Abstract
Autophagy, a lysosomal degradation pathway for cellular constituents and organelles, is an adaptive and essential process required for cellular homeostasis. Although autophagy functions as a survival mechanism in response to cellular stressors such as nutrient or growth factor deprivation, it can also lead to a non-apoptotic form of programmed cell death (PCD) called autophagy-induced cell death or autophagy-associated cell death (type II PCD). Current evidence suggests that cell death through autophagy can be induced as an alternative to apoptosis (type I PCD), with therapeutic purpose in cancer cells that are resistant to apoptosis. Thus, modulating autophagy is of great interest in cancer research and therapy. Natural polyphenolic compounds that are present in our diet, such as rottlerin, genistein, quercetin, curcumin, and resveratrol, can trigger type II PCD via various mechanisms through the canonical (Beclin-1 dependent) and non-canonical (Beclin-1 independent) routes of autophagy. The capacity of these compounds to provide a means of cancer cell death that enhances the effects of standard therapies should be taken into consideration for designing novel therapeutic strategies. This review focuses on the autophagy- and cell death-inducing effects of these polyphenolic compounds in cancer.
Facts
Natural polyphenolic compounds that are present in our diet, such as rottlerin, genistein, quercetin, curcumin, and resveratrol can alter the effects of signaling pathways and induce cell death not only via apoptosis but also via autophagy. Thus, these compounds could be used as a co-therapy with standard therapies in cancer.
These compounds can trigger type II PCD via various mechanisms through the canonical (Beclin-1 dependent) and non-canonical (Beclin-1 independent) routes of autophagy.
Rottlerin or its related analogs may be used in the development of novel agents for the induction of autophagic cell death as it has been proven, pharmacodynamically in a mice xenograft model, to be efficiently absorbed in cells and tissues against pancreatic cancer.
Genistein induced autophagy due to changes in apoptotic signaling, which is beneficial against chemoresistance usually seen in cancer cells.
Quercetin induced extensive autophagy and subsequent death in cancer cells mediated by the inhibition of proteasomal activity and mTOR signaling.
Curcumin induced G2/M arrest and autophagy in malignant glioma cells through the inhibition of the Akt/mTOR/p70S6K and activation of the extracellular signal-regulated kinase (ERK)1/2 pathways, which implied that cell death via autophagy, might be pathway specific.
Resveratrol induced cell death through autophagy in five ovarian cancer cell lines, suggesting that it may be effective treatment in apoptosis-resistant ovarian cancer. Autophagy can be induced with acute exposure to resveratrol, whereas prolonged exposure activates a caspase-mediated cell death pathway.
Open Questions
The induction of cellular senescence was accompanied by autophagy in colon cancer cells with an increase in Beclin-1 and p62/SQSTM1 protein levels. Therefore, the functional link between senescence and autophagy in these curcumin-treated cancer cells should be further investigated.
Autophagy inhibitors may have the potential to enhance resveratrol antitumor efficacy.
The ability of natural polyphenolic compounds to induce autophagic cell death that enhances the effects of standard therapies should be taken into consideration for designing novel therapeutic strategies.
Combining FDA (food and drug activation)-approved drugs with these polyphenolic compounds such as rottlerin, genistein, quercetin, curcumin, and resveratrol may provide novel therapeutic strategies in the treatment of cancer.
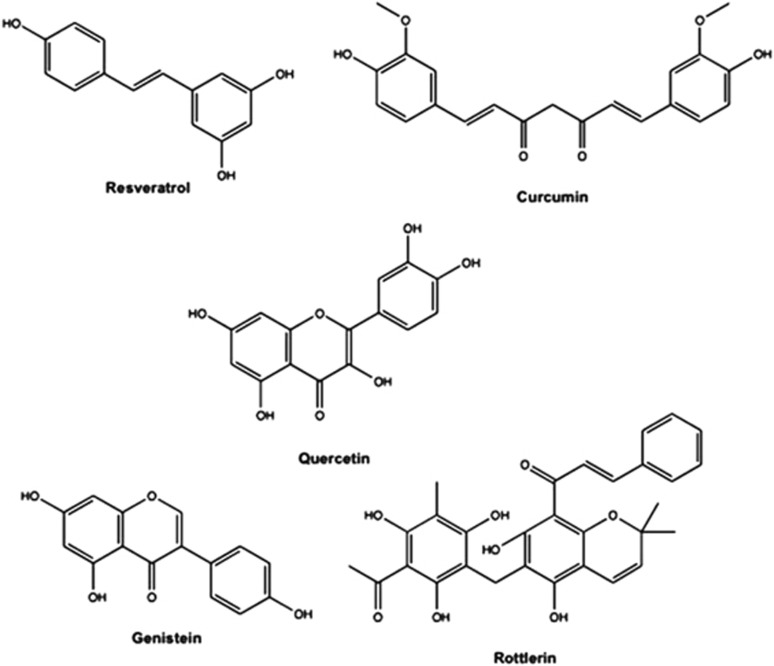
Natural plant-derived polyphenols are chemical substances characterized by the presence of more than one phenol unit per molecule. They are present in some foods and have been shown to exert anticancer properties. Some important examples are rottlerin, genistein, quercetin, curcumin, and resveratrol, all of which have been shown to induce autophagy death in various cancer cells (Figures 1,2,3 and Table 1).
Figure 1.
Chemical structures of autophagy-inducing polyphenolic compounds
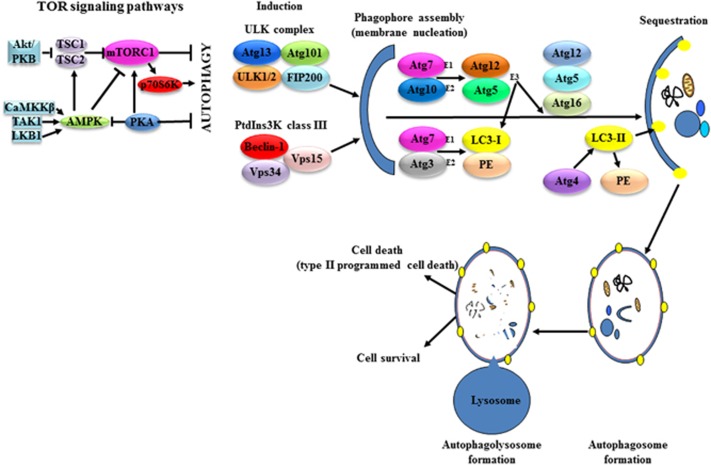
Figure 2.
The major signaling pathway regulation and core machinery of autophagy. Various pathways regulate autophagy either positively or negatively. Most of these pathways, including AMPK and PKA, merge at mTORC1. PKA directly activates mTORC1, inactivating both AMPK and autophagy. AMPK negatively regulates mTORC1 in two ways, either directly or by activating the TSC2 protein. The mTORC1 substrate p70S6K is a positive regulator of autophagy. Another important upstream factor is Akt/PKB, a negative regulator of the TSC1/2 complex. The core machinery includes the processes of induction, phagophore assembly (membrane nucleation), sequestration, autophagosome formation, and autophagolysosome formation. The process of induction involves initiation of the UNC-51-like kinase complex members ULK 1/2, Atg13, Atg101, and FIP200. Activation of the PtdIns3K complex (Beclin-1, Vps34, and Vps15) is an essential step in phagophore assembly (membrane nucleation). The E1-like enzyme Atg7 activates both Atg12 and LC3-I, and the E2-like enzyme activates Atg10 (for Atg12) and Atg3 (for LC3-I). Atg12 is conjugated to Atg5 and the Atg12-Atg5 complex acts as an E3 ubiquitin ligase to catalyze the conjugation of LC3-I to the lipid phosphatidylethanolamine (PE) in the process of sequestration. The subsequent autophagosome formation is dependent on the Atg12-Atg5-Atg16 complex. After autophagosome completion, the Atg12-Atg5-Atg16 complex dissociates from autophagosomes to allow Atg4 access to LC3-II-PE for deconjugation. When the autophagosome is completed, it fuses with lysosome to form an autophagolysosome in which the cytosolic macromolecules, proteins, and organelles will either be degraded by acid hydrolases in what is known as type II programmed cell death or involved in a survival mechanism to be released back into the cytosol with the help of permeases against cellular stress. Akt/PKB, protein kinase B; AMPK, adenosine monophosphate-activated protein kinase; CaMKKβ, calmodulin kinase kinase β Atg, autophagy-related genes; FIP200, focal adhesion kinase (FAK) family-interacting protein of 200kD; LC3, microtubule-associated protein 1 light chain 3; LKB1, liver kinase B1; mTORC1, mammalian target of rapamycin complex 1; PE, phosphatidylethanolamine; p70S6K, p70S6 kinase; PKA, protein kinase A; PtdIns3K, phosphatidylinositol 3-kinase; TAK1, TGFβ-activated kinase 1; TSC1/2, tuberous sclerosis complex 1/2; ULK 1/2, UNC-51–like kinases 1/2; Vps 15/34, vacuolar protein sorting 15/34
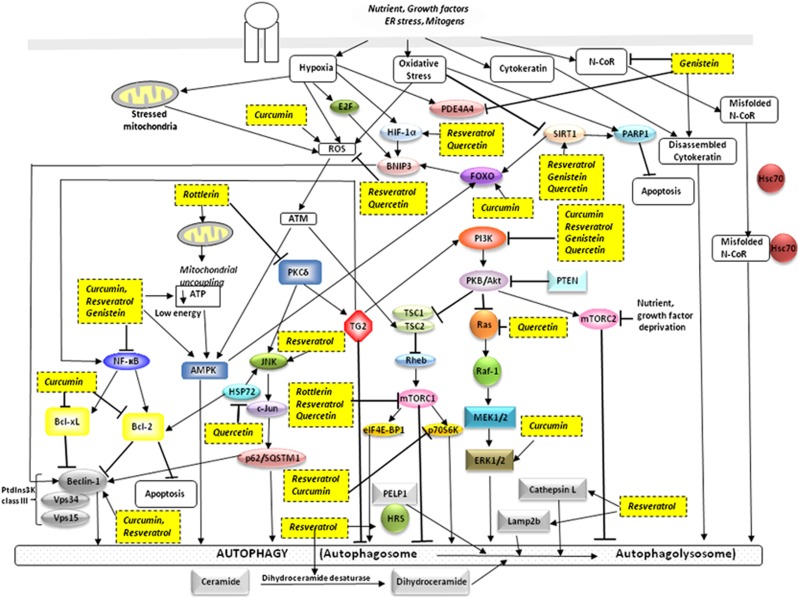
Figure 3.
Polyphenols promote autophagic cell death in cancer cells. The polyphenolic compounds rottlerin, genistein, quercetin, curcumin, and resveratrol induce cell death via autophagy through inhibition or activation of multiple signaling pathways and cellular targets in cancer cells. The common pathways involved are PI3K/Akt/mTOR, NF-κB, MEK/ERK, PKCδ/TG2, JNK/p62/SQSTM1, and AMPK/TSC2. Akt/PKB, protein kinase B; AMPK, adenosine monophosphate-activated protein kinase; ATM, ataxia telangiectasia mutated; ATP, adenosine triphosphate; Bcl-2, B-cell lymphoma 2; Bcl-XL, B cell lymphoma-extra large; BNIP3, Bcl-2 and nineteen KD interacting protein-3; eIF4E-BP1, eukaryotic translation initiation factor 4E-binding protein 1; ERK1/2, extracellular signal-regulated kinase 1/2; FOXO, forkhead transcription factors; HIF-1α, Hypoxia-inducible factor 1-alpha; HRS, hepatocyte growth factor-regulated tyrosine kinase substrate; Hsc70, 70-kDa heat shock protein family expressed constitutively; HSP72, 70-kDa heat shock protein family stress- and heat shock-induced; JNK, c-Jun N-terminal kinase; Lamp2b, lysosome-associated membrane proteins 2b; MEK1/2, mitogen-activated protein kinase 1/2; mTORC1/2, mammalian target of rapamycin complex 1/2; N-CoR, putative corepressor; NF-κB, nuclear factor-kappa B; PARP1, poly(ADP-ribose) polymerase 1; PDE4A4, cyclic AMP phosphodiesterase-4A4; PELP1; proline-, glutamic acid-, and leucine-rich protein-1; PI3K, phosphatidylinositol 3-kinase; PKCδ, protein kinase C-delta; PTEN, phosphatase and tensin homolog deleted on chromosome ten; p62/SQSTM1, p62 protein /sequestosome 1; p70S6K, p70 ribosomal protein S6 kinase; Raf-1, oncoprotein activated by Ras; Ras, oncoprotein; Rheb, Ras homolog enriched in brain; ROS, reactive oxygen species; SIRT1, sirtuin 1; TG2, transglutaminase 2; TSC1/2, tuberous sclerosis complex 1/2; Vps 15/34, vacuolar protein sorting 15/34
Table 1. Effect of polyphenols on induction of autophagy in cancer.
| Treatment | Cancer cell/tissue | Mechanism |
Dose |
Reference | |
|---|---|---|---|---|---|
| In vitro (μM) | In vivo (mg/kg) | ||||
| Rottlerin | Pancreatic | PKCδ/TG2 | 2–4 | — | 12, 13 |
| Fibrosarcoma | PKCδ-indep | 0.5–10 | — | 131 | |
| Breast | mTORC1 | 3 | — | 77 | |
| Genistein | Rat hepatocytes | Cytokeratin | — | 100 | 64, 65 |
| Ovarian | Akt | 25–100 | — | 82 | |
| Ovarian | PDE4A4& p62/SQSTM1 | 10 | — | 83 | |
| Lung | N-CoR/Hsc70 | 25–50 | — | 85 | |
| Quercetin | Colon | Ras | 20 | — | 66 |
| Fibroblast-breast | ROS | 10 | — | 91 | |
| Gastric | Akt/mTOR&HIF-1α | 10–160 | 50 | 90 | |
| Rat mesothelial | HSP72/JNK&Beclin-1 | — | 100 | 93 | |
| Breast, Cervical, Ovarian | mTOR/eIF4E-BP1/p70S6K | 30–90 | — | 92 | |
| Curcumin | Brain | Akt/mTOR/p70S6K | 10–50 | 100 | 62, 96 |
| ERK1/2 | |||||
| CML | Bcl-2 | 5–20 | — | 102 | |
| Bladder | Akt | 40 | — | 98 | |
| Prostate | Bcl-xL | 10–50 | — | 103 | |
| Colon | ROS | 10–40 | — | 104 | |
| Brain | PI3K/Akt/mTOR | 2 | 300 | 97 | |
| Mesothelioma | ND | 10–50 | — | 100 | |
| Oral | ROS | 10 | — | 105 | |
| Colon | Beclin-1&p62/SQSTM1 | 10 | — | 01 | |
| Endothelial | PI3K/Akt/mTOR&FOXO1 | 1–10 | — | 99 | |
| Resveratrol | Ovarian | ND | 50 | — | 3 |
| Salivary gland | PELP1/HRS | 50–100 | — | 110 | |
| Ovarian | Akt/mTOR/ p70S6K | 25–100 | — | 108 | |
| Lung | PELP1/HRS | 50–100 | — | 111 | |
| Colorectal | PI3K/Beclin-1/Lamp2b | 100 | — | 112 | |
| Breast | Akt/PKB/mTOR/p70S6K | 64 | — | 132 | |
| Cervical | Cathepsin L | 100 | — | 115 | |
| Gastric | Dihydroceramide desaturase | 50 | — | 116 | |
| Brain | Beclin-1 | 150 | — | 117 | |
| Fibroblast, Cervical | p70S6K | 50–200 | — | 118 | |
| CML | JNK/p62, AMPK/mTOR | 10–50 | — | 133 | |
| Lung | SIRT1/PARP-1 | 10 | — | 120 | |
| Hepatoma | ND | 20 | — | 125 | |
| Liver | SIRT1, AMPK, HIF-1α | — | 20 | 123 | |
| Colon | SIRT1 | 100 | — | 122 | |
| Brain | ND | 30 | — | 126 | |
| Cervical | ATAD3A | 30–100 | — | 124 | |
| Osteosarcoma, melanoma, cervical, breast | WIPI-1 | 64 | — | 128 | |
| Colon | ROS | 25–150 | — | 113 | |
| Skin | mTORC2 (Rictor) | 50 | — | 129 | |
Abbreviations: Akt/PKB, protein kinase B; AMPK, adenosine monophosphate-activated protein kinase; ATAD3A, ATPase family AAA domain containing 3A; Bcl-2, B-cell lymphoma 2; eIF4E-BP1, eukaryotic initiation factor 4E binding protein 1; ERK 1 / 2, extracellular signal-regulated kinases 1 / 2; HIF-1α, hypoxia-inducible factors -1 alpha; HRS, hepatocyte growth factor-regulated tyrosine kinase substrate; HSP72, heat shock protein 72; JNK, c-Jun N-terminal kinases; Lamp 2b, lysosome associated membrane proteins 2b; mTOR, mammalian target of rapamycin; mTORC 1 / 2, mTOR complex 1 / 2; N-CoR, nuclear receptor corepressor; ND, not determined; p62/SQSTM1, the ubiquitin-binding protein p62 or sequestosome 1; p70S6K, p70S6 kinase; PARP-1, Poly [ADP-ribose] polymerase 1; PDE4A4, cyclic AMP phosphodiesterase-4A4; PELP1, proline-, glutamic acid-, and leucine-rich protein-1; PI3K, phosphatidylinositol 3-kinases; PKCδ, protein kinase C delta; Ras, rat sarcoma protein; ROS, reactive oxygen species; SIRT1, Sirtuin 1; Tcf-4, transcription factor -4; TG2, tissue transglutaminase; TSC1/2, tuberous sclerosis 1 / 2; Vps 34/ Class III PI 3-kinase; WIPI, WD-repeat protein interacting with phosphoinosides; Wnt, wingless integration signaling pathway. Induction of autophagic death by the five dietary polyphenolic compounds rottlerin, genistein, quercetin, curcumin and resveratrol in various cancer types, the regulatory mechanisms involved and the effective doses for in vitro as well as in vivo analyses
Macroautophagy (hereafter called autophagy) is a lysosomal catabolic process conserved through evolution in eukaryotes for degrading long-lived proteins, macromolecules, and organelles from the cytoplasm.1, 2 Autophagy has different roles in normal and cancer cells, especially in the tumor microenvironment. Although genetic evidence indicates that autophagy functions as a tumor suppressor in normal cells, it can promote survival of established tumors in the presence of cellular stress factors, including nutrient deprivation, hypoxia, metabolic, and therapy-induced stress.3 If excessively induced, autophagy can lead to a non-apoptotic form of programmed cell death (type II PCD) which is caspase independent. Autophagy may be seen in cells that have a high threshold for induction of apoptosis or defective apoptotic machinery, such as inappropriate regulation of pro- and antiapoptotic Bcl-2 family member proteins.4
Autophagy requires the sequestration of cytoplasmic content or organelles through the formation of double-membrane vesicles, controlled by autophagy-related genes (ATG) and their protein products. Beclin-1, one of the major Atg proteins (its counterpart in yeast is known as Atg6), controls vital steps in the autophagic pathway. It is interesting that autophagy can be regulated by various canonical and non-canonical pathways, some involving Beclin-1,5, 6 others Beclin-1 dependent and Atg5/Atg7 independent,7 Beclin-1/Vps34 independent,8, 9 or Beclin-1 independent and Vps34 dependent.10, 11 Studies characterizing metabolic cascades known to have a role in autophagy involved common pathways, such as PI3K/Akt/mTOR, NF-κB, MAPK/MEK/ERK, protein kinase C delta (PKCδ)/ transglutaminase 2 (TG2), JNK/p62 /SQSTM1, and AMPK/TSC2, are reviewed here to gain insight into the regulation of autophagy (Table 1 and Figure 3).
In this review, which is an update to previous reports by our group,3, 12, 13 we look into the effects of five common dietary polyphenols – rottlerin, genistein, quercetin, curcumin, and resveratrol – on autophagy in several cancer types and their regulation of the various metabolic cascades that leads to the induction or inhibition of this alternative cell death mechanism.
Autophagic pathways
Autophagy is a multistep process involving induction, phagophore formation, sequestration, formation of autophagosome, and finally its fusion with lysosome to form autophagolysosome, which then induces either the death or survival pathway (Figure 2). The autophagic pathway dependent on Beclin-1 defines the canonical pathway; the other, independent of Beclin-1, is accepted as non-canonical autophagy. The presence of non-canonical autophagy means that autophagy can be triggered in cells in which the expression of Beclin-1 is too low to induce canonical autophagy.
Canonical/classic pathway
Beclin-1, an autophagy-promoting gene, was determined to function as a tumor suppressor after it was shown to be deleted in breast, ovarian, and prostate cancers.5 Beclin-1 forms a core complex known as PtdIns3K class III with Vps34 and Vps15 regulatory subunits (Figures 2 and 3), and this complex has been shown to promote autophagy by mediating autophagosome nucleation.14 Within the Beclin-1-dependent autophagy are two alternative downstream pathways, the more common, Atg5/Atg7 dependent and the other, Atg5/Atg7 independent.7
Non-canonical/non-classic pathway
In contrast to canonical, non-canonical autophagy is a process that does not require the entire set of Atg proteins to form the autophagosome. This alternative form of autophagy, which is independent of Beclin-1 and, typically Vps34, has been described elsewhere.8, 15 It is important to note that Beclin-1-independent autophagy does not always imply independence from Vps34.10, 11 A common hallmark of this Beclin-1 independent autophagy is its dependence on the activity of the UNC-51 like kinase 1/2 complex to induce autophagy and LC3 for phagophore formation (Figures 2 and 3). The stimuli that trigger these forms of autophagy and the various interacting proteins involved in the formation and maturation of autophagosomes are not completely understood.
Autophagy and cancer
Defects in autophagy alter cells' metabolic state and their capacity for protein degradation, and are associated with various disease conditions.16, 17, 18 Malignant cells often display defective autophagic activities compared with their normal counterparts. However, the role of autophagy in cancer is paradoxical in normal and malignant cells, and available data suggest that autophagy can promote tumor growth by helping tumor cells survive and inhibit tumor growth through its tumor-suppressor function in normal cells.19 Removal of damaged proteins and organelles may prevent cancer initiation while allowing established tumors to adapt to nutrient-deprived or hypoxic conditions during cancer progression.20
Initial genetic evidence in mice suggested that Beclin-1 functions as a haplo-insufficient tumor suppressor and that its mono-allelic deletion leads to spontaneous tumors and upon re-expression, it restores autophagy and suppresses tumorigenesis.21, 22 Although Beclin-1 and LC3 are important mediators of autophagy, other molecules and signaling pathways (for example, p53, PI3K/AKT/mTOR) require critical examination to determine their roles in cells' autophagic capacity toward cell death or survival following various triggers.
On the other hand, the antiapoptotic Bcl-2 family members from the endoplasmic reticulum and not the mitochondria, such as Bcl-2 and Bcl-xL, inhibit autophagy by binding to the BH3 domain of Beclin-1.23 Furthermore, recent data suggest that the oncogenic effect of Bcl-2 arises from its ability to inhibit autophagy but not apoptosis. Inhibition of Bcl-2 leads to autophagic cell death in MCF7 breast cancer cells.24 Therefore these evidences suggest that modulating autophagy may be important in designing anticancer therapies.22 Finally, studies suggest that the genetic makeup of cells will determine its fate in terms of undergoing autophagy and response to standard or novel therapies.
Signaling pathways regulating autophagy
Disruption of the normal balance between pro- and anti-autophagic signaling pathways is linked to cancer and other diseases. Several signaling pathways kinases, such as mTOR, AMPK, PI3Ks, MAPKs (ERK and JNK), and PKC, respond to various external factors and are often dysregulated in cancer (Figure 3). These kinases may be part of an energy-sensing mechanism and stress response. For example, the mTOR kinase is an important repressor of autophagy and controller of cell growth and proliferation. Therefore, it is not surprising that it is regulated by multiple signaling pathways; it is activated by the Akt/PKB proto-oncogenic pathway and often upregulated in various cancer types.25
There are two mTOR complexes, mTORC1 and mTORC2, which are regulated differently even though they are both induced in response to nutrient starvation, stress, and reduced growth factor signaling. TSC 1/2, an inhibitor of mTOR, is the gateway by which other signaling pathways influence mTOR activity. Alexander et al.26 provided evidence that the ATM protein signals to TSC2 via the AMPK metabolic pathway in the cytoplasm to repress mTORC1 in response to reactive oxygen species (ROS), thereby inducing autophagy. Alternatively, the mTOR kinase can be activated by Akt independently of TSC 1/2 and can be regulated by MAPKs. mTORC2 is rapamycin insensitive and activates Akt by phosphorylation, to contribute to another route in the regulation of autophagy.25, 27
Wong et al.28 observed the activation of ERK and JNK as upstream effectors controlling autophagy induced by ROS production. Wang et al.28 reported a non-canonical MEK signaling pathway positioned downstream of AMPK and upstream of TSC that mediates autophagy via regulation of Beclin-1.6 Puissant and Auberger29 observed the activation of AMPK/mTOR- and JNK-mediated p62/SQSTM1 in triggering autophagic cell death. AMPK, which promotes autophagy by activating TSC 1/2 and suppressing mTOR, has been implicated in an energy check point because of its roles in phosphorylating p53 and inducing cell cycle arrest following energy deprivation.30 We previously reported that PKCδ positively regulates the expression of TG2,13, 14 which leads to the suppression of autophagy through various downstream pathways, including NF-κB, PI3K/Akt/mTOR/p70S6K and Bcl-2 anti-autophagic/apoptotic protein.3, 12 Hence, studies suggest that activated or inhibited signaling pathways that are involved in the regulation of autophagy will direct it in cancer cells.
Clinical significance of autophagy and its modulation for treatment of cancer
The observations in clinical trials showed that Beclin-1 expression is altered in some cancers, overexpressed in others (intrahepatic cholangiocarcinoma and gastric tumors), whereas underexpressed in several solid tumors (breast, ovarian, cervical, lung, brain, liver, esophageal, gastric, and pancreatic) and osteosarcomas.22, 31, 32, 33, 34, 35, 36 Beclin-1 gene is found to be deleted in ovarian, breast, and prostate cancers, suggesting that reduced autophagic capacity is usual in some commonly diagnosed cancers.37, 38 Apparently, the expression of LC3 and Beclin-1 is associated with cancer stage, low expression levels being associated with poorly differentiated tumors and more advanced clinical stage of disease. Examples of this correlation of Beclin-1 level with clinical cancer stage include ovarian,32 esophageal squamous cell carcinoma,39 chondrosarcoma,40 and lymphomas,41, 42 whereas no such connection has been found in cervix,43 nasopharynx44 or adenoid cystic carcinoma.44 More importantly, many chemotherapeutic agents, such as cisplatin, plant alkaloids, antimetabolites,45, 46 tyrosine kinase inhibitors,47, 48 and radiotherapy49 induce autophagy. However, it remains to be determined whether the effectiveness of these therapies is dependent on hyper or under functional autophagy.20
Currently, >30 clinical trials are investigating the effects of autophagy in combination with cytotoxic chemotherapy or targeted agents in various human cancers.50 For example, chloroquine and hydroxychloroquine, routinely used for the treatment of diverse diseases, inhibit lysosomal acidification and prevent autophagy. In cancer treatment, chloroquine is often used in combination with chemotherapeutic drugs, such as cisplatin or PI3K inhibitor, LY294002 or the mTOR inhibitor, rapamycin. With all these agents, however, sensitization occurs independently of autophagy inhibition, and was not mimicked by Atg12, Beclin-1 knockdown, or bafilomycin treatment, and occurred even in the absence of Atg12.51 The results of these clinical trials are critical for better understanding the process and role of autophagy in tumor biology and to validate the strategy of targeting autophagy to enhance therapeutic benefits to patients.
Cell death by autophagy
Apoptosis (type I PCD) and necrosis (type III PCD) are well-known mechanisms of cell death induced by anticancer therapies. Recent studies have shown a non-apoptotic form of programmed death called autophagy, which is termed type II PCD and is often caspase independent.52 In both apoptosis and autophagy, the degraded cells are disposed by phagocytosis without an inflammatory response, in contrast to the extensive cellular disintegration and subsequent inflammation that occur in necrosis.
Studies have shown that there is a complex interplay between the apoptotic and autophagic processes. Autophagy may precede apoptosis or be induced simultaneously depending on the genetic context and cellular background of the cells. Oligomerized caspase-8 binds autophagosome membrane, leading to its activation and creating a mechanism for transition from apoptosis to autophagy.53 Experimental findings suggest that activated caspase-3 can cleave Beclin-1, producing a fragment that translocates to mitochondria and induces apoptosis.54
Other studies indicate that the Bcl-2 family of proteins not only regulates apoptosis but also controls cell death that depends on the autophagy genes.24 Cytotoxic signals can induce autophagy in apoptosis resistant cells, such as, those expressing high levels of Bcl-2 or Bcl-xL, lacking Bax and Bak, or exposed to pan-caspase inhibitors, such as, zVAD-fmk, suggesting autophagy to act as a default mechanism leading to cell death.4 For instance, although embryonic fibroblasts from Bax/Bak double-knockout mice are resistant to apoptosis, they can undergo autophagic death after stimulation.4 This non-apoptotic cell death was suppressed by autophagy inhibitors, such as, 3-MA, bafilomycin, or hydroxyl-choloroquine and or genetic silencing of autophagic genes (for example, ATG5, ATG7, or Beclin-1).4 This indicates that the autophagic process has a significant role in caspase-independent cell death. Further studies supported these findings when apoptosis-resistant Bax−/− and Bak−/− knockout fibroblast cells underwent autophagic death following induction, such as, starvation, growth factor withdrawal, chemotherapy (etoposide), or radiation.55 In addition, the knockdown of ATG5 or Beclin-1 in cancer cells has shown marked reduction in cell death and autophagic effects in response to death stimuli, with no sign of apoptosis.12
Autophagy has important roles in both, maintenance of cellular homeostasis under regular growth conditions and protection of cell viability under stress. In the housekeeping pathway, autophagy removes sources of ROS such as damaged or aggregated proteins and organelles to prevent tumor initiation via suppression of oxidative and genotoxic stress.56, 57 Under stress, however, autophagy supports tumor cell survival by providing substrates for mitochondrial metabolism.58 In situations of defective mitophagy, autophagy-deficient cells would accumulate damaged mitochondria and deregulate ROS levels, which have been suggested to contribute to induction of tumors.59 Kaminskyy et al.60 showed that autophagy suppression led to the inhibition of proliferation of non-small cell lung carcinoma cells and sensitized them to cisplatin-induced caspase-dependent and -independent apoptosis by stimulation of ROS formation.
Autophagy leading to cell death is induced in various cancer cells in response to treatment by several polyphenolic compounds,61 including rottlerin (Figure 4),12 curcumin,62 resveratrol,63 genistein,64, 65 and quercetin,66 as well as some chemotherapeutic agents, such as cytosine arabinoside,67 etoposide,4 and staurosporine,68 and growth factor deprivation.67
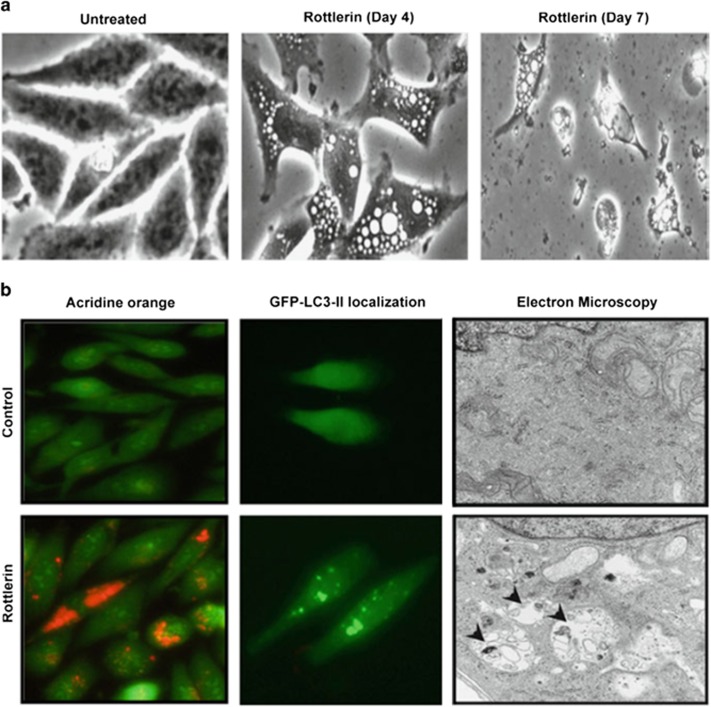
Figure 4.
Rottlerin induced massive autophagy and cell death in pancreatic cancer cells. (a) Morphological changes and formation of autophagic vacuoles in MDA-Panc28 pancreatic cancer cells that were treated with 4 μM rottlerin. Microphotographs were taken using a phase-contrast microscope ( × 300 magnification). Left panel, untreated cells at 48 h; center panel, cells treated for 4 days; and right panel, cells treated for 7 days. (b) MDA-Panc28 cells were treated with Rottlerin for 48 h and then were stained with acridine orange for detection of acidic vesicular organelles by fluorescent microscopy (left panels) and localization of LC3-II at autophagosomes in cells after transfection with GFP-LC3 plasmid (middle panels).12, 13 Green fluorescence indicates cytoplasm and nucleus, whereas red fluorescence shows acidic vesicular organelles. Right panels, electron micrographs showing the ultrastructure of rottlerin-treated cells (4 M). Numerous autophagic vacuoles, indicated by arrows, were observed in the rottlerin-treated cells
Polyphenolic compounds and autophagic cell death
Polyphenols are a structural class of natural organic chemicals characterized by the presence of large multiples of phenol structural units. These compounds found in foods possess anticancer activities in their ability to alter the effects of signaling pathways and induce cell death not only via apoptosis but also autophagy.69, 70
Rottlerin
Rottlerin (5, 7-dihydroxy-2, 2-dimethyl-6-(2, 4, 6-trihydroxy-3-methyl-5-acetylbenzyl)-8-cinnamoyl-1, 2-chromine), also called mallotoxin, is isolated from Mallotus phillippinensis (the monkey-faced tree). Rottlerin induces autophagy via three distinct mechanisms, that is, through PKCδ/TG2, PKCδ-independent, and mTORC1 pathways, in pancreatic cancer, fibrosarcoma, and breast cancer, respectively (Table 1and Figure 3).
Rottlerin displayed antioxidant properties and inhibitory effect on NF-κB in breast and colon cancer cells.71 In fact, in these cells, expressions of PKCδ and TG2 led to activation of NF-κB,72, 73 whereas inhibition led to induction of autophagy death.74 Importantly, rottlerin is accepted widely as a PKCδ-selective inhibitor.
The first evidence of massive autophagy induction leading to death only in cells with PKCδ via induction of TG2 was reported in pancreatic cancer cells.12, 13 However, recent studies suggested that rottlerin can also induce apoptosis through PKCδ-independent mechanisms in fibrosarcoma cells.75 It was suggested that the early autophagy might serve as a survival mechanism against late apoptosis in this cancer type.76
Rottlerin was able to inhibit mTORC1 signaling, via its negative regulator, TSC2 to induce autophagosome accumulation in breast cancer cells in nutrient-rich conditions.77
Rottlerin inhibition of NF-κB was able to induce AMPK induction, which led to significantly reduced cellular ATP levels and induction of autophagy in cancer cells.78 AMPK can also activate the cyclin-dependent kinase inhibitor, p27, by a mechanism involving the SIRT1/FOXO pathway to induce autophagy.79
Overall, rottlerin-induced autophagy may involve multiple signaling pathways and cellular mechanisms for induction of autophagy and eventual cell death. However, the most important factor determining the fate of cells is probably the cellular context, increased apoptotic threshold/resistance and activated/inhibited signaling pathways. Thus rottlerin or its related analogs may be used in the development of novel agents for induction of autophagic cell death as it has been proven pharmacodynamically in a mice xenograft model to be efficiently absorbed in cells and tissues against pancreatic cancer.80
Genistein
Genistein (4′, 5, 7-trihydroxyisoflavone), a naturally occurring isoflavonoid found in soy products, has been shown to have anticancer properties. It has the capacity to induce cell death through both apoptosis81 and autophagy.82 Thus, genistein may be beneficial against chemoresistance owing to changes in apoptotic signaling (Table 1).
It has been shown to completely protect the cytokeratin network in stress, nutrient- and growth factor-deprived environments. Various studies showed its ability to overcome the disruptive effects of okadaic acid, a strong inhibitor of autophagy, on the organization of the cytoskeleton and cytokeratin in rat hepatocytes.64, 65 This is of consequence, as the cytokeratin filaments are involved in the development of autophagy.
Genistein has been shown to be cytotoxic in ovarian cancer cells with mechanism of death involving not only apoptosis but also autophagy.82 Treatment markedly inhibited glucose uptake in these cells, and methyl pyruvate, the substrate for oxidative phosphorylation and fatty acid synthesis, could rescue cells from genistein-induced autophagy. Gossner et al.82 also showed that treatment reduced levels of phosphorylated Akt. This may have contributed to limiting glucose utilization, which suggested that a starvation-like signaling response would eventually lead to autophagy death. Christian et al.83 reported its ability to inhibit both PKC and ERK inhibitors via inhibition of PDE4A4 aggregate formation to activate autophagy in ovarian cancer cells. These authors suggested that, as PDE4A4 aggregates are neither autophagosomes nor aggresomes and constitutively co-immunoprecipitated with the p62 protein (SQSTM1). Therefore, inhibiting their formation would be beneficial in ensuring induction of autophagy, as p62 interacts with LC3, which is critical for membrane encapsulation in autophagosomes.84
Ali et al.85 reported evidence for genistein inhibition of N-CoR misfolding, an important component in the activation of the oncogenic survival pathway in non-small cell lung carcinoma, which was found to be associated with Hsc70, a molecular chaperone in autophagy (Figure 3).
Although, genistein can induce autophagy and apoptotic death in cancer cells, ADME studies revealed that it has intrinsically low oral bioavailability because of metabolic enzyme and efflux transporter. This should be further investigated to improve its efficacy in the treatment of apoptotic resistant cancers.86
Quercetin
Quercetin (3, 3′, 4′, 5, 7-pentahydroxyflavone), a natural flavonoid molecule found in fruits, vegetables, leaves, and grains, has anticancer effects linked to its capacity for targeting key molecules, organelles, and tumorigenic pathways (Table 1).87, 88, 89 To confirm the involvement of autophagy, Psahoulia et al.66 treated RAS-transformed colon cells with 3-MA at the early stages, which resulted in the inhibition of vacuolization. Moreover, zVAD-FMK also failed to inhibit vacuolization, showing that the autophagy induced was caspase independent. Treatment of gastric cancer cells induced the vital stages that initiated autophagy progression.90 Administration of the inhibitor chloroquine or selective ablation of Atg5 or Beclin-1 using siRNA increased apoptotic cell death, suggesting that autophagy has a protective role against quercetin-induced apoptosis. Functional studies revealed that the activated autophagy is modulated via the Akt-mTOR and HIF-1α signaling. Therefore, these xenograft models provided in vivo evidence for quercetin-induced apoptosis and autophagy.90
Martinez-Outschoorn et al.91 presented evidence that treatment with quercetin promoted the removal of defective mitochondria from cancer-associated fibroblasts by autophagy /mitophagy that was induced by oxidative stress. As a consequence, the ‘reverse Warburg effect,' whereby these fibroblasts provided nutrients to stimulate mitochondrial biogenesis and oxidative metabolism in adjacent cancer cells, was proposed.
Quercetin treatment induced extensive intracellular vacuolization and phagolysosome formation with accumulation of autophagic biomarkers in epithelial cancer cells, which led to cell cycle arrest and induction of apoptosis.92 Before the formation of autophagosomes, inhibition of mTOR activity was observed, accompanied by a marked reduction in the phosphorylation of its substrates, the ribosomal S6 subunit via p70S6 kinase and the eIF4 via its inhibitor 4E-BP1. Inhibition of proteasome activity by quercetin was also observed with accumulation of polyubiquitinated protein aggregates, suggesting that proteasome inhibition is a major cause of the cancer cell death. Therefore, quercetin induces extensive autophagy and subsequent death in cancer cells mediated by the inhibition of proteasomal activity and mTOR signaling.92
Li et al.93 investigated whether autophagy contributes to HSP72-mediated cytoprotection in lipopolysaccharide-induced peritonitis. This is because HSP72 is known to induce autophagy but provide protection against apoptosis. The initial exposure of cultured peritoneal mesothelial cells to lipopolysaccharide resulted first in cell death via autophagy, with subsequent death seen to occur via apoptosis. Therefore, the activation of autophagy acted as a prosurvival mechanism. When autophagy was inhibited by 3-MA or Beclin-1 siRNA, the cells were sensitized to apoptosis, and the antiapoptotic effect of HSP72 was abolished. Also, overexpression of HSP72 enhanced autophagy through JNK phosphorylation and Beclin-1 upregulation. The suppression of JNK activity reversed HSP72-mediated Beclin-1 upregulation and autophagy, which indicated that HSP72-mediated autophagy, is JNK dependent. In the in vivo rat model of lipopolysaccharide-induced peritonitis, autophagy was seen to occur prior to apoptosis. When HSP72 was upregulated by geranylacetone, autophagy was increased, whereas apoptosis was inhibited and peritoneal injury reduced. These effects were reversed by downregulation of HSP72 with quercetin. When Li et al.93 blocked autophagy by chloroquine, there was induction of apoptosis and increased peritoneal dysfunction. Thus, they concluded that HSP72 protects peritoneum from lipopolysaccharide-induced mesothelial cell injury by inducing JNK activation-dependent autophagy and inhibiting apoptosis.
In a study to characterize the bioavailability and metabolic pharmacokinetics of quercetin in rats, 93.8% of the dose was circulating as its sulfates and glucuronides in the bloodstream when administered intravenously and 53% when given orally. These metabolites were seen to be responsible for the in vivo effects of quercetin. In both instances, the parent form of quercetin was not detected.94
Therefore, treatment with quercetin has numerous anticancer effects, including not only the induction of cell cycle arrest and apoptosis but also of autophagy through modulation of important autophagy signaling pathways such as Akt-mTOR and HIF-1α (Figure 3).
Curcumin
Curcumin (diferuloylmethane), an active ingredient of the spice turmeric Curcuma longa, has a potent anticancer effect on cancer cells.95 Curcumin targets mainly the PI3K/Akt/mTOR signaling pathway and NF-κB-regulated proteins (Table 1).
Various studies provided evidence that curcumin induced G2/M arrest and autophagy in malignant glioma cells through inhibition of the Akt/mTOR/p70S6K and activation of the ERK1/2 pathways, which implied that cell death via autophagy, might be pathway specific.62, 96, 97 In a xenograft glioma model, Aoki et al.96 observed that curcumin induced autophagy and inhibited tumor growth significantly. Shinojima et al.62 observed that inhibition of NF-κB, which is the main anticancer target of curcumin, does not have a major role in the death of malignant glioma cells. Therefore, autophagy and not NF-κB, is the causal factor of the anticancer effects seen in these cells. Chadalapaka et al.98 showed that curcumin decreased expression of Sp proteins, whose overexpression in gastric and pancreatic cancers correlates with poor survival and tumor aggressiveness. Downregulation of EGFR (a Sp-regulated gene that suppresses autophagy) and decreased phosphorylation of Akt led to the induction of LC3 and cell death in bladder cancer. Curcumin protected HUVECs cells from oxidative stress by inducing autophagy via pI3K/Akt/mTOR and FOXO pathways to interact with Atg7; the involvement of FOXO (a mediator of autophagy) was confirmed in a knockdown study in which autophagy was inhibited by an siRNA.99
Curcumin induced autophagy in mesothelioma cells as indicated by increased conversion of LC3-I to LC3-II and formation of autophagosomes, which were reduced by RNA silencing of Atg5.100 Mosieniak et al.101 showed that induction of cellular senescence was accompanied by autophagy in colon cancer cells with an increase in Beclin-1 and p62/SQSTM1 protein levels. Interestingly, the inhibition of autophagy due to diminished expression of Atg5 by RNA interference also decreased the number of cells induced into senescence by curcumin, but did not lead to increased cell death. This study revealed a possible functional link between senescence and autophagy in curcumin-treated cells, a finding that should be investigated further.
Jia et al.102 presented evidence that curcumin is able to induce both autophagy and apoptosis in a chronic myeloid leukemia cell line via downregulation of the Bcl-2 protein. These effects were confirmed when the potent inhibitor of autophagosome–lysosome fusion, bafilomycin A1 and pan-caspase inhibitor zVAD-FMK suppressed the cell death. In a study on prostate cancer cells, curcumin was shown to induce autophagy cell death through downregulation of another important Bcl-2 family member, Bcl-xL. Curcumin treatment did not induce the cleavage of procaspase-8, -9, -3, or -7 or PARP but led to appearance of the LC3B-II isoform and increased number of autophagosomes.103
Treatment of human colon cancer cells with curcumin induced the conversion of LC3-I to LC3-II and degradation of SQSTM1 and autophagosomes.104 The autophagic changes induced by curcumin were almost completely blocked in the presence of the antioxidant NAC, indicating that treatment led to ROS production, autophagosome formation, and autolysosomal degradation. The reduction of SQSTM1 degradation by bafilomycin, further confirmed the activation of autophagy cell death.104 Kim et al.105 reported anticancer activity involving ROS in oral squamous cell carcinoma via both apoptosis and autophagy, which was confirmed when NAC blocked autophagic vacuole formation.
Curcumin has been proven to not only induce apoptosis on its own but to also have synergistic effects with various FDA-approved drugs via major inflammatory biomarkers and oncoproteins.106 As curcumin promotes autophagy similarly via most of these proteins, therefore this compound is effective in inducing both apoptosis and autophagy in cancer therapy.
However, a major restriction on the use of curcumin as an anticancer agent is its poor absorption, biodistribution, metabolism, and bioavailability. When 400 mg of curcumin were fed to rats, about 60% were found to be absorbed. To address these problems, several formulations have been used, which include nanoparticles, liposomes, micelles, and phospholipid complexes, all with limited success. Also curcumin undergoes metabolism to form various metabolites, such as, glucuronide, sulfate, tetrahydrocurcumin, hexahydrocurcumin, octahydrocurcumin, and hexahydrocurcuminol after oral administration. However, all these metabolites displayed anticancer effects. 107
Resveratrol
Resveratrol (3,5,4-trihydroxystilbene), a natural phytoalexin present in grapes, nuts, and red wine, has chemopreventive properties and multiple mechanisms of action, which may be activated depending on the specific cell type and cellular environment (Table 1).
Opipari et al.63 provided the first evidence that resveratrol induces cell death through autophagy in five ovarian cancer cell lines, which suggested it as an effective treatment in apoptosis-resistant cells. As autophagy is an adaptive response to nutrient starvation and resveratrol's ability to induce a starvation-like signaling response, that is, reducing the levels of phosphorylated Akt and mTOR to initiate autophagy, is observed in ovarian cancer cells.108
Resveratrol was shown to bind to a novel estrogen receptor coactivator, PELP1, and induce its accumulation in autophagosomes.109 PELP1 was identified for the first time by a trafficking molecule, hepatocyte growth factor-regulated tyrosine kinase substrate, which binds to it.110 The role of hepatocyte growth factor-regulated tyrosine kinase substrate in facilitating the transport of cytoplasmic proteins to autophagosomes for their selective degradation was confirmed in another study involving resveratrol treatment of lung cancer cells.111
Puissant et al.29 presented evidence of resveratrol triggering autophagic death in chronic myeloid leukemia cells via both JNK-mediated p62/SQSTM1 overexpression and AMPK/mTOR activation.108 Also resveratrol enhanced the expression of several tubulin subunits, which is important for movement of autophagosomes inside the cell.
Trincheri et al.112 reported that autophagy can be induced with acute exposure to resveratrol, whereas prolonged exposure activated a caspase-mediated cell death pathway. They observed that genetic inactivation of PI3K, Beclin-1, and Lamp2b inhibited resveratrol toxicity. Beclin-dependent autophagy was confirmed when supplementing the cells with asparagine or knocking down Beclin-1 by RNA interference, abrogated the effect. The effect of Lamp2b was confirmed when its silencing inhibited the fusion of autophagosomes with lysosomes and induced cell viability. Interestingly, zVAD-FMK inhibited cell death but not autophagy. This study uncovered a novel pathway of resveratrol cytotoxicity in which autophagy has two roles, that is, a prosurvival stress response that later in the process, changes to a caspase-dependent apoptotic response. In another study, resveratrol increased ROS level with induction of caspase-8 and caspase-3 cleavage and elevation of LC3-II expression in colon cancer cells; these effects were diminished by NAC.113
An interesting non-canonical autophagic process characterized by a Beclin-1/Vps34-independent pathway was observed in MCF-7 cancer cells in response to resveratrol treatment. Overexpression of Bcl-2, which is known to block canonical starvation-induced autophagy by binding to Beclin-1, was unable to reverse the non-canonical autophagy triggered by resveratrol in these breast cancer cells.8, 114
SCCA 1, an endogenous cathepsin L inhibitor, is expressed widely in uterine cervical cells. Hsu et al.115 showed that the cathepsin L–SCCA 1 lysosomal pathway and autophagy were involved in resveratrol-induced cytotoxicity in cervical cancer cells. Inhibition of the autophagic response by wortmannin or asparagine resulted in decreased autophagic death.
Dihydroceramide, an immediate precursor of the apoptotic mediator ceramide in the de novo sphingolipid synthesis pathway, was accumulated when resveratrol-induced autophagy occurred as a result of inhibition of dihydroceramide desaturase activity in gastric cancer cells. These effects of resveratrol were mimicked by a dihydroceramide desaturase inhibitor.116
Resveratrol-induced autophagy in human glioma cells has the ability to inhibit resveratrol-induced apoptosis.117 Autophagy and apoptosis seemed to have different roles, apoptosis causing these cells' death, whereas autophagy delayed apoptosis and protected the cells from death. This suggests that autophagy inhibitors may have the potential to enhance resveratrol antitumor efficacy.117
In a large-scale in vitro kinase screen, p70 S6 kinase (S6K1) was identified as a target of resveratrol. Blocking S6K1 activity by expression of a dominant-negative mutant or RNA interference was sufficient to disrupt autophagy to an extent similar to resveratrol. Also, co-administration of resveratrol with S6K1 knockdown did not produce an additive effect, which indicated that S6K1 is important for the full induction of autophagy and some of the beneficial effects of resveratrol are due to modulation of S6K1 activity.118
SIRT1, one of the best characterized targets of resveratrol, upon activation by this compound induced both autophagy and apoptosis.119 However Armour et al.118 observed that resveratrol decreased autophagy in response to nutrient limitation in multiple cell lines through a pathway independent of SIRT1. Resveratrol is able to induce autophagy in lung cells with cigarette smoke-mediated oxidative stress via regulation of SIRT1 and PARP.120, 121, 122 Resveratrol prevented the decline in ATP concentration and SIRT1 expression, as well as the increase in HIF-1α expression and autophagy, in the livers of endotoxin-challenged wild-type mice but not in the liver of SIRT1 knockout mice.123 These findings provided an insight to the potential roles of SIRT1 and HIF-1α expression in systemic inflammation.
Persistent human papillomavirus infection may stabilize ATAD3A (an anti-autophagy factor), inhibit cell autophagy and apoptosis and to increase drug resistance in uterine cervical cancer. Resveratrol's anticancer effects were confirmed by its capacity for reducing ATAD3A expression, increasing abrasion of the mitochondrial outer membrane, and increasing the numbers of autophagosomes.124
Resveratrol was shown to trigger autophagic cell death through increased expression of Atg5, 7, 9, and 12 proteins in a human hepatitis C-induced hepatoma cell line.125 Also, Filippi-Chiela et al.126 observed that resveratrol induced the formation of autophagosomes through upregulation of Atg5, Beclin-1, and LC3-II in glioblastoma cells. The PtdIns(3)P effectors, WIPI-1, and WIPI-2 were shown to function downstream during initiation of autophagosome formation. Localization of WIPI-1 at the endoplasmic reticulum and the plasma membranes upon the induction of autophagy confirmed its involvement with the autophagosomal membrane.127 In another study, WIPI-1 specifically bound PtdIns(3)P, accumulated at the phagophore, and become a membrane protein of the autophagosome generated. WIPI-1 was observed to function upstream of both Atg7 and 5 and stimulated an increase of LC3-II upon nutrient starvation. These findings constituted evidence that resveratrol-mediated autophagy was via the non-canonical pathway, Beclin-1 independent but Atg7 and 5 dependent.128
As seen with curcumin, resveratrol can induce premature senescence that is associated with a blockade of autolysosome formation, as assessed by the absence of colocalization of important markers of autophagosomes and lysosomes, LC3 and Lamp-2, respectively. Resveratrol was also able to downregulate the level of Rictor, a vital component of the mTORC2 complex, which led to decreases in RhoA-GTPase activity, altered actin cytoskeleton network and increases in senescence-associated β-gal activity. Therefore, resveratrol has the ability to attenuate the autophagic process via downregulation of Rictor, which may be the mechanism of tumor suppression associated with premature senescence.129
Resveratrol upon administration is readily absorbed and metabolized as glucuronides and sulfates and the oral bioavailability of resveratrol is almost zero owing to its rapid metabolism activity.130 However, as resveratrol has shown versatility in inducing autophagy in numerous cancer types and targeting a wide array of autophagy-associated proteins, further clinical investigations are important to fully evaluate the efficacy and bioactivity of resveratrol in the killing of cancer cells via autophagy.
Conclusion
One of the most important unsolved problems in cancer therapy is increased tumor resistance to treatment, be it chemotherapy, radiotherapy, or any of the targeted therapies. This increased resistance is a direct effect of defects in apoptosis. An alternative form of cell death, namely, autophagy, may be the ultimate solution for this problem. The evidence presented here (Table 1 and Figure 2) demonstrates that several phytochemical polyphenolic compounds found in foods are able to mediate both canonical and non-canonical autophagy via multiple pathways targeting important proteins in a number of cancer types. Understanding their mechanisms is of vital importance in ensuring cell death versus survival. These polyphenolic compounds have the ability to induce both apoptosis and autophagy, thereby maximizing death of cancer cells. As they are food compounds, furthermore, they may offer greater safety, both through their inherently lower toxicity and through allowing reduction of doses and side effects as compared with synthetic drugs. Combining FDA-approved drugs with known polyphenolic compounds such as rottlerin, genistein, quercetin, curcumin, and resveratrol may provide novel therapeutic strategies in the treatment of cancer to combat the substantial problem of drug resistance in cancer therapy.
Acknowledgments
This work was supported by the Centre for Research in Biotechnology for Agriculture (CEBAR) University Malaya RU Operation Grant (RU005C-2014). The funder had no role in decision to publish or preparation of the paper.
Glossary
- 3-MA
3-methyl adenine
- 4E-BP1
eIF4E binding protein 1
- ADME
absorption, distribution, metabolism and excretion
- AMPK
AMP-dependent protein kinase
- ATAD3A
ATPase family AAA domain containing 3A
- ATG
autophagy regulated genes
- ATM
ataxia telangiectasia mutated
- CML
chronic myeloid leukemia
- eIF4
eukaryotic initiation factor of protein biosynthesis
- ER
endoplasmic reticulum
- ERK
extracellular signal-regulated kinase
- FDA
Food and Drug Administration
- FOXO
forkhead box class O
- HIF-1α
hypoxia-induced factor 1α
- HRS
hepatocyte growth factor–regulated tyrosine kinase substrate
- HSP72
heat shock protein 72
- HUVECs
human umbilical vein endothelial cells
- JNK c-Jun
N-terminal kinase
- Lamp2b
lysosome-associated membrane proteins 2b
- MAPK
mitogen-activated protein kinase
- MDC
monodansylcadaverin
- mTOR
mammalian target of rapamycin
- NAC
N-acetylcysteine
- N-CoR
nuclear receptor co-repressor
- NF-κB
nuclear factor kappa B
- NSCLC
non-small cell lung carcinoma
- PARP
poly(ADP ribose) polymerase
- PCD
programmed cell death
- PELP1
proline-, glutamic acid–, and leucine-rich protein-1
- PDE4A4
cyclic AMP phosphodiesterase-4A4
- PI3K/Akt
phosphatidylinositide 3-kinase
- PKB
protein kinase B
- PKCδ
protein kinase C delta
- ROS
reactive oxygen species
- SCCA
squamous cell carcinoma antigen
- siRNA
small interfering RNA
- SIRT1
sirtuin 1
- SQSTM1
p62/sequestome 1
- TG2
transglutaminase 2
- TSC2
tuberous sclerosis complex 2
- WIPI-1/2
Wd-repeat proteins interacting with phosphoinositides
- zVAD-fmk
benzyloxycarbonyl-Val–Ala–Asp fluoromethylketone
The authors declare no conflict of interest.
Footnotes
Edited by M Diederich
References
- Klionsky DJ, Ohsumi Y. Vacuolar import of proteins and organelles from the cytoplasm. Annu Rev Cell Dev Biol. 1999;15:1–32. doi: 10.1146/annurev.cellbio.15.1.1. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290:1717–1721. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalby KN, Tekedereli I, Lopez-Berestein G, Ozpolat B. Targeting the prodeath and prosurvival functions of autophagy as novel therapeutic strategies in cancer. Autophagy. 2010;6:322–329. doi: 10.4161/auto.6.3.11625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu S, Kanaseki T, Mizushima N, Mizuta T, Arakawa-Kobayashi S, Thompson CB, et al. Role of Bcl-2 family proteins in a non-apoptotic programmed cell death dependent on autophagy genes. Nat Cell Biol. 2004;6:1221–1228. doi: 10.1038/ncb1192. [DOI] [PubMed] [Google Scholar]
- Aita VM, Liang XH, Murty VV, Pincus DL, Yu W, Cayanis E, et al. Cloning and genomic organization of beclin 1, a candidate tumor suppressor gene on chromosome 17q21. Genomics. 1999;59:59–65. doi: 10.1006/geno.1999.5851. [DOI] [PubMed] [Google Scholar]
- Wang J, Whiteman MW, Lian H, Wang G, Singh A, Huang D, et al. A non-canonical MEK/ERK signaling pathway regulates autophagy via regulating Beclin 1. J Biol Chem. 2009;284:21412–21424. doi: 10.1074/jbc.M109.026013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida Y, Arakawa S, Fujitani K, Yamaguchi H, Mizuta T, Kanaseki T, et al. Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature. 2009;461:654–658. doi: 10.1038/nature08455. [DOI] [PubMed] [Google Scholar]
- Scarlatti F, Maffei R, Beau I, Ghidoni R, Codogno P. Non-canonical autophagy: an exception or an underestimated form of autophagy. Autophagy. 2008;4:1083–1085. doi: 10.4161/auto.7068. [DOI] [PubMed] [Google Scholar]
- Juenemann K, Reits EA. Alternative macroautophagic pathways. Int J Cell Biol. 2012;2012:189794. doi: 10.1155/2012/189794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P, Bauvy C, Souquere S, Tonelli G, Liu L, Zhu Y, et al. The Bcl-2 homology domain 3 mimetic gossypol induces both Beclin 1-dependent and Beclin 1-independent cytoprotective autophagy in cancer cells. J Biol Chem. 2010;285:25570–25581. doi: 10.1074/jbc.M110.118125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DM, Patel S, Raffoul F, Haller E, Mills GB, Nanjundan M. Arsenic trioxide induces a beclin-1-independent autophagic pathway via modulation of SnoN/SkiL expression in ovarian carcinoma cells. Cell Death Differ. 2010;17:1867–1881. doi: 10.1038/cdd.2010.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akar U, Ozpolat B, Mehta K, Fok J, Kondo Y, Lopez-Berestein G. Tissue transglutaminase inhibits autophagy in pancreatic cancer cells. Mol Cancer Res. 2007;5:241–249. doi: 10.1158/1541-7786.MCR-06-0229. [DOI] [PubMed] [Google Scholar]
- Ozpolat B, Akar U, Mehta K, Lopez-Berestein G. PKC delta and tissue transglutaminase are novel inhibitors of autophagy in pancreatic cancer cells. Autophagy. 2007;3:480–483. doi: 10.4161/auto.4349. [DOI] [PubMed] [Google Scholar]
- Funderburk SF, Wang QJ, Yue Z. The Beclin 1-VPS34 complex—at the crossroads of autophagy and beyond. Trends Cell Biol. 2010;20:355–362. doi: 10.1016/j.tcb.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JH, Horbinski C, Guo F, Watkins S, Uchiyama Y, Chu CT. Regulation of autophagy by extracellular signal-regulated protein kinases during 1-methyl-4-phenylpyridinium-induced cell death. Am J Pathol. 2007;170:75–86. doi: 10.2353/ajpath.2007.060524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ. The molecular machinery of autophagy: unanswered questions. J Cell Sci. 2005;118 (Pt 1:7–18. doi: 10.1242/jcs.01620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469:323–335. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy JM, Thorburn A. Targeting autophagy during cancer therapy to improve clinical outcomes. Pharmacol Ther. 2011;131:130–141. doi: 10.1016/j.pharmthera.2011.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swampillai AL, Salomoni P, Short SC. The role of autophagy in clinical practice. Clin Onco. 2012;24:387–395. doi: 10.1016/j.clon.2011.09.010. [DOI] [PubMed] [Google Scholar]
- Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci USA. 2003;100:15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Akar U, Chaves-Reyez A, Barria M, Tari A, Sanguino A, Kondo Y, et al. Silencing of Bcl-2 expression by small interfering RNA induces autophagic cell death in MCF-7 breast cancer cells. Autophagy. 2008;4:669–679. doi: 10.4161/auto.6083. [DOI] [PubMed] [Google Scholar]
- Bhaskar PT, Hay N. The two TORCs and Akt. Dev Cell. 2007;12:487–502. doi: 10.1016/j.devcel.2007.03.020. [DOI] [PubMed] [Google Scholar]
- Alexander A, Cai SL, Kim J, Nanez A, Sahin M, MacLean KH, et al. ATM signals to TSC2 in the cytoplasm to regulate mTORC1 in response to ROS. Proc Natl Acad Sci USA. 2010;107:4153–4158. doi: 10.1073/pnas.0913860107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- Wong CH, Iskandar KB, Yadav SK, Hirpara JL, Loh T, Pervaiz S. Simultaneous induction of non-canonical autophagy and apoptosis in cancer cells by ROS-dependent ERK and JNK activation. PLoS One. 2010;5:e9996. doi: 10.1371/journal.pone.0009996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puissant A, Robert G, Fenouille N, Luciano F, Cassuto JP, Raynaud S, et al. Resveratrol promotes autophagic cell death in chronic myelogenous leukemia cells via JNK-mediated p62/SQSTM1 expression and AMPK activation. Cancer Res. 2010;70:1042–1052. doi: 10.1158/0008-5472.CAN-09-3537. [DOI] [PubMed] [Google Scholar]
- Hardie DG. AMP-activated/SNF1 protein kinases: conserved guardians of cellular energy. Nat Rev Mol Cell Biol. 2007;8:774–785. doi: 10.1038/nrm2249. [DOI] [PubMed] [Google Scholar]
- Karantza-Wadsworth V, White E. Role of autophagy in breast cancer. Autophagy. 2007;3:610–613. doi: 10.4161/auto.4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Li DD, Wang LL, Deng R, Zhu XF. Decreased expression of autophagy-related proteins in malignant epithelial ovarian cancer. Autophagy. 2008;4:1067–1068. doi: 10.4161/auto.6827. [DOI] [PubMed] [Google Scholar]
- Miracco C, Cosci E, Oliveri G, Luzi P, Pacenti L, Monciatti I, et al. Protein and mRNA expression of autophagy gene Beclin 1 in human brain tumours. Int J Oncol. 2007;30:429–436. [PubMed] [Google Scholar]
- Daniel F, Legrand A, Pessayre D, Borrega-Pires F, Mbida L, Lardeux B, et al. Beclin 1 mRNA strongly correlates with Bcl-XLmRNA expression in human hepatocellular carcinoma. Cancer Invest. 2007;25:226–231. doi: 10.1080/07357900701206323. [DOI] [PubMed] [Google Scholar]
- Ahn CH, Jeong EG, Lee JW, Kim MS, Kim SH, Kim SS, et al. Expression of beclin-1, an autophagy-related protein, in gastric and colorectal cancers. APMIS. 2007;115:1344–1349. doi: 10.1111/j.1600-0463.2007.00858.x. [DOI] [PubMed] [Google Scholar]
- Lebovitz CB, Bortnik SB, Gorski SM. Here, there be dragons: charting autophagy-related alterations in human tumors. Clin Cancer Res. 2012;18:1214–1226. doi: 10.1158/1078-0432.CCR-11-2465. [DOI] [PubMed] [Google Scholar]
- Eccles DM, Cranston G, Steel CM, Nakamura Y, Leonard RC. Allele losses on chromosome 17 in human epithelial ovarian carcinoma. Oncogene. 1990;5:1599–1601. [PubMed] [Google Scholar]
- Chen N, Karantza-Wadsworth V. Role and regulation of autophagy in cancer. Biochim Biophys Acta. 2009;1793:1516–1523. doi: 10.1016/j.bbamcr.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Lu Y, Lu C, Zhang L. Beclin-1 expression is a predictor of clinical outcome in patients with esophageal squamous cell carcinoma and correlated to hypoxia-inducible factor (HIF)-1alpha expression. Pathol Oncol Res. 2009;15:487–493. doi: 10.1007/s12253-008-9143-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Ma Q, Ma X, Liu Z, Liu X. Association of elevated HIF-2alpha levels with low Beclin 1 expression and poor prognosis in patients with chondrosarcoma. Ann Surg Oncol. 2011;18:2364–2372. doi: 10.1245/s10434-011-1587-5. [DOI] [PubMed] [Google Scholar]
- Huang JJ, Li HR, Huang Y, Jiang WQ, Xu RH, Huang HQ, et al. Beclin 1 expression: a predictor of prognosis in patients with extranodal natural killer T-cell lymphoma, nasal type. Autophagy. 2010;6:777–783. doi: 10.4161/auto.6.6.12784. [DOI] [PubMed] [Google Scholar]
- Huang JJ, Zhu YJ, Lin TY, Jiang WQ, Huang HQ, Li ZM. Beclin 1 expression predicts favorable clinical outcome in patients with diffuse large B-cell lymphoma treated with R-CHOP. Hum Pathol. 2011;42:1459–1466. doi: 10.1016/j.humpath.2010.12.014. [DOI] [PubMed] [Google Scholar]
- Zhu W, Pan X, Li F, Zhang Y, Lu X. Expression of Beclin 1 and LC3 in FIGO stage I-II cervical squamous cell carcinoma and relationship to survival. Tumour Biol. 2012;33:1653–1659. doi: 10.1007/s13277-012-0421-4. [DOI] [PubMed] [Google Scholar]
- Wan SB, Yang H, Zhou Z, Cui QC, Chen D, Kanwar J, et al. Evaluation of curcumin acetates and amino acid conjugates as proteasome inhibitors. Int J Mol Med. 2010;26:447–455. doi: 10.3892/ijmm_00000484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi G, Hu X, Wu B, Jiang H, Young CY, Pang Y, et al. Autophagy inhibition promotes paclitaxel-induced apoptosis in cancer cells. Cancer Lett. 2011;307:141–148. doi: 10.1016/j.canlet.2011.03.026. [DOI] [PubMed] [Google Scholar]
- Li J, Hou N, Faried A, Tsutsumi S, Kuwano H. Inhibition of autophagy augments 5-fluorouracil chemotherapy in human colon cancer in vitro and in vivo model. Eur J Cancer. 2010;46:1900–1909. doi: 10.1016/j.ejca.2010.02.021. [DOI] [PubMed] [Google Scholar]
- Bellodi C, Lidonnici MR, Hamilton A, Helgason GV, Soliera AR, Ronchetti M, et al. Targeting autophagy potentiates tyrosine kinase inhibitor-induced cell death in Philadelphia chromosome-positive cells, including primary CML stem cells. J Clin Invest. 2009;119:1109–1123. doi: 10.1172/JCI35660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabretta B, Salomoni P. Inhibition of autophagy: a new strategy to enhance sensitivity of chronic myeloid leukemia stem cells to tyrosine kinase inhibitors. Leuk Lymph. 2011;52 (Suppl 1:54–59. doi: 10.3109/10428194.2010.546913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H, Daido S, Kanzawa T, Kondo S, Kondo Y. Radiation-induced autophagy is associated with LC3 and its inhibition sensitizes malignant glioma cells. Int J Oncol. 2005;26:1401–1410. [PubMed] [Google Scholar]
- http://www.cancer.gov/clinicaltrials/search/results?protocolsearchid=10999187 .
- Maycotte P, Aryal S, Cummings CT, Thorburn J, Morgan MJ, Thorburn A. Chloroquine sensitizes breast cancer cells to chemotherapy independent of autophagy. Autophagy. 2012;8:200–212. doi: 10.4161/auto.8.2.18554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirisako T, Baba M, Ishihara N, Miyazawa K, Ohsumi M, Yoshimori T, et al. Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J Cell Biol. 1999;147:435–446. doi: 10.1083/jcb.147.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou W, Han J, Lu C, Goldstein LA, Rabinowich H. Autophagic degradation of active caspase-8: a crosstalk mechanism between autophagy and apoptosis. Autophagy. 2010;6:891–900. doi: 10.4161/auto.6.7.13038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang R, Zeh HJ, Lotze MT, Tang D. The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ. 2011;18:571–580. doi: 10.1038/cdd.2010.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti L, Attia A, Kim KW, Lu B. Crosstalk between Bak/Bax and mTOR signaling regulates radiation-induced autophagy. Autophagy. 2007;3:142–144. doi: 10.4161/auto.3607. [DOI] [PubMed] [Google Scholar]
- Karantza-Wadsworth V, Patel S, Kravchuk O, Chen G, Mathew R, Jin S, et al. Autophagy mitigates metabolic stress and genome damage in mammary tumorigenesis. Genes & development. 2007;21:1621–1635. doi: 10.1101/gad.1565707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew R, Kongara S, Beaudoin B, Karp CM, Bray K, Degenhardt K, et al. Autophagy suppresses tumor progression by limiting chromosomal instability. Genes Dev. 2007;21:1367–1381. doi: 10.1101/gad.1545107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JY, Chen HY, Mathew R, Fan J, Strohecker AM, Karsli-Uzunbas G, et al. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev. 2011;25:460–470. doi: 10.1101/gad.2016311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kongara S, Karantza V. The interplay between autophagy and ROS in tumorigenesis. FrontOncol. 2012;2:171. doi: 10.3389/fonc.2012.00171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminskyy VO, Piskunova T, Zborovskaya IB, Tchevkina EM, Zhivotovsky B. Suppression of basal autophagy reduces lung cancer cell proliferation and enhances caspase-dependent and -independent apoptosis by stimulating ROS formation. Autophagy. 2012;8:7. doi: 10.4161/auto.20123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singletary K, Milner J. Diet autophagy, and cancer: a review. Cancer Epidemiol, Biomarkers Prevention. 2008;17:1596–1610. doi: 10.1158/1055-9965.EPI-07-2917. [DOI] [PubMed] [Google Scholar]
- Shinojima N, Yokoyama T, Kondo Y, Kondo S. Roles of the Akt/mTOR/p70S6K and ERK1/2 signaling pathways in curcumin-induced autophagy. Autophagy. 2007;3:635–637. doi: 10.4161/auto.4916. [DOI] [PubMed] [Google Scholar]
- Opipari AW, Jr., Tan L, Boitano AE, Sorenson DR, Aurora A, Liu JR. Resveratrol-induced autophagocytosis in ovarian cancer cells. Cancer Res. 2004;64:696–703. doi: 10.1158/0008-5472.can-03-2404. [DOI] [PubMed] [Google Scholar]
- Gordon PB, Holen I, Seglen PO. Protection by naringin and some other flavonoids of hepatocytic autophagy and endocytosis against inhibition by okadaic acid. J Biol Chem. 1995;270:5830–5838. doi: 10.1074/jbc.270.11.5830. [DOI] [PubMed] [Google Scholar]
- Blankson H, Holen I, Seglen PO. Disruption of the cytokeratin cytoskeleton and inhibition of hepatocytic autophagy by okadaic acid. Exp Cell Res. 1995;218:522–530. doi: 10.1006/excr.1995.1187. [DOI] [PubMed] [Google Scholar]
- Psahoulia FH, Moumtzi S, Roberts ML, Sasazuki T, Shirasawa S, Pintzas A. Quercetin mediates preferential degradation of oncogenic Ras and causes autophagy in Ha-RAS-transformed human colon cells. Carcinogenesis. 2007;28:1021–1031. doi: 10.1093/carcin/bgl232. [DOI] [PubMed] [Google Scholar]
- Xue L, Fletcher GC, Tolkovsky AM. Autophagy is activated by apoptotic signalling in sympathetic neurons: an alternative mechanism of death execution. Mol Cell Neurosci. 1999;14:180–198. doi: 10.1006/mcne.1999.0780. [DOI] [PubMed] [Google Scholar]
- Zhang L, Shimizu S, Sakamaki K, Yonehara S, Tsujimoto Y. A caspase-8-independent signaling pathway activated by Fas ligation leads to exposure of the Bak N terminus. J Biol Chem. 2004;279:33865–33874. doi: 10.1074/jbc.M403499200. [DOI] [PubMed] [Google Scholar]
- Tan W, Lu J, Huang M, Li Y, Chen M, Wu G, et al. Anti-cancer natural products isolated from chinese medicinal herbs. Chinese Med. 2011;6:27. doi: 10.1186/1749-8546-6-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiyer HS, Warri AM, Woode DR, Hilakivi-Clarke L, Clarke R. Influence of berry polyphenols on receptor signaling and cell-death pathways: implications for breast cancer prevention. J Agri Food Chem. 2012;60:5693–5708. doi: 10.1021/jf204084f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maioli E, Greci L, Soucek K, Hyzdalova M, Pecorelli A, Fortino V, et al. Rottlerin inhibits ROS formation and prevents NFkappaB activation in MCF-7 and HT-29 cells. J Biomed Biotech. 2009;2009:742936. doi: 10.1155/2009/742936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh A, Gukovskaya AS, Nieto JM, Cheng JH, Gukovsky I, Reeve JR, Jr, et al. PKC-delta and -epsilon regulate NF-kappaB activation induced by cholecystokinin and TNF-alpha in pancreatic acinar cells. Am J Physiol Gastrointest Liver Physiol. 2004;287:G582–G591. doi: 10.1152/ajpgi.00087.2004. [DOI] [PubMed] [Google Scholar]
- Mann AP, Verma A, Sethi G, Manavathi B, Wang H, Fok JY, et al. Overexpression of tissue transglutaminase leads to constitutive activation of nuclear factor-kappaB in cancer cells: delineation of a novel pathway. Cancer Res. 2006;66:8788–8795. doi: 10.1158/0008-5472.CAN-06-1457. [DOI] [PubMed] [Google Scholar]
- Fabre C, Carvalho G, Tasdemir E, Braun T, Ades L, Grosjean J, et al. NF-kappaB inhibition sensitizes to starvation-induced cell death in high-risk myelodysplastic syndrome and acute myeloid leukemia. Oncogene. 2007;26:4071–4083. doi: 10.1038/sj.onc.1210187. [DOI] [PubMed] [Google Scholar]
- Soltoff SP. Rottlerin: an inappropriate and ineffective inhibitor of PKCdelta. Trends PharmacolSci. 2007;28:453–458. doi: 10.1016/j.tips.2007.07.003. [DOI] [PubMed] [Google Scholar]
- Song KS, Kim JS, Yun EJ, Kim YR, Seo KS, Park JH, et al. Rottlerin induces autophagy and apoptotic cell death through a PKC-delta-independent pathway in HT1080 human fibrosarcoma cells: the protective role of autophagy in apoptosis. Autophagy. 2008;4:650–658. doi: 10.4161/auto.6057. [DOI] [PubMed] [Google Scholar]
- Balgi AD, Fonseca BD, Donohue E, Tsang TC, Lajoie P, Proud CG, et al. Screen for chemical modulators of autophagy reveals novel therapeutic inhibitors of mTORC1 signaling. PLoS One. 2009;4:e7124. doi: 10.1371/journal.pone.0007124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cacicedo JM, Yagihashi N, Keaney JF, Jr, Ruderman NB, Ido Y. AMPK inhibits fatty acid-induced increases in NF-kappaB transactivation in cultured human umbilical vein endothelial cells. Biochem Biophys Res Commun. 2004;324:1204–1209. doi: 10.1016/j.bbrc.2004.09.177. [DOI] [PubMed] [Google Scholar]
- Dijkers PF, Medema RH, Pals C, Banerji L, Thomas NS, Lam EW, et al. Forkhead transcription factor FKHR-L1 modulates cytokine-dependent transcriptional regulation of p27(KIP1) Mol Cell Biol. 2000;20:9138–9148. doi: 10.1128/mcb.20.24.9138-9148.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu QY, Zhang L, Lugea A, Moro A, Edderkaoui M, Eibl G, et al. determination of rottlerin, a natural protein kinases c inhibitor, in pancreatic cancer cells and mouse xenografts by rp-hplc method. J Chromatogr Sep Tech. 2013;4:1. [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Li W, Liu F. Inhibition of proliferation and induction of apoptosis by genistein in colon cancer HT-29 cells. Cancer Lett. 2004;215:159–166. doi: 10.1016/j.canlet.2004.06.010. [DOI] [PubMed] [Google Scholar]
- Gossner G, Choi M, Tan L, Fogoros S, Griffith KA, Kuenker M, et al. Genistein-induced apoptosis and autophagocytosis in ovarian cancer cells. Gynecol Oncol. 2007;105:23–30. doi: 10.1016/j.ygyno.2006.11.009. [DOI] [PubMed] [Google Scholar]
- Christian F, Anthony DF, Vadrevu S, Riddell T, Day JP, McLeod R, et al. p62 (SQSTM1) and cyclic AMP phosphodiesterase-4A4 (PDE4A4) locate to a novel, reversible protein aggregate with links to autophagy and proteasome degradation pathways. Cell Signal. 2010;22:1576–1596. doi: 10.1016/j.cellsig.2010.06.003. [DOI] [PubMed] [Google Scholar]
- Houslay MD. p62 (SQSTM1) forms part of a novel, reversible aggregate containing a specific conformer of the cAMP degrading phosphodiesterase, PDE4A4. Autophagy. 2010;6:1198–1200. doi: 10.4161/auto.6.8.13479. [DOI] [PubMed] [Google Scholar]
- Ali AB, Nin DS, Tam J, Khan M. Role of chaperone mediated autophagy (CMA) in the degradation of misfolded N-CoR protein in non-small cell lung cancer (NSCLC) cells. PLoS One. 2011;6:e25268. doi: 10.1371/journal.pone.0025268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Kulkarni K, Zhu W, Hu M. Bioavailability and pharmacokinetics of genistein: mechanistic studies on its ADME. Anticancer Agents Med Chem. 2012;12:1264–1280. doi: 10.2174/187152012803833107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avila MA, Velasco JA, Cansado J, Notario V. Quercetin mediates the down-regulation of mutant p53 in the human breast cancer cell line MDA-MB468. Cancer Res. 1994;54:2424–2428. [PubMed] [Google Scholar]
- Stavric B. Quercetin in our diet: from potent mutagen to probable anticarcinogen. Clin Biochem. 1994;27:245–248. doi: 10.1016/0009-9120(94)90025-6. [DOI] [PubMed] [Google Scholar]
- Ranelletti FO, Maggiano N, Serra FG, Ricci R, Larocca LM, Lanza P, et al. Quercetin inhibits p21-RAS expression in human colon cancer cell lines and in primary colorectal tumors. Int J Cancer. 2000;85:438–445. [PubMed] [Google Scholar]
- Wang K, Liu R, Li J, Mao J, Lei Y, Wu J, et al. Quercetin induces protective autophagy in gastric cancer cells: involvement of Akt-mTOR- and hypoxia-induced factor 1alpha-mediated signaling. Autophagy. 2011;7:966–978. doi: 10.4161/auto.7.9.15863. [DOI] [PubMed] [Google Scholar]
- Martinez-Outschoorn UE, Balliet RM, Rivadeneira DB, Chiavarina B, Pavlides S, Wang C, et al. Oxidative stress in cancer associated fibroblasts drives tumor-stroma co-evolution: A new paradigm for understanding tumor metabolism, the field effect and genomic instability in cancer cells. Cell Cycle. 2010;9:3256–3276. doi: 10.4161/cc.9.16.12553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klappan AK, Hones S, Mylonas I, Bruning A. Proteasome inhibition by quercetin triggers macroautophagy and blocks mTOR activity. Histochem Cell Biol. 2012;137:25–36. doi: 10.1007/s00418-011-0869-0. [DOI] [PubMed] [Google Scholar]
- Li S, Zhou Y, Fan J, Cao S, Cao T, Huang F, et al. Heat shock protein 72 enhances autophagy as a protective mechanism in lipopolysaccharide-induced peritonitis in rats. Am J Pathol. 2011;179:2822–2834. doi: 10.1016/j.ajpath.2011.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CY, Hsiu S. L, Wen K. C, Lin S. P, Hou S. Y, Chao Y.C, et al. Bioavailability and metabolic pharmacokinetics of Rutin and Quercetin in rats. J Food Drug Anal. 2005;13:244–250. [Google Scholar]
- Aggarwal BB, Kumar A, Bharti AC. Anticancer potential of curcumin: preclinical and clinical studies. Anticancer Res. 2003;23 (1A:363–398. [PubMed] [Google Scholar]
- Aoki H, Takada Y, Kondo S, Sawaya R, Aggarwal BB, Kondo Y. Evidence that curcumin suppresses the growth of malignant gliomas in vitro and in vivo through induction of autophagy: role of Akt and extracellular signal-regulated kinase signaling pathways. Mol Pharmacol. 2007;72:29–39. doi: 10.1124/mol.106.033167. [DOI] [PubMed] [Google Scholar]
- Zhuang W, Long L, Zheng B, Ji W, Yang N, Zhang Q, et al. Curcumin promotes differentiation of glioma-initiating cells by inducing autophagy. Cancer Sci. 2012;103:684–690. doi: 10.1111/j.1349-7006.2011.02198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadalapaka G, Jutooru I, Burghardt R, Safe S. Drugs that target specificity proteins downregulate epidermal growth factor receptor in bladder cancer cells. Mol Cancer Res. 2010;8:739–750. doi: 10.1158/1541-7786.MCR-09-0493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Pan XY, Xu Y, Xiao Y, An Y, Tie L, et al. Curcumin induces autophagy to protect vascular endothelial cell survival from oxidative stress damage. Autophagy. 2012;8:5. doi: 10.4161/auto.19471. [DOI] [PubMed] [Google Scholar]
- Yamauchi Y, Izumi Y, Asakura K, Hayashi Y, Nomori H. Curcumin Induces Autophagy in ACC-MESO-1 Cells. Phytother Res. 2012;26:1779–1783. doi: 10.1002/ptr.4645. [DOI] [PubMed] [Google Scholar]
- Mosieniak G, Adamowicz M, Alster O, Jaskowiak H, Szczepankiewicz AA, Wilczynski GM, et al. Curcumin induces permanent growth arrest of human colon cancer cells: Link between senescence and autophagy. Mech Age Dev. 2012;133:444–455. doi: 10.1016/j.mad.2012.05.004. [DOI] [PubMed] [Google Scholar]
- Jia YL, Li J, Qin ZH, Liang ZQ. Autophagic and apoptotic mechanisms of curcumin-induced death in K562 cells. J Asian Nat Prod Res. 2009;11:918–928. doi: 10.1080/10286020903264077. [DOI] [PubMed] [Google Scholar]
- Teiten MH, Gaascht F, Cronauer M, Henry E, Dicato M, Diederich M. Anti-proliferative potential of curcumin in androgen-dependent prostate cancer cells occurs through modulation of the Wingless signaling pathway. Int J Oncology. 2011;38:603–611. doi: 10.3892/ijo.2011.905. [DOI] [PubMed] [Google Scholar]
- Lee YJ, Kim NY, Suh YA, Lee C. Involvement of ROS in curcumin-induced autophagic cell death. Korean J Physiol Pharmacol. 2011;15:1–7. doi: 10.4196/kjpp.2011.15.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JY, Cho TJ, Woo BH, Choi KU, Lee CH, Ryu MH, et al. Curcumin-induced autophagy contributes to the decreased survival of oral cancer cells. Arch Oral Biol. 2012;57:1018–1025. doi: 10.1016/j.archoralbio.2012.04.005. [DOI] [PubMed] [Google Scholar]
- Hasima N, Aggarwal BB. Cancer-linked targets modulated by curcumin. Int J Biochem Mol Biol. 2012;3:328–351. [PMC free article] [PubMed] [Google Scholar]
- Prasad S, Tyagi AK, Aggarwal BB. Recent developments in delivery, bioavailability, absorption and metabolism of curcumin: the golden pigment from golden spice. Cancer Res Treat. 2014;46:2–18. doi: 10.4143/crt.2014.46.1.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kueck A, Opipari AW, Jr., Griffith KA, Tan L, Choi M, Huang J, et al. Resveratrol inhibits glucose metabolism in human ovarian cancer cells. Gynecol Oncol. 2007;107:450–457. doi: 10.1016/j.ygyno.2007.07.065. [DOI] [PubMed] [Google Scholar]
- Bowers JL, Tyulmenkov VV, Jernigan SC, Klinge CM. Resveratrol acts as a mixed agonist/antagonist for estrogen receptors alpha and beta. Endocrinology. 2000;141:3657–3667. doi: 10.1210/endo.141.10.7721. [DOI] [PubMed] [Google Scholar]
- Ohshiro K, Rayala SK, Kondo S, Gaur A, Vadlamudi RK, El-Naggar AK, et al. Identifying the estrogen receptor coactivator PELP1 in autophagosomes. Cancer Res. 2007;67:8164–8171. doi: 10.1158/0008-5472.CAN-07-0038. [DOI] [PubMed] [Google Scholar]
- Ohshiro K, Rayala SK, El-Naggar AK, Kumar R. Delivery of cytoplasmic proteins to autophagosomes. Autophagy. 2008;4:104–106. doi: 10.4161/auto.5223. [DOI] [PubMed] [Google Scholar]
- Trincheri NF, Follo C, Nicotra G, Peracchio C, Castino R, Isidoro C. Resveratrol-induced apoptosis depends on the lipid kinase activity of Vps34 and on the formation of autophagolysosomes. Carcinogenesis. 2008;29:381–389. doi: 10.1093/carcin/bgm271. [DOI] [PubMed] [Google Scholar]
- Miki H, Uehara N, Kimura A, Sasaki T, Yuri T, Yoshizawa K, et al. Resveratrol induces apoptosis via ROS-triggered autophagy in human colon cancer cells. Int J Oncol. 2012;40:1020–1028. doi: 10.3892/ijo.2012.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarlatti F, Maffei R, Beau I, Codogno P, Ghidoni R. Role of non-canonical Beclin 1-independent autophagy in cell death induced by resveratrol in human breast cancer cells. Cell Death Differ. 2008;15:1318–1329. doi: 10.1038/cdd.2008.51. [DOI] [PubMed] [Google Scholar]
- Hsu KF, Wu CL, Huang SC, Wu CM, Hsiao JR, Yo YT, et al. Cathepsin L mediates resveratrol-induced autophagy and apoptotic cell death in cervical cancer cells. Autophagy. 2009;5:451–460. doi: 10.4161/auto.5.4.7666. [DOI] [PubMed] [Google Scholar]
- Signorelli P, Munoz-Olaya JM, Gagliostro V, Casas J, Ghidoni R, Fabrias G. Dihydroceramide intracellular increase in response to resveratrol treatment mediates autophagy in gastric cancer cells. Cancer Lett. 2009;282:238–243. doi: 10.1016/j.canlet.2009.03.020. [DOI] [PubMed] [Google Scholar]
- Li J, Qin Z, Liang Z. The prosurvival role of autophagy in Resveratrol-induced cytotoxicity in human U251 glioma cells. BMC Cancer. 2009;9:215. doi: 10.1186/1471-2407-9-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armour SM, Baur JA, Hsieh SN, Land-Bracha A, Thomas SM, Sinclair DA. Inhibition of mammalian S6 kinase by resveratrol suppresses autophagy. Aging (Albany NY) 2009;1:515–528. doi: 10.18632/aging.100056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S, Yao H, Caito S, Hwang JW, Arunachalam G, Rahman I. Regulation of SIRT1 in cellular functions: role of polyphenols. Arch Biochem Biophys. 2010;501:79–90. doi: 10.1016/j.abb.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang JW, Chung S, Sundar IK, Yao H, Arunachalam G, McBurney MW, et al. Cigarette smoke-induced autophagy is regulated by SIRT1-PARP-1-dependent mechanism: implication in pathogenesis of COPD. Arch Biochem Biophys. 2010;500:203–209. doi: 10.1016/j.abb.2010.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caito S, Hwang JW, Chung S, Yao H, Sundar IK, Rahman I. PARP-1 inhibition does not restore oxidant-mediated reduction in SIRT1 activity. Biochem Biophys Res Commun. 2010;392:264–270. doi: 10.1016/j.bbrc.2009.12.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morselli E, Marino G, Bennetzen MV, Eisenberg T, Megalou E, Schroeder S, et al. Spermidine and resveratrol induce autophagy by distinct pathways converging on the acetylproteome. J Cell Biol. 2011;192:615–629. doi: 10.1083/jcb.201008167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Lowry SF, Guarente L, Haimovich B. Roles of SIRT1 in the acute and restorative phases following induction of inflammation. J Biol Chem. 2010;285:41391–41401. doi: 10.1074/jbc.M110.174482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen TC, Hung YC, Lin TY, Chang HW, Chiang IP, Chen YY, et al. Human papillomavirus infection and expression of ATPase family AAA domain containing 3A, a novel anti-autophagy factor, in uterine cervical cancer. Int J Mol Med. 2011;28:689–696. doi: 10.3892/ijmm.2011.743. [DOI] [PubMed] [Google Scholar]
- Liao PC, Ng LT, Lin LT, Richardson CD, Wang GH, Lin CC. Resveratrol arrests cell cycle and induces apoptosis in human hepatocellular carcinoma Huh-7 cells. J Med Food. 2010;13:1415–1423. doi: 10.1089/jmf.2010.1126. [DOI] [PubMed] [Google Scholar]
- Filippi-Chiela EC, Villodre ES, Zamin LL, Lenz G. Autophagy interplay with apoptosis and cell cycle regulation in the growth inhibiting effect of resveratrol in glioma cells. PLoS One. 2011;6:e20849. doi: 10.1371/journal.pone.0020849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proikas-Cezanne T, Codogno P. Beclin 1 or not Beclin 1. Autophagy. 2011;7:671–672. doi: 10.4161/auto.7.7.14877. [DOI] [PubMed] [Google Scholar]
- Mauthe M, Jacob A, Freiberger S, Hentschel K, Stierhof YD, Codogno P, et al. Resveratrol-mediated autophagy requires WIPI-1-regulated LC3 lipidation in the absence of induced phagophore formation. Autophagy. 2011;7:1448–1461. doi: 10.4161/auto.7.12.17802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Back JH, Zhu Y, Calabro A, Queenan C, Kim AS, Arbesman J, et al. Resveratrol-Mediated Downregulation of Rictor Attenuates Autophagic Process and Suppresses UV-Induced Skin Carcinogenesis(dagger) Photochem Photobiol. 2012;88:1165–1172. doi: 10.1111/j.1751-1097.2012.01097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel E, Somoza V. Metabolism and bioavailability of trans-resveratrol. Mol Nutr Food Res. 2005;49:472–481. doi: 10.1002/mnfr.200500010. [DOI] [PubMed] [Google Scholar]
- Pan W, Yang H, Cao C, Song X, Wallin B, Kivlin R, et al. AMPK mediates curcumin-induced cell death in CaOV3 ovarian cancer cells. Oncol Rep. 2008;20:1553–1559. [PubMed] [Google Scholar]
- Scarlatti F, Maffei R, Beau I, Codogno P, Ghidoni R. Role of non-canonical Beclin 1-independent autophagy in cell death induced by resveratrol in human breast cancer cells. Cell Death Different. 2008;15:1318–1329. doi: 10.1038/cdd.2008.51. [DOI] [PubMed] [Google Scholar]
- Puissant A, Auberger P. AMPK - and p62/SQSTM1-dependent autophagy mediate Resveratrol-induced cell death in chronic myelogenous leukemia. Autophagy. 2010;6:5. doi: 10.4161/auto.6.5.12126. [DOI] [PubMed] [Google Scholar]