Significance
A prominent epigenetic mechanism for gene regulation is methylation of cytosine bases in DNA. TET enzymes facilitate DNA demethylation by converting 5-methylcytosine (5mC) to oxidized methylcytosines (oxi-mCs). We show that oxi-mCs are generated by conserved TET/JBP enzymes encoded in the genome of the model organism Coprinopsis cinerea and present a method for simultaneous mapping of the three different species of oxi-mCs at near–base-pair resolution. We observe that centromeres and transposable elements exhibit distinctive patterns of 5mC and oxi-mC, and show that gene body 5mC and oxi-mC mark silent paralogous multicopy genes. Our study describes a method to map three species of oxi-mC simultaneously and reveals the colocation of 5mC and oxi-mC at functional elements throughout the C. cinerea genome.
Keywords: TET, 5mC, 5fC, 5caC, SMRT-seq
Abstract
TET/JBP enzymes oxidize 5-methylpyrimidines in DNA. In mammals, the oxidized methylcytosines (oxi-mCs) function as epigenetic marks and likely intermediates in DNA demethylation. Here we present a method based on diglucosylation of 5-hydroxymethylcytosine (5hmC) to simultaneously map 5hmC, 5-formylcytosine, and 5-carboxylcytosine at near–base-pair resolution. We have used the method to map the distribution of oxi-mC across the genome of Coprinopsis cinerea, a basidiomycete that encodes 47 TET/JBP paralogs in a previously unidentified class of DNA transposons. Like 5-methylcytosine residues from which they are derived, oxi-mC modifications are enriched at centromeres, TET/JBP transposons, and multicopy paralogous genes that are not expressed, but rarely mark genes whose expression changes between two developmental stages. Our study provides evidence for the emergence of an epigenetic regulatory system through recruitment of selfish elements in a eukaryotic lineage, and describes a method to map all three different species of oxi-mCs simultaneously.
The discovery that oxidative modifications of DNA bases are catalyzed by the TET/JBP family of 2-oxoglutarate and iron-dependent dioxygenases (1–4) opened a major area of research into the epigenetics of various eukaryotic lineages (reviewed in refs. 5–7). In metazoans, the TET/JBP family is represented by TET proteins, which are present in all animals that are known to possess DNA cytosine methylation (3, 8, 9). The three TET paralogs of vertebrates have been shown to catalyze the oxidation of 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC) (2), which is progressively oxidized to 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC) (10–12). The discovery of these oxidized methylcytosine (oxi-mC) modifications triggered a flurry of studies on the mammalian TET paralogs; the reports increasingly point toward important roles for these oxidative modifications as intermediates in the enigmatic process of DNA demethylation, and also as epigenetic marks in their own right (13, 14) (reviewed in refs. 5–7). TET proteins have roles in diverse biological processes, including epigenetic regulation of gene transcription, embryonic development, stem cell function, and cancer (5), but the mechanisms underlying their biological activities are still poorly defined.
Several methods have been developed to profile individual oxi-mC species at base resolution in genomic DNA (reviewed in ref. 5). However, none of these methods can simultaneously map all three oxi-mC species—5hmC, 5fC, and 5caC—at the same time; rather, they rely on chemical or enzymatic conversion of individual oxi-mCs followed by bisulfite sequencing. Two recent sequencing technologies, single-molecule, real-time (SMRT) sequencing and protein nanopore sequencing, are capable of recognizing modified bases in unamplified genomic DNA (15–18). In SMRT sequencing, 5mC and 5hmC are barely detectable in unmodified DNA, whereas 5fC and 5caC yield a robust kinetic signature (16). As 5hmC is an abundant oxi-mC modification in mammalian DNA, we sought a method for mapping 5hmC together with 5fC and 5caC by using the SMRT sequencing technique.
We applied the method to the genome of Coprinopsis cinerea, an organism that has been used as a fungal model to study DNA methylation (19–21). C. cinerea was chosen for these studies because of its small genome size (∼36 Mb, ∼1/100th the size of the human genome), a fully sequenced and assembled genome (22, 23), and the presence of multiple copies of DNA transposons with genes encoding TET/JBP proteins (1, 8, 9, 22). We show that oxi-mCs mark TET/JBP transposons and other repetitive elements in C. cinerea, and are also enriched at centromeres. There is an overall correlation between the distribution of 5mC and oxi-mC in C. cinerea, as in mammals (24); however, whereas 5hmC is enriched in the gene bodies of highly expressed genes in mammals (14, 25, 26), oxi-mC modifications mark genes that are silent or poorly expressed in C. cinerea, and tend to be excluded from highly expressed genes and genes whose expression is altered between oidia (haploid spores) and haploid mycelia.
Results
TET/JBP-Coding Transposons and oxi-mCs.
The C. cinerea genome contains a total of 47 TET/JBP genes (Table S1), most of which are part of Kyakuja transposons (8). At least 32 of the TET/JBP genes are predicted to encode catalytically active proteins, and 29 of them belong to “complete” Kyakuja elements defined as possessing the three core genes, encoding a transposase, the TET/JBP enzyme, and a protein with a divergent HMG domain (8) (Fig. 1A and Tables S1 and S2). The C. cinerea genome has two distinct paralogs of DNA cytosine methyltransferase 1 (DNMT1) (CC1G_01237 and CC1G_00579) that are present across all Agaricomycetes (mushrooms), but not other fungi (8). C. cinerea also encodes a third predicted DNA cytosine methylase (CC1G_00872, also called DNMT5) with a distinct architecture (8) which has been recently shown to methylate cytosines with a bias towards those in internucleosomal linker regions (27). The modified cytosines in C. cinerea are likely to arise as a result of the interplay between these DNA methylases and the active TET/JBP proteins.
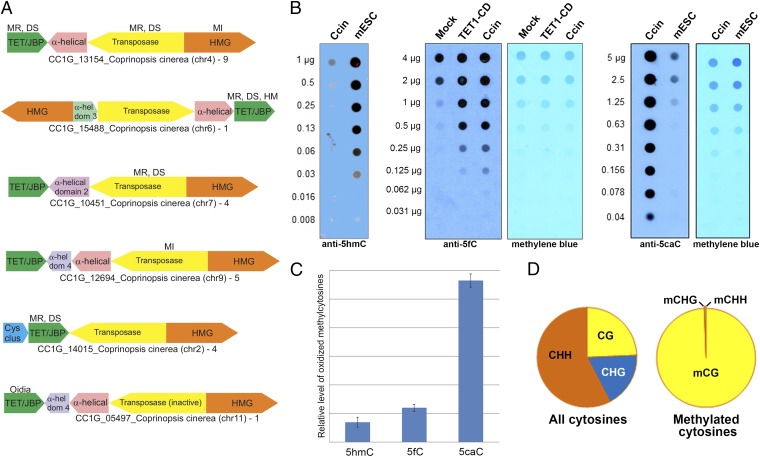
Fig. 1.
TET/JBP-coding transposons and oxi-mCs. (A) Genomic organization of the Kyakuja transposons in C. cinerea. Genes are depicted as arrows pointing from the 5′ to the 3′ direction of the coding sequence. Gene neighborhoods are labeled with the name of the TET/JBP gene, species name, and the sequence identification (gi) number with the chromosome number in brackets. The number of complete elements with a particular organization in the genome is shown next to the label. Five of the depicted transposons represent the more prevalent Kyakuja-1 element, which contains a gene coding for a predominantly α-helical protein between the TET/JBP and transposase genes that may also be fused to the transposase; the sixth is a less frequent Kyakuja-2 element that contains a gene for a cysteine-rich protein with conserved cysteine residues (Cys-clus) 5′ to the TET/JBP gene. In most Kyakuja elements, the TET/JBP gene is encoded on the opposite strand of DNA relative to the gene for the transposase, with the two in a head-to-head orientation; the TET/JBP gene and the gene encoding the HMG domain-containing protein (HMG) are usually on the same strand. The GenBank entry for CC1G_15488 and the transposase associated with CC1G_05497 fuses several distinct domains on the same strand. These are depicted as separate genes. α-hel dom, α-helical domain; DS, dikaryon-specific; HM, expressed in haploid mycelium; MI, meiotically induced; MR, meiotically repressed; MS, meiosis-specific; oidia, expressed in oidia. Stage-specific gene expression information is from Burns et al. (50). (B) Dot blots showing the relative abundance of 5hmC (Left), 5fC (Middle), and 5caC (Right) in C. cinerea (Ccin) compared with mouse embryonic stem cell DNA. TET1CD, DNA from HEK293 cells transfected with the TET1 catalytic domain. (C) Relative levels of 5hmC, 5fC, and 5caC in C. cinerea estimated by MS. (D) All ∼18.5 million cytosines (on both strands) in the 13 assembled chromosomes of the C. cinerea genome were categorized into three groups according to their sequence context (H = A, C, or T). Although only one fourth of all cytosines occur in the CpG context, more than 99% of all methylated cytosines (or hydroxymethylated cytosines, as bisulfite sequencing cannot distinguish between 5mC and 5hmC) are observed at CpGs.
The C. cinerea genome contains all three oxi-mCs 5hmC, 5fC, and 5caC (Fig. 1B). The relative levels of 5hmC in C. cinerea DNA are lower, and the relative levels of 5caC higher, than in mouse embryonic stem cell DNA, as judged by MS and DNA dot blot (Fig. 1 B and C). Bisulfite analysis showed, as previously observed (27), that the majority of methylated cytosines were located in the CpG context [we note that some fraction of these may be hydroxymethylated, as bisulfite sequencing cannot distinguish between 5mC and 5hmC (28)] (Fig. 1D).
Enhancing the Kinetic Signature of 5hmC in SMRT Sequencing.
In an effort to identify all three oxi-mC species simultaneously in the C. cinerea DNA, we turned to SMRT sequencing (29–31). 5mC and 5hmC are only weakly detected by SMRT sequencing (15), whereas 5caC yields a strong signal (16). To enhance the kinetic signature of 5hmC, we converted 5hmC to a diglucosylated adduct by using two T-even bacteriophage enzymes in succession (Fig. 2A). Tests on a synthetic SMRTbell DNA template (30) with 5hmC incorporated at two positions (Fig. 2B) confirmed that the interpulse duration (IPD) ratio of 5hmC to unmodified C was small (∼2) (15) (Fig. 2C, Top). Addition of a single glucose using T4 phage β-glucosyltransferase (BGT) increased the IPD ratio to between 2 and 10 (Fig. 2C, Middle); diglucosylation by successive treatment with BGT followed by T6 phage β-glucosyl α-glucosyl transferase (BGAGT) (32) increased the IPD ratio to between 10 and 29 (Fig. 2C, Bottom). The magnitude and pattern of the observed signal showed a clear dependence on the sequence context (Fig. 2 C and D and Fig. S1A).
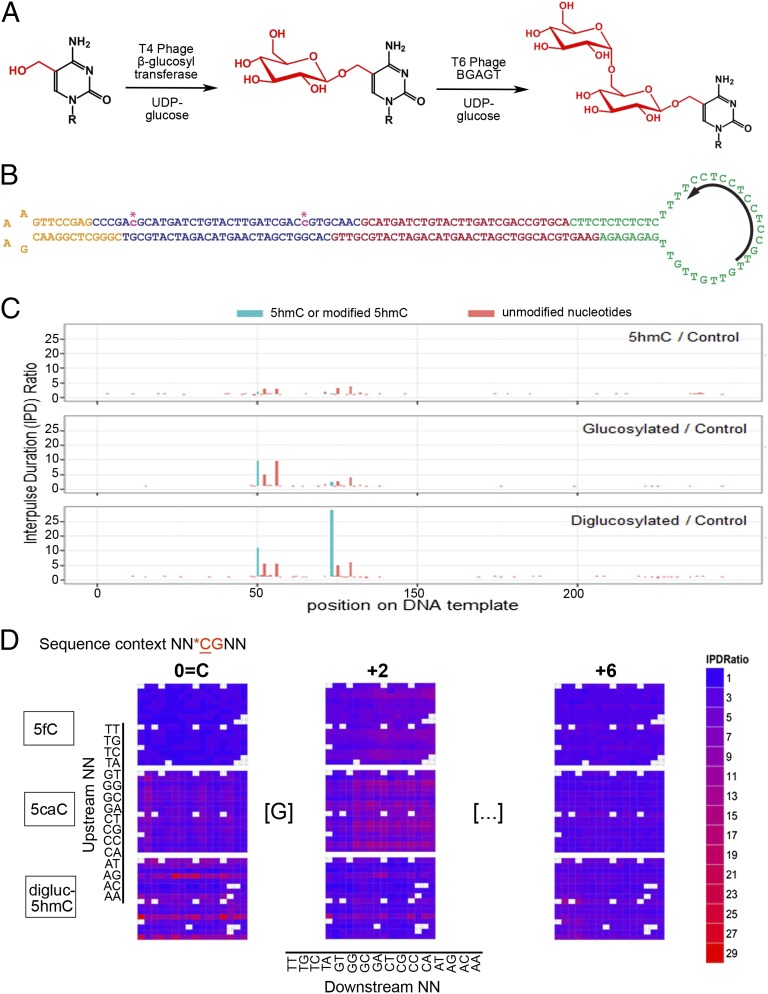
Fig. 2.
Enhancing the kinetic signature of 5hmC in SMRT sequencing. (A) Conversion of 5hmC to a diglucosylated adduct using two T-even bacteriophage enzymes in succession. (B) Synthetic SMRTbell template with 5hmC incorporated at two CpG positions (asterisks). (C) IPD ratio signatures of 5hmC (Top), glucosylated 5hmC (Middle), and diglucosylated 5hmC (Bottom). (D) Heat map showing SMRT sequencing IPD ratio signatures for 5fC, 5caC, and diglucosylated 5hmC at SMRTbell templates containing these modified cytosines in a randomized NNCGNN sequence context. Shown are the IPD ratios at the modified CpG, and at the +2 bp and +6 bp positions downstream.
We compared signals from 5mC, 5hmC, diglucosylated 5hmC, 5fC, and 5caC by sequencing SMRTbells containing these modified cytosines in a degenerate sequence context, in which the two nucleotides immediately 5′ and 3′ of the CpG dinucleotide were randomized (NNCGNN; Fig. 2D and Fig. S1A). For 5fC, high IPD ratios were observed at position +2 with respect to the CpG cytosine (Fig. 2D and Fig. S1A); for 5caC (16) and diglucosylated 5caC, high IPD ratios were observed at the CpG cytosine (position 0), at position +2 and, to a lesser extent, at position +6 (Fig. 2D and Fig. S1A). For 5mC, close spacing of modified cytosines had a cumulative effect: when we sequenced a SMRTbell template that was methylated at defined closely spaced positions, successive 5mC residues yielded progressively increasing IPD ratios, albeit not in a linear fashion (Fig. S1B). Thus, SMRT sequencing on native DNA yields the positions of 5fC and 5caC, whereas SMRT sequencing on diglucosylated DNA yields the positions of all three oxi-mCs (Fig. 2D) (16).
Mapping the Genomic Locations of oxi-mCs in the C. cinerea Genome.
A major advantage of SMRT sequencing is the ability to sequence long stretches of genomic DNA without PCR amplification. For diglucosylated C. cinerea DNA obtained from oidia (19, 33), we performed SMRT sequencing on two SMRTbell templates, one containing DNA sheared to a length of 400–500 bp and another sheared to ∼6 kb; for native DNA, we sequenced only ∼6-kb fragments of DNA. The longer insert libraries permitted unambiguous assignment of reads to specific repetitive genomic regions that are shorter than the long reads. Both samples were sequenced to ∼120× total coverage. To identify statistically significant kinetic variants indicative for modified bases, we calculated log-transformed P values by applying t tests to the sample IPDs against an in silico control at every genomic position [Qmod values, calculated as −10log(P value); SI Materials and Methods].
To confirm the results of SMRT sequencing of diglucosylated DNA (digluc-SMRT-seq), we used cytosine 5-methylenesulphonate (CMS) immunoprecipitation (IP) as a validated method of mapping 5hmC (34–36). Treatment with sodium bisulfite converts 5hmC to CMS, a highly immunogenic adduct (34). We immunoprecipitated bisulfite-treated C. cinerea DNA obtained from oidia (19, 33) with a specific and sensitive antiserum against CMS (34, 36) and mapped 5hmC-enriched regions of the genome (HERGs) by using a statistical model that explicitly considers biological and technical replicates for calculating overdispersion (37, 38) (SI Materials and Methods). In pairwise comparisons, the correlation between technical replicates was 99%, and that between biological replicates was >95% (Fig. S2A). The number of reads reached saturation as judged by multiple random sampling of different numbers of reads (Fig. S2B), and coverage was high, with >99% of all CpGs in the C. cinerea genome covered by extended (250-bp) reads (Fig. S2C). However, standard mapping considering only uniquely mapped reads (Fig. 3A, unique mappers) revealed a substantial number of repetitive genomic regions where CMS enrichment could not be calculated, as neither input nor CMS-IP data could be mapped to a unique position in the genome. To address this problem, we included reads that mapped to as many as 100 different genomic regions in input and CMS-IP analyses, and then calculated HERGs (Fig. 3A, CMS-IP, multiple mappers; details and mapping statistics are provided in SI Materials and Methods and Tables S3 and S4). The HERGs obtained by multiple mapping were merged with the original HERGs in which we considered only uniquely mapped reads, to obtain “extended” CMS-IP HERGs (Fig. 3A, Ext. HERGs) used in subsequent analyses.
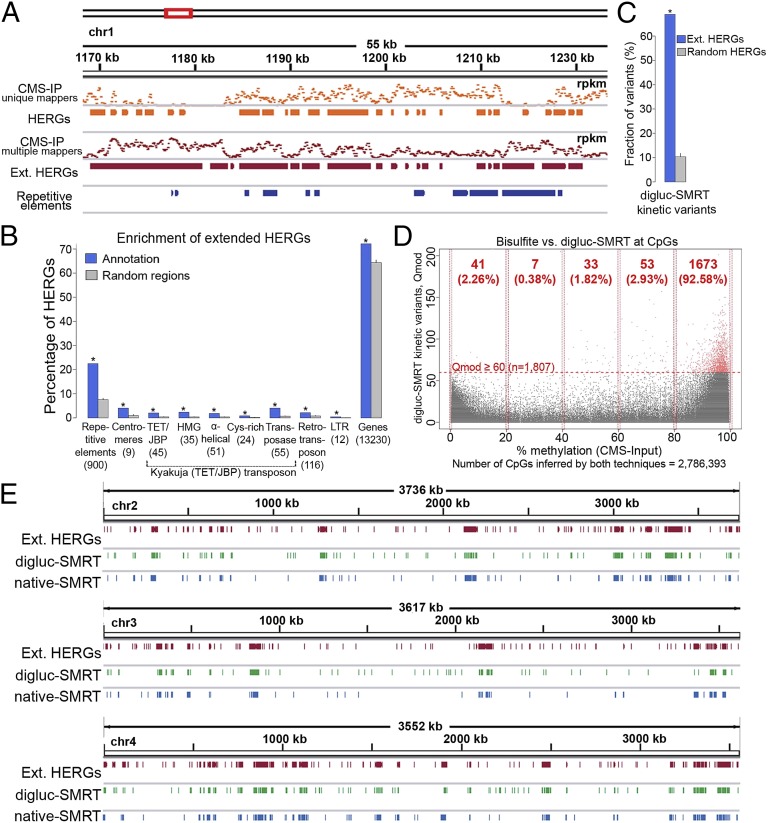
Fig. 3.
Mapping oxi-mCs in the C. cinerea genome. (A) Anti-CMS IP coverage [in reads per kilobase per million (rpkm)] of short reads mapping to only one position in the genome (unique mappers) or as many as 100 positions in the genome (multiple mappers). By considering only uniquely mapped reads, a substantial number of repetitive genomic regions cannot be covered by input or CMS-IP data. To address this problem, we also considered reads that mapped to as many as 100 different genomic regions in input and CMS-IP analyses (SI Materials and Methods). Extended HERGs are obtained by merging HERGs identified by the two alternative approaches. (B) To estimate the enrichment of HERGs at different annotated genomic regions, we performed 100 random samplings of the same number of genomic regions with the same size distribution (SI Materials and Methods). This analysis showed substantial enrichment of HERGs at repetitive elements and Kyakuja transposons (P ≤ 0.001, empirical test). A substantial amount of HERGs (72%) accumulate at genes and 4% of HERGs mapped to annotated centromeres. (C) An analogous analysis shows substantial enrichment of digluc-SMRT kinetic variants at extended HERGs (P ≤ 0.001), suggesting a high agreement between methods. (D) Single base-level comparison of cytosines in a CpG context covered by bisulfite (CMS input, x axis) and digluc-SMRT (y axis) sequencing (n = 2,786,393). Horizontal dashed red line represents a Qmod 60 threshold applied for defining digluc-SMRT kinetic variants. Bisulfite methylation shows a common bimodal distribution, and 5hmC is detected by digluc-SMRT essentially only in the high range of bisulfite methylation that is incorrectly considered as highly methylated by bisulfite sequencing. (E) Chromosome-wide views of extended HERGs (CMS IP), digluc-SMRT, and native SMRT sequencing data shows high correspondence among the distributions of oxi-mC species detected by these different technologies.
An advantage of CMS-IP is that the input DNA has been bisulfite-treated, and hence can be used to infer the sum of 5mC and 5hmC at each CpG (28). Single-base analysis of the bisulfite-treated input DNA confirmed that the majority of methylated/hydroxymethylated cytosines were present in a CpG context (Fig. 1D). In total, there are 371,971 CpGs with a C-to-T conversion rate of less than 20% in the bisulfite-treated input DNA, whereas the lambda control sequences have a mean C-to-T conversion rate of 98.8% (SI Materials and Methods), indicating that 8.26% of all cytosines genome wide (or 9.3% of all covered cytosines) are highly methylated/hydroxymethylated (>80%; Table S5).
Almost all repetitive elements and Kyakuja (TET/JBP) transposons overlap with HERGs, indicating that they are strongly marked by oxi-mCs; in contrast, only a small fraction (13.8%) of genes overlap with HERGs (Fig. S2D). To estimate the enrichment of HERGs at different annotated genomic regions, we performed 100 random samplings of the same number of genomic regions with the same size distribution (SI Materials and Methods). This analysis showed substantial enrichment of HERGs at repetitive elements and Kyakuja transposons (P ≤ 0.001, empirical test; Fig. 3B). Although only a small fraction of annotated genes are marked by hydroxymethylation (Fig. S2D), a substantial amount of HERGs (72%) accumulate at these genes, a number slightly but significantly greater than the enrichment calculated for random genomic sequences of similar length (Fig. 3B). Finally, 4% of HERGs mapped to annotated centromeres (Fig. 3B), whereas almost 100% of the annotated centromeres overlap with HERGs (Fig. S2D; discussed further later).
Signals obtained by digluc-SMRT-seq were significantly enriched in extended CMS-IP HERGs (Fig. 3C). Consequently, digluc-SMRT-seq signals are also significantly enriched at repetitive elements, Kyakuja transposons, and centromeres (P ≤ 0.001, empirical test). The presence of 5hmC correlated with the presence of 5mC: ∼92% of all diglucosylation signals from SMRT sequencing with a Qmod value ≥60 corresponded to CpG cytosines that are highly (80–100%) methylated (5mC/5hmC) as determined by bisulfite sequencing (Fig. 3D). The approximate genome-wide fraction of oxi-mCs (inferred by 1,807 digluc-SMRT–derived kinetic variants at CpGs with Qmod value ≥60) compared with highly methylated (>80%) CpGs inferred by bisulfite sequencing is 0.5% (Fig. 3D and Table S5). There was a strong overall correspondence among the three techniques, anti–CMS-IP, digluc-SMRT-seq, and native SMRT sequencing, particularly evident in chromosome-wide views (Fig. 3E and Fig. S2E).
oxi-mCs Mark DNA Transposons, Centromeres, and Retrotransposons.
Kyakuja transposons containing predicted active TET/JBP enzymes were major foci for 5mC oxidation, suggesting that TET/JBP proteins might regulate the methylation status of their own transposons. In each case, an extensive region including not only the Kyakuja transposon itself but also nearby repetitive elements was enriched for oxi-mC, with the length of the enriched region ranging from ∼30 kb to ∼90 kb in the examples shown (Fig. 4A and Fig. S3A). In addition, we found significant enrichment of oxi-mC at other mobile selfish elements in the C. cinerea genome. Among these are the DNA transposons Dileera and Zisupton, whose transposases are related to those of the Kyakuja elements (1, 8). There are 14 copies of Dileera and 8 copies of Zisupton transposons in the C. cinerea genome, and the majority of them (57%) contain oxi-mC (an example is shown in Fig. S3B). As also observed in the Kyakuja transposons, oxi-mCs are distributed within the bodies of the transposase genes of these elements.
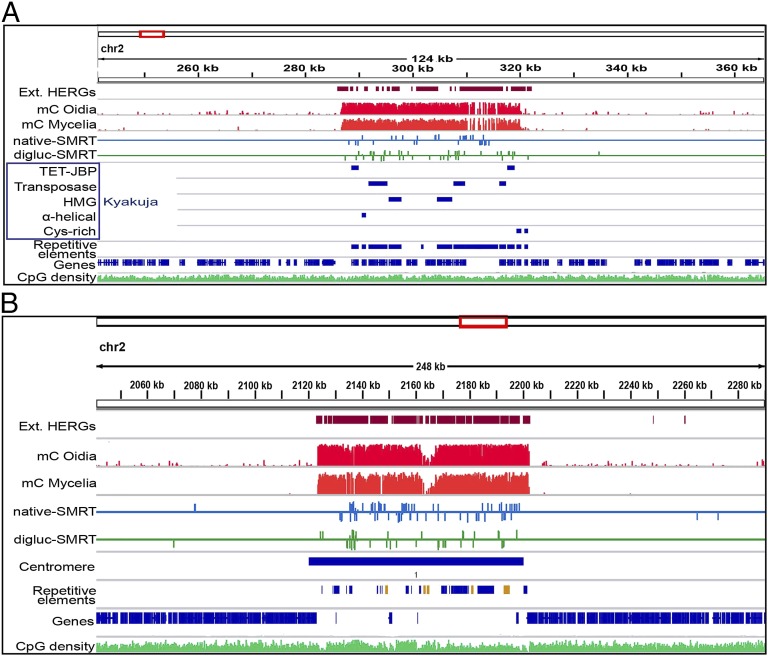
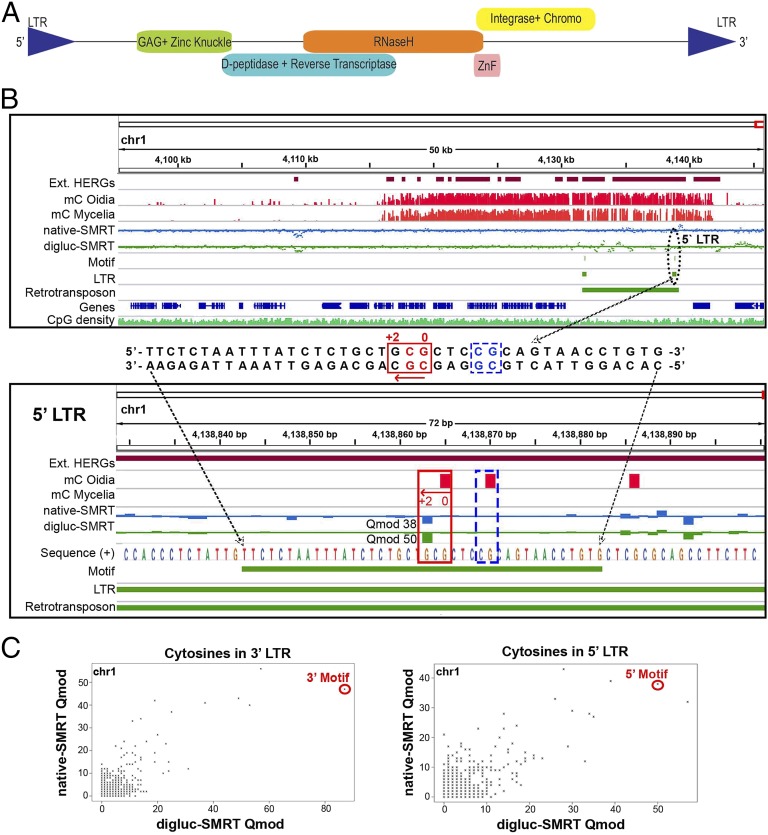
Fig. 4.
oxi-mCs mark TET/JBP transposons and centromeres. (A) TET/JBP transposons are embedded in regions with high abundance of oxi-mC, supporting the model that they regulate their own methylation status. Units are non–strand-specific fractions (as percentages) of the sum of 5mC and 5hmC compared with all sequenced cytosines at each position deduced by bisulfite sequencing for mC oidia and mC mycelia, and strand-specific Qmod values for digluc-SMRT and native SMRT sequencing. (B) Example of the distinct pattern of DNA modifications as observed at 8 of 13 centromeres (units are as described above).
There was also a strong correspondence of extended regions of methylcytosine oxidation with centromeres (Fig. 4B). The positions of 9 of 13 cytologically defined centromeres (39) are highly correlated with a cluster of transposon-related sequences, which are otherwise rare in the genome except for their occurrence near telomeres (22). Repeated sequences have previously been shown to be a primary target of cytosine methylation in C. cinerea (27), as confirmed in this work; however, instead of observing uniform methylation across the presumed centromere regions, we observed a distinctive pattern of DNA modification at 8 of 13 centromeres (chr 1, 2, 3, 4, 6, 7, 10, 12), consisting of local hypomethylated islands in regions of elevated CpG density surrounded by a large region of densely modified sequences that contained 5mC, 5hmC, and other oxi-mCs (Fig. 4B and Fig. S3 C–E). These local regions of hypomethylation were distinguished by the presence of Copia-like transposon fragments (shown in orange in the “repetitive element” tracks in Fig. 4B and Fig. S3 C–E), and were flanked by regions enriched for oxi-mC (Fig. 4B and Fig. S3 C–E). The Copia elements were also present in the flanking pericentric regions; however, the density of 5mC differed dramatically in otherwise identical Copia sequences, with <5% methylated within the presumed centromere core region in contrast to >18% methylated within the flanking pericentric region. The pattern on chromosome 10 occurred at a location other than the cytologically annotated centromere (Fig. S4E), which may reflect a strain difference: numerous chromosome-length polymorphisms have been described previously in C. cinerea (40). For these eight chromosomes, the new correlation between centromeres and hypomethylated/copia regions was even higher than the previous correlation (22) based simply on the presence of transposon clusters (R2 = 0.93 vs. R2= 0.89).
A search for conserved sequences with oxi-mC signals recovered an extended repetitive motif with a single oxi-mC (5′-CACAGGTTACTGCGGAGCGCAGCAGAGATAAATTAGAGAA-3′) that was considerably expanded in the C. cinerea genome (more than 30 copies). The motifs tended to be found in pairs, with the individual motifs in a pair typically ∼7.9 kb apart (Fig. 5B and Fig. S4), and comparison of signals from native and digluc-SMRT-seq suggested that it was 5fC or 5caC (Fig. 5C). Further analysis showed that the motifs were located in the LTRs of a group of mushroom retrotransposons related to the Ty/gypsy-like elements of ascomycete fungi and included among the clusters of repetitive elements previously annotated to Gypsy and Copia families (22). The complete versions of the Ty/gypsy-like elements are retrotransposons that encode polyproteins with serial GAG, Zn-Knuckle, aspartyl peptidase, reverse transcriptase, RNase H, ZnF, integrase and chromodomain (Fig. 5A). The C. cinerea genome has six complete copies of these Ty/gypsy-like retrotransposons on chromosomes 1, 4, 7, 9, 11, and 12, of which at most three (on chromosomes 1, 9, and 11) are likely to be transcribed, based on the presence of ESTs mapping to them; the remaining retrotransposon copies are all fragments at different stages of degeneration. For active copies and inactive fragments, there is a very widespread correlation of the retrotransposon with regions with a high frequency of oxi-mC, implying that TET/JBP enzymes specifically target these elements for modification (Fig. 5 B and C and Fig. S4). In addition, we found three copies of a DIRS1-family retrotransposon in C. cinerea (41, 42), which also contained oxi-mC in the body of the element. Overall, our data indicate that large genomic regions surrounding Kyakuja transposons as well as other DNA transposons and retrotransposons are heavily modified with oxi-mC.
Fig. 5.
oxi-mCs identify LTRs of Ty/gypsy-like retrotransposons. (A) Schematic view of a complete retrotransposon distantly related to the Ty-like elements of ascomycete fungi, encoding a polyprotein with serial GAG, Zn-Knuckle, aspartyl peptidase, reverse transcriptase, RNase H, ZnF, integrase, and chromodomain. (B) Regional view (Top) of a complete copy of the retrotransposon showing its location in a genomic region free of annotated genes but with high abundance of 5mC toward the end of chromosome 1. A conserved DNA sequence (Middle) has been identified in the LTRs of the retrotransposons as a result of a single modified cytosine as highlighted in a local view (Bottom) of the motif in the 5′ LTR of the retrotransposon shown above. The presence of elevated signals in native-SMRT (strand specific blue track, Qmod value 38) and digluc-SMRT (strand-specific green track, Qmod value 50) sequencing, suggests that the cytosine in the CpG context located two bases upstream of the actual signal at the minus strand reflects a population of 5hmC and either 5fC or 5caC. Units are non–strand-specific fractions (as percentages) of the sum of 5mC and 5hmC compared with all sequenced cytosines at each position deduced by bisulfite sequencing for mC oidia and mC mycelia, and strand-specific Qmod values for digluc-SMRT and native SMRT sequencing. (C) Direct correlation of digluc-SMRT (x axis) and native SMRT (y axis) sequencing-derived Qmod values of all cytosines in the 5′ (Top) and 3′ (Bottom) LTR of the retrotransposon shown in Fig. 5B. Highlighted by red circles are the single modified cytosines in the LTR motif.
A C. cinerea TET/JBP Is Transcribed at the Oidial Stage and Is Catalytically Active.
Although both DNMT1 paralogues (CC1G_01237, CC1G_00579) and the third DNA cytosine methylase (CC1G_00872, DNMT5) were expressed at the RNA level at the oidial stage (Fig. S5B and Dataset S1), only a single TET/JBP (CC1G_05497) was expressed at this stage (Fig. S5A and Dataset S1). To confirm the catalytic activity of this C. cinerea TET protein (here termed CcinTET), we expressed a FLAG-tagged version in mammalian HEK293 cells and compared its activity with that of FLAG-tagged human TET2 (Fig. 6 A and B). Unlike hTET2, which is completely localized to the nucleus, a significant proportion of CcinTET was localized to the cytoplasm in HEK293 cells cultured at 37 °C (Fig. 6A and Fig. S5C), but the enzyme still generated copious amounts of 5hmC as judged by immunocytochemistry, comparable to the amounts generated by hTET2 (Fig. 6B). However, in contrast to a previous report using a different C. cinerea TET/JBP protein (43), we were unable to detect significant 5caC in cells expressing CcinTET, even though hTET2 clearly generated 5caC over the background level observed in untransfected cells (Fig. S5D).
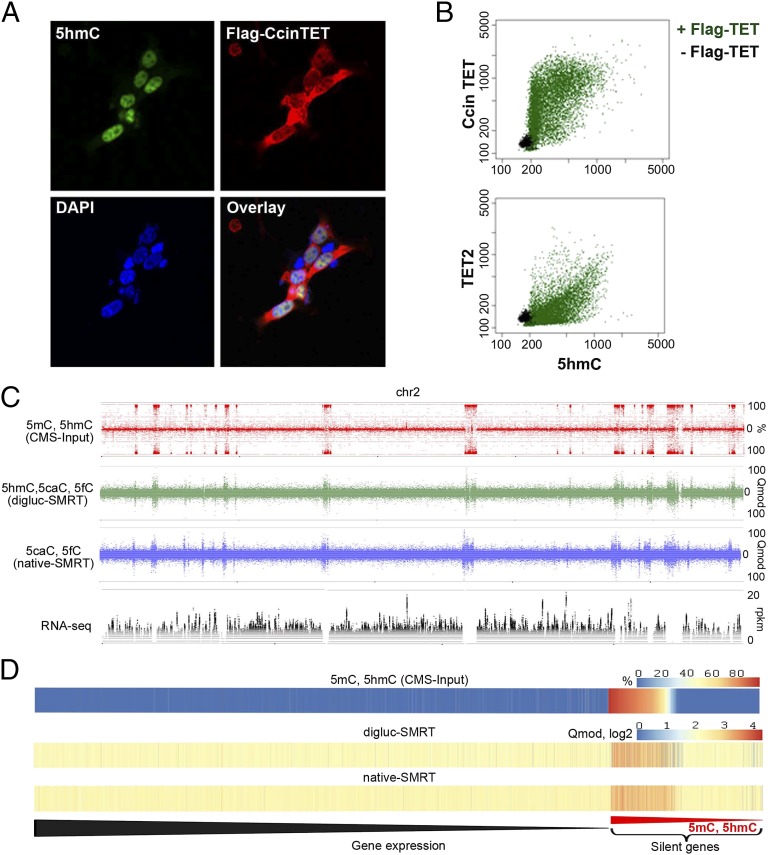
Fig. 6.
Catalytically active TET/JBP gene and relation of DNA modifications to gene expression. (A) Immunostaining of FLAG-CcinTET and 5hmC. HEK293T cells were transfected with pEF-Flag-CcinTET. 5hmC and CcinTET levels were detected by rabbit anti-5hmC (active motif) and mouse anti-Flag (Sigma) antibodies, followed by Alexa Fluor 488 goat anti-rabbit IgG (green) and Alexa Fluor 647 goat anti-mouse IgG antibodies (red). The nuclei were stained with DAPI (blue). (B) Use of IXM high-content imaging system to examine the catalytic activity of CcinTET in HEK293T cells. Each dot indicates the intensity of 5hmC and CcinTET expression (Flag-positive cells) of a single cell (22,248 cells analyzed). Green indicates HEK293T cell expressed with pEF-Flag-CcinTET; black indicates HEK293T cell expressed with empty vector. (C) Complete strand separated data representation of (from top to bottom) bisulfite (CMS-Input)-derived modifications (5mC and 5hmC), digluc-SMRT, native SMRT sequencing (assessed by Qmod values), and RNA-seq data coverage at 10-bp windows along the full chromosome 2. Note the complete coincidence of oxi-mC and 5mC-enriched regions and the strong suppression of transcription in regions of heavy 5mC/oxi-mC. (D) Heat maps of mean 5mC/5hmC (CMS-Input), digluc-SMRT, and native-SMRT signals per gene in oidia, sorted by gene expression in oidia from high to low. Elevated mC/oxi-mC signals are predominantly observed at silenced genes (Right). Silenced genes are sorted from high (at left) to low (at right) 5mC/5hmC level.
We examined the correlation of 5mC and oxi-mC distribution with gene transcription at the oidial stage. There is a complete coincidence of oxi-mC and 5mC-enriched regions using the different methods (shown for chromosome 2 in Fig. 6C), but, at this stage of development, there are no large genomic regions that show enrichment for oxi-mC over 5mC. As expected, transcription was strongly suppressed in regions of heavy 5mC, which correlated well with regions enriched for oxi-mC (Fig. 6 C and D).
Paralogous Multicopy Genes Are Less Expressed in Oidia and Show Higher oxi-mC.
C. cinerea has an efficient mechanism to detect DNA duplications introduced by DNA-mediated transformation and methylate them during the sexual cycle, a process called methylation induced premeiotically, or MIP (21). We confirmed that methylation/hydroxymethylation occurred primarily at sequences present in multiple copies, by inserting two additional copies of the tryptophan synthetase gene (trp1) at the endogenous locus (CC1G_13871; Fig. S6A). Bisulfite sequencing of 20 haploid meiotic segregants revealed that all CpG sequences within the 451-bp promoter region of this tandem triplication were methylated (or hydroxymethylated) to varying degrees (Fig. S6B, Left); no other cytosines were modified. The endogenous locus of WT C. cinerea is not methylated (Fig. S6B, Right), indicating that DNA cytosine methylation/hydroxymethylation is acquired only when duplications are introduced.
This finding led us to investigate the cytosine modification status of endogenous duplicated genes. The 13,236 protein-coding genes on the assembled chromosomes in the C. cinerea genome were previously classified into orphan genes with no obvious homologs in related species such as Laccaria bicolor (orphans, n = 3,689), single-copy genes with at least one ortholog in a related species (orthologs, n = 5,830), and paralogous multicopy genes that are primarily distributed in regions with average or high rates of meiotic recombination (paralogs, n = 3,717) (22) (Fig. S6C). Digluc-SMRT sequencing showed that oxi-mCs were significantly enriched at paralogous multicopy genes, strongly depleted at conserved single-copy genes (i.e., orthologs), and slightly depleted at nonconserved single copy genes (i.e., orphans; Fig. S6D). In total, we observed 5hmC, 5fC, and/or 5caC modifications at 1,043 of the 13,236 annotated genes (7.88%) based on digluc-SMRT sequencing: in detail, 634 (17%) paralogs, 238 (6.3%) orphans, and 177 (3%) orthologs are modified with oxi-mC.
RNA-sequencing (RNA-seq) showed that 98% of all orthologous genes but only 77% of orphan genes and 63% of paralogous genes are expressed at the oidial stage. Strikingly, oxi-mC almost exclusively marks nonexpressed orphan and nonexpressed paralogous genes (Fig. S6E). Even expressed paralogous genes tend to be less expressed than single-copy orthologous genes (Fig. S6F and Dataset S1). Thus, gene body 5mC and oxi-mC appear to be modifications associated with low gene expression in C. cinerea at the oidial stage.
Oxi-mC Modifications Are Excluded from Oidial Genes That Change in Expression from the Oidial to the Hyphal Stage.
To examine the relation of oxi-mC to changes in gene expression during the C. cinerea life cycle, we obtained RNA from haploid mycelia and examined gene transcription by RNA-seq. An M (log ratios, y axis) vs. A [mean average (MA), x axis] plot comparing gene expression in oidia and hyphae shows a large number of differentially expressed genes between these two developmental stages, with approximately equal numbers of genes showing increased and decreased expression (Fig. S6G). Only 30% of multicopy paralogous genes, but ∼40% of nonconserved single-copy genes (orphans) and ∼65% of conserved single-copy genes (orthologs), showed significant changes in expression between the oidial and hyphal stages. Notably, these differentially expressed genes were almost devoid of oxi-mC modifications [Fig. S6H, red (up-regulated) and green (down-regulated) bars; genes unchanged in their expression are shown in blue; dark bars indicate genes devoid of oxi-mC modifications]; in contrast, oxi-mC modifications marked a substantial fraction of multicopy paralogous genes and nonconserved (orphan) single-copy genes whose expression was unchanged between the oidial and hyphal stages (Fig. S6H, light blue bars). These findings suggest the existence of mechanisms that limit epigenetic cytosine modifications at genes whose expression is dynamically altered during the C. cinerea life cycle.
Discussion
We have devised a method to simultaneously map the three different species of oxi-mC (5hmC, 5fC, and 5caC) at near–base-pair resolution by using diglucosylated SMRT sequencing, and have used it to report, to our knowledge, the first whole-genome analysis of oxidative cytosine modifications in the genome of the model fungus C. cinerea, a mushroom that attracted the attention of geneticists and cytologists because it can be cultured on defined media in the laboratory (19, 33). The sequences of the 13 chromosomes of C. cinerea have been assembled through shotgun sequencing (22), and the DNA methylation status of its genome has been mapped by whole-genome bisulfite sequencing (27). The small genome size of this organism (∼36 Mb) facilitated our analysis, and SMRT sequencing of long DNA fragments (∼6 kb) allowed us to map repetitive sequences unambiguously. The technologies and analysis methods we have used can be modified for extension to larger genomes.
Although digluc-SMRT sequencing can be used for rapid analysis of oxi-mC modifications, it has some potential limitations. (i) Given the strong dependence of the IPD ratio on sequence context and modification density, certain modified methylcytosines are likely to be missed. (ii) Although 5fC/5caC, which give signals in native DNA, can be distinguished from 5hmC, which provides a robust signal only after diglucosylation, the three oxi-mC species are not readily distinguishable if only diglucosylated DNA is sequenced. (iii) It may be challenging to identify the exact oxi-mC modification in regions that are rich in modified CpGs. (iv) For larger genomes, the low throughput of the instrument requires enrichment of desired DNA regions before SMRT sequencing.
The presence of all three oxi-mCs in C. cinerea DNA, and the fact that the C. cinerea genome encodes at least 32 TET/JBP paralogs predicted computationally to be catalytically active, suggests that methylcytosine oxidation is a dynamic regulatory modification in this organism. We find that, like 5mC, oxi-mCs are enriched at Kyakuja transposons, at other transposons and repetitive elements, and at annotated centromeres. Moreover, 5mC and oxi-mC are excluded from most orthologous single-copy genes and all orphan and paralogous genes whose expression changes between two stages of the C. cinerea life cycle. The data suggest that as for DNA methylation, a primary function of methylcytosine oxidation in C. cinerea is to limit gene expression at repetitive elements and paralogous gene arrays. In contrast, several groups have reported selective enrichment of 5hmC at gene bodies of highly expressed genes in differentiated mammalian cells (14, 25, 26). Potentially, this difference could reflect a difference in the functions of 5hmC and 5fC/5caC, the oxi-mC species present at high levels in mammalian cells and C. cinerea, respectively (Fig. 1B). TET2 in mammals is reported to associate with the SET1/COMPASS complex, which travels with RNA polymerase II (44), and would therefore mark gene bodies of highly expressed genes (25); in contrast, 5fC/ 5caC have been reported, by using in vitro assays, to diminish the processivity of RNA polymerase II. A similar function has been attributed to base J (β-d-glucosyl-hydroxymethyluracil): a fraction of base J in Leishmania is found at transcription termination sites where two polycistronic transcription units that are transcribed in opposite directions converge, and the loss of base J is accompanied by a dramatic increase in read-through transcription at these sites (45).
Unexpectedly, we found that C. cinerea centromeres could be defined not only by methylcytosine oxidation, but also by a central hypomethylated region. The hypomethylation patterns are similar in DNAs extracted from nondividing cells (oidia) and dividing cells (mycelia), suggesting that hypomethylation and the presence of oxi-mC may be important components of the epigenetic mechanism that marks the centromere. It will be of interest to determine if this hypomethylation pattern and enrichment for oxi-mC also characterize centromeres in other organisms.
Notably, only one of the 47 TET/JBP proteins is active in oidia; in hyphae, one TET/JBP is expressed at moderate levels whereas several others are expressed at extremely low levels (Dataset S1). Other TET/JBPs are expressed at other stages of the life cycle (Fig. 1A). We have shown experimentally that the TET/JBP expressed in oidia (CC1G_05497) is enzymatically active. However, this protein produces 5hmC but not 5caC under our experimental conditions, despite the fact that the majority of oxi-mC in oidia is 5caC; in contrast, one of the Naegleria Tet proteins whose structure was recently determined (46) yields 5caC as the end product. There are several reasons why the only TET protein expressed in oidia does not generate 5caC efficiently when expressed in mammalian cells: (i) because oidia are spores, a different TET/JBP protein might deposit oxi-mC before the oidial stage; (ii) our experimental conditions might not mimic those in the oidia; or (iii) the high level of 5caC in C. cinerea may reflect low activity of thymine DNA glycosylase, which is known to excise 5fC and 5caC in mammalian DNA (11, 47–49). Although expressed at low levels in the cell types examined to date (refs. 22, 50 and present report), the C. cinerea TDG homolog (encoded by CC1G_09247) retains the residues necessary to specifically recognize and excise oxi-mC (3).
In summary, we have developed the digluc-SMRT-seq approach to simultaneously map the oxi-mC distributions at near-base resolution and have generated genome-wide maps of oxi-mC in the model organism C. cinerea. Our findings emphasize the generality of oxi-mC species as potential epigenetic marks across eukaryotic lineages, and provide a genome-scale snapshot of an epigenetic regulatory system that appears to have emerged via recruitment of selfish elements (8, 51). Although the functional significance of the fungal systems and modifications described here remain to be explored in detail, they open unexpected leads for understanding gene regulation and chromatin structure, not only in basidiomycete fungi but in other eukaryotes as well. Given the prevalence of 5caC despite the presence of a predicted active TDG, C. cinerea could serve as a model system for studying the functions of oxidized cytosine marks.
Materials and Methods
Purification of T6 Phage BGAGT.
An ORF encoding the BGAGT gene was amplified by PCR from T6 phage DNA and cloned into the inducible bacterial expression vector pRSETB (Invitrogen) to give pRSETB-BGAGT. BL21 DE3 pLysS competent cells (Stratagene) were transformed with pRSETB-BGAGT and induced with IPTG during log-phase growth to induce expression of His-tagged BGAGT. Bacterial pellets were resuspended in lysis buffer (50 mM Tris, pH 8.0, 300 mM NaCl, 5 mM β-mercaptoethanol) plus 10 mM imidazole, and sonicated on ice for four intervals of 30 s each. Lysates were clarified by centrifugation at 13,800 × g for 20 min. His-tagged BGAGT was purified on a HisTrap FF column (GE Healthcare) as follows: 5 mL column was equilibrated with 50 mL lysis buffer plus 10 mM imidazole. Lysate was applied to the column, and the column was washed with 50 mL lysis buffer plus 40 mM imidazole. His-tagged BGAGT was eluted in one step using 10 mL lysis buffer plus 250 mM imidazole. Purified product was dialyzed by using a Slide-a-lyzer Dialysis Cassette (MWCO 3500; Pierce) to 50 mM Tris, pH 8.0, 100 mM NaCl, and stored at −80 °C in aliquots after addition of 50% (vol/vol) glycerol.
Diglucosylation of 5hmC.
Genomic DNA was isolated from C. cinerea oidia protoplast cells by using DNeasy plant maxi-kit (Qiagen) following the manufacturer’s instructions. Purified genomic DNA was sheared to ∼400–500 bp fragments by using Covaris S2. A maximum of 1 µg fragmented DNA was treated with 8 U T4-BGT (M0357; NEB) and 200 ng recombinant BGAGT in 100 µL of 100 mM Tris, pH 7.4, 50 µM UDP-glucose (NEB), 40 mM β-mercaptoethanol, and 25 mM MgCl2 for 2 h at 30 °C. DNA was purified using the MinElute PCR purification kit (Qiagen) and then converted into a SMRTbell library for sequencing using the PacBio RS system. Control experiments with a purified 5hmC-containing oligonucleotide (35) showed that both glucosylation steps catalyzed by T4-BGT and T6-BGAGT resulted in essentially complete modification at each step.
Preparation of C. cinerea Genomic DNA.
For preparation of C. cinerea genomic DNA before dot blot estimation of 5hmC, 5fC, and 5caC, 107 oidia were lysed in 1.5 mL 40 mM Tris, pH 8, 40 mM EDTA, 0.5% SDS containing 100 mg RNAseA/mL and incubated for 30 min at 37 °C. Proteinase K (10 µL, 10 mg/mL) was added, and sample was incubated at 65 °C for 5 h. An extra 10 µL proteinase K (10 mg/mL) was added, and samples were incubated overnight at 65 °C. One-tenth volume 5 M KOAc was added and sample was place on ice for 15 min and then centrifuged at 21,000 × g for 20 min at 4 °C. The supernatant was collected, and genomic DNA was precipitated by the addition of two volumes of 100% ethanol and washed twice with 70% (vol/vol) ethanol. Genomic DNA was resuspended in 10 mM Tris, pH 8, 1 mM EDTA containing RNaseA and left at 4 °C overnight. Genomic DNA was extracted with equal volumes of phenol, phenol:chloroform:isoamyl alcohol (25:24:1), and chloroform:isoamyl alcohol (24:1), and then precipitated by the addition of one-tenth volume 3 M NaOAc and two volumes of 100% ethanol, washed twice with 70% (vol/vol) ethanol, and then resuspended in 10 mM Tris, 0.1 mM EDTA, pH 8.0, and allowed to resuspend overnight at 32 °C.
Analysis of 5fC and 5caC Levels by Dot Blot.
C. cinerea genomic DNA samples were denatured by heating to 95 °C for 10 min in 10 mM Tris, 6.25 mM EDTA, pH 8.0. Samples were neutralized by the addition of ammonium acetate, pH 7, to a final concentration of 770 mM. Twofold serial dilutions were spotted onto a nitrocellulose membrane in an assembled Bio-Dot apparatus (Bio-Rad). The membrane was washed briefly in 2× SSC, air-dried, vacuum-baked, blocked in 1× Dulbecco PBS solution, 0.05% Tween-20, and 5% (wt/vol) milk solids for 1 h at room temperature.
5caC.
Membranes were incubated with rabbit anti-5caC (diluted to 1:10,000, no. 61225; Active Motif) overnight at 4 °C, washed, and then incubated with goat anti-rabbit IgG-HRP. To ensure equal loading of genomic DNA, the same blot was stained with 0.02% methylene blue in 0.3 M sodium acetate (pH 5.2).
5fC.
5fC levels were compared by using the same procedure with the following modifications. Genomic DNA samples were denatured by heating to 95 °C in 10 mM Tris, 62.5 mM NaOH, 6.25 mM EDTA, pH 8.0, for 10 min. Membranes were incubated with rabbit anti-5fC (no. 61223; Active Motif) diluted to 1:10,000 overnight at 4 °C.
MS.
DNA digestion, separation of nucleosides, and liquid chromatography/tandem MS (i.e., LC-MS/MS/MS) analysis were performed as described previously (52).
CMS-IP.
C. cinerea genomic DNA (5 µg) was sheared by using Covaris to yield a majority of fragments of ∼250 bp, then purified by using a Qiagen MiniElute column. Fragmented DNA was end-repaired, 3′ adenylated, and ligated to methylated Illumina adaptors using an Illumina TruSeq library preparation kit. The libraries were then bisulfite-converted by using a Methylcode bisulfite conversion kit (Invitrogen) according to the manufacturer’s instructions. 5hmC is converted to CMS after bisulfite treatment as reported previously (36). The bisulfite-converted genomic DNA library was denatured in 0.4 M NaOH, 10 mM EDTA for 10 min at 95 °C, neutralized by addition of an equal volume of cold 2M ammonium acetate, pH 7.0, incubated with anti-CMS antiserum in 1× IP buffer (10 mM sodium phosphate pH 7.0, 140 mM NaCl, 0.05% Triton X-100) overnight at 4 °C, then precipitated with Protein G beads (34–36). Precipitated DNA was washed three times with 1× IP buffer and eluted with Proteinase K, then purified with phenol-chloroform. The immunoprecipitated library DNA was amplified by using 12 cycles of PCR (KAPA HiFi Uracil+ polymerase; KAPAbiosystems) and DNA sequencing was performed by using an Illumina Genome AnalyzerII (GAII) (34–36).
SMRT Sequencing.
SMRTbell templates for sequencing were prepared as described previously (16). For the diglucosylated sample, SMRTbell templates from ∼500-bp and ∼6-kb insert libraries were sequenced using C2/C2 chemistry and 2 × 45 min and 1 × 90 min movie acquisition modes, respectively. For the native sample, ∼6-kb SMRTbell templates were sequenced by using the same chemistry and 1 × 90 min movie acquisition mode. All samples were sequenced to achieve ∼120× total sequencing coverage.
Library Preparation for RNA-Seq.
C. cinerea oidia mRNA was isolated using Qiagen RNeasy plant mini kit. mRNA (5 µg) was used for two rounds of polyA selection following the manufacturer’s instructions [Poly(A)Purist MAG kit; Life Technologies]. For oidia, RNA library preparation was done by using SOLiD Total RNA-seq kit (Life Technologies), and, for hyphae, RNA library preparation was done after polyA selection by using an NEB NEBNext Ultra RNA Library Prep Kit for Illumina. Library quality was analyzed by using Agilent 2100 Bioanalyzer platform.
Immunocytochemistry and ImageXpress Micro Quantification.
Actively transcribed TET/JBP (CC1G_05497) at oidia stage of C. cinerea was cloned into pEF-V5 vector with Flag-tag and transfected into HEK293T cell by using Lipofectamine 2000. For immunocytochemistry experiments, transfected cells were seeded on cover class and fixed with 4% (wt/vol) paraformaldehyde for 15 min followed by permeabilization in PBS solution/0.2% Triton-X100 for 15 min. Then DNA was denatured with 2N HCl for 30 min and neutralized with 100 mM Tris⋅Cl (pH 8.5) for 10 min. Next, cells were blocked with PBS solution containing 1% BSA and 0.05% Tween 20 for 1 h. Cells were then incubated with primary antibodies [anti-Flag (Sigma) and anti-5hmC or 5fC or 5caC (Active Motif)] for 1 h at room temperature and followed by incubation with secondary antibodies (Alexa Fluor 488 goat anti-rabbit IgG and Alexa Fluor 647 goat anti-mouse IgG) and DAPI for 1 h. Cells were washed with PBS solution three times after primary and secondary antibody incubation. Images were taken by using an Olympus FV1000 confocal microscope. For ImageXpress Micro (IXM) quantification experiments, transfected cells were cultured on Corning Costar assay plates and stained with anti-Flag and anti-5hmC or 5fC or 5caC antibodies as described earlier. Images were taken by using an IXM imaging system, and data were analyzed by MetaXpress Software Application Modules.
Supplementary Material
Acknowledgments
This work was supported by NIH Grants R01 HD065812 (to A.R.), R01 AI44432 (to A.R.), R01 CA151535 (to A.R.), K08 HL089150 (to S.A.), and R01 CA101864 (to Y.W.); California Institute for Regenerative Medicine Grant RM1-01729 (to A.R.); Leukemia Society of America Translational Research Program Award 6187-12 (to A.R.); a grant to University of North Carolina Chapel Hill from the Howard Hughes Medical Institute through the Undergraduate Science Education Program (to P.J.P.); a pilot grant from Harvard Catalyst, the Harvard Clinical and Translational Science Center [NIH Grant 1 UL1 RR 025758-02 (to S.A.)]; a postdoctoral fellowship from the Leukemia and Lymphoma Society (to Y.H.); a predoctoral fellowship from the National Science Foundation (to W.A.P.); a Feodor Lynen Research Fellowship from the Alexander von Humboldt Foundation (to L.C.); and intramural funds of the National Library of Medicine, NIH (L.A. and L.M.I.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The 5hmC mapping and RNA sequencing data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE46965), and the native and digluc single-molecule, real-time sequencing data have been deposited to the Sequence Read Archive (SRA), www.ncbi.nlm.nih.gov/sra (accession no. SRP041464).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1419513111/-/DCSupplemental.
References
- 1.Iyer LM, Tahiliani M, Rao A, Aravind L. Prediction of novel families of enzymes involved in oxidative and other complex modifications of bases in nucleic acids. Cell Cycle. 2009;8(11):1698–1710. doi: 10.4161/cc.8.11.8580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tahiliani M, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324(5929):930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iyer LM, Abhiman S, Aravind L. Natural history of eukaryotic DNA methylation systems. Prog Mol Biol Transl Sci. 2011;101:25–104. doi: 10.1016/B978-0-12-387685-0.00002-0. [DOI] [PubMed] [Google Scholar]
- 4.Cliffe LJ, et al. JBP1 and JBP2 proteins are Fe2+/2-oxoglutarate-dependent dioxygenases regulating hydroxylation of thymidine residues in trypanosome DNA. J Biol Chem. 2012;287(24):19886–19895. doi: 10.1074/jbc.M112.341974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pastor WA, Aravind L, Rao A. TETonic shift: Biological roles of TET proteins in DNA demethylation and transcription. Nat Rev Mol Cell Biol. 2013;14(6):341–356. doi: 10.1038/nrm3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu H, Zhang Y. Reversing DNA methylation: Mechanisms, genomics, and biological functions. Cell. 2014;156(1-2):45–68. doi: 10.1016/j.cell.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kohli RM, Zhang Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature. 2013;502(7472):472–479. doi: 10.1038/nature12750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iyer LM, et al. Lineage-specific expansions of TET/JBP genes and a new class of DNA transposons shape fungal genomic and epigenetic landscapes. Proc Natl Acad Sci USA. 2014;111(5):1676–1683. doi: 10.1073/pnas.1321818111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iyer LM, Zhang D, Burroughs AM, Aravind L. Computational identification of novel biochemical systems involved in oxidation, glycosylation and other complex modifications of bases in DNA. Nucleic Acids Res. 2013;41(16):7635–7655. doi: 10.1093/nar/gkt573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ito S, et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333(6047):1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He YF, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333(6047):1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pfaffeneder T, et al. The discovery of 5-formylcytosine in embryonic stem cell DNA. Angew Chem Int Ed Engl. 2011;50(31):7008–7012. doi: 10.1002/anie.201103899. [DOI] [PubMed] [Google Scholar]
- 13.Spruijt CG, et al. Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell. 2013;152(5):1146–1159. doi: 10.1016/j.cell.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 14.Mellén M, Ayata P, Dewell S, Kriaucionis S, Heintz N. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell. 2012;151(7):1417–1430. doi: 10.1016/j.cell.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flusberg BA, et al. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat Methods. 2010;7(6):461–465. doi: 10.1038/nmeth.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clark TA, et al. Enhanced 5-methylcytosine detection in single-molecule, real-time sequencing via Tet1 oxidation. BMC Biol. 2013;11:4. doi: 10.1186/1741-7007-11-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wallace EV, et al. Identification of epigenetic DNA modifications with a protein nanopore. Chem Commun (Camb) 2010;46(43):8195–8197. doi: 10.1039/c0cc02864a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li WW, Gong L, Bayley H. Single-molecule detection of 5-hydroxymethylcytosine in DNA through chemical modification and nanopore analysis. Angew Chem Int Ed Engl. 2013;52(16):4350–4355. doi: 10.1002/anie.201300413. [DOI] [PubMed] [Google Scholar]
- 19.Pukkila PJ. Coprinopsis cinerea. Curr Biol. 2011;21(16):R616–R617. doi: 10.1016/j.cub.2011.05.042. [DOI] [PubMed] [Google Scholar]
- 20.Zolan ME, Pukkila PJ. Inheritance of DNA methylation in Coprinus cinereus. Mol Cell Biol. 1986;6(1):195–200. doi: 10.1128/mcb.6.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Freedman T, Pukkila PJ. De novo methylation of repeated sequences in Coprinus cinereus. Genetics. 1993;135(2):357–366. doi: 10.1093/genetics/135.2.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stajich JE, et al. Insights into evolution of multicellular fungi from the assembled chromosomes of the mushroom Coprinopsis cinerea (Coprinus cinereus) Proc Natl Acad Sci USA. 2010;107(26):11889–11894. doi: 10.1073/pnas.1003391107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taylor JW, Ellison CE. Mushrooms: Morphological complexity in the fungi. Proc Natl Acad Sci USA. 2010;107(26):11655–11656. doi: 10.1073/pnas.1006430107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lister R, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462(7271):315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang Y, et al. Distinct roles of the methylcytosine oxidases Tet1 and Tet2 in mouse embryonic stem cells. Proc Natl Acad Sci USA. 2014;111(4):1361–1366. doi: 10.1073/pnas.1322921111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song CX, et al. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat Biotechnol. 2011;29(1):68–72. doi: 10.1038/nbt.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zemach A, McDaniel IE, Silva P, Zilberman D. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science. 2010;328(5980):916–919. doi: 10.1126/science.1186366. [DOI] [PubMed] [Google Scholar]
- 28.Huang Y, et al. The behaviour of 5-hydroxymethylcytosine in bisulfite sequencing. PLoS ONE. 2010;5(1):e8888. doi: 10.1371/journal.pone.0008888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clark TA, et al. Characterization of DNA methyltransferase specificities using single-molecule, real-time DNA sequencing. Nucleic Acids Res. 2012;40(4):e29. doi: 10.1093/nar/gkr1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eid J, et al. Real-time DNA sequencing from single polymerase molecules. Science. 2009;323(5910):133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- 31.Fang G, et al. Genome-wide mapping of methylated adenine residues in pathogenic Escherichia coli using single-molecule real-time sequencing. Nat Biotechnol. 2012;30(12):1232–1239. doi: 10.1038/nbt.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kornberg SR, Zimmerman SB, Kornberg A. Glucosylation of deoxyribonucleic acid by enzymes from bacteriophage-infected Escherichia coli. J Biol Chem. 1961;236:1487–1493. [PubMed] [Google Scholar]
- 33.Kües U. Life history and developmental processes in the basidiomycete Coprinus cinereus. Microbiol Mol Biol Rev. 2000;64(2):316–353. doi: 10.1128/mmbr.64.2.316-353.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ko M, et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010;468(7325):839–843. doi: 10.1038/nature09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pastor WA, et al. Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature. 2011;473(7347):394–397. doi: 10.1038/nature10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang Y, Pastor WA, Zepeda-Martínez JA, Rao A. The anti-CMS technique for genome-wide mapping of 5-hydroxymethylcytosine. Nat Protoc. 2012;7(10):1897–1908. doi: 10.1038/nprot.2012.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chavez L, et al. Computational analysis of genome-wide DNA methylation during the differentiation of human embryonic stem cells along the endodermal lineage. Genome Res. 2010;20(10):1441–1450. doi: 10.1101/gr.110114.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lienhard M, Grimm C, Morkel M, Herwig R, Chavez L. MEDIPS: Genome-wide differential coverage analysis of sequencing data derived from DNA enrichment experiments. Bioinformatics. 2014;30(2):284–286. doi: 10.1093/bioinformatics/btt650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Holm PB, Rasmussen SW, Zickler D, Lu BC, Sage J. Chromosome pairing, recombination nodules and chiasma formation in the basidiomycete Coprinus cinereus. Carlsberg Res Commun. 1981;46(5):305–346. [Google Scholar]
- 40.Zolan ME, Heyler NK, Stassen NY. Inheritance of chromosome-length polymorphisms in Coprinus cinereus. Genetics. 1994;137(1):87–94. doi: 10.1093/genetics/137.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Iyer LM, Aravind L. ALOG domains: Provenance of plant homeotic and developmental regulators from the DNA-binding domain of a novel class of DIRS1-type retroposons. Biol Direct. 2012;7:39. doi: 10.1186/1745-6150-7-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Piednoël M, Gonçalves IR, Higuet D, Bonnivard E. Eukaryote DIRS1-like retrotransposons: An overview. BMC Genomics. 2011;12:621. doi: 10.1186/1471-2164-12-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang L, et al. A TET homologue protein from Coprinopsis cinerea (CcTET) that biochemically converts 5-methylcytosine to 5-hydroxymethylcytosine, 5-formylcytosine, and 5-carboxylcytosine. J Am Chem Soc. 2014;136(13):4801–4804. doi: 10.1021/ja500979k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Deplus R, et al. TET2 and TET3 regulate GlcNAcylation and H3K4 methylation through OGT and SET1/COMPASS. EMBO J. 2013;32(5):645–655. doi: 10.1038/emboj.2012.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Luenen HG, et al. Glucosylated hydroxymethyluracil, DNA base J, prevents transcriptional readthrough in Leishmania. Cell. 2012;150(5):909–921. doi: 10.1016/j.cell.2012.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hashimoto H, et al. Structure of a Naegleria Tet-like dioxygenase in complex with 5-methylcytosine DNA. Nature. 2014;506(7488):391–395. doi: 10.1038/nature12905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maiti A, Drohat AC. Thymine DNA glycosylase can rapidly excise 5-formylcytosine and 5-carboxylcytosine: Potential implications for active demethylation of CpG sites. J Biol Chem. 2011;286(41):35334–35338. doi: 10.1074/jbc.C111.284620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Song CX, et al. Genome-wide profiling of 5-formylcytosine reveals its roles in epigenetic priming. Cell. 2013;153(3):678–691. doi: 10.1016/j.cell.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shen L, et al. Genome-wide analysis reveals TET- and TDG-dependent 5-methylcytosine oxidation dynamics. Cell. 2013;153(3):692–706. doi: 10.1016/j.cell.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Burns C, et al. Analysis of the Basidiomycete Coprinopsis cinerea reveals conservation of the core meiotic expression program over half a billion years of evolution. PLoS Genet. 2010;6(9):e1001135. doi: 10.1371/journal.pgen.1001135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aravind L, Anantharaman V, Zhang D, de Souza RF, Iyer LM. Gene flow and biological conflict systems in the origin and evolution of eukaryotes. Front Cell Infect Microbiol. 2012;2:89. doi: 10.3389/fcimb.2012.00089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu S, et al. Quantitative assessment of Tet-induced oxidation products of 5-methylcytosine in cellular and tissue DNA. Nucleic Acids Res. 2013;41(13):6421–6429. doi: 10.1093/nar/gkt360. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.