Abstract
Glucocorticoids (GCs) induce apoptosis in lymphocytes and are commonly used to treat hematologic malignancies. However, they are also associated with significant adverse effects and their molecular mechanism of action is not fully understood. GC treatment induces expression of the mTORC1 inhibitor Regulated in Development and DNA Damage Response 1 (REDD1), also known as DNA-Damage Inducible Transcript 4 (DDIT4), and mTORC1 inhibition may distinguish GC-sensitive from GC-resistant acute lymphoblastic leukemia (ALL) cells. Interestingly, REDD1 induction was impaired in GC-resistant ALL cells and inhibition of mTORC1 using rapamycin restored GC sensitivity. These data suggest that REDD1 may be essential for the response of ALL cells to glucocorticoids. To further investigate the role of REDD1, we evaluated the effects of glucocorticoids on primary thymocytes from wild-type and REDD1-deficient mice. GC-mediated apoptosis was blocked by a GC receptor antagonist and by an inhibitor of transcription, which interfered with REDD1 induction and mTORC1 inhibition. However, REDD1 ablation had no effect on GC-induced mTORC1 inhibition and apoptosis in thymocytes ex vivo. Overall, these data not only demonstrate the contextual differences of downstream signaling following GC treatment but also provide a better mechanistic understanding of the role of REDD1.
Keywords: REDD1, glucocorticoid, apoptosis, mTOR, thymocyte
Introduction
Glucocorticoid hormones, which are produced by the adrenal cortex, affect a diverse number of cell types and physiological processes in the body, including development, brain function (cognition, memory, mood, etc.), energy metabolism, and immune response. In the immune system, glucocorticoids (GCs) attenuate immune activity, and are very effective and commonly prescribed anti-inflammatory drugs (1). GCs induce apoptotic cell death in a number of cell types, including T and B lymphocytes, and thymocytes represent one of the first reported examples of apoptosis (2). For several decades now, GCs have been a mainstay component of treatment regimens for a variety of hematological malignancies, including leukemia and lymphoma (3, 4). GC-induced apoptotic cell death in these cells requires the transactivation domain of the glucocorticoid receptor (5), a transcription factor of the nuclear hormone receptor family (6) seemingly expressed in every cell in the body, with the exception of non-nucleated red blood cells (7). More recently it was reported that dexamethasone, a synthetic steroid hormone, can also induce autophagy in particular cell types, including lymphocytes (8–10), and that autophagy may play a role in the GC-induced cell death resistance phenotype acquired by some cell types (9).
The Redd1 gene (for Regulated in Development and DNA Damage Response 1) is highly conserved from flies to humans. It encodes a protein with no known functional domains. A recent crystal structure revealed that REDD1 forms an α/β sandwich and identified a highly conserved surface that is required for activity and might interact with effector proteins (11). Also known as DDIT4, Dig2, and RTP801, Redd1 was identified in a screen for genes that are upregulated by hypoxia (12), and was subsequently shown to be upregulated in response to a variety of other cellular stresses, including DNA damage, endoplasmic reticulum (ER) stress, and energy stress, as well as glucocorticoid treatment (13–16). Using microarray analysis, Wang et al. identified Dig2 (dexamethasone-induced gene 2) as a gene that was upregulated in murine T-cell lymphoma cell lines and primary thymocytes (16). REDD1 is an inhibitor of the mammalian target of rapamycin complex 1 (mTORC1) (17–19), which integrates upstream inputs and responds by regulating cell growth and cell proliferation (20). In most cell types examined, the hypoxia-induced inhibition of mTOR signaling requires REDD1 as well as the TSC1/TSC2 complex (17). However, this requirement is cell-type specific, as we found that REDD1 was not required for hypoxia-induced mTORC1 inhibition in hepatocytes (21).
CEM cell lines are established human T-cell acute lymphoblastic leukemia (ALL) cell lines that are commonly used to investigate molecular mechanisms and signaling pathways involved in GC-induced cell death. Resistance to GCs can arise in some patients undergoing long-term treatment with GCs, and analysis of GC-sensitive versus GC-resistant CEM cell lines is being exploited to investigate molecular differences that might play a role in the resistance phenotype (22–24). Interestingly, recent reports have shown that rapamycin, an mTORC1 inhibitor, can sensitize GC-resistant CEM cells to dexamethasone indicating that inhibiting mTORC1 signaling may be sufficient to bypass resistance (25, 26). Inasmuch as REDD1 is induced by dexamethasone (27–29) and that forced overexpression of REDD1 is sufficient to inhibit mTORC1 (17), we hypothesized that differential sensitivity of CEM cells to GCs may be dependent on REDD1.
Herein we sought to clarify the role of REDD1 in dexamethasone-induced cell death in CEM cells and primary thymocytes. While a recent report implicated a pro-survival role for Redd1 in glucocorticoid treated lymphocytes and primary thymocytes in vivo (27), here we show that Redd1 ablation had no effect on GC-induced apoptosis in thymocytes ex vivo. Additionally, overexpression of REDD1 in GC-resistant ALL cells could not overcome the resistance phenotype. Our data demonstrate that REDD1 is neither necessary nor sufficient for GC-induced cell death.
Materials and Methods
Isolation and Treatment of Primary Thymocytes
Thymocytes were harvested as described in Wolff et al. (21). Briefly, four- to six-week-old mice were sacrificed with an isofluorane overdose followed by cervical dislocation, and removal of the thymus. Cells were dispersed with a pestle in a 1.5-ml tube in DMEM containing 10% FCS and 1% Pen-Strep. After dilution in additional medium, cells were counted using a 1:1 TURKS solution (3% glacial acetic acid, 0.1% crystal violet in H2O) and plated at 2.5 × 106 cells/ml for experiments. Wild-type and Redd1βgeo/βgeo thymocytes were treated with 1 μM dexamethasone (Sigma), 800 nM actinomycin D (Sigma), or 1 μM dexamethasone + 800 nM actinomycin D.
CEM Cells
CEM cell lines were obtained from Dr. E. Brad Thompson at University of Texas Medical Branch, Galveston. These lymphoblastic cells were derived from a child with acute lymphoblastic leukemia (ALL)(30). As described in Medh et al. (31) and Miller et al. (32), dexamethasone-sensitive (CEM-C7–14) and dexamethasone-resistant (CEM-C1–15) cell clones were subcloned from the original C7 and C1 clones without selective pressure. The CEM-C1–6 line is a dexamethasone-sensitive spontaneous revertant derived from the dexamethasone-resistant C1 parent. The GC-sensitive (S) cell lines (CEM-C1-6 and CEM-C7-14) and GC-resistant (R) cell line (CEM-C1-15) were seeded at a density of 4 × 105 cells/ml, and treated as described in figure legends.
Propidium Iodide Staining and Fluorescence-Activated Cell Sorting (FACS)
Cells were pelleted and resuspended in PBS containing 2% FCS, and 20 ug/mL propidium iodide (Sigma), filtered through 70 μm mesh, placed on ice, and analyzed by FACS immediately at 620 nm.
RNA Extraction and Northern Blotting
RNA was extracted and Northern blots performed as described previously (21). Briefly, RNA was extracted using either Trizol (Invitrogen) or RNA minikit (Qiagen) and resuspended in RNase-free H2O. Twenty μg of total RNA were resuspended using sample loading buffer (Ambion), incubated at 65°C for 15 min, run on a 1.2% agarose–formaldehyde denaturing gel, transferred by capillary action to a nylon membrane (Amersham), cross-linked using UV light, and prehybridized at 68°C for 1 hour in ExpressHyb solution (Clontech). The membrane was hybridized with heat-denatured probes in ExpressHyb solution for 1 hour at 68°C, washed, and exposed to film. Probes were generated using High Prime DNA labeling kit (Roche) and [α-32P]dCTP according to the manufacturer’s instructions. Templates were generated by PCR using the following primers:
5′ – TTATCGATGAGCGTGGTGGTTATGC (p114) and 3′-GCGCGTACATCGGGCAAATAATATC (p115) for βgeo,
5′-CGGCTTCTGTGCGCCTTCAT (p125) and 3′ - CTCCGGCCCGAAGCCACTGT (p126) for Redd1 (exon 2),
5′ - ACTCCTCATACCTGGATGGGG (p127) and 3′ – TTAACAGCCCCTGGATCTTG (p111) for Redd1 (exon 3),
5′ – AGGCCGTGATTCAGTACAGG (p611) and 3′ – GAACGACTCTGAGGCTTTGG (P612) for Fkbp5 (exon 12)
5′ - TGCTCCTCCTGAGCGCAAGTACTC (p190) and 3′-CTCAGACCTGGGCCATTCAGAAAT for Actin.
Growth Curves
CEM, NAMALWA, and Raji cells were plated (at 4 × 105 cells/ml) in triplicate. At the indicated time points, the cells were resuspended and 50 μL removed for counting (mixed with 50 μL of trypan blue solution). Cells were counted on a hemocytometer and both live and dead (trypan blue-positive) cells were scored.
Reagents
Antibodies against cleaved caspase 3 and REDD1 were from Bethyl. Antibodies against phosphorylated S6K (T389), phosphorylated S6 (S235/236), and phosphorylated 4E-BP1(T37/42) were from Cell Signaling. Antibody against tubulin was from Sigma. Anti-β-galactosidase was from ICN. Dexamethasone was from Sigma-Aldrich. Rapamycin (LC Laboratories) was dissolved in methanol. Vehicle control was methanol.
Immunoblotting
Cells were washed in ice-cold PBS and lysed in ice-cold lysis buffer (50 mM Tris-HCl pH 7.4, 250 mM NaCl, 0.5% Igepal CA-630 containing protease (0.1 μM aprotinin, 0.02 mM leupeptin, 0.01 mM pepstatin, 0.5 mM benzamidine, 0.5 mM PMSF, 0.01 M NaF) and phosphatase (2 mM imidazole, 1.15 mM sodium molybdate, 1 mM sodium orthovanadate, 5 nM microcystin) inhibitors for 10 min. Lysates were cleared by centrifugation and protein concentration was measured using Bradford’s method (Bio-Rad). The protein samples were supplemented with 3x loading buffer (6.7% SDS, 33.3% glycerol, 300 mM DTT and bromophenol blue), heated to 100°C for 10 min, resolved by SDS-PAGE, and transferred to nitrocellulose membranes (Bio-Rad). After blocking in 5% milk in TBS-T (10 mM Tris-HCl pH 8, 150 mM NaCl, 0.1% Tween-20), membranes were incubated at 4°C overnight with the desired primary antibodies (see above) diluted in blocking buffer followed by appropriate secondary antibodies conjugated to HRP. The signal was detected by chemiluminescence by mixing 1:1 solution 1 – 2.5 mM luminol (Sigma), 0.4 mM pCoumaric acid (Sigma), 0.1 M Tris-HCl) and solution 2 (0.015% H2O2, 0.1 M Tris-HCl).
REDD1 Overexpression
The C-terminal HA-tagged REDD1 construct (lab database plasmid #177) was generated as described previously (17). For retroviral transduction, Phoenix cells were transfected with control pBabe/HA (lab database plasmid #332) or pBabe/HA-REDD1 (Rd1-HA) (lab database plasmid #177) using Lipofectamine Plus (Invitrogen). Supernatant was collected and used to infect exponentially growing CEM, NAMALWA, and Raji cells following a “spinfection” protocol as follows. Cells were seeded at 2 × 106 cells/well in a 6-well dish in the presence of 2 ml viral supernatant, 4 μg/ml polybrene, and 0.5 ml FCS, spun at 2500 rpm (1,113 × g) for 1.5 hours at 37°C. Cells were resuspended in fresh media (RPMI + 1x P/S + 10% FCS) and incubated overnight. Spinfection protocol was repeated twice more and transduced cells were selected in media containing 2 μg/ml puromycin for 6 days.
Results
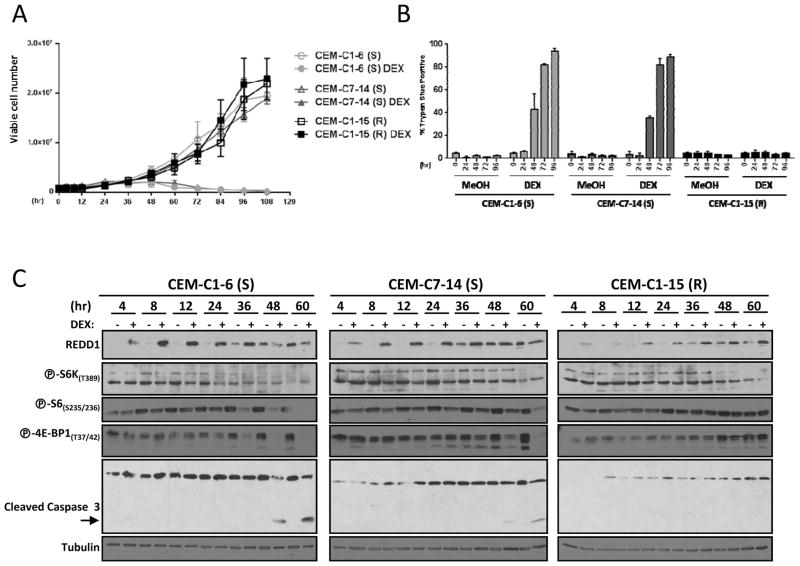
Two GC-sensitive (S) cell lines (CEM-C1-6 and CEM-C7-14) and one GC-resistant (R) cell line (CEM-C1-15) were treated with either vehicle control or 1 μM dexamethasone for up to 110 hours. Cells were harvested at different intervals for trypan blue exclusion to measure cell viability (Fig. 1A), or trypan blue staining to measure nonviable cells (Fig. 1B). As expected, the GC-sensitive CEM-C1-6 (S) and CEM-C7-14 (S) cells did not proliferate upon treatment with dexamethasone, while dexamethasone-treated GC-resistant CEM-C1-15 (R) cells grew at the same rate as vehicle-treated control cells (Fig. 1A). Cell death measurements were inversely correlated with these data, with the dexamethasone-treated CEM-C1-6 (S) and CEM-C7-14 (S) cells showing increased levels of cell death over time, while CEM-C1-15 (R) cells had very low levels of cell death upon dexamethasone treatment (Fig. 1B).
Figure 1. GC-resistant ALL cells have reduced upregulation of REDD1 and fail to inhibit mTORC1 upon dexamethasone treatment.
A) CEM-C1-6 (S), CEM-C7-14 (S), and CEM-C1-15 (R) cells were seeded at a density of 4 × 105 cells/ml and treated with 1 μM dexamethasone (DEX) or vehicle control (MeOH) for up to 110 hours. At the indicated intervals, cell growth/viability was measured by counting trypan blue excluded cells. B) CEM-C1-6 (S), CEM-C7-14 (S), and CEM-C1-15 (R) cells were treated with vehicle control or 1 μM dexamethasone for 0, 24, 48, 72, or 96 hours, then analyzed for cell death by trypan blue staining-cells. C) Immunoblots of protein lysates from CEM-C1-6 (S), CEM-C7-14 (S), and CEM-C1-15 (R) cells treated with 1 μM dexamethasone (DEX) or vehicle control (−) for 4, 8, 12, 24, 36, 48, or 60 hours. Blots were probed for REDD1, phospho-S6K(T389), phospho-S6(S235/236), phospho-4E-BP1(T37/42), caspase 3, and tubulin (loading control). Although separate gels/blots were performed, protein loading and exposures are comparable. (S) = GC-sensitive; (R) = GC-resistant.
Immunoblot analysis of protein lysates from these cells showed the expected increase in cleaved caspase 3 (indicative of increased apoptosis) in CEM-C1-6 (S) and CEM-C7-14 (S) cells. Next, we asked whether dexamethasone treatment had differential effects on mTORC1 in GC-sensitive versus GC-resistant cell lines. For these experiments, we evaluated the phosphorylation state of two substrates of mTORC1, S6K and 4E-BP1, as well as ribosomal protein S6 (a substrate of S6K). Compared to vehicle treated samples collected at the same time, we observed a progressive inhibition of mTORC1 over time in both sensitive cell lines (Fig. 1C). Interestingly, no mTORC1 inhibition was detected in CEM-C1-15 (R) cells over the course of the experiment (Fig. 1C). Thus, a correlation was observed between sensitivity to dexamethasone and mTORC1 inhibition. These data suggest that mTORC1 inhibition may contribute to dexamethasone sensitivity.
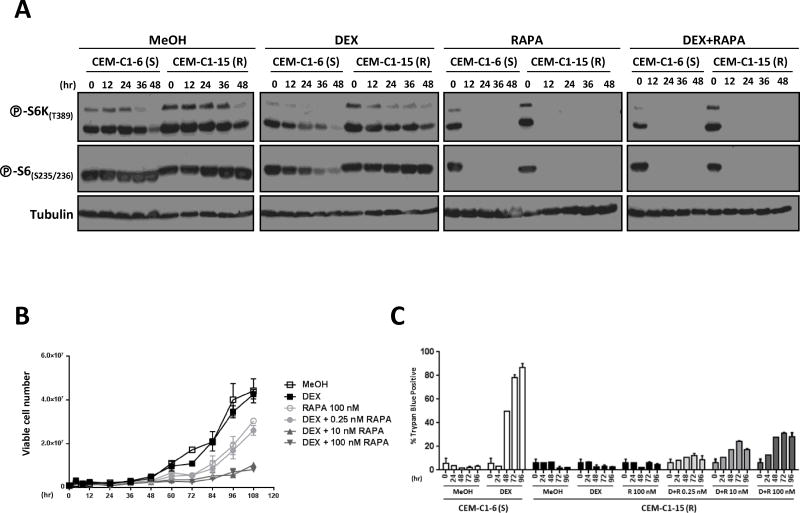
To determine whether mTORC1 inhibition was implicated in dexamethasone sensitization, we treated CEM-C1-15 (R) cells with rapamycin, an mTORC1 inhibitor. While dexamethasone alone failed to inhibit mTORC1 in these cells, mTORC1 was profoundly inhibited by treatment with rapamycin (Fig. 2A). Having restored mTORC1 inhibition in CEM-C1-15 (R) cells by treatment with rapamycin, we evaluated the effects of dexamethasone/rapamycin combination treatment on cell growth and apoptosis. The addition of rapamycin had a synergistic effect with dexamethasone in these cells and suppressed cell proliferation in a manner dependent on rapamycin concentration (Fig. 2B), which is consistent with previous results (26, 33). (The lack of difference between the Dex+10 nM rapa and Dex+100 nM rapa is most likely due to the fact that cell viability is already greatly reduced in the Dex+10 nM rapa-treated cells.) In contrast, rapamycin alone inhibited cell proliferation to a much lesser extent (Fig. 2B). Thus, despite that rapamycin by itself was sufficient to inhibit mTORC1 (Fig. 2A), the suppression of cell proliferation required both rapamycin and dexamethasone (Fig. 2B). The suppression of cell proliferation by the rapamycin/dexamethasone combination was due, at least in part, to increased cell death (Fig. 2C). However, even at the highest rapamycin concentration used, cell death in CEM-C1-15 (R) cells was lower than in dexamethasone-treated CEM-C1-6 (S) cells (Fig. 2C). Given the very profound effect of the combination treatment on cell proliferation (Fig. 2B), rapamycin may synergize with dexamethasone in more than one way.
Figure 2. Rapamycin sensitizes GC-resistant cells to GC-mediated cell death.
A) Immunoblots of protein lysates from CEM-C1-6 (S) and CEM-C1-15 (R) cells treated with vehicle control, 1 μM dexamethasone, 100 nM rapamycin, or 1 μM dexamethasone + 100 nM rapamycin for 0, 12, 24, 36, or 48 hours were probed as indicated. B) CEM-C1-15 (R) cells were seeded at a density of 4 × 105 cells/ml and treated with vehicle control (MeOH), 1 μM dexamethasone (DEX), 100 nM rapamycin (RAPA), or 1 μM dexamethasone plus either 0.25 nM, 10 nM, or 100 nM rapamycin for up to 108 hours. Cell growth and viability were measured by trypan blue staining. C) CEM-C1-6 (S) cells were treated with vehicle control or 1 μM dexamethasone, and CEM-C1-15 (R) cells were treated with vehicle control, 1 μM dexamethasone, and 1 μM dexamethasone plus either 0.25 nM, 10 nM, or 100 nM rapamycin for 0, 24, 48, 72, or 96 hours. Cells were then analyzed for cell death by trypan blue staining. (S) = GC-sensitive; (R) = GC-resistant.
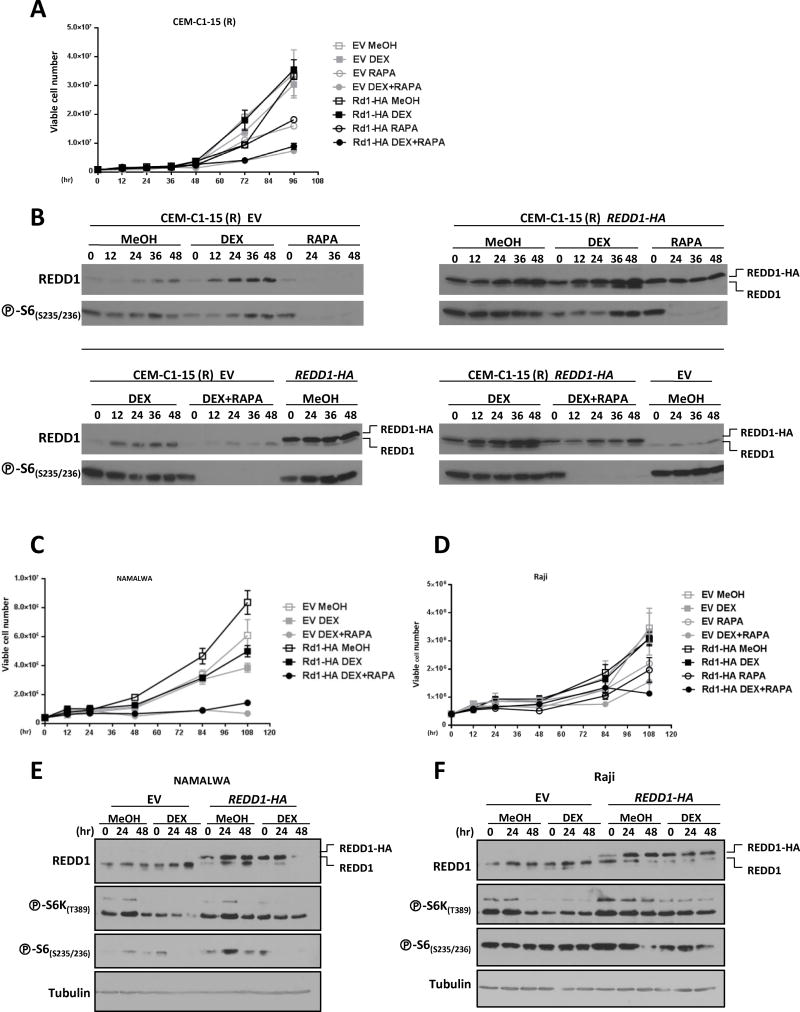
Overall, these data show that mTORC1 inhibition can sensitize resistant ALL cells to dexamethasone. To obtain insight into the mechanism whereby mTORC1 is normally inhibited in response to dexamethasone, we evaluated REDD1 expression. REDD1 is induced by GCs (16, 27, 28), and is sufficient to inhibit mTORC1 (17). In the two sensitive ALL cell lines, REDD1 expression was upregulated within a few hours and remained elevated throughout the time course (Fig. 1C). The parallel, but lesser, increase in REDD1 in the controls may be due to a progressive increase in cell density in the cultures. REDD1 was also induced in resistant CEM-C1-15 (R), but to a lesser extent (Fig. 1C). We hypothesized that lower levels of REDD1 may explain the lack of mTORC1 inhibition in dexamethasone-treated CEM-C1-15 (R) cells. We therefore asked whether, as for rapamycin, forced expression of REDD1 could overcome GC resistance in CEM-C1-15 (R) cells. CEM-C1-15 (R) cells were transduced with an HA-tagged REDD1 (or an empty vector control), and then treated with dexamethasone or vehicle (Fig. 3A). We found that REDD1 overexpression did not sensitize the CEM-C1-15 (R) cells to dexamethasone, nor did it further sensitize the cells to rapamycin treatment (Fig. 3A). These results might be explained by the fact that despite ectopic REDD1 overexpression, there was very little effect on mTORC1 inhibition, as evidenced by phospho-S6 levels (Fig. 3B). Overexpression of REDD1 is therefore not sufficient to overcome the GC-resistance phenotype of CEM-C1-15 (R) cells.
Figure 3. REDD1 overexpression does not sensitize GC-resistant cells to dexamethasone.
A) CEM-C1-15 (R) cells transduced with either an empty vector control (EV) or an HA-tagged REDD1 cDNA (Rd1 HA) were treated with either vehicle control (MeOH), 1 μM dexamethasone (DEX), 100 nM rapamycin (RAPA), or 1 μM dexamethasone + 100 nM rapamycin for up to 100 hours. Cell growth/viability was measured by trypan blue staining. B) Immunoblot analysis of cells as in (A). The upper REDD1 band is the HA-tagged REDD1 protein, which has a slightly higher molecular weight than endogenous REDD1 protein. C and D) NAMALWA (C) and Raji (D) cells transduced with either an empty vector control or an HA-tagged REDD1 cDNA were treated with vehicle control, 1 μM dexamethasone, 100 nM rapamycin, or 1 μM dexamethasone + 100 nM rapamycin for 100 hours. Cell growth/viability was measured by trypan blue staining. E and F) Immunoblot analysis of NAMALWA cells (E) and Raji cells (F) treated as in (C) and (D), respectively, to examine REDD1 protein expression, and phospho-S6K(T389), phospho-S6(S235/236), and tubulin (loading control) levels.
We next looked at whether higher levels of ectopic REDD1 expression might be achieved in other GC-resistant cell lines. The NAMALWA and Raji cell lines are human Burkitt’s lymphoma cell lines that have been shown to be resistant to GC treatment-induced cell death (Fig. 3, C and D) (33, 34). Similar to CEM-C1-15 (R) GC-resistant cells, these cell lines could also be sensitized to dexamethasone-induced cell death by treatment with rapamycin (Fig. 3, C and D) (26). We overexpressed HA-tagged REDD1 in these two cell lines. Modest levels of REDD1 expression were achieved (Fig. 3, E and F), and neither the NAMALWA cells nor the Raji cells showed increased sensitivity to GC treatment upon REDD1 overexpression (Fig. 3, C and D). Additionally, overexpression of REDD1 did not lead to increased inhibition of mTORC1 signaling upon GC treatment (Fig. 3, E and F). Overall, experiments in these GC-resistant leukemia/lymphoma cell lines show that REDD1 overexpression is not sufficient for GC-induced cell death.
While experiments in the resistant cell lines show that REDD1 induction is not sufficient to cause cell death, it remains to be determined whether REDD1 is necessary for GC-mediated cell death in sensitive cells. For these experiments, CEM sensitive cell lines may be subjected to REDD1 depletion by shRNA, and exposed to dexamethasone. However, shRNA-mediated depletion is seldom complete, and despite the use of multiple shRNAs, confounding off-target effects can never be completely ruled out. For these reasons, we turned to a better experimental system. We tried to answer the question using primary thymocytes from wild-type and Redd1-deficient mice.
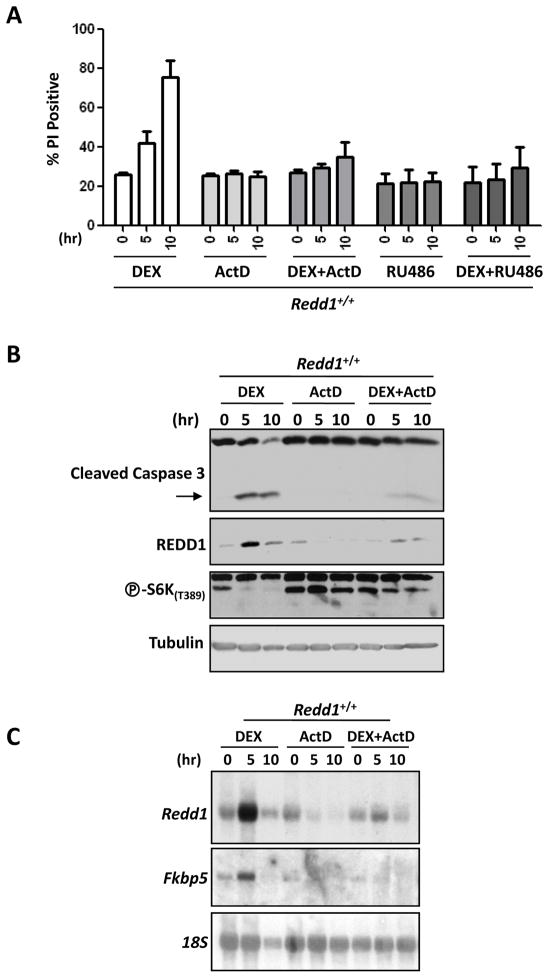
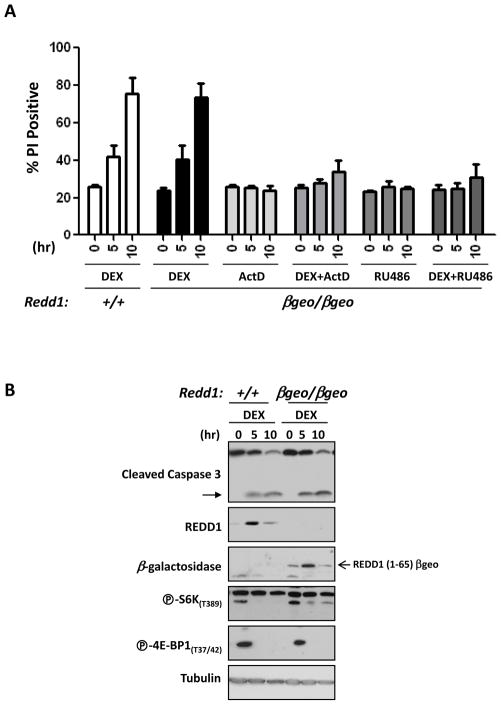
Primary thymocytes have long been known to be sensitive to GC treatment (35, 36), and it has been reported that REDD1 is upregulated in thymocytes in response to treatment with dexamethasone (27, 29). Primary thymocytes were harvested from 4- to 6-week-old mice and treated for 0, 5, or 10 hours with 1 μM dexamethasone, and then stained with PI and subjected to FACS to measure percentage of dead cells. As expected, the percentage of PI-positive cells increased over time, demonstrating an increase in cell death upon dexamethasone treatment (Fig. 4A). After 10 hours, roughly 80% of thymocytes were dead. This induction of cell death was blocked by treatment with actinomycin D (ActD), indicating a requirement for new transcription, as well as by treatment with RU486, a GC receptor antagonist, demonstrating that ligand binding and activation through the GC receptor is also required (Fig. 4A).
Figure 4. Dexamethasone treatment of wild-type primary thymocytes upregulates REDD1 expression, inhibits mTORC1 signaling, and induces cell death.
A) Primary thymocytes were isolated from wild-type (Redd1+/+) mice and treated with 1 μM dexamethasone (DEX), 800 nM actinomycin D (ActD), 1 μM dexamethasone + 800 nM actinomycin D (DEX+ActD), 1 μM RU486, or 1 μM dexamethasone + 1 μM RU486 (DEX+RU486). Cells were harvested at 0, 5, and 10 hours following treatment and analyzed for cell death by propidium iodide (PI) staining and FACS for two independent experiments. All cells were harvested simultaneously 10 hours after extraction from the mice and plating. B) Immunoblots of protein lysates from primary thymocytes treated with DEX, ActD, or DEX+ActD for 0, 5, or 10 hours were probed for caspase 3, REDD1, phospho-S6K(T389), and tubulin (loading control). C) Northern blot analysis of cell samples treated as above. Blots were probed for Redd1, Fkbp5 (positive control for dexamethasone induction), and 18S rRNA (loading control). The lower levels of REDD1 protein and Redd1 mRNA seen in the 10-hour dexamethasone-treated cell samples compared to the 5-hour samples (Fig. 4B and C) is likely due to the considerable cell death (almost 80% of the cells – Fig. 4A) that occurs by this time point.
Dexamethasone treatment of primary thymocytes induced caspase 3 cleavage, and this could be blocked by treatment with ActD (Fig. 4B). The induction of caspase 3 cleavage was paralleled by an induction of REDD1 and the inhibition of mTORC1 (Fig. 4B). Expression of the canonical GC-response gene Fkbp5 (37) was blocked by ActD (Fig. 4C). Redd1 induction (at both the mRNA and protein level), as well as mTORC1 inhibition, was also blocked by ActD (Fig. 4B and C) indicating that new transcription following dexamethasone treatment is required for these observed changes.
To evaluate the role of Redd1 in GC induction of apoptosis, we utilized a Redd1 gene trap mouse strain in which a βgeo cassette is inserted into the second intron of the Redd1 locus. We previously used this gene trap line to analyze the role of Redd1 in mTORC1 regulation by hypoxia, and demonstrated that Redd1βgeo is a null allele (21). The insertion of βgeo results in a fusion transcript and precludes the expression of exon 3, which encodes most of the protein and is required for Redd1 function. Redd1βgeo/βgeo mice develop normally, with no obvious phenotypic abnormalities (21). Primary thymocytes were harvested from wild-type mice and from Redd1βgeo/βgeo littermate mice, treated with dexamethasone, and analyzed for percentage of dead cells. Surprisingly, we found no difference in the percentage of dead cells between dexamethasone-treated wild-type and Redd1βgeo/βgeo thymocytes (Fig. 5A). At either 5 hours or 10 hours of dexamethasone treatment there was no appreciable difference in cell death. Primary thymocytes from the Redd1βgeo/βgeo mice were also treated with ActD, dexamethasone + ActD, RU486, or dexamethasone + RU486. As seen with the wild-type primary thymocytes, treatment with either actinomycin D or RU486 inhibited dexamethasone-induced cell death (Fig. 5A).
Figure 5. Redd1 is dispensable for dexamethasone-induced mTORC1 inhibition and cell death in primary thymocytes.
Primary thymocytes were isolated from Redd1βgeo/βgeo mice and treated with 1 μM dexamethasone (DEX), 800 nM actinomycin D (ActD), 1 μM dexamethasone + 800 nM actinomycin D (DEX+ActD), 1 μM RU486, or 1 μM dexamethasone + RU486 (DEX+RU486). Cells were harvested at 0, 5, and 10 hours following treatment and analyzed for cell death by propidium iodide (PI) staining and FACS. The percentage of PI-positive cells was compared to wild-type primary thymocytes (Redd1+/+) treated with DEX for 0, 5, and 10 hours, for two independent experiments. All cells were harvested simultaneously 10 hours after extraction from the mice and plating. B) Immunoblots of protein lysates from wild-type primary thymocytes and Redd1βgeo/βgeo thymocytes treated with 1 μM dexamethasone (DEX) for 0, 5, or 10 hours were probed for caspase 3, REDD1, β-galactosidase, phospho-S6K(T389), phospho-4E-BP1(T37/42), and tubulin (loading control).
We next asked whether loss of Redd1 had an effect on GC-induced mTORC1 inhibition in thymocytes. Surprisingly, we found that loss of Redd1 did not affect mTORC1 inhibition by dexamethasone (Fig. 5B). Overall, phospho-S6K and phospho-4E-BP1 levels were very similar in dexamethasone-treated Redd1βgeo/βgeo thymocytes compared to wild-type (Fig. 5B). Immunoblot analysis showed upregulation of REDD1 in wild-type cells upon dexamethasone treatment, and confirmed that the REDD1(1-65)-β-galactosidase chimeric reporter protein, but not endogenous REDD1 protein, was expressed in the Redd1βgeo/βgeo cells (Fig. 5B). Cleaved caspase 3 levels were also similar between the wild-type and Redd1βgeo/βgeo cells demonstrating similar levels of apoptosis (Fig. 4B). Thus, these data show that Redd1 is dispensable for GC-induced cell death in thymocytes.
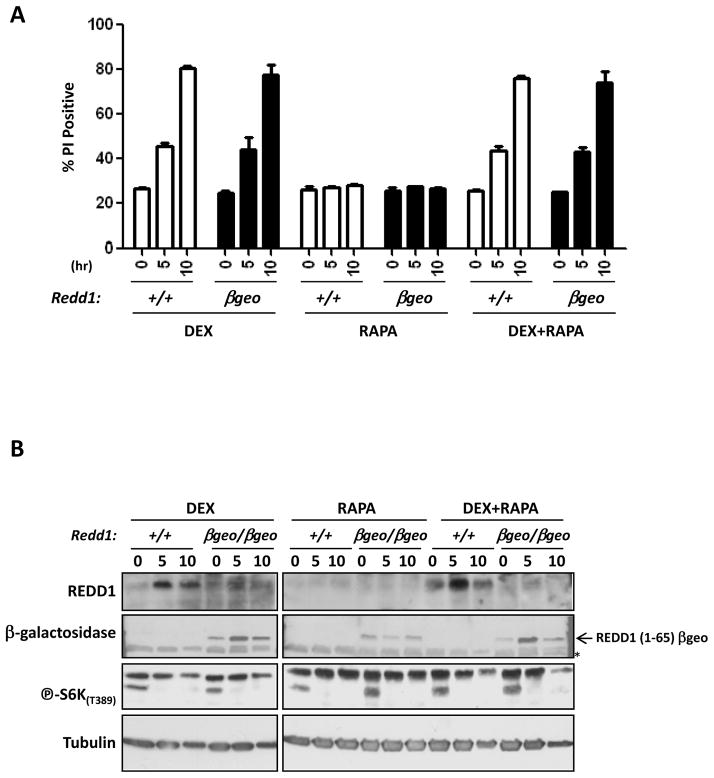
We reasoned that Redd1 might be dispensable because it is not necessary to inhibit mTORC1 in response to GC treatment, and asked whether mTORC1 inhibition alone would be sufficient to induce apoptosis in primary thymocytes. We found that rapamycin treatment alone did not induce cell death in either Redd1βgeo/βgeo or wild-type thymocytes, nor did it enhance the cell death induced by dexamethasone treatment (Fig. 6A). Immunoblot analysis confirmed the upregulation of REDD1 in wild-type cells and the absence of endogenous REDD1 protein in the Redd1βgeo/βgeo cells (Fig. 6B). It also confirmed that rapamycin treatment effectively inhibited mTORC1 signaling as evidenced by the decrease in phospho-S6K levels (Fig. 6B).
Figure 6. Rapamycin treatment of either wild-type or Redd1βgeo/βgeo primary thymocytes does not induce cell death or synergize with dexamethasone.
Primary thymocytes were isolated from wild-type (+/+) or Redd1βgeo/βgeo (βgeo) mice and treated with 1 μM dexamethasone (DEX), 25 μM rapamycin (RAPA), or 1 μM dexamethasone + 25 uM rapamycin (DEX+RAPA). Cells were harvested at 0, 5, and 10 hours following treatment and analyzed for cell death by PI staining and FACS. B) Immunoblots of protein lysates from wild-type primary thymocytes (+/+) or Redd1βgeo/βgeo (βgeo/βgeo) thymocytes treated with 1 μM dexamethasone (DEX), 25 μM rapamycin (RAPA), or dexamethasone + 25 μM rapamycin (DEX+RAPA) for 0, 5, or 10 hours were probed for REDD1, β-galactosidase, phospho-S6K(T389), and tubulin (loading control).
Discussion
Our lab has had a long-standing interest in Redd1 and its involvement in mTORC1 signaling. Here, using a variety of cell types, both tumor-derived and primary cells, we characterize the role of REDD1 and mTORC1 in GC-induced cell death. We show that Redd1 is neither necessary nor sufficient for dexamethasone-induced cell death. In addition, Redd1 is dispensable for mTORC1 inhibition by dexamethasone in thymocytes. However, whereas mTORC1 inhibition does not affect dexamethasone-induced cell death in thymocytes, rapamycin, via its mTORC1 inhibitory action, or perhaps by other mechanisms, sensitizes glucocorticoid-resistant leukemia and lymphoma cells to dexamethasone-induced cell death. In contrast, rapamycin has no effect on glucocorticoid-induced cell death in primary thymocytes.
Our results are consistent with the notion that GC resistance in leukemia and lymphoma cells may be reverted by treatment with an mTORC1 inhibitor: rapamycin synergized with dexamethasone in CEM-C1-15 (R), NAMALWA, and Raji cells. Despite this, and that forced overexpression of REDD1 is sufficient to inhibit mTORC1 in most cell lines examined (17, 18, 20), in these resistant cells, ectopic expression of REDD1 was not enough. This may be explained by the modest levels of REDD1 expression in all three cell lines. These cell lines are difficult to manipulate and experiments were performed in stable cells in which REDD1 was constitutively expressed. Thus, it is possible that higher REDD1-expressing cells were not recovered because of lower fitness. This is consistent with the notion that rapamycin reduced, though not very significantly, the growth rates in CEM and Raji cells. Alternatively, REDD1 overexpression may not be sufficient to inhibit mTORC1 in these cell types. We have previously shown that REDD1-mediated mTORC1 inhibition is TSC1/TSC2-dependent (17), and disruption of this complex blocks REDD1 effects. Interestingly, REDD1 is constitutively upregulated in renal cancer, where it also fails to induce mTORC1 inhibition (38). Overall, these data reveal a cell context-dependency of REDD1-mediated inhibition of mTORC1. Understanding the determinants of permissiveness may shed insight into fundamental mechanisms of mTORC1 regulation by REDD1.
REDD1 has been previously implicated as having a protective role against apoptosis. While focusing on the role of Redd1 in autophagy, Molitoris et al. reported that Redd1 null thymocytes have increased apoptosis compared to wild-type thymocytes in response to dexamethasone treatment (27). These authors reported a 50% increase in thymocyte cell death in the absence of Redd1. However, our data show that the induction of Redd1 in dying cells undergoing apoptosis does not necessarily imply a role for REDD1 (protective or otherwise) in cell death. We show that Redd1 is induced in response to dexamethasone in primary thymocytes and that this treatment results in 80% cell death by 10 hours. This process is accompanied by the induction of Redd1 and requires de novo transcription (as established by ActD experiments). However, we found that REDD1 ablation has no effect on GC-induced cell death in primary thymocytes, and that levels of GC-induced apoptosis are the same in wild-type and Redd1−/− thymocytes. One possible explanation for this difference is that in the Molitoris et al. study, they treated Redd1−/− mice with dexamethasone by intraperitoneal injection, and then harvested the thymus and isolated thymocytes for analysis 4 hours later (27), while we harvested the thymocytes from untreated mice and treated them ex vivo with dexamethasone. In addition, these investigators observed that Redd1 was required for the inhibition of S6K by dexamethasone. While the in vivo treatment is more physiological, the results are open to confounding factors arising from the effects of dexamethasone treatment of other cell populations. Our results with thymocytes in vitro show that Redd1 is dispensable for dexamethasone-induced mTORC1 inhibition and cell death in a strictly cell autonomous fashion. Thus, our findings complement those of Molitoris et al. by reporting the cell autonomous response of thymocytes to the GC treatment. Furthermore, we also found that Redd1 is dispensable for mTORC1 inhibition by dexamethasone. These data suggest that dexamethasone inhibits mTORC1 through, at least in part, Redd1-independent mechanisms. Inasmuch as the effect of dexamethasone can be blocked by ActD, the data are consistent with the notion that GCs induce several genes that inhibit mTORC1. As for Redd1, inactivation of this putative second mechanism may not be sufficient to restore mTORC1 in GC-treated cells, and simultaneous loss with Redd1 may be required.
It is interesting that rapamycin synergized with dexamethasone in the tumor-derived cell lines, but not in the primary thymocytes, and that rapamycin alone, while inhibiting the proliferation of tumor cells, had no effect on thymocyte apoptosis. However, primary thymocytes do not survive well in vitro and these experiments involved different assays. Thus, we cannot exclude that under conditions allowing the expansion of primary thymocytes in vitro, rapamycin may have similarly inhibited cell growth.
GC therapy is a common component in the chemotherapeutic treatment of hematological cancers, including lymphomas and leukemias, due to their ability to induce cell death in these cells. Yet, while GCs are beneficial, they are also associated with undesirable side effects such as hyperglycemia, osteoporosis, cardiovascular disease, cataracts and glaucoma. A better understanding of the molecular mechanisms underlying GC action may help in designing novel, more efficacious, and less toxic analogues (39). While much has been learned about the downstream genes and pathways that mediate GC hormone-induced cell death, their precise mechanism of action is still not fully understood. Therefore, it is critical that the signaling pathways involved be fully investigated.
Implications.
These molecular findings underlying GC action and the role of REDD1 are fundamental for the design of novel, more efficacious, and less toxic analogues.
Acknowledgments
Grant Support: This work was supported by the following NIH grants to J.B.: K08NS051843 and RO1CA129387. J.B. is a Virginia Murchison Linthicum Scholar in medical research at UT Southwestern Medical Center.
We thank the members of the Brugarolas lab for helpful discussions. We thank Dr. Ellen Vitetta at UT Southwestern Medical Center, Dallas for the generous gift of the NAMALWA and Raji cells, and Dr. E. Brad Thompson at UT Medical Branch, Galveston for the generous gift of the CEM cell lines.
Footnotes
Disclosure of Potential Conflicts of Interest: No potential conflicts of interest were disclosed by the authors.
Authors’ Contributions
Conception and Design: J. Brugarolas, N.C. Wolff
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): N.C. Wolff, J. Brugarolas
Analysis and interpretation of data: N.C. Wolff, R.M. McKay, J. Brugarolas
Writing, review and/or revision of the manuscript: R.M. McKay, N.C. Wolff, J. Brugarolas
Administrative, technical, or material support (i.e. reporting or organizing data, constructing databases): N.C. Wolff, R.M. McKay
References
- 1.Barnes PJ. Corticosteroids: the drugs to beat. Eur J Pharmacol. 2006;533(1–3):2–14. doi: 10.1016/j.ejphar.2005.12.052. [DOI] [PubMed] [Google Scholar]
- 2.Wyllie AH. Glucocorticoid-induced thymocyte apoptosis is associated with endogenous endonuclease activation. Nature. 1980;284(5756):555–6. doi: 10.1038/284555a0. [DOI] [PubMed] [Google Scholar]
- 3.Greenstein S, Ghias K, Krett NL, Rosen ST. Mechanisms of glucocorticoid-mediated apoptosis in hematological malignancies. Clin Cancer Res. 2002;8(6):1681–94. [PubMed] [Google Scholar]
- 4.Inaba H, Pui CH. Glucocorticoid use in acute lymphoblastic leukaemia. Lancet Oncol. 2010;11(11):1096–106. doi: 10.1016/S1470-2045(10)70114-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Distelhorst CW. Recent insights into the mechanism of glucocorticosteroid-induced apoptosis. Cell Death Differ. 2002;9(1):6–19. doi: 10.1038/sj.cdd.4400969. [DOI] [PubMed] [Google Scholar]
- 6.Giguere V, Hollenberg SM, Rosenfeld MG, Evans RM. Functional domains of the human glucocorticoid receptor. Cell. 1986;46(5):645–52. doi: 10.1016/0092-8674(86)90339-9. [DOI] [PubMed] [Google Scholar]
- 7.Bloomfield CD. Glucocorticoid receptors in leukemia and lymphoma. J Clin Oncol. 1984;2(4):323–8. doi: 10.1200/JCO.1984.2.4.323. [DOI] [PubMed] [Google Scholar]
- 8.Laane E, Tamm KP, Buentke E, Ito K, Kharaziha P, Oscarsson J, et al. Cell death induced by dexamethasone in lymphoid leukemia is mediated through initiation of autophagy. Cell Death Differ. 2009;16(7):1018–29. doi: 10.1038/cdd.2009.46. [DOI] [PubMed] [Google Scholar]
- 9.Swerdlow S, McColl K, Rong Y, Lam M, Gupta A, Distelhorst CW. Apoptosis inhibition by Bcl-2 gives way to autophagy in glucocorticoid-treated lymphocytes. Autophagy. 2008;4(5):612–20. doi: 10.4161/auto.5920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xia X, Kar R, Gluhak-Heinrich J, Yao W, Lane NE, Bonewald LF, et al. Glucocorticoid-induced autophagy in osteocytes. J Bone Miner Res. 2010;25(11):2479–88. doi: 10.1002/jbmr.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vega-Rubin-de-Celis S, Abdallah Z, Kinch L, Grishin NV, Brugarolas J, Zhang X. Structural analysis and functional implications of the negative mTORC1 regulator REDD1. Biochemistry. 2010;49(11):2491–501. doi: 10.1021/bi902135e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shoshani T, Faerman A, Mett I, Zelin E, Tenne T, Gorodin S, et al. Identification of a novel hypoxia-inducible factor 1-responsive gene, RTP801, involved in apoptosis. Mol Cell Biol. 2002;22(7):2283–93. doi: 10.1128/MCB.22.7.2283-2293.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ellisen LW, Ramsayer KD, Johannessen CM, Yang A, Beppu H, Minda K, et al. REDD1, a developmentally regulated transcriptional target of p63 and p53, links p63 to regulation of reactive oxygen species. Mol Cell. 2002;10(5):995–1005. doi: 10.1016/s1097-2765(02)00706-2. [DOI] [PubMed] [Google Scholar]
- 14.Protiva P, Hopkins ME, Baggett S, Yang H, Lipkin M, Holt PR, et al. Growth inhibition of colon cancer cells by polyisoprenylated benzophenones is associated with induction of the endoplasmic reticulum response. Int J Cancer. 2008;123(3):687–94. doi: 10.1002/ijc.23515. [DOI] [PubMed] [Google Scholar]
- 15.Sofer A, Lei K, Johannessen CM, Ellisen LW. Regulation of mTOR and cell growth in response to energy stress by REDD1. Mol Cell Biol. 2005;25(14):5834–45. doi: 10.1128/MCB.25.14.5834-5845.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Z, Malone MH, Thomenius MJ, Zhong F, Xu F, Distelhorst CW. Dexamethasone-induced gene 2 (dig2) is a novel pro-survival stress gene induced rapidly by diverse apoptotic signals. J Biol Chem. 2003;278(29):27053–8. doi: 10.1074/jbc.M303723200. [DOI] [PubMed] [Google Scholar]
- 17.Brugarolas J, Lei K, Hurley RL, Manning BD, Reiling JH, Hafen E, et al. Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. Genes Dev. 2004;18(23):2893–904. doi: 10.1101/gad.1256804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Corradetti MN, Inoki K, Guan KL. The stress-inducted proteins RTP801 and RTP801L are negative regulators of the mammalian target of rapamycin pathway. J Biol Chem. 2005;280(11):9769–72. doi: 10.1074/jbc.C400557200. [DOI] [PubMed] [Google Scholar]
- 19.DeYoung MP, Horak P, Sofer A, Sgroi D, Ellisen LW. Hypoxia regulates TSC1/2-mTOR signaling and tumor suppression through REDD1-mediated 14-3-3 shuttling. Genes Dev. 2008;22(2):239–51. doi: 10.1101/gad.1617608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brugarolas J. mTORC1 signaling and hypoxia. mTOR pathway and mTOR inhibitors in cancer therapy. In: Polunovsky VAA, Houghton PJJ, editors. Cancer drug discovery and development. New York, NY: Humana Press; 2010. [Google Scholar]
- 21.Wolff NC, Vega-Rubin-de-Celis S, Xie XJ, Castrillon DH, Kabbani W, Brugarolas J. Cell-type-dependent regulation of mTORC1 by REDD1 and the tumor suppressors TSC1/TSC2 and LKB1 in response to hypoxia. Mol Cell Biol. 2011;31(9):1870–84. doi: 10.1128/MCB.01393-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bhadri VA, Trahair TN, Lock RB. Glucocorticoid resistance in paediatric acute lymphoblastic leukaemia. J Paediatr Child Health. 2012;48(8):634–40. doi: 10.1111/j.1440-1754.2011.02212.x. [DOI] [PubMed] [Google Scholar]
- 23.Kofler R, Schmidt S, Kofler A, Ausserlechner MJ. Resistance to glucocorticoid-induced apoptosis in lymphoblastic leukemia. J Endocrinol. 2003;178(1):19–27. doi: 10.1677/joe.0.1780019. [DOI] [PubMed] [Google Scholar]
- 24.Schlossmacher G, Stevens A, White A. Glucocorticoid receptor-mediated apoptosis: mechanisms of resistance in cancer cells. J Endocrinol. 2011;211(1):17–25. doi: 10.1530/JOE-11-0135. [DOI] [PubMed] [Google Scholar]
- 25.Gu L, Zhou C, Liu H, Gao J, Li Q, Mu D, et al. Rapamycin sensitizes T-ALL cells to dexamethasone-induced apoptosis. J Exp Clin Cancer Res. 2010;29:150. doi: 10.1186/1756-9966-29-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guo X, Zhou CY, Li Q, Gao J, Zhu YP, Gu L, et al. Rapamycin sensitizes glucocorticoid resistant acute lymphoblastic leukemia CEM-C1 cells to dexamethasone induced apoptosis through both mTOR suppression and up-regulation and activation of glucocorticoid receptor. Biomed Environ Sci. 2013;26(5):371–81. doi: 10.3967/0895-3988.2013.05.006. [DOI] [PubMed] [Google Scholar]
- 27.Molitoris JK, McColl KS, Swerdlow S, Matsuyama M, Lam M, Finkel TH, et al. Glucocorticoid elevation of dexamethasone-induced gene 2 (Dig2/RTP801/REDD1) protein mediates autophagy in lymphocytes. J Biol Chem. 2011;286(34):30181–9. doi: 10.1074/jbc.M111.245423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang H, Kubica N, Ellisen LW, Jefferson LS, Kimball SR. Dexamethasone represses signaling through the mammalian target of rapamycin in muscle cells by enhancing expression of REDD1. J Biol Chem. 2006;281(51):39128–34. doi: 10.1074/jbc.M610023200. [DOI] [PubMed] [Google Scholar]
- 29.Schmidt S, Rainer J, Riml S, Ploner C, Jesacher S, Achmuller C, et al. Identification of glucocorticoid-response genes in children with acute lymphoblastic leukemia. Blood. 2006;107(5):2061–9. doi: 10.1182/blood-2005-07-2853. [DOI] [PubMed] [Google Scholar]
- 30.Foley GE, Lazarus H, Farber S, Uzman BG, Boone BA, McCarthy RE. Continuous Culture of Human Lymphoblasts from Peripheral Blood of a Child with Acute Leukemia. Cancer. 1965;18:522–9. doi: 10.1002/1097-0142(196504)18:4<522::aid-cncr2820180418>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 31.Medh RD, Webb MS, Miller AL, Johnson BH, Fofanov Y, Li T, et al. Gene expression profile of human lymphoid CEM cells sensitive and resistant to glucocorticoid-evoked apoptosis. Genomics. 2003;81(6):543–55. doi: 10.1016/s0888-7543(03)00045-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller AL, Komak S, Webb MS, Leiter EH, Thompson EB. Gene expression profiling of leukemic cells and primary thymocytes predicts a signature for apoptotic sensitivity to glucocorticoids. Cancer Cell Int. 2007;7:18. doi: 10.1186/1475-2867-7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wei G, Twomey D, Lamb J, Schlis K, Agarwal J, Stam RW, et al. Gene expression-based chemical genomics identifies rapamycin as a modulator of MCL1 and glucocorticoid resistance. Cancer Cell. 2006;10(4):331–42. doi: 10.1016/j.ccr.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 34.Joncas J, Boucher J, Boudreault A, Granger-Julien M. Effect of hydrocortisone on cell viability, Epstein-Barr virus genome expression, and interferon synthesis in human lymphoblastoid cell lines. Cancer Res. 1973;33(9):2142–8. [PubMed] [Google Scholar]
- 35.Cifone MG, Migliorati G, Parroni R, Marchetti C, Millimaggi D, Santoni A, et al. Dexamethasone-induced thymocyte apoptosis: apoptotic signal involves the sequential activation of phosphoinositide-specific phospholipase C, acidic sphingomyelinase, and caspases. Blood. 1999;93(7):2282–96. [PubMed] [Google Scholar]
- 36.Claman HN. Corticosteroids and lymphoid cells. N Engl J Med. 1972;287(8):388–97. doi: 10.1056/NEJM197208242870806. [DOI] [PubMed] [Google Scholar]
- 37.Vermeer H, Hendriks-Stegeman BI, van der Burg B, van Buul-Offers SC, Jansen M. Glucocorticoid-induced increase in lymphocytic FKBP51 messenger ribonucleic acid expression: a potential marker for glucocorticoid sensitivity, potency, and bioavailability. J Clin Endocrinol Metab. 2003;88(1):277–84. doi: 10.1210/jc.2002-020354. [DOI] [PubMed] [Google Scholar]
- 38.Kucejova B, Pena-Llopis S, Yamasaki T, Sivanand S, Tran TA, Alexander S, et al. Interplay between pVHL and mTORC1 pathways in clear-cell renal cell carcinoma. Mol Cancer Res. 2011;9(9):1255–65. doi: 10.1158/1541-7786.MCR-11-0302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schacke H, Schottelius A, Docke WD, Strehlke P, Jaroch S, Schmees N, et al. Dissociation of transactivation from transrepression by a selective glucocorticoid receptor agonist leads to separation of therapeutic effects from side effects. Proc Natl Acad Sci U S A. 2004;101(1):227–32. doi: 10.1073/pnas.0300372101. [DOI] [PMC free article] [PubMed] [Google Scholar]