Abstract
Bacterial gut symbiont communities are critical for the health of many insect species. However, little is known about how microbial communities vary among host species or how they respond to anthropogenic disturbances. Bacterial communities that differ in richness or composition may vary in their ability to provide nutrients or defenses. We used deep sequencing to investigate gut microbiota of three species in the genus Bombus (bumble bees). Bombus are among the most economically and ecologically important non-managed pollinators. Some species have experienced dramatic declines, probably due to pathogens and land-use change. We examined variation within and across bee species and between semi-natural and conventional agricultural habitats. We categorized as ‘core bacteria' any operational taxonomic units (OTUs) with closest hits to sequences previously found exclusively or primarily in the guts of honey bees and bumble bees (genera Apis and Bombus). Microbial community composition differed among bee species. Richness, defined as number of bacterial OTUs, was highest for B. bimaculatus and B. impatiens. For B. bimaculatus, this was due to high richness of non-core bacteria. We found little effect of habitat on microbial communities. Richness of non-core bacteria was negatively associated with bacterial abundance in individual bees, possibly due to deeper sampling of non-core bacteria in bees with low populations of core bacteria. Infection by the gut parasite Crithidia was negatively associated with abundance of the core bacterium Gilliamella and positively associated with richness of non-core bacteria. Our results indicate that Bombus species have distinctive gut communities, and community-level variation is associated with pathogen infection.
Keywords: Bombus, Gilliamella, gut microbiota, land-use change, pyrosequencing
Introduction
Diverse communities of bacteria inhabit insect guts and provide critical functions for their hosts, such as digestion (Warnecke et al., 2007), pathogen defense (Dillon et al., 2005) and insecticide resistance (Kikuchi et al., 2012). Most work on functional importance of gut microbiota has focused on one or a few species of bacteria. However, community-level factors such as composition and richness may also impact insect health and survival through mechanisms, such as colonization resistance (Dillon et al., 2005). For example, particular bacterial communities are better able to resist invasion by pathogens in humans (Berg, 1996). Therefore broad-scale surveys of gut microbial communities coupled with studies of functional consequences for hosts, as has been done in human systems (Frank et al., 2007), will further the understanding of the role of gut microbiota in insect health, ecology and evolution (Hamdi et al., 2011).
When taking a community approach to understanding the insect microbiome, it is useful to differentiate between ‘core' taxa that are repeatedly found in individuals of a particular host species or cluster of closely related host species and ‘non-core' taxa that often occur in non-host-associated environments. Whereas some insect species possess erratic gut communities consisting of environmental bacteria that vary widely among individuals (Engel and Moran, 2013; Wong et al., 2013), other insect species have gut bacteria that are completely or largely restricted to the guts of their hosts (Warnecke et al., 2007; Martinson et al., 2011; Anderson et al., 2012; Sudakaran et al., 2012). Dominance by such core bacterial species may be more common in social insects as interactions among host individuals facilitate transmission (Martinson et al., 2011). Core bacteria may have evolved in close association with hosts and are potentially commensal or beneficial (Berg, 1996; Martinson et al., 2011; Koch et al., 2013). However, this assumption has rarely been tested. Although they are facultative, environmentally acquired bacteria sometimes provide fitness benefits to their hosts (Kikuchi et al., 2012; McFrederick et al., 2012; Ridley et al., 2012). High richness or abundance of non-core bacterial may also reflect dysbioses associated with disease and pathogens (Sartor, 2008; Hamdi et al., 2011).
In this study, we examine the composition of gut microbial communities in Bombus species (bumble bees). Bombus are among the most important wild pollinators in natural and agricultural habitats (Hegland and Totland, 2008; Garibaldi et al., 2013). Furthermore, some Bombus species have undergone dramatic recent declines. Some of these population declines may result from land-use change and pathogens (Goulson et al., 2008; Grixti et al., 2009; Williams and Osborne, 2009; Cameron et al., 2011), but the mechanisms underlying the declines of some Bombus species remain unknown. The few studies of Bombus gut microbiota reveal a simple yet distinctive bacteria community (Mohr and Tebbe, 2006; Martinson et al., 2011; Koch and Schmid-Hempel, 2011b). Bombus and Apis share many of the same groups of core bacterial taxa, including species within Alphaproteobacteria, Betaproteobacteria, Gammaproteobacteria and Lactobacillales (Martinson et al., 2011; Koch et al., 2012). However, previous studies of Bombus gut bacteria have mostly been targeted to specific taxa, and the few broad sequence-based surveys suggest more variation in Bombus gut communities (Martinson et al., 2011).
One critical role of gut bacteria could be pathogen defense, and pathogens are likely an important cause of declines in Bombus populations. Nosema bombi, an intracellular microsporidian that initially invades midgut epithelial cells, is suspected to negatively impact some North American Bombus species (Cameron et al., 2011), and Crithida bombi, a trypanosome that infect Bombus guts, may also lead to declines (Brown et al., 2003; Otterstatter and Thomson, 2008; but see Cordes et al., 2012). Using experimental manipulations, Koch and Schmid-Hempel (2011a, 2012) found that gut colonization by core bacteria, including Gilliamella (Gammaproteobacteria) and Snodgrassella (Betaproteobacteria) species, lowered Crithidia infection in an European bumble bee. Although the mechanism is unknown, these bacteria produce a biofilm on the hindgut wall that may impede infection by gut pathogens (Engel et al., 2012, Martinson et al., 2012).
Here we use deep sequencing to examine the gut microbial communities of three native, wild Bombus species (B. bimaculatus Cresson, B. impatiens Cresson and B. griseocollis DeGeer) collected from semi-natural and agricultural habitats. We sought to first determine whether microbial communities differ among bee species and, second, whether microbial communities are influenced by anthropogenic change. We focus on agricultural land-use change as it represents the dominant form of anthropogenic change globally (Pereira et al., 2010). Work in non-Bombus systems has shown that gut microbial communities can be influenced by host genotype, diet, geography and environmental factors (Zouache et al., 2010; Huang and Zhang, 2013). However, no studies have examined the effect of land-use change on the gut communities of Bombus. Agrochemical inputs can have dramatic negative consequences for managed and native bees (Whitehorn et al., 2012; Di Prisco et al., 2013; Pettis et al., 2013). A recent analysis conducted in our New Jersey study region found that cranberry agriculture (the habitat type examined here) used more types of fungicides than other crops and that fungicide exposure was associated with increased infection of honey bees (Apis mellifera) by the gut pathogen Nosema (Pettis et al., 2013).
Specifically, we investigated three questions: (1) How do gut microbial community metrics such as richness (number of operational taxonomic units (OTUs)), beta diversity and composition differ among three common Bombus species and between semi-natural and agricultural habitats? (2) Do core and non-core gut microbiota differ among species and between semi-natural and agricultural habitats? (3) Do core and non-core gut microbiota have different associations with (a) the infection of Bombus by two important gut parasites: Crithidia sp. and Nosema sp., and (b) the overall abundance of gut bacteria of the host?
Materials and methods
Study design and field data collection
To assess whether agricultural land-use affects Bombus gut microbiota, specimens were collected from six agricultural and six semi-natural sites in Burlington County, NJ, USA in May and June 2011 (Supplementary Figure S1). Agricultural sites were conventionally managed cranberry farms with high fungicide use implicated to have negative consequences for honey bee health (Pettis et al., 2013). Semi-natural habitats were abandoned cranberry bogs that had not been managed as farms for at least 5 years before collection and are reverting to natural wetlands. In all the 12 sites, the dominant flowering plant was cranberry (Vaccinium macrocarpon Aiton: Ericaceae); the use of a focal study plant limited between-treatment differences in diet that might confound effects of land-use and agrochemical input. Sites were separated by a mean of 19.9 km (4.2–38.4 km) to minimize the probability of collecting specimens from the same colony at different sites. In addition, sites of semi-natural habitats were located a mean of 20.2 km (6.1–38.4 km) from agricultural habitats to ensure that collected bees did not forage in areas of active agriculture. A Mantel test using our primary outcome variable (proportion of core bacterial species) indicated no spatial autocorrelation among sites (Mantel r=0.19; P=0.11).
At least three individual workers from each species were collected at each site. Specimens were identified in the field using morphological characteristics, placed into vials with 50 ml of 100% ethanol and held at 4 °C until further processing.
DNA preparation
DNA from whole guts was extracted and quantified using protocols described in Moran et al. (2012), and DNA extracts were normalized to⩽50 ng μl−1 for further procedures. Process controls included DNA of both Escherichia coli K-12 and A. mellifera microbiota (sample AZ125.3); the latter was characterized previously using the same pyrotag approach used in Moran et al. (2012). The presence of microbial DNA was verified by PCR using universal primers: 27F′-HT and 1492R′-HT (Tyson et al., 2004). Any specimens for which DNA failed to amplify were replaced by preparations from alternate specimens. Bombus species identifications were verified by amplifying and sequencing the Cytochrome Oxidase I gene using LepF1 and LepR1 primers (Hebert et al., 2004). Cleaned Cytochrome Oxidase I amplicons were sequenced at Yale Science Hill DNA Analysis Facility and returned sequences were submitted to the Barcode of Life Database (http://www.boldsystems.org) for taxonomic assignment (Ratnasingham and Herbert, 2007). Sequences from specimens field-identified as B. griseocollis contained several polymorphic regions, and taxonomic assignments for B. griseocollis specimens were based on manually trimmed monomorphic segments. Based on these sequences, one specimen was incorrectly identified in the field and removed from analysis. Primers and reaction conditions are listed in Supplementary Table S1.
PCR, pyrosequencing and pyrotag analysis
To amplify 16S rRNA gene sequences, primers 338GF_TBF and 1492R_TBRn were designed with 8 bp barcodes in BARCRAWL (Frank, 2009) with Titanium adaptors and Lib L keys (Supplementary Tables S1 and S2). Approximately 50 ng DNA from three individuals per species per site was amplified in triplicate along with a negative control. The triplicate reactions were pooled, and products were screened on 1% agarose gels. Reactions that failed to amplify or for which a negative control amplified were repeated. DNA from specimens that failed three attempts at amplification were replaced with alternate specimens. Any of the specimens of a species at a particular site that failed to amplify were pulled from the data set (n=7). Pooled amplicon was cleaned using Agencourt Ampure beads (Beckman Coulter, Brea, CA, USA) and quantified using the Quant-it Picogreen dsDNA quantification kit (Invitrogen, Life Technologies, Grand Island, NY, USA) on a Victor X3 plate reader (Perkin Elmer, Waltham, MA, USA). Equimolar amounts of cleaned amplicon were pooled and sent to the University of Georgia Genomics Facility for pyrosequencing via GS FLX Titanium XL+ (454 Life Sciences, Branford, CT, USA). Initially, a 1/8 pilot plate was sequenced followed by a full plate. Reads were analyzed using tools in QIIME versions 1.3 and 1.4 (Caporaso et al., 2010). A summary of read-processing steps and read-related data is in Supplementary Table S3, and a mapping file may be found in Supplementary Table S4.
Relative abundance of microbiota in individual samples was visualized by using ‘high abundance' OTUs binned taxonomically (Supplementary Table S5 and Supplementary Figure S2). To assign ‘high abundance' OTUs to specific taxonomic groups, BLASTn (Altschul et al., 1997) searches against the GenBank nr database (accessed 20 September 2012) were performed through the National Center for Biotechnology Information sequence search function of Geneious (Drummond et al., 2011). High abundance OTUs were defined as those constituting>1% of any one sample. These 76 OTUs (from a total of 352) contained 98.95% of total reads. The top 10 BLAST hits for each OTU were reviewed, and the best BLAST hit was chosen as a representative assignment for that OTU. Best BLAST hits were chosen based on the highest bit score. When multiple hits had equivalent ranking, sequences derived from bees, floral samples or other insect hosts were chosen. OTUs were binned at the taxonomic level of Order for analysis (Supplementary Table S5). Previously observed bacterial species found in corbiculate bees (for example, Gilliamella apicola and Snodgrassella alvi) are noted down to genus in the taxonomic and phylogenetic analyses. These taxonomical bins were used in order to visualize microbial community structure within samples (Supplementary Figure S2) while statistical analyses utilized ungrouped but taxonomically assigned OTUs. Control samples and one Bombus sample with few reads (<200) were removed. A total of 99 Bombus specimens were analyzed in the pyrotag data set comprised of 30 B. bimaculatus, 36 B. impatiens and 33 B. griseocollis.
Characterization of core vs non-core bacteria
Characterization of OTUs as either core or non-core was determined by phylogenetic placement for representative genomic sequences. OTUs were classified as core if they were monophyletic with other bacteria consistently and primarily associated with Apis and Bombus (major groups of corbiculate bees). This analysis was performed by retrieving full-length and nearly full-length 16S rRNA sequences of closely related taxa from GenBank. Representative sequences of bee-associated and other closely related taxa from previously published studies of Bombus and Apis microbiota were used (Cox-Foster et al., 2007; Martinson et al., 2011; Koch and Schmid-Hempel, 2011a). All sequences were aligned via web submission to Infernal aligner of RDP 2.2 (Wang et al., 2007). The alignments were used to reconstruct maximum-likelihood trees with bootstrap support via RAxML v7.4.2 (Stamatakis, 2006) in the CIPRES web portal (Miller et al., 2009). A GTRgamma base substitution model was used with 1000 iterations. Resulting Newick files were visualized with Figtree 1.4 (http://tree.bio.ed.ac.uk/software/figtree/).
The Lactobacillus tree was inferred by a different route because of the complexity and size of the genus and ambiguities within 16S rRNA gene alignments (Claesson et al., 2008). We used an alignment based on a secondary structure model used in a previous study of host specificity between hymenopterans and Lactobacilli (McFrederick et al., 2013). We obtained the alignment from TreeBASE (Morell, 1996), trimmed representative OTU sequences to 636 bp (5′ bases were ambiguously aligned in initial attempts) and then added them to the alignment using the PyNAST tool within QIIME. This full alignment was used to reconstruct maximum-likelihood trees and bin core associates as described above. All resulting trees are presented in Supplementary Figure S3. Sequences are deposited in the Sequence Read Archive under BioProject ID PRJNA217796.
Crithidia and Nosema infection
Infection status with the eukaryotic parasites Crithidia and Nosema was detected by PCRs specific for either parasite using DNA extracted from guts as template. For Crithidia, a part of the ITS region was amplified using the Crithidia-specific primers CrithITS1f and CrithITS1r (Schmid-Hempel and Tognazzo, 2010). To detect infections with Nosema, we used the Nosema-specific primer pair SSUrRNA-f1 and SSUrRNA-r1b (Li et al., 2012) targeting the microsporidian small subunit rRNA gene. In addition, all samples were screened with the N. bombi-specific primer pair N.b.af and N.b.ar (Erler et al., 2012). Primers and reaction conditions are listed in Supplementary Table S1. Positive amplification and product sizes were verified on a 1.5% agarose gel. Products were sequenced directly from positive PCR reactions with the respective forward primer for confirmation. Nosema SSU sequences were also sequenced with the reverse primer and assembled in MacVector (12.7). Nosema sequences were aligned in ClustalW (http://www.genome.jp/tools/clustalw/) (Thompson et al., 1994). We included sequences representing the known microsporidian parasites of bees and additional close matches in GenBank. A maximum-likelihood phylogenetic tree was computed with PhyML (Guindon et al., 2010) using a GTR+G+I model and 500 bootstrap replicates.
Quantification of bacterial abundance
To determine differences in abundance of bacteria among individual bees, absolute copy numbers of 16S rRNA genes in each sample were assessed using real-time qPCR. DNA samples were diluted to 1:10, and 1 μl was amplified in triplicate with 16S rRNA primers 27F and 355R (Castillo et al., 2006). Absolute gene copy numbers were determined with a standard curve using previously described methods (Martinson et al., 2012). Primers and reaction conditions are listed in Supplementary Table S1.
Statistical analysis
The total number of bacterial reads per sample was rarefied to the smallest number of reads for a given sample (n=2500). Statistical tests were performed using this subset.
Chao1 estimates of alpha diversity (number of bacteria OTUs) were calculated in QIIME and were used to examine differences in bacteria between the three species and habitat by performing analysis of variance in JMP10 (SAS, Cary, NC, USA). We use the number of OTUs as our measure of richness. Rarefaction curves of the Chao1 estimate were constructed in QIIME at 10 subsamplings of every 100 reads and were used to verify adequate depth of sampling (Supplementary Figures S4a and b). We used a beta-dispersion test to examine differences in beta diversity between habitats and bee species using a Bray–Curtis dissimilarity matrix (Anderson, 2006). This test determines whether the turnover of bacterial communities among individual bees differed among groups (species or habitat). We used an NMDS (nonmetric multidimensional scaling) to visualize differences in bacterial community composition among species and habitat and assessed differences statistically using a PERMANOVA for Bray–Curtis distances in community composition among groups (Anderson, 2001). Factors in the PERMANOVA were bee species, habitat and their interaction. All community composition analyses were done using the vegan package in R (Oksanen et al., 2013).
To determine whether core and non-core bacteria varied between habitat types or among bee species, a generalized linear mixed model was applied using the lmer package in R (Pinheiro et al., 2013). The response variable was proportion of core bacteria per bee. This parameter indicates how much of the total gut microbiota is composed of core bacterial groups. Bee species, habitat and their interaction were fixed effects. Collection site was a random effect. We used a Gaussian distribution as residuals were normally distributed. For this and all following generalized linear mixed models, non-significant interactions and fixed effects were removed from the model (P>0.05). To determine which core bacterial OTUs best characterized the gut microbiota communities as a function of bee species, habitat and pathogen infection status, we used indicator species analysis in labdsv package in R (Roberts, 2012). Indicator species values are based on how specific and widespread an OTU is within a particular group and are independent of the relative abundances of other bacteria (Dufrêne and Legendre, 1997).
A generalized linear mixed model was used to determine whether core and non-core gut microbiota differ in their association with infection by Crithidia and Nosema. Separate models were conducted for each pathogen. The presence or absence of Crithidia or Nosema was the response variable; therefore, a binomial error distribution was used. Bee species, absolute number of 16S rRNA gene copies, richness of core bacteria, richness of non-core bacteria and habitat were fixed effects. Site was a random effect.
Finally, we used a generalized linear mixed model to determine whether absolute number of 16S rRNA gene copies (an index of total bacterial counts per bee) was influenced by gut community richness. The response variable was the number of 16S rRNA genes, and it was log-transformed to control for overdispersion. As the residuals were normally distributed following transformation, we used a Gaussian error structure. Fixed effects were richness of core bacteria, richness of non-core bacteria, bee species and habitat.
Results
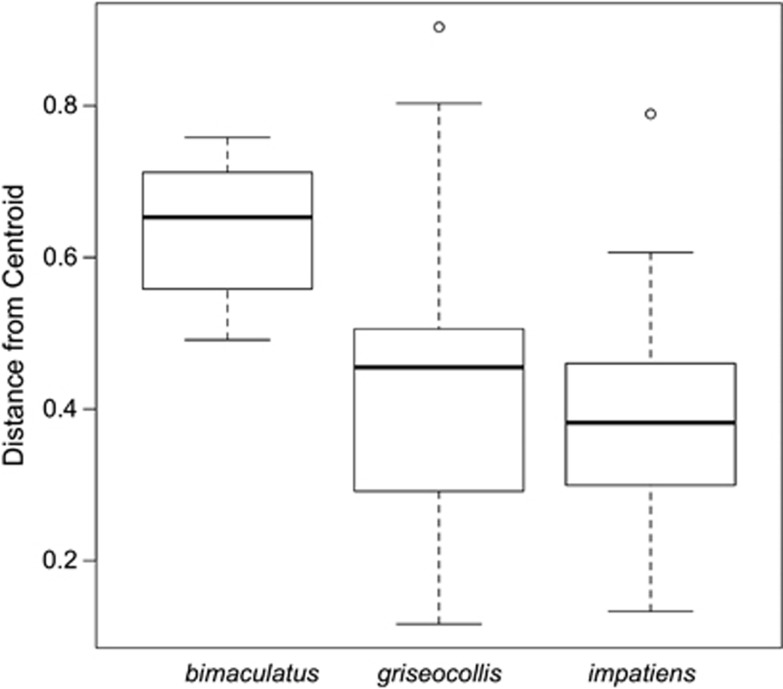
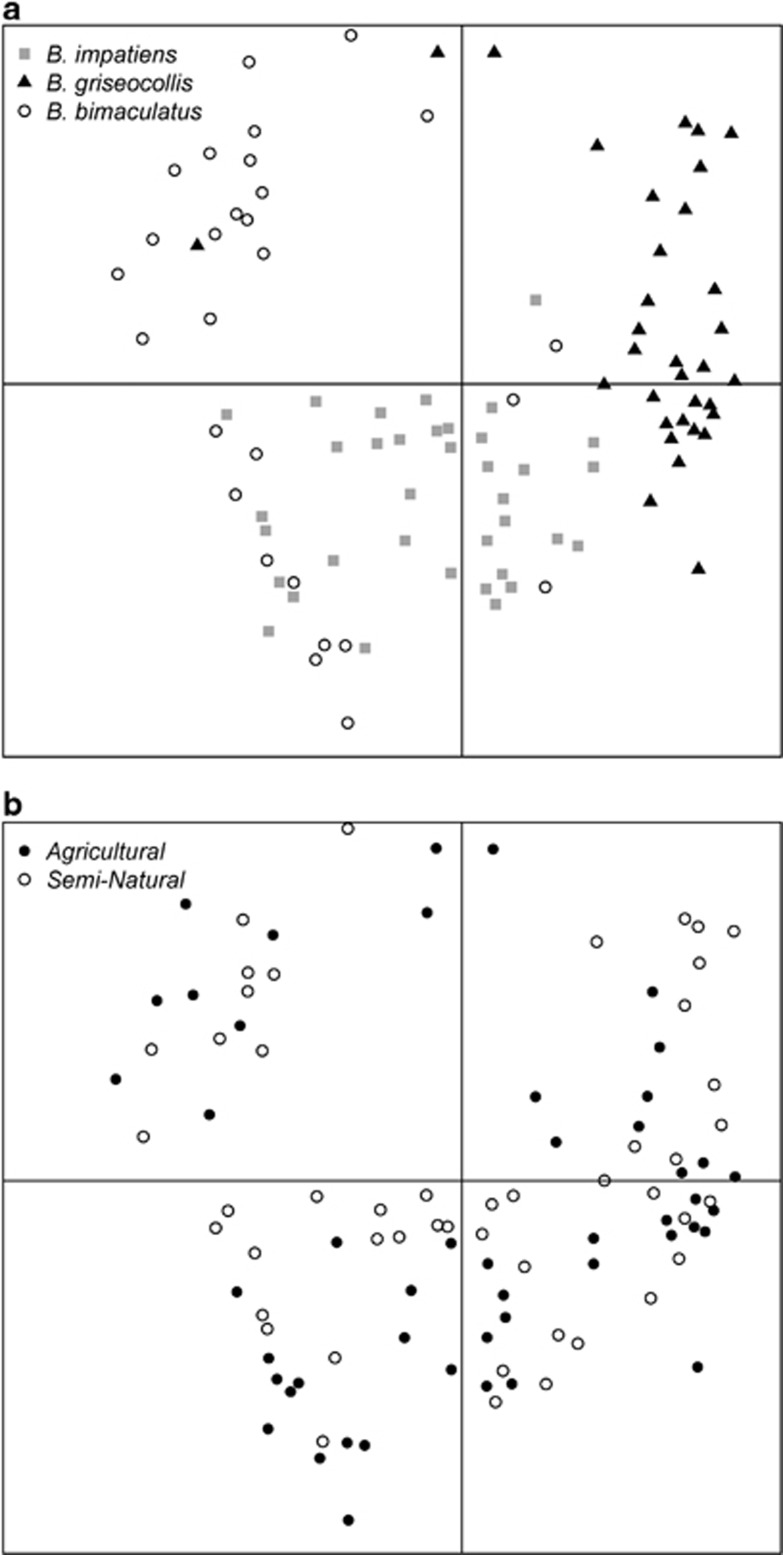
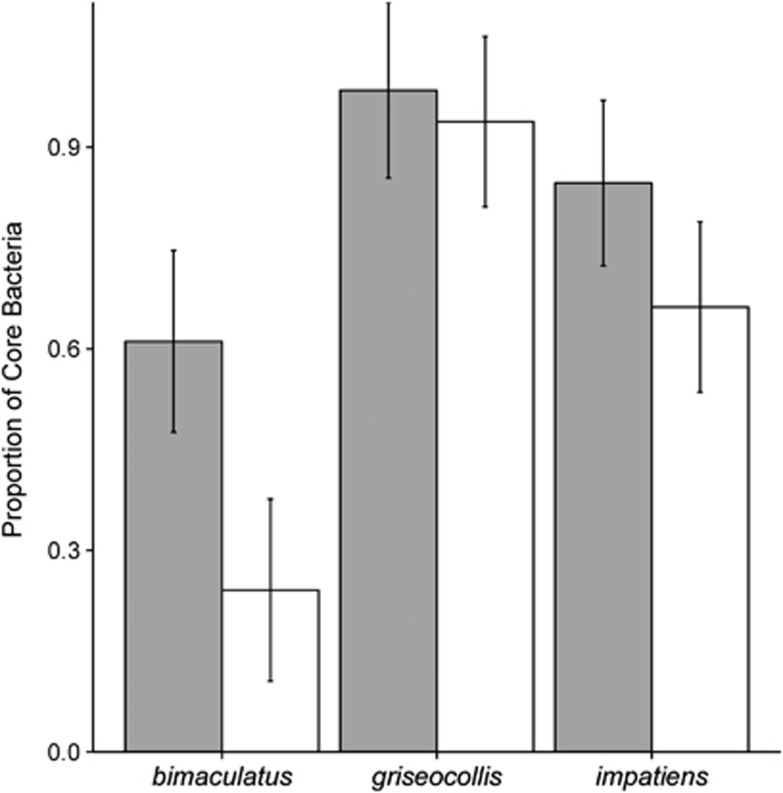
Overall community-level measures differed among Bombus species but not among habitat types. B. bimaculatus and B. impatiens (45.0±4.0 s.e., 47.1±3.6 s.e., respectively) had a richer gut microbiota than B. griseocollis (33.6±3.8, Supplementary Figure S4a). Rarefaction curves of Chao1 alpha diversity estimates showed that 2500 reads reflected a saturated sampling depth (Supplementary Figure S4b). Chao1 alpha diversity estimates for Bombus gut microbiota did not differ between semi-natural and agricultural habitats for any species (Supplementary Table S6). B. griseocollis had lower alpha diversity than the other two bee species (Supplementary Table S6). Beta diversity analysis indicated that variability in gut microbial communities among B. bimaculatus samples was higher than the other two species (beta-dispersion test: F2,93=17.52; P<0.001; Figure 1). There was no difference in beta diversity between habitat types (F1,97=0.62; P=0.43). Similarly, microbial community composition differed among species (PERMANOVA R2=0.27; F2,93=17.7; P<0.001; Figure 2a) but was not affected by habitat types (F1,93=1.29; P=0.23; Figure 2b) or the interaction between habitat type and species (F2,93=1.29; P=0.22). Comparing across bee species, the gut microbiota of B. bimaculatus had the lowest proportion of core bacteria (0.61±0.07 s.e., Figure 3), followed by B. impatiens (0.74±0.09 s.e., Figure 3) with B. griseocollis having the highest (0.98±0.09 s.e.; Figure 3). For B. bimaculatus, the proportion of core bacteria was lower in semi-natural than agricultural habitats (0.24±0.09 s.e. vs 0.61±0.07 s.e.), but this proportion did not vary for the other two species (Figure 3).
Figure 1.
Results of multivariate dispersion test. Gut communities among B. bimaculatus are more variable than those among B. impatiens and B. griseocollis.
Figure 2.
Results of NMDS (nonmetric multidimensional scaling) performed on bacterial gut communities, using individual Bombus specimens as the replicates. Effects of (a) Bombus species and (b) habitat type.
Figure 3.
Estimates (means) from mixed model output showing the proportion of gut microbiota made up of core OTUs. Grey bars represent agricultural habitats while white bars represent semi-natural habitats. Error bars represent±s.e.m.
Indicator species analysis restricted to core OTUs revealed that different bacterial OTUs dominate gut communities of the three Bombus species. The OTUs classified as core in the B. bimaculatus gut microbiota are best characterized by two Acetobacteraceae (Order Rhodospirillales), corresponding to taxa previously referred to as ‘Alpha-2.2' (Mohr and Tebbe, 2006; Cox-Foster et al., 2007; Martinson et al., 2011, Table 1, Supplementary Table S5). We categorized Alpha-2.2 OTUs as core bacteria, because they have been found repeatedly in Apis and Bombus guts in previous studies. However, unlike other core bacteria, the Alpha 2.2 OTUs are also common in floral products, including nectar and pollen. Thus, the finding that they are characteristic of B. bimaculatus is consistent with the conclusion that this host species is dominated by gut bacteria acquired from environmental sources (Table 1). In contrast, B. impatiens and B. griseocollis are best characterized by core bacteria that have only been sampled from the guts of Apis and Bombus (Table 1, Martinson et al., 2011).
Table 1. Results of indicator species analysis showing the association between OTUs and host species.
| Indicator OTUs | Association (GenBank) | B. bimaculatus Ind Val | B. griseocollis Ind Val | B. impatiens Ind Val | P-Value |
|---|---|---|---|---|---|
| Acetobacteraceae, Alpha-2.2 (418) | Floral | 0.56 | — | — | 0.003 |
| Acetobacteraceae, Alpha-2.2 (384) | Floral | 0.44 | — | — | 0.002 |
| Gilliamella (22) | Apis mellifera | — | 0.67 | — | 0.001 |
| Snodgrassella (167) | Bombus bimaculatus | — | 0.64 | — | 0.001 |
| Unclass Gammaproteobacteria (258) | Bombus terrestris | 0.61 | — | 0.001 | |
| Lactobacillaceae (362) | Bombus terrestris | — | 0.49 | — | 0.001 |
| Lactobacillaceae (4) | Bombus terrestris | — | 0.45 | — | 0.001 |
| Gilliamella (424) | Bombus terrestris | — | — | 0.87 | 0.001 |
| Gilliamella (342) | Bombus impatiens | — | — | 0.77 | 0.001 |
| Unclass Bacteroidetes (366) | Bombus terrestris | — | — | 0.74 | 0.001 |
| Snodgrassella (16) | Bombus vagans | — | — | 0.48 | 0.001 |
| Snodgrassella (394) | Apis mellifera | — | — | 0.47 | 0.002 |
| Snodgrassella (447) | Bombus vagans | — | — | 0.46 | 0.001 |
| Snodgrassella (72) | Apis mellifera | — | — | 0.45 | 0.001 |
| Gilliamella (105) | Bombus impatiens | — | — | 0.35 | 0.003 |
In the indicator OTU column, names represent taxonomic affiliation of OTU based on GenBank BLAST search. Numbers in parentheses are study-specific OTU identifiers. Results are from only those OTUs classified as core based on GenBank searches and having indicator values >0.25 and significant at P <0.05.
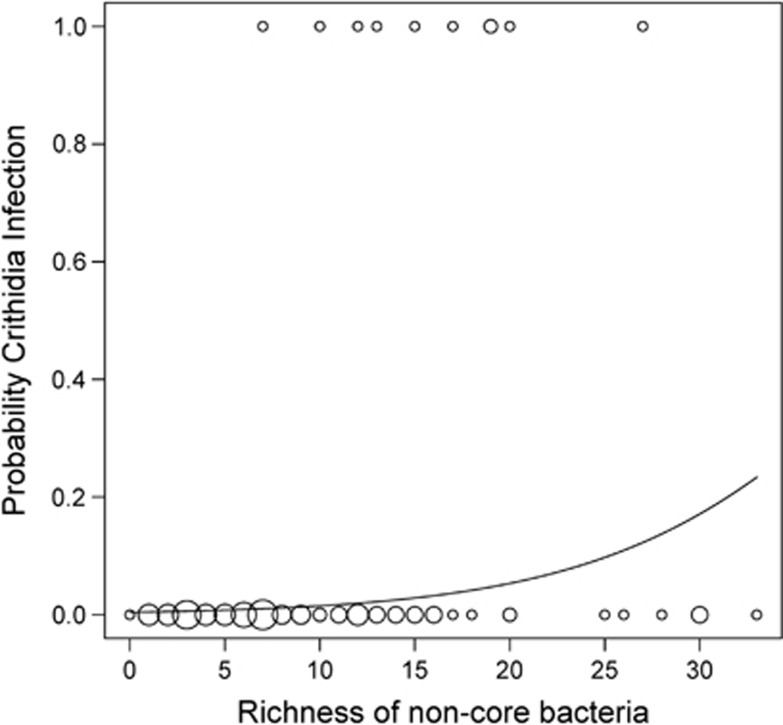
We found that gut communities differed between individuals infected and uninfected by pathogens. Ten of the 99 specimens we analyzed were infected with Crithidia (four impatiens, one griseocollis and five bimaculatus). Indicator species analysis demonstrated that gut microbiota of individuals infected by Crithidia were characterized primarily by the ‘Alpha-2.2'-related OTUs (Table 2), and those not infected were characterized by a Gillamella OTU (no. 424; Table 2). Furthermore, logistic regression analysis revealed that Crithidia infection increased as the richness of non-core bacteria increased (β=0.13±0.06 s.e., P=0.024, Figure 4). Bombus species, number of 16S rRNA gene copies, richness of core bacteria and habitat were not significant and were removed from the analysis.
Table 2. Results of indicator species analysis showing the association of OTUs with the presence or absence of Crithidia and Nosema pathogens.
| Pathogen | Indicator OTUs | Association (GenBank) | Pathogen Absent Ind Val | Pathogen Present Ind Val | P-Value |
|---|---|---|---|---|---|
| Crithidia | Gilliamella (424) | Bombus terrestris | 0.70 | — | 0.040 |
| Crithidia | Acetobacteraceae, Alpha-2.2 (418) | Pollen | — | 0.60 | 0.020 |
| Crithidia | Acetobacteraceae, Alpha-2.2 (384) | Floral | — | 0.54 | 0.002 |
| Crithidia | Acetobacteraceae. Alpha-2.2 (355) | Apis dorsata | — | 0.45 | 0.001 |
| Crithidia | Acetobacteraceae. Alpha-2.2 (62) | Apis dorsata | — | 0.36 | 0.001 |
| Crithidia | Acetobacteraceae, Alpha-2.2 (267) | Pollen | — | 0.27 | 0.013 |
| Nosema | Snodgrassella (72) | Apis mellifera | — | 0.50 | 0.027 |
In the indicator OTUs column, names represent taxonomic affiliations of OTU based on GenBank BLAST searches. Numbers in parentheses are study-specific OTU identifiers. Results are based on OTUs with indicator values >0.25 and significant at P <0.05.
Figure 4.
Results of logistic regression analysis. X axis represents the richness of non-core gut microbiota. Y axis represents the probability of infection by Crithidia. Different size points represent the number of samples at a given coordinate.
Nosema was found in 11 specimens (five impatiens, four griseocollis and two bimaculatus). We found no difference in gut microbial communities between hosts with and without Nosema. Interestingly, based on a maximum-likelihood tree using accessions from GenBank and the ones found in this study, most infections are from a novel strain of Nosema not previously represented in GenBank (Supplementary Figure S5), with an additional three individuals infected with N. ceranae. None of the specimens contained N. bombi. Indicator species analysis showed that one OTU (Snodgrassella no. 72) characterized Bombus infected by Nosema. Mixed model analysis of Nosema infection did not reveal a significant relationship for Bombus species, habitat type or proportion of core bacteria.
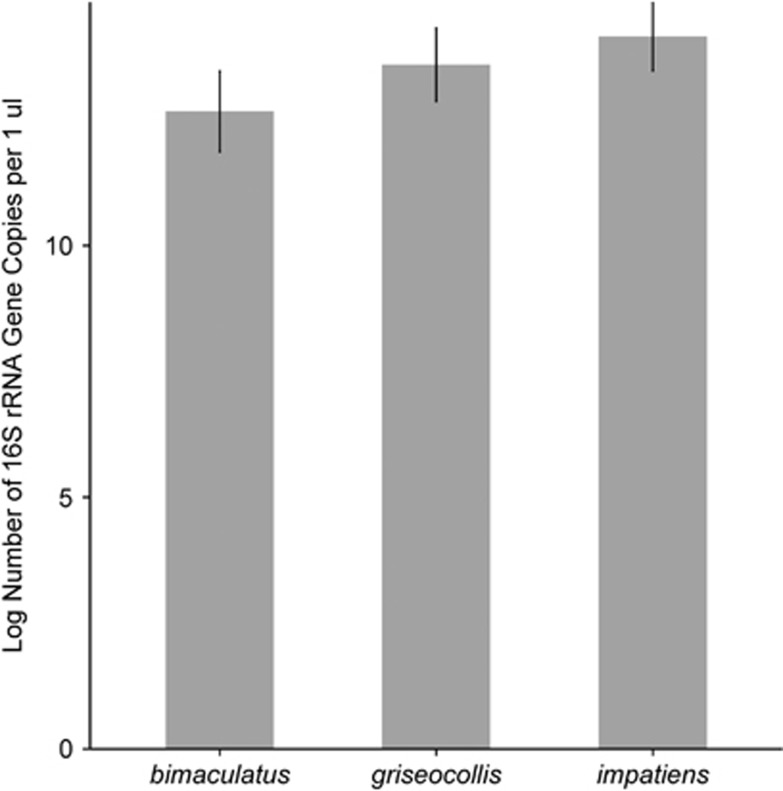
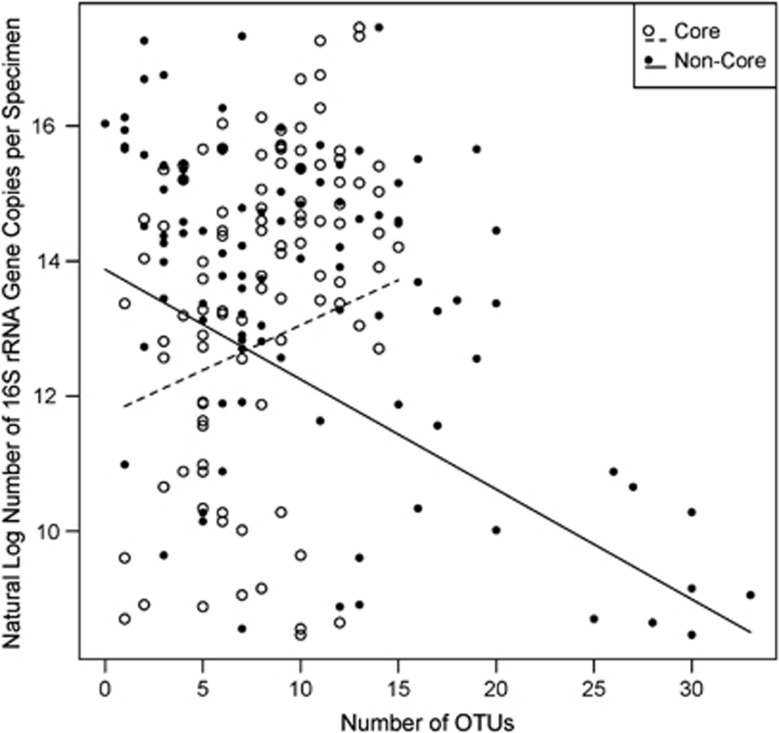
The absolute number of gut bacteria per bee, measured as the number of bacterial rRNA gene copies, varied among Bombus species. Results from the generalized linear mixed model indicated that the log copy number was highest in B. impatiens (14.37±0.21 s.e., Figure 5), followed by B. griseocollis (14.3±0.35 s.e., Figure 5) and B. bimaculatus (11.7±0.4 5 s.e., Figure 5). The average 30-fold difference in size of gut bacterial communities between B. impatiens and B. bimaculatus does not correspond to a difference in worker body size, which is similar for the three species. The number of bacterial rRNA gene copies was negatively associated with richness of non-core OTUs and positively associated with richness of core OTUs (Figure 6). Habitat type was not significant and was removed from the final model.
Figure 5.
Results of mixed model analysis showing the mean number of bacterial 16S rRNA gene copies per specimen. Error bars represent±s.e.m.
Figure 6.
Results of mixed model analysis showing that the richness of non-core (closed circles and solid line) bacteria OTUs is negatively associated with total number of 16S rRNA gene copies per bee specimen, whereas richness of core (open circles and dashed line) bacteria OTUs is positively associated.
Discussion
We used deep sequencing to characterize the composition of gut communities of three Bombus species in two habitat types and to test for associations between core and non-core gut bacteria, pathogen infection and overall bacteria abundance. We defined core bacteria as OTUs corresponding to species primarily associated with Apis and Bombus hosts in previous studies (Martinson et al., 2011; Moran et al., 2012). OTU richness of core bacteria had a positive association with absolute numbers of bacteria. In contrast, richness of non-core bacteria was negatively associated with the number of bacteria per bee and positively associated with Crithidia presence. Non-core bacterial taxa may be less adapted to the bee gut environment and thus less able to proliferate there, with the result that Bombus individuals with a high proportion of non-core bacteria may have lower numbers of gut bacteria. However, this mechanistic explanation remains untested. Also, in hosts with lower proportions of core bacteria and lower overall numbers of bacteria, non-core bacteria will be sampled more deeply, resulting in higher richness estimates due to sampling effects.
Although overall microbiota richness has been assumed to be important in microbial gut function, few studies have differentiated core from non-core bacteria. Experimental work in locusts demonstrated that increasing diversity of gut bacteria reduces susceptibility to a pathogen (Dillon et al., 2005). In contrast, Koch et al. (2012) found that microbiota richness at the colony level was positively associated with Crithidia infection in bumble bees. Our work may reconcile these seemingly disparate results, as richness of core taxa may be associated with healthy hosts while richness of non-core taxa may represent dysbiosis.
We found that specific OTUs varied in their association with pathogen infection. In particular, an OTU corresponding to Gilliamella was positively associated with bees uninfected by Crithidia. In honey bees, Gilliamella produces a biofilm on the ileum wall (Martinson et al., 2012; Kwong and Moran, 2013) and may provide a barrier to attachment or entry of gut pathogens such as Crithidia. Further, Gilliamella had a weak, negative association with Crithidia infection in B. terrestris in Europe (Koch and Schmid-Hempel, 2011a). The results presented here provide further evidence that Gilliamella may confer protective benefits. We found no evidence of a protective association of gut bacteria against infection by Nosema, consistent with previous studies (Koch and Schmid-Hempel, 2011a). In fact, one OTU of the core bacterium Snodgrassella had a positive association with Nosema infection. We note that our study cannot determine whether associations are causal. In particular, positive associations could result if transmission occurs through common routes, causing both a gut bacterium and pathogen to be transferred together. Finally, five other core bacterial OTUs, previously referred to as ‘Alpha-2.2' within the family Acetobacteraceae, had a positive associations with Crithidia infection. Although we categorized this group as core based on repeated retrieval from Apis and Bombus (Martinson et al., 2011; Moran et al., 2012), these bacteria are also common in nectar (Jojima et al., 2004) and solitary bees that otherwise lack core bacteria found in Apis and Bombus (Martinson et al., 2011; Koch et al., 2013). Therefore, these Acetobacteraceae OTUs may be repeatedly introduced from nectar, as suggested by studies on honey bees (Vojvodic et al., 2013).
We found the largest differences in microbial communities among the three Bombus species. The most dramatic difference involved B. bimaculatus, which had the highest variability among individuals and the lowest proportion and richness of core bacteria. In indicator species analysis, B. bimaculatus was characterized only by the Acetobacteraceae OTUs, which are also found in nectar (Jojima et al., 2004; Vojvodic et al., 2013). The distinctness of B. bimaculatus communities does not reflect phylogenetic distance, as it is in the same subgenus as B. impatiens (Pyrobombus) while B. griseocollis is in the subgenus Cullumanobombus. Samples of gut microbiota from multiple Bombus species coupled with host traits and local ecological conditions are needed to uncover mechanisms that drive variation within and among species. In A. mellifera workers, the proportion of core bacteria in individual guts ranges from 0.95 to >0.99 (Moran et al., 2012), with one species having similar (B. griseocollis mean=0.96) and the other two Bombus species having lower (B. bimaculatus mean=0.48, B. impatiens mean=0.76) representation of core taxa, as compared with A. mellifera. Potentially, these differences among species reflect differences in gut physiology or morphology, or differences in colony life cycle, that impact transmission between hosts.
Despite the fact that bees were sampled from farms with high agro-chemical use (Pettis et al., 2013), we found little effect of habitat type on any metric of gut microbial communities. High fungicide applications are reported to increase risk of Nosema infection in honey bees (Pettis et al., 2013). However, the only habitat effect evident in our study was that B. bimaculatus had a greater proportion of core bacteria when collected from farms. We may have failed to detect a larger effect of habitat on gut communities, because cultivated cranberry is a temporary resource with bloom lasting for approximately 6 weeks, whereas the Bombus species we studied are active for up to 6 months per year. Potentially, these core gut bacteria are evolutionarily conserved in host lineages and little affected by local environment. In a study of 35 Bombus species collected world-wide, Koch et al. (2013) found that specific strains of core bacteria in the Snodgrassella group were consistently associated with particular phylogenetic groups within Bombus, independently of geographic location. However, that study examined only presence/absence and did not exclude an effect of environmental conditions on relative or absolute abundance of particular bacterial strains.
The health of wild Bombus species is important to understand as they are among the most important pollinators for many native plants as well as a number of crops (Hegland and Totland, 2008; Garibaldi et al., 2013). For example, native bees are important pollinators at the cranberry farms studied here, as Bombus provide>70% of total pollination from native, wild bees (Cariveau et al., 2013). Several Bombus species have recently undergone steep population declines, with pathogen infection being a potential cause (Otterstatter and Thomson, 2008; Cameron et al., 2011). Our results suggest that the community structure of gut microbiota may be important factors in, or indicators for, the health of wild Bombus.
Acknowledgments
We thank L Fenner and Z Portman for help in collecting Bombus, J Klein for DNA extractions and P Degnan for assistance with bioinformatics. This research was funded by an NSF-Dimensions in Biodiversity Grant No. 1046153 to NAM (PI), J Evans (Co-PI) and RW (Senior Investigator) and Swiss National Science Foundation Grants 140157 and 147881 to HK.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson KE, Russell JA, Moreau CS, Kautz S, Sullam KE, Hu YI, et al. Highly similar microbial communities are shared among related and trophically similar ant species. Mol Ecol. 2012;21:2282–2296. doi: 10.1111/j.1365-294X.2011.05464.x. [DOI] [PubMed] [Google Scholar]
- Anderson MJ. Distance-based tests for homogeneity of multivariate dispersions. Biometrics. 2006;62:245–253. doi: 10.1111/j.1541-0420.2005.00440.x. [DOI] [PubMed] [Google Scholar]
- Anderson MJ. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 2001;26:32–46. [Google Scholar]
- Berg RD. The indigenous gastrointestinal microflora. Trends Microbiol. 1996;4:430–435. doi: 10.1016/0966-842x(96)10057-3. [DOI] [PubMed] [Google Scholar]
- Brown MJF, Schmid-Hempel R, Schmid-Hempel P. Strong context-dependent virulence in a host-parasite system: reconciling genetic evidence with theory. J Anim Ecol. 2003;72:994–1002. [Google Scholar]
- Cameron SA, Lozier JD, Strange JP, Koch JB, Cordes N, Solter LF, et al. Patterns of widespread decline in North American bumble bees. Proc Natl Acad Sci USA. 2011;108:662–667. doi: 10.1073/pnas.1014743108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics. 2010;26:266–267. doi: 10.1093/bioinformatics/btp636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cariveau DP, Williams NM, Benjamin FE, Winfree R. Response diversity to land use occurs but does not consistently stabilise ecosystem services provided by native pollinators. Ecol Lett. 2013;16:903–911. doi: 10.1111/ele.12126. [DOI] [PubMed] [Google Scholar]
- Castillo M, Martin-Orue SM, Manzanilla EG, Badiola I, Martin M, Gasa J. Quantification of total bacteria, enterobacteria and lactobacilli populations in pig digesta by real-time PCR. Vet Microbiol. 2006;114:165–170. doi: 10.1016/j.vetmic.2005.11.055. [DOI] [PubMed] [Google Scholar]
- Claesson MJ, van Sinderen D, O'Toole PW. Lactobacillus phylogenomics - towards a reclassification of the genus. Int J Syst Evol Microbiol. 2008;58:2945–2954. doi: 10.1099/ijs.0.65848-0. [DOI] [PubMed] [Google Scholar]
- Cordes N, Huang W-F, Strange JP, Cameron SA, Griswold TL, Lozier JD, et al. Interspecific geographic distribution and variation of the pathogens Nosema bombi and Crithidia species in United States bumble bee populations. J Invertebr Pathol. 2012;109:209–216. doi: 10.1016/j.jip.2011.11.005. [DOI] [PubMed] [Google Scholar]
- Cox-Foster DL, Conlan S, Holmes EC, Palacios G, Evans JD, Moran NA, et al. A metagenomic survey of microbes in honey bee colony collapse disorder. Science. 2007;318:283–287. doi: 10.1126/science.1146498. [DOI] [PubMed] [Google Scholar]
- Di Prisco G, Cavaliere V, Annoscia D, Varricchio P, Caprio E, Nazzi F, et al. Neonicotinoid clothianidin adversely affects insect immunity and promotes replication of a viral pathogen in honey bees. Proc Natl Acad Sci USA. 2013;110:18466–18471. doi: 10.1073/pnas.1314923110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon RJ, Vennard CT, Buckling A, Charnley AK. Diversity of locust gut bacteria protects against pathogen invasion. Ecol Lett. 2005;8:1291–1298. [Google Scholar]
- Drummond AJ, Ashton B, Buxton S, Cheung M, Cooper A, Duran C, et al. 2011. Geneious v5.4 website. Available from http://www.geneious.com Accessed 5 Jan 2012.
- Dufrêne M, Legendre P. Species assemblages and indicator species: the need for a flexible asymmetrical approach. Ecol Monogr. 1997;67:345–366. [Google Scholar]
- Engel P, Moran NA. The gut microbiota of insects—diversity in structure and function. FEMS Microbiol Rev. 2013;37:699–735. doi: 10.1111/1574-6976.12025. [DOI] [PubMed] [Google Scholar]
- Engel P, Martinson VG, Moran NA. Functional diversity within the simple gut microbiota of the honey bee. Proc Natl Acad Sci USA. 2012;109:11002–11007. doi: 10.1073/pnas.1202970109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erler S, Lommatzsch S, Lattorff HMG. Comparative analysis of detection limits and specificity of molecular diagnostic markers for three pathogens (Microsporidia, Nosema spp.) in the key pollinators Apis mellifera and Bombus terrestris. Parasitol Res. 2012;110:1403–1410. doi: 10.1007/s00436-011-2640-9. [DOI] [PubMed] [Google Scholar]
- Frank DN. BARCRAWL and BARTAB: software tools for the design and implementation of barcoded primers for highly multiplexed DNA sequencing. BMC Bioinformatics. 2009;10:362. doi: 10.1186/1471-2105-10-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci USA. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garibaldi LA, Steffan-Dewenter I, Winfree R, Aizen MA, Bommarco R, Cunningham SA, et al. Wild pollinators enhance fruit set of crops regardless of honey bee abundance. Science. 2013;339:1608–1611. doi: 10.1126/science.1230200. [DOI] [PubMed] [Google Scholar]
- Goulson D, Lye GC, Darvill B. Decline and conservation of bumble bees. Annu Rev Entomol. 2008;53:191–208. doi: 10.1146/annurev.ento.53.103106.093454. [DOI] [PubMed] [Google Scholar]
- Grixti JC, Wong LT, Cameron SA, Favret C. Decline of bumble bees (Bombus) in the North American Midwest. Biol Conserv. 2009;142:75–84. [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Hamdi C, Balloi A, Essanaa J, Crotti E, Gonella E, Raddadi N, et al. Gut microbiome dysbiosis and honeybee health. J Appl Entomol. 2011;135:524–533. [Google Scholar]
- Hebert PDN, Penton EH, Burns JM, Janzen DH, Hallwachs W. Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator. Proc Natl Acad Sci USA. 2004;101:14812–14817. doi: 10.1073/pnas.0406166101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegland SJ, Totland Ø. Is the magnitude of pollen limitation in a plant community affected by pollinator visitation and plant species specialisation levels. Oikos. 2008;117:883–891. [Google Scholar]
- Huang S, Zhang H. The impact of environmental heterogeneity and life stage on the hindgut microbiota of Holotrichia parallela larvae (Coleoptera: Scarabaeidae) PLoS One. 2013;8:e57169. doi: 10.1371/journal.pone.0057169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jojima Y, Mihara Y, Suzuki S, Yokozeki K, Yamanaka S, Fodu R. Saccharibacter floricola gen. nov., sp. nov., a novel osmophilic acetic acid bacterium isolated from pollen. Int J Syst Evol Microbiol. 2004;54:2263–2267. doi: 10.1099/ijs.0.02911-0. [DOI] [PubMed] [Google Scholar]
- Kikuchi Y, Hayatsu M, Hosokawa T, Nagayama A, Tago K, Fukatsu T. Symbiont-mediated insecticide resistance. Proc Natl Acad Sci USA. 2012;109:8618–8622. doi: 10.1073/pnas.1200231109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch H, Schmid-Hempel P. Socially transmitted gut microbiota protect bumble bees against an intestinal parasite. Proc Natl Acad Sci USA. 2011;108:19288–19292. doi: 10.1073/pnas.1110474108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch H, Schmid-Hempel P. Bacterial communities in Central European bumblebees: low diversity and high specificity. Microb Ecol. 2011;62:121–133. doi: 10.1007/s00248-011-9854-3. [DOI] [PubMed] [Google Scholar]
- Koch H, Schmid-Hempel P. Gut microbiota instead of host genotype drive the specificity in the interaction of a natural host-parasite system. Ecol Lett. 2012;15:1095–1103. doi: 10.1111/j.1461-0248.2012.01831.x. [DOI] [PubMed] [Google Scholar]
- Koch H, Abrol DP, Li J, Schmid-Hempel P. Diversity and evolutionary patterns of bacterial gut associates of corbiculate bees. Mol Ecol. 2013;22:2028–2044. doi: 10.1111/mec.12209. [DOI] [PubMed] [Google Scholar]
- Koch H, Cisarovsky G, Schmid-Hempel P. Ecological effects on gut bacterial communities in wild bumblebee colonies. J Anim Ecol. 2012;81:1202–1210. doi: 10.1111/j.1365-2656.2012.02004.x. [DOI] [PubMed] [Google Scholar]
- Kwong WK, Moran NA. Cultivation and characterization of the gut symbionts of honey bees and bumble bees: Snodgrassella alvi gen. nov., sp. nov., a member of the Neisseriaceae family of the Betaproteobacteria; and Gilliamella apicola gen. nov., sp. nov., a member of Orbaceae fam. nov., Orbales ord. nov., a sister taxon to the Enterobacteriales order of the Gammaproteobacteria. Int J Syst Evol Microbiol. 2013;63:2008–2018. doi: 10.1099/ijs.0.044875-0. [DOI] [PubMed] [Google Scholar]
- Li J, Chen W, Wu J, Peng W, An JD, Schmid-Hempel P, et al. Diversity of Nosema associated with bumblebees (Bombus spp.) from China. Int J Parasitol. 2012;42:49–61. doi: 10.1016/j.ijpara.2011.10.005. [DOI] [PubMed] [Google Scholar]
- Martinson VG, Danforth BN, Minckley RL, Rueppell O, Tingek S, Moran NA. A simple and distinctive microbiota associated with honey bees and bumble bees. Mol Ecol. 2011;20:619–628. doi: 10.1111/j.1365-294X.2010.04959.x. [DOI] [PubMed] [Google Scholar]
- Martinson VG, Moy J, Moran NA. Establishment of characteristic gut bacteria during development of the honeybee worker. Appl Environ Microbiol. 2012;78:2830–2840. doi: 10.1128/AEM.07810-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFrederick QS, Cannone JJ, Gutell RR, Kellner K, Plowes RM, Mueller UG. Specificity between lactobacilli and hymenopteran hosts is the exception rather than the rule. Appl Environ Microbiol. 2013;79:1803–1812. doi: 10.1128/AEM.03681-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFrederick QS, Wcislo WT, Taylor DR, Ishak HD, Dowd SE, Mueller UG. Environment or kin: whence do bees obtain acidophilic bacteria. Mol Ecol. 2012;21:1754–1768. doi: 10.1111/j.1365-294X.2012.05496.x. [DOI] [PubMed] [Google Scholar]
- Miller MA, Holder MT, Vos R, Midford PE, Liebowitz T, Chan L, et al. (eds). (2009. The CIPRES Portals. CIPRES. URL http://www.phylo.org/sub_sections/portal .
- Mohr KI, Tebbe CC. Diversity and phylotype consistency of bacteria in the guts of three bee species (Apoidea) at an oilseed rape field. Environ Microbiol. 2006;8:258–272. doi: 10.1111/j.1462-2920.2005.00893.x. [DOI] [PubMed] [Google Scholar]
- Moran NA, Hansen AK, Powell JE, Sabree ZL. Distinctive gut microbiota of honey bees assessed using deep sampling from individual worker bees. PLoS One. 2012;7:e36393. doi: 10.1371/journal.pone.0036393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morell V. TreeBASE: The roots of phylogeny. Science. 1996;273:569. [Google Scholar]
- Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O'Hara RB, et al. Vegan: community ecology package. R package version 2.0-7 2013.
- Otterstatter MC, Thomson JD. Does pathogen spillover from commercially reared bumble bees threaten wild pollinators. PLoS One. 2008;3:e2771. doi: 10.1371/journal.pone.0002771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettis JS, Lichtenberg AM, Andree M, Stitzinger J, Rose R, vanEngelsdorp D. Crop pollination exposes honey bees to pesticides which alters their susceptibility to the gut pathogen Nosema ceranae. PLoS One. 2013;8:e70182. doi: 10.1371/journal.pone.0070182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira HM, Leadley PW, Proenca V, Alkemade R, Scharlemann JPW, Fernandez-Manjarres JF, et al. Scenarios for global biodiversity in the 21st century. Science. 2010;330:1496–1501. doi: 10.1126/science.1196624. [DOI] [PubMed] [Google Scholar]
- Pettis JS, Lichtenberg EM, Andree M, Stitzinger J, Rose R, vanEngelsdorp D. Crop pollination exposes honey bees to pesticides which alters their susceptibility to the gut pathogen Nosema ceranae. PLoS One. 2013;8:e70182. doi: 10.1371/journal.pone.0070182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinheiro J, Bates D, DebRoy S, Sarkar D, and the R Development Core Team.2013nlme: Linear and Nonlinear Mixed Effects ModelsR Package version 3.1-109.
- Ratnasingham S, Herbert PD.2007BOLD: The Barcode of Life Data System ( www.barcodinglife.org ). Mol Ecol Notes 7355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley EV, Wong AC-N, Westmiller S, Douglas AE. Impact of the resident microbiota on the nutritional phenotype of Drosophila melanogaster. PLoS One. 2012;7:e36765. doi: 10.1371/journal.pone.0036765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts DW. labdsv: Ordination and Multivariate Analysis for Ecology. R package version. 2012;1:5–0. [Google Scholar]
- Sartor RB. Therapeutic correction of bacterial dysbiosis discovered by molecular techniques. Proc Natl Acad Sci USA. 2008;105:16413–16414. doi: 10.1073/pnas.0809363105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid-Hempel R, Tognazzo M. Molecular divergence defines two distinct lineages of Crithidia bombi (Trypanosomatidae), parasites of bumblebees. J Eukaryot Microbiol. 2010;57:337–345. doi: 10.1111/j.1550-7408.2010.00480.x. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Sudakaran S, Salem H, Kost C, Kaltenpoth M. Geographical and ecological stability of the symbiotic mid-gut microbiota in European firebugs, Pyrrhocoris apterus (Hemiptera, Pyrrhocoridae) Mol Ecol. 2012;21:6134–6151. doi: 10.1111/mec.12027. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyson GW, Chapman J, Hugenholtz P, Allen EE, Ram RJ, Richardson PM, et al. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature. 2004;428:37–43. doi: 10.1038/nature02340. [DOI] [PubMed] [Google Scholar]
- Vojvodic S, Rehan SM, Anderson KE. Microbial gut diversity of Africanized and European honey bee larval instars. PLoS One. 2013;8:e72106. doi: 10.1371/journal.pone.0072106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Garrity GM, Tiedje JM, Cole JR. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnecke F, Luginbühl P, Ivanova N, Ghassemian M, Richardson TH, Stege JT, et al. Metagenomic and functional analysis of hindgut microbiota of a wood-feeding higher termite. Nature. 2007;450:560–565. doi: 10.1038/nature06269. [DOI] [PubMed] [Google Scholar]
- Whitehorn PR, O'Connor S, Wackers FL, Goulson D. Neonicotinoid pesticide reduces bumble bee colony growth and queen production. Science. 2012;336:351–352. doi: 10.1126/science.1215025. [DOI] [PubMed] [Google Scholar]
- Williams PH, Osborne JL. Bumblebee vulnerability and conservation world-wide. Apidologie. 2009;40:367–387. [Google Scholar]
- Wong AC-N, Chaston JM, Douglas AE. The inconstant gut microbiota of Drosophila species revealed by 16S rRNA gene analysis. ISME J. 2013;7:1922–1932. doi: 10.1038/ismej.2013.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zouache K, Raharimalala FN, Raquin V, Tran-Van V, Raveloson LHR, Ravelonandro P, et al. Bacterial diversity of field-caught mosquitoes, Aedes albopictus and Aedes aegypti, from different geographic regions of Madagascar. FEMS Microbiol Ecol. 2010;75:377–389. doi: 10.1111/j.1574-6941.2010.01012.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.