Abstract
All aspects of biological diversification ultimately trace to evolutionary modifications at the cellular level. This central role of cells frames the basic questions as to how cells work and how cells come to be the way they are. Although these two lines of inquiry lie respectively within the traditional provenance of cell biology and evolutionary biology, a comprehensive synthesis of evolutionary and cell-biological thinking is lacking. We define evolutionary cell biology as the fusion of these two eponymous fields with the theoretical and quantitative branches of biochemistry, biophysics, and population genetics. The key goals are to develop a mechanistic understanding of general evolutionary processes, while specifically infusing cell biology with an evolutionary perspective. The full development of this interdisciplinary field has the potential to solve numerous problems in diverse areas of biology, including the degree to which selection, effectively neutral processes, historical contingencies, and/or constraints at the chemical and biophysical levels dictate patterns of variation for intracellular features. These problems can now be examined at both the within- and among-species levels, with single-cell methodologies even allowing quantification of variation within genotypes. Some results from this emerging field have already had a substantial impact on cell biology, and future findings will significantly influence applications in agriculture, medicine, environmental science, and synthetic biology.
Keywords: evolutionary cell biology, cell biology, adaptive evolution, random genetic drift, cellular evolution
The origin of cells constituted one of life’s most important early evolutionary transitions, simultaneously enabling replicating entities to corral the fruits of their catalytic labor and providing a unit of inheritance necessary for further evolutionary refinement and diversification. The centrality of cellular features to all aspects of biology motivates the focus of cell biology on the biophysical/biochemical aspects of a broad swath of traits that include gene expression, metabolism, intracellular transport and communication, cell–cell interactions, locomotion, and growth. No one questions the rich contributions that have resulted from this focus on how cells work. However, with an emphasis on maximizing experimental consistency in a few well-characterized model systems, cell biologists have generally eschewed the variation that motivates most questions in evolutionary biology.
Because all evolutionary change ultimately requires modifications at the cellular level, questioning and understanding how cellular features arise and diversify should be a central research venue in evolutionary biology. However, if there is one glaring gap in this field, it is the absence of widespread cell-biological thinking. Despite the surge of interest at the molecular, genomic, and developmental levels, much of today’s study of evolution is only moderately concerned with cellular features, perhaps due to lack of appreciation for their wide variation among taxa. However, a full mechanistic understanding of evolutionary processes will never be achieved without an elucidation of how cellular features become established and modified.
The time is ripe for bridging the gap between the historically disconnected fields of cell biology and evolutionary biology and integrating them with the principles of biophysics and biochemistry into a formal field of evolutionary cell biology. Recent advances in cell-biological analysis and the acquisition of ’omic-scale datasets have broadened the opportunities for research on nonstandard model organisms, thereby facilitating the incorporation of phylogenetic diversity into cell-level studies. Our vision for this synthesis is motivated by the growing realization in both communities that an intellectual merger will yield dramatic increases in our understanding of cell-biological structures, functions, and processes, as well as insights into the cellular basis for evolutionary change. Although not an exhaustive list, the following questions motivate and illustrate the potential for this new field.
Why Are Cells the Way They Are, and Why Aren’t They Perfect?
Although it is easy to marvel about the refined features of cells and their robustness to perturbations (1), the field of bioengineering imagines and even implements more efficient cellular mechanisms in extant organisms. What, then, limits the levels of molecular/cellular refinements that have been achieved by natural selection?
To What Extent Is Cell Biology Beholden to Historical Contingency?
We have learned an enormous amount about the genetic mechanisms of evolution since Darwin, and it remains true that evolution is an opportunistic process of “descent with modification,” working with the resources made available in previous generations. Once established, useful features cannot be easily dismantled and reassembled de novo unless there is an intermediate period of redundancy.
One remarkable example of how history continues to influence today’s cell biology is the near universal use of ATP synthase as a mechanism for energy generation (2). Embedded in the surface membranes of bacteria and organellar membranes of eukaryotes, this complex molecular machine uses the potential energy of a proton gradient to generate a rotational force that converts ADP to ATP, much like a turbine converts the potential energy of a water gradient into electricity. However, the proton gradient does not come for free: cells first use energy derived from metabolism to pump protons out of membrane-bound compartments, creating the gradient necessary for reentry through ATP synthase. Even assuming that ATP production is an essential requirement for the origin of life, it is by no means clear that the path chosen for ADP-to-ATP conversion is the only possibility.
Rather, the universal reliance of all of life on this mechanism of energy conversion may be a historical relic of the exploitable energy source present at the time of life's foundation: e.g., a precellular period in which energy acquisition derived from a natural proton gradient between overlying low-pH marine waters and the alkaline interiors of vent mounds (3, 4). Despite the central significance of ATP synthase to bioenergetics across the Tree of Life and the invariance of the basic mechanism of ATP regeneration, many examples are known in which the structure of the complex has been modified with respect to the numbers and types of subunits (2, 5, 6).
How Is Cell Biology Constrained by the Laws of Physics and Chemistry?
Although cataloging and explaining biodiversity are central themes of evolutionary biology, deciphering the roles by which biophysical/biochemical barriers channel cellular characteristics into a limited range of alternatives is equally important. Like the near-universal genetic code, the laws of physics endow cells with specific properties, but, unlike the nucleotide sequences of genes, these laws are immutable and have potential impacts at all levels of biological organization.
Examples of relevant organizing principles at the molecular scale include the role of the hydrophobic effect in protein folding and assembly and constraints imposed by intracellular molecular crowding. For example, rather than operating as monomers, the majority of proteins self-assemble into higher-order structures such as dimers, tetramers, etc. Remarkably, however, unlike the strong, general trend toward dramatic increases in gene structural complexity from prokaryotes to unicellular eukaryotes to multicellular species (7), higher-order structural complexity of proteins does not noticeably scale with organismal complexity across the Tree of Life (8). Comparative biochemical and protein-structural analysis within a phylogenetic framework has great potential to address many outstanding questions in this area, including whether variation in the multimeric states of proteins is a simple consequence of stochastic mutations of adhesive interface residues, with minimal effects on catalytic efficiency.
Similar questions arise about the biophysical properties of supermolecular structures, such as microtubules, actin filaments, and the endomembrane systems of eukaryotic cells (9). The self-assembly of lipid bilayers emerges spontaneously from the biophysical properties of amphiphilic molecules, and recent origin-of-life research suggests that some of the key first steps in the origin of life, such as the assembly and division of vesicles, are inevitable consequences of the behavior of organic molecules in water (10, 11).
Finally, general biophysical phenomena are undoubtedly involved in the patterning of phenotypes at the whole-cell level. For example, constraints on surface:volume scaling may have been involved in the establishment of internal membranes and their above-noted associations with bioenergetics (12). Such constraints may also have played a central role in the evolution of cell size and features of the nuclear envelope (13). The emergence of the nuclear envelope may have, in turn, had secondary evolutionary consequences, such as the establishment of a permissive environment for intron proliferation (7), which requires efficient pretranslational splicing of transcripts.
Although the preceding observations suggest that the emergence and diversification of numerous cellular features may be predictable on biophysical grounds alone, the imposition of constraints on a complex trait need not preclude substantial opportunities for modifying the underlying components, as previously discussed with respect to ATP synthase. For example, although there are common organizational principles in diverse regulatory, signal-transduction, and metabolic pathways, dramatic cases of rewiring have been revealed with the expansion of molecular and cell biological investigations to multiple species. Such examples include aspects of mating-type specification (14, 15), meiosis (16), cell cycle (17, 18), biosynthetic pathways (19–23), protein transport (24), nuclear organization (25), and ribosome production (26, 27). These kinds of observations imply that there are often numerous degrees of freedom for reorganizing the underlying determinants of otherwise constant cellular processes.
How Much of Cellular Complexity Is the Result of Adaptation?
A commonly held but incorrect stance is that essentially all of evolution is a simple consequence of natural selection. Leaving no room for doubt on the process, this narrow view leaves the impression that the only unknowns in evolutionary biology are the identities of the selective agents operating on specific traits. However, population-genetic models make clear that the power of natural selection to promote beneficial mutations and to remove deleterious mutations is strongly influenced by other factors. Most notable among these factors is random genetic drift, which imposes noise in the evolutionary process owing to the finite numbers of individuals and chromosome architecture. Such stochasticity leads to the drift-barrier hypothesis for the evolvable limits to molecular refinement (28, 29), which postulates that the degree to which natural selection can refine any adaptation is defined by the genetic effective population size. One of the most dramatic examples of this principle is the inverse relationship between levels of replication fidelity and the effective population sizes of species across the Tree of Life (30). Reduced effective population sizes also lead to the establishment of weakly harmful embellishments such as introns and mobile-element insertions (7). Thus, rather than genome complexity being driven by natural selection, many aspects of the former actually arise as a consequence of inefficient selection.
Indeed, many pathways to greater complexity do not confer a selective fitness advantage at all. For example, due to pervasive duplication of entire genes (7) and their regulatory regions (31) and the promiscuity of many proteins (32), genes commonly acquire multiple modular functions. Subsequent duplication of such genes can then lead to a situation in which each copy loses a complementary subfunction, channeling both down independent evolutionary paths (33). Such dynamics may be responsible for the numerous cases of rewiring of regulatory and metabolic networks noted in the previous section (34, 35). In addition, the effectively neutral acquisition of a protein–protein-binding interaction can facilitate the subsequent accumulation of mutational alterations of interface residues that would be harmful if exposed, thereby rendering what was previously a monomeric structure permanently and irreversibly heteromeric (8, 36–39). Finally, although it has long been assumed that selection virtually always accepts only mutations with immediate positive effects on fitness, it is now known that, in sufficiently large populations, trait modifications involving mutations with individually deleterious effects can become established in large populations when the small subset of maladapted individuals maintained by recurrent mutation acquire complementary secondary mutations that restore or even enhance fitness (40, 41).
One goal of evolutionary cell biology should be to determine whether these general principles involving effectively neutral paths of molecular evolution extend to even higher-order biological features, such as intracellular architecture (37). Is natural selection a sufficient or even a necessary explanation for the evolution of the complex features of the ribosome, the spliceosome, the nuclear-pore complex, and the Golgi apparatus? Or is a march toward increased, and potentially irreversible, cellular complexity an inevitable outcome of mutation pressure and the inefficiencies of selection processes in finite populations?
The points raised above are not meant to suggest that structures as complex as ribosomes or ATP synthase are maladaptive. Certainly, today’s cells cannot survive without such molecular machines. However, the existence of complex cellular features need not imply that each of the myriad of changes that sculpted such structures over evolutionary time was adaptive at the time of establishment. The determination of whether it is even feasible for a cellular innovation to have been promoted by purely adaptive processes cannot be made in the absence of information about the population-genetic environment: i.e., the magnitudes of the power of mutation, recombination, and random genetic drift. All three features vary by orders of magnitude across the Tree of Life and can only roughly be inferred for ancestral species. Uncertainty in this area is a major challenge for evolutionary cell biology (30, 42).
How Do Cellular Innovations Arise?
For practical reasons, cell biology has historically focused on the average features of the members of large populations of genetically uniform cells. However, natural selection does not operate directly on population means but on variation among individuals. Moreover, the evolutionary response to selection on a trait is not a simple matter of variation, but a function of the fraction of variation that has a genetic basis. Estimation of these key parameters is now within reach as new technologies allow assays of single cells in a high-throughput manner. Applications of these methods to genetically uniform populations reveal substantial cell-to-cell variation in gene-specific numbers of transcripts and proteins in all domains of life (43–45), and such variation (intrinsic cellular noise) seems to be a natural outcome of biophysical features of interactions between transcription factors and their binding sites, which can be quantified in mechanistic terms (46, 47). These kinds of observations, which can be extended to other intracellular traits (48), are essential to understanding the limits to the evolvability of cellular features. This is because environmental variance (intracellular noise) reduces the ability of a population to respond to selection by overshadowing the heritable genetic component of variation (49).
Although conceptually straightforward, resolving the degree to which variation (and covariation) of phenotypes in populations of cells is a consequence of genetic vs. environmental causes will require large-scale experimental designs including genetically variable isolates. When applied in this way, single-cell phenotyping down to the level of individual molecules has the potential to revolutionize the field of quantitative genetics by elucidating the precise sources of variation underlying the expression of higher-order cellular features. Notably, the statistical framework of quantitative genetics is also fully equipped to address the evolutionary consequences of transient epigenetic effects (49), whose influences are dissipated over time with various levels of reinforcement (e.g., refs. 50–52).
Where Do Cellular Innovations Map onto the Tree of Life?
A first step in nearly all studies in evolutionary biology is the elucidation of phylogenetic patterns of variation. Although a purely historical perspective cannot reveal the mechanisms by which evolution proceeds, it does clarify what needs to be explained. Traditional cell biology is largely devoid of comprehensive comparative analyses, but recent studies demonstrate the power of such approaches, as illustrated by the following three examples.
The first example addresses the evolutionary origins of the network of organelles and underlying molecular features by which membrane trafficking emerged in eukaryotes. The sorting of proteins and lipids among the intracellular compartments of eukaryotic cells is mediated in part by a family of protein complexes called adaptins. Although it had been accepted for over a decade that there are only four adaptin complexes in eukaryotes, comparative genomics suggested the presence of a fifth highly divergent adaptin-like complex across eukaryotes (53). Subsequent characterization of the protein in human cells identified its cellular location and function, thereby fundamentally altering our basic understanding of vesicle-transport systems and the likely order of evolutionary events leading to their diversification. An even more recent phylogenetic analysis suggests the existence of a sixth form of adaptor complex (54), raising the possibility that still more remain to be discovered, perhaps with some complexes being restricted to a subset of taxa.
A second striking example of the power of comparative analysis to inform our basic understanding of cell biology involves the discovery of an evolutionary relationship between what were considered two very different kinds of membrane-deformation proteins. Cargo transport in eukaryotic cells involves the use of diverse pathways initiating with membrane-coated vesicles supported by clathrin, and the cage forming proteins of cytoplasmic coat protein complexes I and II (COPI and COPII). Although these proteins are lacking in amino acid sequence similarity, comparative structural analysis suggests a common molecular architecture that is also related to the membrane-curving proteins involved in both the nuclear-pore complex (NPC) (55) and the adaptins discussed above. The structural and functional insights emerging from these observations guided the development of a mechanistic understanding of the NPC (56) and yielded a novel evolutionary proposal—the “protocoatomer” hypothesis, which postulates that many vesicle-coating complexes and the NPC arose by descent with modification (55). Among other things, this concept has provided a potential explanation for how the diverse body plans of eukaryotic cells could have arisen from a simpler prokaryote-like ancestor.
In a third example, an integration of molecular and morphological phylogenetic analysis has led to the identification of novel components of centrioles and cilia, as well as to evolutionary hypotheses for how their coordinated biogenesis and functions in different cellular contexts have been achieved through duplication and divergence of an ancestral gene set (57, 58).
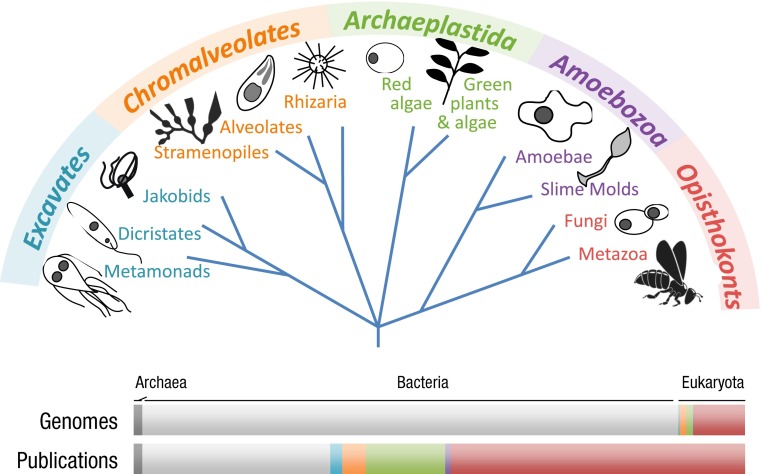
This small set of examples illustrates the considerable potential for more elaborate comparative analyses to elucidate the evolutionary foundations of the most basic eukaryotic cellular features. Of course, ascertainment of where cell-biological innovations map onto the Tree of Life and inference of phylogenetic points of gain and loss of various modifications will require a substantial increase in taxonomic sampling of cellular diversity. Of the estimated 5–100 million extant species, only ∼1.5 million have been described at even a rudimentary level, and most of these taxa are heavily biased toward plants, animals, fungi, and microbes with direct human impact (59) (Fig. 1). Future studies of biodiversity are likely to continue to extend to the discovery of novel phyla for quite some time (e.g., refs. 60–62). These issues, together with the fact that typically about a third of predicted protein-coding genes per sequenced genome are undefined and/or restricted to narrow taxonomic groupings, make clear that we are still missing immense swaths of information on cellular diversity. This “missing phylogeny” is likely of high value to applied research efforts in medicine, agriculture, and environmental science.
Fig. 1.
Taxonomic distribution of research articles and sequenced genomes. Modern taxonomy identifies five major eukaryotic supergroups: the Excavates (turquoise), Chromalveolates (orange), Archaeplastida (green), Amoebozoa (purple), and Opisthokonts (red). Although the total number of species on earth remains unknown, it is clear that there are far more unicellular eukaryotes than the combined total of all animals (Metazoa, an Opisthokont lineage), fungi (also Opisthokonts), and plants (Archaeplastida). However, research activity displays considerable taxonomic bias. As of January 2014, the National Center for Biotechnology Information taxonomy browser (www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi) lists 338 Archaeal genomes (dark gray), 20,709 Eubacteria (light gray), 769 Metazoa, 1,201 Fungi, 251 green plants/algae, and 336 genomes from all other eukaryotic taxa (13% of eukaryotic genomes). The taxonomic distribution of PubMed citations is as follows: Archaea, 19,000; Eubacteria, 397,000; Metazoa, 576,000; Fungi, 135,000; green plants/algae, 168,000; and all other eukaryotes combined, 97,000 (<9% of publications on Eukaryotes).
Unfortunately, parts lists inferred from genome information alone can take us only so far. Although results from transcriptomics, metabolomics, etc. can provide additional information, such work must ultimately be coupled to detailed studies of individual gene products in diverse taxa. To this end, we envision the need for a new grand challenge in biology, such as the proposed Atlas of the Biology of Cells (www.nsf.gov/publications/pub_summ.jsp?ods_key=bio12009). The fundamental idea here is to develop a database for cellular/subcellular features for a judiciously chosen, phylogenetically broad set of organisms, with the goal of sampling the functional diversity of metabolic and cellular morphological traits in the fullest possible sense. To be maximally productive, such an enterprise will require the further development of automated, generalizable, and high-throughput cell-biological methods. Significant support for appropriate phylogenetic sampling, development of reliable culture methods, and standardized measurement methodology will also be necessary. Most importantly, the latter will require the establishment of not only controlled vocabularies and ontologies to provide a conceptual framework for data comparison, but also quantitative metrics for defining, comparing, and predicting cell-biological structures and processes.
The payoffs of such an organized research program are likely to be substantial. As an analogy to where evolutionary cell biology is and where it might lead, consider that whole-genome sequencing was barely a dream 25 y ago but, in the past decade, has revolutionized virtually every aspect of biology, vastly increasing our understanding of human-genetic disorders, methods for disease control, energy production, and ecosystem function. Such advances continue to inspire the development of new ’omics technologies with enormous increases in accuracy and efficiency, as well as the emergence of novel computational technologies for storage, integration, and analysis that facilitate the rapid transformation of data into knowledge.
How Can Effective Implementation of Lessons from Evolutionary Cell Biology Be Ensured?
Cell biology textbooks traditionally focus on structures and pathways perceived to be common to all cells, only occasionally addressing specializations in individual phylogenetic lineages, and even more rarely mentioning their modes of diversification. In effect, we have built up a sort of canonical molecular and cell biology based on a few serendipitously selected model organisms. How things work in Escherichia coli, Saccharomyces cerevisiae, Drosophila melanogaster, and mouse cells is all too often viewed as the “normal” mode of biology, with differences observed in other organisms often being viewed as little more than amusing oddities. Imagine what today’s biology might look like if our models had been Nanoarchaeum (archaebacterium), Paramecium (ciliate), Ceratium (dinoflagellate), and Pinus (gymnosperm).
The view that intracellular structures are essentially invariant in diverse organisms engenders the false impression that an evolutionary biologist has little to gain by pursuing studies at the cellular level. Moreover, the few statements about evolution that can be found in cell-biology textbooks and journal articles frequently speculate on the adaptive significance of cellular features, oversimplifying and obscuring our understanding of evolutionary mechanisms (42, 63). This outmoded view of evolutionary processes still gives rise to major misunderstandings, with substantial implications (64).
In summary, we have attempted to highlight why bridging the conceptual gap between cell biology and evolutionary biology is likely to enrich our understanding of virtually all biological processes. For example, although the natural spatial delimitation of cell biology resides at the cell membrane, an understanding of the evolutionary roots of various cellular features is of central relevance to evolutionary developmental biologists concerned with the origin of cell types (65). Evolutionary cell biology has a particularly high potential for informing a variety of practical matters with ecological, economic, and health benefits. Such applications include the facilitation of drug development and the elucidation of the mechanisms of drug sensitivity and resistance, and of the identification of the mechanisms of nutrient fluxes through the environment and their dependence on species-specific features. The removal of real and perceived conceptual and communication barriers (including those engendered by the use of specialized vocabularies) and the design and implementation of cross-disciplinary educational initiatives are central keys to building an interactive community of scientists essential for igniting an effective field of evolutionary cell biology.
Acknowledgments
We thank W. Ford Doolittle for helpful comments. This paper was, in part, inspired by the National Science Foundation-sponsored Workshop on Evolutionary Cell Biology (Grant MCB-1228570), and we acknowledge the many insightful discussions among the participants (for details, see www.nsf.gov/publications/pub_summ.jsp?ods_key=bio12009). We are grateful for support from National Science Foundation Grants IOS-1051962 (to S.S.), MCB-1050161 (to M.L.), MCB-1051985 (to A.P.T.), and MCB-1244593 (to H.V.G.), National Institutes of Health Grants R01-GM036827 (to M.L.), R01-105783 (to A.P.T.), R01-GM74108 (to H.S.M.), and R01-AI49301 (to D.S.R.), US Army Research Office Grant W911NF-09-1-0444 (to M.L.), and Fundação para a Ciência e Tecnologia Grant PTDC/EBB-BIO/119006/2010 (to J.B.P.-L.). H.S.M. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Wagner A. Robustness and Evolvability in Living Systems. Princeton Univ Press, Princeton; 2007. [Google Scholar]
- 2.Walker JE. The ATP synthase: The understood, the uncertain and the unknown. Biochem Soc Trans. 2013;41(1):1–16. doi: 10.1042/BST20110773. [DOI] [PubMed] [Google Scholar]
- 3.Martin W, Baross J, Kelley D, Russell MJ. Hydrothermal vents and the origin of life. Nat Rev Microbiol. 2008;6(11):805–814. doi: 10.1038/nrmicro1991. [DOI] [PubMed] [Google Scholar]
- 4.Lane N, Allen JF, Martin W. How did LUCA make a living? Chemiosmosis in the origin of life. BioEssays. 2010;32(4):271–280. doi: 10.1002/bies.200900131. [DOI] [PubMed] [Google Scholar]
- 5.Balabaskaran Nina P, et al. Highly divergent mitochondrial ATP synthase complexes in Tetrahymena thermophila. PLoS Biol. 2010;8(7):e1000418. doi: 10.1371/journal.pbio.1000418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lapaille M, et al. Atypical subunit composition of the chlorophycean mitochondrial F1FO-ATP synthase and role of Asa7 protein in stability and oligomycin resistance of the enzyme. Mol Biol Evol. 2010;27(7):1630–1644. doi: 10.1093/molbev/msq049. [DOI] [PubMed] [Google Scholar]
- 7.Lynch M. The Origins of Genome Architecture. Sinauer Associates; Sunderland, MA: 2007. [Google Scholar]
- 8.Lynch M. Evolutionary diversification of the multimeric states of proteins. Proc Natl Acad Sci USA. 2013;110(30):E2821–E2828. doi: 10.1073/pnas.1310980110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karsenti E. Self-organization in cell biology: A brief history. Nat Rev Mol Cell Biol. 2008;9(3):255–262. doi: 10.1038/nrm2357. [DOI] [PubMed] [Google Scholar]
- 10.Budin I, Szostak JW. Physical effects underlying the transition from primitive to modern cell membranes. Proc Natl Acad Sci USA. 2011;108(13):5249–5254. doi: 10.1073/pnas.1100498108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen IA. GE Prize-winning essay: The emergence of cells during the origin of life. Science. 2006;314(5805):1558–1559. doi: 10.1126/science.1137541. [DOI] [PubMed] [Google Scholar]
- 12.Lane N, Martin W. The energetics of genome complexity. Nature. 2010;467(7318):929–934. doi: 10.1038/nature09486. [DOI] [PubMed] [Google Scholar]
- 13.Cavalier-Smith T. Nuclear volume control by nucleoskeletal DNA, selection for cell volume and cell growth rate, and the solution of the DNA C-value paradox. J Cell Sci. 1978;34:247–278. doi: 10.1242/jcs.34.1.247. [DOI] [PubMed] [Google Scholar]
- 14.Baker CR, Tuch BB, Johnson AD. Extensive DNA-binding specificity divergence of a conserved transcription regulator. Proc Natl Acad Sci USA. 2011;108(18):7493–7498. doi: 10.1073/pnas.1019177108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singh DP, et al. Genome-defence small RNAs exapted for epigenetic mating-type inheritance. Nature. 2014;509(7501):447–452. doi: 10.1038/nature13318. [DOI] [PubMed] [Google Scholar]
- 16.Kohl KP, Sekelsky J. Meiotic and mitotic recombination in meiosis. Genetics. 2013;194(2):327–334. doi: 10.1534/genetics.113.150581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cross FR, Buchler NE, Skotheim JM. Evolution of networks and sequences in eukaryotic cell cycle control. Philos Trans R Soc Lond B Biol Sci. 2011;366(1584):3532–3544. doi: 10.1098/rstb.2011.0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harashima H, Dissmeyer N, Schnittger A. Cell cycle control across the eukaryotic kingdom. Trends Cell Biol. 2013;23(7):345–356. doi: 10.1016/j.tcb.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 19.Koonin EV, Mushegian AR, Bork P. Non-orthologous gene displacement. Trends Genet. 1996;12(9):334–336. [PubMed] [Google Scholar]
- 20.Omelchenko MV, Galperin MY, Wolf YI, Koonin EV. Non-homologous isofunctional enzymes: A systematic analysis of alternative solutions in enzyme evolution. Biol Direct. 2010;5:31. doi: 10.1186/1745-6150-5-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Allen AE, et al. Evolution and functional diversification of fructose bisphosphate aldolase genes in photosynthetic marine diatoms. Mol Biol Evol. 2012;29(1):367–379. doi: 10.1093/molbev/msr223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liapounova NA, et al. Reconstructing the mosaic glycolytic pathway of the anaerobic eukaryote Monocercomonoides. Eukaryot Cell. 2006;5(12):2138–2146. doi: 10.1128/EC.00258-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oria-Hernández J, Riveros-Rosas H, Ramírez-Sílva L. Dichotomic phylogenetic tree of the pyruvate kinase family: K+ -dependent and -independent enzymes. J Biol Chem. 2006;281(41):30717–30724. doi: 10.1074/jbc.M605310200. [DOI] [PubMed] [Google Scholar]
- 24.Raposo G, Marks MS. The dark side of lysosome-related organelles: Specialization of the endocytic pathway for melanosome biogenesis. Traffic. 2002;3(4):237–248. doi: 10.1034/j.1600-0854.2002.030401.x. [DOI] [PubMed] [Google Scholar]
- 25.Devos DP, Gräf R, Field MC. Evolution of the nucleus. Curr Opin Cell Biol. 2014;28:8–15. doi: 10.1016/j.ceb.2014.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lavoie H, et al. Evolutionary tinkering with conserved components of a transcriptional regulatory network. PLoS Biol. 2010;8(3):e1000329. doi: 10.1371/journal.pbio.1000329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tanay A, Regev A, Shamir R. Conservation and evolvability in regulatory networks: The evolution of ribosomal regulation in yeast. Proc Natl Acad Sci USA. 2005;102(20):7203–7208. doi: 10.1073/pnas.0502521102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lynch M. The lower bound to the evolution of mutation rates. Genome Biol Evol. 2011;3:1107–1118. doi: 10.1093/gbe/evr066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lynch M. Evolutionary layering and the limits to cellular perfection. Proc Natl Acad Sci USA. 2012;109(46):18851–18856. doi: 10.1073/pnas.1216130109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sung W, Ackerman MS, Miller SF, Doak TG, Lynch M. Drift-barrier hypothesis and mutation-rate evolution. Proc Natl Acad Sci USA. 2012;109(45):18488–18492. doi: 10.1073/pnas.1216223109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Force A, et al. The origin of subfunctions and modular gene regulation. Genetics. 2005;170(1):433–446. doi: 10.1534/genetics.104.027607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tokuriki N, Tawfik DS. Protein dynamism and evolvability. Science. 2009;324(5924):203–207. doi: 10.1126/science.1169375. [DOI] [PubMed] [Google Scholar]
- 33.Force A, et al. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151(4):1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tuch BB, Li H, Johnson AD. Evolution of eukaryotic transcription circuits. Science. 2008;319(5871):1797–1799. doi: 10.1126/science.1152398. [DOI] [PubMed] [Google Scholar]
- 35.Lynch M. The evolution of genetic networks by non-adaptive processes. Nat Rev Genet. 2007;8(10):803–813. doi: 10.1038/nrg2192. [DOI] [PubMed] [Google Scholar]
- 36.Stoltzfus A. On the possibility of constructive neutral evolution. J Mol Evol. 1999;49(2):169–181. doi: 10.1007/pl00006540. [DOI] [PubMed] [Google Scholar]
- 37.Lukeš J, Archibald JM, Keeling PJ, Doolittle WF, Gray MW. How a neutral evolutionary ratchet can build cellular complexity. IUBMB Life. 2011;63(7):528–537. doi: 10.1002/iub.489. [DOI] [PubMed] [Google Scholar]
- 38.Finnigan GC, Hanson-Smith V, Stevens TH, Thornton JW. Evolution of increased complexity in a molecular machine. Nature. 2012;481(7381):360–364. doi: 10.1038/nature10724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lynch M. The evolution of multimeric protein assemblages. Mol Biol Evol. 2012;29(5):1353–1366. doi: 10.1093/molbev/msr300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lynch M, Abegg A. The rate of establishment of complex adaptations. Mol Biol Evol. 2010;27(6):1404–1414. doi: 10.1093/molbev/msq020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weissman DB, Feldman MW, Fisher DS. The rate of fitness-valley crossing in sexual populations. Genetics. 2010;186(4):1389–1410. doi: 10.1534/genetics.110.123240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lynch M. The frailty of adaptive hypotheses for the origins of organismal complexity. Proc Natl Acad Sci USA. 2007;104(Suppl 1):8597–8604. doi: 10.1073/pnas.0702207104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Newman JR, et al. Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature. 2006;441(7095):840–846. doi: 10.1038/nature04785. [DOI] [PubMed] [Google Scholar]
- 44.Schwanhäusser B, et al. Global quantification of mammalian gene expression control. Nature. 2011;473(7347):337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 45.Taniguchi Y, et al. Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science. 2010;329(5991):533–538. doi: 10.1126/science.1188308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shahrezaei V, Swain PS. Analytical distributions for stochastic gene expression. Proc Natl Acad Sci USA. 2008;105(45):17256–17261. doi: 10.1073/pnas.0803850105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thattai M, van Oudenaarden A. Intrinsic noise in gene regulatory networks. Proc Natl Acad Sci USA. 2001;98(15):8614–8619. doi: 10.1073/pnas.151588598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Farhadifar R, Needleman D. Automated segmentation of the first mitotic spindle in differential interference contrast microcopy images of C. elegans embryos. Methods Mol Biol. 2014;1136:41–45. doi: 10.1007/978-1-4939-0329-0_3. [DOI] [PubMed] [Google Scholar]
- 49.Lynch M, Walsh JB. Genetics and Analysis of Quantitative Traits. Sinauer Associates; Sunderland, MA: 1998. [Google Scholar]
- 50.Azimzadeh J, Marshall WF. Building the centriole. Curr Biol. 2010;20(18):R816–R825. doi: 10.1016/j.cub.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Colby DW, Prusiner SB. Prions. Cold Spring Harb Perspect Biol. 2011;3(1):a006833. doi: 10.1101/cshperspect.a006833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shimi T, Butin-Israeli V, Adam SA, Goldman RD. Nuclear lamins in cell regulation and disease. Cold Spring Harb Symp Quant Biol. 2010;75:525–531. doi: 10.1101/sqb.2010.75.045. [DOI] [PubMed] [Google Scholar]
- 53.Hirst J, et al. The fifth adaptor protein complex. PLoS Biol. 2011;9(10):e1001170. doi: 10.1371/journal.pbio.1001170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hirst J, et al. Characterization of TSET, an ancient and widespread membrane trafficking complex. eLife. 2014;3:e02866. doi: 10.7554/eLife.02866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Devos D, et al. Components of coated vesicles and nuclear pore complexes share a common molecular architecture. PLoS Biol. 2004;2(12):e380. doi: 10.1371/journal.pbio.0020380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Devos D, et al. Simple fold composition and modular architecture of the nuclear pore complex. Proc Natl Acad Sci USA. 2006;103(7):2172–2177. doi: 10.1073/pnas.0506345103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Carvalho-Santos Z, Azimzadeh J, Pereira-Leal JB, Bettencourt-Dias M. Evolution: Tracing the origins of centrioles, cilia, and flagella. J Cell Biol. 2011;194(2):165–175. doi: 10.1083/jcb.201011152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Carvalho-Santos Z, et al. Stepwise evolution of the centriole-assembly pathway. J Cell Sci. 2010;123(Pt 9):1414–1426. doi: 10.1242/jcs.064931. [DOI] [PubMed] [Google Scholar]
- 59.Mora C, Tittensor DP, Adl S, Simpson AG, Worm B. How many species are there on Earth and in the ocean? PLoS Biol. 2011;9(8):e1001127. doi: 10.1371/journal.pbio.1001127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim E, et al. Newly identified and diverse plastid-bearing branch on the eukaryotic tree of life. Proc Natl Acad Sci USA. 2011;108(4):1496–1500. doi: 10.1073/pnas.1013337108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Keeling PJ, et al. The Marine Microbial Eukaryote Transcriptome Sequencing Project (MMETSP): Illuminating the functional diversity of eukaryotic life in the oceans through transcriptome sequencing. PLoS Biol. 2014;12(6):e1001889. doi: 10.1371/journal.pbio.1001889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Karsenti E, et al. Tara Oceans Consortium A holistic approach to marine eco-systems biology. PLoS Biol. 2011;9(10):e1001177. doi: 10.1371/journal.pbio.1001177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gould SJ, Lewontin RC. The spandrels of San Marco and the Panglossian paradigm: a critique of the adaptationist programme. Proc R Soc Lond B Biol Sci. 1979;205(1161):581–598. doi: 10.1098/rspb.1979.0086. [DOI] [PubMed] [Google Scholar]
- 64.Graur D, et al. On the immortality of television sets: “Function” in the human genome according to the evolution-free gospel of ENCODE. Genome Biol Evol. 2013;5(3):578–590. doi: 10.1093/gbe/evt028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Arendt D. The evolution of cell types in animals: emerging principles from molecular studies. Nat Rev Genet. 2008;9(11):868–882. doi: 10.1038/nrg2416. [DOI] [PubMed] [Google Scholar]