Significance
The ability to design and build nonnative DNA sequences conferring specific and useful engineered functions on cells defines the field of synthetic biology. Here, we describe a synthetic biology device built for Saccharomyces cerevisiae. The telomerator inducibly and precisely linearizes circular DNA molecules in vivo, yielding derivatives that encode functional telomeres at each end. The telomerator provides the means to engineer artificial chromosomes, typically constructed as circular molecules, because telomerator-mediated linearization permits specific positioning of genes to modulate their expression. The telomerator also provides a tool to probe spatial relationships between functional elements on chromosomes such as telomeres, centromeres, and replication origins. In the future, the telomerator may be applied to engineer artificial chromosomes in other organisms.
Keywords: telomerator, chromosome engineering, telomeric silencing, Sc2.0, synthetic chromosome
Abstract
Chromosome engineering is a major focus in the fields of systems biology, genetics, synthetic biology, and the functional analysis of genomes. Here, we describe the “telomerator,” a new synthetic biology device for use in Saccharomyces cerevisiae. The telomerator is designed to inducibly convert circular DNA molecules into mitotically stable, linear chromosomes replete with functional telomeres in vivo. The telomerator cassette encodes convergent yeast telomere seed sequences flanking the I-SceI homing endonuclease recognition site in the center of an intron artificially transplanted into the URA3 selectable/counterselectable auxotrophic marker. We show that inducible expression of the homing endonuclease efficiently generates linear molecules, identified by using a simple plate-based screening method. To showcase its functionality and utility, we use the telomerator to circularly permute a synthetic yeast chromosome originally constructed as a circular molecule, synIXR, to generate 51 linear variants. Many of the derived linear chromosomes confer unexpected phenotypic properties. This finding indicates that the telomerator offers a new way to study the effects of gene placement on chromosomes (i.e., telomere proximity). However, that the majority of synIXR linear derivatives support viability highlights inherent tolerance of S. cerevisiae to changes in gene order and overall chromosome structure. The telomerator serves as an important tool to construct artificial linear chromosomes in yeast; the concept can be extended to other eukaryotes.
Chromosome engineering is the study of genetic modifications that affect large segments of chromosomes. Top-down approaches start with preexisting chromosomes and modify them in vivo by introducing, for instance, deletions, translocations, or duplications. Bottom-up approaches involve design and construction of chromosomes de novo. To facilitate successful chromosome engineering efforts, we need a strong understanding of chromosomal features that confer mitotic stability such as centromeres, telomeres, and replication origins. Moreover, it is important to appreciate the effects of the spatial relationships between such elements and other critical features such as genes.
The Saccharomyces cerevisiae genome is an excellent platform to develop tools for eukaryotic chromosome engineering given its ease of genetic manipulation. The S. cerevisiae genome is composed of 12 Mb of DNA organized as 16 linear chromosomes ranging in size from 230 kb to more than 2 Mb (1). Three important cis elements are required to maintain chromosome stability through mitosis and meiosis: Compact point centromeres (∼125 bp) ensure faithful segregation of sister chromatids (2) and homologs in meiosis I, replication origins are necessary for genome duplication before cell division (3, 4), and conserved telomere sequences protect chromosome ends ensuring maintenance of chromosome length during replication (5, 6). With these elements intact, many lines of evidence indicate that budding yeast tolerate a high degree of chromosomal modification without affecting viability. For instance: (i) the largest yeast chromosome (IV) can be subdivided into 11 separate minichromosomes (7); (ii) more than 500 kb, including 247 nonessential genes, can be deleted in a single haploid strain (8); (iii) any of the 16 chromosomes can be individually destabilized in a diploid cell to generate a chromosomal complement of 2n−1 (9); (iv) synthetic chromosomes such as synIII (10) and synIXR (11), built to the designer specifications of the Sc2.0 Yeast Genome Project, power growth of budding yeast in the absence of the corresponding native chromosomes (10, 11).
With the goal of systematically and specifically perturbing the order and orientation of genetic elements on chromosomes in S. cerevisiae, we developed the telomerator, a genetic tool that can linearize circular DNA molecules in vivo on demand. Importantly, the linear derivatives generated via the telomerator encode functional telomeres and are thus mitotically stable. We used the telomerator to circularly permute the synthetic yeast chromosome, synIXR, encoded in the form of a bacterial artificial chromosome (BAC; herein referred to as synIXR BAC). In 51 viable linear permutants, we discovered substantial phenotypic diversity that depends on changes in expression of genes required for growth, mediated by telomere position effects. Further, our results support the conclusion that telomerator-induced linearization generates linear chromosomes with functional telomeres on which heterochromatin is fully established.
Results
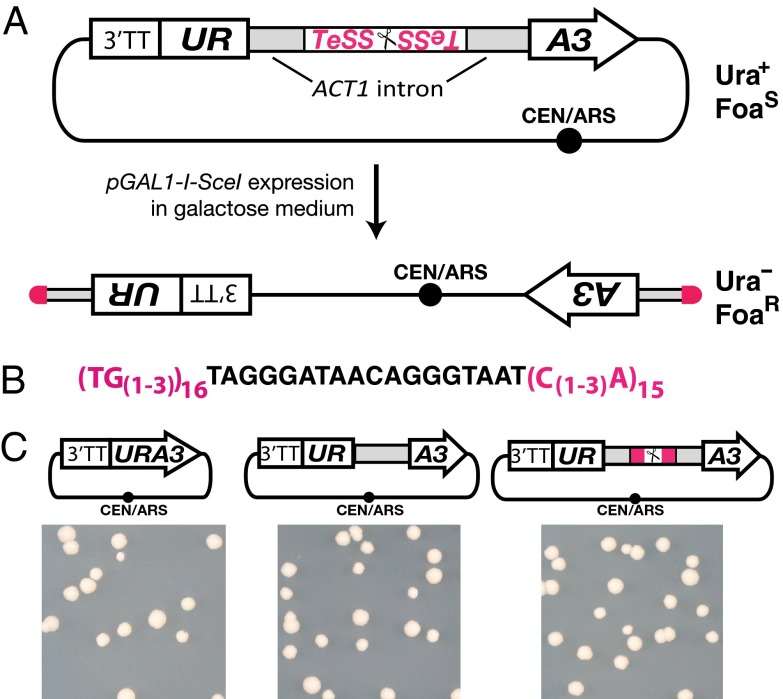
The design of the telomerator includes several different elements for its function (Fig. 1A). Centrally, the telomerator encodes an 18-bp recognition sequence for the I-SceI homing endonuclease (12, 13). This site is flanked by a pair of convergent telomere seed sequences (TeSSs) that encode ∼40 bp of conserved yeast telomere repeats [(TG(1-3))16…(C(1-3)A)15] previously shown to serve as a minimal telomeric repeat (14) (Fig. 1B). The entire TeSS–I-SceI recognition sequence-TeSS cassette is encoded within an intron that interrupts the S. cerevisiae URA3 auxotrophic selectable marker coding sequence. Previous work showed that transplantation of the ACT1 gene intron into the middle of the URA3 coding sequence did not disrupt complementation of gene function as measured by growth of a ura3 mutant on medium lacking uracil. This result indicates the mRNA was expressed and the nonnative intron correctly spliced before translation of the URA3 gene product (15). To facilitate inducible linearization, I-SceI expression can be placed under control of the inducible GAL1 promoter and, thus, linearization specifically induced by growth in medium containing galactose. Under standard growth conditions in glucose medium, the telomerator cassette remains intact with cells expressing a functional Ura3 protein, complementing growth of a ura3Δ0 strain on medium lacking uracil. Following growth in galactose, cells containing the linearization product can be selected on 5-fluoroorotic acid (Foa) medium, which counterselects URA3+ cells (16). A circular telomerator-containing molecule grows on medium lacking uracil and dies on FOA, but a cell harboring a linearized molecule, in which the URA3 gene component of the telomerator is literally split in two, display the opposite growth phenotype [Ura–, FoaR (uracil requiring and resistant to Foa)] (Fig. 1A). Once converted to a linear molecule, the newly terminally exposed telomere seed sequences should be extended by native telomerase to generate functional telomeres and confer mitotic stability.
Fig. 1.
Telomerator design and function. (A) The telomerator encodes a selectable URA3 gene sequence interrupted by an intron (ACT1 intron) that harbors a homing endonuclease recognition site (depicted by scissors). The recognition site is flanked by telomere seed sequences (TeSS). The URA3 promoter sequence also encodes a strong 3′ transcriptional terminator sequence (3′TT). The telomerator cassette is encoded on a circular DNA molecule yielding cells that grow on medium lacking uracil (Ura+) but are sensitive to the uracil counterselection drug Foa (FoaS), assuming the circular DNA molecule encodes at least one essential genetic part. Expression of I-SceI, under the control of the GAL1 promoter (pGAL1), is induced by growth in galactose medium. Cells harboring newly linearized molecules should no longer grow on plates lacking uracil (Ura–) and should be resistant to Foa (FoaR). (B) Detailed sequence information for the TeSSs (pink) flanking the I-SceI recognition sequence (black) of the telomerator. (C) The telomerator complements yeast cell growth on medium lacking uracil. Plasmids encoding a native URA3 gene (Left), a URA3 gene interrupted by the intron of ACT1 (Middle), or a URA3 gene interrupted by a telomerator sequence encoded within the ACT1 intron (Right) were transformed into a yeast strain (BY4741) incapable of growing on medium lacking uracil (SC–Ura). Growth of the resulting transformed colonies on SC–Ura was indistinguishable, indicating normal expression of the URA3 gene product from the telomerator-encoding construct.
Whereas expression of the URA3 gene interrupted by the ACT1 intron was shown to complement growth of ura3 mutants on medium lacking uracil (15), the effect on complementation of further encoding the TeSS–I-SceI–TeSS sequence within the intron was unknown. To evaluate the effect of the TeSS–I-SceI–TeSS insertion, we first constructed the telomerator cassette in pRS413, a centromere-based yeast shuttle vector encoding the HIS3 selectable marker (17). The telomerator-containing construct was transformed into BY4741, a yeast strain with a complete URA3 deletion (ura3Δ0) (18), and plated on medium lacking uracil. Growth of the resulting transformants was compared with cells transformed with pRS416, another centromere-based yeast shuttle vector encoding a wild-type URA3 gene, or a plasmid encoding the URA3exon1-intronACT1-URA3exon2 cassette (15), identical to the telomerator plasmid but lacking the TeSS–I-SceI–TeSS sequence. Growth of cells transformed with any of the three constructs on medium lacking uracil was indistinguishable (Fig. 1C). Thus, the presence of the TeSS–I-SceI–TeSS sequence within the ACT1 intron interrupting the URA3 gene does not interfere with growth on medium lacking uracil.
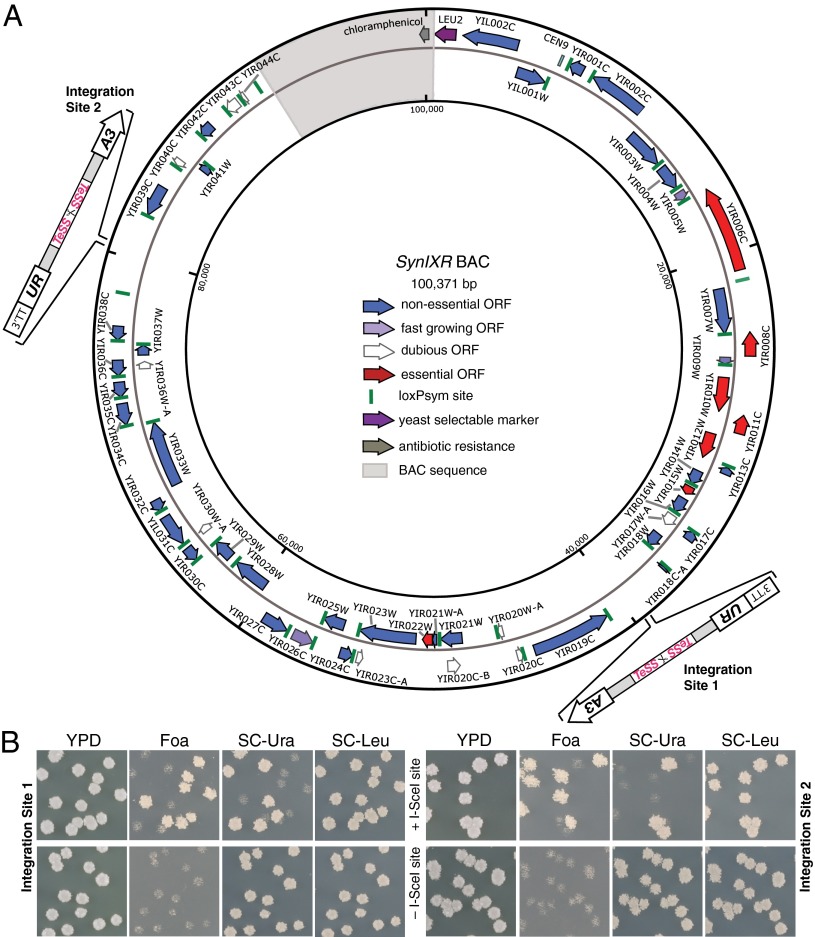
Chromosome length has important consequences for mitotic stability in S. cerevisiae because linear chromosomes shorter than 90 kb exhibit markedly decreased stability (19). Therefore, we tested the capacity of the telomerator to linearize circular DNA molecules in vivo by using synIXR BAC, a circular synthetic yeast chromosome arm (11, 20) ∼100 kb in size. SynIXR BAC encodes all 52 genes from the right arm of chromosome 9 (YIR001C-YIR044C), 2 genes from the left arm (YIL001W, YIL002C), the native chromosome 9 centromere (CEN9), a yeast selectable marker (LEU2), plus ∼10 kb of BAC vector sequence including a chloramphenicol resistance gene and bacterial replication origin (Fig. 2A). SynIXR, which encodes seven essential genes, was shown to be mitotically stable in vivo and power yeast cell growth at wild-type levels in the absence of the wild-type chromosome arm IXR (11). Two “gene-free” regions of synIXR BAC were chosen as initial candidate integration sites for the telomerator cassette, which was then introduced by homologous recombination to generate two new derivative circular chromosomes designed for subsequent linearization. Additionally, the URA3exon1-intronACT1-URA3exon2 cassette (15), lacking the TeSS–I-SceI–TeSS sequence, was integrated at the same two positions to generate auxotrophically equivalent, but nonlinearizeable control strains. All four strains were transformed with a plasmid expressing pGAL1–I-SceI, grown in galactose medium for 24 h, and finally ∼200 cells were plated on YPD, permissive yeast medium containing glucose. These plates were replica-plated onto medium containing Foa and uracil, or on medium lacking uracil [synthetic complete (SC)–Ura] or leucine (SC–Leu) (Fig. 2B). At either integration site 1 or 2, in cells expressing the functional telomerator cassette (+ I-SceI), ∼50% of cells became resistant to growth on Foa and correspondingly lost ability to grow on SC–Ura, consistent with successful linearization (Fig. 2B, Top). The remaining half of cells that were sensitive to Foa was able to grow on SC-Ura, suggesting that in those cells, the synIXR BAC had not been linearized. When comparing cells harboring circular or putative linear synIXR molecules, there were no apparent changes to colony size or shape, suggesting the linearized construct was mitotically stable. In cells lacking the I-SceI recognition site (–I-SceI), the FoaR phenotype was not observed, demonstrating that linearization depends on this sequence (Fig. 2B, Bottom). Taken together, these results suggest the telomerator efficiently linearizes circular DNA molecules in vivo upon induction.
Fig. 2.
The telomerator drives in vivo linearization of a synthetic yeast chromosome arm, synIXR BAC. (A) synIXR BAC is a ∼100-kb circular chromosome arm that can power a yeast cell. In independent integration experiments, the telomerator cassette was introduced by homologous recombination at two gene-free regions of synIXR BAC as indicated (integration site 1, ∼40 kb; integration site 2, ∼80 kb). (B) Following 24 h of growth in galactose-containing medium to induce expression of I-SceI, single cells from integration site 1 (Left) and integration site 2 (Right) were plated on YPD medium, and then replica plated onto SC medium supplemented with 5′-fluoroorotic acid (Foa), and SC medium lacking uracil or leucine (SC–Ura and SC–Leu, respectively). Only in cell populations encoding an intact I-SceI recognition sequence as part of the telomerator cassette (+ I-SceI; Top) did linearization occur, as assayed by appearance of colonies displaying growth on Foa and nongrowth on SC–Ura.
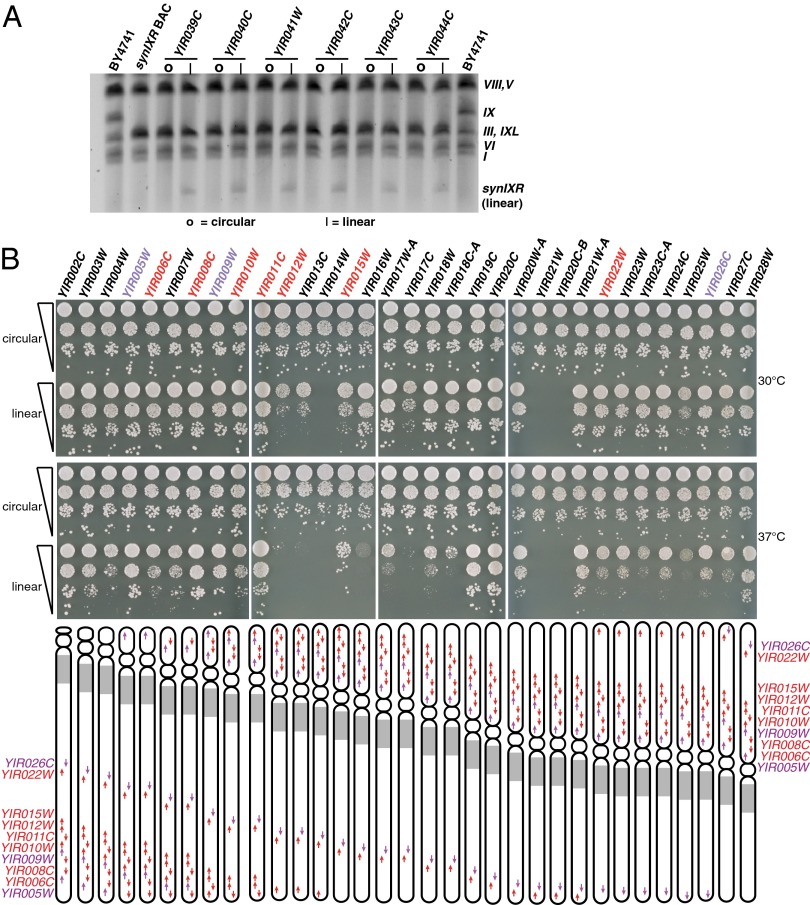
The telomerator enables precise control over the location at which a circular DNA molecule is linearized. Thus, it represents a tool to probe the function of gene order, orientation, and chromosomal structure in vivo. For instance, if integrated between two genes, telomerator-driven linearization will yield novel telomere-promixal gene positioning that could lead to subtelomeric silencing. Furthermore, depending on the orientation of each gene with respect to the telomerator cassette, linearization could disrupt either 5′ or 3′ regulatory elements that drive their transcription. To test these ideas, we integrated the telomerator cassette 3 bp downstream of all chromosome IX genes on the synIXR BAC, generating an array of 54 permutable strains. We chose this position because it was previously shown to tolerate the insertion of synthetic sequences (11). Indeed, the synIXR BAC encodes a 34-bp site-specific recombination site (loxPsym) at this position downstream of every nonessential gene on the BAC with no effect on cell fitness (Fig. 2A). Moreover, the promoter region of the telomerator-encoded URA3 contains a terminator sequence for polyadenylation of incoming transcripts (21). Integration of the telomerator 3′ downstream of any gene, while disrupting native 3′ end forming sequences, provides a built-in, nonnative terminator sequence for the resulting transcript. For the telomerator cassette integration, at each of the nonessential genes, we “overwrote” the preexisting loxPsym site, and for the seven essential genes (which lacked loxPsym sites), we inserted the cassette 3 bp downstream of the stop codon. In all cases, the telomerator was encoded on the same DNA strand as the targeted gene. Integration at the correct locus was confirmed for all 54 permutable strains by PCR using a gene-specific primer in combination with a telomerator-specific primer (Fig. S1A). Next, the pGAL1–I-SceI construct was transformed into the 54 strains, which were grown in galactose medium for 24 h to induce linearization. Foa resistant cells were selected, enabling isolation of the permuted array of linearized synIXR-containing strains (Fig. 3A). Each permutant resulted in relocation of the targeted gene into the subtelomere, positioning the stop codon ∼800 bp upstream of a TeSS (Fig. 3). To directly investigate whether the synIXR BACs had been linearized, full-length chromosomes from permuted strains, prelinearization and postlinearization, were separated by pulsed field gel electrophoresis. Only the telomerator strains subjected to growth in galactose and subsequent selection on Foa exhibited a band on the gel corresponding to the linearized form of synIXR (Fig. 4A and Fig. S1B). Also, consistent with linearization, we found that FoaR/Ura– isolates from the entire panel of viable permutants no longer yielded PCR amplicons spanning the TeSS–I-SceI–TeSS junction (Fig. S1A). In the circular conformation, we found that simply integrating the telomerator caused no noticeable change in fitness at any of the 54 loci (Fig. 4B). Of the 54 sites into which the telomerator had been integrated, 51 could be linearized and give rise to viable FoaR colonies (Fig. 4B). The three positions at which linearization of synIXR was lethal were 3′ of YIR014W, YIR021W, and YIR020C-B (Fig. 4B). YIR014W encodes a putative protein of unknown function and is located directly upstream of the essential YIR015W; linearization 3′ of YIR014W repositioned the promoter of YIR015W ∼800 bp downstream of a telomere. YIR021W (MRS1) encodes a splicing factor for mitochondrial group I introns and YIR020C-B is a dubious ORF that overlaps YIR021W; linearization 3′ of either of these two genes repositioned the essential YIR022W subtelomerically. Although not lethal, many other linear chromosome derivatives resulted in modest growth defects at 30 °C (Fig. 4B). At 37 °C, almost half of all permuted strains grew slowly. Together, the data indicate that telomerator-induced linearization provides a new way to generate diverse growth phenotypes.
Fig. 3.
Circular permutation of a synthetic eukaryotic chromosome, synIXR BAC. Schematic depicting 54 expected synIXR linear chromosome structures achieved via telomerator-mediated linearization downstream of each yeast genes on the chromosome. The systematic yeast gene names denote both the gene downstream of which the telomerator was integrated plus the final chromosomal positioning of that gene in the linear structure.
Fig. 4.
Characterization of strain encoding linear permutations of synIXR. (A) Pulsed field gel showing appearance of linear synIXR after galactose induction in six representative strains (the telomerator was integrated downstream of YIR039C, YIR040C, YIR041W, YIR042C, YIR043C, YIR044C), compared with a wild-type strain encoding a native chromosome IX (BY4741), or the parental strain used for telomerator integrations (synIXR BAC). The circular form of synIXR BAC is not detectable on pulsed field gel. |, linear; O, circular. (B) Fitness assays of permuted strains. Growth of strains, prelinearization (circular) and postlinearization (linear), was assessed at 30 °C (Top) or 37 °C (Bottom) on rich medium. Shown here are 10-fold serial dilutions after 2 d growth. Permuted strains not pictured here did not exhibit growth defects.
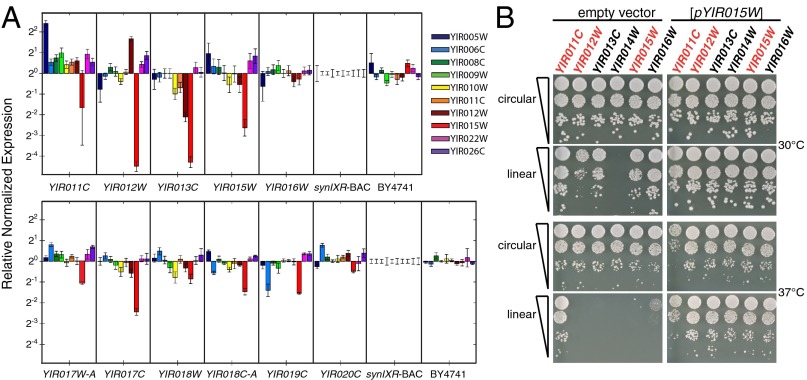
We hypothesized that telomeric silencing could underlie the fitness defects observed in many of the linear permutations of synIXR (Fig. 4B). Telomeric silencing, or the transcriptional repression of genes in proximity to telomeres, is an epigenetic phenomenon relying on the establishment of heterochromatin that spreads inward from the ends of chromosomes. Linearization-dependent relocation of essential genes into subtelomeric regions and the subsequent reduction in expression could have a major impact on cell fitness and/or survival. Indeed, mapping the position of the seven essential genes onto the array of linearized permutations of synIXR suggests that fitness defects tend to occur in strains in which one or more essential genes are relocated to within 10 kb of a telomere (Fig. 3). To study the effect of linearization on gene expression, we tested the expression level of the seven essential genes (YIR006C, YIR008C, YIR010W, YIR011C, YIR012W, YIR015W, YIR022W) plus three genes required for fast growth (YIR005W, YIR009W, YIR026C) by quantitative PCR in 11 linearly permuted strains of synIXR BAC (linearized 3′ of YIR011–YIR020C; Fig. 5A). These particular strains were selected because they are located on either side of one of the positions at which linearization was found to be lethal (3′ of YIR014W), several exhibit slow growth at 30 °C, and many are dead or barely alive at 37 °C (Fig. 4B). In cells growing at 30 °C, we discovered changes in expression of many of these 10 genes (Fig. 5A). However, most consistently we found a reduction in expression of the essential gene YIR015W, an observation that largely correlated with decreased fitness at 30 °C and/or 37 °C in the strains (Figs. 5A and 4B). YIR015W (RPR2) encodes an essential subunit of the nuclear RNase P complex that cleaves tRNA precursors to generate mature 5′ ends and facilitates turnover of nuclear RNAs (22, 23). That yeast cells may be exquisitely sensitive to reduced expression of YIR015W is consistent with the observation that telomerator-driven linearization 3′ of YIR014W, which repositions the YIR015W promoter adjacent to a telomere, cannot be tolerated. To specifically test this hypothesis, we cloned the YIR015W gene into a CEN vector and transformed it, or the corresponding empty vector, into six strains harboring the telomerator cassette integrated at unique positions (3′ of YIR011C–YIR016W) on the synIXR BAC. The linear permutations of synIXR were generated by growth in galactose in the presence of the pGAL1–I-SceI construct, and FoaR/Ura– cells were identified. We discovered that the plasmid-borne copy of YIR015W rescued both slow growth and lethality at 30 and 37 °C in all strains examined, including linearization downstream of YIR014W, whereas cells harboring an empty vector displayed growth defects/lethality as described previously (Figs. 4B and 5B). Interestingly, increased expression of YIR015W in a linear permutant in which expression of this gene was not altered (linearization 3′ of YIR011C; Fig. 5A) resulted in slow growth in both the circular and linear format at 37 °C, suggesting that both gain- and loss-of-function of YIR015W inhibit yeast cell growth. This result is consistent with the fact that the YIR015W gene product, Rpr2, is a member of a protein complex (22) whose function may be disrupted by imbalanced stoichiometry among subunits.
Fig. 5.
SynIXR linear permutations display changes in expression of essential genes and genes required for fast growth. (A) The relative normalized expression of 10 different genes (listed in Inset legend) was determined for 11 different synIXR permutations (YIR011C, YIR012W, YIR013C, YIR015W, YIR016W, YIR017W-A, YIR017C, YIR018W, YIR018C-A, YIR019C, YIR020C) compared with a strain encoding synIXR BAC. Expression of the same 10 genes was also determined relative to synIXR BAC for BY4741, which encodes native chromosome IX. (B) Fitness assays of permuted strains expressing YIR015 episomally [pYIR015W] or an empty vector. Shown here are 10-fold serial dilutions after 2 d growth on selective medium.
The process of telomeric silencing is mediated in part by the lysine deacetylase enzyme Sir2 (24), which deacetylates histone H4, forming a heterochromatin-like structure (25). To globally test the hypothesis that silencing of telomere-proximal genes impacts fitness across the entire array of linearly permuted strains, we deleted SIR2 from each of the 54 linearizable synIXR BAC strains. Following transformation of the pGAL1–I-SceI construct and growth in galactose, cells harboring linearized synIXR molecules were selected on medium containing Foa. We discovered that disruption of telomeric silencing by SIR2 deletion rescued nearly all of the growth defects associated with linear permutations of synIXR (Fig. 6). Notably, deletion of SIR2 allowed linearization where it was not observed in the wild type, namely 3′ of YIR014W (Fig. 3). Importantly, these results are consistent with the conclusion that telomerator-encoded 40-bp telomere seed sequences generate functional telomeres complete with the potential for the appropriate establishment of heterochromatin. In native chromosomes, subtelomeric sequences can buffer the effects of gene silencing, as can insulators. Future telomerator designs could incorporate such elements.
Fig. 6.
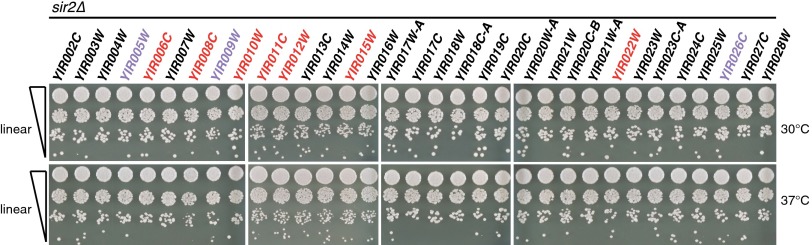
Deletion of SIR2 rescues growth defects of synIXR BAC circular permutations. The SIR2 gene was deleted in the each of the 54 unique telomerator-containing synIXR BAC strains. Following transformation of the pGAL1–I-SceI construct and growth in galactose, linear permutations were isolated on Foa medium. Shown here are 10-fold serial dilutions after 2 d growth on rich medium (YPD) at the indicated temperatures. Strains correspond to those shown above in Fig. 4B.
Next, to investigate the stability of “telomerated” linear chromosomes, we tested their propensity to recircularize. Here, we asked whether six high fitness synIXR linear permutations (linearized 3′ of YIR029W, YIR030C, YIR030W-A, YIR031C, YIR032C, and YIR033W) could regain the ability to grow on medium lacking uracil, a selective pressure favoring the action of DNA repair mechanism(s) rejoining the two halves of the ACT1 intron at the termini of the linearized chromosome to reinstate URA3 expression. The six linear permutants were grown to saturation in YPD medium, washed once with water, and ∼2 × 107 cells were plated on synthetic medium lacking uracil. After 5 d, one to five robustly growing Ura+ colonies arose on three of the six plates. Colonies from each of the three plates were selected for sequence analysis. In all cases examined, we discovered the genetic basis underlying the Ura+ phenotype was likely due to gene conversion templated by the native ACT1 intron locus (Fig. S2). The small fraction of Ura+ colonies recovered in this experiment suggests that telomerator-generated linear chromosomes and their associated nascent telomeres are not only functional but also highly stable.
Discussion
The telomerator represents a new, convenient, and precise way to linearize circular DNA molecules in vivo in S. cerevisiae. It can be applied to the construction and expression of artificial chromosomes encoding nonnative pathways that could confer a multitude of new functions to the cell. At least two previous methods have been used to construct artificial linear chromosomes in yeast. First, nonnative chromosomes (i.e., YACs) were generated by in vitro ligation of synthetic telomere sequences onto linear DNA followed by transformation and selection in yeast (26, 27). This approach overcomes the complicating factor that linear DNA molecules cannot be replicated in Escherichia coli and can therefore be difficult to construct. Second, a circular derivative of chromosome three (28) was linearized by integrating into it synthetic telomere sequences; over time these sequences resolved in vivo to generate a linear molecule (29). Together these studies uncovered important features that underlie mitotic stability of S. cerevisiae chromosomes. In terms of chromosome engineering, the telomerator now presents many new advantages: (i) the use of telomere seed sequences derived from native yeast telomeres; (ii) the ease with which a telomerator construct can be PCR amplified and, thus, integrated at any number of specifically targeted loci; (iii) recovery of virtually 100% correct clones with no requirement for cumbersome screening approaches; and (iv) inducible control over the timing of linearization in vivo.
Here, by circularly permuting synIXR BAC with the telomerator, we generated 51 linear chromosome structures that supported viability. With the exception of effects on gene expression at some positions (see below), the panel of permuted synIXR variants displays surprisingly little variation in behavior, suggesting that the relative placement of telomeres and centromeres in S. cerevisiae is flexible. For instance, linearization at either YIL001W or YIR001C produces telocentric versions of synIXR with only ∼1 kb separating the centromere from a telomere (Fig. 3); that these permutations exhibit no apparent growth defect is consistent with previous work suggesting telocentric chromosomes in budding yeast are mitotically stable (19, 29).
In several instances, either outright lethality or severe growth defects were revealed upon linearization, at least in a SIR2+ background. We performed a variety of experiments to better understand these exceptional cases. Our data suggest that the slow growth or lethality associated with linearization in the cases examined depends on expression changes of essential genes. Moreover, all observed fitness defects were mediated by Sir2-dependent telomeric silencing. In the three cases where linearization proved intolerable, an essential gene was repositioned into a subtelomeric region such that its transcription occurred inwards toward the centromere. Specifically, linearization 3′ of YIR014W repositioned the start codon of the essential gene YIR015W 0.9 kb from a TeSS, and linearization 3′ of YIR021W or YIR021C-B placed the start codon of the essential gene YIR022W 0.9 kb and 2.1 kb from the TeSS, respectively. Interestingly, linearization 3′ of either of these essential genes themselves, which repositioned the YIR015W or YIR022W adjacent to a TeSS but pointing toward it (either 1.1 or 1.3 kb from start to TeSS), was not lethal. This observation suggests that transcriptional elongation toward the telomere may sufficiently counteract the inward spread of heterochromatin to allow production of enough mRNA to support viability.
At one position, 3′ of YIR021W-A, we were surprised to successfully integrate the telomerator cassette into synIXR BAC. Targeting the telomerator to this position should have disrupted the promoter of the neighboring essential gene YIR022W, whose start codon is just 6 bases downstream of the YIR021W-A stop codon (Fig. S3). Although we generated Ura+ transformants and confirmed integration of the telomerator cassette 3′ of YIR021W-A by PCR, over time the Ura+ phenotype could be lost; this result was not observed in any other telomerator-integrated derivatives of the synIXR BAC. This finding suggests the presence of two copies of synIXR BAC, one in which the telomerator was integrated and the other presumably required for expression of YIR022W. Indeed, testing expression of essential genes and genes required for fast growth postlinearization 3′ of YIR021W-A revealed an ∼twofold increase in expression of all genes except YIR022W, whose expression is identical to the circular parental synIXR BAC (Fig. S3). These observations suggest duplication of the synIXR molecule as a mechanism to compensate for reduced expression of one or more genes. Although we did not see evidence for duplication of linearized synIXR in any of the linear derivatives examined by qPCR (i.e., pervasive ∼twofold increase in gene expression) (Fig. 5A), in rare cases we were able to isolate FoaR/Ura– isolates of synIXR linearized 3′ of YIR021W or YIR020C-B, two of the positions at which linearization was lethal. We predict that duplication of the linearized synIXR derivative underlies the growth phenotype in these rare events.
In principle, the telomerator technology can be extended to engineering both native and artificial chromosomes in S. cerevisiae and other eukaryotic organisms. Although native eukaryotic chromosomes are typically linear, instances of circular chromosomes have been reported. For example, in addition to synIXR BAC (11), S. cerevisiae chromosome III can also exist in a ring structure with no effect on fitness (28). These results suggest that other native yeast chromosomes could easily be circularized, for instance, by homologous recombination using a marker gene flanked by the appropriate targeting sequences, thus providing a template for telomerator-driven linearization at any position of interest. Ring chromosomes have also been documented in plants, insects, rodents, and humans, usually the result of fusion of two broken chromosome ends and in human patients often associated with severe pathologies or cancer (30–32). However, rather than native chromosomes, we envision a major application of the telomerator to be engineering of artificial chromosomes, often constructed initially as circular molecules to take advantage of facile cloning methods in both E. coli and S. cerevisiae. Modifying gene expression of heterologous pathways in microorganisms is a major interest in the field of metabolic engineering, and the telomerator could provide a new tool to facilitate this goal. In addition to S. cerevisiae, the telomerator technology should be readily expandable to other industrially relevant yeast species (e.g., Pichia pastoris, high-level secreted protein production; Ashbya gossypii, riboflavin production) and other eukaryotic microbes such as algae. Applying the telomerator to chromosome engineering in mammalian cells will be more challenging because “regional” mammalian centromeres can extend for hundreds of kilobases and include repetitive sequences. Artificially generated human minichromosomes, existing as either linear or circular constructs, have been described and they may serve as a starting point for mammalian artificial chromosome engineering (33–39). We ultimately envision such artificial chromosomes as valuable platforms for gene targeting that will allow delivery of large DNA sequences to recipient cells and provide a means by which to investigate the incorporation of complex segments of DNA encoding networks and pathways. The linearization function of the telomerator can be achieved with a variety of homing endonucleases (40), CRISPR/Cas systems (41), zinc finger nucleases (42), or TALENs (43) in addition to I-SceI demonstrated here. To build plant artificial chromosomes, endonuclease-dependent linearization of a circular plant artificial chromosome encoding convergent telomere-like sequences has been attempted in vitro before bombardment into maize immature embryos (44); however, it is unclear whether the resulting minichromosomes arose from telomere truncation (45). For in vivo linearization, one important consideration is that the recognition sequence of the enzyme must be sufficiently rare within the sequence of the host genome (i.e., no or few existing sites) to work without causing “collateral damage” in the rest of the genome. In this regard, the I-SceI site is not present in yeast nuclear genome (46). A variety of selectable/counterselectable markers could also be used.
Materials and Methods
Construction of the Telomerator.
The TeSS–I-SceI–TeSS-I sequence was introduced by one-step isothermal assembly (47) into the unique XhoI site encoded centrally within the ACT1 intron that had been previously transplanted into URA3 (15). Specifically, the TeSS–I-SceI recognition sequence-TeSS sequence (Fig. 1B) flanked by 40 bp corresponding to sequence on either side of the XhoI site was ordered from IDT, Inc. (Coralville, IA) as an Ultramer. This sequence was exponentially amplified by using external primers [Forward (For): ATCCCATTTAACTGTAAGAAGAATTGC3′; Reverse (Rev) GGAGAGTGAAAAATAGTAAAAAAAGGT). Expression of the URA3-intronACT1-TeSS-I-Sce-I-TeSS-intronACT1-URA3 cassette was verified by functional complementation assay as described in the text (Fig. 1C).
Integration of Telomerator into synIXR BAC.
Primers were designed to bind upstream of the URA3 promoter (For: CCCGGGGGATCCGGTGATTG3) and downstream of the URA3 terminator (Rev: CCAAAGCTGGAGCTCCACCG3) on the telomerator construct. Fifty base pairs corresponding to either side of each unique integration site was then appended to these two primers. Standard PCR conditions were used to amplify the telomerator, which was then transformed into a synIXR BAC containing strain (yJS587; ref. 11). Integration was confirmed with a location-specific primer in combination with a primer internal to the URA3 promoter sequence.
Linearization of synIXR.
The pGAL1–I-SceI sequence was subcloned from a preexisting plasmid (48) into the yeast shuttle vector pRS413 (17) at the SalI recognition site. This construct was transformed into strains to be linearized and selected on synthetic complete medium lacking histidine (SC–His). Linearization was induced in liquid culture in synthetic medium lacking histidine with 2% galactose for 24 h. Strains were then transferred onto synthetic complete solid medium supplemented with Foa and extra uracil. Each strain was subjected to a single round of colony purification on YPD medium, and single isolates that had lost the pGAL1–I-Sce-I construct, confirmed by replica-plating on SC–His, were selected for further analysis. Circular versus linear status of synIXR was tested by PCR using the primers originally designed to amplify the TeSS–I-Sce-I recognition sequence-TeSS ultramer (Fig. S1A).
Pulsed Field Gel Electrophoresis.
Full-length yeast chromosomes were prepared in agarose plugs as described (49). Chromosomes were separated by clamped homogeneous electric field (CHEF) gel electrophoresis by using the CHEF-DR III Pulsed Field Electrophoresis Systems (Bio-Rad) with the following settings: 6 V/cm, switch time 60–120 s over 24 h, 14 °C, 0.5× Tris-borate-EDTA buffer, and 1% gel prepared with low melting point agarose (Lonza, 50100). Gels were stained with 5 μg/mL ethidium bromide in water postelectrophoresis, destained in water, and then imaged.
Dot Assays.
Strains were inoculated into 150 μL of appropriate medium in sterile 96-well plates and incubated overnight at 30 °C. Ten-fold serial dilutions were carried out in water, and the dilutions corresponding to 10−2 to 10−6 spotted on the appropriate agar plates. For strain carrying the YIR015W construct or corresponding empty vector, YPD medium was supplemented with 200 μg/mL G418. Incubations were carried out at 30 °C or 37 °C for 2 or 3 d.
Gene Expression.
Eleven linearly permuted strains of synIXR BAC (linearized 3′ of YIR011C-YIR020C) plus two control strains [the parental wild-type strain to synIXR BAC, BY4741, and synIXR BAC (yJS587) without integration of the telomerator cassette] were grown overnight at 30 °C in 5-mL YPD culture. RNA was prepared from 250 μL of culture by using the RNeasy Minikit (Qiagen; 74106) as per the manufacturer’s instructions. In brief, cells were lysed enzymatically, and eluted RNA was treated with DNase (Qiagen; 79254) in solution before passage over a second column and elution in 30 μL of water. Complete digestion of gDNA was confirmed in a control qPCR experiment lacking reverse transcriptase by using 100 ng of prepared RNA and two different primer pairs corresponding to the reference genes (UBC6 and TAF10; ref. 50). Primers were designed by using the PrimeQuest Design tool on the IDT website and are listed in Table S1. Primers were selected to anneal outside PCRTag sequences (10, 11) and, thus, amplify both synthetic and native DNA. cDNA was prepared from ∼5 μg of RNA by using SuperScript III Reverse Transcriptase (Invitrogen; 18080044) as per the manufacturer’s instructions. qPCR reactions were carried out in a final volume of 4 μL by using SsoAdvanced Universal SYBR Green Supermix (Bio-Rad; 172-5272). Each reaction included 75 nL of cDNA and 50 nL of forward/reverse primers mix (50 μM each) introduced into each well of a Hard-Shell Thin-Wall 384-Well Skirted PCR Plate (Bio-Rad; HSP-3905) by using the Echo (LabCyte). Gene expression analysis was performed by using the CFX Software Manager (Bio-Rad). This experiment was performed in two batches with independently grown control strains (synIXR BAC and BY4741). For the first batch (linearized 3′ of YIR011C, YIR012W, YIR013C, YIR015W, YIR016W), qPCR was performed on technical quadruplicates and data analyzed by using a single reference gene (UBC6). For the second batch (linearized 3′ of YIR017W-A, YIR017C, YIR018W, YIR018C-A, YIR019C, YIR020C), qPCR was performed on technical duplicates and two reference genes (TAF10 and UBC6) were used for data analysis.
Cloning YIR015W.
YIR015W was amplified from synIXR BAC gDNA by using primers designed to anneal ∼500 bp upstream and ∼200 bp downstream of the start and stop codon, respectively. Further, to each primer was appended ∼30 bases of sequence corresponding to the terminal ends of an EcoRI/BamHI digested, modified version of a pRS CEN vector (17) encoding resistance to G418 (pLM200). The primers sequences are as follows (underlined portion anneals to yeast gDNA); Forward: cctcgaggtcgacggtatcgataagcttgatatcggggaatttacttgtccggtaccc; Reverse: gctccaccgcggtggcggccgctctagaactagtgggcaagacggaatcctacattacc. The final construct was built by using one-step isothermal assembly (47) and a single digestion-verified construct selected for transformation into yeast.
SIR2 Gene Deletion.
The sir2∆kanMX locus was amplified from the deletion mutant collection strain (51) by using primers ∼500 bp flanking the gene deletion and subsequently transformed into the array of permutable strains by selection on YPD supplemented with 200 µg/mL geneticin. SIR2 gene deletion was confirmed functionally by testing for the loss of mating.
Recircularization.
Six linear permutation strains (linearized 3′ of YIR029W, YIR030C, YIR030W-A, YIR031C, YIR032C, and YIR034C) were inoculated into 5 mL of YPD medium and incubated with rotation for 24 h at 30 °C. Cells from 1 mL of each culture (∼2 × 107) were collected by centrifugation, washed once with water, and spread onto synthetic medium lacking uracil (SC–Ura) plates. After 5 d of incubation at 30 °C, four Ura+ colonies from the YIR029W plate, two colonies from the YIR031C plate, and two colonies from the YIR034C plate were restreaked for single colonies on SC–Ura medium. Genomic DNA was prepared from a single colony growing for each of the eight Ura+ strains and primers designed to anneal within URA3, upstream and downstream of the ACT1 intron (For: AGCGGTTTGAAGCAGGCGG; Rev: CCAGTAGATAGGGAGCCCTTGC), were used to amplify the across the intron with a high fidelity polymerase (Phusion; NEB). Amplicons were subcloned by using the Zero Blunt TOPO PCR cloning Kit (Life Technologies; 45-0245), transformed into E. coli, and inserts were sequenced with M13F and M13R.
Supplementary Material
Acknowledgments
We thank Rodney Rothstein and Abram Gabriel for plasmids and Gabrielle Rieckhof for comments on the manuscript. This work was supported in part by National Science Foundation Grant MCB-0718846 and Defense Advanced Research Projects Agency Contract N66001-12-C-4020 (to J.D.B.). L.A.M. was funded by a postdoctoral fellowship from the Natural Sciences and Engineering Research Council of Canada.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1414399111/-/DCSupplemental.
References
- 1.Goffeau A, et al. Life with 6000 genes. Science. 1996;274(5287):546, 563–547. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- 2.Clarke L, Carbon J. Isolation of a yeast centromere and construction of functional small circular chromosomes. Nature. 1980;287(5782):504–509. doi: 10.1038/287504a0. [DOI] [PubMed] [Google Scholar]
- 3.Chan CS, Tye BK. Autonomously replicating sequences in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1980;77(11):6329–6333. doi: 10.1073/pnas.77.11.6329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kearsey S. Analysis of sequences conferring autonomous replication in baker’s yeast. EMBO J. 1983;2(9):1571–1575. doi: 10.1002/j.1460-2075.1983.tb01626.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shampay J, Szostak JW, Blackburn EH. DNA sequences of telomeres maintained in yeast. Nature. 1984;310(5973):154–157. doi: 10.1038/310154a0. [DOI] [PubMed] [Google Scholar]
- 6.Walmsley RW, Chan CS, Tye BK, Petes TD. Unusual DNA sequences associated with the ends of yeast chromosomes. Nature. 1984;310(5973):157–160. doi: 10.1038/310157a0. [DOI] [PubMed] [Google Scholar]
- 7.Ueda Y, et al. Large-scale genome reorganization in Saccharomyces cerevisiae through combinatorial loss of mini-chromosomes. J Biosci Bioeng. 2012;113(6):675–682. doi: 10.1016/j.jbiosc.2012.01.013. [DOI] [PubMed] [Google Scholar]
- 8.Murakami K, et al. Large scale deletions in the Saccharomyces cerevisiae genome create strains with altered regulation of carbon metabolism. Appl Microbiol Biotechnol. 2007;75(3):589–597. doi: 10.1007/s00253-007-0859-2. [DOI] [PubMed] [Google Scholar]
- 9.Reid RJ, et al. Chromosome-scale genetic mapping using a set of 16 conditionally stable Saccharomyces cerevisiae chromosomes. Genetics. 2008;180(4):1799–1808. doi: 10.1534/genetics.108.087999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Annaluru N, et al. Total synthesis of a functional designer eukaryotic chromosome. Science. 2014;344(6179):55–58. doi: 10.1126/science.1249252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dymond JS, et al. Synthetic chromosome arms function in yeast and generate phenotypic diversity by design. Nature. 2011;477(7365):471–476. doi: 10.1038/nature10403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Plessis A, Perrin A, Haber JE, Dujon B. Site-specific recombination determined by I-SceI, a mitochondrial group I intron-encoded endonuclease expressed in the yeast nucleus. Genetics. 1992;130(3):451–460. doi: 10.1093/genetics/130.3.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Colleaux L, D’Auriol L, Galibert F, Dujon B. Recognition and cleavage site of the intron-encoded omega transposase. Proc Natl Acad Sci USA. 1988;85(16):6022–6026. doi: 10.1073/pnas.85.16.6022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davis AP, Symington LS. RAD51-dependent break-induced replication in yeast. Mol Cell Biol. 2004;24(6):2344–2351. doi: 10.1128/MCB.24.6.2344-2351.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu X, Gabriel A. Patching broken chromosomes with extranuclear cellular DNA. Mol Cell. 1999;4(5):873–881. doi: 10.1016/s1097-2765(00)80397-4. [DOI] [PubMed] [Google Scholar]
- 16.Boeke JD, Trueheart J, Natsoulis G, Fink GR. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- 17.Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122(1):19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brachmann CB, et al. Designer deletion strains derived from Saccharomyces cerevisiae S288C: A useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14(2):115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 19.Surosky RT, Newlon CS, Tye BK. The mitotic stability of deletion derivatives of chromosome III in yeast. Proc Natl Acad Sci USA. 1986;83(2):414–418. doi: 10.1073/pnas.83.2.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blake WJ, et al. Pairwise selection assembly for sequence-independent construction of long-length DNA. Nucleic Acids Res. 2010;38(8):2594–2602. doi: 10.1093/nar/gkq123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yarger JG, Armilei G, Gorman MC. Transcription terminator-like element within a Saccharomyces cerevisiae promoter region. Mol Cell Biol. 1986;6(4):1095–1101. doi: 10.1128/mcb.6.4.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chamberlain JR, Lee Y, Lane WS, Engelke DR. Purification and characterization of the nuclear RNase P holoenzyme complex reveals extensive subunit overlap with RNase MRP. Genes Dev. 1998;12(11):1678–1690. doi: 10.1101/gad.12.11.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marvin MC, et al. Accumulation of noncoding RNA due to an RNase P defect in Saccharomyces cerevisiae. RNA. 2011;17(8):1441–1450. doi: 10.1261/rna.2737511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aparicio OM, Billington BL, Gottschling DE. Modifiers of position effect are shared between telomeric and silent mating-type loci in S. cerevisiae. Cell. 1991;66(6):1279–1287. doi: 10.1016/0092-8674(91)90049-5. [DOI] [PubMed] [Google Scholar]
- 25.Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403(6771):795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 26.Dani GM, Zakian VA. Mitotic and meiotic stability of linear plasmids in yeast. Proc Natl Acad Sci USA. 1983;80(11):3406–3410. doi: 10.1073/pnas.80.11.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murray AW, Szostak JW. Construction of artificial chromosomes in yeast. Nature. 1983;305(5931):189–193. doi: 10.1038/305189a0. [DOI] [PubMed] [Google Scholar]
- 28.Haber JE, Thorburn PC, Rogers D. Meiotic and mitotic behavior of dicentric chromosomes in Saccharomyces cerevisiae. Genetics. 1984;106(2):185–205. doi: 10.1093/genetics/106.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murray AW, Szostak JW. Construction and behavior of circularly permuted and telocentric chromosomes in Saccharomyces cerevisiae. Mol Cell Biol. 1986;6(9):3166–3172. doi: 10.1128/mcb.6.9.3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ferree PM. Mitotic misbehavior of a Drosophila melanogaster satellite in ring chromosomes: Insights into intragenomic conflict among heterochromatic sequences. Fly (Austin) 2014 doi: 10.4161/fly.29488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gisselsson D, et al. The structure and dynamics of ring chromosomes in human neoplastic and non-neoplastic cells. Hum Genet. 1999;104(4):315–325. doi: 10.1007/s004390050960. [DOI] [PubMed] [Google Scholar]
- 32.Schulz-Schaeffer Jr . Cytogenetics, Plants, Animals, Humans. Springer; New York: 1980. pp xiii. [Google Scholar]
- 33.Kazuki Y, et al. Refined human artificial chromosome vectors for gene therapy and animal transgenesis. Gene Ther. 2011;18(4):384–393. doi: 10.1038/gt.2010.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saffery R, et al. Construction of neocentromere-based human minichromosomes by telomere-associated chromosomal truncation. Proc Natl Acad Sci USA. 2001;98(10):5705–5710. doi: 10.1073/pnas.091468498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harrington JJ, Van Bokkelen G, Mays RW, Gustashaw K, Willard HF. Formation of de novo centromeres and construction of first-generation human artificial microchromosomes. Nat Genet. 1997;15(4):345–355. doi: 10.1038/ng0497-345. [DOI] [PubMed] [Google Scholar]
- 36.Ebersole TA, et al. Mammalian artificial chromosome formation from circular alphoid input DNA does not require telomere repeats. Hum Mol Genet. 2000;9(11):1623–1631. doi: 10.1093/hmg/9.11.1623. [DOI] [PubMed] [Google Scholar]
- 37.Ikeno M, et al. Construction of YAC-based mammalian artificial chromosomes. Nat Biotechnol. 1998;16(5):431–439. doi: 10.1038/nbt0598-431. [DOI] [PubMed] [Google Scholar]
- 38.Moralli D, Vagnarelli P, Bensi M, De Carli L, Raimondi E. Insertion of a loxP site in a size-reduced human accessory chromosome. Cytogenet Cell Genet. 2001;94(3-4):113–120. doi: 10.1159/000048801. [DOI] [PubMed] [Google Scholar]
- 39.Iida Y, et al. Human artificial chromosome with a conditional centromere for gene delivery and gene expression. DNA Res. 2010;17(5):293–301. doi: 10.1093/dnares/dsq020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marcaida MJ, Muñoz IG, Blanco FJ, Prieto J, Montoya G. Homing endonucleases: From basics to therapeutic applications. Cell Mol Life Sci. 2010;67(5):727–748. doi: 10.1007/s00018-009-0188-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim YG, Cha J, Chandrasegaran S. Hybrid restriction enzymes: Zinc finger fusions to Fok I cleavage domain. Proc Natl Acad Sci USA. 1996;93(3):1156–1160. doi: 10.1073/pnas.93.3.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bogdanove AJ, Voytas DF. TAL effectors: Customizable proteins for DNA targeting. Science. 2011;333(6051):1843–1846. doi: 10.1126/science.1204094. [DOI] [PubMed] [Google Scholar]
- 44.Ananiev EV, et al. Artificial chromosome formation in maize (Zea mays L.) Chromosoma. 2009;118(2):157–177. doi: 10.1007/s00412-008-0191-3. [DOI] [PubMed] [Google Scholar]
- 45.Gaeta RT, et al. In vivo modification of a maize engineered minichromosome. Chromosoma. 2013;122(3):221–232. doi: 10.1007/s00412-013-0403-3. [DOI] [PubMed] [Google Scholar]
- 46.Noël AJ, Wende W, Pingoud A. DNA recognition by the homing endonuclease PI-SceI involves a divalent metal ion cofactor-induced conformational change. J Biol Chem. 2004;279(8):6794–6804. doi: 10.1074/jbc.M311372200. [DOI] [PubMed] [Google Scholar]
- 47.Gibson DG, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6(5):343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 48.Lisby M, Mortensen UH, Rothstein R. Colocalization of multiple DNA double-strand breaks at a single Rad52 repair centre. Nat Cell Biol. 2003;5(6):572–577. doi: 10.1038/ncb997. [DOI] [PubMed] [Google Scholar]
- 49.Schwartz DC, Cantor CR. Separation of yeast chromosome-sized DNAs by pulsed field gradient gel electrophoresis. Cell. 1984;37(1):67–75. doi: 10.1016/0092-8674(84)90301-5. [DOI] [PubMed] [Google Scholar]
- 50.Teste MA, Duquenne M, François JM, Parrou JL. Validation of reference genes for quantitative expression analysis by real-time RT-PCR in Saccharomyces cerevisiae. BMC Mol Biol. 2009;10:99. doi: 10.1186/1471-2199-10-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Winzeler EA, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285(5429):901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.