Abstract
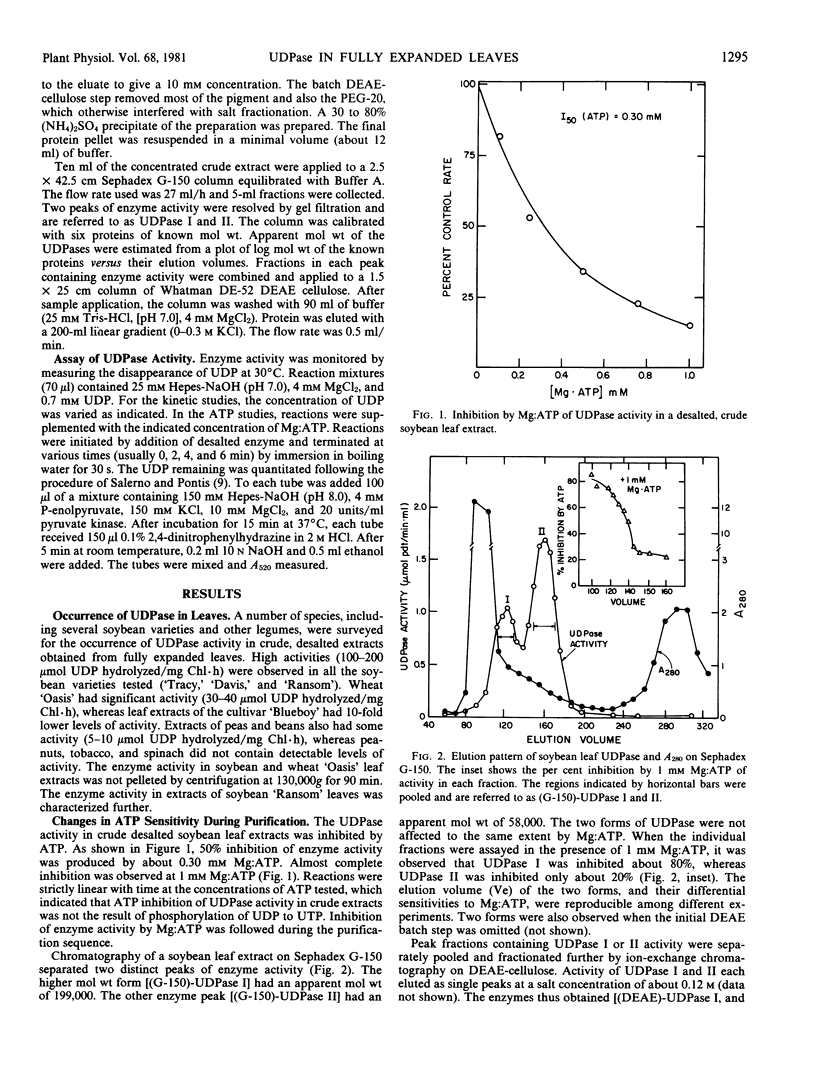
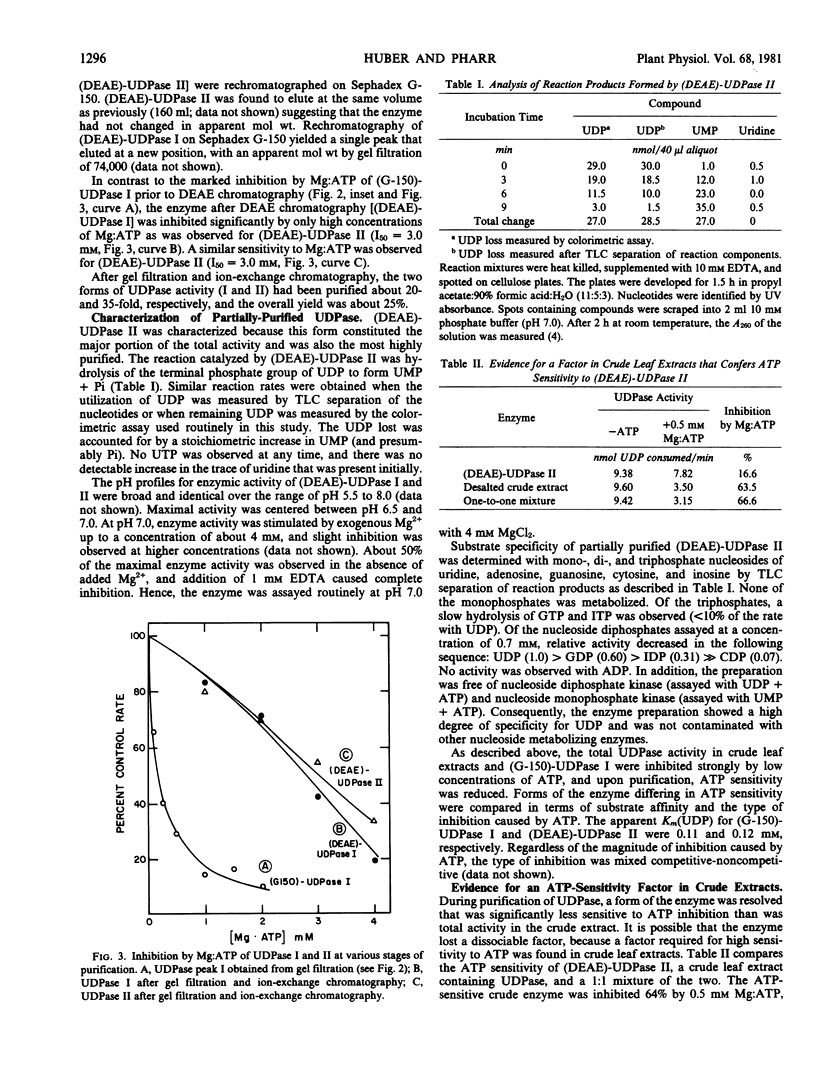
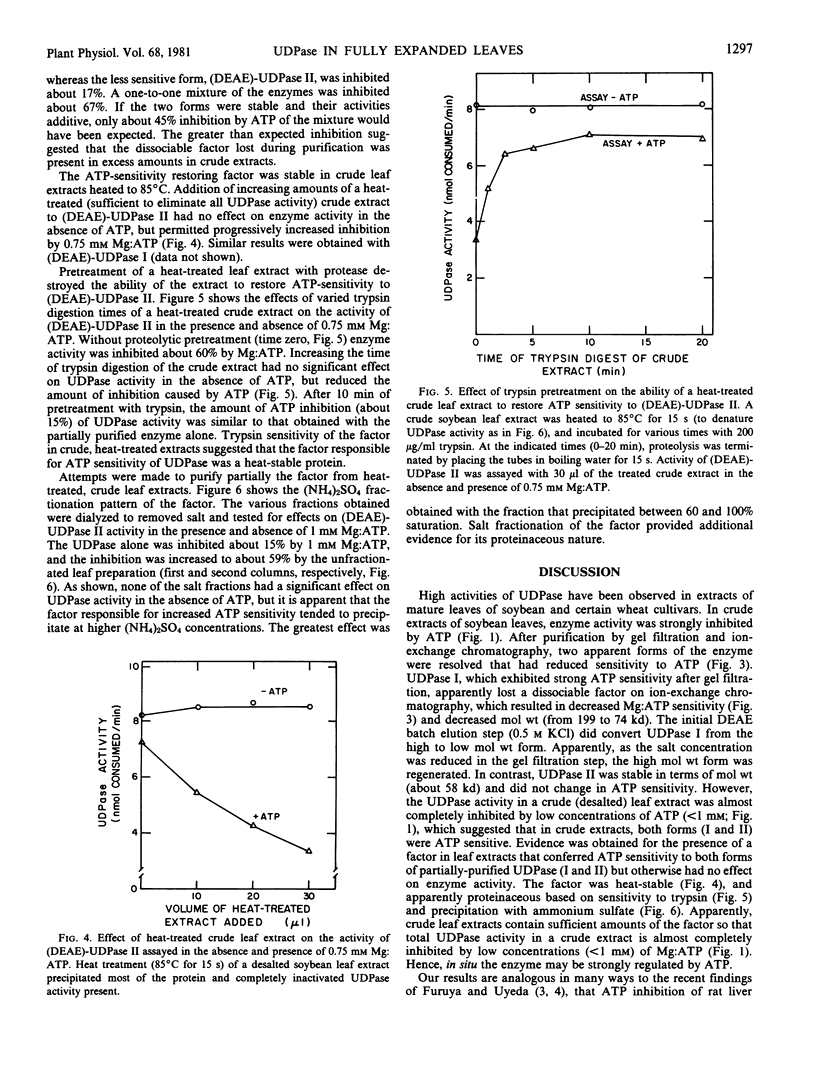
High activities (100-200 micromoles UDP hydrolyzed per milligram chlorophyll per hour) of uridine-5′ diphosphatase (UDPase) have been identified in extracts of fully expanded soybean (Glycine max Merr.) leaves. In desalted crude extracts, UDPase activity was strongly inhibited by low concentrations of Mg:ATP (I50 = 0.3 millimolar). Two forms of the enzyme were resolved by gel filtration on Sephadex G-150. The higher molecular weight form (UDPase I, about 199 kilodaltons by gel filtration) retained ATP sensitivity (I50 = 0.3 millimolar), whereas the major, lower molecular weight form (UDPase II, about 58 kilodaltons) was markedly less sensitive to ATP inhibition (I50 = 2.7-3.0 millimolar). Subsequent purification of UDPase I by ion-exchange chromatography on DEAE cellulose produced a lower molecular weight enzyme (about 74 kilodaltons by gel filtration) that had reduced ATP sensitivity similar to UDPase II. Ion-exchange chromatography of UDPase II did not alter molecular weight or ATP sensitivity. UDPase II, after the DEAE-cellulose step, was specific for nucleoside diphosphates. Maximum reaction velocity decreased in the following sequence; UDP > GDP > CDP. ADP was not a substrate for the enzyme. The reaction catalyzed was hydrolysis of the terminal-P of UDP to form UMP. The enzyme was stimulated by Mg2+ and the pH optimum was centered between pH 6.5 and 7.0. In a survey of various species, soybean cultivars had highest activities of apparent UDPase and other species ranged in apparent activity from 0 to 30 micromoles hydrolyzed per milligram chlorophyll per hour.
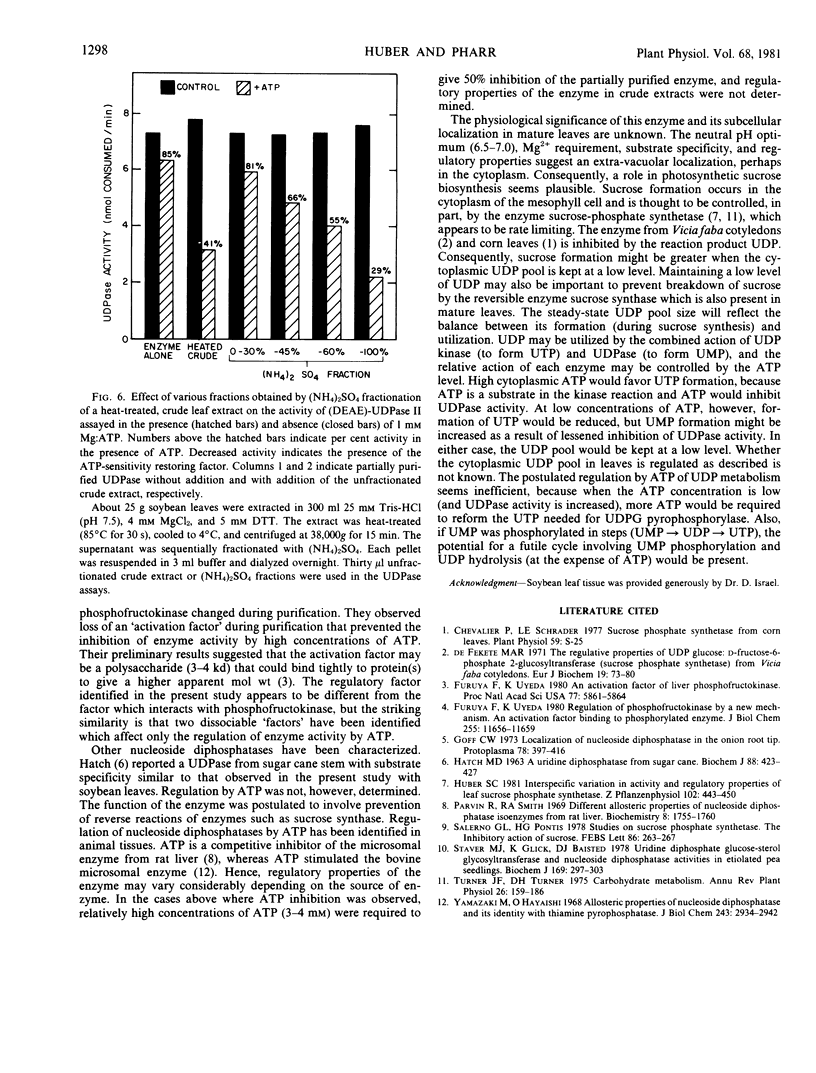
A heat-stable proteinaceous factor was identified in desalted crude leaf extracts that increased ATP sensitivity of the partially purified enzyme. Apparently, during purification a dissociable factor, that is required for maximum sensitivity to low concentrations (<1 millimolar) of Mg:ATP, is lost from the enzyme.
Full text
PDF
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Furuya E., Uyeda K. An activation factor of liver phosphofructokinase. Proc Natl Acad Sci U S A. 1980 Oct;77(10):5861–5864. doi: 10.1073/pnas.77.10.5861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuya E., Uyeda K. Regulation of phosphofructokinase by a new mechanism. An activation factor binding to phosphorylated enzyme. J Biol Chem. 1980 Dec 25;255(24):11656–11659. [PubMed] [Google Scholar]
- Goff C. W. Localization of nucleoside diphosphatase in the onion root tip. Protoplasma. 1973;78(4):397–416. doi: 10.1007/BF01275775. [DOI] [PubMed] [Google Scholar]
- Hatch M. D. A uridine diphosphatase from sugar cane. Biochem J. 1963 Sep;88(3):423–427. doi: 10.1042/bj0880423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvin R., Smith R. A. Different allosteric properties of nucleoside diphosphatase isoenzymes from rat liver. Biochemistry. 1969 Apr;8(4):1755–1760. doi: 10.1021/bi00832a059. [DOI] [PubMed] [Google Scholar]
- Salerno G. L., Pontis H. G. Studies on sucrose phosphate synthetase. The inhibitory action of sucrose. FEBS Lett. 1978 Feb 15;86(2):263–267. doi: 10.1016/0014-5793(78)80576-6. [DOI] [PubMed] [Google Scholar]
- Staver M. J., Glick K., Baisted D. J. Uridine diphosphate glucose-sterol glucosyltransferase and nucleoside diphosphatase activities in etiolated pea seedlings. Biochem J. 1978 Feb 1;169(2):297–303. doi: 10.1042/bj1690297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki M., Hayaishi O. Allosteric properties of nucleoside diphosphatase and its identity with thiamine pyrophosphatase. J Biol Chem. 1968 Jun 10;243(11):2934–2942. [PubMed] [Google Scholar]
- de Fekete M. A. The regulative properties of UDPglucose: D-fructose-6-phosphate 2-glucosyltransferase (sucrose phosphate synthetase) from Vicia faba cotyledons. Eur J Biochem. 1971 Mar 1;19(1):73–80. doi: 10.1111/j.1432-1033.1971.tb01289.x. [DOI] [PubMed] [Google Scholar]


