Abstract
Ammonium/ammonia is the sole energy substrate of ammonia oxidizers, and is also an essential nitrogen source for other microorganisms. Ammonia oxidizers therefore must compete with other soil microorganisms such as methane-oxidizing bacteria (MOB) in terrestrial ecosystems when ammonium concentrations are limiting. Here we report on the interactions between nitrifying communities dominated by ammonia-oxidizing archaea (AOA) and Nitrospira-like nitrite-oxidizing bacteria (NOB), and communities of MOB in controlled microcosm experiments with two levels of ammonium and methane availability. We observed strong stimulatory effects of elevated ammonium concentration on the processes of nitrification and methane oxidation as well as on the abundances of autotrophically growing nitrifiers. However, the key players in nitrification and methane oxidation, identified by stable-isotope labeling using 13CO2 and 13CH4, were the same under both ammonium levels, namely type 1.1a AOA, sublineage I and II Nitrospira-like NOB and Methylomicrobium-/Methylosarcina-like MOB, respectively. Ammonia-oxidizing bacteria were nearly absent, and ammonia oxidation could almost exclusively be attributed to AOA. Interestingly, although AOA functional gene abundance increased 10-fold during incubation, there was very limited evidence of autotrophic growth, suggesting a partly mixotrophic lifestyle. Furthermore, autotrophic growth of AOA and NOB was inhibited by active MOB at both ammonium levels. Our results suggest the existence of a previously overlooked competition for nitrogen between nitrifiers and methane oxidizers in soil, thus linking two of the most important biogeochemical cycles in nature.
Keywords: ammonia-oxidizing archaea, methane-oxidizing bacteria, nitrification, methanotrophy, mixotrophy, stable-isotope probing
Introduction
Ammonia-oxidizing archaea and bacteria (AOA and AOB, respectively) play a key role in the global nitrogen cycle. They convert relatively immobile ammonium into more mobile nitrite that is then further oxidized into nitrate by nitrite-oxidizing bacteria (NOB). Thereby, these two functional microbial guilds (that is, AOA/AOB and NOB), each referred to as nitrifiers, produce a reactive form of inorganic nitrogen with numerous environmental consequences (Gruber and Galloway, 2008). In nitrogen-limited soils, AOA and AOB must compete with ammonium-assimilating organisms such as plants and heterotrophic microorganisms, and it has been shown that AOB are poorer competitors for limiting amounts of ammonium in grassland soils (Verhagen et al., 1995). Only recently, indications of interactions between AOA and other microorganisms, which are relevant to both structure and activity of archaeal ammonia oxidizer communities, have been predicted in the ocean and described for a geothermal spring (Boyett et al., 2013; Hamilton et al., 2014). However, the ability of AOA to compete with ammonium-assimilating organisms in ammonium-limited, terrestrial environments has not been studied.
The necessity of AOA to compete for ammonium may vary between species, as the cultured soil AOA ‘Ca. Nitrososphaera gargensis' and ‘Ca. Nitrosotalea devanaterra' have a preference for low ammonium levels, whereas Nitrososphaera viennensis EN76 and ‘Ca. Nitrosoarchaeum koreensis' seem to be adapted to substantially higher ammonium concentrations (Hatzenpichler et al., 2008; Jung et al., 2011; Lehtovirta-Morley et al., 2011; Tourna et al., 2011). Considering that the archaeal ammonia-monooxygenase (AMO) is reported to have a higher affinity for its substrate than known for most other microbial enzymes (Martens-Habbena et al., 2009; Jung et al., 2011; Lehtovirta-Morley et al., 2011; Kim et al., 2012) and ammonium uptake is likely facilitated by high densities of NH4+/NH3 transporters (Urakawa et al., 2011), AOA are possibly strong competitors for ammonium. However, it is currently unclear whether their competitive strength and ammonia oxidation activity varies depending on the availability of ammonium. Field and microcosm studies show either enhanced abundance and activity of AOA or inhibition in response to nitrogen (N) fertilization (Di et al., 2009; Stopnisek et al., 2010; Pratscher et al., 2011; Verhamme et al., 2011; Levicnik-Hofferle et al., 2012; Ke et al., 2013).
Interactions between nitrogen- and methane-cycling microbes are very likely, as the process of methane oxidation and the growth of methane-oxidizing bacteria (MOB) are influenced by ammonium (Bodelier and Laanbroek, 2004; Bodelier, 2011). Furthermore, MOB share many physiological, structural and ecological features with AOA such as dependence on monooxygenase reactions catalyzed by enzymes of the copper-containing membrane-bound monooxygenase superfamily, intracellular membrane systems, sensitivity to the same inhibitors, possession of hydroxylamine oxidoreductase systems, reliance on copper and the occupancy of oxic environments (Stein et al., 2012).
Whether MOB are inhibited or stimulated by ammonium in soil largely depends on the ratio of methane-C to ammonium-N and on the genetic potential of the MOB community to deal with ammonia directly as well as indirectly via the ammonia oxidation products, hydroxylamine and nitrite (Stein et al. 2012). Given a steady methane availability enabling strong microbial growth, such as in wetlands and other high methane environments, ammonium is more likely to serve as a nutrient than as an inhibitor to methanotrophy (Bodelier and Laanbroek, 2004). In environments with such conditions, the regulation of carbon (C) and N cycling is tightly linked to nitrifiers and MOB competing for mineral N as a source of growth and/or metabolic activity. The balance in activity between these three functional guilds (that is, ammonia, nitrite and methane oxidizers) will determine whether N will be lost through gaseous N oxides and mineral leaching (Lipschultz et al., 1981; Freitag and Bock, 1990; Stein and Klotz, 2011; Jung et al., 2013) and whether the soil is a source or sink of methane. Moreover, competition for ammonium may be an important modulator of niche differentiation and the selection of nitrifier and MOB ecotypes in oxic, N-limited soils. Despite important ecological implications for ecosystem functioning that are tied to the coupling of C and N cycles in soil, we have a rather limited understanding of the involved microorganisms and how they interact in situ.
Hypothesizing that ammonium-assimilating MOB inhibit nitrification by competition with AOA depending on the amount of generally available ammonium, we performed a microcosm study with two controlled levels of ammonium. By incubating a soil with limited ammonium availability and in which no AOB could be detected (Daebeler et al., 2012) we utilized an ideal model system to study the importance of AOA for nitrification and their (competitive) interactions with other microorganisms. The nitrifier and MOB communities were targeted by a combination of stable-isotope probing (SIP) and deep sequencing. This approach allowed us to determine the autotrophically growing nitrifiers and active methanotrophs and enabled us to study (competitive) interactions and niche adaptation of nitrifiers and methanotrophs to different ammonium levels.
Materials and methods
Soil microcosms
The effects of ammonium concentration and methane availability on nitrification, methane oxidation and the respective ammonia- and methane-oxidizer community dynamics were determined in microcosms containing grassland soil taken from Grændalur valley (64° 1′ 7′′ N, 21° 11′ 20′′ W), Iceland, in August 2012. The soil is a Histic Andosol with two tephra layers in the top 30 cm, has a pH of 6.7±0.1 (as determined in replicated 1:5 soil/demineralized water solutions on day 0 of the incubation) and an in situ ammonium content of 0.5–1.1 μg NH4+-N per g dry soil (determined in August 2010 and May 2012 as described below). It has experienced stable, elevated temperatures of 33 °C through geothermal activity from at least 2005 and is cooling down since an earthquake nearby in 2009 with a recorded temperature of 14 °C in August 2012. Details of sampling and determination of water content and water-holding capacity can be found in the Supplementary Information.
The incubation was conducted in triplicate microcosms consisting of sterile 250 ml bottles containing 14 g of moist, sieved soil (corresponding to 5 g of dry soil). Loosely capped bottles were preincubated for 14 days to reduce the flux of CO2 released from soil due to respiration. Conditions of the preincubation are given in the Supplementary Information.
On day 14 of preincubation, microcosms were amended with ammonium sulfate solutions to final additions of 15 (further referred to as LAT or low ammonium treatment) and 150 μg NH4+-N per g dry soil (further referred to as HAT or high ammonium treatment). We determined the ammonia losses due to adsorption to soil particles with a 15N-tracing approach (see Supplementary Information) and could therefore estimate that ammonia availability was elevated by the ammonium treatments by at least 4.5 and 45 μg NH4+-N per g dry soil in LAT and HAT microcosms, respectively, after the first addition.
Microcosms were sealed by rubber stoppers and aluminum caps and incubated for 28 days in the dark at 25 °C. HAT and LAT were refreshed weekly with ammonium sulfate solutions by addition of 7.5 and 75 μg NH4+-N per g dry soil to the LAT and HAT microcosms, respectively, on days 7, 14 and 21 by injection through the rubber stopper. Soil moisture content was monitored by weight loss and kept constant at 70% water-holding capacity by addition of demineralized water or respective volumes of ammonium sulfate solutions.
The soil microcosms were amended with 5% (vol/vol) 12C-CO2 or 13C-CO2 (99% at excess, Campro Scientific BV, Veenendaal, The Netherlands) by injection through the rubber stopper on days 0, 7, 14 and 21. Before every gas addition microcosms were flushed with synthetic air (21% O2 in N2) for 5 min to ensure oxic conditions. Half of the bottles were additionally amended with 1% (10 000 p.p.m.v.) 12C-CH4 or 13C-CH4 (99% at excess, Campro Scientific BV) after 14 days of incubation. For an overview of the amendments made to the microcosms, see Supplementary Table A in the Supplementary Information. Microcosms receiving both 13C-CH4 and 13C-CO2 from day 14 were amended with 12C-CO2 only until day 14 to ensure that a 13C-signal in genomic DNA would only be found in microbes active in the presence of methane. Concentrations of methane and carbon dioxide in the headspace were monitored regularly as described in the Supplementary Information.
Triplicate LAT and HAT microcosms with either 12C-CO2 or 13C-CO2 were sampled destructively after 0, 14, 21 and 28 days of incubation. In the same manner, triplicate LAT and HAT microcosms with either 12C-CO2 and 12C-CH4 or 13C-CO2 and 13C-CH4 were sampled at day 28. At sampling, soil for extraction and analysis of DNA was mixed and immediately stored at −20 °C. The remaining soil of each microcosm (∼12 g) was taken for measurements of inorganic nitrogen as described in the Supplementary Information.
Nucleic acid extraction and SIP gradient fractionation
Total nucleic acids were extracted from 0.5 g soil according to Lueders et al. (2004) and the concentration was determined using a NanoDrop ND 1000 (Thermo Fisher Scientific, Wilmington, DE, USA) before storing at −20 °C until further analysis of DNA.
Density gradient centrifugation and gradient fractionation was performed as described in Lu and Jia (2013) with 6 μg DNA extracted from duplicate LAT and HAT microcosms that had been incubated with 13CO2 for 14 and 28 days without methane as well as from 13CO2 +13CH4 and 12CO2 +12CH4 microcosms amended with methane on day 14 and sampled on day 28.
Quantitative PCR
Quantification of archaeal amoA genes was performed as described previously (Daebeler et al., 2012). Primers used for all quantitative PCR (qPCR) assays as well as details of cycling conditions and standards used can be found in the Supplementary Information. For quantification of AOB and MOB, the bacterial amoA and the pmoA genes were targeted. NOB-based qPCR assays were not performed because of polyphyly of NOB, phylogenetically nonconserved copy numbers of the nitrite-reductase gene cluster and nonspecificity of primers. For both the archaeal and the bacterial amoA assays, the detection limit of qPCR was 2.85 × 103 per g dry soil; for pmoA it was 4.35 × 103 per g dry soil.
Pyrosequencing and sequence analysis
To characterize microbial communities in the gradient fractions from 13C and from 12C-control microcosms obtained after density gradient centrifugation, we carried out barcoded pyrosequencing of fractions 3 to 12 of each gradient targeting the 16S rRNA gene using a Roche 454 GS FLX Titanium sequencer (Roche Diagnostics Corporation, Branford, CT, USA) as reported previously (Xia et al., 2011; Lu and Jia, 2013). The raw, demultiplexed SFF files were deposited into the EMBL-ENA SRA database (https://www.ebi.ac.uk/ena/) and can be found under study accession number PRJEB5248 at the following site: http://www.ebi.ac.uk/ena/data/view/PRJEB5248.
A total of 1 138 939 raw sequences were processed with mothur v1.31.1 (Schloss et al., 2009) according to the standard operating procedure published by Schloss et al. (2011; detailed description accessible at http://www.mothur.org/wiki/454_SOP#OTUs) until the extraction of representative sequences for each operational taxonomic unit (OTU). Only the alignment was performed differently than described by Schloss et al. (2011). Sequence alignment was generated with SINA (http://www.arb-silva.de/aligner/) and the alignment was then imported back to mothur for subsequent analysis steps according to the mothur standard procedure referenced above. OTUs were clustered with mothur using the average neighbor algorithm at 99% sequence divergence. The OTUs containing <50 sequences over the entire data set were discarded to increase computation speed. Finally, we standardized to 5426 sequences per sample, obtaining a final number of 868 160 sequences of which 254 676 were unique. Details of the phylogenetic analysis are given in the Supplementary Information.
Network analysis was performed using the R-package qgraph (Epskamp et al., 2012) based on a Spearman's rank correlation matrix generated for the 10 most abundant AOA, NOB and MOB phylotypes in samples from LAT and HAT 13/12C microcosms incubated with and without methane. Only correlation pairs with a ρ-value above 0.6 and significance level below 0.05 were used. The network layout was generated using a modified version of the force-directed algorithm proposed by Fruchterman and Reingold (1991) as implemented in qgraph.
Statistical analysis
To test for effects of treatments and time as well as their interaction on AOA and MOB abundance, methane oxidation rate and mineral N concentrations, we performed analysis of variance (ANOVA) followed by Tukey's HSD (honest significant difference) post hoc tests. Abundance data of AOA and MOB genes were log-transformed to meet homoscedasticity assumptions. We calculated the balance between expected and observed nitrification by subtracting the total amount of accumulated NO3−-N on day 28 from all added and initially present NH4+-N. Differences in community compositions of nitrifiers and methane oxidizers based on Bray–Curtis distances of weighted phylotype profiles were assessed using the adonis function of the R package vegan v2.0-8 (Oksanen et al., 2010) that compares dissimilarities of variance in groups and uses permutation tests to assess significance.
Results
Nitrification and methane oxidation in soil microcosms
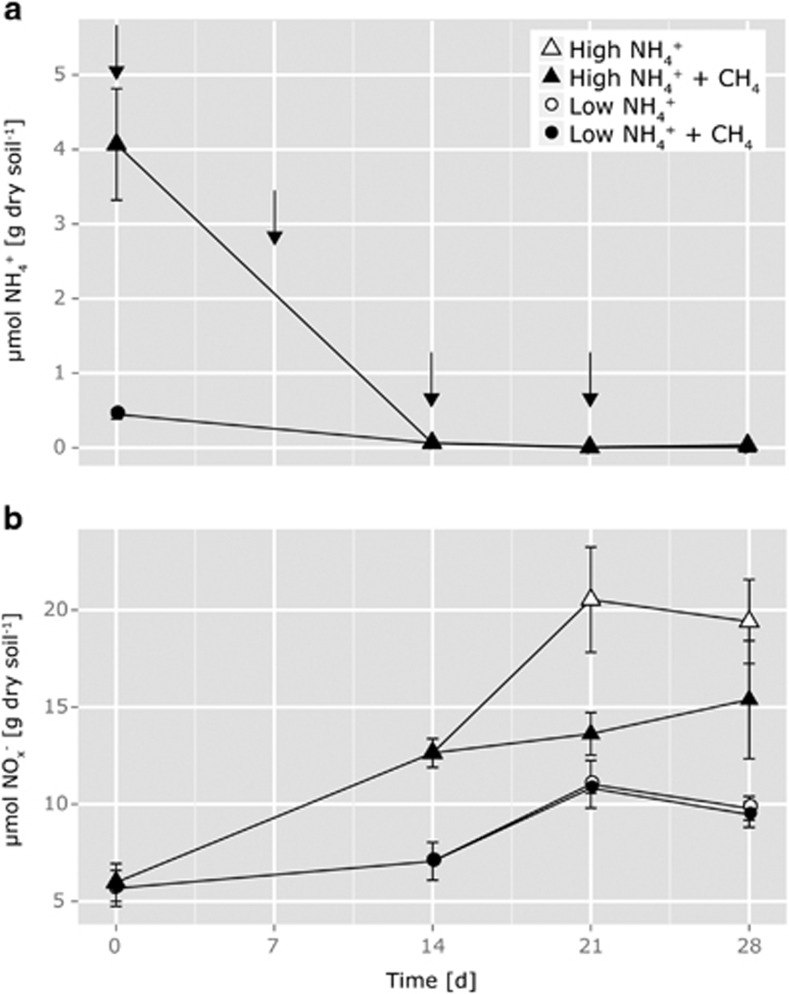
Concentrations of ammonium in the soil microcosms were barely measurable throughout the incubation, except for initial concentrations because of ammonium additions to the LAT and HAT microcosms (Figure 1a). Concentrations close to zero were measured for days 14, 21 and 28 in all microcosms before the weekly reapplication of ammonium took place.
Figure 1.
Changes in ammonium (a) and nitrite plus nitrate (b) concentrations in soil microcosms incubated at 25 °C for 0, 14, 21 or 28 days at low (15 μg NH4+-N per g dry soil added initially; 7.5 μg NH4+-N per g dry soil on days 7, 14 and 21) and high (150 μg NH4+-N per g dry soil added initially; 75 μg NH4+-N per g dry soil on days 7, 14 and 21) levels of ammonium and concentrations of 0% and 1% methane in the headspace. Arrows in (a) indicate ammonium addition. Data plotted are mean values and s.e. from 3 to 6 replicate microcosms destructively sampled at each time point, with some symbols nonvisible because they are masked by others.
Significant net nitrification assessed via production of NO2−+NO3− was only observed in HAT and HAT+CH4 microcosms over the time frame of the entire incubation (Figure 1b; Tukey's HSD, pday0:28<0.001, and 0.033 respectively). Net nitrification was significantly affected by the ammonium treatment (ANOVA, plow:high=0.013) but methane-amended microcosms did not differ from nonmethane-amended microcosms of the same ammonium treatment. Both LAT microcosms without and with methane showed a negative balance between expected nitrification and measured nitrification by −5.3 and −0.9 μg N per g dry soil, respectively. In contrast, the balance between expected and measured nitrification in HAT microcosms without and with methane was positive with 336.2 and 393.0 μg N per g dry soil, respectively.
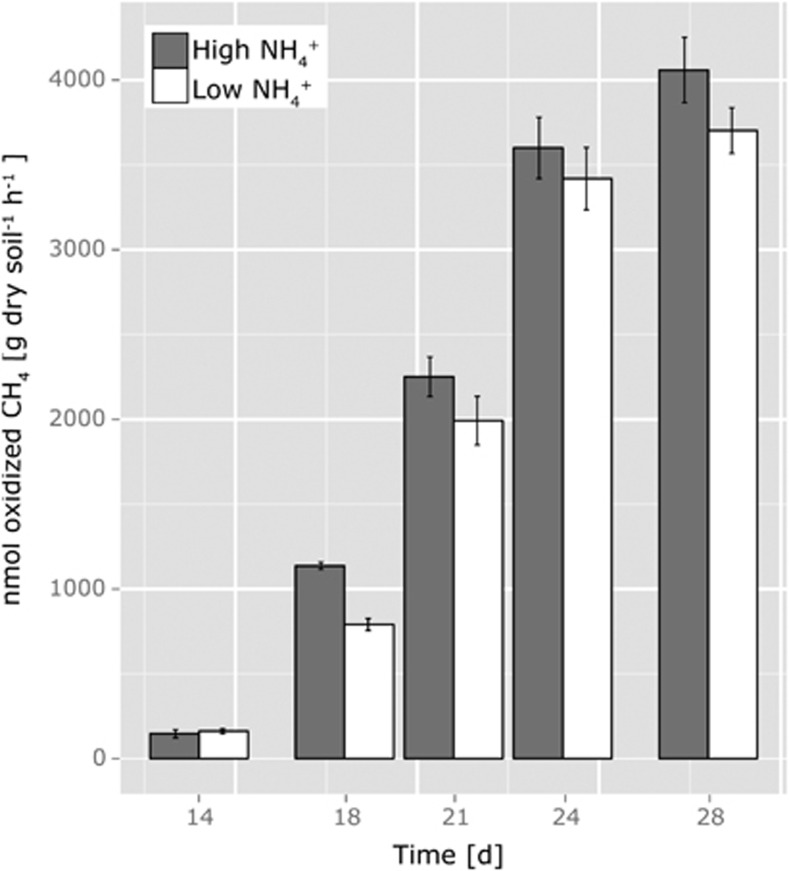
Consumption of methane was observed upon the first addition of methane on day 14 with rates of 161.3 (±13.3) and 146.37 (±23.98) nmol oxidized methane per g dry soil per h after a lag time of 20 h and 50 h in the LAT and HAT microcosms, respectively (Figure 2). With the repeated addition of 1% methane to the headspace, methane oxidation rates increased throughout the 14 subsequent days of incubation and were significantly higher in the HAT than in the LAT microcosms (ANOVA, plow:high=0.003) reaching values of 3702.3 (±134.6) and 4058.5 (±192.4) nmol oxidized methane per g dry soil per h, respectively, at the end of the incubation.
Figure 2.
Methane oxidation rates on days 14, 18, 21, 24 and 28 of microcosms incubated at 25 °C at low (15 μg NH4+-N per g dry soil added initially; 7.5 μg NH4+-N per g dry soil on days 7, 14 and 21) and high (150 μg NH4+-N per g dry soil added initially; 75 μg NH4+-N per g dry soil on days 7, 14 and 21) levels of ammonium and repeated addition of 1% methane to the headspace starting on day 14. Methane addition was repeated every 3 to 4 days. Data plotted are mean values and s.e. from triplicate nonlabeled microcosms.
Quantification of ammonia- and methane-oxidizing communities
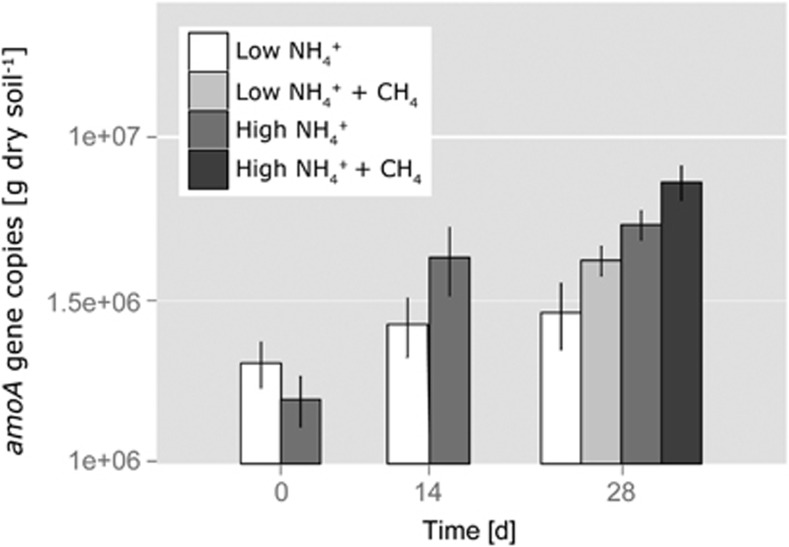
Archaeal amoA abundance increased significantly only in HAT+CH4 microcosms (Tukey's HSD, pday0:28=0.006; Figure 3). The addition of methane significantly elevated archaeal amoA gene abundance (ANOVA, p-CH4:+CH4=0.025) and we found significantly more amoA genes in high than in low ammonium microcosms (ANOVA, phigh:low<0.001). Ammonia-oxidizing bacteria were undetectable by qPCR.
Figure 3.
Changes in archaeal amoA gene copy numbers in soil microcosms incubated at 25 °C for 0, 14 and 28 days at low (15 μg NH4+-N per g dry soil added initially; 7.5 μg NH4+-N per g dry soil on days 7, 14 and 21) and high (150 μg NH4+-N per g dry soil added initially; 75 μg NH4+-N per g dry soil on days 7, 14 and 21) levels of ammonium as well as 0% and 1% methane in the headspace. Repeated methane addition every 3 to 4 days was started on day 14. Data plotted are mean values and s.e. from triplicate nonlabeled microcosms destructively sampled at each time point.
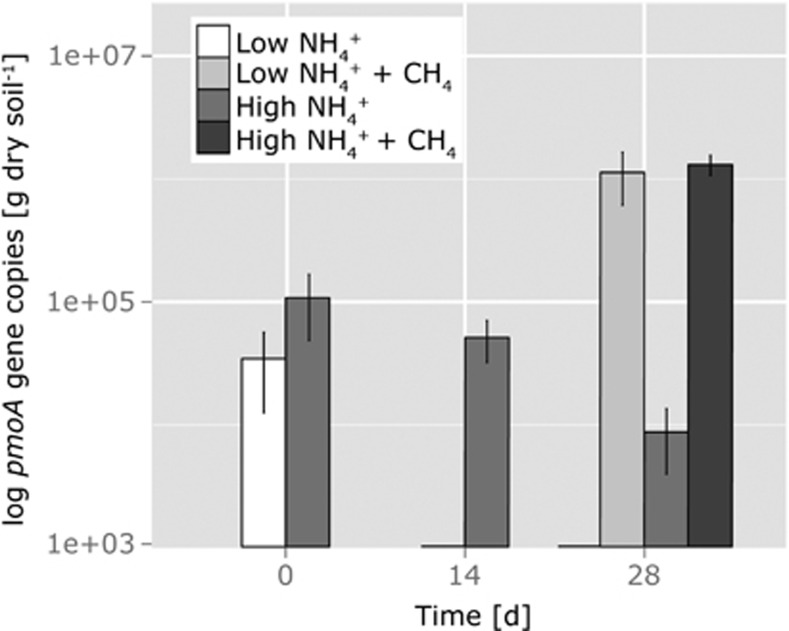
The community size of methane-oxidizing bacteria was not significantly different at the start of the incubation between LAT and HAT microcosms (Figure 4). In the absence of methane, pmoA gene abundance dropped below detection limit in LAT microcosms, whereas the abundance in HAT microcosms on day 28 did not differ significantly from day 0. In the presence of methane, pmoA gene abundance drastically increased within 2 weeks in the LAT microcosms from below detection limit on day 14, the day of the first methane amendment, to 1.13 × 106 copies per g dry soil on day 28. The pmoA gene abundance in HAT+CH4 microcosms reached a rather similar level constituting significant growth upon methane addition (Tukey's HSD, pday14:28=0.039).
Figure 4.
Changes in bacterial pmoA gene copy numbers in soil microcosms incubated at 25 °C for 0, 14 and 28 days at low (15 μg NH4+-N per g dry soil added initially; 7.5 μg NH4+-N per g dry soil on days 7, 14 and 21) and high (150 μg NH4+-N per g dry soil added initially; 75 μg NH4+-N per g dry soil on days 7, 14 and 21) levels of ammonium as well as 0% and 1% methane in the headspace. Repeated methane addition every 3 to 4 days was started on day 14. Data plotted are mean values (n=2 or 3) and s.e. from nonlabeled microcosms destructively sampled at each time point.
DNA-SIP analysis of nitrifying and methane-oxidizing communities
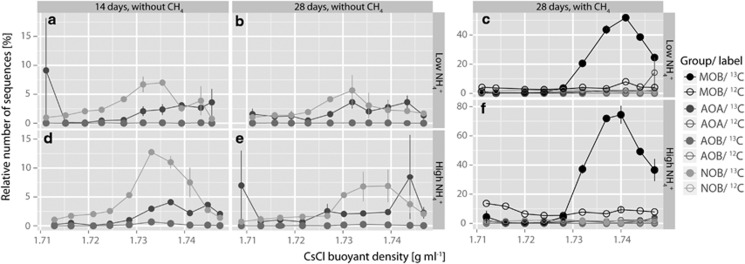
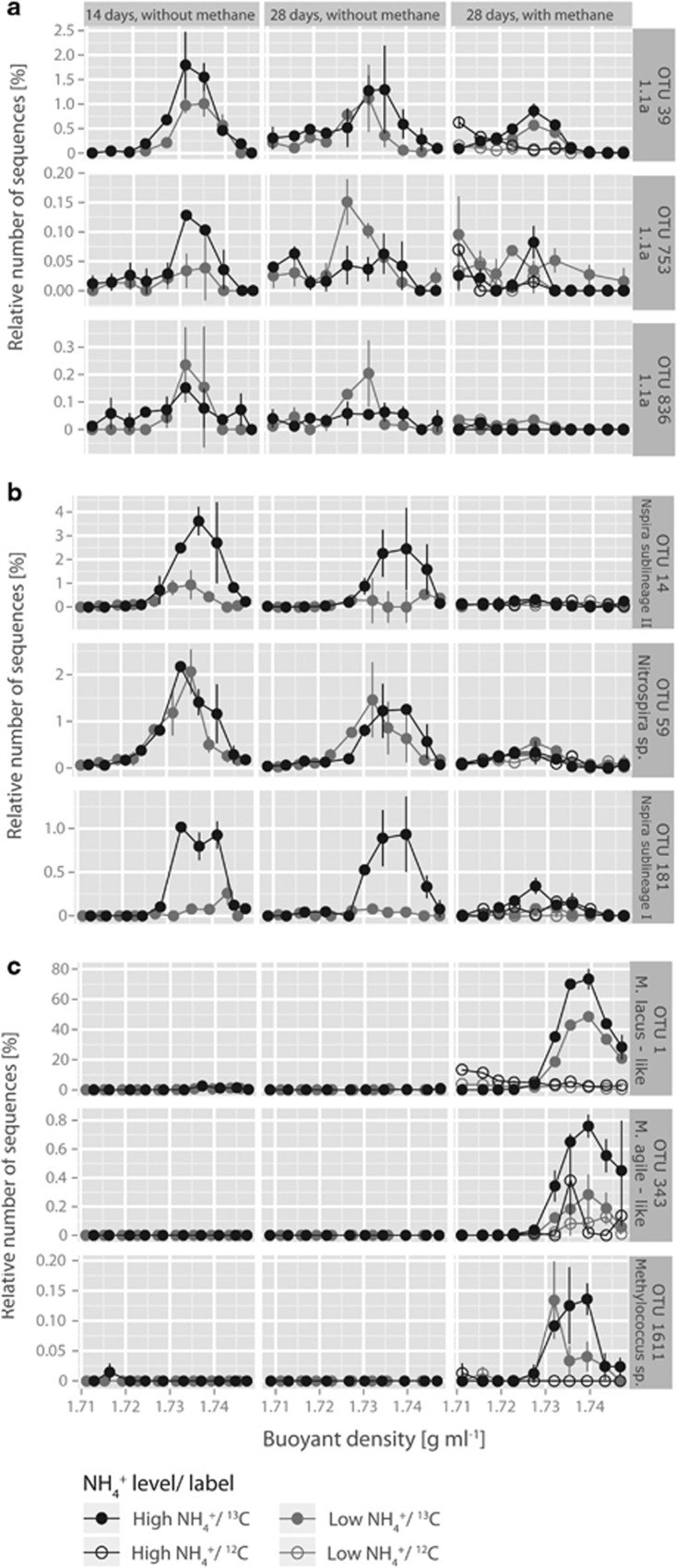
In order to reveal which part of the nitrifying and methane-oxidizing community was actively growing by assimilating CO2/CH4, we separated ‘heavy' (13C-enriched) DNA from ‘light' (12C) DNA by isopycnic, CsCl gradient ultracentrifugation of DNA from microcosms incubated with 13CO2 for 14 and 28 days as well as from microcosms incubated with 13CO2 and 13CH4 from day 14 to 28, and their respective 12C controls (Figures 5 and 6).
Figure 5.
Average relative abundances of AOA, AOB, NOB and MOB 16S rRNA genes in fractionated DNA from microcosms regularly amended with low ((a–c) 15 μg NH4+-N per g dry soil added initially; 7.5 μg NH4+-N per g dry soil on days 7, 14 and 21) and high ((d–f); 150 μg NH4+-N per g dry soil added initially; 75 μg NH4+-N per g dry soil on days 7, 14 and 21) levels of ammonium and 5% 13CO2 sampled on day 14 (a, d) and day 28 (b, c, e, f) of incubation. Microcosms were incubated in the absence (a, b, d, e) and presence (c, f) of 1% 13CH4 from day 14 onwards. The relative frequencies are expressed as the percentage of 16S rRNA genes from the respective functional group to total 16S rRNA genes of each DNA fraction. Data plotted are mean values and s.e. from DNA obtained from gradient fractions of duplicate nonlabeled 12C (open symbols) and labeled 13C (closed symbols) microcosms.
Figure 6.
Changes in the relative frequencies of 16S rRNA gene sequence reads related to the three most abundant and labeled phylotypes of the archaeal ammonia-oxidizing (a), nitrite-oxidizing (b) and methane-oxidizing (c) community in fractionated DNA from microcosms regularly amended with low (15 μg NH4+-N per g dry soil added initially; 7.5 μg NH4+-N per g dry soil on days 7, 14 and 21) and high (150 μg NH4+-N per g dry soil added initially; 75 μg NH4+-N per g dry soil on days 7, 14 and 21) levels of ammonium sampled on days 14 and 28. Microcosms were incubated with and without 1% methane in the headspace for the last 14 days of incubation. The relative frequencies are expressed as the percentage of 16S rRNA genes from the respective OTU to total 16S rRNA genes of each DNA fraction. Data plotted are mean values and s.e. from DNA obtained from gradient fractions of duplicate nonlabeled 12C (open symbols) and labeled 13C (closed symbols) microcosms.
Total active communities
Relative abundances of AOA, AOB, NOB and MOB 16S rRNA genes in purified fractions 3 to 12 of the 15 CsCl gradient fractions were determined by pyrosequencing. The 16S rRNA gene sequences of AOB were mostly not detectable in the gradient fractions and if present never constituted >0.68% of the total number of sequence reads in any fraction, whereas AOA and NOB reads were found in all fractions and comprised up to 9.13% and 12.75% of total reads in a fraction, respectively (Figure 5). Because of the rareness of detected AOB sequences, no specific pattern of distribution along the CsCl gradients could be obtained, and consequently we did not find evidence for the incorporation of CO2 into genomic DNA of AOB.
The relative numbers of AOA-related 16S rRNA genes tended to increase in fractions above 1.73 g CsCl per ml in all microcosms without methane amendment, irrespective of the ammonium level applied. This distribution pattern was rather weak and remained at a low level of AOA-related 16S rRNA gene enrichment in the ‘heavy' end of the CsCl gradient. We did however find a clear enrichment of NOB-related 16S rRNA genes in all ‘heavier' fractions of the CsCl gradients (Figures 5a, b, d and e). In addition, we could detect different distributions of NOB-related 16S rRNA genes between the two ammonium treatment levels and sampling days with a trend to a larger, actively CO2-incorporating NOB community in the HAT microcosms as seen by a peak shift in relative sequence abundance toward the ‘heavy' end of the gradient, especially for day 14 (Figure 5d).
In the presence of methane, we could not detect an increase in relative 16S rRNA gene sequence abundances along the gradient for either of the nitrifiers (Figures 5c and f). In contrast, we observed a large enrichment of relative MOB 16S rRNA gene abundance in the fractions above 1.73 g CsCl per ml from the 13C-labeled microcosms, but not from 12C microcosms (Figures 5c and f), indicative of a very active, methanotrophic community. Furthermore, there was an unmistakable effect of ammonium treatment level on MOB as apparent from an increase of 13C-labeled MOB DNA in the ‘heavy' fractions by nearly one-third in the microcosms with high compared with those with low NH4+ treatment (Figures 5c and f).
In addition, we quantified archaeal amoA and bacterial pmoA genes in purified CsCl fractions 2 to 13 by qPCR which confirmed the pyrosequencing results and is shown in Supplementary Figure A in the Supplementary Information.
Community composition and active nitrifier and methanotroph phylotypes
The most abundant AOA phylotype in all samples fell within the 1.1a-associated Nitrosotalea cluster of the Thaumarchaeal phylum (Supplementary Figures B and E). In addition, we found four phylotypes belonging to the 1.1a cluster as well as five phylotypes of the 1.1b cluster. Generally, the 1.1b phylotypes were of much lower relative abundance than the 1.1a phylotypes. The AOA community composition in ‘heavy' and ‘light' fractions was significantly different (adonis, R2=0.17, p=0.025). In addition, ‘heavy' fractions from day 28 of methane-amended 13C-microcosms were significantly different from 12C-microcosms of the same day (adonis, R2=0.29, p=0.034). We found no significant difference between AOA communities in ‘heavy' fractions of replicates between the two ammonium treatment levels, time points or between methane-amended and nonamended microcosms.
For the majority of AOA phylotypes there was no clear evidence for incorporation of CO2 as they showed similar abundances in the ‘heavy' fractions from 13C and 12C microcosms. Only three phylotypes of the 1.1a cluster (that is, 39, 753 and 836) were enriched in the ‘heavy' end of the CsCl gradient from 13C but not 12C microcosms (Figure 6a), although at a rather low level of maximally 1.80% relative sequence abundance. Their distribution pattern is indicative of active CO2 incorporation and was significantly lower, but not absent in samples from methane-amended microcosms. For all three phylotypes, there was no significant difference of relative abundance in ‘heavy' fractions between the LAT and HAT microcosms, except in the incubations without methane on day 28. Here, the enrichment of phylotypes 753 and 836 in ‘heavy' fractions was larger in samples from LAT microcosms.
The low abundance of AOB in our microcosms was coupled to a low diversity. We only obtained three AOB phylotypes in total. They were affiliated with Nitrosomonas oligotropha and a Nitrosospira-associated cluster (Supplementary Figure F). Community analysis of AOB based on relative abundances was not possible because of their absence from most samples and the extremely low sequence counts in the samples where we did detect them.
Analysis of phylotype-based community composition of NOB showed that the whole community consisted exclusively of the genus Nitrospira. It was foremost a Nitrospira sublineage II-, a Nitrospira sp.- and a Nitrospira sublineage I-like phylotype that were enriched in the ‘heavy' fractions from the 13C microcosms (Figure 6b and Supplementary Figures C and G). These three NOB phylotypes strongly dominated the active autotrophic NOB community (Supplementary Figure C). There was no significant difference in NOB community composition between replicated ‘heavy' fractions, but there was a significant difference between NOB communities in ‘heavy' and ‘light' fractions (adonis, R2=0.33, p=0.001), showing that NOB did not only actively incorporate CO2 in our microcosm incubations, but that the autotrophically active NOB community was different from the nonautotrophically active one. This difference was significantly related to the ammonium treatment level (adonis, R2=0.31, p=0.003), the methane amendment (adonis, R2=0.27, p=0.003) as well as the time of sampling (adonis, R2=0.15, p=0.006). These factors jointly explained 74% of the observed variance. There was no significant difference in NOB communities between ‘heavy' fractions of 12C control and 13C-labeled, methane-amended microcosms on day 28 of the incubation, but a large difference in ‘heavy' fractions between methane-amended and nonamended microcosms of the same day. Moreover, we observed a substantial and significant decrease in relative abundance of the actively CO2-incorporating NOB phylotypes 14, 59 and 181 in the ‘heavy' end of the CsCl gradient in samples from methane-amended microcosms as compared with microcosms without methane (Figure 6b).
The methanotrophic community comprised eight phylotypes with the two most dominant ones closely related to Methylosarcina lacus (Supplementary Figures D and H). Adonis analysis of variances for MOB communities showed that there was no significant difference between replicates, but a significant difference between communities in ‘heavy' fractions from microcosms with and without methane amendment (adonis, R2=0.87, p=0.002) and between communities of 12C and 13C microcosms in the presence of methane on day 28 (adonis, R2=0.84, p=0.014). Furthermore, MOB communities in ‘heavy' fractions differed significantly from the ones in ‘light' fractions in microcosms to which 1% 13C-methane was added (adonis, R2=0.57, p=0.019), whereas there was no difference in community composition related to ammonium level. It was foremost a single M. lacus-like MOB phylotype that was responsible for these results and that was enriched from 0 to up to 73.40% relative abundance in ‘heavy' fractions of samples from methane-amended microcosms (Figure 6c).
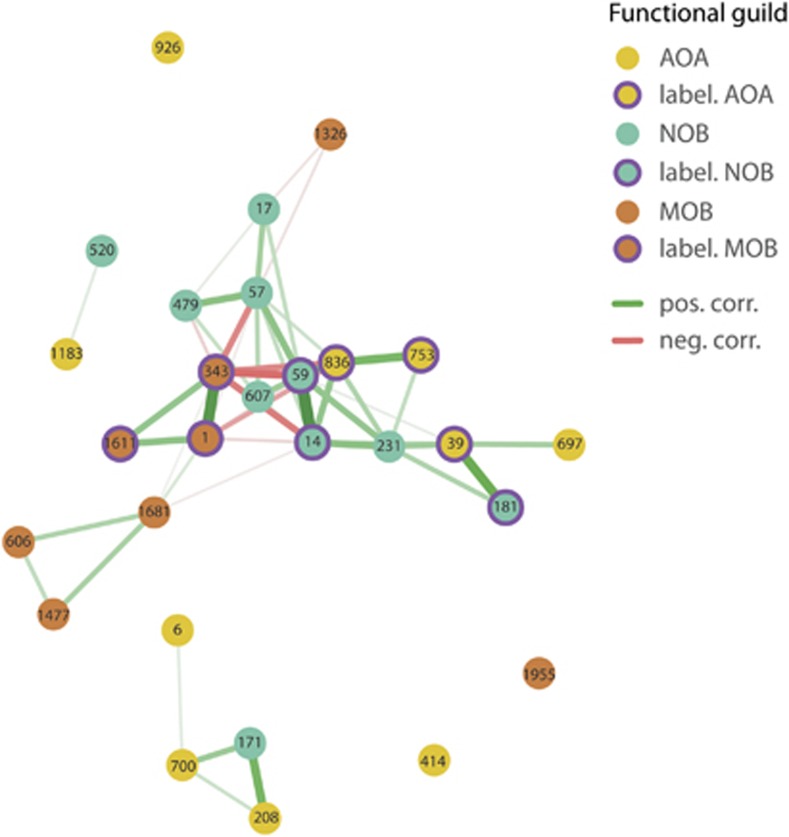
Network analysis
In order to elucidate interactions between AOA, NOB and MOB in our microcosms, we performed a network analysis based on co-occurring phylotypes of the three functional guilds in all incubations. The results show exclusively positive, stronger and more frequent correlations between the AOA and NOB than between MOB and one of the other guilds (Figure 7). The actively CO2-incorporating AOA phylotypes 39, 753 and 836 of the 1.1a cluster (see Figure 6a) were all strongly and positively correlated with NOB phylotypes, among which were the autotrophically growing phylotypes 14, 59 and 181 (see Figure 6b). Interestingly, two of these NOB phylotypes were negatively correlated to OTU 1, which is closely affiliated with M. lacus, the dominant and active methane-oxidizer phylotype (see Figure 6c). Another interesting correlation suggesting negative interactions is the one between the 1.1a AOA phylotype 836, whose autotrophic growth was suppressed in methane-amended microcosms, and the actively methane-oxidizing phylotypes 1 and 343. OTU 6, which fell within the Nitrosotalea cluster and was the most abundant phylotype of AOA in our microcosms, only showed a positive correlation with OTU 700, which is associated with the Nitrososphaera cluster.
Figure 7.
Network analysis of co-occurring phylotypes of AOA, NOB and MOB in microcosms incubated at low (15 μg NH4+-N per g dry soil added initially; 7.5 μg NH4+-N per g dry soil on days 7, 14 and 21) and high (150 μg NH4+-N per g dry soil added initially; 75 μg NH4+-N per g dry soil on days 7, 14 and 21) levels of ammonium for 14 and 28 days as well as with and without 1% methane for the last 14 days of incubation. Nodes represent the 10 most abundant OTUs of each of the three functional guilds and connecting lines stand for strong (Spearman's ρ>0.6) and significant (p-value of <0.05) positive (green) or negative (red) correlations where a thicker and shorter line represents a stronger correlation. Purple circles around nodes mark phylotypes that had actively incorporated the 13C label.
Discussion
Dominance of AOA and Nitrospira among nitrifiers
The ammonia-oxidizing community was clearly dominated by archaea as amoA-targeted qPCR as well as pyrosequencing of the total microbial community could demonstrate. Although we cannot exclude that AOB are a substantial fraction of the rare biosphere, this confirms the finding of a previous study conducted with soils of the same geothermal area (Daebeler et al., 2012).
An adaptation to low substrate best explains the numerical dominance of AOA in this soil that has the typically high cation exchange capacity of an Andosol, resulting in low ammonia availability. AOA of the 1.1a and the Nitrosotalea lineages, which comprise the vast majority of the AOA community detected in our incubations, are often found under oligotrophic conditions and may be adapted to low ammonium environments (Martens-Habbena et al., 2009; Martens-Habbena and Stahl, 2011; French et al., 2012; Nakagawa and Stahl, 2013; Gorman-Lewis et al., 2014). Nitrosotalea-like AOA have been demonstrated to be adapted to low pH environments, but are also abundant among ammonia oxidizers in many other terrestrial ecosystems and not exclusively found in low pH soils (Leininger et al., 2006; Gubry-Rangin et al., 2011; Lehtovirta-Morley et al., 2011; Pester et al., 2011). Their relatively high abundance in our soil may hence reflect their adaptation to low substrate availability rather than to low pH itself. On the other hand, cultivated members of the 1.1b lineage, which were a minority among the AOA observed in our incubations, have been shown to prefer higher substrate concentrations (Jung et al., 2011; Tourna et al., 2011).
Dominance of Nitrospira sp., the proposed k-strategist nitrite oxidizer with high substrate affinity and hence adaptation to low nitrite concentrations (Ehrich et al., 1995; Schramm et al., 1999; Koops and Pommerening-Röser, 2001; Attard et al., 2010), may provide further evidence of selection for low substrate-adapted nitrifiers in this volcanic grassland soil.
Activity and growth of nitrifiers and methane oxidizers
We observed an increase in archaeal amoA gene abundance in HAT microcosms without and with methane supply. This demonstrates that AOA were indeed actively growing despite the fact that evidence for autotrophic growth was only found at a rather low level and only for three specific 1.1a AOA phylotypes. These results of limited autotrophic growth by AOA are similar to those of Jia and Conrad (2009) and Pratscher et al. (2011) who concluded AOB to be more important for ammonia oxidation. However, in our soil AOB represent such low numbers that they are unlikely responsible for the observed ammonia oxidation. Accordingly, net nitrification rates were comparable to those found previously (Zhang et al., 2010; Verhamme et al., 2011), where the authors reported ammonia oxidation to be mainly driven by AOA. The detected CO2 incorporation by 1.1a AOA phylotypes, which jointly represented 23% of the total AOA community, suggests that at least some AOA did indeed grow autotrophically, likely coupled to ammonia oxidation. Such preferential activity of 1.1a AOA confirms findings of previous studies (Tourna et al., 2008; Offre et al., 2009; Zhang et al., 2010).
As no cultured AOA has been found to contain more than one copy of the amoA gene (Hallam et al., 2006; Walker et al., 2010; Blainey et al., 2011; Spang et al., 2012) we may assume that copy numbers of archaeal amoA can be directly related to cell abundance. This is not the case for AOB, as several AOB harbor several copies of the amoC/amoA/amoB gene complex (Chain et al., 2003; Stein et al., 2007; Norton et al., 2008) and it is therefore more reliable to infer AOB cell abundance from 16S rRNA gene quantification. Even if the estimated number of labeled AOA cells (23% of the archaeal amoA abundance) was the same throughout the entire incubation period as on day 28 and if all AOA cells were growing coupled to ammonia oxidation with maximum specific activity rates similar to reported values for 1.1a AOA (Jung et al., 2011), they could have only oxidized a maximum of 0.5 μM NH3. We however measured for example an oxidation of 67.1 μM NH3 in HAT microcosms without methane, suggesting heterotrophic or mixotrophic ammonia oxidation performed by AOA. The increase in amoA gene abundance suggests that the AOA community consisted of 75% newly grown cells by the end of the incubation period. Such growth, if supported autotrophically, would have been detected by DNA SIP to a much larger extent, analogous to previous studies (Zhang et al., 2010; Xia et al., 2011; Lu and Jia, 2013). Mixotrophy has been suggested for AOA in soil before (Jia and Conrad, 2009; Mussmann et al., 2011; Tourna et al., 2011) and through genomic and metagenomic studies, the mechanisms and abilities of lineage 1.1a Thaumarchaeota to utilize small organic substances have been identified (Ouverney and Fuhrman, 2000; Ingalls et al., 2006; Walker et al., 2010; Tully et al., 2012). Nevertheless, it can be imagined that some of the observed ammonia oxidation was not coupled to growth of AOA or AOB, as gene abundance was one and four orders of magnitude lower than expected from relating observed net nitrification to reported per cell activity rates for AOA and AOB, respectively (Prosser, 1989; Laanbroek and Gerards, 1993; Jung et al., 2011).
Of the detected Nitrospira sp. only a minority was assimilating CO2, suggesting a high importance of specific Nitrospira spp. for autotrophic nitrite oxidation which is consistent with previous reports (Freitag et al., 2005; Xia et al., 2011). Differences in autotrophic growth of the Nitrospira sublineage I OTU 181 between HAT and LAT microcosms supports the report on a preference for higher nitrite concentrations of this sublineage (Maixner et al. (2006). In contrast we found the autotrophic activity of a Nitrospira sublineage II phylotype increased upon addition of more NH4+. We may therefore conclude, that substrate-driven niche differentiation exists within sublineage II Nitrospira and that at least some nitrite oxidizers of this sublineage are selected by elevated nitrite concentrations as well.
The methane-oxidizing community in our incubations responded rapidly and vigorously to the availability of CH4, suggesting methanotrophy to be an important metabolic lifestyle in the Icelandic grassland soil studied here. MOB abundance rapidly dropped in LAT but not HAT microcosms without methane. This is indicative of N limitation and nitrogen dependence, but not in relation to methanotrophy. Despite the origin of the soil material used, being top soil from a grassland site, availability of biologically derived methane from deeper layers as a substrate for MOB is highly likely as anoxic microcosm incubations produced significant amounts of methane at a rate of 1000 nmol per g dry soil per d after a lag phase of 2 weeks (A. Daebeler, nonpublished results). Furthermore, steams of geothermal areas in Iceland carry average methane concentrations of 0.44 mmol kg−1 leading to estimated methane emission rates of 1300 t per year from Icelandic geothermal sites (Etiope et al., 2007). In combination with the readily responsive MOB community, this suggests the mitigation of methane emission as an important ecosystem service for these grasslands.
Surprisingly, over 98% of the observed CH4-C assimilation was carried out by only a single phylotype closely related to M. lacus. Although we cannot exclude growth of other MOB through alternative carbon sources (Semrau et al., 2011), the increase in relative abundance of this M. lacus-like MOB under methane-amended conditions leads to the conclusion that the observed growth of the MOB community was to a large extent because of the growth of this organism.
Putative interactions of nitrifiers and MOB
The design of our study allowed for analysis of responses of CO2- and CH4-incorporating microorganisms to low and elevated ammonium availabilities. As only HAT but not LAT microcosms showed more expected than observed nitrification, we conclude that significant amounts of ammonia were immobilized only in HAT microcosms. This further leads to the conclusion that this immobilization was likely not due to adsorption of ammonium to soil particles, but to assimilation by heterotrophic microorganisms. Therefore, we are convinced that the ammonium additions led to conditions of more and less N limitation, resulting in different forces of competition for the common nutrient.
Both nitrifying guilds studied were responsive to ammonium amendment: amoA gene abundances of AOA were elevated and incorporation of CO2-C by NOB was stronger in HAT than in LAT microcosms. In addition, net nitrification was stimulated by ammonium amendment, suggesting that both nitrifying guilds may be N limited under in situ conditions and are adapted to tolerate and utilize higher substrate concentrations. We cannot exclude, however, that specific groups with a higher tolerance and lower affinity for ammonium than the in situ populations were selected by the incubation conditions.
Likely, the AOA will have transmitted the higher substrate availability in HAT microcosms to NOB by providing them with more nitrite. Conclusively, the network analysis showed that phylotypes of AOA and NOB had exclusively positive correlations. This is especially interesting in the case of autotrophically growing NOB and AOA as it may display their occupancy of the same niche space as well as the syntrophic relationship that is apparent from their energy metabolism as nitrifiers. We are therefore convinced that strong and positive links between autotrophically growing AOA of the 1.1a cluster and nitrite-oxidizing Nitrospira exist in the in situ soil environment, comparable to co-occurrence patterns of amoA-encoding archaea and Nitrospina in the ocean (Mincer et al., 2007). In addition, positive co-occurrence patterns between AOA and NOB phylotypes which were not actively incorporating CO2 leave room for the possibility of interrelations of AOA and NOB with metabolic lifestyles other than chemolithoautotrophic ammonia and nitrite oxidation.
In the presence of methane, autotrophic growth of AOA and NOB ceased at both ammonium levels, more so in case of the NOB, and the network analysis showed exclusively negative correlations between active MOB and autotrophically growing nitrifiers. Interestingly, net nitrification was not significantly affected by the addition of methane, possibly suggesting a metabolic switch of nitrifiers. However, it is possible that only nitrite oxidation was in fact inhibited by the presence of active MOB. Unfortunately, the absence of data on nitrite accumulation separately from nitrate failed to resolve this possibility. Little is known about interactions between nitrifiers and methanotrophs, but our data show inhibition of autotrophic ammonia and nitrite oxidation by an active, methane-oxidizing community coupled to a shift in community composition of AOA and NOB. This result supports our hypothesis of a competition for common substrates between active methane oxidizers and nitrifiers. Perhaps, the suppression of autotrophic nitrification by active MOB was further a direct consequence of oxygen consumption by MOB causing suboxic conditions which limited the growth of strictly autotrophic NOB, but not to the same extent the growth of AOA who are known to tolerate low oxygen concentrations (Martens-Habbena et al., 2009; Park et al., 2010; Jung et al., 2011; Martens-Habbena and Stahl, 2011). When assuming a 1:1 CH4/O2 stoichiometry for methane oxidation, MOB must have consumed ∼0.51 and 0.55 mmol oxygen in the LAT and HAT microcosms, respectively, within the last week of the incubation and may thus have created oxygen-limited conditions within parts of the soil matrix. Under such low oxygen conditions, MOB equipped to use nitrite as electron acceptor alternative to oxygen as reported by Nyerges et al. (2010) would likely have competed with Nitrospira for nitrite, resulting in detrimental circumstances for the nitrite oxidizers. This effect would be more unfavorable in HAT microcosms and may have caused the observed pattern of higher methane oxidation, ceased NOB activity, but less affected AOA growth with no detectable decrease in oxidized mineral N.
Summary
In the Icelandic grassland soil studied, syntrophic communities of autotrophic 1.1a AOA and autotrophic Nitrospira likely drove chemolithotrophic nitrification, and mixotrophic AOA may also have contributed to nitrification. Our results challenge the perception of archaeal ammonia oxidizers and nitrite oxidizers as physiologically uniform guilds and hint at metabolic plasticity of AOA and NOB, such as active selection for different modes of growth as well as uncoupling of metabolic activity from growth. Both nitrifying guilds showed positive responses to inorganic N supply and we found indications of niche differentiation within the genus of Nitrospira as a result of adaptation to N availability. Likely because of strongly nitrogen-limited conditions in the soil, bacterial ammonia oxidizers constituted a negligible fraction of the microbial community and remained unresponsive throughout the incubation period. The availability of mineral nitrogen played a pivotal role not only for the regulation of nitrification and abundance activity patterns of soil nitrifiers, but also for the extent to which the soil was capable of methane oxidation. Our results show that nitrogen stimulation of active methanotrophs can lead to suppression of autotrophic nitrification and of autotrophic growth of AOA and NOB, perhaps through competition for mineral nitrogen and oxygen. This previously unrecognized competition between nitrifiers and methanotrophs may therefore cause uncoupling of the N cycle and will have to be accounted for when addressing the impacts of N deposition.
Acknowledgments
We thank Bjarni D Sigurdsson and the Agricultural University of Iceland for access to the sampling site and Bas Dingemans for collecting the soil samples. We also thank Roos Keijzer and Marion Meima-Franke for excellent technical assistance. The Schure-Beijerinck-Popping fund is acknowledged for funding the stay of AD as a guest researcher in the ZJ group. This study was partially supported by the Ministry of Science and Technology of China (2010DFA22770). This is publication number 5603 of The Netherlands Institute of Ecology (NIOO-KNAW).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Attard E, Poly F, Commeaux C, Laurent F, Terada A, Smets BF, et al. Shifts between Nitrospira- and Nitrobacter-like nitrite oxidizers underlie the response of soil potential nitrite oxidation to changes in tillage practices. Environ Microbiol. 2010;12:315–326. doi: 10.1111/j.1462-2920.2009.02070.x. [DOI] [PubMed] [Google Scholar]
- Blainey PC, Mosier AC, Potanina A, Francis CA, Quake SR. Genome of a low-salinity ammonia-oxidizing archaeon determined by single-cell and metagenomic analysis. PLoS One. 2011;6:e16626. doi: 10.1371/journal.pone.0016626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodelier PLE, Laanbroek HJ. Nitrogen as a regulatory factor of methane oxidation in soils and sediments. FEMS Microbiol Ecol. 2004;47:265–277. doi: 10.1016/S0168-6496(03)00304-0. [DOI] [PubMed] [Google Scholar]
- Bodelier PLE. Interactions between nitrogenous fertilizers and methane cycling in wetland and upland soils. Curr Opin Environ Sustainability. 2011;3:379–388. [Google Scholar]
- Boyett MR, Tavakkoli A, Sobolev D. Mathematical modeling of competition for ammonium among Bacteria, Archaea and cyanobacteria within cyanobacterial mats: can ammonia-oxidizers force nitrogen fixation. Ocean Sci J. 2013;48:269–277. [Google Scholar]
- Chain P, Lamerdin J, Larimer FW, Regala W, Lao V, Land M, et al. Complete genome sequence of the ammonia-oxidizing bacterium and obligate chemolithoautotroph Nitrosomonas europaea. J Bacteriol. 2003;185:2759–2773. doi: 10.1128/JB.185.9.2759-2773.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daebeler A, Abell GCJ, Bodelier PLE, Bodrossy L, Frampton DMF, Hefting MM, et al. Archaeal dominated ammonia-oxidizing communities in Icelandic grassland soils are moderately affected by long-term N fertilization and geothermal heating. Fronti Microbiol. 2012;3:352. doi: 10.3389/fmicb.2012.00352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di HJ, Cameron KC, Shen JP, Winefield CS, O'Callaghan M, Bowatte S, et al. Nitrification driven by bacteria and not archaea in nitrogen-rich grassland soils. Nat Geosci. 2009;2:621–624. [Google Scholar]
- Ehrich S, Behrens D, Lebedeva E, Ludwig W, Bock E. A new obligately chemolithoautotrophic, nitrite-oxidizing bacterium,Nitrospira moscoviensis sp. nov. and its phylogenetic relationship. Arch Microbiol. 1995;164:16–23. doi: 10.1007/BF02568729. [DOI] [PubMed] [Google Scholar]
- Epskamp S, Cramer AOJ, Waldorp LJ, Schmittmann VD, Borsboom D. qgraph: network visualizations of relationships in psychometric data. J Stat Softw. 2012;48:1–18. [Google Scholar]
- Etiope G, Fridriksson T, Italiano F, Winiwarter W, Theloke J. Natural emissions of methane from geothermal and volcanic sources in Europe. J Volcanol Geoth Res. 2007;165:76–86. [Google Scholar]
- Freitag A, Bock E. Energy conservation in Nitrobacter. FEMS Microbiol Lett. 1990;66:157–162. [Google Scholar]
- Freitag TE, Chang L, Clegg CD, Prosser JI. Influence of inorganic nitrogen management regime on the diversity of nitrite-oxidizing bacteria in agricultural grassland soils. Appl Environ Microbiol. 2005;71:8323–8334. doi: 10.1128/AEM.71.12.8323-8334.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French E, Kozlowski JA, Mukherjee M, Bullerjahn G, Bollmann A. Ecophysiological characterization of ammonia-oxidizing archaea and bacteria from freshwater. Appl Environ Microbiol. 2012;78:5773–5780. doi: 10.1128/AEM.00432-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fruchterman TMJ, Reingold EM. Graph drawing by force-directed placement. Softw Pract Exp. 1991;21:1129–1164. [Google Scholar]
- Gorman-Lewis D, Martens-Habbena W, Stahl DA. Thermodynamic characterization of proton-ionizable functional groups on the cell surfaces of ammonia-oxidizing bacteria and archaea. Geobiology. 2014;12:157–171. doi: 10.1111/gbi.12075. [DOI] [PubMed] [Google Scholar]
- Gruber N, Galloway JN. An Earth-system perspective of the global nitrogen cycle. Nature. 2008;451:293–296. doi: 10.1038/nature06592. [DOI] [PubMed] [Google Scholar]
- Gubry-Rangin C, Hai B, Quince C, Engel M, Thomson BC, James P, et al. Niche specialization of terrestrial archaeal ammonia oxidizers. Proc Natl Acad Sci USA. 2011;108:21206–21211. doi: 10.1073/pnas.1109000108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallam SJ, Konstantinidis KT, Putnam N, Schleper C, Watanabe Y-i, Sugahara J, et al. Genomic analysis of the uncultivated marine crenarchaeote Cenarchaeum symbiosum. Proc Natl Acad Sci USA. 2006;103:18296–18301. doi: 10.1073/pnas.0608549103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton TL, Koonce E, Howells A, Havig JR, Jewell T, de la Torre JR, et al. Competition for ammonia influences the structure of chemotrophic communities in geothermal springs. Appl Environ Microbiol. 2014;80:653–661. doi: 10.1128/AEM.02577-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzenpichler R, Lebedeva EV, Spieck E, Stoecker K, Richter A, Daims H, et al. A moderately thermophilic ammonia-oxidizing crenarchaeote from a hot spring. Proc Natl Acad Sci USA. 2008;105:2134–2139. doi: 10.1073/pnas.0708857105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingalls AE, Shah SR, Hansman RL, Aluwihare LI, Santos GM, Druffel ERM, et al. Quantifying archaeal community autotrophy in the mesopelagic ocean using natural radiocarbon. Proc Natl Acad Sci USA. 2006;103:6442–6447. doi: 10.1073/pnas.0510157103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Z, Conrad R. Bacteria rather than Archaea dominate microbial ammonia oxidation in an agricultural soil. Environ Microbiol. 2009;11:1658–1671. doi: 10.1111/j.1462-2920.2009.01891.x. [DOI] [PubMed] [Google Scholar]
- Jung M-Y, Park S-J, Min D, Kim J-S, Rijpstra WIC, Sinninghe Damsté JS, et al. Enrichment and characterization of an autotrophic ammonia-oxidizing archaeon of mesophilic crenarchaeal group I.1a from an agricultural soil. Appl Environ Microbiol. 2011;77:8635–8647. doi: 10.1128/AEM.05787-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung M-Y, Well R, Min D, Giesemann A, Park S-J, Kim J-G, et al. Isotopic signatures of N2O produced by ammonia-oxidizing archaea from soils. ISME J. 2013;8:1115–1125. doi: 10.1038/ismej.2013.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke X, Angel R, Lu Y, Conrad R. Niche differentiation of ammonia oxidizers and nitrite oxidizers in rice paddy soil. Environ Microbiol. 2013;15:2275–2292. doi: 10.1111/1462-2920.12098. [DOI] [PubMed] [Google Scholar]
- Kim J-G, Jung M-Y, Park S-J, Rijpstra WIC, Sinninghe Damsté JS, Madsen EL, et al. Cultivation of a highly enriched ammonia-oxidizing archaeon of thaumarchaeotal group I.1b from an agricultural soil. Environ Microbiol. 2012;14:1528–1543. doi: 10.1111/j.1462-2920.2012.02740.x. [DOI] [PubMed] [Google Scholar]
- Koops H-P, Pommerening-Röser A. Distribution and ecophysiology of the nitrifying bacteria emphasizing cultured species. FEMS Microbiol Ecol. 2001;37:1–9. [Google Scholar]
- Laanbroek H, Gerards S. Competition for limiting amounts of oxygen between Nitrosomonas europaea and Nitrobacter winogradskyi grown in mixed continuous cultures. Arch Microbiol. 1993;159:453–459. [Google Scholar]
- Lehtovirta-Morley LE, Stoecker K, Vilcinskas A, Prosser JI, Nicol GW. Cultivation of an obligate acidophilic ammonia oxidizer from a nitrifying acid soil. Proc Natl Acad Sci USA. 2011;108:15892–15897. doi: 10.1073/pnas.1107196108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leininger S, Urich T, Schloter M, Schwark L, Qi J, Nicol GW, et al. Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature. 2006;442:806–809. doi: 10.1038/nature04983. [DOI] [PubMed] [Google Scholar]
- Levicnik-Hofferle S, Nicol GW, Ausec L, Mandic-Mulec I, Prosser JI. Stimulation of thaumarchaeal ammonia oxidation by ammonia derived from organic nitrogen but not added inorganic nitrogen. FEMS Microbiol Ecol. 2012;80:114–123. doi: 10.1111/j.1574-6941.2011.01275.x. [DOI] [PubMed] [Google Scholar]
- Lipschultz F, Zafiriou OC, Wofsy SC, McElroy MB, Valois FW, Watson SW. Production of NO and N2O by soil nitrifying bacteria. Nature. 1981;294:641–643. [Google Scholar]
- Lu L, Jia Z. Urease gene-containing Archaea dominate autotrophic ammonia oxidation in two acid soils. Environ Microbiol. 2013;15:1795–1809. doi: 10.1111/1462-2920.12071. [DOI] [PubMed] [Google Scholar]
- Lueders T, Manefield M, Friedrich MW. Enhanced sensitivity of DNA- and rRNA-based stable isotope probing by fractionation and quantitative analysis of isopycnic centrifugation gradients. Environ Microbiol. 2004;6:73–78. doi: 10.1046/j.1462-2920.2003.00536.x. [DOI] [PubMed] [Google Scholar]
- Maixner F, Noguera DR, Anneser B, Stoecker K, Wegl G, Wagner M, et al. Nitrite concentration influences the population structure of Nitrospira-like bacteria. Environ Microbiol. 2006;8:1487–1495. doi: 10.1111/j.1462-2920.2006.01033.x. [DOI] [PubMed] [Google Scholar]
- Martens-Habbena W, Berube PM, Urakawa H, de la Torre JR, Stahl DA. Ammonia oxidation kinetics determine niche separation of nitrifying Archaea and Bacteria. Nature. 2009;461:976–979. doi: 10.1038/nature08465. [DOI] [PubMed] [Google Scholar]
- Martens-Habbena W, Stahl DA. Nitrogen metabolism and kinetics of ammonia-oxidizing archaea. Method Enzymol. 2011;496:465–487. doi: 10.1016/B978-0-12-386489-5.00019-1. [DOI] [PubMed] [Google Scholar]
- Mincer TJ, Church MJ, Taylor LT, Preston C, Kar DM, DeLong EF. Quantitative distribution of presumptive archaeal and bacterial nitrifiers in Monterey Bay and the North Pacific Subtropical Gyre. Environ Microbiol. 2007;9:1162–1175. doi: 10.1111/j.1462-2920.2007.01239.x. [DOI] [PubMed] [Google Scholar]
- Mussmann M, Brito I, Pitcher A, Damste JSS, Hatzenpichler R, Richter A, et al. Thaumarchaeotes abundant in refinery nitrifying sludges express amoA but are not obligate autotrophic ammonia oxidizers. Proc Natl Acad Sci USA. 2011;108:16771–16776. doi: 10.1073/pnas.1106427108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Stahl DA. Transcriptional response of the archaeal ammonia oxidizer Nitrosopumilus maritimus to low and environmentally relevant ammonia concentrations. Appl Environ Microbiol. 2013;79:6911–6916. doi: 10.1128/AEM.02028-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norton JM, Klotz MG, Stein LY, Arp DJ, Bottomley PJ, Chain PSG, et al. Complete genome sequence of nitrosospira multiformis, an ammonia-oxidizing bacterium from the soil environment. Appl Environ Microbiol. 2008;74:3559–3572. doi: 10.1128/AEM.02722-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyerges G, Han S-K, Stein LY. Effects of ammonium and nitrite on growth and competitive fitness of cultivated methanotrophic bacteria. Appl Environ Microbiol. 2010;76:5648–5651. doi: 10.1128/AEM.00747-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Offre P, Prosser JI, Nicol GW. Growth of ammonia-oxidizing archaea in soil microcosms is inhibited by acetylene. FEMS Microbiol Ecol. 2009;70:99–108. doi: 10.1111/j.1574-6941.2009.00725.x. [DOI] [PubMed] [Google Scholar]
- Oksanen J, Blanchet G, Kindt R, Legendre P, O'Hara RG, Simpson GL, et al. 2010. Vegan: community ecology package. R package version 1.17-1. Available at: : http://CRAN.R-project.org/package=vegan .
- Ouverney CC, Fuhrman JA. Marine planktonic archaea take up amino acids. Appl Environ Microbiol. 2000;66:4829–4833. doi: 10.1128/aem.66.11.4829-4833.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park B-J, Park S-J, Yoon D-N, Schouten S, Sinninghe Damste JS, Rhee S-K. Cultivation of autotrophic ammonia-oxidizing archaea from marine sediments in coculture with sulfur-oxidizing bacteria. Appl Environ Microbiol. 2010;76:7575–7587. doi: 10.1128/AEM.01478-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pester M, Rattei T, Flechl S, Gröngröft A, Richter A, Overmann J, et al. amoA-based consensus phylogeny of ammonia-oxidizing archaea and deep sequencing of amoA genes from soils of four different geographic regions. Environ Microbiol. 2011;14:525–539. doi: 10.1111/j.1462-2920.2011.02666.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratscher J, Dumont MG, Conrad R. Ammonia oxidation coupled to CO2 fixation by archaea and bacteria in an agricultural soil. Proc Natl Acad Sci USA. 2011;108:4170–4175. doi: 10.1073/pnas.1010981108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosser JI. Autotrophic nitrification in bacteria. Adv Microb Physiol. 1989;30:125–181. doi: 10.1016/s0065-2911(08)60112-5. [DOI] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Gevers D, Westcott SL. Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PLoS One. 2011;6:e27310. doi: 10.1371/journal.pone.0027310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramm A, de Beer D, van den Heuvel JC, Ottengraf S, Amann R. Microscale distribution of populations and activities of Nitrosospira and Nitrospira spp. along a macroscale gradient in a nitrifying bioreactor: quantification by in situ hybridization and the use of microsensors. Appl Environ Microbiol. 1999;65:3690–3696. doi: 10.1128/aem.65.8.3690-3696.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semrau JD, Dispirito AA, Vuilleumier S. Facultative methanotrophy: false leads, true results, and suggestions for future research. FEMS Microbiol Lett. 2011;323:1–12. doi: 10.1111/j.1574-6968.2011.02315.x. [DOI] [PubMed] [Google Scholar]
- Spang A, Poehlein A, Offre P, Zumbragel S, Haider S, Rychlik N, et al. The genome of the ammonia-oxidizing Candidatus Nitrososphaera gargensis: insights into metabolic versatility and environmental adaptations. Environ Microbiol. 2012;14:3122–3145. doi: 10.1111/j.1462-2920.2012.02893.x. [DOI] [PubMed] [Google Scholar]
- Stein LY, Daniel JA, Paul MB, Patrick SGC, Loren H, Mike SMJ, et al. Whole-genome analysis of the ammonia-oxidizing bacterium, Nitrosomonas eutropha C91: implications for niche adaptation. Environ Microbiol. 2007;9:2993–3007. doi: 10.1111/j.1462-2920.2007.01409.x. [DOI] [PubMed] [Google Scholar]
- Stein LY, Klotz MG. Nitrifying and denitrifying pathways of methanotrophic bacteria. Biochem Soc Trans. 2011;39:1826–1831. doi: 10.1042/BST20110712. [DOI] [PubMed] [Google Scholar]
- Stein LY, Roy R, Dunfield PF. Aerobic Methanotrophy and Nitrification: Processes and Connections. eLS. John Wiley & Sons, Ltd; 2012. [Google Scholar]
- Stopnisek N, Gubry-Rangin C, Hofferle S, Nicol GW, Mandic-Mulec I, Prosser JI. Thaumarchaeal ammonia oxidation in an acidic forest peat soil is not influenced by ammonium amendment. Appl Environ Microbiol. 2010;76:7626–7634. doi: 10.1128/AEM.00595-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourna M, Freitag TE, Nicol GW, Prosser JI. Growth, activity and temperature responses of ammonia-oxidizing archaea and bacteria in soil microcosms. Environ Microbiol. 2008;10:1357–1364. doi: 10.1111/j.1462-2920.2007.01563.x. [DOI] [PubMed] [Google Scholar]
- Tourna M, Stieglmeier M, Spang A, Konneke M, Schintlmeister A, Urich T, et al. Nitrososphaera viennensis, an ammonia oxidizing archaeon from soil. Proc Natl Acad Sci USA. 2011;108:8420–8425. doi: 10.1073/pnas.1013488108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tully BJ, Nelson WC, Heidelberg JF. Metagenomic analysis of a complex marine planktonic thaumarchaeal community from the Gulf of Maine. Environ Microbiol. 2012;14:254–267. doi: 10.1111/j.1462-2920.2011.02628.x. [DOI] [PubMed] [Google Scholar]
- Urakawa H, Martens-Habbena W, Stahl DA.2011Physiology and genomics of ammonia-oxidizing archaeaIn: Ward BB, Arp DJ, Klotz MG (eds)Nitrification ASM Press: Washington, DC; 117–155. [Google Scholar]
- Verhagen FJM, Laanbroek HJ, Woldendorp JW. Competition for ammonium between nitrifying bacteria and plant roots in soil pots; effects of grazing by flagellates and fertilization. Plant Soil. 1995;170:241–250. [Google Scholar]
- Verhamme DT, Prosser JI, Nicol GW. Ammonia concentration determines differential growth of ammonia-oxidising archaea and bacteria in soil microcosms. ISME J. 2011;5:1067–1071. doi: 10.1038/ismej.2010.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker CB, de la Torre JR, Klotz MG, Urakawa H, Pinel N, Arp DJ, et al. Nitrosopumilus maritimus genome reveals unique mechanisms for nitrification and autotrophy in globally distributed marine crenarchaea. Proc Natl Acad Sci USA. 2010;107:8818–8823. doi: 10.1073/pnas.0913533107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia WW, Zhang CX, Zeng XW, Feng YZ, Weng JH, Lin XG, et al. Autotrophic growth of nitrifying community in an agricultural soil. ISME J. 2011;5:1226–1236. doi: 10.1038/ismej.2011.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang LM, Offre PR, He JZ, Verhamme DT, Nicol GW, Prosser JI. Autotrophic ammonia oxidation by soil thaumarchaea. Proc Natl Acad Sci USA. 2010;107:17240–17245. doi: 10.1073/pnas.1004947107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.