Abstract
The role of methylotrophic bacteria in the fate of the oil and gas released into the Gulf of Mexico during the Deepwater Horizon oil spill has been controversial, particularly in relation to whether organisms such as Methylophaga had contributed to the consumption of methane. Whereas methanotrophy remains unqualified in these organisms, recent work by our group using DNA-based stable-isotope probing coupled with cultivation-based methods has uncovered hydrocarbon-degrading Methylophaga. Recent findings have also shown that methylotrophs, including Methylophaga, were in a heightened state of metabolic activity within oil plume waters during the active phase of the spill. Taken collectively, these findings suggest that members of this group may have participated in the degradation of high-molecular-weight hydrocarbons in plume waters. The discovery of hydrocarbon-degrading Methylophaga also highlights the importance of considering these organisms in playing a role to the fate of oil hydrocarbons at oil-impacted sites.
Keywords: Deepwater Horizon, Methylophaga, degradation, Gulf of Mexico, hydrocarbons, marine environment
The Deepwater Horizon disaster of 20 April 2010 is possibly the worst accidental maritime oil spill in the history of the oil and gas industry. Approximately 7 × 105 tonnes of crude oil and gas were released into the Gulf of Mexico over a period of 83 days (Reddy et al., 2012). The spill was unprecedented by virtue of the depth at which it occurred and formation of a deepwater oil plume that became entrained at ∼1000–1300 m within the water column and which spanned an impressive 35 × 2 × 0.2 km (Camilli et al., 2010; Diercks et al., 2010). The earliest sampling of the plume (end of May 2010) revealed the dominance of a specific cluster within the Oceanospirillales—designated DWH Oceanospirillales—that constituted up to 90% of total bacterial 16S rRNA gene clone (Hazen et al., 2010) and pyrosequencing (Yang et al., 2014) libraries. By early June, the plume community had become dominated by members affiliated to Cycloclasticus and Colwellia (Redmond and Valentine, 2012; Yang et al., 2014). Collectively, these microbial community ‘snap-shots', thanks to opportune research cruises that ventured into the Gulf during the spill, have hitherto provided a temporal timeline on the plume microbial dynamics. A contentious and largely unresolved issue, however, relates to the role that methylotrophs had played in the fate of the oil and gas.
During the active phase of the spill (20 April to 15 July), methylotrophic bacteria were not detected near the leaky wellhead or plume (Hazen et al., 2010; Valentine et al., 2010; Yang et al., 2014), whereas these organisms were reported to account for at least 5% of the total bacterial community after the spill (Kessler et al. (2011) reported 5–36% in September 2010; Yang et al. (2014) reported 0.23–6% and 2% in September 2010 and October 2010, respectively). Kessler et al. (2011) postulated that methylotrophs, mainly Methylophaga, had contributed to the consumption of methane released during the spill; their involvement was inferred in part from the high numbers of methylotrophs detected in plume waters almost 3 months after capping of the leaky well, and which were thought to be remnants of a July bloom of methane-consuming bacteria (Kessler et al., 2011). This interpretation, however, was severely contested by Joye et al. (2011), in part by the fact that the role of Methylophaga in methane oxidation remains unsubstantiated as these organisms are not recognized for carrying out methane oxidation or being capable of growth on methane as a carbon and energy source. The enrichment of methylotrophs detected during this post-spill period (September to October 2010) appears more likely associated with an eukaryotic phytoplankton bloom that was detected during August 2010 in the vicinity of the spill site (Hu et al., 2011). Phytoplankton blooms can produce large quantities of extracellular high-molecular-weight dissolved organic matter (Biddanda and Benner, 1997) that acts as a rich source of methylated sugars (Panagiotopoulos et al., 2013) exploitable by planktonic bacteria (Baines and Pace, 1991) such as methylotrophs (McCarren et al., 2010). The observed enrichment of these organisms near the Deepwater Horizon spill site during the late summer of 2010 may have thus been associated with a role as terminal degraders of phytoplankton-derived dissolved organic matter (Yang et al., 2014).
Whereas a few studies have reported the enrichment of Methylophaga spp. in oil-contaminated field samples and laboratory experiments with oil (Vila et al., 2010 and references therein), the ability of any member of this genus to degrade hydrocarbons has hitherto remained unsubstantiated. The known substrate spectrum for the Methylophaga group is confined exclusively to C1 sources (methanol, methylamine, dimethylsulfide) as sole carbon and energy sources, with the exception of some species that are also capable of utilizing fructose (Janvier and Grimont, 1995). Therefore, the notion of these organisms to have contributed to the degradation of high-molecular-weight hydrocarbons during the Deepwater Horizon spill has, understandably, been brushed aside—at least until now.
Recent work by our group using DNA-based stable-isotope probing with uniformly labeled [13C]n-hexadecane, coupled with cultivation-based methods, has uncovered for the first time members of Methylophaga with the ability to utilize n-hexadecane (Mishamandani et al., 2014; Figure 1). Although Methylophaga were detected only in very low abundance within the plume (<0.1% of total 16S rRNA gene sequences in pyrosequencing libraries), these organisms were actually not metabolically ‘procrastinating'. A recent report assessing the transcriptional response of water column bacterial communities to the spill revealed that about 1/3 (out of 50) taxa identified in the plume had significantly higher numbers of transcripts (up to 2 orders of magnitude) relative to their levels in non-plume waters (Rivers et al., 2013). All these taxa belonged to the Gammaproteobacteria and included Methylophaga (represented by three operational taxonomic units; Table 1) that were reported in a heightened state of metabolic activity, possibly linked to their consumption of methanol produced from methanotrophic oxidation of methane (Rivers et al., 2013). During crude oil enrichment experiments with natural seawater (Mishamandani et al., 2014) and with a non-axenic cosmopolitan marine diatom (Mishamandani et al., unpublished results), we had observed a rapid (within 72 h) and short-lived bloom of Methylophaga. Taken collectively, we hypothesize that a bloom of Methylophaga with hydrocarbon-degrading qualities may have occurred, but had been missed, during the initial stages (first couple weeks) following the DWH blowout.
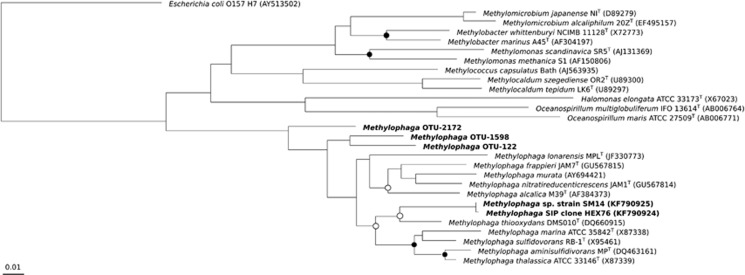
Figure 1.
Phylogenetic tree of Methylophaga operational taxonomic units (OTUs) 122, 1598 and 2172 identified in oil-contaminated plume waters at Deepwater Horizon (Rivers et al., 2013) and the hydrocarbon-degrading SIP clone HEX76 and isolated strain SM14 identified in a North Carolina surface seawater sample (Mishamandani et al., 2014). These sequences are shown in bold along with related type strains of Methylophaga and other methylotrophs in the class Gammaproteobacteria from the SILVA SSU NR 111 NR database (Quast et al., 2013). GenBank accession numbers are in parentheses. The tree was constructed using the neighbour-joining algorithm. Nodes with bootstrap support of at least 65% (○) and 90% (●) are marked (1000 replications). Escherichia coli 0157 H7 (AY513502) was used as the outgroup. The scale bar indicates the difference of number of substitutions per site.
Table 1. Methylophaga OTUs 122, 1598 and 2172 in plume waters during the active phase of the Deepwater Horizon oil spill (Rivers et al., 2013) and their similarity, based on 16S rRNA gene sequencing, to Methylophaga strain SM14.
| OTU | Closest BLASTn matcha | % ID to strain SM14b |
|---|---|---|
| 122 | Methylophaga nitratireducenticrescens (97% GU567814) | 96.0 |
| 1598 | Methylophaga thalassica (96% NR036802) | 96.3 |
| 2172 | Methylophaga frappieri (96% GU567815) | 94.5 |
Abbreviation: OTU, operational taxonomic unit.
Results are to the closest type strain; percentage similarity and accession number shown in parentheses.
% ID, percentage identity based on 16S rRNA gene sequencing.
The discovery of Methylophaga with the capacity to degrade hydrocarbons not only expands the paradigm beyond C1-specific metabolism for this group of methylotrophs, but also adds another dimension to the role these organisms may have had in the fate of the oil during the Gulf spill—specifically the possibility that these organisms had contributed to a hydrocarbon-degradation cascade.
Acknowledgments
This work was supported by a Marie Curie International Outgoing Fellowship (PIOF-GA-2008-220129) within the seventh European Community Framework Programme. Partial support was also provided through the US National Institute of Environmental Health Sciences, grant 5 P42ES005948.
The authors declare no conflict of interest.
References
- Baines SB, Pace ML. The production of dissolved organic matter by phytoplankton and its importance to bacteria: Patterns across marine and freshwater systems. Limnol Oceanogr. 1991;36:1078–1090. [Google Scholar]
- Biddanda B, Benner R. Carbon, nitrogen, and carbohydrate fluxes during the production of particulate and dissolved organic matter by marine phytoplankton. Limnol Oceanogr. 1997;42:506–518. [Google Scholar]
- Camilli R, Reddy CM, Yoerger DR, Van Mooy BAS, Jakuba MV, Kinsey JC, et al. Tracking hydrocarbon plume transport and biodegradation at deepwater horizon. Science. 2010;330:201–204. doi: 10.1126/science.1195223. [DOI] [PubMed] [Google Scholar]
- Diercks A-R, Highsmith RC, Asper VL, Joung DJ, Zhou Z, Guo L, et al. Characterization of subsurface polycyclic aromatic hydrocarbons at the Deepwater Horizon wellhead site. Geophys Res Lett. 2010;37:L20602. [Google Scholar]
- Hazen TC, Dubinsky EA, DeSantis TZ, Andersen GL, Piceno YM, Singh N, et al. Deep-sea oil plume enriches indigenous oil-degrading bacteria. Science. 2010;330:204–208. doi: 10.1126/science.1195979. [DOI] [PubMed] [Google Scholar]
- Hu C, Weisberg RH, Liu Y, Zheng L, Daly KL, English DC, et al. Did the northeastern Gulf of Mexico become greener after the Deepwater Horizon oil spill. Geophys Res Lett. 2011;38:L09601. [Google Scholar]
- Janvier M, Grimont PAD. The genus Methylophaga, a new line of descent within phylogenetic branch γ of Proteobacteria. Res Microbiol. 1995;146:543–550. doi: 10.1016/0923-2508(96)80560-2. [DOI] [PubMed] [Google Scholar]
- Joye SB, Leifer I, MacDonald IR, Chanton JP, Meile CD, Teske AP, et al. Technical comment on ‘A persistent oxygen anomaly reveals the fate of spilled methane in the deep Gulf of Mexico'. Science. 2011;332:1033–1034. doi: 10.1126/science.1203307. [DOI] [PubMed] [Google Scholar]
- Kessler JD, Valentine DL, Redmond MC, Du M, Chan EW, Mendes SD, et al. A persistent oxygen anomaly reveals the fate of spilled methane in the deep Gulf of Mexico. Science. 2011;331:312–315. doi: 10.1126/science.1199697. [DOI] [PubMed] [Google Scholar]
- McCarren J, Becker JW, Repeta DJ, Shi Y, Young CR, Malmstrom RR, et al. Microbial community transcriptomes reveal microbes and metabolic pathways associated with dissolved organic matter turnover in the sea. Proc Natl Acad Sci USA. 2010;107:16420–16427. doi: 10.1073/pnas.1010732107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishamandani S, Gutierrez T, Aitken MD. DNA-based stable isotope probing coupled with cultivation methods implicates Methylophaga in hydrocarbon degradation. Front Microbiol. 2014;5:76. doi: 10.3389/fmicb.2014.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panagiotopoulos C, Repeta DJ, Mathieu L, Rontani J-F, Sempéré Molecular level characterization of methyl sugars in marine high molecular weight dissolved organic matter. Mar Chem. 2013;154:34–45. [Google Scholar]
- Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy CM, Arey JS, Seewald JS, Sylva SP, Lemkau KL, Nelson RK, et al. Composition and fate of gas and oil released to the water column during the Deepwater Horizon oil spill. Proc Natl Acad Sci USA. 2012;109:20229–20234. doi: 10.1073/pnas.1101242108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redmond MC, Valentine DL. Natural gas and temperature structured a microbial community response to the Deepwater Horizon oil spill. Proc Natl Acad Sci USA. 2012;109:20292–20297. doi: 10.1073/pnas.1108756108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivers AR, Sharma S, Tringe SG, Martin J, Joye SB, Moran MA. Transcriptional response of bathypelagic marine bacterioplankton to the Deepwater Horizon oil spill. ISME J. 2013;7:2315–2329. doi: 10.1038/ismej.2013.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentine DL, Kessler JD, Redmond MC, Mendes SD, Heintz MB, Farwell C, et al. Propane respiration jump-starts microbial response to a deep oil spill. Science. 2010;330:208–211. doi: 10.1126/science.1196830. [DOI] [PubMed] [Google Scholar]
- Vila J, Nieto JM, Mertens J, Springael D, Grifoll M. Microbial community structure of a heavy fuel oil-degrading marine consortium: linking microbial dynamics with polycyclic aromatic hydrocarbon utilization. FEMS Microbiol Ecol. 2010;73:349–362. doi: 10.1111/j.1574-6941.2010.00902.x. [DOI] [PubMed] [Google Scholar]
- Yang T, Nigro LM, Gutierrez T, D'Ambrosio L, Joye SB, Highsmith R, et al. 2014Pulsed blooms and persistent oil-degrading bacterial populations in the water column during and after the Deepwater Horizon blowout Deep-Sea Res IIdoi: 10.1016/j.dsr2.2014.01.014 [DOI]