fig. 3 E and F).
Fig. 3.
Actin-containing filopodia extensions bridge the cell body with substrate in front of the leading edge. Filopodia are often difficult to detect with phase contrast microscopy (B, Inset), but become visible, upon staining with fluorescent phalloidin, as thin projections from the cell body (A, Inset). Similar structures are found extending from cells that have already occupied one small island (C and D, Insets). (Scale bar, 10 μm.) Scanning electron microscope images show protrusions that extend from the cell body above the underlying hydrogel to land on an adjacent small island (E). Enlarged view shows a filopodia protrusion along the top edge of the cell (F).
Cdc42 and formins are known to regulate filopodia function (28–30). Inhibition of Cdc42 or formins with inhibitor ML-141 or small molecule inhibitor of formin homology 2 domains (SMIFH2), respectively, caused a similar decrease in the rate of probing by thin extensions (Fig. 2D). Unexpectedly, both agents caused an increase in cell spreading onto small islands on testing substrates, allowing more cells to cross the rigidity border and at a higher rate than the spreading of untreated cells (Fig. S4 A and B). These results suggest that probing of filopodia may be involved in generating a retraction signal against cell spreading on soft substrates.
Nascent Protrusions Detect Substrate Rigidity Outside the Cell Border Based on Substrate Strain.
The results above suggest that thin extensions such as filopodia are responsible for probing substrate rigidity, which then activates a retraction response if the substrate is soft. Otherwise spreading represents the default response when the substrate is rigid or when the sensing mechanism is defective. A plausible mechanism may involve the response to substrate movement associated with the strain caused by traction forces.
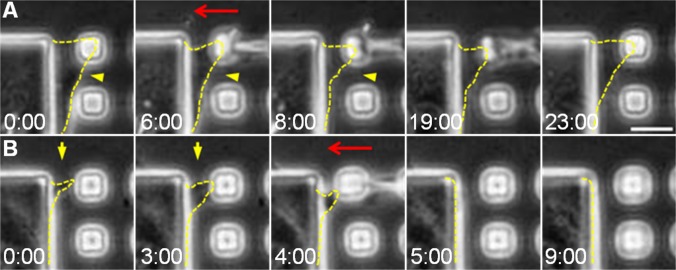
To test this hypothesis, we identified small islands on a rigid underlying gel that have recently been occupied by a cell extension, and used a microneedle to push the island toward the cell body to simulate the deformation of soft substrates by cellular probing forces (Fig. 4 A and B). We found that nascent extensions, defined as filopodia or fine protrusions that became detectable in phase contrast optics for less than 10 min, readily retracted after the induced centripetal movement (91% retracted from the island; Fig. 4B and Movie S5). More mature protrusions, which have typically expanded sufficiently to occupy a small island (Fig. 4A, first panel), showed a reduction in size but persisted without releasing from the island (87.5% remained associated with the island; Fig. 4A and Movie S6). These results suggest that, whereas both nascent and mature protrusions respond to mechanical input, only nascent protrusions and the associated nascent adhesions are able to retract completely from soft substrates. The sensitivity to rigidity decreases once the lamellipodium has spread over the adhered area.
Fig. 4.
Deformation of the substrate by micromanipulation causes retraction of nascent but not mature protrusions. A microneedle was used to push a small island toward the cell body to simulate substrate deformation as a result of probing by traction forces (red arrows). Mature protrusions that adhere to the small island respond by decreasing their size without fully releasing from the island (A, yellow arrowheads). In contrast, nascent protrusions, which have become visible for less than 10 min, retract beyond detection in response to the same manipulation (B, yellow arrows). Cell protrusions are outlined by yellow dashed lines. Numbers at the bottom of each image indicate the lapse time in minutes and seconds. 0:00 is an arbitrary time that indicates the start of the time-lapse imaging. (Scale bar, 10 μm.)
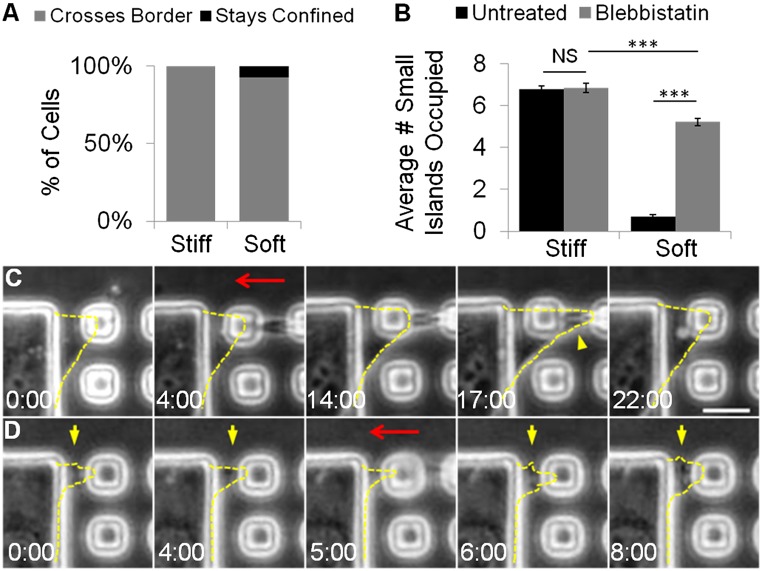
Myosin II Is Required for both the Probing Forces and the Subsequent Retraction from Soft Substrates.
The previous results suggest that rigidity sensing involves pulling forces of filopodia or other thin extensions to probe the deformability of the substrate. We hypothesized that myosin II is responsible for generating these probing forces, such that cells should become insensitive to substrate rigidity and show the default behavior of spreading upon the inhibition of myosin II. Indeed, cells treated with 10 μM blebbistatin readily crossed the rigidity border on testing substrates (Fig. 5A and histogram shown in Fig. S5A) to occupy more small islands than untreated cells (Fig. 5B). In addition, as reported previously, blebbistatin-treated cells adopted a more irregular shape regardless of the stiffness of the substrate (19). There was still a small but significant difference in the average number of small islands occupied on testing substrates versus control substrates, similar to the difference in spread area on unpatterned gels (Fig. S5B). Nevertheless, these observations support the notion that, when a cell becomes unable to probe and deform the substrate, it would interpret all of the substrates as being rigid.
Fig. 5.
Myosin II is required for the cellular response to substrate rigidity. NIH 3T3 cells treated with 10 μM blebbistatin are no longer confined to the large island on testing substrates with soft gels (A, right bar), such that there is a significant increase in number of small islands taken by treated cells (B, gray bar on right, n = 192) than by untreated cells (B, black bar on right). On rigid gels cells are able to occupy small islands to a similar extent regardless of blebbistatin treatment (B, left bars, n = 51). Neither mature (C) nor nascent protrusions (D, yellow arrows) visibly respond to the deformation of the substrate (red arrows show the direction of microneedle manipulation). Some mature protrusions even continue to spread beneath the microneedle during the micromanipulation (C, yellow arrowhead). Protrusions are outlined by yellow dashed lines. Numbers at the bottom of each image indicate the lapse time in minutes and seconds. 0:00 is an arbitrary time that indicates the start of the time-lapse imaging. (Scale bar, 10 μm.) Error bars represent SEM ***P < 0.001.
If the sole function of myosin II in rigidity sensing is to provide contractile probing forces for deforming the substrate, then one may expect that artificial deformation of the substrate, as induced by a microneedle, described earlier, should cause cell protrusions to retract regardless of myosin II inhibition. However, neither nascent nor mature protrusions of blebbistatin-treated cells retracted upon microneedle-induced centripetal movement of small islands (Fig. 5 C and D and Movies S7 and S8). Some of these cells even spread beneath the microneedle during the pushing action (Fig. 5C, yellow arrow head, and Movie S8). These results may be explained if myosin II activity is required not only for probing but also for the subsequent retraction in response to substrate deformation. In addition, the results are consistent with the notion that spreading represents the default response to substrate adhesion in the absence of substrate deformation and associated reactions.
Discussion
Rigidity sensing has emerged as an important factor in many biological processes. Various hydrogel-based experimental approaches, including spatial borders and temporal modulation of rigidity, have been used to study rigidity sensing (18, 31–33). However, many of these systems require lengthy time-lapse recording over a wide area to capture the brief moment when a cell encounters a rigidity border. Whereas some recent studies have developed substrates with micropatterned rigidity (15–17, 34), the present composite substrate has several distinct advantages. First, it traps single cells at a simulated rigidity border, which allows prolonged observation of cell behavior. Second, it allows a large number of cells to be studied under the same condition, which provides reproducible results and allows efficient screening of conditions that affect rigidity sensing. Third, the presence of nonadhesive polyacrylamide gels between adhesive islands allows the analysis of cellular responses to substrate rigidity at a distance from the cell border.
NIH 3T3 cells exhibited strikingly different behavior on testing substrates (with a soft underlying gel to create a rigidity border) versus control substrates (with a rigid underlying gel to create a nondeformable surface across). On control substrates, cells occupied the majority of small islands within 8 h. In contrast, on testing substrates, 74% of the cells remained completely confined to the rigid domain after 16 h. In the present experiment, this transition in response took place at an underlying hydrogel rigidity of 1000 Pa. However, this value cannot be directly compared with the transition of rigidity response for cells plated on homogeneous substrates, where a filopodium would adhere to and move an area not much larger than its cross-section during probing. In contrast, a filopodium in the present configuration would attach to an island much larger than its cross-section and must move the entire island during probing. Therefore, the equivalent rigidity threshold on homogeneous substrates may be one to two orders of magnitude higher or between 10 and 100 kPa.
Our observations with phalloidin staining and scanning electron microscopy suggested that actin-containing filopodia in front of the leading edge are responsible for rigidity sensing. This is complementary to other studies suggesting that filopodia play a role in probing the extracellular environment during axonal guidance and in the sensing of substrate topography, and that fluctuations in protrusive activities can bias directional cell migration (35–37). Time-lapse recordings further indicated a probing process mediated by filopodia followed by the spreading of lamellipodia. Although cells on both soft and rigid substrates exhibited a similar probing rate, there was a dramatic difference in the rate of subsequent spreading across soft or rigid substrates. On the control substrate, it took ∼3 h for cells to spread onto the first island and another 5 h onto the remaining seven islands. In contrast, on testing substrates it took over 10 h for a small fraction of cells to spread across soft hydrogels onto the first island. This difference was due to the differential stability of filopodia extensions following initial adhesion and probing. Extensions onto soft materials tended to release and retract, whereas extensions onto rigid materials were stable to allow continuous spreading. These results also suggest that fibroblasts are able to detect substrate rigidity at a distance in front of their leading edge through nascent adhesions, which have been previously shown to generate strong traction stress likely for the purpose of probing substrates (38). Supporting this hypothesis, we have observed minute deflections of small islands on soft gels just before extensions became visible in phase contrast optics. This finding is also supported by earlier observations that filopodia were able to exert forces on the surrounding substrate (39, 40).
One may consider two alternative mechanisms for rigidity sensing—through either the promotion of spreading on rigid substrates, or the promotion of retraction on soft substrates. Inhibition of Cdc42 or formins, two positive filopodia regulators, reduced the appearance of thin probing extensions while promoting cell spreading onto soft substrates. In addition, consistent with the previous suggestion that actomyosin-dependent traction forces are crucial for rigidity sensing (18, 19), inhibition of myosin II also caused cells to spread onto soft substrates (Fig. 5A). These results suggest that filopodia and associated traction forces are required for probing substrate rigidity and that spreading is the default response when the substrate fails to deform or when the downstream signal transduction mechanism is defective. One may then expect cell retraction when the substrate was pushed artificially toward the cell body to simulate the effect of traction forces, and that this retraction response be inhibited by blebbistatin. Both predictions were supported by experiments.
Using a similar composite substrate, we previously suggested that rigidity sensing is based on long-range strain of the substrate in response to cellular traction forces (21). The present results indicate that this range is defined by the length of filopodia extensions, which may be as long as 20–30 μm (28, 41, 42), rather than the tip-to-tail length of the cell. A related question concerns rigidity sensitivity of mature focal adhesions, which was suggested by the retraction of cell lamellipodia and shrinkage of mature focal adhesions when a spread region was pushed toward the cell body to reduce tension (10). Based on their localization behind the leading edge, we suspect that mature focal adhesions play a secondary role in guiding cell migration, such as during durotaxis. They may nevertheless facilitate the responses to dynamic changes in substrate rigidity, as demonstrated upon softening of the anterior substrate using a UV-sensitive hydrogel substrate (32).
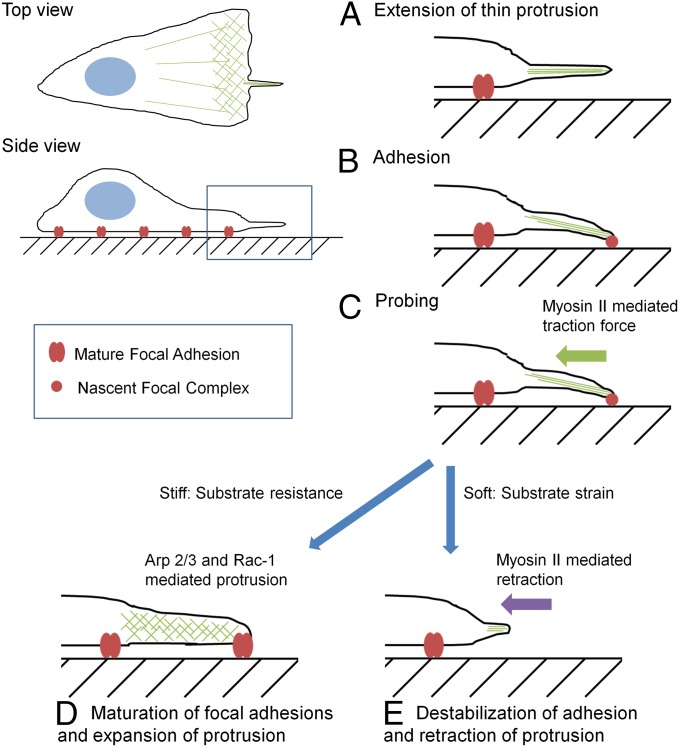
Fig. 6 depicts a plausible rigidity sensing mechanism, where myosin II-driven pulling forces are exerted at nascent adhesions at the tip of filopodia to induce substrate deformation (Fig. 6). Resistance of rigid substrates causes tension to build up at nascent adhesions, which promotes maturation of focal adhesions and allows the default response of active extension/spreading of the cell possibly by maintaining the small GTPase Rac (43, 44) and Arp 2/3 mediated actin polymerization (25, 45). On soft materials, the strain of the substrate inhibits the buildup of tension at nascent adhesions and keeps them from maturation into focal adhesions, which then promotes the retraction of filopodia through myosin II-dependent contractile forces. In the absence of these mechanically active filopodia, or in the absence of proper downstream responses to substrate deformation, cells exhibit the default response of spreading.
Fig. 6.
Schematic of rigidity sensing in front of the leading edge. Actin-containing filopodia extend in front of the leading edge (A), and establish nascent adhesions at a distance in front of the leading edge (B). Myosin II-mediated traction forces drive the filopodia probing process by pulling on the nascent adhesions (C). The resulting strain then determines the cellular response. Resistance from rigid substrates causes nascent adhesions to mature into focal adhesions and protrusions to expand via Arp 2/3- and Rac1-mediated actin polymerization (D). Large strain from soft substrates causes nascent adhesions to disassemble and protrusions to retract via a myosin II-dependent process (E).
Sensing mediated by filopodia in front of the leading edge provides an efficient mechanism for durotaxis and possibly other responses to physical properties of the substrate, by allowing cells to determine physical characteristics before moving over an area and to avoid the formation of mature focal adhesions on soft substrates to avoid backtracking. Although the present study was conducted with cells in 2D culture, the mechanism may also allow cells to maintain a similar sensitivity in physiologically relevant 3D environments, where they are unable to form broad lamellipodia or large focal adhesions but maintain the ability to form filopodia and other types of extensions.
Methods
Preparation of composite materials was performed as described previously (21, 22); complete details of the preparation and characterization of composite substrates are given in SI Methods. All experiments were performed using NIH 3T3 cells; complete details of cell culture conditions, reagents used, and microscopy are given in SI Methods. Image analysis was performed using ImageJ; complete details are given in SI Methods.
Supplementary Material
Acknowledgments
We thank Joe Suhan at the Electron Microscopy Facility at Carnegie Mellon University for assisting with the scanning electron microscopy. This work was supported by NIH Grant GM-32476 (to Y.-L.W.) and by NIH Predoctoral Training Grant T32EB003392 (to S.W.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1412285111/-/DCSupplemental.
References
- 1.Wang HB, Dembo M, Wang YL. Substrate flexibility regulates growth and apoptosis of normal but not transformed cells. Am J Physiol Cell Physiol. 2000;279(5):C1345–C1350. doi: 10.1152/ajpcell.2000.279.5.C1345. [DOI] [PubMed] [Google Scholar]
- 2.Discher DE, Janmey P, Wang Y-L. Tissue cells feel and respond to the stiffness of their substrate. Science. 2005;310:1139–43. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- 3.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126(4):677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 4.Butcher DT, Alliston T, Weaver VM. A tense situation: Forcing tumour progression. Nat Rev Cancer. 2009;9(2):108–122. doi: 10.1038/nrc2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paszek MJ, et al. Tensional homeostasis and the malignant phenotype. Cancer Cell. 2005;8(3):241–254. doi: 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 6.Tilghman RW, et al. Matrix rigidity regulates cancer cell growth and cellular phenotype. PLoS ONE. 2010;5(9):e12905. doi: 10.1371/journal.pone.0012905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ulrich TA, de Juan Pardo EM, Kumar S. The mechanical rigidity of the extracellular matrix regulates the structure, motility, and proliferation of glioma cells. Cancer Res. 2009;69(10):4167–4174. doi: 10.1158/0008-5472.CAN-08-4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levy-Mishali M, Zoldan J, Levenberg S. Effect of scaffold stiffness on myoblast differentiation. Tissue Eng Part A. 2009;15(4):935–944. doi: 10.1089/ten.tea.2008.0111. [DOI] [PubMed] [Google Scholar]
- 9.Mason BN, Califano JP, Reinhart-King CA. In: Engineering Biomaterials for Regenerative Medicine. Bhatia SK, editor. Springer; New York: 2012. pp. 19–38. [Google Scholar]
- 10.Lo CM, Wang HB, Dembo M, Wang Y-L. Cell movement is guided by the rigidity of the substrate. Biophys J. 2000;79(1):144–152. doi: 10.1016/S0006-3495(00)76279-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pelham RJ, Jr, Wang Yl. Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc Natl Acad Sci USA. 1997;94(25):13661–13665. doi: 10.1073/pnas.94.25.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tan JL, et al. Cells lying on a bed of microneedles: An approach to isolate mechanical force. Proc Natl Acad Sci USA. 2003;100(4):1484–1489. doi: 10.1073/pnas.0235407100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang G, Huang AH, Cai Y, Tanase M, Sheetz MP. Rigidity sensing at the leading edge through alphavbeta3 integrins and RPTPalpha. Biophys J. 2006;90(5):1804–1809. doi: 10.1529/biophysj.105.072462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Plotnikov SV, Pasapera AM, Sabass B, Waterman CM. Force fluctuations within focal adhesions mediate ECM-rigidity sensing to guide directed cell migration. Cell. 2012;151(7):1513–1527. doi: 10.1016/j.cell.2012.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gray DS, Tien J, Chen CS. Repositioning of cells by mechanotaxis on surfaces with micropatterned Young’s modulus. J Biomed Mater Res A. 2003;66(3):605–614. doi: 10.1002/jbm.a.10585. [DOI] [PubMed] [Google Scholar]
- 16.Nemir S, Hayenga HN, West JL. PEGDA hydrogels with patterned elasticity: Novel tools for the study of cell response to substrate rigidity. Biotechnol Bioeng. 2010;105(3):636–644. doi: 10.1002/bit.22574. [DOI] [PubMed] [Google Scholar]
- 17.Kawano T, Kidoaki S. Elasticity boundary conditions required for cell mechanotaxis on microelastically-patterned gels. Biomaterials. 2011;32(11):2725–2733. doi: 10.1016/j.biomaterials.2011.01.009. [DOI] [PubMed] [Google Scholar]
- 18.Trichet L, et al. Evidence of a large-scale mechanosensing mechanism for cellular adaptation to substrate stiffness. Proc Natl Acad Sci USA. 2012;109(18):6933–6938. doi: 10.1073/pnas.1117810109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raab M, et al. Crawling from soft to stiff matrix polarizes the cytoskeleton and phosphoregulates myosin-II heavy chain. J Cell Biol. 2012;199(4):669–683. doi: 10.1083/jcb.201205056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Breckenridge MT, Desai RA, Yang MT, Fu J, Chen CS. Substrates with engineered step changes in rigidity induce traction force polarity and durotaxis. Cell Mol Bioeng. 2013;7:26–34. doi: 10.1007/s12195-013-0307-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoffecker IT, Guo WH, Wang YL. Assessing the spatial resolution of cellular rigidity sensing using a micropatterned hydrogel-photoresist composite. Lab Chip. 2011;11(20):3538–3544. doi: 10.1039/c1lc20504h. [DOI] [PubMed] [Google Scholar]
- 22.Wong S, Guo W, Hoffecker IT, Wang Y. Preparation of a micropatterned rigid-soft composite substrate for probing cellular rigidity sensing. Methods in Cell Biology. 2014;121:3–15. doi: 10.1016/B978-0-12-800281-0.00001-4. [DOI] [PubMed] [Google Scholar]
- 23.Rape AD, Guo W-H, Wang Y-L. The regulation of traction force in relation to cell shape and focal adhesions. Biomaterials. 2011;32(8):2043–2051. doi: 10.1016/j.biomaterials.2010.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen CS, Alonso JL, Ostuni E, Whitesides GM, Ingber DE. Cell shape provides global control of focal adhesion assembly. Biochem Biophys Res Commun. 2003;307(2):355–361. doi: 10.1016/s0006-291x(03)01165-3. [DOI] [PubMed] [Google Scholar]
- 25.Suraneni P, et al. The Arp2/3 complex is required for lamellipodia extension and directional fibroblast cell migration. J Cell Biol. 2012;197(2):239–251. doi: 10.1083/jcb.201112113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nolen BJ, et al. Characterization of two classes of small molecule inhibitors of Arp2/3 complex. Nature. 2009;460(7258):1031–1034. doi: 10.1038/nature08231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Steffen A, et al. Rac function is crucial for cell migration but is not required for spreading and focal adhesion formation. J Cell Sci. 2013;126(Pt 20):4572–4588. doi: 10.1242/jcs.118232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nobes CD, Hall A. Rho, rac, and cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell. 1995;81(1):53–62. doi: 10.1016/0092-8674(95)90370-4. [DOI] [PubMed] [Google Scholar]
- 29.Jaiswal R, et al. The formin Daam1 and fascin directly collaborate to promote filopodia formation. Curr Biol. 2013;23(14):1373–1379. doi: 10.1016/j.cub.2013.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mellor H. The role of formins in filopodia formation. Biochim Biophys Acta. 2010;1803(2):191–200. doi: 10.1016/j.bbamcr.2008.12.018. [DOI] [PubMed] [Google Scholar]
- 31.Wong JY, Velasco A, Rajagopalan P, Pham Q. Directed movement of vascular smooth muscle cells on gradient-compliant hydrogels. Langmuir. 2003;19:1908–1913. [Google Scholar]
- 32.Frey MT, Wang Y-L. A photo-modulatable material for probing cellular responses to substrate rigidity. Soft Matter. 2009;5:1918–1924. doi: 10.1039/b818104g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sunyer R, Jin AJ, Nossal R, Sackett DL. Fabrication of hydrogels with steep stiffness gradients for studying cell mechanical response. PLoS ONE. 2012;7(10):e46107. doi: 10.1371/journal.pone.0046107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Choi YS, et al. The alignment and fusion assembly of adipose-derived stem cells on mechanically patterned matrices. Biomaterials. 2012;33(29):6943–6951. doi: 10.1016/j.biomaterials.2012.06.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dalby MJJ, Gadegaard N, Riehle MO, Wilkinson CDW, Curtis ASG. Investigating filopodia sensing using arrays of defined nano-pits down to 35 nm diameter in size. Int J Biochem Cell Biol. 2004;36(10):2005–2015. doi: 10.1016/j.biocel.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 36.Davenport RW, Dou P, Rehder V, Kater SB. A sensory role for neuronal growth cone filopodia. Nature. 1993;361(6414):721–724. doi: 10.1038/361721a0. [DOI] [PubMed] [Google Scholar]
- 37.Caballero D, Voituriez R, Riveline D. Protrusion fluctuations direct cell motion. Biophys J. 2014;107(1):34–42. doi: 10.1016/j.bpj.2014.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beningo KA, Dembo M, Kaverina I, Small JV, Wang YL. Nascent focal adhesions are responsible for the generation of strong propulsive forces in migrating fibroblasts. J Cell Biol. 2001;153(4):881–888. doi: 10.1083/jcb.153.4.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Albuschies J, Vogel V. The role of filopodia in the recognition of nanotopographies. Sci Rep. 2013;3 doi: 10.1038/srep01658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chan CE, Odde DJ. Traction dynamics of filopodia on compliant substrates. Science. 2008;322:1687–1691. doi: 10.1126/science.1163595. [DOI] [PubMed] [Google Scholar]
- 41.Lehnert D, et al. Cell behaviour on micropatterned substrata: Limits of extracellular matrix geometry for spreading and adhesion. J Cell Sci. 2004;117(Pt 1):41–52. doi: 10.1242/jcs.00836. [DOI] [PubMed] [Google Scholar]
- 42.Partridge MA, Marcantonio EE. Initiation of attachment and generation of mature focal adhesions by integrin-containing filopodia in cell spreading. Mol Biol Cell. 2006;17(10):4237–4248. doi: 10.1091/mbc.E06-06-0496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ridley AJ, Hall A. The small GTP-binding protein rho regulates the assembly of focal adhesions and actin stress fibers in response to growth factors. Cell. 1992;70:389–99. doi: 10.1016/0092-8674(92)90163-7. [DOI] [PubMed] [Google Scholar]
- 44.Waterman-Storer CM, Worthylake RA, Liu BP, Burridge K, Salmon ED. Microtubule growth activates Rac1 to promote lamellipodial protrusion in fibroblasts. Nat Cell Biol. 1999;1(1):45–50. doi: 10.1038/9018. [DOI] [PubMed] [Google Scholar]
- 45.Wu C, et al. Arp2/3 is critical for lamellipodia and response to extracellular matrix cues but is dispensable for chemotaxis. Cell. 2012;148(5):973–987. doi: 10.1016/j.cell.2011.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.