Abstract
Circadian clocks are cell autonomous, transcriptionally-based, molecular mechanisms that confer the selective advantage of anticipation, enabling cells/organs to respond to environmental factors in a temporally appropriate manner. Critical to circadian clock function are two transcription factors, CLOCK and BMAL1. The purpose of the present study was to reveal novel physiologic functions of BMAL1 in the heart, as well as determine the pathologic consequences of chronic disruption of this circadian clock component. In order to address this goal, we generated cardiomyocyte-specific Bmal1 knockout (CBK) mice. Following validation of the CBK model, combined microarray and in silico analyses were performed, identifying 19 putative direct BMAL1 target genes, which included a number of metabolic (e.g., β-hydroxybutyrate dehydrogenase 1 [Bdh1]) and signaling (e.g., the p85α regulatory subunit of phosphatidylinositol 3-kinase [Pik3r1]) genes. Results from subsequent validation studies were consistent with regulation of Bdh1 and Pik3r1 by BMAL1, with predicted impairments in ketone body metabolism and signaling observed in CBK hearts. Furthermore, CBK hearts exhibited depressed glucose utilization, as well as a differential response to a physiologic metabolic stress (i.e., fasting). Consistent with BMAL1 influencing critical functions in the heart, echocardiographic, gravimetric, histologic, and molecular analyses revealed age-onset development of dilated cardiomyopathy in CBK mice, which was associated with a severe reduction in lifespan. Collectively, our studies reveal that BMAL1 influences metabolism, signaling, and contractile function of the heart.
Keywords: Chronobiology, circadian, metabolism, signaling, transcriptome
INTRODUCTION
Marked time-of-day-dependent oscillations in cardiovascular function are observed in mammals, including increased blood pressure, heart rate, and cardiac output during the awake period (Degaute, et al. 1994; Degaute, et al. 1991; Delp, et al. 1991; Richards, et al. 1986). Adverse cardiovascular events (e.g., myocardial infarction, sudden cardiac death) also exhibit a time-of-day-dependence in humans, with regards to their onset (Carson, et al. 2000; Muller, et al. 1989), while various behaviors known to adversely impact normal daily rhythms (e.g., night shift work) significantly increase cardiovascular disease risk (Harma and Ilmarinen 1999; Knutsson, et al. 1986; Koller 1983). A host of neurohumoral factors oscillate over the course of the day (e.g., autonomic, adrenergic, and sympathetic stimulation), many of which likely contribute towards daily rhythms in physiologic and pathologic cardiovascular parameters (Muller et al. 1989; Prinz, et al. 1979; Richards et al. 1986). However, it is becoming increasingly clear that cardiovascular components exhibit time-of-day-dependent oscillations in their responsiveness to extrinsic factors, which in turn likely influence outcomes following pathologic events (e.g., myocardial infarction) (Collins and Rodrigo 2010; Durgan, et al. 2010; Durgan, et al. 2006; Durgan, et al. 2011b; Durgan and Young 2010; Sachan, et al. 2011). Currently, the relative contribution of extrinsic versus intrinsic factors towards rhythms in cardiovascular function and dysfunction remains undefined. Recently, multiple studies by Scheer & Shea highlighted that time-of-day-dependent oscillations in epinephrine, norepinephrine, fibrinolytic activity, heart rate and blood pressure are mediated, at least in part, by an intrinsic circadian system in humans (Scheer, et al. 2010; Scheer, et al. 2011; Shea, et al. 2011). One intrinsic mechanism that has emerged as a significant modulator of the responsiveness of cardiovascular-relevant cells is the circadian clock (Rudic and Fulton 2009; Young 2009).
Circadian clocks are cell autonomous molecular mechanisms that confer the selective advantage of anticipation, facilitating responsiveness of a cell/organ to an extrinsic stimulus/stress in a temporally appropriate manner (Edery 2000). The circadian clock is a transcriptionally-based mechanism that has been identified in all mammalian cells investigated to date (Dibner, et al. 2010; Takahashi, et al. 2008). At the core of the mechanism are two transcription factors, CLOCK and BMAL1, which, upon heterodimerization, bind to E-boxes in the promoters of target genes (Gekakis, et al. 1998; Hogenesch, et al. 1998). Ubiquitous genetic disruption of circadian clock genes, as well as clock-controlled genes, often precipitates cardiovascular dysfunction. For example, heterozygous tau hamsters (that harbor a mutation in CK1ɛ, resulting in a 22 hour circadian clock) develop hypertrophic cardiomyopathy (Martino, et al. 2008). A cardiomyopathic phenotype is also observed when the clock output genes Dbp, Hlf, and Tef are genetically deleted (Wang, et al. 2010). Interestingly, targeted ablation of the core clock component Bmal1 in a ubiquitous manner results in severe cardiac dysfunction, which is associated with increased mortality (Kondratov, et al. 2006; Lefta, et al. 2012). Collectively, these observations provide evidence that functional circadian clocks are essential for normal cardiovascular function.
In an attempt to define roles for the cardiomyocyte-specific circadian clock, we have previously targeted the transcription factor CLOCK in a dominant negative manner. Using cardiomyocyte-specific Clock mutant (CCM) mice, we have highlighted critical roles for this transcription factor in both cardiac physiology (e.g., transcription, metabolism, contractile function) and pathophysiology (e.g., ischemia/reperfusion tolerance, hypertrophic growth) (Bray, et al. 2008; Durgan et al. 2010; Durgan et al. 2006; Durgan, et al. 2011a; Durgan et al. 2011b; Tsai, et al. 2010). In contrast, the role of BMAL1 in the cardiomyocyte remains undefined. To address this deficiency, we recently generated a cardiomyocyte-specific Bmal1 knockout (CBK) mouse model (Durgan et al. 2011b). Through combined microarray and in silico analyses, the present study identified 19 putative direct BMAL1 target genes in the hearts, many of which influence metabolism and signaling. Validation studies confirmed that Bdh1 and Pik3r1 are regulated by BMAL1, and that their chronic repression in CBK hearts is associated with impaired ketone body metabolism and signaling, respectively. In addition, CBK hearts exhibit abnormalities in glucose utilization in the fed state, as well as abnormal metabolic responses to acute fasting (i.e., a physiologic metabolic stress). CBK mice also exhibit an age-onset cardiomyopathy, which is associated with early mortality. Collectively, these observations highlight critical roles of BMAL1 in the heart.
MATERIALS AND METHODS
Mice
The present study utilized: 1) CBK (BMAL1flox/flox/α-MHC-CRE+/−) and littermate controls (BMAL1flox/flox/α-MHC-CRE−/−) on the C57Bl/6J background; 2) α-MHC-CRE (α-MHC-CRE+/−) and littermate wild-types (α-MHC-CRE−/−) on the C57Bl/6J background; and 3) CCM (α-MHC-dnCLOCK+/−) and littermate wild-types (α-MHC-dnCLOCK−/−) on the FVB/N background; both CBK and CCM mice have been described previously (Durgan et al. 2006; Durgan et al. 2011b). All experimental mice were male, and were housed at the Center for Comparative Medicine at the University of Alabama at Birmingham (UAB), under temperature-, humidity-, and light- controlled conditions. A strict 12-hour light/12-hour dark cycle regime was enforced (lights on at 6AM; zeitgeber time [ZT] 0); the light/dark cycle was maintained throughout these studies, facilitating elucidation of the potential roles for cardiomyocyte circadian clock components under physiological conditions. As such, diurnal variations were investigated in mice (as opposed to circadian rhythms). Mice received food and water ad libitum, unless otherwise stated. Mice were housed in standard micro-isolator cages, with the exception of the whole body behavioral/metabolic analyses and fasting studies, during which time mice were housed within CLAMS (Comprehensive Laboratory Animal Monitoring System) and wire bottom cages, respectively. All animal experiments were approved by the Institutional Animal Care and Use Committee of the University of Alabama at Birmingham.
Whole body behavioral and metabolic monitoring
Twenty-four hour patterns of physical activity, food intake, and energy expenditure (indirect calorimetry) were measured in mice using a CLAMS (Columbus Instruments Inc., Columbus, OH), as described previously (Bray, et al. 2012).
Echocardiographic analysis
Left ventricular function was determined in vivo through echocardiography, at an established UAB core facility. Mice were anesthetized using 1.5% isofluorane with 95% oxygen; heart rate and body temperature were kept constant. M-mode echocardiography was performed using the Visualsonics imaging/processing system as described previously (Bray et al. 2008). Control versus CBK assessments were performed in 2011–2012 on a Vevo 2100 (Toronto, Canada), whereas wild-type versus α-MHC-Cre assessments were performed in 2013 by a different technician on a Vevo 7700 (Toronto, Canada). As such, direct comparisons are possible only within each distinct sub-study.
Humoral factor analysis
Plasma glucose, triglyceride, non-esterified fatty acids (NEFA), insulin, leptin, and adiponectin concentrations were measured with commercially available kits (Stanbio Laboratory, Boerne, TX; EMD Millipore, Billerica, MA; TOSOH Bioscience, San Francisco, CA; Wako Chemicals, Richmond, VA) using a Sirrus Clinical Chemistry Analyzer (Stanbio Laboratory, Boerne, TX).
Histologic Assessment
Cross sections from the medial heart were taken immediately upon removal of heart and flash-frozen in OCT. Laminin staining was utilized for measurement of myocyte cross-sectional area; at least 100 myocytes were assessed per heart using ImagePro Plus software (Media Cybernetics, Inc., Rockville, MD), as described previously (Durgan et al. 2011b). Picrosirius Red staining of collagen fibers was utilized for semi-quantitative measurement of left ventricular fibrosis, using ImagePro Plus software (Media Cybernetics, Inc., Rockville, MD), as described previously (Durgan et al. 2011b).
Quantitative RT-PCR
RNA was extracted from hearts using standard procedures (Chomczynski and Sacchi 1987). Candidate gene expression analysis was performed by quantitative RT-PCR, using previously described methods (Gibson, et al. 1996; Heid, et al. 1996). For quantitative RT-PCR, specific Taqman assays were designed for each gene from mouse sequences available in GenBank. Primer and probe sequences utilized are listed in Supplemental Table 1. All quantitative RT-PCR data are presented as fold change from littermate control.
Microarray Analysis
Microarray analysis was performed using the mouse Ref-8 BeadChips and the BeadStation System (Illumina, Inc., San Diego, CA) according to the manufacturer’s guidelines, as described previously (Bray et al. 2008). Ventricular tissue was collected from CBK and littermate controls every three hours for a period of 24 hours (n=4 samples per time point; 64 samples total), and RNA was extracted. Genes/transcripts were defined as being significantly expressed above background when the 75th percentile of each gene's detection score met or exceeded 0.9. The expression data were normalized within centiles of the distribution of gene expression values. Time-of-day-independent differentially expressed genes between CBK and littermate control samples were identified by 2-way ANOVA (i.e., genotype main effect). Time-of-day-dependent differential expression was performed by initially identify genes exhibiting significant oscillations in control samples using cosinor analysis, as described previously (Bray et al. 2008); those genes that oscillate in control samples and either do not oscillate in CBK samples or oscillated with a decreased amplitude (assessed by two-way ANOVA) were identified as being differentially expressed in a time-of-day-dependent manner. Normalized data have been submitted to GEO archive and are available at http://www.ncbi.nlm.nih.gov/geo/ (GSE43073).
To identify a comprehensive set of putative direct BMAL1 gene targets, we performed a systems level bioinformatics analysis. To do this we compared the most highly induced and repressed genes identified from our CBK microarray data (using fold-change cut-offs of 2.0 and 0.5, respectively, for the time-of-day-independent differentially expressed genes; Supplemental Table 3) against a genome-wide map of murine BMAL1 binding targets identified through functional assays by chromatin immunoprecipitation (ChIP) coupled with deep sequencing (Rey, et al. 2011). In addition, we performed an in silico search within the 5’-regulatory regions of identified genes for putative circadian E-box sequences (CANNTG), the canonical palindromic circadian E-box motif (CACGTG), and E-box like sequences (E’-box) (CACGTT), as described previously (Tsimakouridze, et al. 2012). The 1,000 base pair regions upstream of the coding sequences were retrieved from UCSC Genome Bioinformatics (http://genome.ucsc.edu/cgi-bin/hgTables). Moreover, we interrogated the circadian section of the mammalian Promoter/Enhancer DataBase (PEDB, http://promoter.cdb.riken.jp/circadian_home.html) (Kumaki, et al. 2008) for genes containing high scoring putative E-boxes conserved in the noncoding region of both the human and mouse.
Western blotting
Qualitative analysis of protein expression was performed using standard Western blotting techniques, as described previously (Durgan et al. 2011a). Lysates (10ug) were separated on a 7.5% bis-acrylamide gel by sodium dodecyl sulfate–polyacrylamide gel electrophoresis, transferred to a PVDF membrane and probed with anti-BMAL1 (Millipore; catalogue number AB2298), anti-BDH1 (Abcam; catalogue number AB93931), anti-PIK3R1 (Abcam; catalogue number AB22653), anti-AKT (Millipore; catalogue number 05–591), anti-phospho-Thr308-AKT (Cell Signaling; catalogue number CS-9275), anti-GSK3β (Santa Cruz; catalogue number sc-7291), anti-phospho-Ser9-GSK3β (Cell Signaling; catalogue number CS-9336), or GAPDH (Cell Signaling; catalogue number CS-2118) antibodies. Membranes were incubated with either goat anti-rabbit or anti-mouse horse radish peroxidase-conjugated secondary antibody (Santa Cruz; catalogue numbers sc-2004 and sc-2005, respectively). Bands were visualized with Immunstar Western C detection kit (Biorad) on X-ray film, scanned and quantified using Scion Image version 4.0.3.2, and normalized to Calsequestrin (Abcam; catalogue number AB3516). For phosphorylation to total comparisons, membranes were incubated with either AlexaFluor 680-conjugated secondary donkey anti-rabbit (Invitrogen; catalogue number A10043) or IRDYE800-conjugated secondary donkey anti-mouse (LI-COR; catalogue number 926–32212). Bands were visualized with the LI-COR Odyssey CLx Infrared Imager and quantified using Image Studio version 4.0.21 software.
Enzymatic activities
β-Hydroxybutyrate dehydrogenase (BDH) and citrate synthase (negative control) activities were measured in freshly isolated hearts using previously described assays (Grinblat, et al. 1986; Srere 1969). BDH activity was assessed by following the conversion rate of NAD+ to NADH in the presence of β-hydroxybutyrate. Citrate synthase activity was assessed by following the conversion rate of 5,5'-dithiobis-(2-nitrobenzoic acid) to 2-nitro-5-thiobenzoate in the presence of oxaloacetate and acetyl-CoA. Enzymatic activities were normalized to protein content (determined by BCA assay kit; Pierce).
Working mouse heart perfusions
Myocardial substrate utilization was measured ex vivo through isolated working mouse heart perfusions, as described previously (Bray et al. 2008; Durgan et al. 2011a; Tsai et al. 2010; Tsai, et al. 2013). All hearts were perfused in the working mode (non-recirculating manner) for 40 minutes with a preload of 12.5mmHg and an afterload of 50mmHg. Standard Krebs–Henseleit buffer was supplemented with 8mM glucose, 0.4mM oleate conjugated to 3% BSA (fraction V, fatty acid-free; dialyzed), 10µU/ml insulin (basal/fasting concentration), 2mM β-hydroxybutyrate, 0.2mM acetoacetate, 0.05mM L-carnitine, and 0.13mM glycerol. Metabolic flux were assessed through the use of distinct radiolabeled tracers: 1) [U-14C]-β-hydroxybutyrate (0.04mCi/L; β-hydroxybutyrate oxidation); 2) [U-14C]-glucose (0.12mCi/L; glycolysis, glucose oxidation); and 3) [9,10-3H]-oleate (0.067mCi/L;β-oxidation). Measures of cardiac metabolism (i.e., β-hydroxybutyrate oxidation and oxygen consumption) and function (i.e., cardiac power and rate pressure product) were determined as described previously (Bray et al. 2008; Durgan et al. 2011a; Tsai et al. 2010; Tsai et al. 2013). At the end of the perfusion period, hearts were snap-frozen in liquid nitrogen and stored at −80°C prior to analysis. Data are presented as steady state values (i.e., values during the last 10 minutes of the perfusion protocol).
Isolated Adult Cardiomyocyte Studies
Viable adult mouse and rat cardiomyocytes were isolated as described previously (Durgan, et al. 2005; Marsh, et al. 2011; Shan, et al. 2008). Adult mouse cardiomyocytes were lysed immediately, for subsequent Western blot analysis. Adult rat cardiomyocytes were cultured overnight (equilibration period) followed by challenge with 50% serum for a period of 2 hours (i.e., serum shock); cells were terminated at 4 hour intervals thereafter. We have previously reported circadian clock gene oscillations in these samples (Durgan et al. 2005).
DNA Precipitation Assays
DNA precipitation assays were performed as described previously (Yang and Chow 2012). Briefly, COS cells were transfected with expression vectors for Bmal1 and Clock, and subsequently harvested in Trition-lysis buffer (20 mM Tris [pH 7.4], 134 mM NaCl, 2 mM EDTA, 25 mM β-glycerophosphate, 2 mM NaPPi, 10% glycerol, 1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride, 1 mM benzamidine, and 10 µg/ml leupeptin). Double-stranded biotinylated oligonucleotides (−573 bp E box, 5’-Biotin-CCTGAGTGAGACACTTGGCAGAGAGCACCTGTACTCTGCGTT-3’; −619 bp E box, 5’-Biotin- GAGATTACATCACCTTCCTTAAAGCAGCTAGCAATGAGATG-3’; −1828 bp E box, 5’-Biotin-GGGCTGGGACCACTTGGTAGAAGAACAGCTTCTTTGAA-3’) were incubated with BMAL1 and CLOCK cell extracts for 12 h before precipitation with 20 µl of streptavidin-Sepharose for 2 h. After three washes in Triton-lysis buffer, precipitated DNA and its associated proteins were separated by SDS-PAGE, and an immunoblot was performed to detect BMAL1.
Luciferase Reporter Assays
The Pik3r1 promoter was amplified from mouse genomic DNA and subcloned into the pGL3 basic luciferase reporter plasmid using MluI and XhoI sites. Expression vectors for Bmal1 and Clock (50 ng) were co-transfected with the Pik3r1 luciferase reporter plasmids (100 ng) and control plasmid pRSV β-galactosidase (25 ng) into COS cells. Transfected cells were serum shocked (with 20% serum) for 2 hr, and subsequently maintained in media supplemented with 2% serum for 24 hr before harvest. The data are presented as relative luciferase activity, calculated as the ratio of luciferase activity to the activity of β-galactosidase.
Statistical analysis
Statistical analyses were performed using student t-tests, two-way ANOVA, and cosinor analysis, as described previously (Bray et al. 2008; Bray et al. 2012). Briefly, Excel 2010 was used to compare CBK/CCM to wild-type littermate controls (only when two experimental groups were investigated). Stata version IC10.0 (Stata Corp., San Antonio, TX) or IBM SPSS Statistics 22.0 (IBM, Inc., Armonk, NY) was used to perform two-way ANOVA (with repeated measures, when appropriate) to investigate main effects of genotype, time, feeding status, and/or age (indicated in Figures), followed by Bonferroni post hoc analyses for pair-wise comparisons (only when a significant interaction was observed). Assumptions of normality and homogeneity of variances were met and verified with Shapiro Wilk and Levene’s tests. For repeated measures ANOVA, sphericity (for repeated measures) was examined with Mauchly’s test and degrees of freedom were modified using the Greenhouse-Geisser adjustment when appropriate. Survival data were analyzed by using a Kaplan–Meier survival analysis with a log rank method of statistics. Cosinor analysis was conducted using nonlinear regression with Stata version IC10.0 and SPSS 22. The cosinor analyses were performed based on the assumption that the free-running period is fixed at 24 hours. Data were considered rhythmic if the p-value of the R2 of the linearized cosinor function of f(t) = M + Cos(2πt/24) + Sin(2πt/24) was less than 0.05 (indicated in Supplemental Table 2). In cases in which both comparison groups significantly fit the cosine function, individual parameter estimates for mesor, amplitude, and phase were compared using t-tests determined from the pooled variances. In all analyses, the null hypothesis of no model effects was rejected at p<0.05.
RESULTS
Cardiomyocyte-specific Bmal1 ablation selectively impacts the circadian clock in the heart
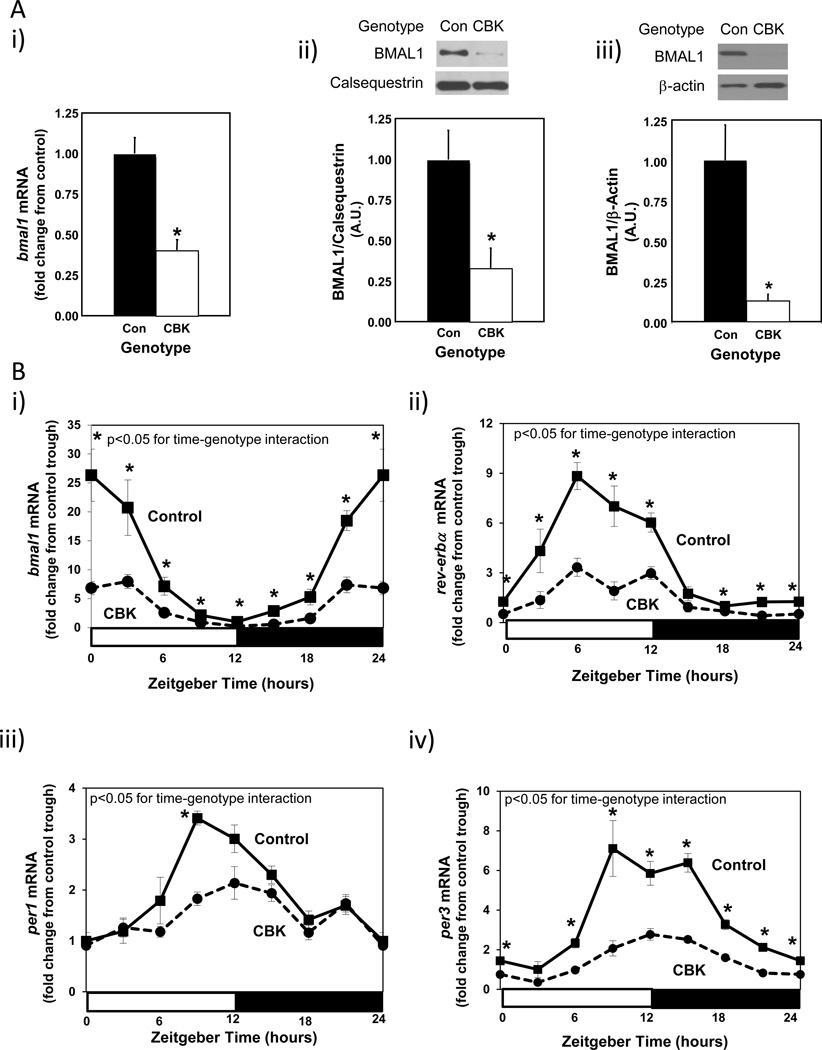
In order to initially validate the CBK model, Bmal1 gene and protein expression were assessed. Figure 1Ai-ii shows decreased bmal1 gene (60%) and BMAL1 protein (67%) expression in intact hearts isolated from 12 week old CBK mice at ZT6 (relative to littermate controls). Importantly, BMAL1 protein was almost undetectable in cardiomyocytes isolated from CBK hearts (Figure 1Aiii), suggesting that residual expression in intact CBK hearts is due primarily to the presence of non-cardiomyocyte cell types (e.g., vascular smooth muscle cells, endothelial cells, fibroblasts). Decreased BMAL1 expression in CBK hearts was associated with a dramatic attenuation in time-of-day-dependent rhythmic oscillations for core circadian clock components (i.e., bmal1, rev-erbα, per1, and per3) and output (i.e., dbp and e4bp4) genes; in the case of clock output genes (considered a marker of clock function), the amplitude of dbp mRNA oscillations were decreased by 73% in CBK hearts, while those of e4bp4 mRNA were completely abolished (Figure 1B and Supplemental Table 2). In contrast, circadian clock function in extra-cardiac peripheral tissues remained intact as expected, as evidenced by identical bmal1 and dbp mRNA oscillations in skeletal muscle (gastrocnemius), liver, kidney, and adipose (epididymal fat) isolated from CBK versus littermate control mice (Supplemental Figure 1A and Supplemental Table 2). Similarly, no significant differences in humoral (i.e., glucose, triglyceride, leptin, adiponectin; Supplemental Figure 1B and Supplemental Table 2) or whole body behavioral (i.e., physical activity and food intake; Supplemental Figure 1C and Supplemental Table 2) factor diurnal variations were observed between CBK and littermate control mice. Whole body energy expenditure also did not significantly differ between CBK and littermate controls (Supplemental Figure 1D and Supplemental Table 2). Collectively, these data are consistent with the cardiac-specific nature of the CBK model.
Figure 1.
Decreased bmal1 mRNA levels in intact CBK hearts (Ai), decreased BMAL1 protein levels in intact CBK hearts (Aii), and in cardiomyocytes isolated from CBK hearts (Aiii), as well as attenuated time-of-day-dependent gene expression oscillations in circadian clock components (bmal1, rev-erbα, per1, and per3) and clock output genes (dbp and e4bp4) in CBK hearts (B). Mice were housed in a 12-hr light:12-hr dark cycle (lights on at ZT0). Hearts were isolated from 12 week old male CBK mice and age-matched littermate controls. For data presented in A, hearts were isolated at ZT6. Note that ZT0 and ZT24 are identical data. Values are expressed as mean ± SEM (n=4–12; sample size range varies dependent on the parameter investigated). *, p<0.05 for CBK versus littermate control at a distinct ZT (post-hoc pair-wise comparison).
Identification of putative direct BMAL1 target genes in the heart
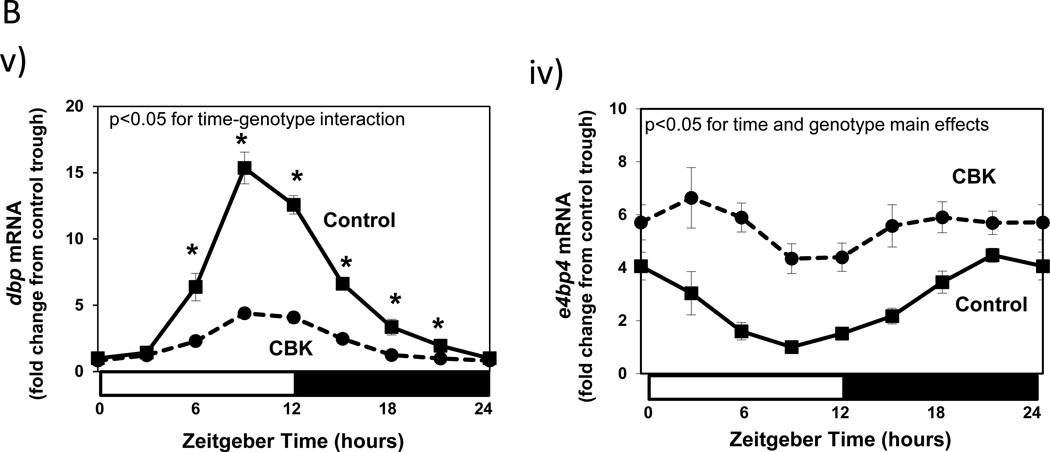
Given that BMAL1 is a transcription factor, we rationalized that investigating the impact of cardiomyocyte-specific genetic ablation of this protein on the cardiac transcriptome might identify novel roles for BMAL1 in the heart. Accordingly, ventricles were collected from 12 week old CBK and littermate control mice at 3 hours intervals over the course of the 24-hour light/dark cycle. Following RNA isolation, an unbiased microarray approach was taken. Microarray data were analyzed in two manners. First, genes chronically induced or repressed in a time-of-day-independent manner in CBK hearts, relative to their littermate controls, were identified through ANOVA; a total of 2037 genes were identified through this analysis, of which 1002 were induced while 1035 were repressed in CBK hearts (Supplemental Table 3). Second, genes that significantly oscillate in a time-of-day-dependent manner (with a periodicity of approximately 24 hours) in control hearts, and that the amplitudes of these oscillations were significantly attenuated/abolished in CBK hearts, were identified through cosinor analysis; a total of 1267 genes were identified by this analysis (Supplemental Table 4). Figure 2 illustrates attenuation (i.e., decreased amplitude) and/or abolition of gene expression oscillations in CBK, relative to littermate control, hearts. It is interesting to note that CBK hearts appear to exhibit a similar temporal suspension as previously described in CCM hearts (Bray et al. 2008), wherein genes that oscillate in control hearts are generally suspended at the beginning of the light/sleep phase in CBK hearts (Figure 2). Collectively, these observations suggest that genetic deletion of BMAL1 in the cardiomyocyte results in marked alterations in the cardiac transcriptome, in both time-of-day- independent and dependent manners.
Figure 2.
Heat map illustrating genes that oscillate in a time-of-day-dependent manner in control hearts (periodicity of 24 hours; i), while oscillations of these genes are significantly attenuated/abolished in CBK hearts (ii). Green represents low level of expression; red represents high level of expression. Data are taken from Supplemental Table 5 (n=4).
Next, in an attempt to identify putative candidate genes for which BMAL1 directly regulates in the heart, we systematically compared our microarray gene expression data to systems-level transcriptional regulatory databases (Kumaki et al. 2008; Rey et al. 2011; Tsimakouridze et al. 2012). A gene was defined as a putative direct BMAL1 target gene if it: 1) was highly induced (fold change of 2 or more) or repressed (fold change of 0.5 or less) in CBK hearts; 2) the 5’-regulatory region of the gene contained a putative BMAL1/CLOCK binding site (i.e., E-box sequences [CANNTG], the canonical palindromic circadian E-box motif [CACGTG], or E-box like sequences [CACGTT]); and 3) the gene was included in a genome-wide map of BMAL1 binding targets in mouse livers identified through functional assays by chromatin immunoprecipitation coupled with deep sequencing (Rey et al. 2011). It should be noted that the latter step likely underestimates the number of BMAL1 target genes in the heart, as previous microarray studies have suggested a high level of tissue specificity in time-of-day-dependent regulation of gene expression (Storch, et al. 2002). This analysis revealed a total of 19 putative direct BMAL1 targets genes (Table 1), 16 of which were previously identified as CLOCK regulated genes through transcriptome analysis of CCM hearts (Bray et al. 2008); of these 16 genes, 15 (i.e., 94%) were differentially expressed in CCM and CBK hearts in an identical manner. Among the 19 putative BMAL1 target genes, the expression of 9 oscillate significantly in wild-type hearts with a periodicity of approximately 24 hours (while 10 do not; Table 1). Based on known biologic function, these 19 genes appeared to fall into three main categories, namely clock function, metabolism, and signaling (Table 1).
Table 1.
Putative direct BMAL1 target genes in the heart. Table lists the most highly induced and repressed genes (fold change cut-offs of 2.0 and 0.5, respectively) that were identified as putative direct BMAL1 target genes (see text). Differential expression of these genes in CCM versus littermate control hearts is included, based on previously published microarray analyses [30]. Not determined is defined as a gene that was not present in the previously published microarray study [30].
| Gene Symbol | Gene Name | Accession Number | CBK versus Control (Fold Change) |
CCM versus Control (Fold Change) |
Role/Function |
|---|---|---|---|---|---|
| Expression does not oscillate significantly in control hearts (with a periodicity of approximately 24 hours) | |||||
| Aldob | Aldolase B, fructose-bisphosphate | NM_144903.2 | Repressed (0.06) | Repressed (0.49) | Carbohydrate Metabolism |
| Bdh1 | 3-Hydroxybutyrate dehydrogenase, type 1 |
NM_175177.3 | Repressed (0.13) | Repressed (0.07) | Ketone Body Metabolism |
| Acsm5 | Acyl-CoA synthetase medium-chain family member 5 | NM_178758.3 | Repressed (0.16) | Not Determined | Fatty Acid Metabolism |
| Gm129 | Gene model 129 | NM_001033302.1 | Repressed (0.17) | Not Determined | Unknown |
| Prlr | Prolactin receptor | NM_011169.3 | Repressed (0.28) | Repressed (0.22) | Signaling |
| Dcakd | Dephospho-CoA kinase domain containing | NM_026551.3 | Repressed (0.43) | Not Determined | Coenzyme A Biosynthesis |
| Coq10b | Coenzyme Q10 homolog B | NM_001039710.1 | Repressed (0.45) | Not Determined | Respiratory Chain Function |
| Lrrc4b | Leucine rich repeat containing 4B | NM_198250.1 | Repressed (0.47) | Not Significant | Excitatory Synapse Formation |
| Per3 | Period 3 | NM_011067 | Repressed (0.50) | Repressed (0.65) | Clock Component |
| Mal | Myelin and lymphocyte protein | NM_010762.4 | Induced (2.02) | Induced (1.43) | Signaling/Myelin Biosynthesis |
| Expression oscillates significantly in control hearts (with a periodicity of approximately 24 hours) | |||||
| Nr1d2 | Nuclear receptor subfamily 1, group D, member 2 |
NM_011584.2 | Repressed (0.26) | Repressed (0.00) | Clock Component |
| Tef | Thyrotrophic embryonic factor | NM_153484.2 | Repressed (0.17) | Repressed (0.43) | Clock Output |
| Dbp | Albumin D-box binding protein | NM_016974.1 | Repressed (0.36) | Repressed (0.36) | Clock Output |
| Nampt | Nicotinamide phosphoribosyltransferase |
NM_021524.1 | Repressed (0.23) | Repressed (0.46) | NAD Metabolism |
| Dgat2 | Diacylglycerol O-acyltransferase 2 | NM_026384.3 | Repressed (0.35) | Repressed (0.55) | Lipid Metabolism |
| Usp2 | Ubiquitin specific peptidase 2 | NM_198091.2 | Repressed (0.48) | Repressed (0.57) | Protein Turnover |
| Pik3r1 | Phosphoinositide-3-kinase, regulatory subunit 1 |
NM_001024955.1 | Repressed (0.50) | Repressed (0.80) | Signaling |
| NFil3 | Nuclear factor, interleukin 3 regulated |
NM_017373.3 | Induced (2.34) | Induced (1.46) | Clock Output |
| 2310022B05Rik | Uncharacterized | NM_175149.3 | Induced (2.18) | Induced (1.45) | Unknown |
BMAL1 regulates Bdh1 in a time-of-day-independent manner
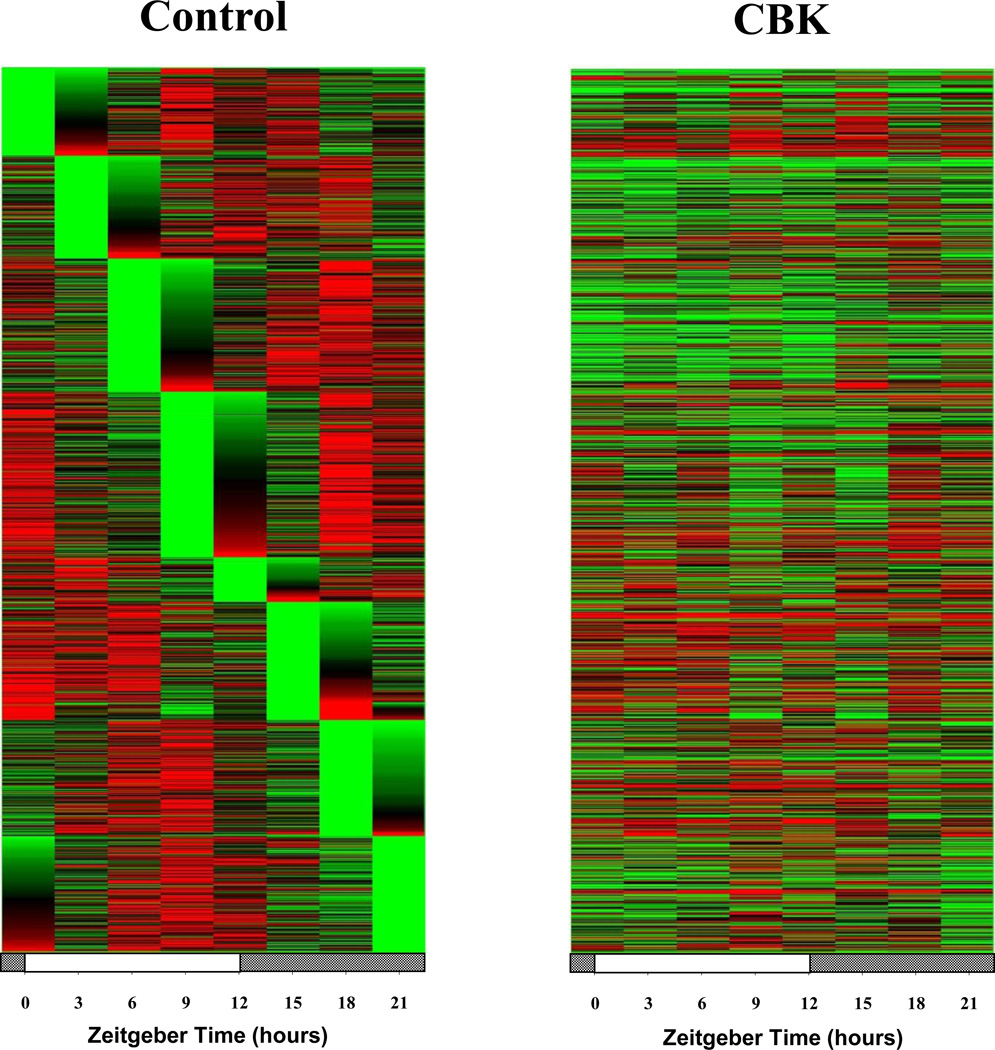
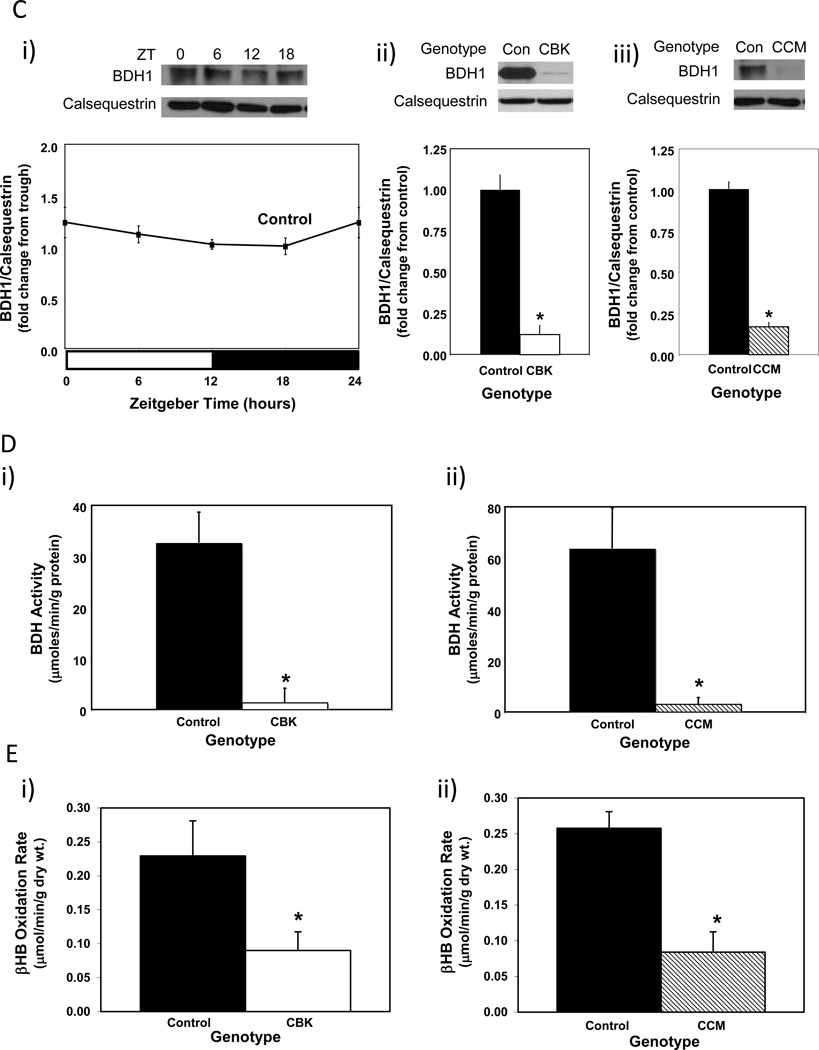
We next sought to validate our combined microarray and in silico analyses. Through the use of CCM mice, we have previously reported that two of the putative direct BMAL1 target genes listed in Table 1 that are known to influence metabolism (namely Dgat2 and Nampt) are regulated by the cardiomyocyte circadian clock (Durgan et al. 2011a; Tsai et al. 2010). One metabolism-related gene identified in Table 2 that has not been investigated previously, in term of circadian clock regulation, encodes for β-hydroxybutyrate dehydrogenase 1 (Bdh1). RT-PCR validation confirmed that bdh1 mRNA levels in CBK hearts (relative to wild-type littermates) were decreased (83%) in a time-of-day-independent manner (Figure 3Ai-ii and Supplemental Table 2). Decreased bdh1 expression was not due to the expression of Cre alone, as α-MHC-Cre mouse hearts exhibit normal bdh1 mRNA levels (1.00 ± 0.17 versus 0.94 ± 0.26 A.U. for α-MHC-Cre and wild-type littermate hearts isolated at ZT15, respectively; p>0.05). bdh1 mRNA levels are also significantly reduced (94%) in CCM hearts in a time-of-day-independent manner (Figure 3Aiii and Supplemental Table 2). Consistent with the lack of significant oscillation of bdh1 mRNA levels in control hearts, this transcript did not significantly oscillate in synchronized adult rat cardiomyocytes (Figure 3B; p>0.05 for cosine analysis). BDH1 protein levels also did not oscillate in a time-of-day-dependent manner in control hearts (although BDH1 protein levels are lower in both CBK (87%) and CCM (85%) hearts; Figure 3C and Supplemental Table 2). Interestingly, decreased BDH1 protein levels in CBK and CCM hearts are associated with decreased BDH enzymatic activity (95% and 91% decrease, respectively; Figure 3D), in the absence of genotype-dependent alterations in citrate synthase activity (Supplemental Figure 2A). Similarly, β-hydroxybutyrate oxidation rates were decreased in ex vivo perfused CBK and CCM hearts (61% and 67% decrease, respectively; Figure 3E), in the absence of genotype-dependent alterations in myocardial oxygen consumption or contractile function (Supplemental Figure 2B). Note that BDH enzymatic activity and β-hydroxybutyrate oxidation rates were assessed at only one time of the day (ZT6), as BDH1 protein levels do not oscillate. Collectively, our findings suggest that Bdh1 is likely influenced by both BMAL1 and CLOCK, in a time-of-day-independent manner.
Table 2.
Gravimetric, histologic, and transcriptional measures in both 12 and 36 week of age old CBK and littermate control mice. All parameters were measured at ZT6. Gene expression was determined in isolated ventricles.
| Parameter | 12 Weeks Old | 36 Weeks Old | ||
|---|---|---|---|---|
| Control | CBK | Control | CBK | |
| Body Weight (g) | 28.0 ± 0.6 | 27.7 ± 0.5 | 27.2 ± 2.8 | 24.51 ± 1.06# |
| BVW (mg) | 124.6 ± 6.5 | 133.5 ± 5.3 | 119.6 ± 8.2 | 162.3 ± 15.0#* |
| TL (mm) | 17.3 ± 0.1 | 17.4 ± 0.1 | 17.7 ± 0.1# | 17.6 ± 0.3 |
| BVW/Body Weight (x1000) | 4.4 ± 0.2 | 4.8 ± 0.2 | 4.5 ± 0.3 | 6.8 ± 0.9#* |
| BVW/TL (mg/mm) | 7.2 ± 0.4 | 7.7 ± 0.3 | 6.7 ± 0.4 | 9.1 ± 0.9* |
| Lung Weight (mg) | 143.7 ± 2.0 | 148.3 ± 1.0 | 168.9 ± 4.5# | 207.6 ± 19.8#* |
| Myocyte X-Sectional Area (mm2) |
423.2± 37.4 | 467.5 ± 23.6 | 370.04 ± 40.5 | 450.2 ± 45.2 |
| Fibrosis (A.U.) | 1.00 ± 0.26 | 1.00 ± 0.22 | 1.04 ± 0.13 | 1.83 ± 0.33#* |
| col3a1 mRNA (A.U.) | 1.00 ± 0.03 | 1.63 ± 0.20* | 0.51 ± 0.08# | 2.23 ± 0.91* |
| col4a1 mRNA (A.U.) | 1.00 ± 0.08 | 1.35 ± 0.09* | 0.67 ± 0.05# | 2.35 ± 0.83* |
| anf mRNA (A.U.) | 1.00 ± 0.35 | 1.72 ± 0.68 | 0.47 ± 0.25 | 3.78 ± 0.76* |
| mhcb mRNA (A.U.) | 1.00 ± 0.09 | 0.82 ± 0.12 | 3.21 ± 0.91 | 34.86 ± 7.29#* |
| serca2a mRNA (A.U.) | 1.00 ± 0.07 | 0.88 ± 0.06 | 0.61 ± 0.10 | 0.34 ± 0.03#* |
| cyclophilin mRNA (A.U.) | 1.00 ± 0.06 | 0.89 ± 0.03 | 1.01 ± 0.12 | 1.36 ± 0.14# |
BVW, biventricular weight; TL, tibia length.
Values are expressed as mean ± SEM (n=5–7).
p<0.05 for 12 weeks old versus 36 weeks old within a genotype.
p<0.05 for CBK compared to age-matched control.
Figure 3.
Alterations in bdh1 mRNA in CBK (microarray [i], RT-PCR [ii]) and CCM (RT-PCR [iii]) hearts, compared to littermate controls (A). Lack of bdh1 mRNA oscillations in serum shocked adult rat cardiomyocytes (i.e., challenged with 50% serum for 2 hours [shaded area]; B). Lack of diurnal variations in BDH1 protein levels in control hearts (Ci). Decreased BDH1 protein levels in CBK (Cii) and CCM (Ciii) hearts at ZT6. Decreased BDH activity in CBK (Di) and CCM (Dii) hearts at ZT6. Decreased β-hydroxybutyrate oxidation in ex vivo perfused CBK (Ei) and CCM (Eii) hearts at ZT6. Mice were housed in a 12-hr light:12-hr dark cycle (lights on at ZT0). Hearts were isolated from 12 week old male CBK, CCM, and age-matched littermate controls. Note that ZT0 and ZT24 are identical data. Values are expressed as mean ± SEM (n=3–7; sample size range varies dependent on the parameter investigated). *, p<0.05 for CBK/CCM versus littermate control at a distinct ZT (post-hoc pair-wise comparison).
BMAL1 regulates Pik3r1 in a time-of-day-dependent manner
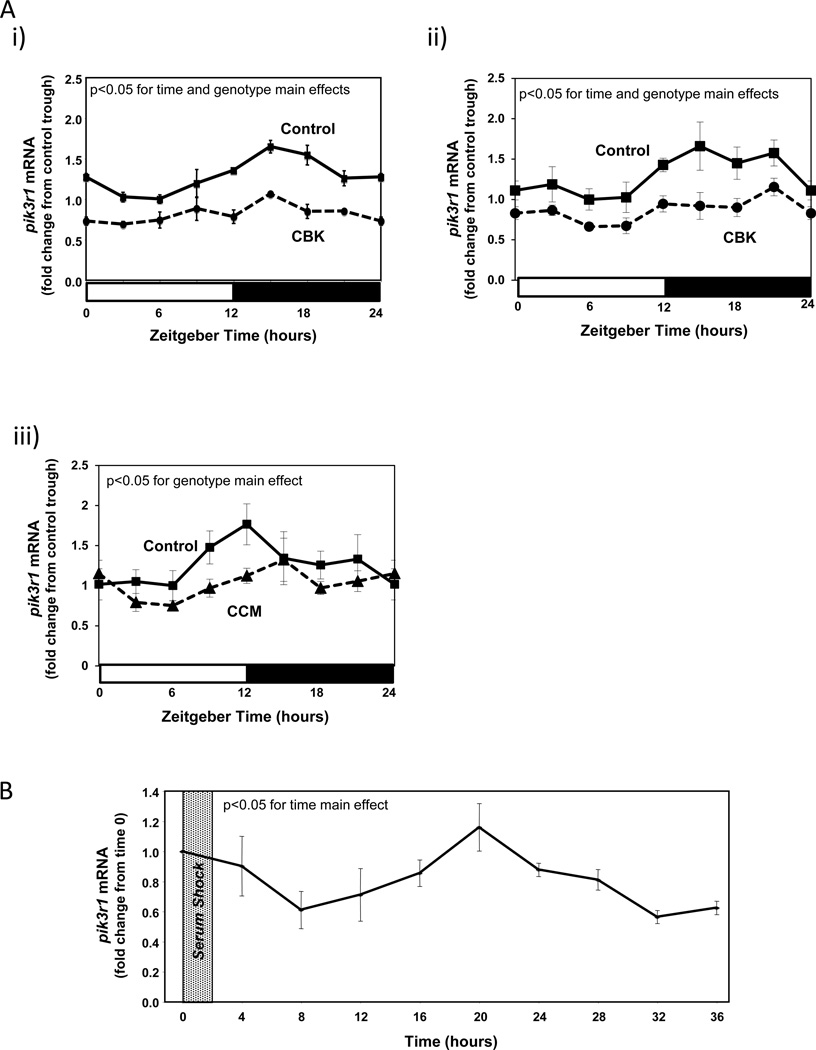
An additional putative BMAL1 target gene identified in Table 1 is Pik3r1, encoding for the p85α regulatory subunit of phosphatidylinositol 3-kinase (PI3K). This gene is of potential interest because: 1) it is a component of the insulin signaling cascade, which impacts both cardiac metabolism and contractile function; 2) insulin sensitivity had been shown to be under circadian control in extra-cardiac tissues; and 3) our previous studies suggest that components downstream of PI3K in the insulin signaling cascade, namely AKT and GSK3β, are regulated by the cardiomyocyte circadian clock (Abel 2004; Durgan et al. 2010; Leighton, et al. 1988). Quantitative RT-PCR confirmed that pik3r1 mRNA oscillates in control hearts in a time-of-day-dependent manner (with a periodicity of approximately 24 hours), and that this oscillation is attenuated/abolished in CBK and CCM hearts (Figure 4A and Supplemental Table 2). Decreased pik3r1 mRNA levels in CBK hearts does not appear to be due to expression of Cre alone, as α-MHC-Cre mouse hearts exhibit similar pik3r1 expression relative to wild-type littermates (1.00 ± 0.10 versus 1.06 ± 0.16 A.U. for α-MHC-Cre and wild-type littermate hearts isolated at ZT15, respectively; p>0.05). Cosinor analysis revealed a significant (p<0.05) circadian oscillation in pik3r1 mRNA levels in isolated cardiomyocytes following serum shock-induced synchronization (Figure 4B).
Figure 4.
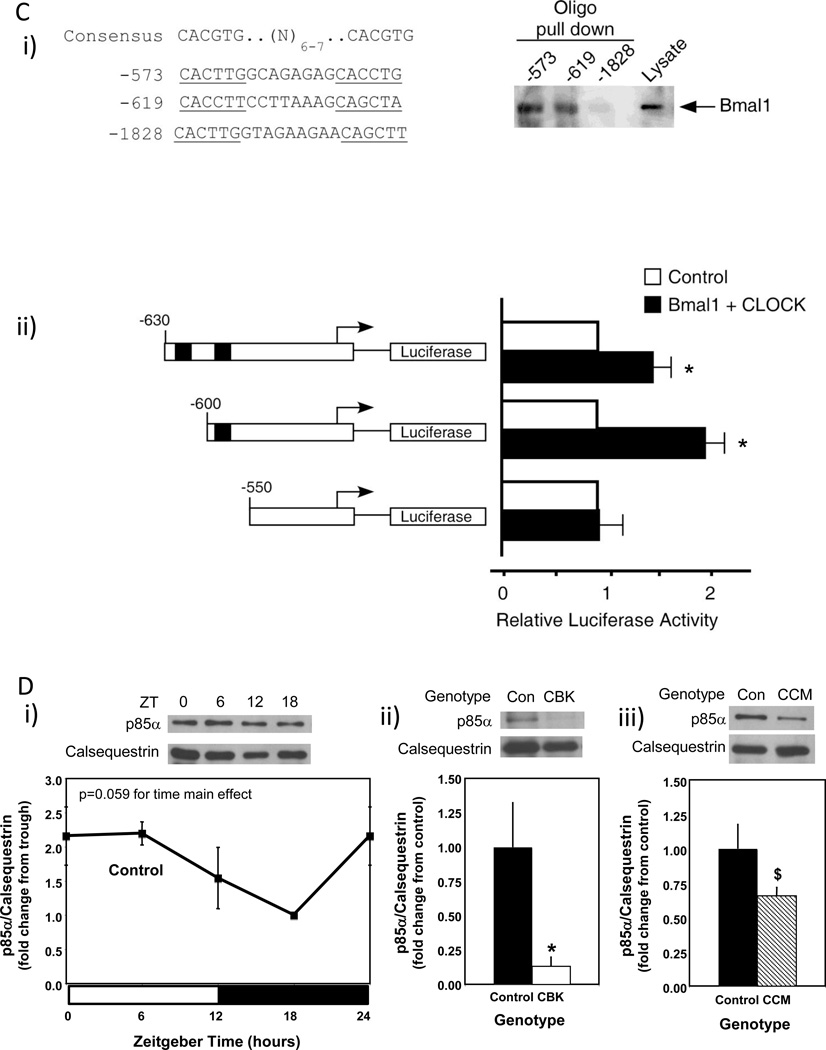
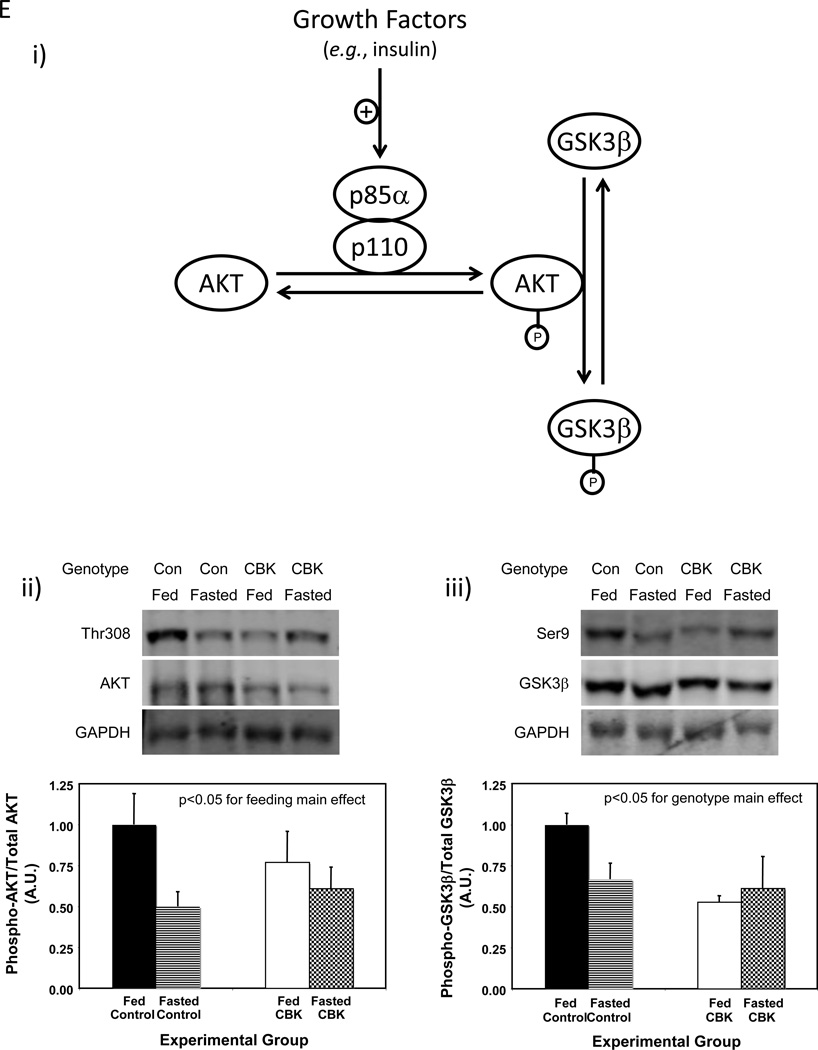
Alterations in time-of-day-dependent oscillations in pik3r1 mRNA in CBK (microarray [i], RT-PCR [ii]) and CCM (RT-PCR [iii]) hearts, compared to littermate controls (A). Persistence of circadian oscillations in pik3r1 mRNA in isolated adult rat cardiomyocytes following serum shock induced synchronization (i.e., challenged with 50% serum for 2 hours [shaded area]; B). Association of BMAL1 with two E-box consensus sequences in the Pik3R1 promoter (Ci); COS cells were transfected with expression vectors for Bmal1 and Clock, followed by DNA precipitation assay with biotinylated oligonucleotides (as described in the Methods section). BMAL1/CLOCK-mediated transcription of the Pik3r1 promoter (Cii); expression vectors for Bmal1 and Clock were co-transfected with Pik3r1 luciferase reporter and control plasmids into COS cells, as described in the Methods section (black boxes represent E-boxes at positions −619 and −573 in the Pik3r1 promoter). Diurnal variations in p85α protein levels in control hearts (Di). Decreased p85α protein levels in CBK (Dii) and CCM (Diii) hearts at ZT6. Relationship between PI3K, AKT, and GSK3β (Ei). CBK hearts exhibit phospho-Thr308-AKT (Eii) and phospho-Ser9-GSK3β (Eiii) levels that are comparable to fasted wild-type hearts. Mice were housed in a 12-hr light:12-hr dark cycle (lights on at ZT0). Hearts were isolated from 12 week old male CBK, CCM, and age-matched littermate controls. Note that ZT0 and ZT24 are identical data. Values are expressed as mean ± SEM (n=3–13; sample size range varies dependent on the parameter investigated). *, p<0.05 for CBK/CCM versus littermate control (post-hoc pair-wise comparison). $, p=0.076 for CCM versus control.
To investigate whether Bmal1 directly regulates Pik3r1, we examined the upstream regulatory sequence of the Pik3r1 promoter. We found several putative E-box sequences on the Pik3r1 promoter that could potentially recruit Bmal1. The putative E-boxes are located at −573, −619, and −1828 bp upstream of the Pik3r1 promoter. Notably, the putative E boxes located at −573 and −619 bp match very closely with the preferred BMAL1 binding motif, including their spacing and location immediately upstream of known Bmal1-regulated promoters (Rey et al. 2011). DNA precipitation assays using biotinylated oligonucleotides were performed, to investigate whether BMAL1 binds directly to any of these three putative E-boxes. Figure 4Ci shows that BMAL1 was precipitated by the E-boxes located at −573 and −619 bp upstream of the Pik3r1 promoter. However, BMAL1 was not detected in the DNA precipitates using the E-box located at −1828 bp upstream of the Pik3r1 promoter. Next, we examined the function of the E-boxes located at −573 and −619 bp upstream of the Pik3r1 promoter using reporter gene assays (Figure 4Cii). Co-transfection of BMAL1 and CLOCK increased Pik3r1 promoter activity. Deletion of E-boxes to −550 bp abrogated BMAL1/CLOCK-mediated increases in Pik3r1 promoter activity. Collectively, these observations suggest that Pik3r1 is directly regulated by the BMAL1/CLOCK heterodimer.
Pik3r1 encodes for p85α, the regulatory subunit of PI3K. Whether oscillations in pik3r1 mRNA observed in control hearts were associated with time-of-day-dependent fluctuations in p85α protein levels was next investigated; Figure 4D (and Supplemental Table 2) shows that p85α significantly oscillates in control hearts, peaking approximately 4 hours into the light phase. In addition, p85α protein levels were decreased in both CBK and CCM hearts (90% and 34% decrease, respectively; Figure 4D). We next investigated whether BMAL1-mediated regulation of Pik3r1 was associated with a functional impact on the downstream signaling components (illustrated in Figure 4Ei), namely AKT and GSK3β. Hearts were isolated during the dark phase (ZT20) from fed and 16-hour fasted (fasted between ZT4 to ZT20) control and CBK mice. The purpose of investigating fed versus fasted mice was to compare basal versus insulin-stimulated states; data presented in Supplemental Figure 3 confirms higher plasma insulin levels in fed mice, independent of genotype. Consistent with the dramatic decreased in p85α levels in CBK hearts (90% lower compared to littermate controls; Figure 4D), AKT phosphorylation at Thr-308 tended to be lower in CBK hearts (Figure 4Eii). Importantly, phosphorylation of the AKT target, glycogen synthase kinase 3 beta (GSK3β), at Ser-9 was significantly reduced in hearts isolated from CBK mice (significant genotype main effect; Figure 4Eiii); since the genotype-feeding status interaction term did not reach statistical significance (p=0.06), pairwise comparisons between experimental groups was not possible. Collectively, our findings suggest that the cardiomyocyte circadian clock regulates both p85α and downstream signaling.
Physiologic role of BMAL1 in the adaptation of the heart to fasting
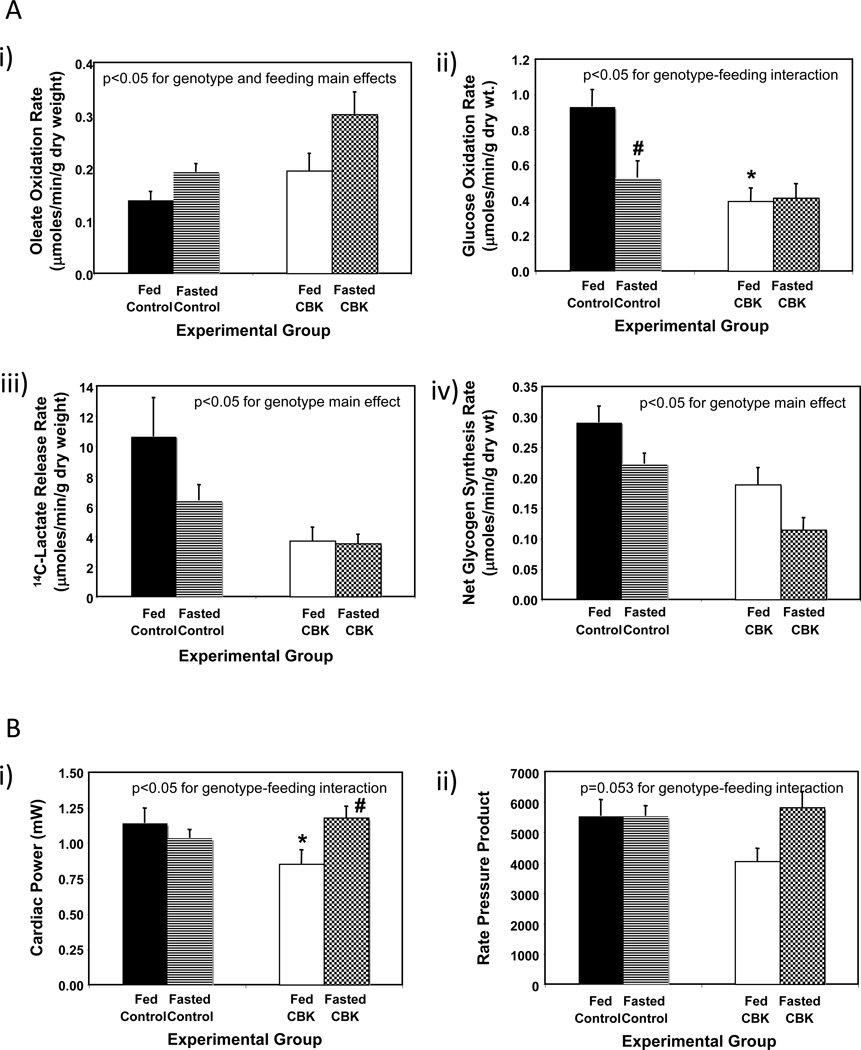
Based on our combined transcriptome and in silico analyses (summarized in Table 1) and subsequent validation studies (Figures 3 and 4), it is clear that BMAL1 influences both myocardial metabolism (e.g., β-hydroxybutyrate oxidation) and signaling (e.g., the PI3K/AKT/GSK3β axis). In order to gain better insight regarding the extent to which BMAL1 influences these processes, we chose to perform a more in depth analysis of substrate metabolism in CBK and littermate control hearts, in both the fed and fasted states. The scenario in which the animal in the wild is unsuccessful in foraging upon awakening was mimicked, by removing food from mice during their sleep time (light phase; ZT4), to ensure prevention of feeding upon awakening; all hearts were investigated at ZT20. Fasting influenced whole body metabolism (e.g., RER) and humoral (e.g., plasma NEFA and insulin) factors in a similar fashion in CBK and littermate control mice (Supplemental Figure 3). Independent of genotype, fasting increased fatty acid (oleate) oxidation, concomitant with decreased rates of glucose oxidation (i.e., p<0.05 for feeding status main effects; Figure 5A). Independent of feeding status, CBK hearts exhibit increased fatty acid (oleate) oxidation, concomitant with decreased rates of glucose oxidation, glycolysis (14C-lactate release) and net glycogen synthesis (i.e., p<0.05 for genotype main effects; Figure 5A). In addition, fasting decreased glucose oxidation in control, but not CBK, hearts (Figure 5A). Finally, cardiac power was decreased in hearts isolated from fed (but not fasted) CBK mice; a similar trend was observed for a second contractile function parameter (i.e., rate pressure product; Figure 5B). Collectively, these observations reveal a critical role of BMAL1 in myocardial substrate utilization.
Figure 5.
Response of CBK mice to a 16-hour fast (relative to littermate control mice) at the level of substrate utilization (A) and contractile function (B). Mice were housed in a 12-hr light:12-hr dark cycle (lights on at ZT0); for fasted mice, food was removed from the cage at ZT4. All parameters represent values at ZT20. Values are expressed as mean ± SEM (n=6–7; sample size range varies dependent on the parameter investigated). *, p<0.05 for CBK versus littermate control within a distinct feeding status (post-hoc pair-wise comparison). #, p<0.05 for fed versus fasted within a distinct genotype (post-hoc pair-wise comparison).
CBK mice exhibit age-onset cardiomyopathy and early mortality
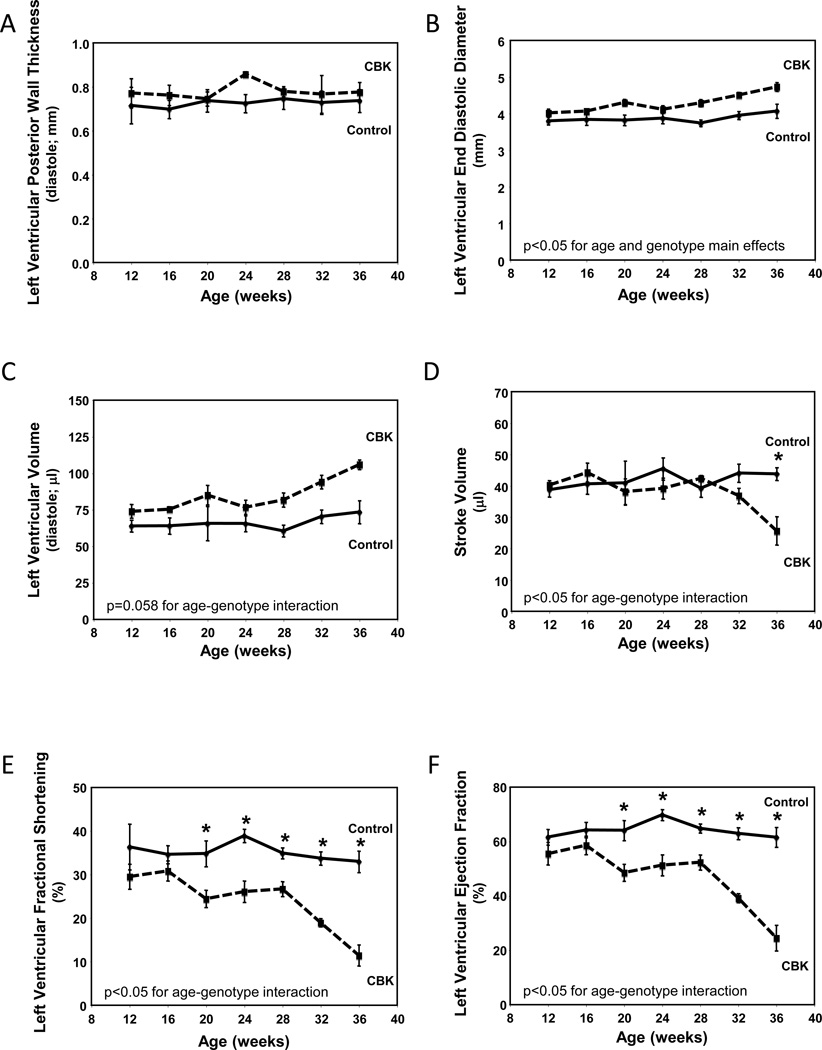
Maintenance of normal metabolic and signaling processes is critical for the heart. Given that these processes are aberrant in CBK hearts, we sought to investigate the long-term consequences of cardiomyocyte-specific BMAL1 deletion. Initially serial echocardiography was performed in CBK versus littermate control mice (at 4-week intervals, between 12 and 36 weeks of age; Figure 6). At 12 weeks of age, echocardiographic parameters were not significantly different between CBK and littermate control mice (Figure 6). By 20 weeks of age, significant differences were observed with respect to genotype for systolic function parameters (fractional shortening and ejection fraction), which worsened with age (Figure 6). Importantly, at 20 and 28 weeks of age, α-MHC-Cre mice do not exhibit differences in systolic function parameters, relative to wild-type littermates (Supplemental Figure 4), suggesting that alone the expression of Cre in cardiomyocytes is not sufficient to mediate the contractile dysfunction observed in CBK hearts.
Figure 6.
Age-dependent depression of cardiac function in CBK, but not littermate control, mice. Left ventricular wall thickness during diastole (A), end diastolic diameter (B), left ventricular volume during diastole (C), stroke volume (D), left ventricular fractional shortening (E), and left ventricular ejection fraction (F). Mice were housed in a 12-hr light:12-hr dark cycle (lights on at ZT0). Serial echocardiography was performed at 4 week intervals, always at ZT6. Values are expressed as mean ± SEM (n=5–7). *, p<0.05 for CBK versus littermate control at a distinct age (post-hoc pair-wise comparison).
The age-dependent cardiomyopathy observed in CBK mice was next characterized at gravimetric, histological, and transcriptional levels. At 12 weeks of age, no significant differences were observed between CBK and littermate control mice for any of the gravimetric parameters investigated (Table 2). In contrast, at 36 weeks of age, biventricular weight, biventricular weight-to-body weight ratio, biventricular weight-to-tibia length ratio, and lung weight were all significantly elevated in CBK mice (relative to age-matched littermate controls; Table 2). Cardiomyocyte cross-sectional area was not significantly different between CBK and littermate control mice (age-independent), although increased fibrosis was observed in hearts isolated from 36 week old CBK mice (Table 2). Collagen isoforms (i.e., col3a1 and col4a1 mRNA) and molecular markers of cardiac dysfunction (i.e., anf and mhcβ mRNA) investigated were significantly induced in hearts isolated from CBK mice at 36 weeks of age, while serca2a mRNA was significantly repressed; cyclophilin mRNA served as a negative control (Table 2). Although col3a1 and col4a1 mRNA were slightly, but significantly, elevated in hearts isolated from CBK mice at 12 weeks of age, molecular markers of cardiac dysfunction were not altered relative to littermate controls at this age (Table 2).
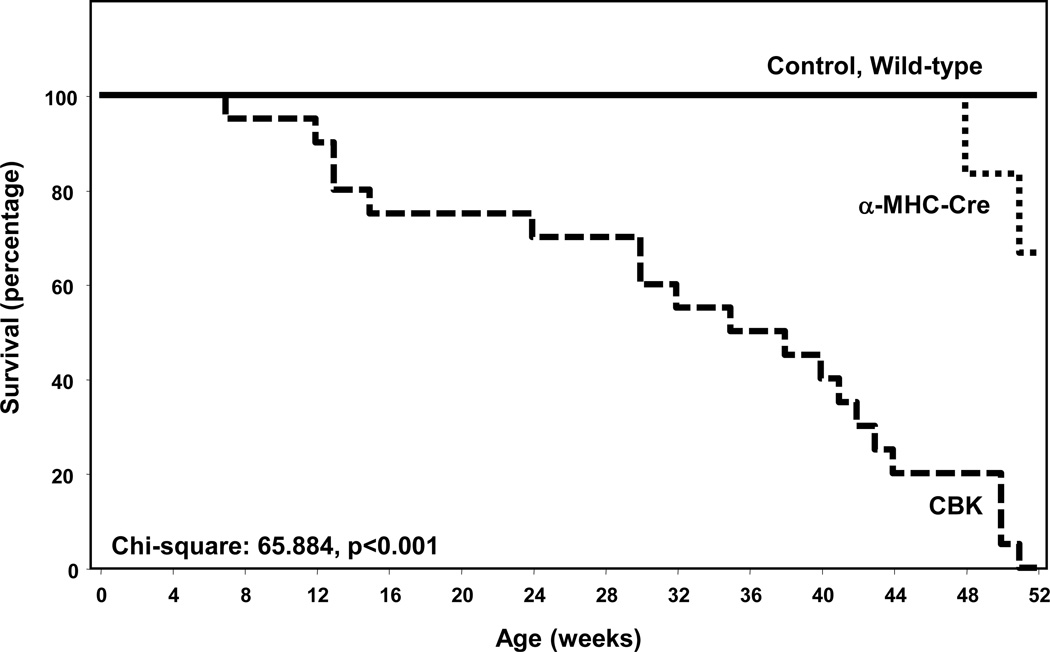
Associated with the development of age-onset cardiomyopathy, CBK mice exhibit a striking decrease in life span. Mean survival age of the CBK mice was 33 ± 3 weeks versus 51 ± 0.7 weeks in MHCα-Cre mice (p<0.0001); no deaths were observed in either littermate control (i.e., BMAL1flox/flox) or wild-type mice during the one year study period (Figure 7). Collectively, these data suggest that chronic genetic deletion of Bmal1 in cardiomyocytes results in cardiomyopathy and decreased lifespan.
Figure 7.
Decreased survival in CBK mice compared to littermate controls. Values are expressed as mean ± SEM (n=20).
DISCUSSION
The purpose of the present study was to investigate the importance of BMAL1 in the heart, through the generation of cardiomyocyte-specific BMAL1 knockout (CBK) mice. Following validation of the cardiac-restricted nature of this model (Figure 1 and Supplemental Figure 1), we identified 19 direct putative BMAL1 target genes, with known functions in metabolism and signaling (Table 1). Subsequent validation studies revealed that aberrant regulation of Bdh1 and Pik3r1 in CBK hearts was associated with impaired ketone body metabolism (Figure 3) and signaling (Figure 4) respectively. We next tested the hypothesis that a physiologic role of BMAL1 is for metabolic adaptation of the heart to fasting; we observed that CBK hearts exhibit a fasting-like metabolic profile (depressed glucose utilization and increased fatty acid oxidation), even in the fed state (Figure 5). Finally, we report that chronic BMAL1 ablation in the heart results in an age-onset cardiomyopathy (Figure 6) and reduced lifespan (Figure 7 and Table 2). Collectively, these studies highlight novel functions for the circadian clock component BMAL1 in the heart.
Animal models wherein circadian clock components are genetically ablated in a ubiquitous fashion have been utilized in attempts to reveal the relative contributions of this molecular mechanism on biology. These studies suggest that a plethora of critical processes, ranging from behavior (e.g., sleep-wake and feeding-fasting cycles), metabolic homeostasis (e.g., insulin secretion and responsiveness), and cellular function (e.g., proliferation), to longevity are under circadian clock control (Bass and Takahashi 2010; Kondratov et al. 2006; Rudic, et al. 2004; Takahashi et al. 2008; Turek, et al. 2005). Two of the best well characterized models, whole-body ClockΔ19 mutant and Bmal1 knockout mice, target the core heterodimer within the mammalian clock mechanism (Bunger, et al. 2000; Vitaterna, et al. 1994). However, despite the fact that CLOCK and BMAL1 function in the circadian clock mechanism in partnership, ClockΔ19 mutant and Bmal1 knockout mice do not always exhibit identical phenotypes. For example, ClockΔ19 mutant mice are obesity prone and exhibit normal lifespan, while Bmal1 knockout mice are lean and exhibit reduced lifespan (Bunger, et al. 2005; Kondratov et al. 2006; Turek et al. 2005). Differences in phenotype have been explained in terms of the extent to which the circadian clock mechanism is impacted (lesser residual circadian clock function in Bmal1 knockout mice) and/or functions of CLOCK and BMAL1 that are independent of the circadian clock mechanism. Appreciation of these reports underscores the need to: 1) generate cell type specific models to identify direct circadian clock functions in distinct organs; and 2) to genetically manipulate multiple clock components, such that circadian clock dependent- and independent- mechanisms can be unveiled. Accordingly, we have generated two cardiomyocyte-specific models, namely CCM and CBK mice (Durgan et al. 2006; Durgan et al. 2011b).
Multiple parameters of cardiac physiology and pathophysiology vary markedly as a function of time-of-day (Durgan and Young 2010; Young 2006). At a molecular level, previous microarray studies have revealed that approximately 10–15% of the cardiac transcriptome oscillates in a time-of-day-dependent fashion in wild-type mice (Martino, et al. 2004; Storch et al. 2002). However, the relative contribution of factors extrinsic (e.g., neurohumoral factors) versus intrinsic (e.g., cell autonomous clock) to the heart is unknown. Through the use of CCM mice, we have previously reported that genetic disruption of CLOCK in the cardiomyocyte impacts a host of genes whose protein products are involved in transcription, cellular signaling, ion homeostasis, and metabolism (Bray et al. 2008). With regards to the latter, we have revealed that wild-type hearts exhibit marked time-of-day-dependent rhythms in both glucose and fatty acid metabolism, which are dependent on cardiomyocyte CLOCK function (Bray et al. 2008; Durgan et al. 2011a; Tsai et al. 2010). The present study identified 19 putative direct BMAL1 target genes in the heart, several of which are involved in metabolism (Table 1). Consistent with identification of the triglyceride synthesis gene Dgat2 (Table 1), we have previously reported attenuated oscillations of this gene, and of triglyceride synthesis, in CCM hearts (Bray et al. 2008). Here, we uncovered a marked dependence of Bdh1 and ketone body metabolism on both CLOCK and BMAL1 (albeit in a time-of-day-independent manner; Figure 3). Importantly, this is the first report suggesting that the circadian clock influences ketone body metabolism. Future studies are required to interrogate fully the links between the cell autonomous circadian clock and ketone body metabolism, as well as the potential implications during conditions of increased ketone body utilization (e.g., starvation, diabetes).
Several putative direct BMAL1 target genes identified in our analysis are known to play a role in cellular signaling (Table 1). These genes are of particular interest, as they might provide insight with regards to which extracellular stimuli/stresses that the cardiomyocyte circadian clock allows to heart to anticipate on a daily basis. One such gene was Pik3r1, which encodes for an insulin signaling component (i.e., the p85α subunit of PI3K). We have previously reported that the phosphorylation status of two kinases that are downstream of PI3K (namely AKT and GSK3β) appear to be under circadian clock control (as evidenced by loss of time-of-dependent oscillations in CCM hearts) (Durgan et al. 2010; Ko, et al. 2011); similar findings regarding clock regulation of AKT phosphorylation have been reported in extra cardiac tissues, using germline Bmal1 null and Per2 mutant mice (Anea, et al. 2009; Carvas, et al. 2012). Consistent with impaired PI3K activity, both AKT and GSK3β phosphorylation were attenuated in CBK hearts (Figure 4E). Collectively, these data are consistent with direct regulation of Pik3r1 and downstream signaling by the cardiomyocyte circadian clock. This concept is supported by previously published studies, reporting that diurnal variations in whole body insulin sensitivity are influenced by circadian clocks (Rudic et al. 2004). Importantly, the current study is among the first to highlight a potential mechanism by which cell autonomous circadian clocks directly impact the PI3K/AKT/GSK3β signaling axis.
A primary goal of the current study was to determine one (or more) physiologic role(s) for BMAL1 in the heart. Given that BMAL1 directly influenced genes involved in metabolism (e.g., ketone body metabolism) and signaling (e.g., insulin signaling), we hypothesized that BMAL1 may play a role in the metabolic adaptation of the heart to fluctuations in feeding status. Consistent with this concept, previous studies have highlighted a role for BMAL1 and/or the circadian clock in both whole body and hepatic responsiveness to fasting (Lamia, et al. 2008; Zhang, et al. 2010). In addition, we have previously observed a differential transcriptional response of CCM hearts to a 12 hour fast (Durgan et al. 2006). In an attempt to mimic the scenario in which the mouse in the wild is unsuccessful in foraging upon awakening, food was removed from mice during the sleep (light) phase (thereby preventing food consumption upon awakening). We observed that even in the fed state, CBK hearts exhibit a fasting-like metabolic phenotype (increased fatty acid oxidation and decreased glucose utilization), which is consistent with the metabolic profile of CCM hearts (Bray et al. 2008; Durgan et al. 2011a). We speculate that depressed PI3K/AKT/GSK3β signaling in CBK hearts (Figure 4E) signals a chronic fasted state. It should be noted that these observations are somewhat in contrast with recent reports that fatty acid oxidation is depressed in isolated mitochondria from germline Bmal1 null mouse livers (Peek, et al. 2013). However, we speculate that investigation of metabolism in an intact organ likely preserves extra-mitochondrial regulatory points, including substrate supply (e.g., control of fatty acyl-CoA uptake into the mitochondrial matrix is a critical β-oxidation regulatory mechanism in intact tissues/cells, which is abolished in isolated mitochondrial studies). Collectively, these observations confirm an important role for BMAL1 in the regulation of myocardial metabolism, as well as in the adaptation of the heart to fasting (i.e., a physiologic stress).
Given that BMAL1 influences a number of important processes in the heart (e.g., metabolism and signaling), we sought to investigate the consequence of chronic genetic ablation of this transcription factor in a cardiomyocyte-specific manner. We have previously reported that both CCM and CBK hearts exhibit a pro-hypertrophic phenotype (Durgan et al. 2011b). Consistent with these observations, current study reports that CBK mice develop an age-dependent cardiomyopathy (Figure 6), which is associated with decreased lifespan (Figure 7). This phenotype is remarkably similar to that reported for germline BMAL1 null mice, wherein systolic dysfunction begins at 20–24 weeks of age (Lefta et al. 2012).
It is important to acknowledge a number of shortcomings and unanswered questions associated with the current study. For example, identification of the precise mechanism(s) by which loss of BMAL1 in the heart results in cardiomyopathy has not been determined. Our analyses clearly reveal that BMAL1 influences a number of critical functions in the heart, leading to the suggestion that the cardiomyopathy observed in CBK mice is potentially multifactorial, as opposed to disruption of a single physiologic process. It is highly likely that the conservative nature of our in silico analysis resulted in a large number of false negatives, in terms of identification of putative BMAL1 target genes. One contributing factor was use of liver ChIP sequencing data to guide the analysis; this may explain why genes such as Klf15 and Scn5a, which have previously been suggested to be regulated by Bmal1 in the heart (Jeyaraj, et al. 2012; Schroder, et al. 2013), were not included in Table 1. It should also be noted that all animal studies were performed under light/dark conditions, raising the possibility that a sub-set of genes oscillate in the heart secondarily to light-stimulation. Finally, the reason why a sub-set of BMAL1- and CLOCK- regulated genes (such as Bdh1) do not oscillate in the heart is also not known; it is possible that this is due to a long half-life for these mRNA species.
In summary, the current study has revealed novel functions for BMAL1 in the heart, including regulation of substrate utilization (e.g., ketone body, fatty acid, and glucose metabolism) and signaling (e.g., the PI3K/AKT/GSK3β signaling axis). In addition, a functional consequence of chronic genetic ablation of BMAL1 in the cardiomyocyte is development of age-onset cardiomyopathy and reduced lifespan. These studies therefore highlight critical functions for BMAL1 in the heart.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by the National Heart, Lung, and Blood Institute (HL-074259, HL-106199, and HL-107709). Bradley W. Peden was supported by an American Heart Association Fellowship (MSRF9960000). Billy-Joe Ammons was supported by the University of Alabama at Birmingham Summer in Biomedical Sciences (SIBS) Undergraduate Research Program. Analysis of microarray data was performed at the UAB Heflin Center for Genomic Sciences. Echocardiography was performed by the UAB Diabetes Research Center Small Animal Physiology Core (DK11015). Humoral factor analysis was performed by the UAB Metabolism Core Laboratory for the Nutrition Obesity Research Center (P30 DK56336, UL 1RR025777, P60DK079626). We would like to thank Alan Akira, Maximiliano H. Grenett, Betty M. Pat, and William F. Ratcliffe for technical assistance.
Footnotes
DISCLOSURES
None declared.
REFERENCES
- Abel ED. Insulin signaling in heart muscle: lessons from genetically engineered mouse models. Curr Hypertens Rep. 2004;6:416–423. doi: 10.1007/s11906-004-0034-4. [DOI] [PubMed] [Google Scholar]
- Anea CB, Zhang M, Stepp DW, Simkins GB, Reed G, Fulton DJ, Rudic RD. Vascular disease in mice with a dysfunctional circadian clock. Circulation. 2009;119:1510–1517. doi: 10.1161/CIRCULATIONAHA.108.827477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass J, Takahashi JS. Circadian integration of metabolism and energetics. Science. 2010;330:1349–1354. doi: 10.1126/science.1195027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray M, Shaw C, Moore M, Garcia R, Zanquetta M, Durgan D, Jeong W, Tsai J, Bugger H, Zhang D, et al. Disruption of the circadian clock within the cardiomyocyte influences myocardial contractile function; metabolism; and gene expression. Am J Physiol Heart Circ Physiol. 2008;294:H1036–H1047. doi: 10.1152/ajpheart.01291.2007. [DOI] [PubMed] [Google Scholar]
- Bray MS, Ratcliffe WF, Grenett MH, Brewer RA, Gamble KL, Young ME. Quantitative analysis of light-phase restricted feeding reveals metabolic dyssynchrony in mice. Int J Obes (Lond) 2012 doi: 10.1038/ijo.2012.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunger MK, Walisser JA, Sullivan R, Manley PA, Moran SM, Kalscheur VL, Colman RJ, Bradfield CA. Progressive arthropathy in mice with a targeted disruption of the Mop3/Bmal-1 locus. Genesis. 2005;41:122–132. doi: 10.1002/gene.20102. [DOI] [PubMed] [Google Scholar]
- Bunger MK, Wilsbacher LD, Moran SM, Clendenin C, Radcliffe LA, Hogenesch JB, Simon MC, Takahashi JS, Bradfield CA. Mop3 is an essential component of the master circadian pacemaker in mammals. Cell. 2000;103:1009–1017. doi: 10.1016/s0092-8674(00)00205-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson PA, O’Connor CM, Miller AB, Anderson S, Belkin R, Neuberg GW, Wertheimer JH, Frid D, Cropp A, Packer M. Circadian rhythm and sudden death in heart failure: results from Prospective Randomized Amlodipine Survival Trial. J Am Coll Cardiol. 2000;36:541–546. doi: 10.1016/s0735-1097(00)00728-2. [DOI] [PubMed] [Google Scholar]
- Carvas JM, Vukolic A, Yepuri G, Xiong Y, Popp K, Schmutz I, Chappuis S, Albrecht U, Ming XF, Montani JP, et al. Period2 gene mutant mice show compromised insulin-mediated endothelial nitric oxide release and altered glucose homeostasis. Front Physiol. 2012;3:337. doi: 10.3389/fphys.2012.00337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Collins HE, Rodrigo GC. Inotropic response of cardiac ventricular myocytes to beta-adrenergic stimulation with isoproterenol exhibits diurnal variation: involvement of nitric oxide. Circ Res. 2010;106:1244–1252. doi: 10.1161/CIRCRESAHA.109.213942. [DOI] [PubMed] [Google Scholar]
- Degaute JP, Van Cauter E, van de Borne P, Linkowski P. Twenty-four-hour blood pressure and heart rate profiles in humans. A twin study. Hypertension. 1994;23:244–253. doi: 10.1161/01.hyp.23.2.244. [DOI] [PubMed] [Google Scholar]
- Degaute JP, van de Borne P, Linkowski P, Van Cauter E. Quantitative analysis of the 24-hour blood pressure and heart rate patterns in young men. Hypertension. 1991;18:199–210. doi: 10.1161/01.hyp.18.2.199. [DOI] [PubMed] [Google Scholar]
- Delp MD, Manning RO, Bruckner JV, Armstrong RB. Distribution of cardiac output during diurnal changes of activity in rats. Am J Physiol. 1991;261:H1487–H1493. doi: 10.1152/ajpheart.1991.261.5.H1487. [DOI] [PubMed] [Google Scholar]
- Dibner C, Schibler U, Albrecht U. The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annu Rev Physiol. 2010;72:517–549. doi: 10.1146/annurev-physiol-021909-135821. [DOI] [PubMed] [Google Scholar]
- Durgan D, Hotze M, Tomlin T, Egbejimi O, Graveleau C, Abel E, Shaw C, Bray M, Hardin P, Young M. The intrinsic circadian clock within the cardiomyocyte. Am J Physiol Heart Circ Physiol. 2005;289:H1530–H1541. doi: 10.1152/ajpheart.00406.2005. [DOI] [PubMed] [Google Scholar]
- Durgan D, Pulinilkunnil T, Villegas-Montoya C, Garvey M, Frangogiannis N, Michael L, Chow C, Dyck J, Young M. Short communication: ischemia/reperfusion tolerance is time-of-day-dependent: mediation by the cardiomyocyte circadian clock. Circ Res. 2010;106:546–550. doi: 10.1161/CIRCRESAHA.109.209346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durgan D, Trexler N, Egbejimi O, McElfresh T, Suk H, Petterson L, Shaw C, Hardin P, Bray M, Chandler M, et al. The circadian clock within the cardiomyocyte is essential for responsiveness of the heart to fatty acids. J Biol Chem. 2006;281:24254–24269. doi: 10.1074/jbc.M601704200. [DOI] [PubMed] [Google Scholar]
- Durgan DJ, Pat BM, Laczy B, Bradley JA, Tsai JY, Grenett MH, Ratcliffe WF, Brewer RA, Nagendran J, Villegas-Montoya C, et al. O-GlcNAcylation, novel post-translational modification linking myocardial metabolism and cardiomyocyte circadian clock. J Biol Chem. 2011a;286:44606–44619. doi: 10.1074/jbc.M111.278903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durgan DJ, Tsai JY, Grenett MH, Pat BM, Ratcliffe WF, Villegas-Montoya C, Garvey ME, Nagendran J, Dyck JR, Bray MS, et al. Evidence suggesting that the cardiomyocyte circadian clock modulates responsiveness of the heart to hypertrophic stimuli in mice. Chronobiol Int. 2011b;28:187–203. doi: 10.3109/07420528.2010.550406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durgan DJ, Young ME. The cardiomyocyte circadian clock: emerging roles in health and disease. Circ Res. 2010;106:647–658. doi: 10.1161/CIRCRESAHA.109.209957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edery I. Circadian rhythms in a nutshell. Physiol Genomics. 2000;3:59–74. doi: 10.1152/physiolgenomics.2000.3.2.59. [DOI] [PubMed] [Google Scholar]
- Gekakis N, Staknis D, Nguyen H, Davis F, Wilsbacher L, King D, Takahashi J, Weitz C. Role of the CLOCK protein in the mammalian circadian mechanism. Science. 1998;280:1564–1569. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- Gibson UE, Heid CA, Williams PM. A novel method for real time quantitative RT-PCR. Genome Res. 1996;6:995–1001. doi: 10.1101/gr.6.10.995. [DOI] [PubMed] [Google Scholar]
- Grinblat L, Pacheco Bolanos LF, Stoppani AO. Decreased rate of ketone-body oxidation and decreased activity of D-3-hydroxybutyrate dehydrogenase and succinyl-CoA:3-oxo-acid CoA-transferase in heart mitochondria of diabetic rats. Biochem J. 1986;240:49–56. doi: 10.1042/bj2400049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harma M, Ilmarinen J. Towards the 24-hour society--new approaches for aging shift workers? Scand J Work Envion Health. 1999;25:610–615. doi: 10.5271/sjweh.488. [DOI] [PubMed] [Google Scholar]
- Heid CA, Stevens J, Livak KJ, Williams PM. Real time quantitative PCR. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- Hogenesch J, Gu Y, Jain S, Bradfield C. The basic-helix-loop-helix-PAS orphan MOP3 forms transcriptionally active complexes with circadian and hypoxia factors. Proc Natl Aad Sci U.S.A. 1998;95:5474–5479. doi: 10.1073/pnas.95.10.5474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeyaraj D, Haldar SM, Wan X, McCauley MD, Ripperger JA, Hu K, Lu Y, Eapen BL, Sharma N, Ficker E, et al. Circadian rhythms govern cardiac repolarization and arrhythmogenesis. Nature. 2012;483:96–99. doi: 10.1038/nature10852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutsson A, Akerstedt T, Jonsson B, Orth-Gomer K. Increased risk of ischaemic heart disease in shift workers. Lancet. 1986;12:89–92. doi: 10.1016/s0140-6736(86)91619-3. [DOI] [PubMed] [Google Scholar]
- Ko ML, Shi L, Tsai JY, Young ME, Neuendorff N, Earnest DJ, Ko GY. Cardiac-specific mutation of Clock alters the quantitative measurements of physical activities without changing behavioral circadian rhythms. J Biol Rhythms. 2011;26:412–422. doi: 10.1177/0748730411414170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koller M. Health risks related to shift work: An example of time-contingent effects of long-term stress. Int Arch Occup Environ Health. 1983;53:59–75. doi: 10.1007/BF00406178. [DOI] [PubMed] [Google Scholar]
- Kondratov RV, Kondratova AA, Gorbacheva VY, Vykhovanets OV, Antoch MP. Early aging and age-related pathologies in mice deficient in BMAL1, the core componentof the circadian clock. Genes Dev. 2006;20:1868–1873. doi: 10.1101/gad.1432206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumaki Y, Ukai-Tadenuma M, Uno KD, Nishio J, Masumoto KH, Nagano M, Komori T, Shigeyoshi Y, Hogenesch JB, Ueda HR. Analysis and synthesis of high-amplitude Cis-elements in the mammalian circadian clock. Proc Natl Acad Sci U S A. 2008;105:14946–14951. doi: 10.1073/pnas.0802636105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamia KA, Storch KF, Weitz CJ. Physiological significance of a peripheral tissue circadian clock. Proc Natl Acad Sci U S A. 2008;105:15172–15177. doi: 10.1073/pnas.0806717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefta M, Campbell KS, Feng HZ, Jin JP, Esser KA. Development of dilated cardiomyopathy in Bmal1-deficient mice. Am J Physiol Heart Circ Physiol. 2012;303:H475–H485. doi: 10.1152/ajpheart.00238.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leighton B, Kowalchuk JM, Challiss RA, Newsholme EA. Circadian rhythm in sensitivity of glucose metabolism to insulin in rat soleus muscle. Am J Physiol. 1988;255:E41–E45. doi: 10.1152/ajpendo.1988.255.1.E41. [DOI] [PubMed] [Google Scholar]
- Marsh SA, Dell’Italia LJ, Chatham JC. Activation of the hexosamine biosynthesis pathway and protein O-GlcNAcylation modulate hypertrophic and cell signaling pathways in cardiomyocytes from diabetic mice. Amino Acids. 2011;40:819–828. doi: 10.1007/s00726-010-0699-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martino T, Arab S, Straume M, Belsham DD, Tata N, Cai F, Liu P, Trivieri M, Ralph M, Sole MJ. Day/night rhythms in gene expression of the normal murine heart. J Mol Med. 2004;82:256–264. doi: 10.1007/s00109-003-0520-1. [DOI] [PubMed] [Google Scholar]
- Martino TA, Oudit GY, Herzenberg AM, Tata N, Koletar MM, Kabir GM, Belsham DD, Backx PH, Ralph MR, Sole MJ. Circadian rhythm disorganization produces profound cardiovascular and renal disease in hamsters. Am J Physiol Regul Integr Comp Physiol. 2008;294:R1675–R1683. doi: 10.1152/ajpregu.00829.2007. [DOI] [PubMed] [Google Scholar]
- Muller J, Tofler G, Stone P. Circadian variation and triggers of onset of acute cardiovascular disease. Circulation. 1989;79:733–743. doi: 10.1161/01.cir.79.4.733. [DOI] [PubMed] [Google Scholar]
- Peek CB, Affinati AH, Ramsey KM, Kuo H-Y, Yu W, Sena LA, Ilkayeva O, Marcheva B, Kobayashi Y, Omura C, et al. Circadian Clock NAD+ Cycle Drives Mitochondrial Oxidative Metabolism in Mice. Science. 2013:342. doi: 10.1126/science.1243417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinz PN, Halter J, Benedetti C, Raskind M. Circadian variation of plasma catecholamines in young and old men: relation to rapid eye movement and slow wave sleep. J Clin Endocrinol Metab. 1979;49:300–304. doi: 10.1210/jcem-49-2-300. [DOI] [PubMed] [Google Scholar]
- Rey G, Cesbron F, Rougemont J, Reinke H, Brunner M, Naef F. Genome-wide and phase-specific DNA-binding rhythms of BMAL1 control circadian output functions in mouse liver. PLoS Biol. 2011;9:e1000595. doi: 10.1371/journal.pbio.1000595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards AM, Nicholls MG, Espiner EA, Ikram H, Cullens M, Hinton D. Diurnal patterns of blood pressrmones in normal man. Clin Exp Hypertens A. 1986;8:153–166. doi: 10.3109/10641968609074769. [DOI] [PubMed] [Google Scholar]
- Rudic RD, Fulton DJ. Pressed for time: the circadian clock and hypertension. J Appl Physiol. 2009;107:1328–1338. doi: 10.1152/japplphysiol.00661.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudic RD, McNamara P, Curtis AM, Boston RC, Panda S, Hogenesch JB, Fitzgerald GA. BMAL1 and CLOCK, two essential components of the circadian clock, are involved in glucose homeostasis. PLoS Biol. 2004;2:e377. doi: 10.1371/journal.pbio.0020377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachan N, Dey A, Rotter D, Grinsfelder DB, Battiprolu PK, Sikder D, Copeland V, Oh M, Bush E, Shelton JM, et al. Sustained hemodynamic stress disrupts normal circadian rhythms in calcineurin-dependent signaling and protein phosphorylation in the heart. Circ Res. 2011;108:437–445. doi: 10.1161/CIRCRESAHA.110.235309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer FA, Hu K, Evoniuk H, Kelly EE, Malhotra A, Hilton MF, Shea SA. Impact of the human circadian system, exercise, and their interaction on cardiovascular function. Proc Natl Acad Sci U S A. 2010;107:20541–20546. doi: 10.1073/pnas.1006749107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer FA, Michelson AD, Frelinger AL, 3rd, Evoniuk H, Kelly EE, McCarthy M, Doamekpor LA, Barnard MR, Shea SA. The human endogenous circadian system causes greatest platelet activation during the biological morning independent of behaviors. PLoS One. 2011;6:e24549. doi: 10.1371/journal.pone.0024549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder EA, Lefta M, Zhang X, Bartos DC, Feng HZ, Zhao Y, Patwardhan A, Jin JP, Esser KA, Delisle BP. The cardiomyocyte molecular clock, regulation of Scn5a, and arrhythmia susceptibility. Am J Physiol Cell Physiol. 2013;304:C954–C965. doi: 10.1152/ajpcell.00383.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan D, Marchase RB, Chatham JC. Overexpression of TRPC3 increases apoptosis but not necrosis in response to ischemia-reperfusion in adult mouse cardiomyocytes. Am J Physiol Cell Physiol. 2008;294:C833–C841. doi: 10.1152/ajpcell.00313.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shea SA, Hilton MF, Hu K, Scheer FA. Existence of an endogenous circadian blood pressure rhythm in humans that peaks in the evening. Circ Res. 2011;108:980–984. doi: 10.1161/CIRCRESAHA.110.233668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srere PA. Citrate Synthase. Methods in Enzymology. 1969;13:3–11. [Google Scholar]
- Storch KF, Lipan O, Leykin I, Viswanathan N, Davis FC, Wong WH, Weitz CJ. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002;417:78–83. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- Takahashi JS, Hong HK, Ko CH, McDearmon EL. The genetics of mammalian circadian order and disorder: implications for physiology and disease. Nat Rev Genet. 2008;9:764–775. doi: 10.1038/nrg2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai JY, Kienesberger PC, Pulinilkunnil T, Sailors MH, Durgan DJ, Villegas-Montoya C, Jahoor A, Gonzalez R, Garvey ME, Boland B, et al. Direct regulation of myocardial triglyceride metabolism by the cardiomyocyte circadian clock. J Biol Chem. 2010;285:2918–2929. doi: 10.1074/jbc.M109.077800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai JY, Villegas-Montoya C, Boland BB, Blasier Z, Egbejimi O, Gonzalez R, Kueht M, McElfresh TA, Brewer RA, Chandler MP, et al. Influence of dark phase restricted high fat feeding on myocardial adaptation in mice. J Mol Cell Cardiol. 2013;55:147–155. doi: 10.1016/j.yjmcc.2012.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsimakouridze EV, Straume M, Podobed PS, Chin H, LaMarre J, Johnson R, Antenos M, Kirby GM, Mackay A, Huether P, et al. Chronomics of pressure overload-induced cardiac hypertrophy in mice reveals altered day/night gene expression and biomarkers of heart disease. Chronobiol Int. 2012;29:810–821. doi: 10.3109/07420528.2012.691145. [DOI] [PubMed] [Google Scholar]
- Turek F, Joshu C, Kohsaka A, Lin E, Ivanova G, McDearmon E, Laposky A, Losee-Olson S, Easton A, Jensen D, et al. Obesity and metabolic syndrome in Clock mutant mice. Science. 2005;308:1043–1045. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitaterna M, King D, Chang A, Kornhauser J, Lowrey P, McDonald J, Dove W, Pinto L, Turek F, Takahashi J. Mutagenesis and mapping of a mouse gene; Clock; essential for circadian behavior. Science. 1994;264:719–725. doi: 10.1126/science.8171325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Maillard M, Schibler U, Burnier M, Gachon F. Cardiac hypertrophy, low blood pressure, and low aldosterone levels in mice devoid of the three circadian PAR bZip transcription factors DBP, HLF, and TEF. Am J Physiol Regul Integr Comp Physiol. 2010;299:R1013–R1019. doi: 10.1152/ajpregu.00241.2010. [DOI] [PubMed] [Google Scholar]
- Yang TT, Chow CW. Elucidating protein: DNA complex by oligonucleotide DNA affinity purification. Methods Mol Biol. 2012;809:75–84. doi: 10.1007/978-1-61779-376-9_5. [DOI] [PubMed] [Google Scholar]
- Young ME. The circadian clock within the heart: potential influence on myocardial gene expression, metabolism, and function. Am J Physiol Heart Circ Physiol. 2006;290:H1–H16. doi: 10.1152/ajpheart.00582.2005. [DOI] [PubMed] [Google Scholar]
- Young ME. Anticipating anticipation: pursuing identification of cardiomyocyte circadian clock function. J Appl Physiol. 2009;107:1339–1347. doi: 10.1152/japplphysiol.00473.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang EE, Liu Y, Dentin R, Pongsawakul PY, Liu AC, Hirota T, Nusinow DA, Sun X, Landais S, Kodama Y, et al. Cryptochrome mediates circadian regulation of cAMP signaling and hepatic gluconeogenesis. Nat Med. 2010;16:1152–1156. doi: 10.1038/nm.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.