Abstract
Detrimental host–pathogen interactions are a normal phenomenon in aquaculture animal production, and have been counteracted by prophylactic use of antibiotics. Especially, the youngest life stages of cultivated aquatic animals are susceptible to pathogen invasion, resulting in disease and mortality. To establish a more sustainable aquatic food production, there is a need for new microbial management strategies that focus on ‘join them' and not the traditional ‘beat them' approaches. We argue that ecological theory could serve as a foundation for developing sustainable microbial management methods that prevent pathogenic disease in larviculture. Management of the water microbiota in aquaculture systems according to ecological selection principles has been shown to decrease opportunistic pathogen pressure and to result in an improved performance of the cultured animals. We hypothesize that manipulation of the biodiversity of the gut microbiota can increase the host's resistance against pathogenic invasion and infection. However, substantial barriers need to be overcome before active management of the intestinal microbiota can effectively be applied in larviculture.
Keywords: aquaculture, ecological theory, host–microbe, intestinal, larviculture, microbe–microbe
Microbiota and disease in aquaculture
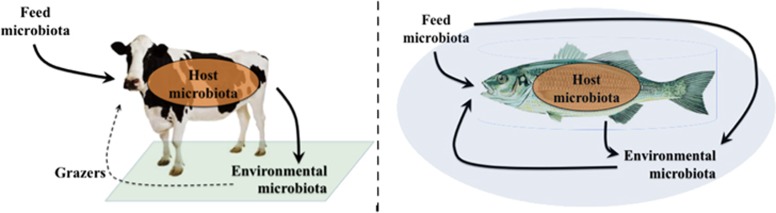
The presence of detrimental microorganisms is a major concern in animal breeding. The life stages shortly after birth or hatching are highly susceptible to disease and mortality, partly because of an immature immune system. Infections are a common problem for broilers (Suzuki, 1994), piglets (Fairbrother et al., 2005) and calves (Lanzas et al., 2008), and also for larvae of various shrimp and fish species (Vadstein et al., 2013). This has resulted in more than 60 years of prophylactic use of antibiotics in animal production (Gustafson and Bowen, 1997). The way aquatic animals live suggests that the association between the host intestinal microbiota and the environmental microbiota is closer than for land-bred animals (Figure 1). In aquaculture systems, microbes and animals have water as a shared environment, and the water is characterized by a high load of organic material supporting microbial growth (Vadstein et al., 2004). This makes the control of proliferation of pathogens extremely challenging.
Figure 1.
Terrestrial animals are cultured on flat surfaces (left). As feed is normally supplied to the animals in cratches, the feed associated-microbiota has little direct interaction with the environmental microbiota as opposed to the host intestinal microbiota. Defecation will modify the environmental microbiota on a local scale, but not necessarily on a regional scale (including the animal's intestine) because of dispersal limitation, or limited active uptake in the case of grazers (Bissett et al., 2010). Aquatically cultured animals live in water that completely surrounds them (right). As feed is dosed directly in the water, the feed microbiota may have a profound effect on the composition of the environmental microbiota. Similarly, the composition of the environmental microbiota may be dependent on the host intestinal microbiota because of defecation in the water. Microorganisms in the water easily spread on a regional scale, including to the animal's intestine because of drinking for osmoregulation, during feeding or by active uptake. The (in)dependency of the host microbiota from both the feed microbiota and the environmental microbiota is thus a relevant question (Wong and Rawls, 2012).
The control over bacterial pathogens in aquaculture systems can be pursued at different levels. Biosecurity measures can be taken to prevent pathogens from entering the rearing facility (Pruder, 2004). However, the implementation of hygienic barriers is not flawless and there is substantial regrowth in the water. Consequently, additional measures should be taken to restrain pathogens within the aquaculture system.
The primary compartment where pathogens should be controlled is the water surrounding the animals. Aquaculture systems are traditionally designed as flow-through (that is, continuous intake and discharge of water) or recirculation systems (that is, internal treatment of the water for removal of nutrients during passage through a biofilter system in order to reuse it). Both are often combined with ozonation or ultraviolet irradiation for removal of harmful microorganisms (Summerfelt et al., 2009). The use of disinfection in aquaculture systems may, however, do more harm than good in terms of disease control when it is considered from a microbial ecological point of view. It may give opportunities for opportunistic pathogens to proliferate (Attramadal et al., 2012a; De Schryver et al., 2014) and should thus be evaluated carefully.
Pathogen control may also be achieved at the level of the host. As the gut is considered a main pathogen transmission route, the current research revolution in aquaculture aims at biological strategies to control pathogenic infections in the gut of the host. The main focus has so far been on the manipulation of the intestinal microbiota using probiotics and to a lesser extent prebiotics, as illustrated by several reviews (Gatesoupe, 1999, 2010; Verschuere et al., 2000; Ringø et al., 2010). We believe, however, that these strategies too often are developed and applied with little rational basis and ignore the ecology of the microbes. It is safe to state that the intestinal microbiota in aquatic animals contributes to nutrition and immune stimulation (Guarner and Malagelada, 2003), and there is a growing body of knowledge describing how gut bacteria interact with their aquatic host (see, for example, Rawls et al., 2004 and Forberg et al., 2012). However, the potential of managing the natural microbiota associated with aquatic animals for disease prevention and control is still mostly unexplored. An ecological perspective would contribute to the development of new prevention and treatment methodologies against pathogenic infections in aquaculture animals. Before efficient gut microbial management based on theoretical microbial ecology comes within reach, there are, however, substantial challenges that must be overcome and that require the combined efforts of scientists from the field of the applied biological science of animal breeding and the field of (theoretical) microbial ecology.
Managing the water microbiota based on ecological theory
The current perception in the aquaculture world is that the rearing water needs to be as low as possible in microbial load, and hence disinfection is a standard water treatment step. As a result, disinfected water in both flow-through and recirculation systems typically contains a perturbed microbial community (Attramadal et al., 2012a). Upon inflow in the rearing unit, the microorganisms arrive in an environment with an increased carrying capacity resulting from the presence of uneaten feed, feces and dead biomass. According to the ecological theory of r/K selection (MacArthur and Wilson, 1967), which also applies to microbial communities (Andrews and Harris, 1986), an environment rich in nutrients per cell, low in competition and with frequent perturbations selects for microorganisms with a high capacity to exploit nutrients and increase in population size, termed fast-growing opportunistic r-strategists. Conversely, a stable environment where there are scarce resources per cell and hence high interspecific competition will select for slow-growing competition specialists—the K-strategists. Current aquaculture practices thus favor the r-selected opportunists (Figure 2). These are often the opportunistic pathogens that attack young and stressed individuals but that are not as able to cause disease in healthy adult individuals (Hajek, 2004).
Figure 2.
Two examples of how current aquaculture practices promote r-selection, increase the probability of opportunistic pathogens and consequently the probability of detrimental infections. (a) Disinfection of intake water is done as a barrier against introduction of pathogens in the system. However, disinfection also decreases bacterial numbers and makes dissolved organic matter more bioavailable that, under normal conditions, results in a bloom of fast-growing r-strategists. (b) Addition of feed to the system will increase the microbial carrying capacity in the rearing tanks directly and indirectly through defecation. This increase in carrying capacity will also result in r-selection. Notice that the water typically resides in the rearing tanks during the r-selection period, and has already left the tanks when K-selection can take place. The figure is a modification of a figure kindly provided by Dr KJK Attramadal.
Based on the principles of r/K selection, the water microbiota may be managed toward a state that decreases the disease risk for the cultured animals. Controlled microbial recolonization of the inflow water with K-strategists counteracts destabilizing forces in the rearing unit (Attramadal et al., 2012b). This microbial maturation can be achieved in a flow-through maturation unit colonized by K-strategists or in recirculating aquaculture systems (Attramadal et al., 2014). In this way, the development of a pioneer community dominated by r-strategists can be avoided in the rearing water, and the contact between opportunistic pathogens and the cultured animals is minimized.
The matured water approach has recently been shown to result in a 65–70% increase in survival of cultured Atlantic cod larvae and to provide temporal stability of the microbial community in the rearing unit (Attramadal et al., 2014). This approach illustrates that an understanding of the ecological principles that are relevant for identified problems can serve as a foundation to act and successfully drive a system in a desired direction. It is thus not always required to know in detail the microbial ecological consequences of measures taken. Nonetheless, it would be interesting to verify whether the change in microbial biodiversity in the water and the larval gut resulting from microbial maturation could be a factor decreasing larval susceptibility to infections.
Managing the gut microbiota toward functionality
The biodiversity of the gut microbiota in aquatic animals can be hypothesized to determine the likelihood of pathogenic invasion and disease. Therefore, a specific ecological state of the intestinal microbial community may be targeted by microbial community management to increase the health status or performance of the host.
Gut biodiversity determines invasibility
Invasion resistance can be defined as the inhibition of exogenous microbes to invade and persist in a local community. It can also apply to native opportunistic bacteria that are present in the community at low population densities (Guarner and Malagelada, 2003; Valéry et al., 2008). Several obligate pathogens cause disease and mortality after invading the gut environment of larval, juvenile and adult aquatic animals. Opportunistic pathogens, however, normally cause microbial interference in larvae (Skjermo and Vadstein, 1999). The commensal microbiota is assumed to act as a barrier against pathogenic invasion and establishment (Stecher and Hardt, 2008). The factors considered to provide protection are: stimulation of the host immune response, production of bacteriocins, competition for nutrients and adhesion sites and alteration of the gut physicochemical environment (McCracken and Lorenz, 2001; Stecher and Hardt, 2008). However, only in rare cases have specific members or mechanisms been shown to cause colonization resistance (Dillon et al., 2005). On the other hand, the community structure of the microbiota has been hypothesized to be related to the functionality of a host's microbiome (Little et al., 2008). Resistance to pathogenic invasion is therefore a community function that may be explained by general ecological principles (Robinson et al., 2010a). This knowledge is of relevance for microbial management in future aquaculture production.
Richness and evenness are considered the most important quantitative measures of biodiversity (Purvis and Hector, 2000). The temporal variation of these variables, the dynamics, is also an interesting community descriptor.
Richness
There is ecological evidence of a positive relationship between diversity and ecosystem functioning and stability (see, for example, Naeem et al., 1994; McGrady-Steed et al., 1997; Ptacnik et al., 2008). In the context of invasion, the diversity resistance hypothesis states that a more diverse community has a higher probability for a protective effect (Kennedy et al., 2002). A possible mechanism behind this diversity–invasibility relationship is niche complementarity (Fargione and Tilman, 2005). Niche opportunities in the form of unused resources and the physical environment that supports the presence of an invader are in theory fewer when a more diverse community inhabits a given environment (Shea and Chesson, 2002; Costello et al., 2012). As resistance to invasion also depends on individual traits, a more diverse community has a higher chance of having a member with an antagonistic trait toward an invader (that is, sampling effect) (Fargione and Tilman, 2005) or is more likely to have species that contribute to each other's invasion resistance (that is, facilitation effect) (Bruno et al., 2003).
Negative richness–invasibility relationships have been found for a variety of ecosystems including plant communities (Knops et al., 1999; Mitchell et al., 2002; Kennedy et al., 2002; Fargione and Tilman, 2005), animal communities (Stachowicz et al., 1999; Johnson et al., 2008) and microbial communities (Cook et al., 2006; van Elsas et al., 2012). The specific case of invasion resistance to pathogens by an animal's gut microbiota has, as far as we know, been the subject of only one study. Dillon et al. (2005) investigated the effects of a bacterial richness gradient in the gut of the desert locust on the invasibility by the pathogenic bacterium Serratia marsescens. They found a significant negative relationship between the number of symbiotic bacterial species and the density of the pathogen inside the gut.
Some authors state that the richness–invasibility relationship is not straight forward in natural ecosystems (Meiners et al., 2004; Bell et al., 2005), and that a high native species richness also can indicate a high risk for invasion (Jiang and Morin, 2004). An ecosystem with a high community richness is indeed more likely to harbor invaders (the community scale), but also implies a higher chance for the presence of an antagonistic or competitive invasion resisting mechanism (the individualistic scale) (Levine, 2000). At the lower levels of species richness, the individual effect is likely to decrease considerably (Bell et al., 2005).
Evenness
Most studies on the ecological aspects of invasion have focused on species richness. However, community evenness may also be of importance for the stability of a variety of functions, including invasion resistance (Emery and Gross, 2007). Evenness usually changes more rapidly than richness after environmental perturbations that may result in functional disturbance (Chapin et al., 2000). Positive evenness–functional stability (Wittebolle et al., 2009) and negative evenness–invasibility (Wilsey and Polley, 2002; De Roy et al., 2013) relationships have been observed for microbial communities. In highly uneven communities, the functionality relies mainly on the traits of the dominating species (Hillebrand et al., 2008). In the short run, under stable conditions, this results in high functionality. The risk for loss of functionality is, however, higher for uneven than for even communities (Balvanera et al., 2005).
Dominance may prevent invasion if the dominant species create the most competitive environment for the invader (Hillebrand et al., 2008). The inhibition effect then depends on the one-to-one interaction between a putative invader and a ‘protector', and is thus specific (Losure et al., 2007). However, there is a risk that the invader outcompetes the protector, or that environmental perturbations cause the protector to become less abundant or even lost from the community. In a more even native community, the protection against invasion could be spread over functionally redundant species or over a wider range of functions that can prevent invasion. This is especially valid for environments subjected to frequent fluctuations (Hillebrand et al., 2008).
Dynamics
Natural microbial communities are not compositionally stable, but are subject to changing selection pressure resulting in a continuous succession of populations (Curtis and Sloan, 2004). In biotechnological applications, it has been shown that dynamic microbial communities can provide functional stability (Cabrol and Malhautier, 2011). This illustrates that on a temporal scale the conservation of traits is vital (Lemon et al., 2012), and presence of functionally redundant species may ensure functional stability at the community level.
The rate at which a microbial community changes may influence its functionality. Healthy gut microbiotas in vertebrates are rather stable (Benskin et al., 2010; Lozupone et al., 2012). In larvae of aquatic animals, however, the gut community is highly dynamic because of the interaction between colonizing bacteria and a rapidly developing gut environment (Rawls et al., 2006; Bakke et al., 2013). The highly dynamic situation in larvae may be a factor contributing to their unpredictable viability as transient communities have populations that are less adapted to the prevailing selection pressure. There are hardly any data on the relationship between community dynamics and invasibility. Robinson et al. (2010b) investigated the resistance of the cabbage white butterfly gut community to invasion. The results suggest that the altered state of the community after perturbation resulted in increased invasibility, but their data were not conclusive with respect to the rate of change in the community (abrupt change vs gradual change). Thus, whether the rate at which microbial communities change in the larval gut affects their susceptibility to (opportunistic) invaders is an open question. It can be hypothesized that the normal changes, representing a succession of functionally redundant community members, minimize opportunities for a pathogen to invade. A gut community should thus be able to quickly adapt to the rapidly changing environment in the larval gut in order to provide invader resistance. More rapidly changing communities then represent a perturbed environment that is more prone to invasions as there is a higher chance for open niches or lack of presence of antagonists during transition toward recovery.
Biodiversity–invasibility applied to aquaculture
Based on the reasoning above, we hypothesize that the following biodiversity–invasibility relationships in the larval gut microbiota can lower the risk for disease caused by pathogens:
(1) Microbial richness should be maximized to lower the risk for pathogen invasion. The relative richness should be considered here, as species richness depends on niche richness. Absolute richness may increase with age of the animal, but the relative richness can go down because the number of niches increases even more. Functionally redundant species that share the same niche also (temporarily) increase richness.
(2) A more evenly organized gut microbiota has a lower risk for pathogen invasion because the probability of instantaneous antagonism is higher and antagonism is partly dependent on population size.
(3) Gut microbial dynamics should be limited to the normal succession of populations in a developing larval gut. In practice, this means that stress on the animals should be avoided, constant environmental conditions should be ensured (physicochemical as well as microbial) and abrupt dietary changes should be limited.
The hypotheses above should be tested experimentally, and the gnotobiotic experimental systems established for aquatic animals are excellent for this purpose (Rawls et al., 2004; Marques et al., 2005; Dierckens et al., 2009; Forberg et al., 2012). Such systems can provide experimental insight into the biodiversity–invasibility relationships in the larval gut under conditions of full control over the microbial community. However, the findings from gnotobiotic systems should be verified under normal culture conditions. The conclusions from such research could serve as a basis for developing gut microbial management strategies.
Managing community assembly of gut microbiota
Securing the establishment of a desired gut microbiota in larvae, and hence proper functionality, is a better microbial management strategy than antibiotics. This is, however, not straight forward. A prerequisite is gaining insight into the ecological mechanisms and processes in the larval gut environment that regulate community composition, that is, the assembly rules. Similar to humans (Costello et al., 2012), the first colonization can be assumed to have long-term consequences for larval health. This motivates the development of management strategies to steer first colonization. This has only recently been acknowledged among aquaculturists.
The randomness of first gut colonization
For aquatic animals, the colonization of the unoccupied gut starts at mouth opening and progresses at the onset of feeding. Recent studies have shown substantial variation in the composition of the microbiota between individuals, even for cohabiting larvae (Fjellheim et al., 2012; Bakke et al., 2013). The same has been observed for terrestrial animals (van der Wielen et al., 2002). In line with theory (Costello et al., 2012), colonization of larvae can be the result of environmental filtering by the host, often termed host selection (that is, similar niches in the gut allow presence of similar assemblies), historical contingency (that is, timing and order of species inflow determines assembly), random sampling (that is, community members are randomly drawn from the species pool) and dispersal limitation (that is, some species gain access to the new habitat and others do not). Dispersal limitation intuitively seems to be of limited importance for the colonization of larvae that live in the same water as the microbes and continuously ingest them at high rates during drinking and by eating prey organisms (Reitan et al., 1998). Analyses of similarities in gut microbiota between cohabiting larvae (Bakke et al., 2013) suggest some degree of selection in the host. However, the interindividual variation is consistent with the involvement of random sampling from the bacterial species pool and the sequence of inflow of functionally redundant species in the colonization process.
Gut biodiversity: niche processes vs neutral processes
The microbial assembly in an animal gut may be determined by the number of available niches and species interactions according to Darwin-based niche theory (Silvertown, 2004). It is then primarily the number of available niches that determine species richness (Wennekes et al., 2012) whereas niche differentiation and interspecies interactions regulate coexistence patterns (Chesson, 2000). Alternatively, neutral theory states that biodiversity is the result of stochasticity in demographic processes such as reproduction, death and dispersal of species that are neutral with respect to competitive ability (Hubbell, 2001).
Several authors have suggested that spatial and temporal changes in biodiversity cannot be explained by either niche or neutral processes (Leibold and McPeek, 2006; Dumbrell et al., 2010; Ofiteru et al., 2010; Jeraldo et al., 2012). It is the interplay of dispersal, diversification and environmental selection that shapes local assemblages (Vellend, 2010). The relative contribution depends on the strength of stabilizing mechanisms (niches) and species fitness equality (neutrality) (Adler et al., 2007). For aquatic animals, the characteristics of the gut environment are age dependent. The immature gut goes through a series of developmental changes from the larval, over juvenile, to the adult stages, and even within the larval stage there is a continuous change (Zambonino-Infante and Cahu, 2001; Elbal et al., 2004; Yang et al,. 2010; He et al., 2012).
The variation in gut microbiota of cohabiting larvae (Fjellheim et al., 2012) raises two questions that are relevant for microbial management: (1) Is the early gut made up out of relatively broad niches that make neutral assembly possible? (2) When the intestine differentiates (Kamisaka and Rønnestad, 2011) will the niches increase in number but become narrower, thus increasing competition and reducing the probability of stochasticity in assembly? Differences in early coexistence patterns may then lead to divergence in microbiota composition between larva as a result of physicochemical and morphological changes in the gut during maturation in combination with competition between the community members (Rogers et al., 2012).
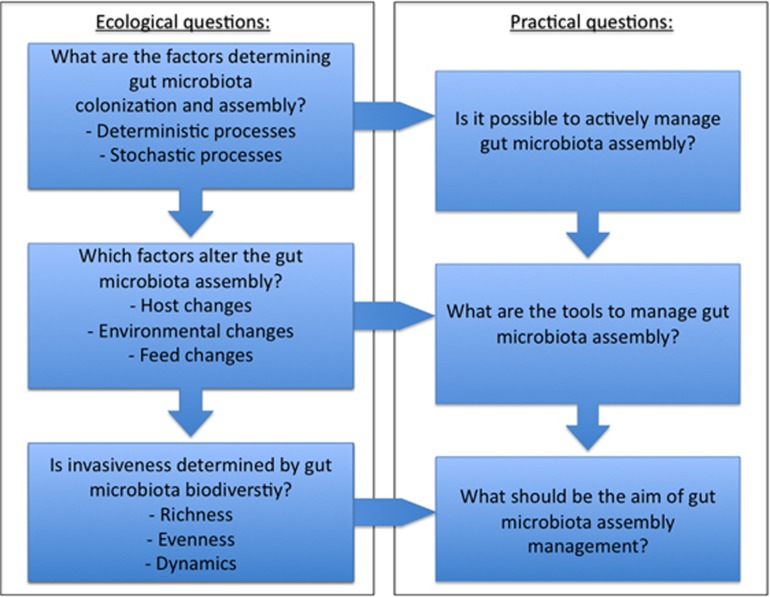
Yan et al. (2012) suggested that assembly in larval zebrafish was deterministic (that is, high interindividual similarity), whereas assembly in juveniles reaching adulthood was stochastic (that is, increased gut turnover rates). The inconsistencies between these data and the aforementioned data indicate the need for more studies that focus on the mechanisms involved in gut microbiota assembly in larvae. Understanding the neutral and/or deterministic processes that determine community assembly can form the basis for strategies that aim at manipulating gut microbial community composition (Marino et al., 2014). An ultimate goal would be the development of predictive models of community assembly in larvae. Quantitative ecology in larvae presents itself, however, as a huge challenge because of the complexity of the system under consideration for which aquaculture requires the input from theoretical microbial ecologists (Figure 3). Fortunately, effective microbial management is not always dependent on such models as illustrated above by the r/K-selection concept.
Figure 3.
Achieving invasion resistance by gut microbiota management in larviculture presents a big challenge. Primarily, knowledge should be gained on the ecological processes that determine the assembly of the microbiota in a developing gut environment. This will allow identification of the opportunities to actively manage the gut microbiota. Second, the development of predictive ecological models should provide information on how factors can be altered to direct assembly of gut microbiota in a desired direction. Even for systems far simpler than a larval gut, this is not an easy task. Finally, information on the biodiversity–invasibility relationship should provide knowledge on which ecological state of the intestinal microbiota is most auspicious to minimize invasion risk. To answer the questions in the figure, use can be made of gnotobiotic model systems that exist for aquatic species such as brine shrimp (Marques et al., 2005), Atlantic cod (Forberg et al., 2012), European sea bass (Dierckens et al., 2009) and zebrafish (Rawls et al., 2004) in combination with synthetic microbial ecosystems as applied in the researches of Wittebolle et al. (2009) and De Roy et al. (2013).
Conclusions
Microbial management offers opportunities for a decrease in diseases and increase in larval viability in aquaculture systems (Vadstein et al., 1993). An ecological perspective would contribute to an understanding of how clinical benefits can be achieved. Active management of the water and the intestinal microbiota as a strategy to control bacterial foes in aquaculture was introduced already in the 1990s (Vadstein et al., 1993; Vadstein, 2000). However, it seems the focus on human health was required for the concept of microbial management to become an acknowledged strategy (Costello et al., 2012). With the increasing ecological knowledge on structure and function of the gut microbiota, the time seems ripe for a reintroduction of the ‘join them' approach in the biological science of animal breeding. Moreover, emerging knowledge on biodiversity–invasibility relationships can form a basis for future research that, at the moment, is still mainly focusing on probiotics and disinfection. This current low richness and evenness in research hampers the progress in knowledge required for establishing sustainable microbial management strategies based on ecological principals in aquaculture.
Acknowledgments
Peter De Schryver is supported as a postdoctoral research fellow by the Research Foundation-Flanders (FWO, Belgium). We thank Dr Ingrid Bakke and Dr Ian Butts for their valuable inputs.
The authors declare no conflict of interest.
References
- Adler PB, HilleRisLambers J, Levine JM. A niche for neutrality. Ecol Lett. 2007;10:95–104. doi: 10.1111/j.1461-0248.2006.00996.x. [DOI] [PubMed] [Google Scholar]
- Andrews JH, Harris JF. r- and K- selection in microbial ecology. Adv Microb Ecol. 1986;9:99–147. [Google Scholar]
- Attramadal KJK, Øie G, Størseth TR, Alver MO, Vadstein O, Olsen Y. The effects of moderate ozonation or high intensity UV-irradiation on the microbial environment in RAS for marine larvae. Aquaculture. 2012;330:121–129. [Google Scholar]
- Attramadal KJK, Salvesen I, Xue RY, Øie G, Størseth TR, Vadstein O, et al. Recirculation as a possible microbial control strategy in the production of marine larvae. Aquacult Eng. 2012;46:27–39. [Google Scholar]
- Attramadal KJK, Truong TM, Bakke I, Skjermo J, Olsen Y, Vadstein O.2014. RAS and microbial maturation as tools for K-selection of microbial communities improve survival in cod larvae. Aquaculture (in press).
- Bakke I, Skjermo J, Vo TA, Vadstein O. Live feed is not a major determinant of the microbiota associated with cod larvae (Gadus morhua) Environ Microbiol Rep. 2013;5:537–548. doi: 10.1111/1758-2229.12042. [DOI] [PubMed] [Google Scholar]
- Balvanera P, Kremen C, Martinez-Ramos M. Applying community structure analysis to ecosystem function: examples from pollination and carbon storage. Ecol Appl. 2005;15:360–375. [Google Scholar]
- Bell T, Newman JA, Silverman BW, Turner SL, Lilley AK. The contribution of species richness and composition to bacterial services. Nature. 2005;436:1157–1160. doi: 10.1038/nature03891. [DOI] [PubMed] [Google Scholar]
- Benskin CMH, Rhodes G, Pickup RW, Wilson K, Hartley IR. Diversity and temporal stability of bacterial communities in a model passerine bird, the zebra finch. Mol Ecol. 2010;19:5531–5544. doi: 10.1111/j.1365-294X.2010.04892.x. [DOI] [PubMed] [Google Scholar]
- Bissett A, Richardson AE, Baker G, Wakelin S, Thrall PH. Life history determines biogeographical patterns of soil bacterial communities over multiple spatial scales. Mol Ecol. 2010;19:4315–4327. doi: 10.1111/j.1365-294x.2010.04804.x. [DOI] [PubMed] [Google Scholar]
- Bruno JF, Stachowicz JJ, Bertness MD. Inclusion of facilitation into ecological theory. Trends Ecol Evol. 2003;18:119–125. [Google Scholar]
- Cabrol L, Malhautier L. Integrating microbial ecology in bioprocess understanding: the case of gas biofiltration. Appl Microbiol Biotechnol. 2011;90:837–849. doi: 10.1007/s00253-011-3191-9. [DOI] [PubMed] [Google Scholar]
- Chapin FS, Zavaleta ES, Eviner VT, Naylor RL, Vitousek PM, Reynolds HL, et al. Consequences of changing biodiversity. Nature. 2000;405:234–242. doi: 10.1038/35012241. [DOI] [PubMed] [Google Scholar]
- Chesson P. Mechanisms of maintenance of species diversity. Annu Rev Ecol Syst. 2000;31:343–366. [Google Scholar]
- Cook KL, Garland JL, Layton AC, Dionisi HM, Levine LH, Sayler GS. Effect of microbial species richness on community stability and community function in a model plant-based wastewater processing system. Microb Ecol. 2006;52:725–737. doi: 10.1007/s00248-006-9105-1. [DOI] [PubMed] [Google Scholar]
- Costello EK, Stagaman K, Dethlefsen L, Bohannan BJM, Relman DA. The application of ecological theory toward an understanding of the human microbiome. Science. 2012;336:1255–1262. doi: 10.1126/science.1224203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis TP, Sloan WT. Prokaryotic diversity and its limits: microbial community structure in nature and implications for microbial ecology. Curr Opin Microbiol. 2004;7:221–226. doi: 10.1016/j.mib.2004.04.010. [DOI] [PubMed] [Google Scholar]
- De Roy K, Marzorati M, Negroni A, Thas O, Balloi A, Fava F, et al. Environmental conditions and community evenness determine the outcome of biological invasion. Nat Commun. 2013;4:1383. doi: 10.1038/ncomms2392. [DOI] [PubMed] [Google Scholar]
- De Schryver P, Defoirdt T, Sorgeloos P. Early mortality syndrome outbreaks: a microbial management issue in shrimp farming. PLoS Pathog. 2014;10:e1003919. doi: 10.1371/journal.ppat.1003919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dierckens K, Rekecki A, Laureau S, Sorgeloos P, Boon N, Van den Broeck W, et al. Development of a bacterial challenge test for gnotobiotic sea bass (Dicentrarchus labrax) larvae. Environ Microbiol. 2009;11:526–533. doi: 10.1111/j.1462-2920.2008.01794.x. [DOI] [PubMed] [Google Scholar]
- Dillon RJ, Vennard CT, Buckling A, Charnley AK. Diversity of locust gut bacteria protects against pathogen invasion. Ecol Lett. 2005;8:1291–1298. [Google Scholar]
- Dumbrell AJ, Nelson M, Helgason T, Dytham C, Fitter AH. Relative roles of niche and neutral processes in structuring a soil microbial community. ISME J. 2010;4:337–345. doi: 10.1038/ismej.2009.122. [DOI] [PubMed] [Google Scholar]
- Elbal MT, Hernandez MPG, Lozano MT, Agulleiro B. Development of the digestive tract of gilthead sea bream (Sparus aurata L.). Light and electron microscopic studies. Aquaculture. 2004;234:215–238. [Google Scholar]
- Emery SM, Gross KL. Dominant species identity, not community evenness, regulates invasion in experimental grassland plant communities. Ecology. 2007;88:954–964. doi: 10.1890/06-0568. [DOI] [PubMed] [Google Scholar]
- Fairbrother JM, Nadeau E, Gyles CL. Escherichia coli in postweaning diarrhea in pigs: an update on bacterial types, pathogenesis, and prevention strategies. Anim Health Res Rev. 2005;6:17–39. doi: 10.1079/ahr2005105. [DOI] [PubMed] [Google Scholar]
- Fargione JE, Tilman D. Diversity decreases invasion via both sampling and complementarity effects. Ecol Lett. 2005;8:604–611. [Google Scholar]
- Fjellheim AJ, Playfoot KJ, Skjermo J, Vadstein O. Inter-individual variation in the dominant intestinal microbiota of reared Atlantic cod (Gadus morhua L.) larvae. Aquac Res. 2012;43:1499–1508. [Google Scholar]
- Forberg T, Vestrum RI, Arukwe A, Vadstein O. Bacterial composition and activity determines host gene-expression responses in gnotobiotic Atlantic cod (Gadus morhua) larvae. Vet Microbiol. 2012;157:420–427. doi: 10.1016/j.vetmic.2012.01.007. [DOI] [PubMed] [Google Scholar]
- Gatesoupe FJ. The use of probiotics in aquaculture. Aquaculture. 1999;180:147–165. [Google Scholar]
- Gatesoupe FJ.2010Probiotics and other microbial manipulations in fish feeds: prospective health benefitsIn: Watson RR, Preedy VR (eds)Bioactive Foods in Promoting Health Academic Press: Oxford; 541–552. [Google Scholar]
- Guarner F, Malagelada JR. Gut flora in health and disease. Lancet. 2003;361:512–519. doi: 10.1016/S0140-6736(03)12489-0. [DOI] [PubMed] [Google Scholar]
- Gustafson RH, Bowen RE. Antibiotic use in animal agriculture. J Appl Microbiol. 1997;83:531–541. doi: 10.1046/j.1365-2672.1997.00280.x. [DOI] [PubMed] [Google Scholar]
- Hajek A. Natural Enemies: An Introduction to Biological Control. Cambridge University Press: Cambridge; 2004. [Google Scholar]
- He T, Xiao ZZ, Liu QH, Ma DY, Xu SH, Xiao YS, et al. Ontogeny of the digestive tract and enzymes in rock bream Oplegnathus fasciatus (Temminck et Schlegel 1844) larvae. Fish Physiol Biochem. 2012;38:297–308. doi: 10.1007/s10695-011-9507-y. [DOI] [PubMed] [Google Scholar]
- Hillebrand H, Bennett DM, Cadotte MW. Consequences of dominance: a review of evenness effects on local and regional ecosystem processes. Ecology. 2008;89:1510–1520. doi: 10.1890/07-1053.1. [DOI] [PubMed] [Google Scholar]
- Hubbell SP. The Unified Neutral Theory of Biodiversity and Biogeography. Princeton University Press: Princeton; 2001. [Google Scholar]
- Jeraldo P, Sipos M, Chia N, Brulc JM, Dhillon AS, Konkel ME, et al. Quantification of the relative roles of niche and neutral processes in structuring gastrointestinal microbiomes. Proc Natl Acad Sci USA. 2012;109:9692–9698. doi: 10.1073/pnas.1206721109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Morin PJ. Productivity gradients cause positive diversity-invasibility relationships in microbial communities. Ecol Lett. 2004;7:1047–1057. [Google Scholar]
- Johnson PTJ, Hartson RB, Larson DJ, Sutherland DR. Diversity and disease: community structure drives parasite transmission and host fitness. Ecol Lett. 2008;11:1017–1026. doi: 10.1111/j.1461-0248.2008.01212.x. [DOI] [PubMed] [Google Scholar]
- Kamisaka Y, Rønnestad I. Reconstructed 3D models of digestive organs of developing Atlantic cod (Gadus morhua) larvae. Mar Biol. 2011;158:233–243. doi: 10.1007/s00227-010-1554-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy TA, Naeem S, Howe KM, Knops JMH, Tilman D, Reich P. Biodiversity as a barrier to ecological invasion. Nature. 2002;417:636–638. doi: 10.1038/nature00776. [DOI] [PubMed] [Google Scholar]
- Knops JMH, Tilman D, Haddad NM, Naeem S, Mitchell CE, Haarstad J, et al. Effects of plant species richness on invasion dynamics, disease outbreaks, insect abundances and diversity. Ecol Lett. 1999;2:286–293. doi: 10.1046/j.1461-0248.1999.00083.x. [DOI] [PubMed] [Google Scholar]
- Lanzas C, Warnick LD, Ivanek R, Ayscue P, Nydam DV, Grohn YT. The risk and control of Salmonella outbreaks in calf-raising operations: a mathematical modeling approach. Vet Res. 2008;39:13. doi: 10.1051/vetres:2008038. [DOI] [PubMed] [Google Scholar]
- Leibold MA, McPeek MA. Coexistence of the niche and neutral perspectives in community ecology. Ecology. 2006;87:1399–1410. doi: 10.1890/0012-9658(2006)87[1399:cotnan]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Lemon KP, Armitage GC, Relman DA, Fischbach MA. Microbiota-targeted herapies: an ecological perspective. Sci Transl Med. 2012;4:137rv5. doi: 10.1126/scitranslmed.3004183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine JM. Species diversity and biological invasions: relating local process to community pattern. Science. 2000;288:852–854. doi: 10.1126/science.288.5467.852. [DOI] [PubMed] [Google Scholar]
- Little AEF, Robinson CJ, Peterson SB, Raffa KE, Handelsman J. Rules of engagement: interspecies interactions that regulate microbial communities. Annu Rev Microbiol. 2008;62:375–401. doi: 10.1146/annurev.micro.030608.101423. [DOI] [PubMed] [Google Scholar]
- Losure DA, Wilsey BJ, Moloney KA. Evenness-invasibility relationships differ between two extinction scenarios in tallgrass prairie. Oikos. 2007;116:87–98. [Google Scholar]
- Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489:220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur RH, Wilson EO. The Theory of Island Biogeography. Princeton University Press: Princeton; 1967. [Google Scholar]
- Marino S, Baxter NT, Huffnagle GB, Petrosino JF, Schloss PD. Mathematical modeling of primary succession of murine intestinal microbiota. Proc Natl Acad Sci USA. 2014;111:439–444. doi: 10.1073/pnas.1311322111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques A, Dinh T, Ioakeimidis C, Huys G, Swings J, Verstraete W, et al. Effects of bacteria on Artemia franciscana cultured in different gnotobiotic environments. Appl Environ Microbiol. 2005;71:4307–4317. doi: 10.1128/AEM.71.8.4307-4317.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken VJ, Lorenz RG. The gastrointestinal ecosystem: a precarious alliance among epithelium, immunity and microbiota. Cell Microbiol. 2001;3:1–11. doi: 10.1046/j.1462-5822.2001.00090.x. [DOI] [PubMed] [Google Scholar]
- McGrady-Steed J, Harris PM, Morin PJ. Biodiversity regulates ecosystem predictability. Nature. 1997;390:162–165. [Google Scholar]
- Meiners SJ, Cadenasso ML, Pickett STA. Beyond biodiversity: individualistic controls of invasion in a self-assembled community. Ecol Lett. 2004;7:121–126. [Google Scholar]
- Mitchell CE, Tilman D, Groth JV. Effects of grassland plant species diversity, abundance, and composition on foliar fungal disease. Ecology. 2002;83:1713–1726. [Google Scholar]
- Naeem S, Thompson LJ, Lawler SP, Lawton JH, Woodfin RM. Declining biodiversity can alter the performance of ecosystems. Nature. 1994;368:734–737. [Google Scholar]
- Ofiteru ID, Lunn M, Curtis TP, Wells GF, Criddle CS, Francis CA, et al. Combined niche and neutral effects in a microbial wastewater treatment community. Proc Natl Acad Sci USA. 2010;107:15345–15350. doi: 10.1073/pnas.1000604107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruder GD. Biosecurity: application in aquaculture. Aquacult Eng. 2004;32:3–10. [Google Scholar]
- Ptacnik R, Solimini AG, Andersen T, Tamminen T, Brettum P, Lepisto L, et al. Diversity predicts stability and resource use efficiency in natural phytoplankton communities. Proc Natl Acad Sci USA. 2008;105:5134–5138. doi: 10.1073/pnas.0708328105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purvis A, Hector A. Getting the measure of biodiversity. Nature. 2000;405:212–219. doi: 10.1038/35012221. [DOI] [PubMed] [Google Scholar]
- Rawls JF, Mahowald MA, Ley RE, Gordon JI. Reciprocal gut microbiota transplants from zebrafish and mice to germ-free recipients reveal host habitat selection. Cell. 2006;127:423–433. doi: 10.1016/j.cell.2006.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawls JF, Samuel BS, Gordon JI. Gnotobiotic zebrafish reveal evolutionarily conserved responses to the gut microbiota. Proc Natl Acad Sci USA. 2004;101:4596–4601. doi: 10.1073/pnas.0400706101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitan KI, Natvik CM, Vadstein O. Drinking rate, uptake of bacteria and microalgae in turbot larvae. J Fish Biol. 1998;53:1145–1154. [Google Scholar]
- Ringø E, Olsen RE, Gifstad TO, Dalmo RA, Amlund H, Hemre GI, et al. Prebiotics in aquaculture: a review. Aquacult Nutr. 2010;16:117–136. [Google Scholar]
- Robinson CJ, Bohannan BJM, Young VB. From structure to function: the ecology of host-associated microbial communities. Microbiol Mol Biol Rev. 2010;74:453–476. doi: 10.1128/MMBR.00014-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson CJ, Schloss P, Ramos Y, Raffa K, Handelsman J. Robustness of the bacterial community in the cabbage white butterfly larval midgut. Microb Ecol. 2010;59:199–211. doi: 10.1007/s00248-009-9595-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers GB, Hoffman LR, Carroll MP, Bruce KD. Interpreting infective microbiota: the importance of an ecological perspective. Trends Microbiol. 2012;21:271–276. doi: 10.1016/j.tim.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shea K, Chesson P. Community ecology theory as a framework for biological invasions. Trends Ecol Evol. 2002;17:170–176. [Google Scholar]
- Silvertown J. Plant coexistence and the niche. Trends Ecol Evol. 2004;19:605–611. [Google Scholar]
- Skjermo J, Vadstein O. Techniques for microbial control in the intensive rearing of marine larvae. Aquaculture. 1999;177:333–343. [Google Scholar]
- Stachowicz JJ, Whitlatch RB, Osman RW. Species diversity and invasion resistance in a marine ecosystem. Science. 1999;286:1577–1579. doi: 10.1126/science.286.5444.1577. [DOI] [PubMed] [Google Scholar]
- Stecher B, Hardt WD. The role of microbiota in infectious disease. Trends Microbiol. 2008;16:107–114. doi: 10.1016/j.tim.2007.12.008. [DOI] [PubMed] [Google Scholar]
- Summerfelt ST, Sharrer MJ, Tsukuda SM, Gearheart M. Process requirements for achieving full-flow disinfection of recirculating water using ozonation and UV irradiation. Aquacult Eng. 2009;40:17–27. [Google Scholar]
- Suzuki S. Pathogenicity of Salmonella enteritidis in poultry. Int J Food Microbiol. 1994;21:89–105. doi: 10.1016/0168-1605(94)90203-8. [DOI] [PubMed] [Google Scholar]
- Vadstein O. Strategies for avoiding bacterial infections: beat them or join them. ASM News. 2000;66:446–447. [Google Scholar]
- Vadstein O, Bergh O, Gatesoupe FJ, Galindo-Villegas J, Mulero V, Picchietti S, et al. Microbiology and immunology of fish larvae. Rev Aquacult. 2013;5:S1–S25. [Google Scholar]
- Vadstein O, Mo TA, Bergh Ø.2004Microbial interactions, prophylaxis and diseasesIn: Moksness E, Kjørsvik E, Olsen Y (eds)Culture of Cold-Water Marine Fishes Blackwell Publishing: Bath; 28–72. [Google Scholar]
- Vadstein O, Øie G, Olsen Y, Salvesen I, Skjermo J, Skjåk-bræk G.1993A strategy to obtain microbial control during larval development of marine fishIn: Reinertsen H, Dahle LA, Jørgensen L, Tvinnereim K (eds)Fish Farming Technology - Proceedings of the First International Conference on Fish Farming Technology Balkema: Rotterdam; 67–75. [Google Scholar]
- Valéry L, Fritz H, Lefeuvre JC, Simberloff D. In search of a real definition of the biological invasion phenomenon itself. Biol Invasions. 2008;10:1345–1351. [Google Scholar]
- van der Wielen P, Keuzenkamp DA, Lipman LJA, van Knapen F, Biesterveld S. Spatial and temporal variation of the intestinal bacterial community in commercially raised broiler chickens during growth. Microb Ecol. 2002;44:286–293. doi: 10.1007/s00248-002-2015-y. [DOI] [PubMed] [Google Scholar]
- van Elsas JD, Chiurazzi M, Mallon CA, Elhottova D, Kristufek V, Salles JF. Microbial diversity determines the invasion of soil by a bacterial pathogen. Proc Natl Acad Sci USA. 2012;109:1159–1164. doi: 10.1073/pnas.1109326109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vellend M. Conceptual synthesis in community ecology. Q Rev Biol. 2010;85:183–206. doi: 10.1086/652373. [DOI] [PubMed] [Google Scholar]
- Verschuere L, Rombaut G, Sorgeloos P, Verstraete W. Probiotic bacteria as biological control agents in aquaculture. Microbiol Mol Biol Rev. 2000;64:655–671. doi: 10.1128/mmbr.64.4.655-671.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wennekes PL, Rosindell J, Etienne RS. The neutral-niche debate: a philosophical perspective. Acta Biotheor. 2012;60:257–271. doi: 10.1007/s10441-012-9144-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilsey BJ, Polley HW. Reductions in grassland species evenness increase dicot seedling invasion and spittle bug infestation. Ecol Lett. 2002;5:676–684. [Google Scholar]
- Wittebolle L, Marzorati M, Clement L, Balloi A, Daffonchio D, Heylen K, et al. Initial community evenness favours functionality under selective stress. Nature. 2009;458:623–626. doi: 10.1038/nature07840. [DOI] [PubMed] [Google Scholar]
- Wong SD, Rawls JF. Intestinal microbiota composition in fishes is influenced by host ecology and environment. Mol Ecol. 2012;21:3100–3102. doi: 10.1111/j.1365-294x.2012.05646.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Q, van der Gast CJ, Yu Y. Bacterial community assembly and turnover within the intestines of developing zebrafish. PLoS One. 2012;7:e30603. doi: 10.1371/journal.pone.0030603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R, Xie C, Fan Q, Gao C, Fang L. Ontogeny of the digestive tract in yellow catfish Pelteobagrus fulvidraco larvae. Aquaculture. 2010;302:112–123. [Google Scholar]
- Zambonino-Infante JL, Cahu CL. Ontogeny of the gastrointestinal tract of marine fish larvae. Comp Biochem Physiol C Toxicol Pharmacol. 2001;130:477–487. doi: 10.1016/s1532-0456(01)00274-5. [DOI] [PubMed] [Google Scholar]