Abstract
Genome architecture of a microbe markedly changes when it transitions from a free-living lifestyle to an obligate symbiotic association within eukaryotic cells. These symbiont genomes experience numerous rearrangements and massive gene loss, which is expected to radically alter gene regulatory networks compared with those of free-living relatives. As such, it remains unclear whether and how these small symbiont genomes regulate gene expression. Here, using a label-free mass-spec quantification approach we found that differential protein regulation occurs in Buchnera, a model symbiont with a reduced genome, when it transitions between two distinct life stages. However, differential mRNA expression could not be detected between Buchnera life stages, despite the presence of a small number of putative transcriptional regulators. Instead a comparative analysis of small RNA expression profiles among five divergent Buchnera lineages, spanning a variety of Buchnera life stages, reveals 140 novel intergenic and antisense small RNAs and 517 untranslated regions that were significantly expressed, some of which have been conserved for ∼65 million years. In addition, the majority of these small RNAs exhibit both sequence covariation and thermodynamic stability, indicators of a potential structural RNA role. Together, these data suggest that gene regulation at the post-transcriptional level may be important in Buchnera. This is the first study to empirically identify Buchnera small RNAs, and we propose that these novel small RNAs may facilitate post-transcriptional regulation through translational inhibition/activation, and/or transcript stability. Ultimately, post-transcriptional regulation may shape metabolic complementation between Buchnera and its aphid host, thus impacting the animal's ecology and evolution.
Introduction
Gene regulation in bacterial genomes relies heavily on operon structure, regulatory proteins and sigma factors. Strikingly, most of these canonical regulatory attributes are lost from reduced genomes, exemplified by obligate intracellular pathogens and symbionts (Dandekar et al., 2000; Shigenobu et al., 2000; Moran and Mira, 2001; Mettenhuber, 2002; Wilcox et al., 2003). Mycoplasma pneumoniae, a host-restricted pathogen with a reduced genome, lacks many of the regulatory elements and genes found in other bacteria (Dandekar et al., 2000). Nevertheless, in this bacterium, complex transcriptional and post-transcriptional gene regulation occurs, resembling a eukaryote more than a prokaryote by utilizing antisense RNAs (asRNAs), alternative-splicing and multifunctional enzymes that interact (Guell et al., 2009; Yus et al., 2009; Kuhner et al., 2009). Presently, it is unknown whether or not this complex type of gene regulation is present in beneficial symbionts with reduced genomes, which unlike Mycoplasma pneumoniae are generally smaller in genome size and uncultivable.
The intracellular bacterial symbiont, Buchnera aphidicola, is a hallmark example of an uncultivable, obligate insect mutualist with a reduced genome (Shigenobu et al., 2000). Buchnera provides its aphid host with essential amino acids that cannot be synthesized de novo by the aphid nor provided in sufficient quantities by the aphid's diet of plant sap (Shigenobu et al., 2000; Hansen and Moran, 2014). Buchnera from the pea aphid has a genome size of 641 kbp and encodes only a few recognizable regulatory proteins including two sigma factors (σ 70: RpoD and σ 32: RpoH; Shigenobu et al., 2000; Wilcox et al., 2003). Despite Buchnera's obligate nutritional role for its insect host, limited evidence has been found for Buchnera's ability to regulate transcription of genes underlying essential amino-acid pathways in response to aphid nutritional demands (Moran et al., 2003, 2005; Reymond et al., 2006; Viñuelas et al., 2011). In addition, a diminished transcriptional response to heat shock was observed in Buchnera during near-lethal heat shock treatments, although Buchnera still encodes a functional heat shock sigma factor (σ 32) and several recognition sequences (Wilcox et al., 2003; Dunbar et al., 2007). Potentially, gene regulation in this intracellular microbe is not important, because of its host-controlled homeostatic environment (Shigenobu et al., 2000). Alternatively, gene regulation may act primarily at the post-transcriptional level, which has not been investigated in Buchnera or any other uncultivable symbionts with reduced genomes. If post-transcriptional gene regulation occurs in Buchnera, it may utilize small RNAs to inhibit translation, as in M. pneumoniae (Guell et al., 2009).
In this study, we evaluate the importance and role of post-transcriptional gene regulation and regulatory small RNAs in Buchnera. First, we determined if transcriptional or post-transcriptional gene regulation occurs across different Buchnera life stages, during which Buchnera transitions from an extracellular, proliferating state to an intracellular non-proliferating one (Koga et al., 2012). Second, we conducted directional RNAseq on different Buchnera life stages isolated from four different aphid species that diverged ∼65 million years ago (Degnan et al., 2011) in order to identify conserved, expressed small RNAs. Together, these data validate a number of computationally predicted regulatory elements and reveal a potential role for small RNAs in regulating gene expression in a symbiont with a reduced genome.
Materials and methods
Proteomic sample preparation and analysis
Two distinct Buchnera life stages were collected to determine if gene regulation in Buchnera occurs at the protein level. Briefly, Buchnera cells were collected from embryos and from maternal bacteriocytes of fourth instar pea aphids (Acyrthosiphon pisum str. LSR1), reared and sampled as described elsewhere (Hansen and Moran, 2011). For embryo samples, entire ovarioles were rapidly dissected out of ∼300 aphids, pooled in buffer A at 4 °C and stored at −80 °C. For the bacteriocyte samples, ∼2.6 g of whole aphids were homogenized in ice-cold buffer, and filtered to enrich for Buchnera cells. Buchnera cells were then isolated on a Percoll density gradient (27%/70%) at 4 °C, and stored at −80 °C (Charles and Ishikawa 1999; Hansen and Moran, 2012). This latter approach preferentially captures Buchnera cells from maternal bacteriocytes. A complete description of the proteomic methods is available in the Supplementary Materials and methods.
Label-free quantification (LFQ) was conducted by the WM Keck Foundation Biotechnology Resource Laboratory at Yale University to determine changes in Buchnera protein expression between three technical replicates of embryo and bacteriocyte samples. Mascot distiller and the Mascot search algorithm were used to identify peptide matches in a database comprising the predicted proteomes of both Buchnera aphidicola (PRJNA59285, PRJNA57805) and its pea aphid host (PRJNA13657, PRJNA29489). The MASCOT search engine confidence level was set to 95% for protein hits based on randomness. Buchnera protein abundances were normalized to a constitutively expressed Buchnera protein, 50S ribosomal subunit protein RplN (BUAP5A_507). The Progenesis LCMS statistical analysis pipeline (Nonlinear Dynamics, LLC, Durham, NC, USA) was used to perform a one-way analysis of variance with the normalized protein abundances. Proteins with a conservative ⩾1.5-fold change between samples and with an analysis of variance P<0.05 were considered significantly up- or downregulated (Blagoev et al., 2004; Babaei et al., 2013). See Supplementary Materials and methods for more detail on proteomic analysis. In addition, a Multidimensional Protein Identification Technology (MudPIT) experiment was conducted on a biological replicate of the maternal bacteriocytes prepared as detailed above for fourth instar A. pisum (str. LSR1) to confirm identified proteins.
Small RNA sample preparation and sequencing
Small RNAs from the aphid symbiont Buchnera aphidicola were prepared as described previously (Hansen and Moran, 2012). Briefly, four aphid species with completely sequenced Buchnera genomes, A. pisum (Buchnera-LSR1 and Buchnera-5A), Acyrthosiphon kondoi (Buchnera-Ak), Uroleucon ambrosiae (UA002, referred to as Buchnera-Ua) and Schizaphis graminum (Buchnera-Sg) were reared in a growth chamber at 20 °C under a 16-h light/8-h dark regime. Insects were maintained on the following host plants: A. pisum and A. kondoi on Vicia faba (fava bean), U. ambrosiae on Tithonia rotundifolia (Mexican sunflower) and S. graminum on Hordeum vulgare (barley). For each strain, 2–3 g of insects were homogenized and Buchnera cells were isolated by filtration and centrifugation. Cells were treated immediately with TRI reagent solution (Ambion, Austin, TX, USA), and total RNA was extracted using the miRNAeasy kit (Qiagen, Valencia, CA, USA) then DNAase treated similar to Hansen and Moran (2012).
Library preparation and sequencing of Buchnera small RNAs for all five aphid strains were conducted by the Yale Center for Genome Analysis. Library preparation of the ⩽200 nt fractions were performed using the Illumina mRNA directional sequencing protocol starting from the phosphatase treatment step. Each library was then loaded and sequenced on an individual Illumina (San Diego, CA, USA) GA IIx, lane, 35-nt sequence reads were generated. As in Hansen and Moran (2012), the CLC Genomic Workbench was used for initial read processing and mapping.
The sequence data for directional RNAseq from all Buchnera samples were submitted to NCBI under study accession PRJNA212118 (5A- SRR935066, LSR1- SRR935070, Ak- SRR935071, Ua- SRR935072 and Sg- SRR935073).
Small RNA identification
Using the initial mapping file generated for each sample with CLC Genomics Workbench, individual reads overlapping known RNAs were removed (ribosomal RNAs, transfer RNAs, rnpB, ssrA and ffs) and the remaining sequences were re-mapped using the Rockhopper software (McClure et al., 2013). In Rockhopper, default parameters were used for strand-specific reads except a final threshold for minimum expression value of untranslated regions (UTRs) and small RNAs (0.3) was chosen based on empirical tests that optimized the agreement between the Buchnera-LSR1 and Buchnera-5A samples.
We then identified and further characterized transcription start sites, UTRs and small RNAs predicted by Rockhopper that were conserved in two or more species. Using a multiple whole-genome alignment generated for the four Buchnera species with Mauve (Darling, 2004), we were able to plot aligned RNAseq coverage curves. This process was aided by the complete collinearity of the Buchnera genomes. Individual coverage plots generated using the ggplot2 module in R (Kahle and Wickham, 2013) and a global plot viewed in Artemis (Rutherford et al., 2000) were manually inspected to determine small RNA boundaries.
Prediction of small RNA structural stability
Putative small RNAs and UTRs were extracted from each genome based on the identified boundaries, and orthologous regions were re-aligned with ClustalW2 (Larkin et al., 2007). RNAalifold was then used to determine a likely secondary structure for the alignment, and of 100 random re-shuffling's of the alignment, which was used as null distribution (Hofacker et al., 2002; Bernhart et al., 2008). Predicted thermodynamic stability and sequence covariation of small RNA secondary structures were measured as the free energy of the thermodynamic ensemble (in units of kcal per mol). Conserved UTRs and predicted small RNA elements were then cross-referenced with existing computational predictions (Degnan et al., 2011).
Reverse transcriptase- quantitative PCR
We compared Buchnera-LSR1 mRNA gene expression for three aphid tissue types (for example, young embryo (YE), old embryo (OE) and maternal bacteriocyte (BAC); Braendle et al., 2003; Koga et al., 2012). As above, YE, OE and BAC samples were rapidly dissected out of fourth instar A. pisum (str. LSR1), pooled in RNA Bacterial Protect (Qiagen) and stored at −80 °C. RNA extractions and DNAase treatment of the samples were conducted as described previously (Hansen and Moran, 2012). Complementary DNA was synthesized from each pooled RNA sample using the iScript cDNA synthesis kit (Bio-Rad, Hercules, CA, USA) and then quantitative PCR was conducted on each sample using a Mastercycler Epgradient Realplex2 (Eppendorf, Hamburg, Germany) in conjunction with the SYBR Fast Universal qPCR reagents (KAPA Biosystems, Woburn, MA, USA). Gene expression values were calculated using the standard curve method for relative quantification (Bookout et al., 2006) and normalized to the housekeeping gene rplN. For statistical analyses, Kal's Z-test (Kal et al., 1999) and multiple comparisons were conducted with a false discovery rate criterion of α=0.05 or less using CLC Genomics Workbench. The oligonucleotide primers used are listed in Supplementary Table S1 and further details are provided in the Supplementary Materials and methods.
Results
Differential protein expression in a symbiont with a reduced genome
To determine if gene regulation occurs at the protein level in Buchnera, we quantitatively compared protein expression profiles of two distinct Buchnera life stages using LFQ. Buchnera experiences a biphasic lifestyle that alternates between a rapidly proliferating extracellular state in aphid ovarioles and a non-proliferating, intracellular state within the maternal bacteriocytes (that is, specialized aphid cells harboring Buchnera; Miura et al., 2003; Braendle et al., 2003; Koga et al., 2012). Using a combination of dissection and filtration techniques to isolate Buchnera-LSR1 from the pea aphid A. pisum str. LSR1, we were able to evaluate protein expression profiles for Buchnera from aphid ovarioles (referred to as embryos) compared with Buchnera from maternal bacteriocytes. In addition, a qualitative protein analysis, MudPIT, was performed on intact Buchnera cells isolated from entire animals to validate the list of proteins identified.
In sum, 355 of 558 proteins encoded in the Buchnera-LSR1 genome were detected (Supplementary Tables S2 and S3). Of these, we identified 80 proteins that were differentially expressed between the developmental stages over 1.5-fold (one-way analysis of variance, P<0.05; Supplementary Table S2). The majority of the differentially enriched proteins was found in the embryos (67/80), whereas only 13 proteins were enriched in the bacteriocytes. The LFQ analysis also detected peptide evidence of ddlB, 1 of 13 putative pseudogenes in the Buchnera-LSR1 genome that harbor single inactivating mutations (for example, homopolymeric frameshifts).
Using gene set enrichment analysis (Subramanian et al., 2005), we found only one Buchnera KEGG pathway was significantly enriched in embryos compared with bacteriocytes (biosynthesis of amino acids, gene set enrichment analysis test statistic=14.4, P=0.034, N=56). The Buchnera proteins, LysA, Fba, ArgA, ThrB, SerC and AroK, were primarily responsible for driving this pattern, exhibiting ⩾1.5-fold enrichment in embryos compared with the bacteriocytes (Table 1). In contrast, HisG, HisC, HisH and IlvH were highly enriched in bacteriocytes compared with embryos (⩾1.5-fold), indicating that not all Buchnera amino-acid pathways are enriched in the embryo (Table 1). Collectively, these results support previous research, which indicated that Buchnera produces essential amino acids for its aphid host (for example, Douglas and Prosser, 1992; Sasaki and Ishikawa, 1995; Shigenobu et al., 2000). However, differential protein expression of essential amino-acid pathways in different aphid tissue types and Buchnera life-cycle stages was unexpected.
Table 1. Differentially regulated proteins involved in essential amino-acid pathways of Buchnera-LSR1.
| Protein name | Pathway | ANOVA P-valuea | Fold change | Transcription factorsb | qRT-PCRc |
|---|---|---|---|---|---|
| Upregulated in bacteriocytes | |||||
| HisG | Histidine | 0.0089 | 1.8 | DksA | NS |
| HisC | Histidine | 0.003 | 1.6 | DksA | NS |
| IlvH | Branched-chain amino acids | 0.0017 | 1.5 | Hns | — |
| Upregulated in embryos | |||||
| AroK | Aromatic amino acids | 0.0063 | 1.6 | *2 | — |
| SerC | Sulfate reduction | 0.0124 | 1.7 | *2 | — |
| ThrB | Threonine | 0.0058 | 1.7 | DksA | NS |
| ArgA | Arginine | 0.0004 | 2.1 | *1 | NS |
| Fba | Glycolysis | 0.0087 | 2.2 | *2 | NS |
| LysA | Lysine | 0.0002 | 2.8 | *2 | NS |
Abbreviations: ANOVA, analysis of variance; LFQ, label-free quantification; NS, not significantly different between treatments; qRT-PCR, quantitative reverse transcriptase-PCR; ‘—', not tested.
LFQ analysis of Buchnera proteins between two Buchnera life stages.
Transcription factors conserved in Buchnera that are critical for E.coli regulation. *N indicates the number of E. coli transcription factors that are missing in Buchnera.
Quantitative reverse transcriptase-PCR was conducted between life stage treatments for the specific gene with differential protein expression.
Other functional classes enriched in the embryo compared with the bacteriocyte include proteins associated with the flagellar apparatus, transfer RNA synthetases, riboflavin biosynthesis and several hypothetical proteins conserved in many bacteria (Supplementary Table S2). In contrast, proteins associated with membrane transport and protein degradation are enriched in the bacteriocyte (Supplementary Table S2).
Limited transcriptional control in Buchnera
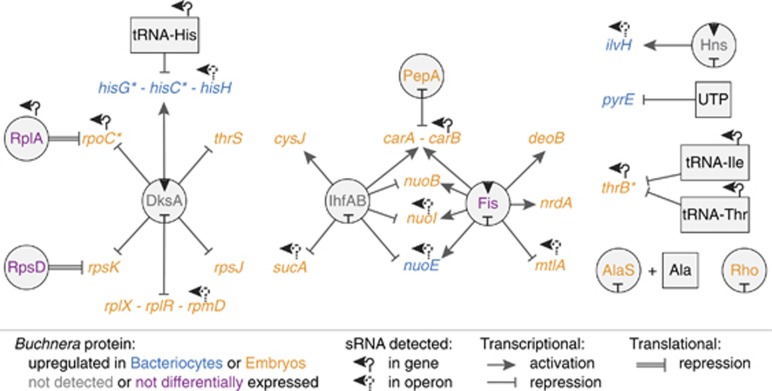
Buchnera's gene repertoire for transcriptional regulation is severely depleted compared with that of its free-living relatives. For example, 78% of transcriptional regulatory proteins present in Escherichia coli, known to regulate orthologs of Buchnera genes whose protein products were differentially expressed in this study are missing in Buchnera; only eight recognizable transcriptional regulatory proteins remain. In E. coli, these 8 proteins have been demonstrated to regulate 26 of the 80 differentially expressed proteins (Figure 1). In addition, approximately half of the operons in Buchnera have been fragmented or radically restructured because of gene loss and genome rearrangements compared with E. coli (Moran and Mira, 2001).
Figure 1.
Genetic regulatory schematic of differentially expressed Buchnera proteins. Only a fraction of proteins differentially expressed by Buchnera bacteriocytes or embryos have conserved transcriptional regulatory factors shared with E. coli. However, several of the genes when tested by quantitative reverse transcriptase-PCR showed no difference between Buchnera life stages (indicated with ‘*'), suggesting post-transcriptional regulation may occur by small RNAs (sRNAs). sRNAs were found associated with nearly half of the genes encoding differentially expressed proteins.
To increase our sensitivity of detecting differential gene expression between Buchnera life stages, we dissected out both early and late embryos in addition to maternal bacteriocytes (that is, three samples in total). Using quantitative reverse transcriptase-PCR, we measured mRNA expression levels of 60 Buchnera-LSR1 genes that showed a range of protein expression responses in our LFQ experiment. Remarkably, we found no difference in the mRNA expression of these genes among the three different samples (Supplementary Table S4). Moreover, four genes that were differentially expressed at the protein but not the mRNA level in this study are generally regulated in free-living relatives at the transcriptional level by the transcription factor DksA. Although DksA is still present in Buchnera-LSR1, the mRNAs of the four transcriptional targets of DksA were not differentially expressed (Table 1), indicating that DksA may have a different regulatory role in Buchnera compared with E. coli. In sum, as the majority of transcriptional regulatory factors and operon structures that are typically responsible for differential protein expression have been lost, and differential mRNA expression could not be detected other regulatory mechanisms, such as small RNAs and regulated protein stability, may underlie Buchnera gene regulation.
RNAseq-based identification of Buchnera small RNAs
Cis- and trans-acting small RNAs are important facilitators of gene regulation at both the transcriptional and post-transcriptional level in free-living bacteria (Waters and Storz, 2009). As Buchnera has lost most of its transcriptional regulators and conserved operons, we determined if other regulatory mechanisms such as those involving small RNAs are present in Buchnera's genome using directional RNAseq on the RNA fraction ⩽200 nt isolated from Buchnera of four distinct aphid species (Table 2). For each sample, small RNAs from all of Buchnera's life stages were pooled to produce a global transcriptome profile for each species (see Materials and methods section). On average, 70% of reads from each sample aligned to Buchnera indicate that the bacterial cell separation and RNA isolation protocol was effective. We focused our subsequent analyses on reads that aligned to regions of the Buchnera genome not containing a known RNA. Approximately half of these directional reads corresponded to predicted protein coding genes or pseudogenes (that is, sense expression). However, quite surprisingly, the other half mapped either within intergenic regions or antisense to predicted protein coding genes or pseudogenes (Table 2). Mean read coverage in the intergenic regions (466–884 reads per bp) was considerably greater than antisense coverage (16–63 reads per bp). To reduce the probability of falsely identifying fragmented intermediates of RNA degradation as small RNA candidates, we used the analysis package Rockhopper (McClure et al., 2013). Rockhopper identifies novel transcripts that are highly expressed (P<0.05), and demarcates boundaries of known transcripts. Rockhopper expression parameters and thresholds were optimized with small RNA transcripts from two clonal Buchnera strains (5A and LSR1) from the same aphid species, A. pisum (see Materials and methods section). We note that the expression profiles for novel small RNA transcripts between the 5A and LSR1 Buchnera strains were highly correlated (Spearman's rho 0.8952, P<0.0001). We identified ‘conserved' small RNAs among Buchnera species using two main criteria: (1) expression of a discreet transcript at a specific location in the genome, and (2) the expression of this transcript is significant based on Rockhopper optimized thresholds as described above. A small RNA candidate was called ‘conserved' if it meets both criteria in two or more species genomes.
Table 2. Directional RNAseq mapping statistics of five Buchnera strains.
|
A. pisum |
A. kondoi |
U. ambrosiae |
S. graminum |
||
|---|---|---|---|---|---|
| Buchnera-5A | Buchnera-LSR1 | Buchnera-Ak | Buchnera-Ua | Buchnera-Sg | |
| Genome features | |||||
| Chromosome length | 642 122 | 641 794 | 615 380 | 641 454 | |
| Protein-coding genes | 558 | 559 | 529 | 548 | |
| Known RNAs | 38 | 38 | 38 | 38 | |
| Disrupted protein-coding genesa | 18 | 18 | 13 | 35 | |
| Directional RNAseq data | |||||
| Total reads in run | 35 487 640 | 35 336 055 | 35 496 905 | 34 314 119 | 33 423 727 |
| Initial trimmed reads mapped | 26 880 929 | 26 358 035 | 26 217 855 | 20 059 556 | 22 712 071 |
| Reads mapping outside of known RNAs | 3 575 290 | 2 921 332 | 3 442 912 | 1 899 567 | 2 833 485 |
| Mapped (sense) to protein-coding genes | 57% | 48% | 43% | 39% | 35% |
| Mapped (antisense) to protein-coding genes | 10% | 10% | 29% | 13% | 12% |
| Mapped to unannotated regions | 33% | 43% | 28% | 48% | 53% |
| Identified small RNAs | |||||
| Total genes with predicted UTRs | 354 | 362 | 306 | 235 | 357 |
| 5′ UTRs | 269 | 269 | 209 | 164 | 263 |
| 3′ UTRs | 218 | 224 | 188 | 132 | 219 |
| Total predicted RNAs (includes >1 element per IGS, gene) | 282 | 275 | 252 | 344 | 269 |
| Intergenic | 46 | 50 | 64 | 51 | 31 |
| Antisense | 236 | 225 | 188 | 293 | 238 |
Abbreviations: IGS, intergenic spacer; UTR, untranslated region.
Includes genes with frameshifts in homopolymeric tracts.
Prevalence of conserved UTRs
Nearly half of all orthologous, annotated Buchnera genes, exhibit significantly expressed UTRs (P<0.05; Table 2). Transcriptional start sites upstream of the identified start codons showed a significant positive length correlation between pairs of genomes (5′ UTRs = Spearman's Rho 0.308–0.774, P<0.01; Supplementary Figure S1). The same was true of 3′ UTRs (Spearman's Rho 0.430–0.8331, P<0.001). We extracted and aligned the 287—5′ and 230—3′ predicted UTRs shared by two or more Buchnera species to identify possible conserved structural elements (as in Degnan et al., 2011). Many of the aligned UTRs were found to have significantly conserved, thermodynamically stable structures (5′ UTRs=137 and 3′ UTRs=140, one-tailed t-tests, d.f.=99, P<0.05; Supplementary Table S5). These calculations are based on alignments of the expressed UTRs and take into account both the thermodynamic stability and sequence covariation, which together are indicators of a potential structural RNA role (Hofacker et al., 2002).
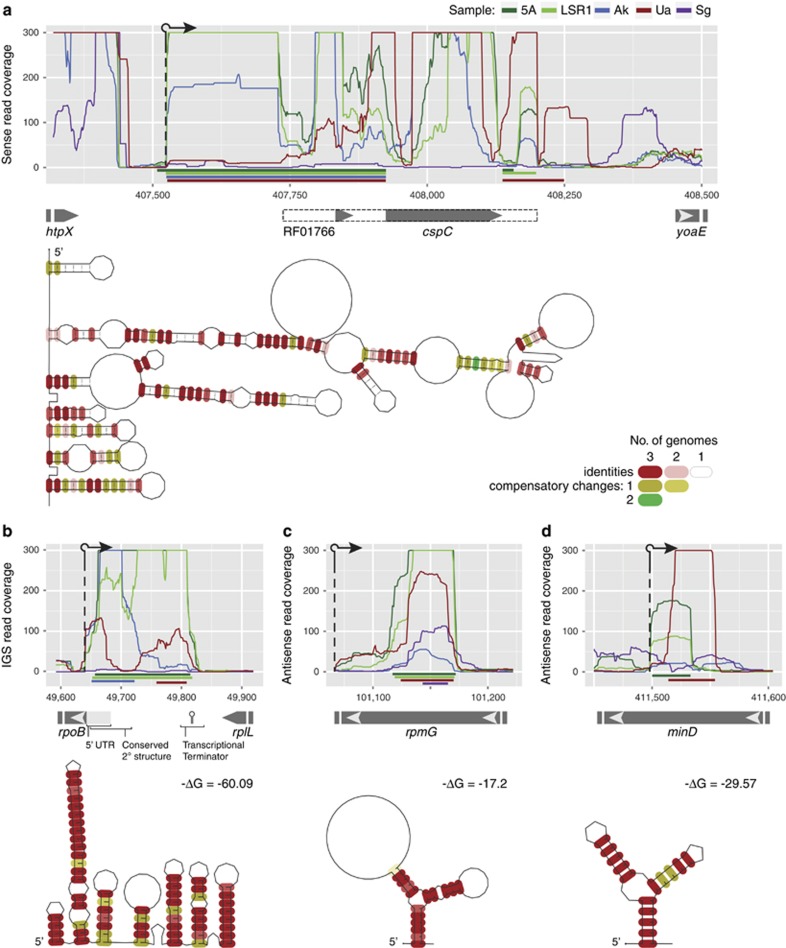
The predicted UTRs are on average longer than those reported for many free-living bacteria (30–60 nt vs 20–40 nt; Table 3); long 5′ UTRs in free-living bacteria often contain regulatory RNA sensor elements in the 5′ UTR region (Caldelari et al., 2013). One of the longest conserved UTRs occurs upstream of cspC in Buchnera-5A, Buchnera-LSR1, Buchnera-Ak and Buchnera-Ua (cspC is absent in Buchnera-Sg; Figure 2a). This region is homologous to the cspA thermoregulatory element characterized in E. coli (RF01766; Giuliodori et al., 2010), although we note that the 5′ UTRs predicted by Rockhopper extend a further 200 nt upstream of the Rfam model (Figure 2a). Within the 5′ UTR of cspC from Buchnera-5A, Buchnera-LSR1 and Buchnera-Ak, we identified a small 36 aa open reading frame, similar to the genetic organization of YobF and CspC in E. coli (Hemm et al., 2009). However, this open reading frame has no sequence similarity to YobF and has two inactivating mutations in Buchnera-Ua, so it remains unclear if this small open reading frame in Buchnera-5A, Buchnera-LSR1 and Buchnera-Ak has any functional significance. The 5′ UTR of cspE, a homolog of cspA, exhibits conserved expression in all four species, but it does not encode a structural match for this thermoregulator.
Table 3. Summary of expressed and structurally conserved UTRs in Buchnera.
| Buchnera-5A | Buchnera-LSR1 | Buchnera-Ak | Buchnera-Ua | Buchnera-Sg | |
|---|---|---|---|---|---|
| 5′ UTRs | |||||
| No. | 184 | 182 | 160 | 113 | 174 |
| Mean±s.d. | 58.6±49.6 | 55.0±46.0 | 49.9±43.7 | 33.3±31.6 | 60.2±66.3 |
| Longest | 229 | 287 | 252 | 226 | 686 |
| No. ⩾100 nt | 102 | 99 | 119 | 150 | 109 |
| 3′ UTRs | |||||
| No. | 211 | 219 | 195 | 150 | 210 |
| Mean±s.d. | 55.4±62.5 | 53.7±53.4 | 46.5±44.6 | 42.9±46.9 | 41.4±40.3 |
| Longest | 330 | 330 | 220 | 348 | 241 |
| No. ⩾100 nt | 83 | 81 | 85 | 122 | 72 |
Abbreviation: UTR, untranslated region.
Figure 2.
Extensive expression and conservation of small RNA elements in Buchnera. Small RNA expression profiles of Buchnera from five aphid strains were compared revealing numerous instances of conserved small RNAs. For example, (a) CspC encodes a particularly long, sequence and structurally conserved 5′ UTR that overlaps a predicted thermoregulatory element (RFAM model RF01766; dashed box). Also discreet small RNAs were identified in 25 intergenic spacers including one between rpoB and rplL (b), which is expressed in the opposite orientation to the flanking genes and ends at the transcriptional terminator predicted for rplL. We also identified over 115 sequence and structurally conserved asRNAs such as those in (c) rmpG and (d) minD. In panels (a–d), RNAseq raw read coverage traces (for example, not normalized) are shown for each Buchnera strain according to the colors indicated in a. Coordinates for the x axes correspond to a global Buchnera genome alignment (see Materials and methods section). Below the read coverage traces, colored horizontal lines indicate the (a) UTR and (b–d) small RNA regions predicted by Rockhopper. Gray and white arrows indicate coding sequence boundaries. Structural conservation diagrams are based on RNAalifold and predicted thermostabilty is indicated when available.
In addition to cspC's putative thermoregulatory element, we found that 77 of the 277 UTRs that were thermodynamically stable correspond to previous computational predictions (5′ UTRs=16 and 3′ UTRs=61; Degnan et al., 2011). The majority of the 3′ UTRs coincide with 45 of 55 predicted Rho-independent transcriptional terminators. As expected transcript coverage ends at or shortly after the 3′ terminus of the predicted element. The remaining conserved UTRs overlap predicted elements that contain highly conserved secondary structures or putative small RNAs (Supplementary Table S5; Degnan et al., 2011).
The widespread occurrence and conservation of such thermodynamically stable UTRs indicate that these UTRs may have an important role in transcriptional or post-transcriptional regulation in Buchnera. For example, 43% of differentially expressed proteins in this study significantly express UTRs that are structurally conserved cis to the gene (Supplementary Table S5). Alternatively, UTRs may simply aid in transcript stability, counteracting the consequences of A+T compositional bias in Buchnera (Lambert and Moran, 1998; Hansen and Moran, 2012).
Discreet small RNAs identified in intergenic spacers
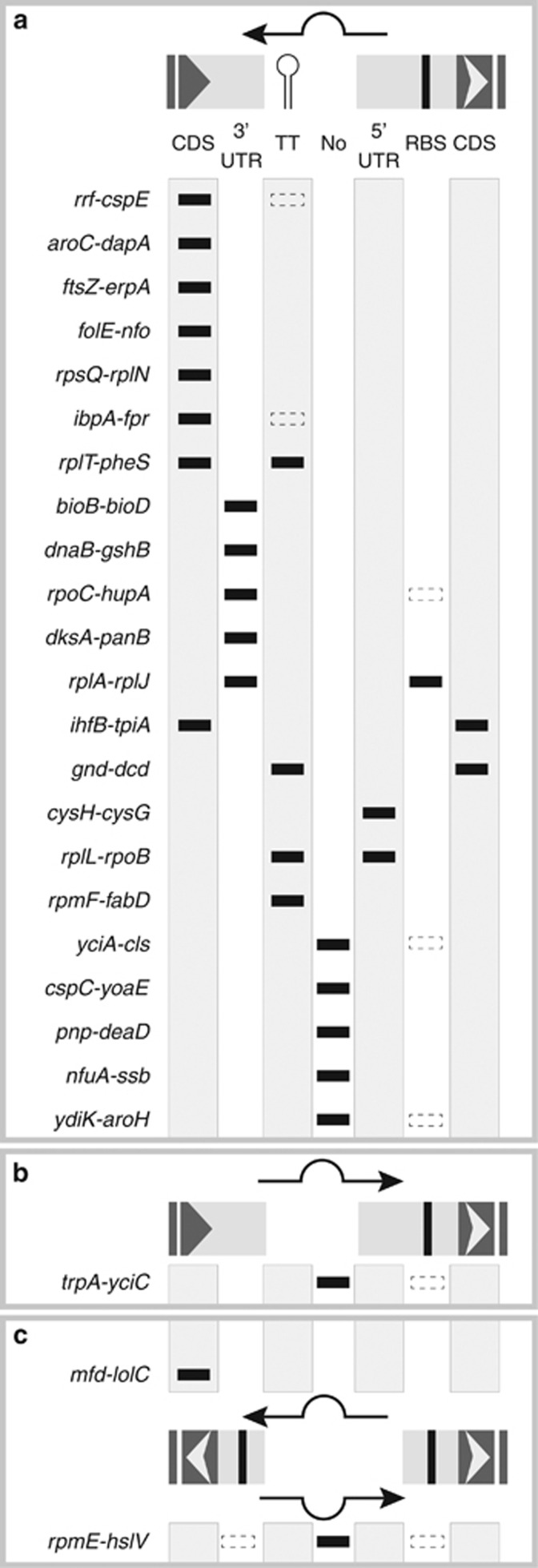
In addition to the UTRs described above, Rockhopper results identified significant expression of 60 possible small RNA elements in orthologous Buchnera intergenic spacers. After manual curation, only 25 putative small RNAs could be readily distinguished as discrete elements (Supplementary Table S6); the remainder were either associated with asRNA expression of transfer RNAs (see Hansen and Moran, 2012) or expressed in the same direction as a possible UTR or co-transcribed operon. Out of 25 discrete intergenic RNAs, 17 have significantly greater predicted thermodynamic stability and sequence covariation than randomized alignments (one-tailed t-test, d.f.=99, P<0.05). Moreover, 15 out of the 25 intergenic spacers with discrete small RNA expression levels were previously predicted as putative conserved functional sequences using computational methods (Degnan et al., 2011) and 73% of these discrete intergenic RNAs overlap the predicted region(s) (Supplementary Table S6). For example, an element that corresponds to a highly conserved region in all four species and is expressed antisense to the 5′ UTR of rpoB may have a potential role in cis-acting gene regulation (Figure 2b). Overall discrete intergenic small RNAs may be important in cis-acting gene regulation because they overlap 5′ or 3′ UTRs, transcription start sites, ribosome-binding sites and/or the coding regions of the flanking genes (Figure 3). We note that four differentially expressed proteins in this study encode significantly expressed discrete intergenic small RNAs cis to the gene (Supplementary Table S6).
Figure 3.
Localization and orientation of Buchnera intergenic small RNAs (sRNAs). Schematic representation of the location of conserved Buchnera sRNAs identified between 25 gene pairs are shown. (a) The vast majority of intergenic sRNAs identified occur antisense to pairs of co-directionally transcribed flanking genes. However, (b) one candidate occurred in the same direction as the flanking genes and (c) two candidates were found between divergently transcribed genes. Filled markers indicate that the sRNA overlaps the indicated element; coding sequence (CDS), 3′ UTR, transcriptional terminator (TT), no conserved element (No), 5′ UTR or a ribosome-binding site (RBS). Dashed markers indicate the presence of a conserved RBS or TT, which is not covered by the sRNA.
Extensive asRNA expression in Buchnera
Ten to nearly 30% of the directional RNAseq reads from all of the Buchnera species were expressed antisense to known gene regions (Table 2). As above, we compared predicted asRNAs from orthologous genes and identified 110 genes with 115 conserved asRNAs present in two or more species (Supplementary Table S7). This indicates that roughly one-fifth of the Buchnera genome experiences significant antisense expression from conserved locations. The majority of the asRNAs also showed significant predicted thermodynamic stability and sequence covariation (80/115, one-tailed t-test, d.f.=99, P<0.05; Figures 2c and d). Examination of the relative location of the asRNA transcripts reveals that ∼40% of asRNAs directly overlap the 5′ or 3′ end of the sense transcript (45/115) with the remainder occurring elsewhere within the transcript. As with the intergenic RNAs, it is possible that these asRNAs contribute to post-transcriptional regulation in Buchnera. For example, 14 proteins differentially expressed in this study display significantly expressed small RNAs antisense to the coding sequence (Supplementary Table S7). Five and two of these asRNAs overlapped the 3′ and 5′ end, respectively, of these differentially expressed genes. Only two of these genes were differentially enriched in the bacteriocytes compared with the embryo (Supplementary Table S7). As such, these asRNAs have the potential to contribute to post-transcriptional regulation of a variety of functional categories including amino-acid biosynthesis (carB and thrB), one of the key functions performed by Buchnera.
Putative translational inhibition by the cis- and trans-acting elements
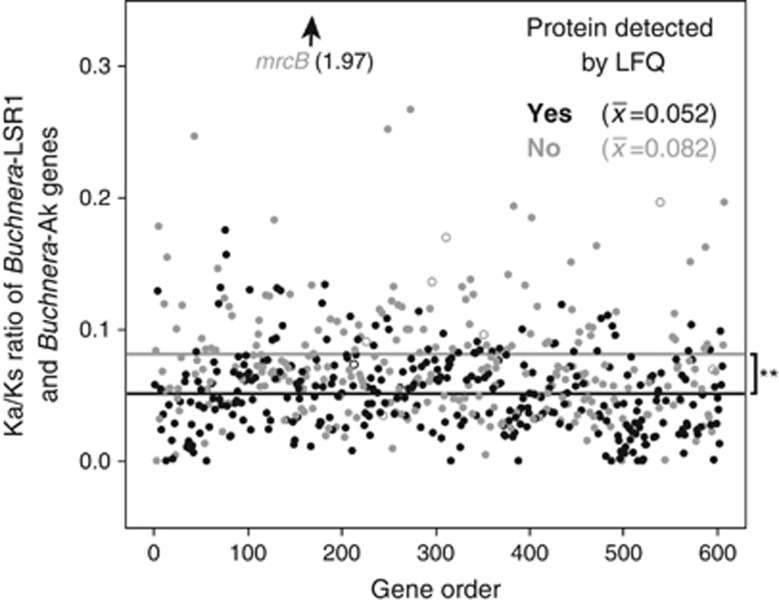
Twenty-eight of the Buchnera-LSR1 genes tested with quantitative reverse transcriptase-PCR flanked intergenic spacers with a small RNA expressed in the opposite direction (Supplementary Figure S2). Although some of the flanking genes were identified as encoding proteins, we found several situations in which proteins corresponding to the upstream or downstream gene were not detected either by LFQ or MudPIT analyses. For example, small RNAs overlapped the 5′ ends of cysG and lolC and the 3′ end of ihfB, dnaB, folE and fpr, and these genes were not detected as proteins (Supplementary Table S6). This may be evidence of translational inhibition by cis-acting elements. A comparison of the Buchnera proteins detected here and those detected by Poliakov et al. (2011) reveals a high degree of correspondence (see Supplementary Materials and methods; Supplementary Figure S3); however, not detecting a protein with LFQ or MudPIT does not necessarily mean that the protein is not expressed. Nevertheless, to indirectly assess the evidence for translational inhibition, we analyzed the relative selective constraint of genes encoding proteins that were detected or not. We calculated the ratio of nonsynonymous to synonymous nucleotide substitutions (Ka/Ks) between Buchnera-LSR1 and a related species Buchnera-Ak. From this analysis, we found that the average Ka/Ks ratio was significantly higher in genes encoding proteins not detected in the LFQ analysis compared with those proteins that were (X=0.082 and 0.052, respectively, Wilcoxon rank sums, P<0.0001; Figure 4). The third codon positions of genes encoding asRNAs may not be strictly neutral leading to the appearance of increased relaxed selection. To test this, we removed regions encoding asRNAs from the sequence alignments and re-estimated Ka/Ks. Nevertheless, after this subsequent analysis the significant pattern persists (X=0.0824 and 0.0518, respectively, Wilcoxon rank sums, P<0.0001) likely due to the fact that the Ka/Ks estimates are averaged across the entire length of the gene and the asRNA sequences are a fraction of overall protein lengths (mean 22%±19%). The pattern also holds when short genes (<150AA) or genes with saturated Ks estimates (⩾3.0) are removed from the analysis. These results are consistent with relaxation of selection (decrease in purifying selection) of the particular genes encoding proteins that were not detected. One consequence of less efficient purifying selection is the accumulation of slightly deleterious amino-acid changes (Moran, 1996). In turn, cis and/or trans-acting small RNAs we identified here may be important inhibitors of these potentially toxic proteins (Kuo and Ochman, 2010). Notably, one-quarter of undetected proteins have cis or antisense small RNAs associated with the gene (52/206) (Supplementary Tables S6 and S7). Buchnera does not encode Hfq, which is generally required to chaperone trans-acting small RNAs in model organisms; however, other candidate chaperone proteins may be involved.
Figure 4.
Expressed Buchnera proteins exhibit a high degree of sequence conservation. Buchnera-LSR1 genes with detected proteins (black) are significantly more conserved than those without detected proteins (grey) (Wilcoxon rank sums, **P<0.001). The degree of sequence conservation is determined as the ratio of nonsynonymous changes per nonsynonymous site (Ka) to synonymous changes per synonymous site (Ks) between orthologous genes of Buchnera-LSR1 and Buchnera-Ak. Ka/Ks ratios for each gene are plotted according to their genome gene order. Pseudogenes are marked with open circles.
Discussion
Our study reveals for the first time that Buchnera, an uncultivable symbiont with a small genome, experiences differential protein expression when it transitions from an extracellular to intracellular life stage. These patterns in differential protein expression are unlikely to be due to ‘typical' transcriptional regulatory mechanisms, because (i) most of its ancestral transcription factors have been eroded from Buchnera's genome, (ii) most conserved operons have been fragmented and (iii) no evidence of differential mRNA expression is evident between life stages. In turn, post-transcriptional processes may be the primary cause of differential gene regulation in Buchnera, similar to what has been widely observed in mitochondria and plastids (Mercer et al., 2011; Cardi et al., 2012).
Small RNAs in free-living bacteria have a role in one type of post-transcriptional gene regulation (Waters and Storz, 2009; Gottesman and Storz, 2011; Storz et al., 2011). Small RNAs are also expressed from human mitochondria, and are predicted to have a post-transcriptional role for organelle gene expression (Mercer et al., 2011). Similarly, we discovered widespread expression of conserved small RNAs in four reduced endosymbiont species that diverged ∼65 million years ago (Figure 5). These conserved small RNAs are expressed in UTRs, intergenic spacers and antisense to coding sequences (asRNA). In addition to having similar expression profiles, most of the small RNAs also have conserved secondary structures. Our findings predict that these novel small RNAs may have important functional roles for Buchnera in transcript stability, regulation and/or inhibition of toxic proteins.
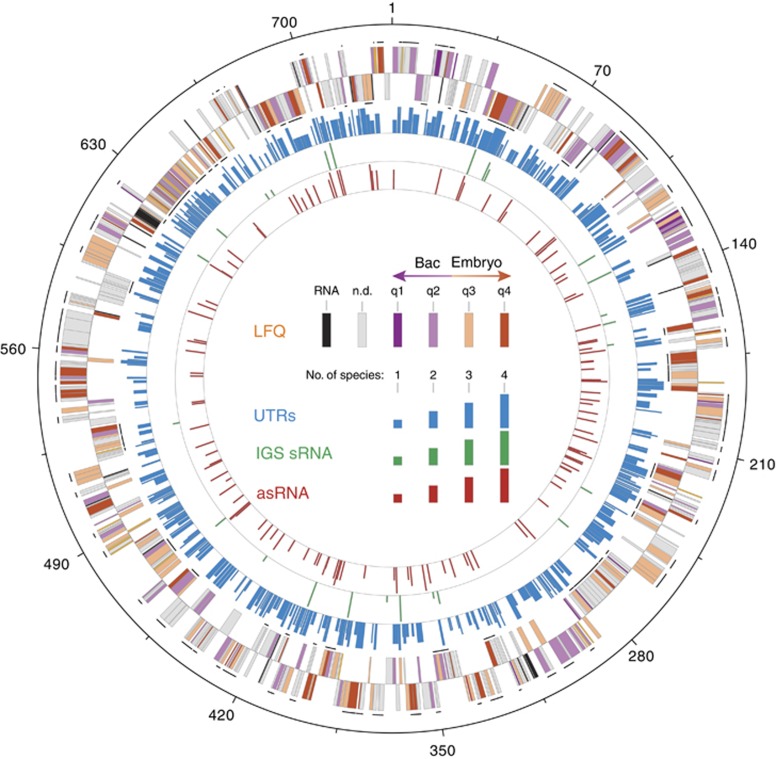
Figure 5.
Genome-wide map of protein and small RNA (sRNA) expression in Buchnera. Protein expression profiles and identified sRNAs are mapped onto the global alignment of the five Buchnera strain genomes. The outermost circles indicate genes on the 5′ and 3′ strands of the genome starting from the origin of replication (nt 1). Genes are colored according to the key as canonical RNAs (black), not detected in the LFQ analysis (gray), or differentially expressed as proteins (dark purple = upregulated in the Bacteriocytes (Bac) to dark orange = upregulated in the Embryos). Predicted transcriptional units with more than one protein are shown as solid black lines. The next circle shows genes that were found to have a 5′ and/or 3′ UTR (blue) and the following two circles denote the locations of the conserved intergenic sRNAs (green) and asRNA (red), respectively. Bar heights correspond to the number of genomes the element was conserved in. For simplicity, Buchnera-LSR1 and Buchnera-5A are represented as the consensus ‘Buchnera-Ap'.
Evolution of base-pairing small RNAs in free-living bacteria is assumed to be rapid. However, closely related bacteria can encode similar trans-encoded small RNAs, they generally do not express the same asRNAs (Raghavan et al., 2012). The widespread maintenance of shared cis and/or trans-encoded small RNAs and asRNAs in the four Buchnera lineages spanning ∼65 million years in divergence is unexpected. Nevertheless, these small RNAs are not conserved in all Buchnera lineages examined in this study, and therefore a subset of small RNAs (for example, conserved in only one or two lineages) are potentially dynamic and rapidly evolving in Buchnera. Potentially in these small genomes, the loss of canonical regulatory proteins and the inability to obtain additional genes through horizontal transfer has resulted in the evolution of alternative regulatory mechanisms, such as small RNAs, which can evolve faster than proteins.
The primary role of Buchnera for its aphid host is to produce essential amino acids (Hansen and Moran, 2014). Consistent with this role, the main KEGG pathway differentially expressed in this study at the protein level was related to the biosynthesis of amino acids, particularly essential amino acids. However, to our surprise we discovered that proteins in different essential amino-acid pathways were differentially regulated depending on Buchnera life stage. These proteins have lost many of the canonical means of transcriptional regulation (Table 1). Nevertheless, most of these genes have either asRNAs identified within their transcriptional unit or small RNAs flanking them (thrB, fba, serC, hisC, hisG and ilvH). Moreover, an additional 16 genes related to amino-acid biosynthesis, but not differentially expressed in this life stage sample comparison, may also be impacted by either asRNAs within their transcriptional unit or a flanking small RNA (Supplementary Tables S2, S6 and S7).
In our study, we found no evidence of differential mRNA expression between Buchnera life stages in spite of a clear signal of differential protein expression, and the retention of only a subset of predicted transcriptional regulators. These results are contrary to previous data from microarrays, which suggested that there were considerable mRNA differences among these life stages (Bermingham et al., 2009). However, reanalysis of these data indicates that, when the necessary false discovery rate criterion and quality controls are applied, there are in fact no significant differences in Buchnera transcription (Supplementary Microarray Reanalysis and Supplementary Figure S4), which is consistent with other studies on Buchnera mRNA regulation (Wilcox et al., 2003; Moran et al., 2003, 2005; Reymond et al., 2006; Viñuelas et al., 2011).
Overall, our study provides evidence of protein regulation in small symbiont genomes and a large number of expressed small RNAs and UTRs that are conserved among divergent lineages of this symbiont (Figure 5). We predict that these small RNA candidates are involved in post-transcriptional regulatory processes. Other post-transcriptional mechanisms such as Buchnera- or aphid-encoded proteases or allosteric regulation may also be important for Buchnera protein regulation. However, if these small genomes rely primarily on small RNAs instead of proteins for gene regulation, they could provide prime examples of how genomes return back to the ‘RNA world' for gene regulation. Furthermore, this work adds new insight and understanding of the model Buchnera-aphid nutritional symbiosis, and thus these findings may have broader implications for other such host–microbe associations (Hansen and Moran, 2014).
Acknowledgments
We thank Nancy Moran for discussions of the experiments and comments on the manuscript. We also thank Cari Vanderpool and three anonymous reviewers for their helpful comments on the manuscript, and Adam Hejmowski and Daniel Phillips for technical assistance. This work was supported by the USDA AFRI postdoctoral fellowship from the United States Department of Agriculture to AKH (2011-67012-30707), and University of Illinois at Urbana-Champaign start-up funds to AKH and PHD.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Babaei F, Ramalingam R, Tavendale A, Liang Y, Yan LS, Ajuh P, et al. Novel blood collection method allows plasma proteome analysis from single zebrafish. J Proteome Res. 2013;12:1580–1590. doi: 10.1021/pr3009226. [DOI] [PubMed] [Google Scholar]
- Bermingham J, Rabatel A, Calevro F, Viñuelas J, Febvay G, Charles H, et al. Impact of host developmental age on the transcriptome of the symbiotic bacterium Buchnera aphidicola in the pea aphid (Acyrthosiphonpisum) Appl Environ Microbiol. 2009;75:7294–7297. doi: 10.1128/AEM.01472-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhart SH, Hofacker IL, Will S, Gruber AR, Stadler PF. RNAalifold: improved consensus structure prediction for RNA alignments. BMC Bioinformatics. 2008;9:474. doi: 10.1186/1471-2105-9-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagoev B, Ong S, Kratchmarova I, Mann M. Temporal analysis of phosphotyrosine-dependent signaling networks by quantitative proteomics. Nature Biotechnol. 2004;22:1139–1145. doi: 10.1038/nbt1005. [DOI] [PubMed] [Google Scholar]
- Bookout AL, Cummins CL, Mangelsdorf DJ, Pesola JM, Kramer MF. Current Protocols in Molecular Biology. John Wiley & Sons Inc.: Hoboken, NJ; 2006. High-throughput real-time quantitative reverse transcription PCR; pp. 15.8.1–15.8.28. [DOI] [PubMed] [Google Scholar]
- Braendle C, Miura T, Bickel R, Shingleton AW, Kambhampati S, Stern DL. Developmental origin and evolution of bacteriocytes in the aphid–Buchnera symbiosis. PloS Biol. 2003;1:e1. doi: 10.1371/journal.pbio.0000021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldelari I, Chao Y, Romby P, Vogel J. RNA-mediated regulation in pathogenic Bacteria. Cold Spring Harb Perspect Med. 2013;3:a010298–a010298. doi: 10.1101/cshperspect.a010298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardi T, Giegé P, Kahlau S, Scotti N. Advances in Photosynthesis and Respiration 35. Springer Netherlands: Houten, The Netherlands; 2012. Expression profiling of organellar genes; pp. 323–355. [Google Scholar]
- Charles H, Ishikawa H. Physical and genetic map of the genome of Buchnera, the primary endosymbiont of the pea aphid Acyrthosiphon pisum. J Mol Evol. 1999;48:142–150. doi: 10.1007/pl00006452. [DOI] [PubMed] [Google Scholar]
- Dandekar T, Huynen M, Regula JT, Ueberle B, Zimmermann CU, Andrade MA, et al. Re-annotating the Mycoplasma pneumoniae genome sequence: adding value, function and reading frames. Nucleic Acids Res. 2000;28:3278–3288. doi: 10.1093/nar/28.17.3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling ACE. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degnan PH, Ochman H, Moran NA. Sequence conservation and functional constraint on intergenic spacers in reduced genomes of the obligate symbiont Buchnera. PLoS Genet. 2011;7:e1002252. doi: 10.1371/journal.pgen.1002252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas AE, Prosser WA. Synthesis of the essential amino acid tryptophan in the pea aphid (Acyrthosiphon pisum) symbiosis. J Insect Physioly. 1992;38:565–568. [Google Scholar]
- Dunbar HE, Wilson AC, Ferguson NR, Moran NA. Aphid thermal tolerance is governed by a point mutation in bacterial symbionts. PLoS Biol. 2007;5:e96. doi: 10.1371/journal.pbio.0050096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuliodori AM, Di Pietro F, Marzi S, Masquida B, Wagner R, Romby P, et al. The cspA mRNA is a thermosensor that modulates translation of the cold-shock protein CspA. Molecular Cell. 2010;37:21–33. doi: 10.1016/j.molcel.2009.11.033. [DOI] [PubMed] [Google Scholar]
- Gottesman S, Storz G. Bacterial small RNA regulators: versatile roles and rapidly evolving variations. Cold Spring Harbor Perspect Biol. 2011;3:a003798–a003798. doi: 10.1101/cshperspect.a003798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guell M, van Noort V, Yus E, Chen WH, Leigh-Bell J, Michalodimitrakis K, et al. Transcriptome complexity in a genome-reduced bacterium. Science. 2009;326:1268–1271. doi: 10.1126/science.1176951. [DOI] [PubMed] [Google Scholar]
- Hansen AK, Moran NA. Aphid genome expression reveals host-symbiont cooperation in the production of amino acids. Proc Natl Acad Sci USA. 2011;108:2849–2854. doi: 10.1073/pnas.1013465108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen AK, Moran NA. Altered tRNA characteristics and 3' maturation in bacterial symbionts with reduced genomes. Nucleic Acids Res. 2012;40:7870–7884. doi: 10.1093/nar/gks503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen AK, Moran NA. The impact of microbial symbionts on host plant utilization by herbivorous insects. Mol Ecol. 2014;23:1473–1496. doi: 10.1111/mec.12421. [DOI] [PubMed] [Google Scholar]
- Hemm MR, Paul BJ, Miranda-Rios J, Zhang A, Soltanzad N, Storz G. Small stress response proteins in Escherichia coli: proteins missed by classical proteomic studies. J Bacteriol. 2009;192:46–58. doi: 10.1128/JB.00872-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofacker IL, Fekete F, Stadler PF. Secondary structure prediction for aligned RNA sequences. J Mol Biol. 2002;319:1059–1066. doi: 10.1016/S0022-2836(02)00308-X. [DOI] [PubMed] [Google Scholar]
- Kahle D, Wickham H. ggmap: spatial visualization with ggplot2. R Journal. 2013;5:144–161. [Google Scholar]
- Kal AJ, van Zonneveld AJ, Benes V, van den Berg M, Koerkamp MG, Albermann K, et al. Dynamics of gene expression revealed by comparison of serial analysis of gene expression transcript profiles from yeast grown on two different carbon sources. Mol Biol Cell. 1999;10:1859–1872. doi: 10.1091/mbc.10.6.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga R, Meng X-Y, Tsuchida T, Fukatsu T. Cellular mechanism for selective vertical transmission of an obligate insect symbiont at the bacteriocyte-embryo interface. Proc Natl Acad Sci USA. 2012;109:E1230–E1237. doi: 10.1073/pnas.1119212109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhner S, van Noort V, Betts MJ, Leo-Macias A, Batisse C, Rode M, et al. Proteome organization in a genome-reduced bacterium. Science. 2009;326:1235–1240. doi: 10.1126/science.1176343. [DOI] [PubMed] [Google Scholar]
- Kuo C-H, Ochman H. The extinction dynamics of bacterial pseudogenes. PLoS Genet. 2010;6:e1001050. doi: 10.1371/journal.pgen.1001050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert JD, Moran NA. Deleterious mutations destabilize ribosomal RNA in endosymbiotic bacteria. Proc Natl Acad Sci USA. 1998;95:4458–4462. doi: 10.1073/pnas.95.8.4458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- McClure R, Balasubramanian D, Sun Y, Bobrovskyy M, Sumby P, Genco CA, et al. Computational analysis of bacterial RNA-Seq data. Nucleic Acids Res. 2013;41:e140–e140. doi: 10.1093/nar/gkt444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Neph S, Dinger ME, Crawford J, Smith MA, Shearwood A-MJ, et al. The human mitochondrial transcriptome. Cell. 2011;146:645–658. doi: 10.1016/j.cell.2011.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittenhuber G. An inventory of genes encoding RNA polymerase sigma factors in 31 completely sequenced eubacterial genomes. J Mol Microbiol Biotechnol. 2002;4:77–91. [PubMed] [Google Scholar]
- Miura T, Braendle C, Shingleton A, Sisk G, Kambhampati S, Stern DL. A comparison of parthenogenetic and sexual embryogenesis of the pea aphid Acyrthosiphon pisum (Hemiptera: Aphidoidea) J Exp Zool B Mol Dev Evol. 2003;295:59–81. doi: 10.1002/jez.b.3. [DOI] [PubMed] [Google Scholar]
- Moran NA. Accelerated evolution and Muller's rachet in endosymbiotic bacteria. Proc Natl Acad Sci USA. 1996;93:2873–2878. doi: 10.1073/pnas.93.7.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran NA, Dunbar HE, Wilcox JL. Regulation of transcription in a reduced bacterial genome: nutrient-provisioning genes of the obligate symbiont Buchnera aphidicola. J Bacteriol. 2005;187:4229–4237. doi: 10.1128/JB.187.12.4229-4237.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran NA, Mira A.2001The process of genome shrinkage in the obligate symbiont Buchneraaphidicola Genome Biol 2RESEARCH0054.1–0054.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran NA, Plague GR, Sandström JP, Wilcox JL. A genomic perspective on nutrient provisioning by bacterial symbionts of insects. Proc Natl Acad Sci USA. 2003;100 (Suppl 2:14543–14548. doi: 10.1073/pnas.2135345100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poliakov A, Russell CW, Ponnala L, Hoops HJ, Sun Q, Douglas AE, van Wijk KJ. Large-scale label-free quantitative proteomics of the pea aphid-Buchnera symbiosis. Molec Cell Proteom. 2011;10:M110.007039. doi: 10.1074/mcp.M110.007039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavan R, Sloan DB, Ochman H. Antisense transcription is pervasive but rarely conserved in enteric bacteria. mBio. 2012;3:e00156–12. doi: 10.1128/mBio.00156-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond N, Calevro F, Viñuelas J, Morin N, Rahbe Y, Febvay G, et al. Different levels of transcriptional regulation due to trophic constraints in the reduced genome of Buchnera aphidicola APS. Appl Environ Microbiol. 2006;72:7760–7766. doi: 10.1128/AEM.01118-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P. Artemis: sequence visulization and annotation. Bioinformatics. 2000;16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- Sasaki T, Ishikawa H. Production of essential amino acids from glutamate by mycetocyte symbionts of the pea aphid Acyrthosiphon pisum. J Insect Physiol. 1995;41:41–46. [Google Scholar]
- Shigenobu S, Watanabe H, Hattori M, Sakaki Y, Ishikawa H. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature. 2000;407:81–86. doi: 10.1038/35024074. [DOI] [PubMed] [Google Scholar]
- Storz G, Vogel J, Wassarman KM. Regulation by small RNAs in Bacteria: expanding frontiers. Mol Cell. 2011;43:880–891. doi: 10.1016/j.molcel.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viñuelas J, Febvay G, Duport G, Colella S, Fayard J-M, Charles H, et al. Multimodal dynamic response of the Buchnera aphidicola pLeu plasmid to variations in leucine demand of its host, the pea aphid Acyrthosiphon pisum. Mol Microbiol. 2011;81:1271–1285. doi: 10.1111/j.1365-2958.2011.07760.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters LS, Storz G. Regulatory RNAs in bacteria. Cell. 2009;136:615–628. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcox JL, Dunbar HE, Wolfinger RD, Moran NA. Consequences of reductive evolution for gene expression in an obligate endosymbiont. Mol Microbiol. 2003;48:1491–1500. doi: 10.1046/j.1365-2958.2003.03522.x. [DOI] [PubMed] [Google Scholar]
- Yus E, Maier T, Michalodimitrakis K, van Noort V, Yamada T, Chen WH, et al. Impact of genome reduction on bacterial metabolism and its regulation. Science. 2009;326:1263–1268. doi: 10.1126/science.1177263. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.