Significance
Understanding the working mechanism of membrane proteins is difficult even when crystal structures are available. One promising approach is ion mobility–mass spectrometry (IM-MS) that detects not only the mass-to-charge ratio but also the area of proteins by measuring the rotationally averaged collision cross-sections (CCS) in the gas phase. We identified detergents that allow the release of membrane proteins at low levels of collisional activation for native MS, thus avoiding denaturing effects. We studied the gating mechanism of an ion channel, which occurs through large conformational changes. Ability to detect several coexisting states during gating with a change as small as 3% will open new avenues for studying dynamic structures of membrane proteins.
Keywords: ion mobility mass spectrometry, MscL, membrane proteins, structure function, ion channel gating
Abstract
Mechanosensitive ion channels are sensors probing membrane tension in all species; despite their importance and vital role in many cell functions, their gating mechanism remains to be elucidated. Here, we determined the conditions for releasing intact mechanosensitive channel of large conductance (MscL) proteins from their detergents in the gas phase using native ion mobility–mass spectrometry (IM-MS). By using IM-MS, we could detect the native mass of MscL from Escherichia coli, determine various global structural changes during its gating by measuring the rotationally averaged collision cross-sections, and show that it can function in the absence of a lipid bilayer. We could detect global conformational changes during MscL gating as small as 3%. Our findings will allow studying native structure of many other membrane proteins.
One of the best candidates to explore the gating of mechanosensitive channels is the mechanosensitive channel of large conductance (MscL) from Escherichia coli. The crystal structure of MscL in its closed/nearly closed state from Mycobacterium tuberculosis revealed this channel as a homopentamer (1). Each subunit has a cytoplasmic N- and C-terminal domain as well as two α-helical transmembrane (TM) domains, TM1 and TM2, which are connected by a periplasmic loop. The five TM1 helices form the pore and the more peripheral TM2 helices interact with the lipid bilayer.
MscL detects changes in membrane tension invoked by a hypoosmotic shock and couples the tension sensing directly to large conformational changes (1, 2). On the basis of a large body of structural and theoretical data, numerous gating models of MscL have been proposed (3–9). These models agree upon (i) the hydrophobic pore constriction of the channel and (ii) the channel opens by an iris-like rotation—i.e., a tilting and outward movement of transmembrane helices that make the channel wider and shorter (5). This mechanism is supported by patch-clamp (10), disulfide cross-linking (11), FRET spectroscopy (12), and site-directed spin labeling EPR experiments (6, 7), as well as computational studies (13–15). So far, direct experimental results have only been observed for short-range local structural changes, and no measure of the overall global structural changes during channel gating have been reported. Because there is no crystal structure available for the open MscL channel, elucidating overall global structural changes from the onset of channel activation is of utmost importance for our understanding of the gating mechanism of mechanosensitive channels. Here, we provide direct experimental evidence for the key areal changes occurring during channel gating by combining our ability to activate MscL in a controlled manner to different subopen states (16) with a native ion mobility-mass spectrometry (IM-MS) approach.
Results and Discussion
Detergents that are Unstable in the Gas Phase Allow Detection of MscL in Its Native Form.
Native mass spectrometry (MS) relies on the gentle ionization and transfer of intact complexes from solution into the gas phase by means of nanoelectrospray ionization (nanoESI) using desolvation voltages and pressures inside the mass spectrometer, which maintain noncovalent interactions (17–21). Together with ion mobility, an adjunct technique that measures collision cross-sections and determines the global (rotationally averaged) size of individual particles, native MS is increasingly used to determine the subunit composition, stoichiometry, size, and shape of biomolecular complexes (22–25). IM-MS has proven exceptionally useful for providing insights into the structure of membrane proteins, such as subunit stoichiometries and phospholipid binding (26, 27). Current methodology depends strongly on the apparently protective capabilities of detergent micelles to transfer membrane proteins intact into the vacuum of the mass spectrometer (26, 28), followed by the removal of these detergents by collisional activation. However, the energy required to rid the membrane protein of its detergent cover often results in the loss of structural integrity (28, 29). To enable analysis of native MscL structure using IM-MS (22–25), we addressed this problem and screened several detergents for their ability to preserve and release oligomeric MscL, hence eliminating the need for collisional activation.
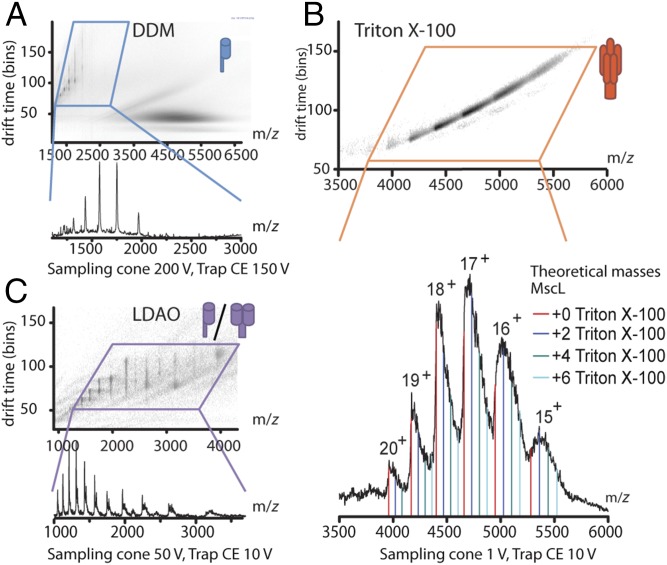
First, we introduced the protein in n-dodecyl-β-d-maltoside (DDM) micelles using nanoESI into the mass spectrometer. DDM is one of the most commonly used and better-studied detergents for native MS of membrane proteins (30). The dissociation of DDM micelles in the gas phase requires moderate to high levels of collisional activation, i.e., 150–200 V. When using IM-MS to study MscL in DDM, we could only detect monomeric MscL, indicating that these conditions were too harsh to keep native oligomeric MscL intact (Fig. 1A and Table S1). Next, we tested Triton X-100. Surprisingly, we found that rather than requiring collisional activation to disassemble detergent–protein complexes, the protein already appears in the spectra at very mild declustering conditions as a series of pentamer peaks centered around 4,750 m/z (Fig. 1B). It appears that Triton X-100 micelles dissociate readily in the gas phase of the mass spectrometer, but we nonetheless detected pentameric MscL with only a few remaining detergent molecules still attached (Fig. 1B, Lower). Upon increasing the collision energy above 50 V to remove these remaining detergents, we found that the energy required to do so was high enough to partially unfold MscL in the gas phase (Fig. S1). We therefore kept the various accelerating voltages inside the mass spectrometer below the threshold for unfolding, accepting broader peaks due to the remaining Triton X-100 detergent molecules but avoiding any artifactual structural change of the protein complex.
Fig. 1.
Comparison of the oligomeric states and collisional activation required to release MscL from detergent micelles in the gas phase. (A) Ion mobility spectrum (Upper) and the excerpt of the mass spectrum in the region where monomeric MscL is observed when released from DDM micelles. Release of MscL from DDM micelles requires high collision energies, which are already enough to disrupt the MscL pentamer, yielding only monomeric subunits. (B) Ion mobility spectrum (Upper) and complementary mass spectrum (Lower) of pentameric MscL released from Triton X-100 micelles. Colored lines indicate calculated m/z values for pentameric MscL with and without several detergent molecules attached (average mass 625 Da for Triton X-100 used). (C) Ion mobility spectrum (Upper) and the excerpt of the mass spectrum in the region where monomeric and dimeric MscL is observed when released from LDAO micelles.
We hypothesize the ease of protein release from Triton X-100 micelles in the gas phase by their greatly reduced hydrogen bond-forming capacity. Detergent micelles form in water largely due to the hydrophobic effect, which is absent in the vacuum of the mass spectrometer. For the maltoside-based detergent DDM, a network of hydrogen bonds in the head group stabilizes the detergent clusters in the gas phase, which then would require considerable additional energy for dissociation. Triton X-100, however, is not able to form such an extensive hydrogen bond network because of its polyethylene oxide head group; its micelles disassemble spontaneously upon transfer into the gas phase, thus revealing the embedded oligomeric protein. To examine this hypothesis, we also tested N,N-dimethyldodecylamine-N-oxide (LDAO), which was not expected to form extensive hydrogen bond networks. LDAO indeed required only 10 V of collision energy to release the protein from its detergent micelles, but yielded only monomeric and dimeric MscL subunits (Fig. 1C). This observation highlights the fact that though LDAO is a suitable detergent for gentle protein release in the gas phase, it does not enable MscL to adopt the correct (oligomeric) form in micelles (31, 32). Our approach, using detergent micelles that are unstable in the gas phase, allowed for the detection of native pentamer of E. coli MscL by IM-MS. Recently, a study on determining the specificity of membrane proteins toward lipids reported that a homologous MscL channel from Mycobacterium tuberculosis (Tb-MscL) also keeps its folded conformation in the gas phase of IM-MS (33). MscL from these two organisms differ in their molecular weight, tension sensitivity, and lipid interactions. Tb-MscL is bigger than E. coli MscL. Indeed, in the study of Laganowsky et al. (33), the molecular mass and collision cross-section (CCS) of M. tuberculosis MscL was measured as 85,578 Da and 5,438.5 Å2, whereas we measured the same parameters for WT E. coli MscL as 78,342 Da and 4,861 Å2 (Table S2).
Ion Mobility-Mass Spectrometry Can Distinguish Individual Heteropentameric MscL Channels by Their Mass-to-Charge Ratio and Size.
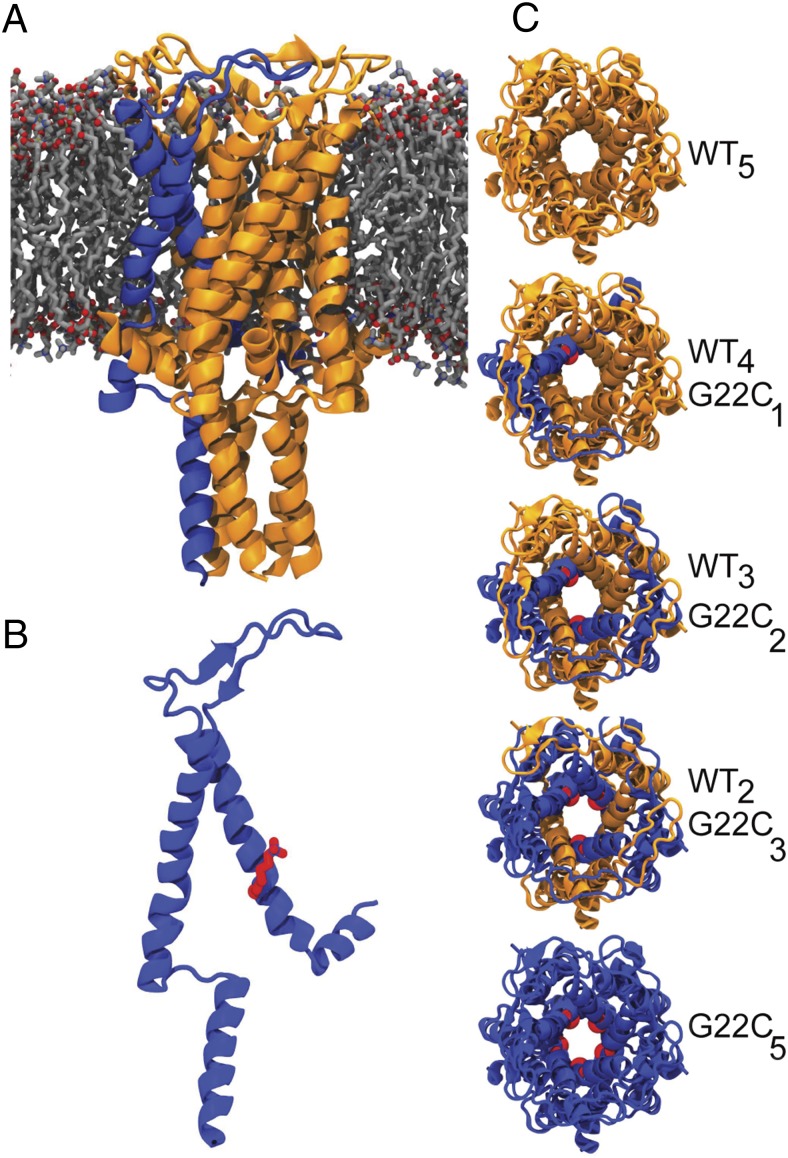
Now able to detect pentameric MscL by IM-MS, we set out to follow a range of large global structural changes occurring during MscL gating, i.e., the opening or closing of the channel. In its closed state, the pore diameter is ∼3.5 Å (1). However, when the channel fully opens, the pore diameter is estimated to become as large as ∼30 Å (34, 35), which can be detected by IM-MS if it were possible to trigger MscL opening in the absence of the lipid bilayer and tension. Here, to open these tension-sensitive channels in our experimental setup, we used heteropentameric MscLs comprised of varying number of WT and cysteine point-mutated subunits (G22C) per pentamer (Fig. 2) (16). Even though in nature, MscL opens in response to tension in the lipid bilayer, attaching charged cysteine-specific compounds to the pore of MscL-G22C homopentamers opens the channel spontaneously (36–38). Recently, based on this principle, we gained control on the degree of MscL opening in the absence of tension. We increased the hydrophilicity of the channel pore one subunit at a time using WT G22C heteropentameric MscLs. As the number of G22C subunits per heteropentamer increased, hence also the number of binding sites for positive charges within the pore constriction, the channels opened further and visited higher subconducting states (16).
Fig. 2.
Structure of MscL. (A) MscL viewed along the membrane plane. (B) MscL monomer highlighting the MTSET bound to the cysteine at position 22 (red). (C) Top view of homo- and heteropentameric MscL with different numbers of WT (yellow) and G22C (blue) subunits. MscL figures were aligned to the E. coli sequence and are based on the crystal structure of pentameric WT MscL from M. tuberculosis (40).
We first tested any conformational differences in individual heteropentameric channels in their closed form using IM-MS. We generated homo- (WT5 and G22C5) and heteropentameric (WT4G22C1, WT3G22C2, WT2G22C3) channels using a duet expression system, as described previously (16). Briefly, two mscL genes (WT-MscL with a StrepII-tag and G22C-MscL with a 6His-tag) were cloned and coexpressed in E. coli PB104, which allowed the production of homo- and heteropentameric channel assemblies in the host cell membrane. Individual heteropentamers were then obtained by using a three-step purification protocol; two-step affinity chromatography to remove the homopentameric MscL, followed by chromatofocusing to separate the individual heteropentameric MscL based on their isoelectric points (Fig. S2). Using IM-MS we determined the mass-to-charge ratio and the rotationally averaged size (CCS) of individual pentamers (Table S2). MscL had a narrow charge distribution (15–20+). In native MS, the lowest-observed significant charge state is known to represent the native conformation best. We chose the 16+ state for CCS determination throughout, because it gave consistent signal quality for all samples. The CCS was found to increase slightly with the number of WT subunits within a given heteropentamer. This observation was attributed to the larger size of the StrepII-tag of WT monomers relative to the size of the 6His-tag. Indeed, IM-MS of the homopentameric WT MscL channels with StrepII-tag gave a larger CCS than the same protein with 6His-tag (Table S2).
MscL Gating Can Be Detected Reversibly by Ion Mobility-Mass Spectrometry.
To see whether the gating of MscL can be detected by IM-MS, we first showed the covalent binding of a positively charged cysteine-specific channel activator [2-(trimethylammonium)ethyl] methanethiosulfonate bromide (MTSET) to cysteines of G22C subunits by tandem MS (Fig. S3). We tested increasing amounts of MTSET with G22C5 homopentamer and showed that MTSET concentrations above 500 µM maximally label the cysteines (beyond 1,000-µM deterioration of the IM-MS signal occurs due to ion suppression). Therefore, for the rest of the experiments, we used 750 µM MTSET for activating MscL channels.
Next, we measured the rotationally averaged collision cross-section of G22C5 homopentamer in the presence of 750 µM MTSET. These MTSET activated homopentamers generated similar narrow charge-state distribution (15–20+) as the closed channels, and we followed the 16+ charge state throughout.
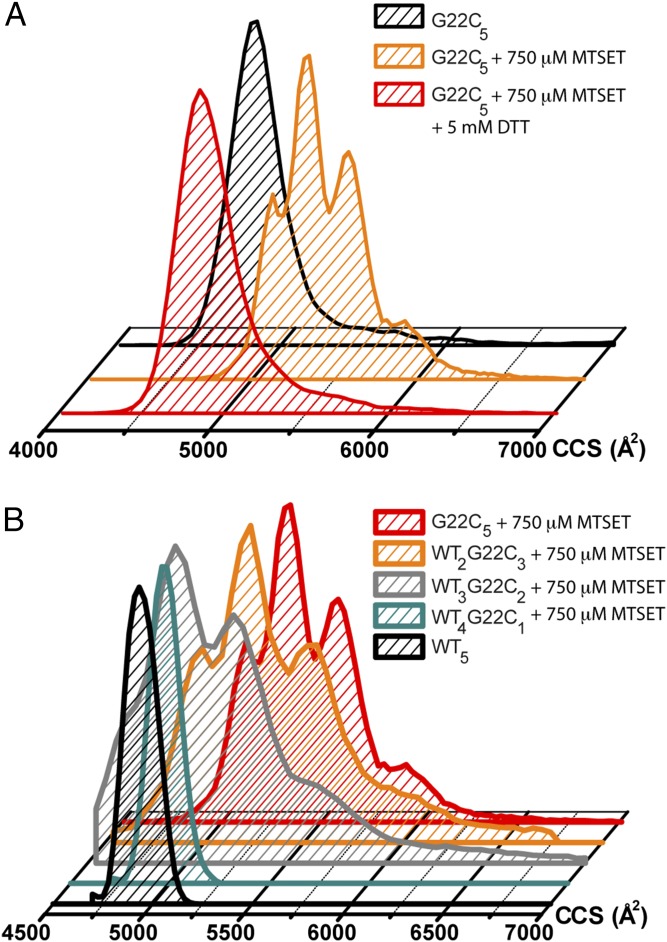
Though the closed channel had a single CCS centered around 4,960 ± 63 Å2 (CCS ± range), after MTSET treatment, the CCS of G22C5 increased and showed four distinct peaks at 5,095 ± 46; 5,330 ± 48; 5,580 ± 56; and 5,890 ± 106 Å2 (CCS ± range), suggesting various subopen conformations of the channel during its gating (Table S2). To prove that the presence of MTSET had opened the channel, we closed these already activated channels by using a reducing agent, DTT, for cleaving MTSET off the protein and showed that the CCS returned back to its original value (Fig. 3A). We also tested the effect of MTSET alone and showed that it did not cause any changes in the CCS of WT5 homopentamer. To further demonstrate that the CCS species of MTSET-activated MscL are different open conformations of the channel, we compared them to collision-induced unfolded MscL (Fig. S1), which shows different states. To our knowledge, this is the first time that ion channel gating is observed by using IM-MS to monitor global conformational changes.
Fig. 3.
Opening of MscL observed by IM-MS. (A) Charge-induced opening of MscL is reversible. The shift and dispersity of the collision cross-section observed for the 16+ charge state of G22C5 MscL (black) upon binding of MTSET indicates opening of the channel (yellow). After adding DTT to the same sample, thus removing the positively charged MTSET from the pore lining, the channel returns to its closed state again (red). (B) Increasing the number of MTSET-labeled cysteines in the pore lining allows MscL to occupy more extended conformations. Reported collision cross-sections are corrected for the different compositions of the purification tags of the heteropentamers of MscL.
The Overall Size of MscL Channels Increases as the Number of Charges in the Pore Increases.
Finally, we could resolve the channel opening to different subopen states by activating heteropentameric MscL with varying numbers of chargeable G22C subunits in the presence of 750 µM MTSET. When WT4G22C1 was activated with MTSET, no detectable change in CCS was observed using IM-MS (Fig. 3B). However, patch-clamp electrophysiology (16), dual-color fluorescence (39), and fluorescence dequenching assays (Fig. S4) showed conductance through the WT4G22C1/MTSET channel pore. The maximum pore opening for this heteropentamer was calculated to be up to 15 Å (39). Introducing a second charge in the pore, using MTSET-activated WT3G22C2 mutant, however, generated two major CCS populations averaged 5,266 ± 64 Å2 and 5,557 ± 98 Å2 (CCS ± range; Fig. 3B and Table S2), indicating that MscL already goes through at least two pronounced conformational states on its journey from its closed to the two-charge activated state. With the addition of the third charge to the pore, i.e., WT2G22C3 activated with MTSET, the channel visited the same extended conformations as WT3G22C2 MscL with higher frequencies (Fig. 3B). Homopentameric G22C5, when activated with MTSET, displayed all intermediate CCSs that were also observed for the heteropentameric forms of MscL, but also showed an additional more compact (5,095 ± 46 Å2) and a more extended (5,890 ± 106 Å2) conformation. These findings on observing increasing CCSs upon addition of MTSET to the pore are in good agreement with the patch-clamp single-channel measurements of individual heteropentamers when activated by MTSET (39). Though WT4G22C1 visits a very low ion-conducting state with 2% probability and stays mainly in the nonconducting closed conformation (P = 98%), as the number of MTSET attached subunits increases, i.e., MTSET attached WT3G22C2, WT2G22C3, WT1G22C4, and G22C5, the channels visit higher subconducting states and also presents a more flickering behavior (39). Homopentameric G22C5, activated with MTSET, spends most of its time in the early substates, whereas it visits the closed form only 25% of the time according to this data (39). Therefore, in a given time, due to its flickering behavior and higher possibility of being in subopen states, MTSET-bound G22C5 channels visits various subopen states frequently, which might explain the smallest conformational state of 5,095 Å2 being visible with 21% relative abundance in G22C5 but not in WT4G22C1. Furthermore, the absence of the fully closed form of activated G22C5 in the CCS distribution but the presence of this very first, distinctly larger 5,095 Å2 species with similar abundance in IM-MS measurements as in patch-clamp measurements, i.e., 21% vs. 25%, respectively, suggests that this population might represent the closed-expanded state of MscL, which could not be differentiated from the closed MscL on ionic conductance-based patch-clamp experiments (39). Overall, IM-MS could detect multiple conformations of MscL during its gating between the closed and the open forms (Fig. 3B).
MTSET-Induced Increase in the Size of MscL Obtained from IM-MS Corresponds with That of Tension-Activated Channel in Molecular Dynamics Simulations.
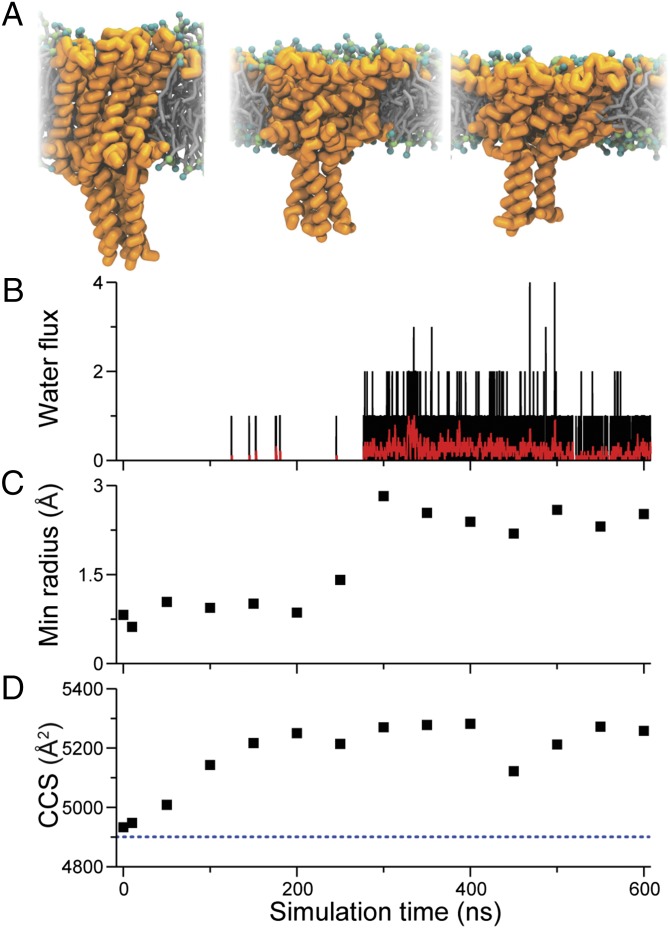
To correlate the observed increase in rotationally averaged collision cross-sections of charge-activated MscL in its very early open states as determined by IM-MS with tension-induced MscL gating, we performed coarse-grained molecular dynamics simulations. Using the closed-state crystal structure of MscL from M. tuberculosis (1, 40), a coarse-grained model (14, 15) was generated and embedded in a 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) bilayer. By applying high tension (65 dyn/cm) on the membrane, we were able to simulate initial channel opening. In the first ∼100 ns following the application of the lateral bilayer tension, the MscL transmembrane helices tilted, extending the extracellular cavity and flattening the channel in the plane of the bilayer (Fig. 4A). The channel took an additional ∼200 ns before the channel pore expanded enough to allow water permeation through the pore (Fig. 4B). The onset of water flux through the channel correlates well with the observed increase in minimum pore radius (Fig. 4C). Fig. 4D shows the calculated CCS for various time points of the simulation. The CCS gradually increased from a value of ∼4,900 Å2 before the tension was applied, to ∼5,200–5,300 Å2 when the channel gates, and reaches its first substates, overall corresponding well with the cross-section values of the closed (4,960 Å2) and first two expanded conformation (5,095 and 5,330 Å2) as determined by the IM-MS experiments (Fig. 3). The absence of water in the pore at CCS of 5,095 Å2 presents additional evidence that this state might indeed be a closed-expanded state at the initiation of MscL gating. These findings further support that the observed very early, hence compact, conformations seen for the charge-activated MscL pass through similar substates as tension-activated MscL in its transitions from the closed to the open conformation (16).
Fig. 4.
MscL gated with applied tension in MD simulations. (A) Snapshots from a Martini CG MD simulation show how the opening of the channel is governed by tilting of the helix bundle in response to flattening of the membrane (from left to right: no tension, with tension after 100 ns, and 300 ns). After applying bilayer tension, the water flux through the channel was monitored (B) showing flux per 0.15 ns (black) and average over 3 ns (red). (C) Minimum pore radius through the pore constriction (residues 12–25). (D) CCS calculated from the MD simulations. The dotted line indicates the CCS before the tension was applied.
Structural Changes During MscL Gating Are Intrinsic to the Protein.
The observed enlargement of heteropentamers with increasing number of MTSET-bound subunits also suggested—for the first time to our knowledge—that MscL could open in the absence of a lipid bilayer. Previously, charge-induced activation of MscL has been shown in intact E. coli cells (11, 41), with spheroplasts (12, 42) and with reconstituted systems (6, 7, 43). In all of these cases, MscL was embedded in a lipid bilayer environment. To support our IM-MS findings, we studied MscL channels in Triton X-100 with and without MTSET using both electron paramagnetic resonance (EPR) spectroscopy and transmission electron microscopy. Together with site-directed spin labeling, EPR provides information on protein structure and dynamics both in detergent and lipid bilayers (44). To make MscL visible for EPR, we partially (50% spin-labeling efficiency) labeled homopentameric G22C mutant with 1-oxyl-2,2,5,5-tetramethylpyrrolin-3-yl)methyl methane thiosulfonate. G22 position of MscL has been previously shown to change its environment when the channel opens (6, 7). We monitored structural changes of MscL in Triton X-100 by following the variations in spin–spin interactions, and the mobility of the spin labels within MscL as a function of MTSET activation of the channels (Fig. S5). If the channel opens, the distance between the spin labels increases and causes a change in the EPR line shape, and an increase in the mobility of the spin labels. The overall line broadening in the absence of MTSET indicated closer proximity between spin labels (i.e., stronger spin–spin interaction) in the closed form (Fig. S5, black trace). However, upon MTSET treatment, broadening disappeared (Fig. S5, red trace), suggesting that the subunits of MscL moved further apart. Furthermore, interaction with MTSET increased the probe mobility (Fig. S5).
Next, we used transmission electron microscopy and image analysis to study the closed (no MTSET) and partially open (750 μM MTSET) states of G22C5 MscL in Triton X-100 detergent micelles. Negatively stained closed and partially open MscL channels appear as bright spots as shown in Fig. S6A, Left and Right, respectively. Using WEB and SPIDER software packages, 1,403 closed and 1,004 open-state, centered, and aligned channel images were averaged. In its closed form, the area of the averaged MscL images in detergent was 5,007 Å2 (Fig. S6B, Left). Upon treatment with MTSET, the channels became flatter and wider and expanded to 6,165 Å2 (Fig. S6B, Right). Even though we took the averaged images as a measure, we were able to classify open and closed channels into different subclasses using the WEB and SPIDER software (Fig. S6 C–F). Whereas Fig. S6 C and D shows the three classes in the closed form of MscL from the top and the side, respectively, Fig. S6 E and F show three classes of the open state from the top and side, respectively. The observed subclasses may represent different subclosed and subopen states; however, it needs further investigation. Furthermore, the EM data on the averaged images of the closed and partially open state with a 27 ± 5% area increase upon MTSET opening is in good agreement with the CCS increase found by IM-MS of 21 ± 3%. Together, EM data strongly supports that our observation with IM-MS shows MscL gating in the absence of detergent or membrane.
In conclusion, detergent micelles that require minimum energy for their disassembly in the gas phase are valuable tools in native IM-MS of integral membrane protein assemblies. Resolving the global shape of the protein in combination with its mass-to-charge ratio made it possible to follow the global structural chances of MscL upon its gating. We could detect conformational changes as small as 3% in the rotationally averaged collision cross-sections of the protein complex from the closed (4,952 Å2) to the first detectable substate of 5,095 Å2, which seem to represent a closed-expanded state at the initiation of MscL gating. Assignment of other CCS species to defined subconducting states of MscL as defined by patch clamp on the basis of ionic conductance is currently difficult. Because a minor increase in a subconducting state, and hence change in pore diameter does not necessarily make a detectable change in the overall conformation of the channel. Indeed, in patch-clamp studies (39) we could detect as many as seven subconducting states of G22C5 when activated by MTSET. However, we could observe four CCS species in IM-MS.
We believe studying membrane proteins in gas phase unstable detergents by employing IM-MS opens new avenues for further discoveries on their dynamic structures.
Methods
Sample Preparation and Native IM-MS.
MscL homo- and heteropentamers were produced as described elsewhere (16) and introduced into the mass spectrometer (SYNAPT G2; Waters), which was carefully tuned for transmission of noncovalent complexes using nanoESI. Homo- and heteropentamer samples were incubated for 5 min at room temperature with 750 µM of MTSET before analysis.
MD Simulations.
Simulations of MscL gating were performed using the Martini coarse-grain (CG) model (45–47) and the GROMACS 4.X simulation package (48) following a similar protocol as described previously (14, 15). In short, the topology of MscL was derived from the crystal structure of the closed state Tb-MscL (PDB ID code 2OAR) (1, 40) using CG Martini 2.0. The channel was solvated in 562 CG DOPC lipids and ∼20,000 CG water beads (corresponding to ∼80,000 water molecules) using the insane.py script. The temperature and pressure were controlled using the Berendsen thermostat (298 K) and barostat (49). The initial system was energy-minimized (steepest descent, 500 steps) and simulated for 1 ns using short time-step 1- to 10-fs simulations and with position restrains on the protein backbone. The restrains were released, the time step set to 30 fs, and the system was equilibrated for 4 μs with 1 bar semiisotropic pressure coupling. Bilayer tension was incrementally applied in seven short (3-ns) simulations to a value of 65 dyn/cm and then simulated for 0.6 μs. In the first ∼100 ns following the application of the lateral bilayer tension and thinning of the bilayer, the MscL transmembrane helixes tilted, extending the extracellular cavity of the channel. The channel hydrophobic gate takes an additional ∼200 ns before expanding and opening the channel. Selected frames from the CG simulation were backmapped into atomistic coordinates using the Wassenaar et al. (50) reverse-transformation method and minimized with the GROMOS 54a7 force field (51). The minimum pore radius was measured for the same backmapped frames using the HOLE 2.0 program (52). The radius was defined as the radius of the largest sphere that could pass through the channel hydrophobic lock (−0.5–1 nm along the channel pore axes starting from the center of mass of residue 18) and interacting with the fully atomistic structure using their van der Waals radii.
All CCS calculations were done using MOBCAL (53, 54) with the projection approximation (PA) algorithm at 298 K and He as a buffer gas. To correct for the use of N2 in the IM-MS experiments and the underestimation of cross-sections by the PA algorithm, all calculated CCS were scaled by the experimentally derived factor of 1.14 (55). Simulations snapshots were generated using the molecular graphics viewer VMD (56).
EPR Spectroscopy.
Continuous-wave EPR measurements were performed using a commercially available MiniScope benchtop X-band EPR spectrometer (MS400 Magnettech GmbH) with a rectangular TE102 resonator. The microwave power was set to 10 mW and the B-field modulation amplitude to 0.20 mT. EPR glass capillaries (0.9 mm inner diameter) were filled with sample volume of 50 µL with a final protein concentration of 285 µM. The microwave and the modulation frequencies were 9.41 GHz and 100 kHz, respectively. Each spectrum corresponds to the accumulation of 36 scans.
Electron Microscopy.
The 0.03-mg/mL MscL in 100 mM ammonium acetate pH 6.8 buffer was applied in its closed or partially open state, i.e., activated by MTSET, to electron microscope grids and stained with 2% uranyl acetate. The specimen was analyzed with a JEOL 2100, LaB6 operating at 200 kV. A Gatan CCD camera was used (4,000 × 4,000; pixel size 15 Å) for collecting the data. Micrographs were recorded at a nominal magnification of 40,000 (pixel size 2.65 Å). The WEB and SPIDER software packages (57) were used for image processing. The 1,403 particles in the closed state and 1,004 particles in the partially open states were picked by hand, windowed, subjected to reference free alignment, and sorted into classes using the K-means clustering (58). Each class contained images of particles viewed from the same orientation.
Chromatofocusing.
Heteropentamers of MscL were separated by chromatofocusing based on their surface pI as described previously (16). The 250-μL fractions were collected and their pH determined at 6 °C. Only the peak fractions were used for efflux and IM-MS experiments.
Fluorescence Dequenching Assay.
Proteins were reconstituted into synthetic liposomes in the presence of a self-quenching fluorescent dye according to Koçer et al. (59). The resulting proteoliposomes were tested for the MscL activity using a fluorescent dequenching assay (59). The data were fitted to the exponential plot, and the first-order rate constants (k) for the fluorescent dye efflux were calculated for each heteropentamer.
The experimental procedures are described in more detail in SI Methods.
Supplementary Material
Acknowledgments
We thank E. Martin, A. Butterer, and J. F. Van Dyck for useful discussions. This work was supported by European Research Council Starting Grant 208814 (to A. Koçer) and a Rubicon grant from the Netherlands Organization for Scientific Research (to H.I.I.). The SYNAPT instrument was funded by the Hercules Foundation, and F.S. holds a Francqui Research Professorship at the University of Antwerp.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. C.V.R. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1413118111/-/DCSupplemental.
References
- 1.Chang G, Spencer RH, Lee AT, Barclay MT, Rees DC. Structure of the MscL homolog from Mycobacterium tuberculosis: A gated mechanosensitive ion channel. Science. 1998;282(5397):2220–2226. doi: 10.1126/science.282.5397.2220. [DOI] [PubMed] [Google Scholar]
- 2.Sukharev SI, Blount P, Martinac B, Kung C. Mechanosensitive channels of Escherichia coli: The MscL gene, protein, and activities. Annu Rev Physiol. 1997;59:633–657. doi: 10.1146/annurev.physiol.59.1.633. [DOI] [PubMed] [Google Scholar]
- 3.Blount P, Moe PC. Bacterial mechanosensitive channels: integrating physiology, structure and function. Trends Microbiol. 1999;7(10):420–424. doi: 10.1016/s0966-842x(99)01594-2. [DOI] [PubMed] [Google Scholar]
- 4.Yoshimura K, Batiza A, Schroeder M, Blount P, Kung C. Hydrophilicity of a single residue within MscL correlates with increased channel mechanosensitivity. Biophys J. 1999;77(4):1960–1972. doi: 10.1016/S0006-3495(99)77037-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sukharev S, Durell SR, Guy HR. Structural models of the MscL gating mechanism. Biophys J. 2001;81(2):917–936. doi: 10.1016/S0006-3495(01)75751-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perozo E, Cortes DM, Sompornpisut P, Kloda A, Martinac B. Open channel structure of MscL and the gating mechanism of mechanosensitive channels. Nature. 2002;418(6901):942–948. doi: 10.1038/nature00992. [DOI] [PubMed] [Google Scholar]
- 7.Perozo E, Kloda A, Cortes DM, Martinac B. Physical principles underlying the transduction of bilayer deformation forces during mechanosensitive channel gating. Nat Struct Biol. 2002;9(9):696–703. doi: 10.1038/nsb827. [DOI] [PubMed] [Google Scholar]
- 8.Liu Z, Gandhi CS, Rees DC. Structure of a tetrameric MscL in an expanded intermediate state. Nature. 2009;461(7260):120–124. doi: 10.1038/nature08277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corry B, et al. An improved open-channel structure of MscL determined from FRET confocal microscopy and simulation. J Gen Physiol. 2010;136(4):483–494. doi: 10.1085/jgp.200910376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sukharev S, Betanzos M, Chiang CS, Guy HR. The gating mechanism of the large mechanosensitive channel MscL. Nature. 2001;409(6821):720–724. doi: 10.1038/35055559. [DOI] [PubMed] [Google Scholar]
- 11.Betanzos M, Chiang C-S, Guy HR, Sukharev S. A large iris-like expansion of a mechanosensitive channel protein induced by membrane tension. Nat Struct Biol. 2002;9(9):704–710. doi: 10.1038/nsb828. [DOI] [PubMed] [Google Scholar]
- 12.Corry B, Rigby P, Liu Z-W, Martinac B. Conformational changes involved in MscL channel gating measured using FRET spectroscopy. Biophys J. 2005;89(6):L49–L51. doi: 10.1529/biophysj.105.072009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gullingsrud J, Schulten K. Gating of MscL studied by steered molecular dynamics. Biophys J. 2003;85(4):2087–2099. doi: 10.1016/s0006-3495(03)74637-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yefimov S, van der Giessen E, Onck PR, Marrink SJ. Mechanosensitive membrane channels in action. Biophys J. 2008;94(8):2994–3002. doi: 10.1529/biophysj.107.119966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Louhivuori M, Risselada HJ, van der Giessen E, Marrink SJ. Release of content through mechano-sensitive gates in pressurized liposomes. Proc Natl Acad Sci USA. 2010;107(46):19856–19860. doi: 10.1073/pnas.1001316107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Birkner JP, Poolman B, Koçer A. Hydrophobic gating of mechanosensitive channel of large conductance evidenced by single-subunit resolution. Proc Natl Acad Sci USA. 2012;109(32):12944–12949. doi: 10.1073/pnas.1205270109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Loo JA. Studying noncovalent protein complexes by electrospray ionization mass spectrometry. Mass Spectrom Rev. 1997;16(1):1–23. doi: 10.1002/(SICI)1098-2787(1997)16:1<1::AID-MAS1>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 18.Sobott F, Hernández H, McCammon MG, Tito MA, Robinson CV. A tandem mass spectrometer for improved transmission and analysis of large macromolecular assemblies. Anal Chem. 2002;74(6):1402–1407. doi: 10.1021/ac0110552. [DOI] [PubMed] [Google Scholar]
- 19.Heck AJR, Van Den Heuvel RHH. Investigation of intact protein complexes by mass spectrometry. Mass Spectrom Rev. 2004;23(5):368–389. doi: 10.1002/mas.10081. [DOI] [PubMed] [Google Scholar]
- 20.Sobott F, McCammon MG, Hernández H, Robinson CV. The flight of macromolecular complexes in a mass spectrometer. Philos Trans A Math Phys Eng Sci. 2005;363(1827):379–389, discussion 389–391. doi: 10.1098/rsta.2004.1498. [DOI] [PubMed] [Google Scholar]
- 21.Hernández H, Robinson CV. Determining the stoichiometry and interactions of macromolecular assemblies from mass spectrometry. Nat Protoc. 2007;2(3):715–726. doi: 10.1038/nprot.2007.73. [DOI] [PubMed] [Google Scholar]
- 22.Ruotolo BT, et al. Evidence for macromolecular protein rings in the absence of bulk water. Science. 2005;310(5754):1658–1661. doi: 10.1126/science.1120177. [DOI] [PubMed] [Google Scholar]
- 23.Ruotolo BT, Benesch JLP, Sandercock AM, Hyung S-J, Robinson CV. Ion mobility-mass spectrometry analysis of large protein complexes. Nat Protoc. 2008;3(7):1139–1152. doi: 10.1038/nprot.2008.78. [DOI] [PubMed] [Google Scholar]
- 24.Uetrecht C, Rose RJ, van Duijn E, Lorenzen K, Heck AJR. Ion mobility mass spectrometry of proteins and protein assemblies. Chem Soc Rev. 2010;39(5):1633–1655. doi: 10.1039/b914002f. [DOI] [PubMed] [Google Scholar]
- 25.Konijnenberg A, Butterer A, Sobott F. Native ion mobility-mass spectrometry and related methods in structural biology. Biochim Biophys Acta. 2013;1834(6):1239–1256. doi: 10.1016/j.bbapap.2012.11.013. [DOI] [PubMed] [Google Scholar]
- 26.Barrera NP, Di Bartolo N, Booth PJ, Robinson CV. Micelles protect membrane complexes from solution to vacuum. Science. 2008;321(5886):243–246. doi: 10.1126/science.1159292. [DOI] [PubMed] [Google Scholar]
- 27.Zhou M, et al. Mass spectrometry of intact V-type ATPases reveals bound lipids and the effects of nucleotide binding. Science. 2011;334(6054):380–385. doi: 10.1126/science.1210148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laganowsky A, Reading E, Hopper JTS, Robinson CV. Mass spectrometry of intact membrane protein complexes. Nat Protoc. 2013;8(4):639–651. doi: 10.1038/nprot.2013.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Borysik AJ, Hewitt DJ, Robinson CV. Detergent release prolongs the lifetime of native-like membrane protein conformations in the gas-phase. J Am Chem Soc. 2013;135(16):6078–6083. doi: 10.1021/ja401736v. [DOI] [PubMed] [Google Scholar]
- 30.Barrera NP, Robinson CV. Advances in the mass spectrometry of membrane proteins: From individual proteins to intact complexes. Annu Rev Biochem. 2011;80:247–271. doi: 10.1146/annurev-biochem-062309-093307. [DOI] [PubMed] [Google Scholar]
- 31.Timmins PA, Hauk J, Wacker T, Welte W. The influence of heptane-1,2,3-triol on the size and shape of LDAO micelles. Implications for the crystallisation of membrane proteins. FEBS Lett. 1991;280(1):115–120. doi: 10.1016/0014-5793(91)80217-q. [DOI] [PubMed] [Google Scholar]
- 32.Dorwart MR, Wray R, Brautigam CA, Jiang Y, Blount P. S. aureus MscL is a pentamer in vivo but of variable stoichiometries in vitro: Implications for detergent-solubilized membrane proteins. PLoS Biol. 2010;8(12):e1000555. doi: 10.1371/journal.pbio.1000555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laganowsky A, et al. Membrane proteins bind lipids selectively to modulate their structure and function. Nature. 2014;510(7503):172–175. doi: 10.1038/nature13419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cruickshank CC, Minchin RF, Le Dain AC, Martinac B. Estimation of the pore size of the large-conductance mechanosensitive ion channel of Escherichia coli. Biophys J. 1997;73(4):1925–1931. doi: 10.1016/S0006-3495(97)78223-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Y, et al. Single molecule FRET reveals pore size and opening mechanism of a mechano-sensitive ion channel. eLife. 2014;3:e01834. doi: 10.7554/eLife.01834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoshimura K, Batiza A, Kung C. Chemically charging the pore constriction opens the mechanosensitive channel MscL. Biophys J. 2001;80(5):2198–2206. doi: 10.1016/S0006-3495(01)76192-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koçer A, Walko M, Meijberg W, Feringa BL. A light-actuated nanovalve derived from a channel protein. Science. 2005;309(5735):755–758. doi: 10.1126/science.1114760. [DOI] [PubMed] [Google Scholar]
- 38.Koçer A, et al. Rationally designed chemical modulators convert a bacterial channel protein into a pH-sensory valve. Angew Chem Int Ed Engl. 2006;45(19):3126–3130. doi: 10.1002/anie.200503403. [DOI] [PubMed] [Google Scholar]
- 39.Mika JT, Birkner JP, Poolman B, Koçer A. On the role of individual subunits in MscL gating: “All for one, one for all?”. FASEB J. 2013;27(3):882–892. doi: 10.1096/fj.12-214361. [DOI] [PubMed] [Google Scholar]
- 40.Steinbacher S, Bass R, Strop P, Rees DC. Structures of the prokaryotic mechanosensitive channels MscL and MscS. Curr Top Membr. 2007;58:1–24. [Google Scholar]
- 41.Bartlett JL, Levin G, Blount P. An in vivo assay identifies changes in residue accessibility on mechanosensitive channel gating. Proc Natl Acad Sci USA. 2004;101(27):10161–10165. doi: 10.1073/pnas.0402040101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bartlett JL, Li Y, Blount P. Mechanosensitive channel gating transitions resolved by functional changes upon pore modification. Biophys J. 2006;91(10):3684–3691. doi: 10.1529/biophysj.106.088062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yoshimura K, Usukura J, Sokabe M. Gating-associated conformational changes in the mechanosensitive channel MscL. Proc Natl Acad Sci USA. 2008;105(10):4033–4038. doi: 10.1073/pnas.0709436105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hubbell WL, Cafiso DS, Altenbach C. Identifying conformational changes with site-directed spin labeling. Nat Struct Biol. 2000;7(9):735–739. doi: 10.1038/78956. [DOI] [PubMed] [Google Scholar]
- 45.Marrink SJ, de Vries AH, Mark AE. Coarse grained model for semiquantitative lipid simulations. J Phys Chem B. 2004;108:750–760. [Google Scholar]
- 46.Marrink SJ, Risselada HJ, Yefimov S, Tieleman DP, de Vries AH. The MARTINI force field: Coarse grained model for biomolecular simulations. J Phys Chem B. 2007;111(27):7812–7824. doi: 10.1021/jp071097f. [DOI] [PubMed] [Google Scholar]
- 47.Monticelli L, et al. The MARTINI coarse-grained force field: Extension to proteins. J Chem Theory Comput. 2008;4:819–834. doi: 10.1021/ct700324x. [DOI] [PubMed] [Google Scholar]
- 48.Hess B, Kutzner C, van der Spoel D, Lindahl E. GROMACS 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput. 2008;4(3):435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 49.Berendsen HJC, Postma JPM, van Gunsteren WF, DiNola A, Haak JR. Molecular dynamics with coupling to an external bath. J Chem Phys. 1984;81:3684–3690. [Google Scholar]
- 50.Wassenaar TA, Pluhackova K, Böckmann RA, Marrink SJ, Tieleman DP. Going backward: A flexible geometric approach to reverse transformation from coarse grained to atomistic models. J Chem Theory Comput. 2014;10(2):676–690. doi: 10.1021/ct400617g. [DOI] [PubMed] [Google Scholar]
- 51.Schmid N, et al. Definition and testing of the GROMOS force-field versions 54A7 and 54B7. Eur Biophys J. 2011;40(7):843–856. doi: 10.1007/s00249-011-0700-9. [DOI] [PubMed] [Google Scholar]
- 52.Smart OS, Goodfellow JM, Wallace BA. The pore dimensions of gramicidin A. Biophys J. 1993;65(6):2455–2460. doi: 10.1016/S0006-3495(93)81293-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mesleh MF, Hunter JM, Shvartsburg AA, Schatz GC, Jarrold MF. Structural information from ion mobility measurements: Effects of the long-range potential. J Phys Chem. 1996;100(40):16082–16086. [Google Scholar]
- 54.Shvartsburg AA, Jarrold MF. An exact hard-spheres scattering model for the mobilities of polyatomic ions. Chem Phys Lett. 1996;261(1-2):86–91. [Google Scholar]
- 55.Hall Z, Politis A, Bush MF, Smith LJ, Robinson CV. Charge-state dependent compaction and dissociation of protein complexes: Insights from ion mobility and molecular dynamics. J Am Chem Soc. 2012;134(7):3429–3438. doi: 10.1021/ja2096859. [DOI] [PubMed] [Google Scholar]
- 56.Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. J Mol Graph. 1996;14(1):33–38, 27–28. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 57.Frank J, et al. SPIDER and WEB: Processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116(1):190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 58.Frank J. Classification of macromolecular assemblies studied as ‘single particles’. Q Rev Biophys. 1990;23(3):281–329. doi: 10.1017/s0033583500005564. [DOI] [PubMed] [Google Scholar]
- 59.Koçer A, Walko M, Feringa BL. Synthesis and utilization of reversible and irreversible light-activated nanovalves derived from the channel protein MscL. Nat Protoc. 2007;2(6):1426–1437. doi: 10.1038/nprot.2007.196. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.