Abstract
CD8 T cells specific for islet autoantigens are major effectors of β cell damage in type 1 diabetes, and measurement of their number and functional characteristics in blood represent potentially important disease biomarkers. CD8 T cell reactivity against glutamic acid decarboxylase 65 (GAD65) in HLA-A*0201 subjects has been reported to focus on an immunogenic region 114–123 (VMNILLQYVV), with studies demonstrating both 114–123 and 114–122 epitopes being targeted. However, the fine specificity of this response is unclear and the key question as to which epitope(s) β cells naturally process and present and, therefore, the pathogenic potential of CD8 T cells with different specificities within this region has not been addressed. We generated human leucocyte antigen (HLA)-A*0201-restricted CD8 T cell clones recognizing either 114–122 alone or both 114–122 and 114–123. Both clone types show potent and comparable effector functions (cytokine and chemokine secretion) and killing of indicator target cells externally pulsed with cognate peptide. However, only clones recognizing 114–123 kill target cells transfected with HLA-A*0201 and GAD2 and HLA-A*0201+ human islet cells. We conclude that the endogenous pathway of antigen processing by HLA-A*0201-expressing cells generates GAD65114–123 as the predominant epitope in this region. These studies highlight the importance of understanding β cell epitope presentation in the design of immune monitoring for potentially pathogenic CD8 T cells.
Keywords: autoimmunity, CD8 T cell clones, GAD65, peptide-processing, type 1 diabetes
Introduction
Type 1 diabetes is a prototypical autoimmune disease involving the specific targeting and destruction of β cells within the islets of Langerhans. The autoimmune processes involved are becoming better defined [1], and there is strong evidence that CD8 T cells play a key role [2–7]. For example, CD8 T cells are the predominant leucocyte present in islet infiltrates close to diagnosis [8,9]. In addition, CD8 T cell clones recognizing preproinsulin peptides kill β cells in vitro and CD8 T cells with the same epitope specificity can be detected in infiltrated islets in patients studied close to diagnosis [3,4,8]. Finally, several studies link disease susceptibility to inheritance of human leucocyte antigen class I (HLAI) genes, with odds ratios second only to those for HLAII genes [10]. Notably, susceptibility is linked to HLA-A*2402 and HLA-B*3906, and while multiple studies have examined HLA-A*0201 subjects, a prevalent allele among European and worldwide populations [11], it is not actually disease-linked [12].
Given this background, and the increasing number of β cell epitopes being reported as targets of autoreactive CD8 T cells [13,14], there is a strong rationale for focusing on these effector cells in the peripheral blood as potential biomarkers of immune-mediated damage. Indeed, recent proof-of-concept for this strategy in the setting of an immunotherapy trial has now been gained [15].
Several technological advances have been highly enabling for this biomarker strategy. These include peptide elution studies [3,4], the wide availability of peptide binding algorithms for common HLAI molecules [16] and the creation of mice transgenic for human HLAI genes [17], each of which facilitates epitope discovery. These have been allied with the development of highly sensitive assays to examine antigen-specific CD8 T cell number (peptide–HLA multimers) [18] and functional responses (enzyme-linked immunoassays) [4]. However, as we have discussed previously [19,20], it is important to appreciate the imperfections in these approaches. Notably, peptides identified via algorithms may bind HLAI and stimulate CD8 T cell responses, but this does not provide evidence that they are generated via endogenous and/or cross-presentation pathways of antigen processing, or that they are displayed on the surface of β cells. In the absence of these features, the disease relevance of epitopes and the CD8 T cells targeting them remains unclear.
In the present study, we focused on CD8 T cell responses targeting glutamic acid decarboxylase (GAD65), a major autoantigen in type 1 diabetes. In a groundbreaking 1995 study, a potentially important, HLA-A*0201-restricted 10-mer peptide of glutamic acid decarboxylase (GAD65) (VMNILLQYVV, representing residues 114–123) was identified as a CD8 T cell target [21]. Short-term T cell lines from patients with type 1 diabetes generated against GAD65114–123 were capable of killing target cell lines (lymphoblastoid B lymphocytes) transfected with GAD2. Although this study provided the first evidence of CD8 T cell targeting directed against any β cell autoantigen, it fell short of demonstrating the pathogenic (i.e. β cell destructive) potential of these responses or their disease specificity. Subsequent studies have highlighted the potential disease association of CD8 T cell responses to both GAD65114–123 and a nested 9-mer peptide, GAD65114–122 [22–25]. The 9-mer peptide was selected for study because of its higher predicted HLA-A*0201 binding than the 10-mer, and because of difficulties encountered in refolding 10-mer-HLA-A*0201 for use in peptide–HLA multimers [22,23]. Given the findings of our recent study, namely that any given αβ T cell receptor exhibits an explicit preference for a single HLAI-peptide length [26], we considered it important to clarify the nature of the cytotoxic CD8 T cell response to the peptides nested in this immunogenic region of GAD65. Specifically, in the present study, we sought to establish whether human β cells process endogenously and present both or either of these target peptides to cytotoxic CD8 T cells, and thereby provide support for the rationale for studying these reactivities ex vivo.
Materials and methods
Cell lines
C1R and K562 transfected with HLA-A*0201 (designated C1R-A2 and K562-A2) were maintained in RPMI-1640 (Gibco, Life Technologies, Paisley, UK) supplemented with 10% fetal calf serum (FCS), 100 IU/ml penicillin and 100 μg/ml streptomycin (Invitrogen, Life Technologies, Carlsbad, CA, USA). GAD65-target cells were generated by cloning GAD2 (Accession number: BC126327; Genome Cube, Source Bioscience, Nottingham, UK) into the retroviral plasmid MigR1-eGFP (gift from Nathan Sherer, Wisconsin University). MigR1-GAD65-eGFP, VSVG plasmid (vesicular stomatitis virus G protein) and cytosine–phosphate–guanosine (CpG) plasmid (encoding gag and pol proteins) were transfected into 293T cells using polyethylenimine (PEI; Sigma-Aldrich, Gillingham, Dorset, UK) to produce non-replicative retrovirus particles (NRRPs). The K562-A2 target cell line was spinoculated at 300 g for 2-h with GAD2-containing NRRPs, and after 48 h GFP-positive clones were sorted and analysed by Western blot for protein expression. Previously generated preproinsulin15–24-specific cloned CD8 T cells (3F2) were used as a positive control in functional assays [4].
GAD65-specific CD8 T cell clone generation and isolation
GAD65-specific CD8 T cell clones were generated using our previously described method and patient collections [4,27]. Cell lines were maintained in X-Vivo 15/5% AB serum/interleukin (IL)-7 (10 ng/m), IL-15 (0·1 ng/ml; Peprotech, Rocky Hill, NJ, USA) and 2·5% Cellkine (ZeptoMetrix, Buffalo, NY, USA). Single-cell clones were isolated by tetramer-guided sorting into wells containing irradiated feeder peripheral blood mononuclear cells (PBMCs) and phytohaemagglutinin (PHA)-M (500 μg/ml; Sigma Aldrich).
Tetramer staining and functional responses of cloned CD8 T cells
Tetramer staining of isolated CD8 T cell clones and measurement of specific human islet-specific cytotoxicity were measured as described previously [28]. Peptide pulsing (10 μg/ml) of target cells was performed for 1 h at 37°C followed by washing off peptide excess. Cytokines and chemokines secreted during 18-h cultures of 2 × 104 CD8 T cell clones in 96-well plates containing 2 × 104 target cells were measured by Luminex (Milliplex Mag Kit; Millipore, Darmstadt, Germany). Intracellular cytokine analysis was performed on 18-h cultures of 2 × 105 CD8 T cell clones and 2 × 105 target cells, using Cytoperm/Cytofix (BD Biosciences, San Jose, CA, USA). ViViD was used to distinguish live populations, co-stained with anti-CD3 and anti-CD8 antibodies. Under identical conditions, degranulation (as a surrogate for cytotoxicity) was examined over 4 h using fluorochrome-conjugated monoclonal anti-CD107a antibody. Non-peptide pulsed target cells, or cells pulsed with an irrelevant peptide, islet-specific glucose-6-phosphatase catalytic subunit-related protein (IGRP265–273), were used as negative controls and ionomycin/phorbol myristate acetate (PMA) as positive controls (1 μg/ml and 50 ng/ml, respectively).
Measuring the thermal stability of HLA-A2–GAD peptide complexes
Thermal stability of HLA-A2 complexes was assessed by circular dichroism (CD) spectroscopy monitoring the change of ellipticities Θ at 218 nm where the CD spectra exhibit a minimum. Data were collected on an Aviv Model 215 spectropolarimeter (Aviv Biomedical Inc., Lakewood, NJ, USA) equipped with a Peltier thermostatted cell holder using a 0·1-cm quartz cell. Proteins were dissolved in 75 mM NaCl, 20 mM PO4−, pH 7·5, at concentrations of 5–7 μM as determined by absorbance at 280 nm using extinction coefficients calculated from the amino acid composition [29]. Melting curves were recorded in 0·5°C intervals from 4°C up to a maximum temperature, when protein aggregation was observed with settings resulting in an average heating rate of ca. 30°C/h. Values were corrected to a calibration curve recorded with the temperature measured in the cell. After subtraction of a buffer baseline, Θ-values were converted to molar ellipticities [Θ]MRW = Θ × MRW/(c × d), where MRW denotes the mean residue weight Mr/(n–1) with n indicating the number of amino acid residues, c the concentration in mg/ml and d the cell path length in mm.
Melting curves were analysed assuming a two-state trimer-to-monomer transition from the native (N) to unfolded (U) conformation N3 ↔ 3U with an equilibrium constant K = [U]3/[N3] = F/[3c2 (1-F)3], where F and c are the degree of folding and protein concentration, respectively. Data were fitted as described [30] using the non-linear least-squares routine of Origin version 7·5 (OriginLab, Northampton, MA, USA). Fitted parameters were the melting temperature Tm, at which 50% of proteins are in the folded and unfolded state, van't Hoff's enthalpy ΔHvH at the transition mid-point and the slope and Θ-intercept of the native baseline assumed as a linear function of the temperature. As all protein complexes aggregated to various degrees upon unfolding, the ellipticity of the unfolded state was set as a constant of −4500 deg cm2 dmol−1 [31]. For all peptides, the coefficient of determination for fitted curves versus measurements was r2 > 0·99.
Results
Generation of GAD65-specific CD8 T cell clones
We successfully isolated and expanded a series of GAD65-specific CD8 T cell clones (Supporting Information, Fig. S1). Two of these are used here to exemplify the specificities we observed and to address processing and presentation of GAD65 by human HLA-A*0201 β cells.
As shown in Fig. 1, we used tetramers to identify clone cells with specificity for 9-mer peptide (two independent clones generated from a single donor: data shown for RK9P9-3; HLA-A*0201–GAD65114–122 tetramer-positive); and clones with specificity that enabled binding to both 9-mer and 10-mer peptide (two independent clones generated from a further donor; data shown for RK9C10-1, HLA-A*0201–GAD65114–122 and HLA-A*0201–GAD65114–123 tetramer-positive). The recognition pattern of these clones for these peptide–HLA-A*0201 complexes was confirmed by measuring tumour necrosis factor (TNF)-α production and degranulation in 18- and 4-h co-cultures, respectively, with peptide-pulsed C1R-A2 target cells. RK9P9-3 produces TNF-α and degranulates when co-cultured with target cells pulsed with 9-mer peptide GAD65114–122 to levels comparable to those achieved with the positive control. In contrast, responses to 10-mer peptide GAD65114–123 are similar to background. By comparison, RK9C10-1 produces TNF-α and degranulates in the presence of both 9-mer (GAD65114–122) and 10-mer (GAD65114–123) pulsed target cells (Fig. 2a,b). Final confirmation that effector function of these clones was restricted in this specific fashion was derived by measuring killing of peptide-pulsed K562-A2 target cells (Fig. 2c). Thus we derived and characterized two CD8 T cell clones specific for the immunogenic region 114–123 region of GAD65. These clones offer distinct specificities that enable their use in examining natural processing and presentation of GAD65. A difference in the level of response by clone RK9C10-1 was observed in assays measuring either cytokine production or killing, in conditions using either GAD9-mer- or GAD10-mer-expressing target cells (Fig. 2). One explanation for the reduced response to 10-mer-expressing targets, compared to 9-mer-pulsed cells, would be reduced stability of 10-mer-bound HLA-A2 complexes, which could contribute to a reduced activation signal. To determine empirically in-silico prediction algorithms reporting weaker binding of HLA-A*0201 by GAD10-mer [32,33], circular dichroism was performed on HLA-A2–GAD10-mer and HLA-A2–GAD9-mer complexes to assess their respective thermostability (Supporting Information, Fig. S2). The results show that GAD9-mer peptide forms a more stable complex with HLA-A2 than GAD10-mer, as it requires more energy to become denatured. The observed difference in GAD–peptide–HLA stability therefore provides an explanation for the different quality of response observed.
Fig. 1.
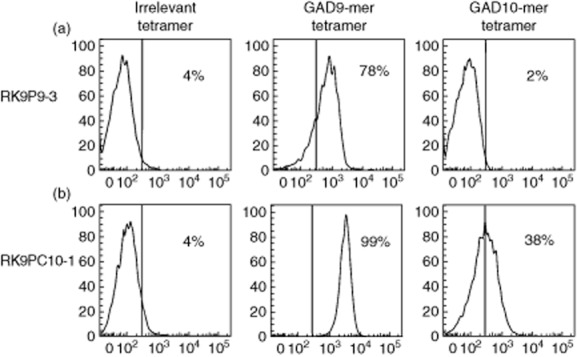
CD8 T cell clones display differential staining with tetramer loaded with either glutamic acid decarboxylase 9 (GAD9)-mer or GAD10-mer peptide. CD3+CD8+ clone cells raised against the immunogenic 114–123 region of GAD65 were stained with an irrelevant human leucocyte antigen (HLA)-A*0201 tetramer (loaded with preproinsulin 15–24), GAD9-mer tetramer (GAD114–122) or GAD10-mer tetramer (GAD114–123). (a) Clone RK9P9-3 stains with the GAD9-mer tetramer only. (b) Clone RK9C10-1 stains with both GAD9-mer and GAD10-mer tetramers. Histograms are displayed as % of maximum for 10 000 live events, the line indicative of positive staining represents the fluorescence-minus-one control.
Fig. 2.
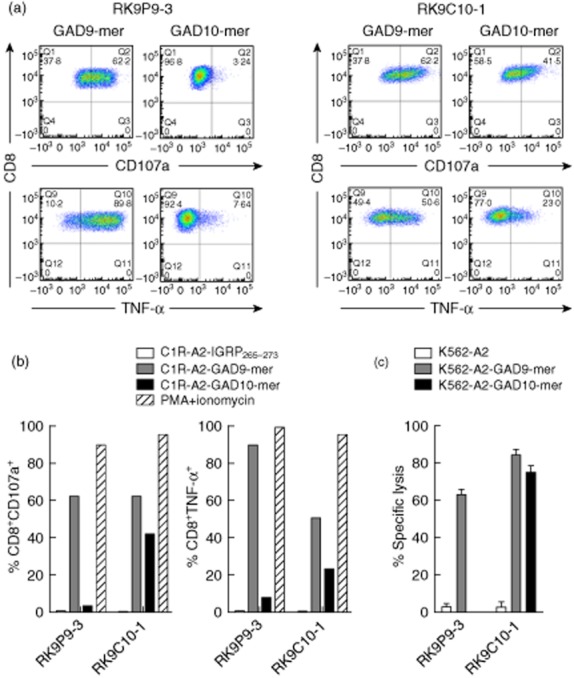
Differential recognition of peptide-pulsed human leucocyte antigen (HLA)-A*0201+ cell lines by CD8 T cell clones. Representative intracellular staining for tumour necrosis factor (TNF)-α and surface staining of CD107a (degranulation marker) of clone cells after 18 h co-culture with peptide-pulsed target cell lines. (a) RK9P9-3 shows a high percentage of events positive for TNF-α production and degranulation following co-culture with glutamic acid decarboxylase 9 (GAD9)-mer-pulsed target cells, but minimal positivity using GAD10-mer-pulsed target cells. In contrast, RK9C10-1 displays a cross-reactive profile to both GAD9-mer- and GAD10-mer peptide-pulsed cell lines. (b) These responses contrasted against a negative control peptide stimulation (IGRP265–273) and the positive control of phorbol myristate acetate/ionomycin stimulation. (c) Specific lysis of GAD9-mer-pulsed K562-A2 target cells by both RK9P9-3 and RK9C10-1 and killing of GAD10-mer-pulsed target cells by RK9C10-1 only; 10 000 live CD3+CD8+ events were recorded for flow cytometry analyses; cytotoxicity and functional assays are representative data performed at an effector : target ratio of 25:1 for 4 h. Data shown are representative single experiments.
GAD9-mer restricted CD8 T cells do not recognize endogenously processed and presented GAD65
K562 cells stably expressing HLA-A*0201 were transduced to express GAD2 (K562-A2–GAD65) and intracellular GAD65 production confirmed by Western blot (not shown). To determine whether these cells processed and presented the 9-mer GAD65114–122 complexed with HLA-A*0201, they were co-cultured with RK9P9-3 (GAD65114–122-specific) and the chemokine response examined in culture supernatants. Clone RK9P9-3 responds to peptide-pulsed K562-A2 cells, but not K562-A2–GAD65 (Fig. 3a). In contrast, RK9C10-1 (9-mer/10-mer; GAD65114–122/123-specific) responds comparably to K562-A2-GAD65 target cells and to K562-A2 target cells pulsed with the 9-mer (GAD65114–122) peptide (Fig. 3b) or 10-mer (data not shown and see Fig. 2b). These data show that clone RK9P9-3 does not engage cognate ligand when cultured with cell lines expressing GAD65 and HLA-A*0201, indicating that the 9-mer peptide GAD65114–122 is not naturally processed and presented by K562 cells. In contrast, clone RK9C10-1 engages cognate ligand when cultured with the same cell lines, indicating that endogenously expressed GAD65 is naturally processed by K562 cells, but that only the 10-mer GAD65114–123 is presented.
Fig. 3.
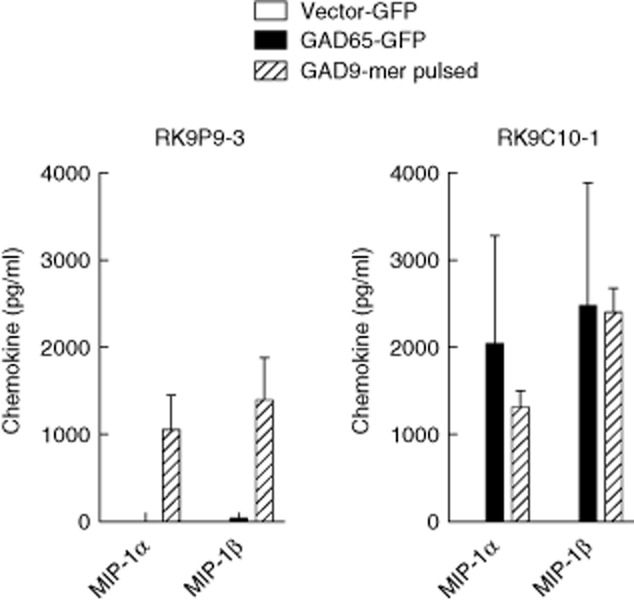
CD8 T cell clone recognition of endogenously presented glutamic acid decarboxylase 65 (GAD65) by transduced target cells. CD8 T cell clones were co-cultured with GAD2 transduced human leucocyte antigen (HLA)-A2 expressing K562 cells (K562-A2-GAD65) for 18 h, and supernatants assessed for macrophage inflammatory protein (MIP)-1α and MIP-1β production. Both RK9P9-3 and RK9C10-1 respond similarly to stimulation with GAD9-mer peptide-pulsed cells. Similarly, neither respond to the empty vector control cell line. RK9C10-1 produced high levels of chemokines when cultured with K562-A2-GAD65 target cells, but RK9P9-3 did not respond. These data indicate that only RK9C10-1 recognizes GAD65 after endogenous processing, and that the naturally processed and presented epitope is GAD65114–123. Cells were incubated at an effector : target ratio of 1:1. Bars show triplicate means and error bars standard errors of the mean of representative experiments.
HLA-A0201+ human islet cell presentation of GAD65
Once characterized for their specificity, these clones provided robust reagents for examining processing and presentation of this immunogenic region of GAD65 by human islet cells. In keeping with our findings in assays performed using K562-A2–GAD65 cells (endogenously expressing GAD2), no reactivity was observed when RK9P9-3 clone was co-cultured with human islet cells (Fig. 4a). In contrast, RK9C10-1 clone cells killed disaggregated human HLA-A*0201 + islet cell preparations (Fig. 4b) and secreted chemokines when co-cultured with these targets over 18 h (Fig. 4a).
Fig. 4.
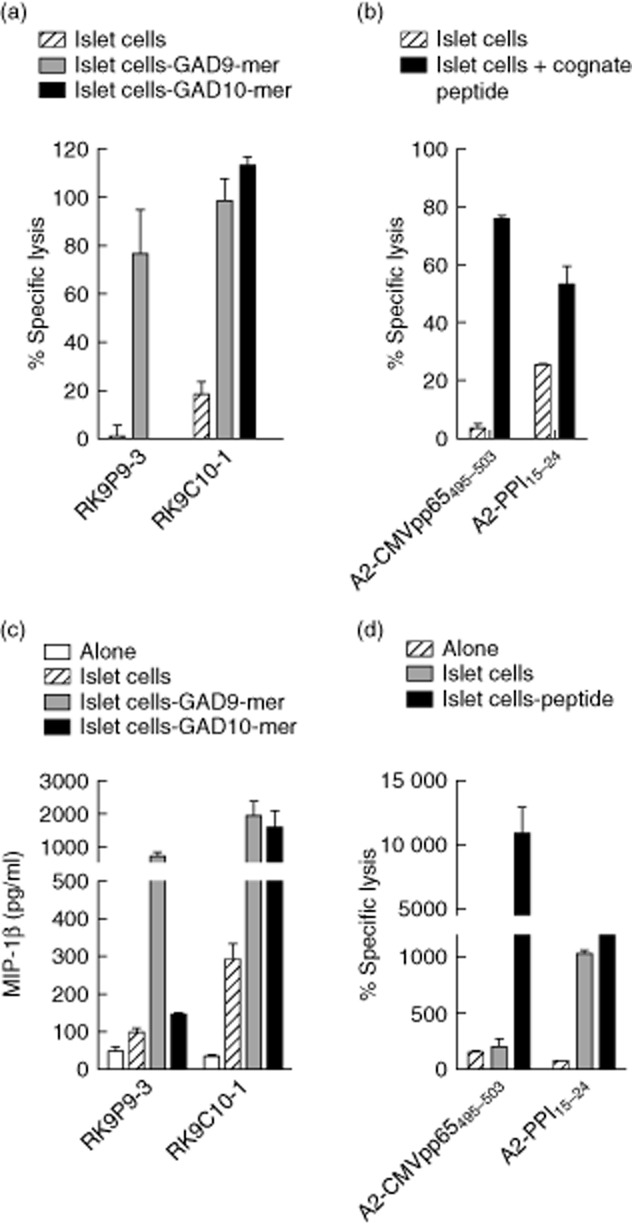
CD8 T cell clone recognizing glutamic acid decarboxylase 65 (GAD65)114–123 recognizes and kills human islet cells. (a) Cytotoxicity assays were performed using RK9P9-3 and RK9C10-1 clone cells and human leucocyte antigen (HLA)-A*0201+ human islet cells as targets. As shown in (a) RK9C10-1 robustly kills islet cells [comparable to the positive control (b), namely killing of islet cells from the same preparation by the preproinsulin (PPI)-specific CD8 T cell clone 3F2 specific for PPI15–24], whereas RK9P9-3 displays background levels of specific lysis in the absence of cognate peptide. (b) Negative control, cytomegalovirus (CMV)-specific clone A2-CMVpp65495–503, which does not kill islet cells in the absence of cognate peptide. (c) Analysis of supernatants of 18-h co-cultures of clone cells and islet cells for secretion of macrophage inflammatory protein (MIP)-1β. RK9C10-1 showed robust production of MIP-1β in response to islet cells, whereas RK9P9-3 production of MIP-1β is similar to background. (d) Negative and positive control responses to the same islet cell preparations. Cytotoxicity assays were performed at an effector : target ratio of 25:1. Bars show means of triplicates and error bars the standard errors of the mean (data are representative of n = 2 experiments using the same islet donor).
Discussion
We describe the generation of CD8 T cell clones specific for an immunogenic region of GAD65, which we have used to examine the natural processing and presentation of endogenously expressed GAD65 in human islet cells. We provide the first evidence that human islet cells can be targeted and destroyed via recognition of GAD65, and show that for the immunogenic region we studied, it is the 10-mer peptide GAD65114–123 that is naturally processed and presented by HLA-A*0201.
Our work was prompted by numerous studies documenting type 1 diabetes-related CD8 T cell responses directed against the region of GAD65 encompassing residues 114–123. Attention to this sequence was drawn originally by Sinigaglia's group, who showed that CD8 T cells with cytotoxic potential could be generated against GAD65114–123, although their work fell short of demonstrating that human islet cells are susceptible to killing through this pathway [21]. Our report is the first to show islet cell killing via targeting of the GAD65 sequence 114–123. Subsequent studies have also provided strong evidence that a focused cytotoxic response against GAD65114–123 may be important in disease pathogenesis, as CD8 T cells with this specificity have been identified in insulitic lesions in type 1 diabetes patients studied close to diagnosis using in-situ tetramer staining [8]. Taken together with our findings, these data indicate that tracking circulating CD8 T cells specific for GAD65114–123 in HLA-A*0201 individuals with, or at risk of, type 1 diabetes has a robust experimental basis as a biomarker strategy.
In contrast, our study of HLA-A*0201-restricted CD8 T cells with specificity for GAD65114–122 presents a more nuanced set of questions. First, we show clearly that this 9-mer peptide is not displayed by target cells transduced to express GAD65, or by human islet cells that express this protein constitutively. This is of interest, as algorithms (http://www.syfpeithi.de/ and http://www.cbs.dtu.dk/services/HLArestrictor/ [32,33]) predict that this shorter peptide binds with higher affinity to HLA-A*0201 than the 10-mer GAD65114–123, the naturally presented species. In our study, this prediction was corroborated by studies using circular dichroism to examine the thermal stability of the relevant peptide–HLA complexes. The reduced stability of HLA-A2–GAD10-mer compared to GAD9-mer complexes most probably explains the reduced response of RK9C10-1 to GAD10-mer expressing target cells. However, the affinity of RK9C10-1 T cell receptor (TCR) for stable peptide–HLA complexes may also be a factor in the observed differential response. It should be noted that the relative instability of HLA–GAD10-mer complexes reported here could also account for the difficulty in tetramer production, and thus the original rationale for investigating GAD-specific CD8 T cell responses with more stable GAD9-mer-containing tetramers [22,23]. As we have stated previously [19,20], this argues for caution in the use of algorithms to predict epitopes, as they promote a focus on high-affinity peptides that may not be generated by the target cell.
However, our findings do not necessarily imply that reactivity against the 9-mer GAD65114–122 lacks disease relevance, rather that it merits further study. First, we provide evidence that TCR clonotypes exist (RK9C10-1) that can target both the 9-mer and 10-mer peptides from this region. Thus, it seems likely that a proportion of CD8 T cells that are identified using HLA-A*0201–GAD65114–122 multimers, or through their functional responses to free 9-mer peptide in PBMC cultures, will have the capacity to respond to endogenously presented GAD65114–123, and thus have a potentially pathogenic role. An additional possibility, which will require addressing in future studies, is that the 9-mer GAD65114–122 is indeed generated by cross-presentation of soluble GAD65 by professional antigen-presenting cells as part of the intra-islet and pancreatic lymph node inflammatory process. As a result, 9-mer-specific CD8 T cells could indeed be induced, disease-related and represent putative biomarkers, in this case of an inflammatory process that may not have islet cell killing as its end-point (‘frustrated autoimmunity’).
Acknowledgments
This work was supported by the Juvenile Diabetes Research Foundation (JDRF) Autoimmunity Centers Consortium (1–2007-1803 to M. P.), a JDRF R&D Grant (type 1 diabetes 217194; 17–2012-352 to A. S., G. D., A. K. S. and M. P.) and the UK Department of Health via the National Institute for Health Research (NIHR) Biomedical Research Centre Award to Guy's & St Thomas' National Health Service Foundation Trust in partnership with King's College London. M. P. receives additional funding via the European Commission Seventh Framework Programme (NAIMIT, PEVNET and EE-ASI). D. K. C. is a Wellcome Trust Career Development Fellow. M. P. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Disclosure
There are no potential conflicts of interest relevant to this article to report.
Author contributions
R. R. K., D. K., M. E. and A. S. designed and performed the experiments and with M. P. analysed data and wrote the manuscript. R. R. K., A. S and M. P. conceived ideas and oversaw research. G. D and A. K. S provided cellular material and input on experimental design. D. K. C. and K. B. performed CD experiments and analysis, and wrote the respective report. M. Z. and G. C. H. performed human islet experiments.
Supporting information
Additional Supporting information may be found in the online version of this article at the publisher's web-site:
Fig. S1. Tetramer staining of glutamic acid decarboxylase 65 (GAD65) peptide-specific clones derived from two donors. CD3+CD8+ clone cells raised against the immunogenic 114–123 region of GAD65 were stained with an irrelevant human leucocyte antigen (HLA)-A*0201 tetramer (loaded with preproinsulin 15–24), GAD9-mer tetramer (GAD114–122) or GAD10-mer tetramer (GAD114–123). Clones RK9P9-3 and RK9C10-1 were used as tools for further studies in this paper to decipher and ascribe relevant importance to putative GAD65 epitopes, due to their differential recognition of GAD peptides from the immunogenic 114–123 region.
Fig. S2. Glutamic acid decarboxylase 65 (GAD65) 10-mer binding to human leucocyte antigen (HLA)-A*0201 is less stable than the 9-mer. Left panel. Circular dichroism thermal denaturation curves recorded at 218 nm are shown for selected samples as indicated. Dots represent measured values fitted assuming a two-state trimer-to-monomer transition, as described in Materials and methods. Open symbols and dashed line for stabilization of GAD65 9-mer and closed circles/solid line for GAD65 10-mer. Right panel: bar graphs of the thermal stability with respect to melting temperature (upper) and van't Hoff's enthalpy of unfolding (lower panel). Error bars show standard deviation of the respective parameter based on fitting each set of measured data.
References
- 1.von Herrath M, Peakman M, Roep B. Progress in immune-based therapies for type 1 diabetes. Clin Exp Immunol. 2013;172:186–202. doi: 10.1111/cei.12085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Knight RR, Kronenberg D, Zhao M, et al. Human beta-cell killing by autoreactive preproinsulin-specific CD8 T cells is predominantly granule-mediated with the potency dependent upon T-cell receptor avidity. Diabetes. 2013;62:205–213. doi: 10.2337/db12-0315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kronenberg D, Knight RR, Estorninho M, et al. Circulating preproinsulin signal peptide-specific CD8 T cells restricted by the susceptibility molecule HLA-A24 are expanded at onset of type 1 diabetes and kill beta-cells. Diabetes. 2012;61:1752–1759. doi: 10.2337/db11-1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Skowera A, Ellis RJ, Varela-Calvino R, et al. CTLs are targeted to kill beta cells in patients with type 1 diabetes through recognition of a glucose-regulated preproinsulin epitope. J Clin Invest. 2008;118:3390–3402. doi: 10.1172/JCI35449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Velthuis JH, Unger WW, Abreu JR, et al. Simultaneous detection of circulating autoreactive CD8+ T-cells specific for different islet cell-associated epitopes using combinatorial MHC multimers. Diabetes. 2010;59:1721–1730. doi: 10.2337/db09-1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fuchs YF, Adler K, Lindner A, et al. IGRP and insulin vaccination induce CD8+ T cell-mediated autoimmune diabetes in the RIP-CD80GP mouse. Clin Exp Immunol. 2014;176:199–206. doi: 10.1111/cei.12263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dolton G, Lissina A, Skowera A, et al. Comparison of peptide–major histocompatibility complex tetramers and dextramers for the identification of antigen-specific T cells. Clin Exp Immunol. 2014;177:47–63. doi: 10.1111/cei.12339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coppieters KT, Dotta F, Amirian N, et al. Demonstration of islet-autoreactive CD8 T cells in insulitic lesions from recent onset and long-term type 1 diabetes patients. J Exp Med. 2012;209:51–60. doi: 10.1084/jem.20111187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Willcox A, Richardson SJ, Bone AJ, Foulis AK, Morgan NG. Analysis of islet inflammation in human type 1 diabetes. Clin Exp Immunol. 2009;155:173–181. doi: 10.1111/j.1365-2249.2008.03860.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nejentsev S, Howson JM, Walker NM, et al. Wellcome Trust Case Control C. Localization of type 1 diabetes susceptibility to the MHC class I genes HLA-B and HLA-A. Nature. 2007;450:887–892. doi: 10.1038/nature06406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gonzalez-Galarza FF, Christmas S, Middleton D, Jones AR. Allele frequency net: a database and online repository for immune gene frequencies in worldwide populations. Nucleic Acids Res. 2011;39:D913–919. doi: 10.1093/nar/gkq1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nejentsev S, Howson JM, Walker NM, et al. Localization of type 1 diabetes susceptibility to the MHC class I genes HLA-B and HLA-A. Nature. 2007;450:887–892. doi: 10.1038/nature06406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scotto M, Afonso G, Larger E, et al. Zinc transporter (ZnT)8(186–194) is an immunodominant CD8+ T cell epitope in HLA-A2+ type 1 diabetic patients. Diabetologia. 2012;55:2026–2031. doi: 10.1007/s00125-012-2543-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li S, Li H, Chen B, et al. Identification of novel HLA-A 0201-restricted cytotoxic T lymphocyte epitopes from Zinc Transporter 8. Vaccine. 2013;31:1610–1615. doi: 10.1016/j.vaccine.2012.12.008. [DOI] [PubMed] [Google Scholar]
- 15.Roep BO, Solvason N, Gottlieb PA, et al. Plasmid-encoded proinsulin preserves C-peptide while specifically reducing proinsulin-specific CD8(+) T cells in type 1 diabetes. Sci Transl Med. 2013;5:191ra82. doi: 10.1126/scitranslmed.3006103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vita R, Zarebski L, Greenbaum JA, et al. The immune epitope database 2·0. Nucleic Acids Res. 2010;38:D854–862. doi: 10.1093/nar/gkp1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boucherma R, Kridane-Miledi H, Bouziat R, et al. HLA-A*01:03, HLA-A*24:02, HLA-B*08:01, HLA-B*27:05, HLA-B*35:01, HLA-B*44:02, and HLA-C*07:01 monochain transgenic/H-2 class I null mice: novel versatile preclinical models of human T cell responses. J Immunol. 2013;191:583–593. doi: 10.4049/jimmunol.1300483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wooldridge L, Lissina A, Cole DK, van den Berg HA, Price DA, Sewell AK. Tricks with tetramers: how to get the most from multimeric peptide-MHC. Immunology. 2009;126:147–164. doi: 10.1111/j.1365-2567.2008.02848.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roep BO, Peakman M. Diabetogenic T lymphocytes in human type 1 diabetes. Curr Opin Immunol. 2011;23:746–753. doi: 10.1016/j.coi.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 20.Di Lorenzo TP, Peakman M, Roep BO. Translational mini-review series on type 1 diabetes: systematic analysis of T cell epitopes in autoimmune diabetes. Clin Exp Immunol. 2007;148:1–16. doi: 10.1111/j.1365-2249.2006.03244.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Panina-Bordignon P, Lang R, van Endert PM, et al. Cytotoxic T cells specific for glutamic acid decarboxylase in autoimmune diabetes. J Exp Med. 1995;181:1923–1927. doi: 10.1084/jem.181.5.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Monti P, Scirpoli M, Rigamonti A, et al. Evidence for in vivo primed and expanded autoreactive T cells as a specific feature of patients with type 1 diabetes. J Immunol. 2007;179:5785–5792. doi: 10.4049/jimmunol.179.9.5785. [DOI] [PubMed] [Google Scholar]
- 23.Monti P, Scirpoli M, Maffi P, et al. Islet transplantation in patients with autoimmune diabetes induces homeostatic cytokines that expand autoreactive memory T cells. J Clin Invest. 2008;118:1806–1814. doi: 10.1172/JCI35197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mallone R, Scotto M, Janicki CN, et al. T-Cell Workshop Committee IoDS. Immunology of diabetes society T-cell workshop: HLA class I tetramer-directed epitope validation initiative T-Cell Workshop Report-HLA Class I Tetramer Validation Initiative. Diabetes Metab Res Rev. 2011;27:720–726. doi: 10.1002/dmrr.1243. [DOI] [PubMed] [Google Scholar]
- 25.Giuliani L, Mele R, Di Giovine M, et al. Detection of GAD65 autoreactive T-cells by HLA class I tetramers in type 1 diabetic patients. J Biomed Biotechnol. 2009;2009:576219. doi: 10.1155/2009/576219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ekeruche-Makinde J, Miles JJ, van den Berg HA, et al. Peptide length determines the outcome of TCR/peptide-MHCI engagement. Blood. 2013;121:1112–1123. doi: 10.1182/blood-2012-06-437202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thompson WS, Pekalski ML, Simons HZ, et al. Multi-parametric flow cytometric and genetic investigation of the peripheral B cell compartment in human type 1 diabetes. Clin Exp Immunol. 2014;177:571–585. doi: 10.1111/cei.12362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang GC, Zhao M, Jones P, et al. The development of new density gradient media for purifying human islets and islet-quality assessments. Transplantation. 2004;77:143–145. doi: 10.1097/01.TP.0000100401.62912.B2. [DOI] [PubMed] [Google Scholar]
- 29.Pace CN, Vajdos F, Fee L, Grimsley G, Gray T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Greenfield NJ. Analysis of circular dichroism data. Met Enzymol. 2004;383:282–317. doi: 10.1016/S0076-6879(04)83012-X. [DOI] [PubMed] [Google Scholar]
- 31.Venyaminov S, Baikalov IA, Shen ZM, Wu CS, Yang JT. Circular dichroic analysis of denatured proteins: inclusion of denatured proteins in the reference set. Anal Biochem. 1993;214:17–24. doi: 10.1006/abio.1993.1450. [DOI] [PubMed] [Google Scholar]
- 32.Rammensee H, Bachmann J, Emmerich NP, Bachor OA, Stevanovic S. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics. 1999;50:213–219. doi: 10.1007/s002510050595. [DOI] [PubMed] [Google Scholar]
- 33.Erup Larsen M, Kloverpris H, Stryhn A, et al. HLA restrictor – a tool for patient-specific predictions of HLA restriction elements and optimal epitopes within peptides. Immunogenetics. 2011;63:43–55. doi: 10.1007/s00251-010-0493-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Tetramer staining of glutamic acid decarboxylase 65 (GAD65) peptide-specific clones derived from two donors. CD3+CD8+ clone cells raised against the immunogenic 114–123 region of GAD65 were stained with an irrelevant human leucocyte antigen (HLA)-A*0201 tetramer (loaded with preproinsulin 15–24), GAD9-mer tetramer (GAD114–122) or GAD10-mer tetramer (GAD114–123). Clones RK9P9-3 and RK9C10-1 were used as tools for further studies in this paper to decipher and ascribe relevant importance to putative GAD65 epitopes, due to their differential recognition of GAD peptides from the immunogenic 114–123 region.
Fig. S2. Glutamic acid decarboxylase 65 (GAD65) 10-mer binding to human leucocyte antigen (HLA)-A*0201 is less stable than the 9-mer. Left panel. Circular dichroism thermal denaturation curves recorded at 218 nm are shown for selected samples as indicated. Dots represent measured values fitted assuming a two-state trimer-to-monomer transition, as described in Materials and methods. Open symbols and dashed line for stabilization of GAD65 9-mer and closed circles/solid line for GAD65 10-mer. Right panel: bar graphs of the thermal stability with respect to melting temperature (upper) and van't Hoff's enthalpy of unfolding (lower panel). Error bars show standard deviation of the respective parameter based on fitting each set of measured data.


