Summary
Vaccines prevent infectious disease largely by inducing protective neutralizing antibodies against vulnerable epitopes. Multiple major pathogens have resisted traditional vaccine development, although vulnerable epitopes targeted by neutralizing antibodies have been identified for several such cases. Hence, new vaccine design methods to induce epitope-specific neutralizing antibodies are needed. Here we show, with a neutralization epitope from respiratory syncytial virus (RSV), that computational protein design can generate small, thermally and conformationally stable protein scaffolds that accurately mimic the viral epitope structure and induce potent neutralizing antibodies. These scaffolds represent promising leads for research and development of a human RSV vaccine needed to protect infants, young children and the elderly. More generally, the results provide proof of principle for epitope-focused and scaffold-based vaccine design, and encourage the evaluation and further development of these strategies for a variety of other vaccine targets including antigenically highly variable pathogens such as HIV and influenza.
Vaccination is a proven, safe, and cost-effective way to protect against infectious disease1,2, but potentially vaccine-preventable illnesses continue to place a heavy burden on the human population. Data from recent epidemiological studies indicate that in 2010, infectious diseases caused 18.5% of all human deaths and 23% of disability-adjusted life years3,4. This burden could be reduced by broader deployment and use of existing vaccines or by other prevention modalities or treatment regimens. However, for maximal, affordable and sustainable gains in global health, new or improved vaccines are needed for multiple major pathogens including: human immunodeficiency virus 1 (HIV)5, malaria6, mycobacterium tuberculosis7, influenza virus8, dengue virus9 and respiratory syncytial virus (RSV)10. One likely impediment to vaccine development in these cases is the limited set of antigen design or presentation methods available to vaccine engineers. For example, current licensed vaccines in the United States11 derive from strategies that have been available for many years – viral vaccines are composed of recombinant viruslike particles or live, live-attenuated, or whole inactivated viruses or subunit vaccines, and bacterial vaccines are composed of bacterial surface proteins, detoxified toxins, or polysaccharides with or without conjugation to a carrier protein.
Epitope-focused vaccine design is a conceptually appealing but unproven method in which immunogens are designed to elicit protective antibody responses against structural epitopes that are defined by protective antibodies isolated from infected patients or animal models12. This strategy, if validated, could offer a potential route to vaccines for many pathogens that have resisted traditional vaccine development, including highly antigenically variable viruses such as HIV, influenza and hepatitis C virus for which broadly-neutralizing antibodies have been discovered and characterized structurally with their target epitopes13. We tested the feasibility of this strategy using an epitope from RSV, a virus that causes lower respiratory tract infections in children and the elderly. In 2010 RSV was estimated to be responsible for 6.7% of all deaths in children of ages one month to one year3. We focused on the epitope targeted by the licensed, prophylactic neutralizing antibody palivizumab (Synagis®, Pali) and an affinity-matured variant, motavizumab (Mota)14. A crystal structure of Mota in complex with its epitope from the RSV Fusion (F) glycoprotein revealed that the antibody-bound epitope attains a helixturn-helix conformation15.
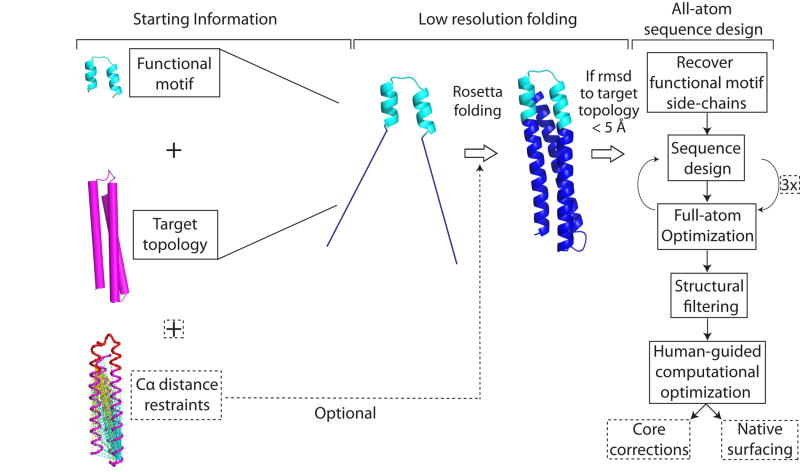
We previously developed “side-chain grafting” and “backbone grafting” methods to transplant continuous or discontinuous epitopes to scaffold proteins of known structure, for epitope conformational stabilization and immune presentation16,17,18,19,20. Epitopescaffold immunogens designed by these methods for epitopes from HIV or RSV (including the Mota epitope) have in some cases induced structure-specific antibodies but have failed to induce neutralizing antibodies16,17,18. Because those methods are restricted to scaffold proteins of predetermined structure, here we developed a new computational method to design scaffold proteins with full backbone flexibility, to allow greater precision in tailoring scaffold structures for particular epitope structures. We used this method to design scaffolds for the Mota epitope, and we found that the scaffolds had favorable biophysical and structural properties and that scaffold immunization of rhesus macaques induced RSV-neutralizing activity (Fig. 1).
Figure 1. A novel computational method to design epitope-focused vaccines, illustrated with a neutralization epitope from RSV.
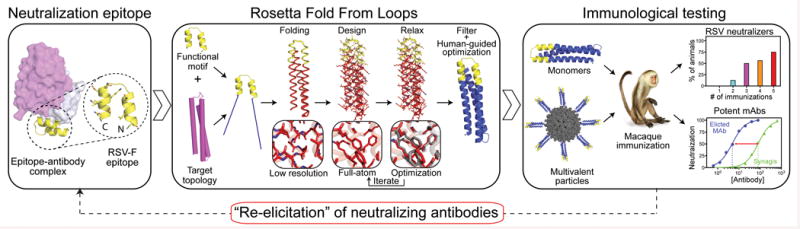
Stages of computational design and immunological evaluation are shown; biophysical and structural evaluation are also important (see text).
Computational Method
Great strides have been made in developing de novo methods to design arbitrary, idealized protein structures21,22, but the resulting proteins have lacked functional activity. We devised a computational method to allow de novo folding and design of scaffold proteins stabilizing functional motifs (Extended Data Fig. 1). This procedure, called Fold from Loops (FFL), has four stages: i) Selection of the functional motif and target topology to be folded around the motif; ii) Ab initio folding to build diverse backbone conformations consistent with the target topology; iii) Iterative sequence design and structural relaxation to select low energy amino acid sequences for the given backbone conformations; iv) Filtering and human-guided optimization, in which the best designs are identified by structural metrics and then subjected to optional human-guided sequence design to correct remaining flaws.
Design of Epitope-Scaffolds
To design scaffolds for the helix-turn-helix conformation of the Mota epitope (PDBid: 3IXT, chain P), we selected a three-helix bundle (PDBid: 3LHP, chain S) as the template topology. Knowing that the template protein folds into thermally stable, soluble monomers16,23, we designed scaffolds of similar length and position-dependent secondary structure. We produced 40000 designs using FFL stages i-iii and then employed multiple structural filters to select eight designs for human-guided optimization. Additional modifications were made to those designs as follows: (i) to optimize solubility, nearly all surface residues outside the epitope were replaced with those from the template protein; and (ii) to optimize side-chain packing in the buried protein core, computational design was used to design larger hydrophobic residues at selected buried positions of most designs (Extended Data Fig. 2). The final eight “FFL” designs had similar but nonidentical backbone conformations (pairwise root mean square deviations (RMSDs) ranging from 0.5 to 3.0 Å) with correspondingly diverse core packing solutions differing from each other by 8 to 42 mutations and from the template by 56 mutations on average (Extended Data Fig. 2). All eight FFL designs had identical surface residues (including non-epitope residues taken from the template, as well as the epitope itself). To create fully artificial scaffolds with different antigenic surfaces, that could be employed in heterologous prime-boost regimens with FFL scaffolds or could be used to map immune responses to FFL scaffolds, we resurfaced23 the FFL_001 design; this produced the “FFL_surf” designs (Extended Data Fig. 2) that differed from FFL_001 by 36 mutations on average and had no significant sequence similarity (BLAST E-value < 10-3) to any known protein except the RSV F protein.
Biophysical and Structural Characterization
Six of eight FFL designs, and three of four FFL_surf designs, could be expressed in Escherichia coli and purified, with yields ranging from 3 to 5 mg/L. These nine scaffolds were monomeric in solution, showed circular dichroism (CD) spectra typical for properly folded helical proteins, and all but one were highly thermally stable with melting temperatures (Tms) greater than 75 °C (Fig. 2a, b, Table 1 and Extended Data Fig. 3). 15N Hetero-nuclear Single-Quantum Coherence (HSQC) spectra were collected for four FFL designs, and these data showed reasonable to good peak dispersion, typical of well behaved, globular proteins with high α-helical content in solution (Fig. 2c, Table 1 and Extended Data Fig. 3).
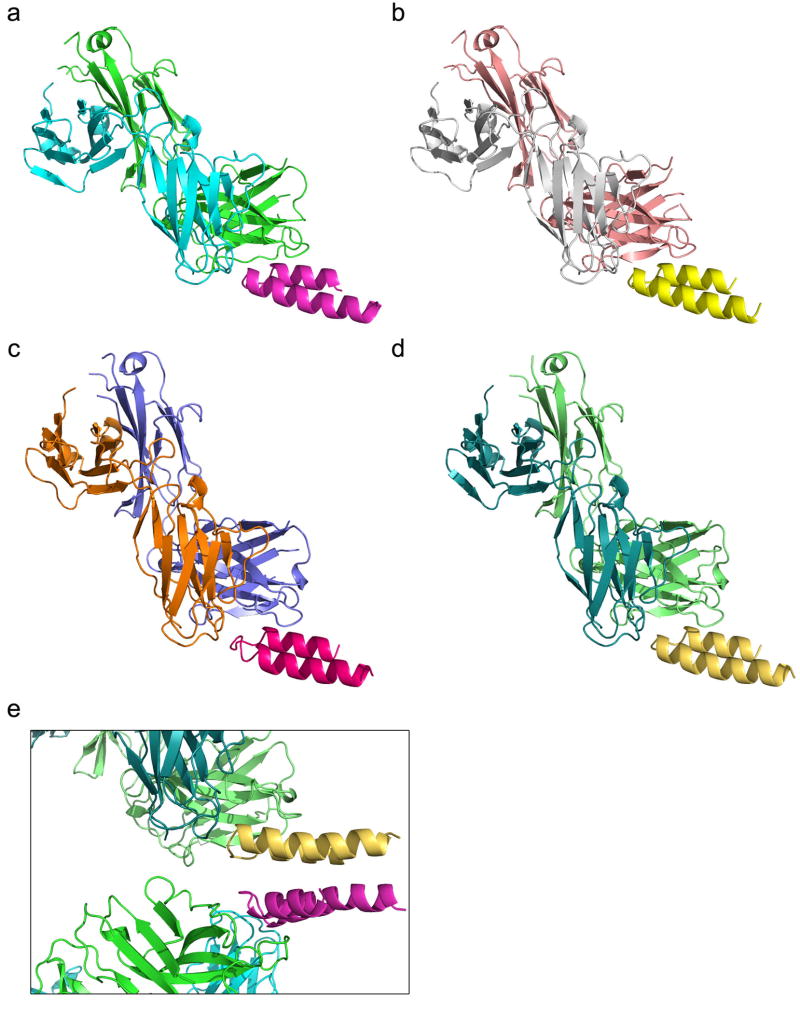
Figure 2. Biophysical and structural characterization of scaffold FFL_001.
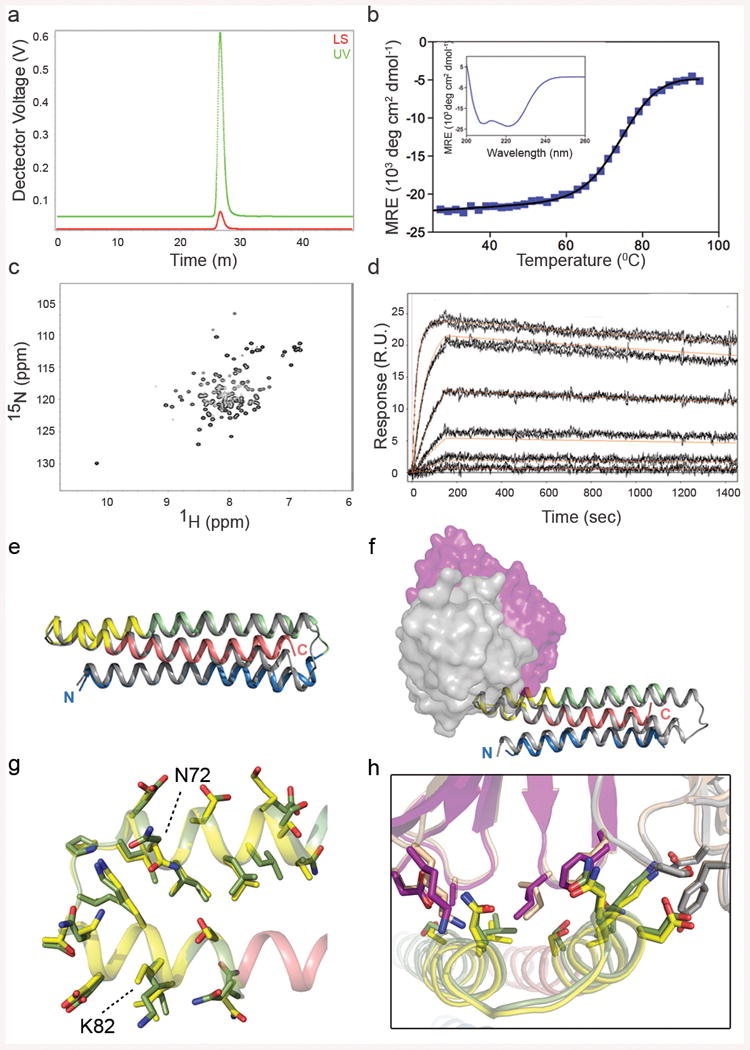
a, Size exclusion chromatography coupled in-line with multi-angle light scattering measured a molecular weight in solution of ∼15 kDa, corresponding to a monomer. b, Circular dichroism data fit with a two state model showed that the protein had a melting temperature of 74 °C. Inset: The wavelength scan at 25 °C exhibited two minima characteristic of an all-helical protein,. c, Two dimensional 1H-15N HSQC spectrum at 25 °C and 600 MHz showed good peak dispersion typical of well-folded, alpha-helical proteins. ppm, parts per million. d, Surface plasmon resonance data and model fits (red lines) of the interaction with Mota Fab analyte, from which the dissociation constant (KD) was measured to be 29.9 pM. e, Crystal structure of unliganded FFL_005 (blue, green, and salmon helices, with yellow epitope), superimposed with the design model (gray with yellow epitope). f, Crystal structure of FFL_001 bound to Mota Fab, superimposed with the design model. Coloring as in e, but with the Fab light and heavy chains in gray or purple, respectively. g, Superposition of the epitope structure from unliganded FFL_005 (yellow) and the complex of peptide (green) bound to Mota from PDBid:3ixt. The positions of escape mutations for Pali (262 and 272) or Mota (272) are noted. h, Superposition of the Mota-liganded structures of FFL_001 and peptide (PDBid:3ixt). The antibody chains of 3ixt are colored in wheat, and the interfacial side-chains of both epitope and antibody are shown in stick representation.
Table 1.
Biophysical properties of scaffolds and scaffold-antibody interactions. ND, not done.
| Molecule | Multimeric State | Tm (°C) |
ΔG (kcal mol-1) | SPR Motavizumab | NMR-HSQC Dispersion | ||
|---|---|---|---|---|---|---|---|
| kon (M-1s-1) | koff (s-1) | koff/kon (pM) | |||||
| FFL_001 | Mon | 76 | ND | 3.99 × 106 | 1.19 × 10-4 | 29.9 | Dispersed |
| FFL_002 | Mon | 49 | ND | 1.56 × 106 | 7.34 × 10-4 | 469.9 | ND |
| FFL_004 | Mon | >85 | ND | 1.05 × 106 | 8.32 × 10-4 | 795.0 | ND |
| FFL_005 | Mon | >100 | 15.0 | 2.97 × 106 | 2.09 × 10-4 | 70.3 | Partially Dispersed |
| FFL_006 | Mon | >85 | ND | 3.57 × 106 | 2.32 × 10-4 | 651.9 | Dispersed |
| FFL_007 | Mon | >85 | ND | 1.45 × 106 | 1.36 × 10-4 | 94.1 | Partially Dispersed |
| FFL_001_surf1 | Mon | 84 | 8.2 | 7.43 × 106 | 4.70 × 10-4 | 6.32 | ND |
| FFL_001_surf2 | Mon | >85 | 8.1 | 5.32 × 106 | 1.58 × 10-4 | 29.6 | ND |
| FFL_001_surf4 | Mon | >85 | 9.0 | 4.80×106 | 1.58 × 10-4 | 32.9 | ND |
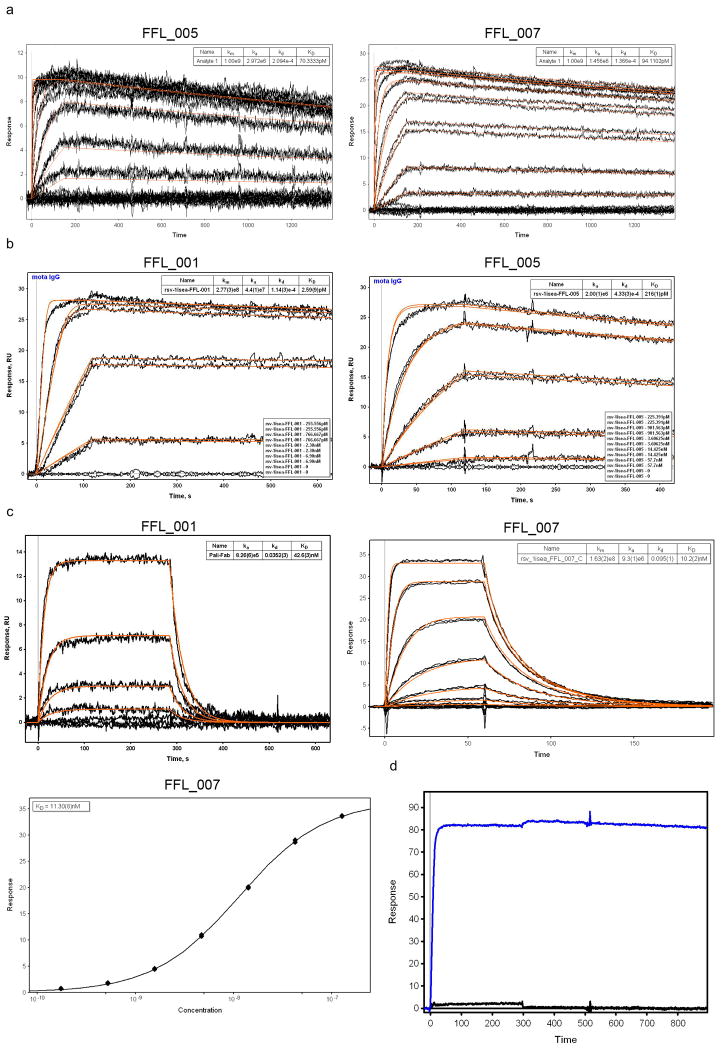
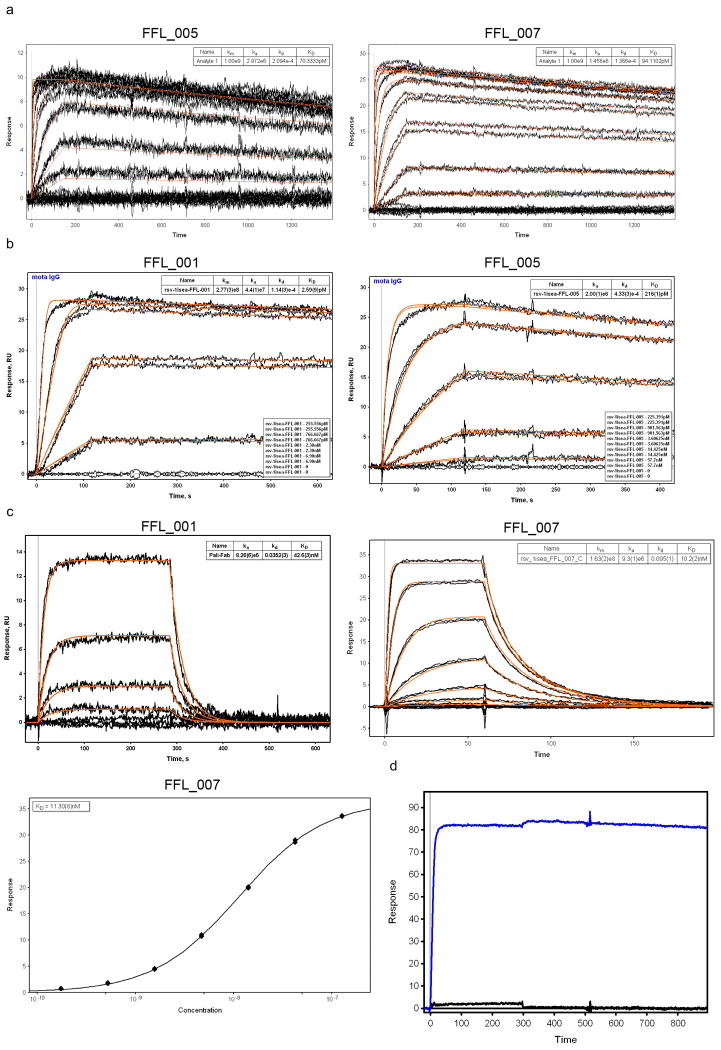
The nine purifiable FFL and FFL_surf scaffolds all had high affinity for Mota, as assessed by surface plasmon resonance (SPR) (KD < 800 pM, Fig. 2d, Table 1 and Extended Data Fig. 4). In particular, six of nine scaffolds had very high Mota affinities (KD = 6 to 94 pM) and slow dissociation rates (koff ∼ 10-4 s-1) comparable to those of the Mota interaction with the RSV F glycoprotein (KD = 35 pM and koff = 0.31 × 10-4 s-1) (ref. 14). The Mota/scaffold interaction was also specific—the point mutation K82E on scaffold FFL_001, analogous to the K272E Mota escape mutation on RSV F24, reduced Mota binding by more than a factor of 1000 (Extended Data Fig. 4). These results suggested that the conformation of the native epitope was reproduced accurately on the scaffolds. The Mota affinities for FFL scaffolds were three to four orders of magnitude higher than the Mota affinities for the free epitope peptide (KD = 210-240 nM18) or for the best side-chain grafting epitope-scaffold previously reported (KD = 90-125 nM18).
To evaluate the degree to which high-resolution structures of the FFL designs recapitulated the design models and the Mota epitope, we solved two crystal structures: unliganded FFL_005 and the complex of FFL_001 bound to Mota Fab (Fig. 2e, f), to resolutions of 2.0 and 2.7 Å, respectively (Extended Data Fig. 5). The crystal structures showed good overall agreement with the design models—the backbone RMSDs over all residues were 1.7 Å for FFL_005 and 1.2 Å for FFL_001 (Fig. 2e, f), and the all-atom RMSDs for the core side-chains were 2.5 Å for FFL_005 and 1.8 Å for FFL_001. Consistent with the biophysical data, both unliganded and Mota-bound structures revealed a high degree of epitope-mimicry. Compared to the structure of peptide in complex with Mota (PDBid: 3IXT), the epitope backbone RMSDs were 0.5 Å for FFL_005 (Fig. 2g) and 0.4 Å for FFL_001. Compared to structures of pre- and postfusion RSV F trimer (PDBids: 4JHW and 3RRR), which were not available at the time the designs were carried out, epitope backbone RMSDs were 0.3 and 0.4 Å for FFL_005, respectively. Furthermore, the interaction of FFL_001 with Mota accurately recapitulated the interaction of Mota with peptide—superposition of the epitope and paratope of both complexes gave an all-atom RMSD of 0.8 Å (Fig. 2h).
Immunological Evaluation
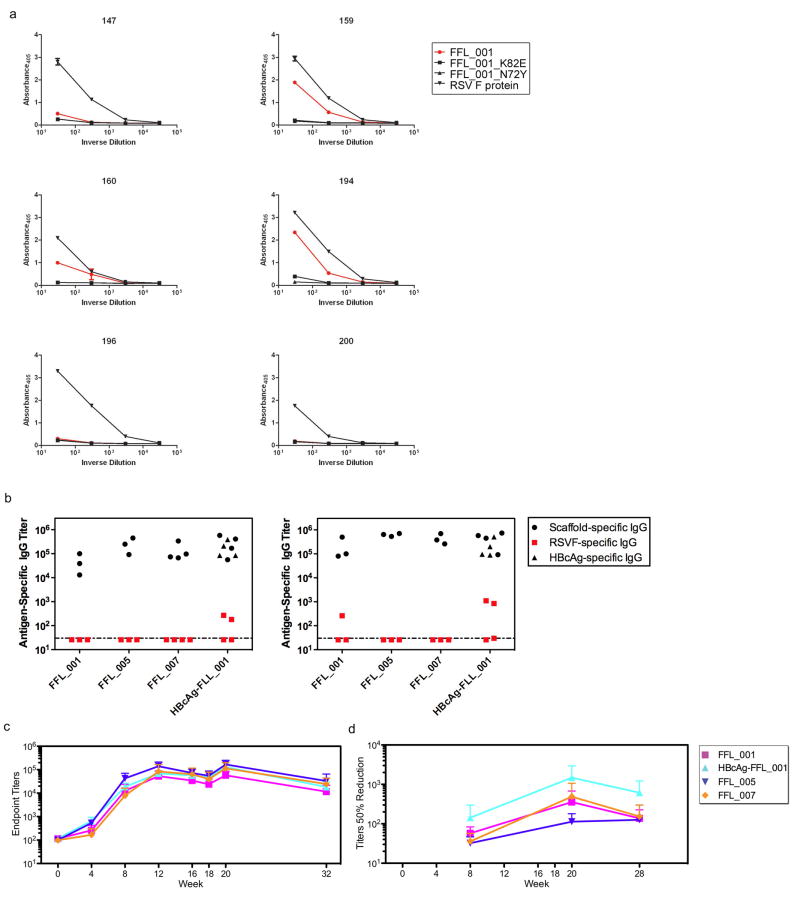
To assess whether humans can make antibodies specific for the RSV epitope structure stabilized on the scaffolds, we tested the binding of sera from six RSV-seropositive humans to RSV F, FFL_001, and FFL_001 variants with two different epitope mutations (N72Y and K82E) corresponding to RSV escape mutations for Pali (N262Y and K272E) and Mota (K272E) (Fig. 2g and Extended Data Fig. 6). While all sera reacted with RSV F and none reacted to the scaffold escape mutants, three sera displayed reactivity to FFL_001. These data confirmed that the FFL scaffolds presented a clinically relevant epitope conformation and illustrated that epitope-scaffolds have promise as reagents to assay levels of epitope-structure-specific antibodies in polyclonal sera.
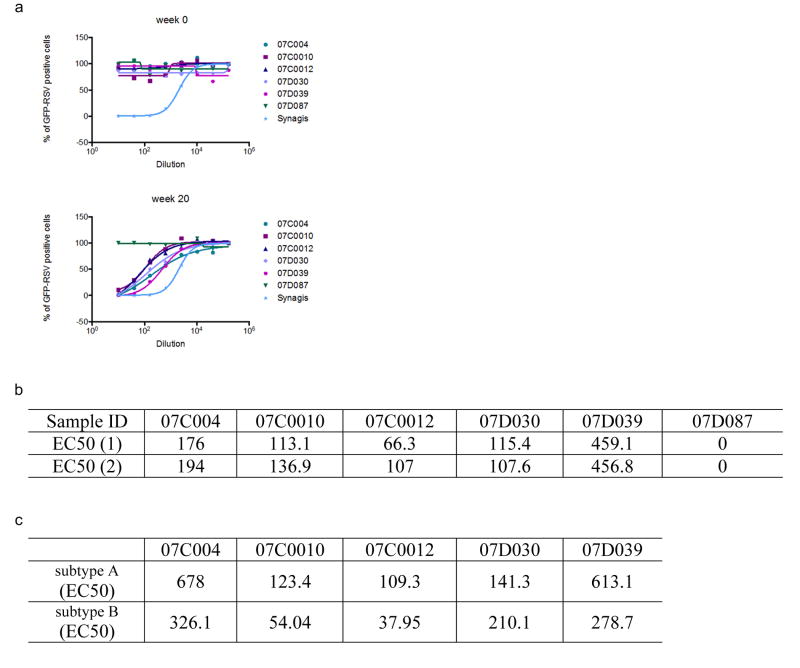
We tested whether the FFL scaffolds could induce RSV-neutralizing antibodies by vaccination, in both BALB/c mice and rhesus macaques. Four immunogens were tested: monomeric scaffolds FFL_001, FFL_005, and FFL_007, and a virus-like particle consisting of hepatitis B core antigen particles conjugated with multiple copies of FFL_00125,26. Mice produced robust binding antibody responses against the autologous antigens, but binding antibody responses against RSV F protein or RSV viral lysate were detected in only a few animals (Extended Data Fig. 6), and neutralizing activity as judged by a plaque reduction assay was not detected (not shown). In contrast to the mouse results, after three immunizations all macaques produced robust binding responses not only against the autologous antigens (Fig. 3a) but also recombinant RSV F protein (Extended Data Fig. 6), and most animals responded to RSV viral lysate (Fig. 3a and Supplementary Table 1). Neutralizing activity was detected by the plaque assay in 7 of 16 macaques after three immunizations and in 12 of 16 macaques after five immunizations (Figs. 1 and 3a and Supplementary Table 2). Neutralizing activities were confirmed at selected timepoints using two different assays (microneutralization and a flow cytometrybased assay) in different laboratories, and included measurement of neutralizing activity against RSV subtype B27 Extended Data Fig. 6, 7 and Supplementary Table 2). To benchmark the neutralization potency, selected macaque sera were tested side-by-side with sera from seropositive human adults, in both the plaque reduction and flow cytometry assays (Fig. 3b, c). The results in both assays demonstrate that the best responding macaques, including two of four animals in the particle group at week 20 and one animal in that group at week 12, have neutralization titers comparable to those induced by natural human infection. This is remarkable given that natural infection exposes multiple epitopes on the RSV F and G glycoproteins, while the scaffolds exposed only one epitope.
Fig 3. Serological analysis of immunized macaques.
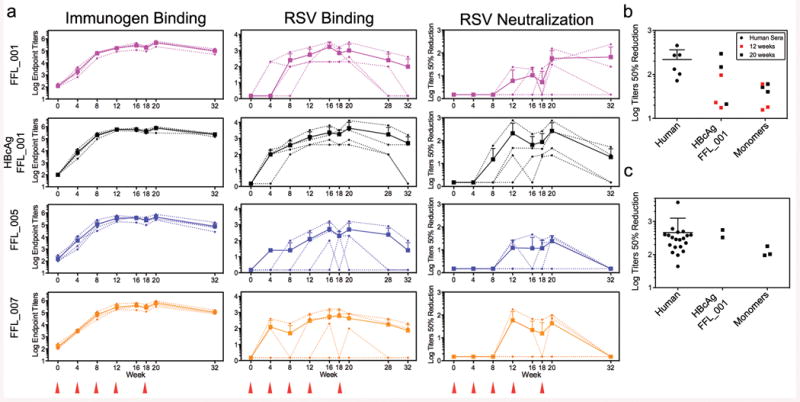
a, ELISA endpoint titers measured against the autologous immunogen (left) or against RSV whole viral lysate (middle), and 50% neutralization titers as determined by the plaque reduction assay (right). The immunization groups are shown at far left, and the schedule is indicated at the bottom. Small symbols connected with dashed lines indicate individual animals. Large symbols connected with solid lines report group averages, with error bars showing standard deviations, measured over the four animals in each group at each time point. b, Comparison of 50% neutralization titers for sera from six RSV-seropositive humans and sera from eight macaques from weeks 12 and 20, measured side-by-side in the plaque reduction assay. Mean ± standard deviation for the human data are 218 ± 145. Two macaque data points at both week 12 and 20 are not visible in the graph because no neutralizing activity was detected. c, Comparison of 50% neutralization titers for sera from twenty RSV-seropositive humans and sera from five macaques from week 20, measured side-by-side in the flow cytometry assay. Mean ± standard deviation for the human data are 462 ± 792.
Monoclonal Antibody Characterization
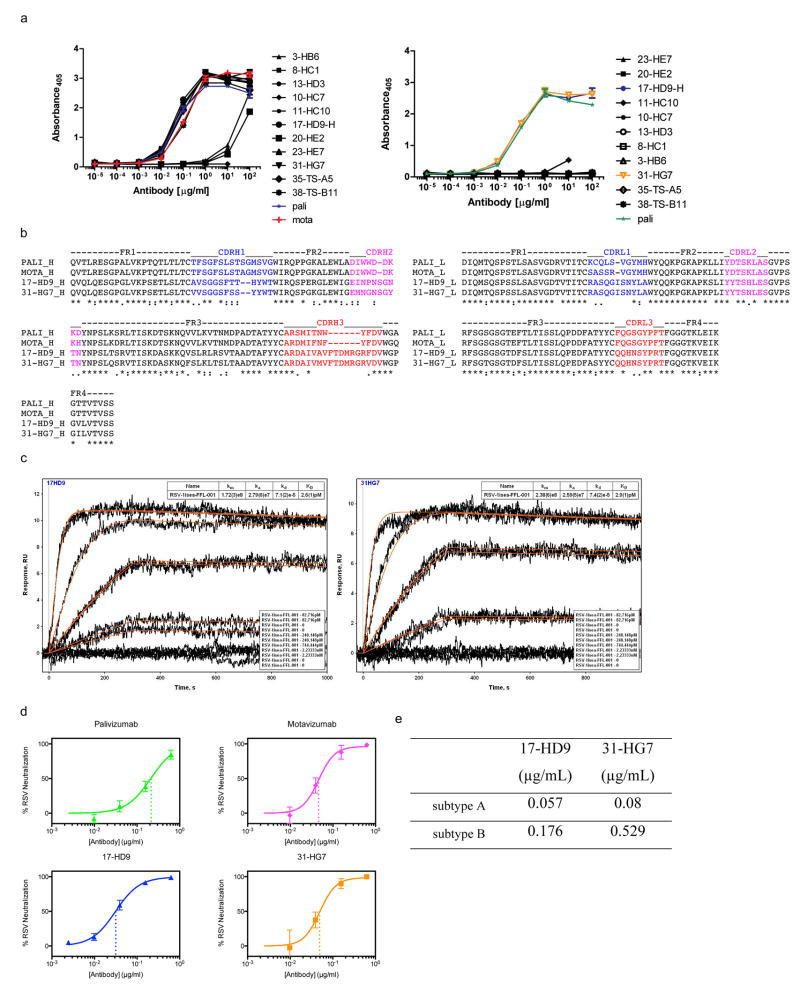
To study the molecular basis for the vaccine-induced neutralizing activity, we used single B cell sorting to isolate epitope-specific monoclonal antibodies (mAbs)28 from memory B cells of one animal from the particle group with potent serum neutralizing activity. We isolated B cells that bound strongly to FFL_001 but not to a double mutant of FFL_001 (FFL_001_N72Y_K82E) containing both Pali escape mutations. Following DNA sequencing of antibody variable genes in those cells, we produced 11 recombinant mAbs, of which eight bound with high avidity to FFL_001 and two (17-HD9 and 31-HG7) bound with high avidity to RSV F protein (Fig. 4a and Extended Data Fig. 8). SPR revealed that these two mAbs, which are clonal relatives, have extremely high affinities (KD ≈ 3 pM) for the scaffold FFL_001 that elicited them when mounted on the particle (Extended Data Fig. 8). Concomitant with high affinities, these two mAbs have neutralization potencies similar to Mota and higher than Pali by nearly an order of magnitude (Fig. 4b and Extended Data Fig. 8).
Fig 4. Analysis of monoclonal antibodies (mAbs) isolated from an immunized macaque.
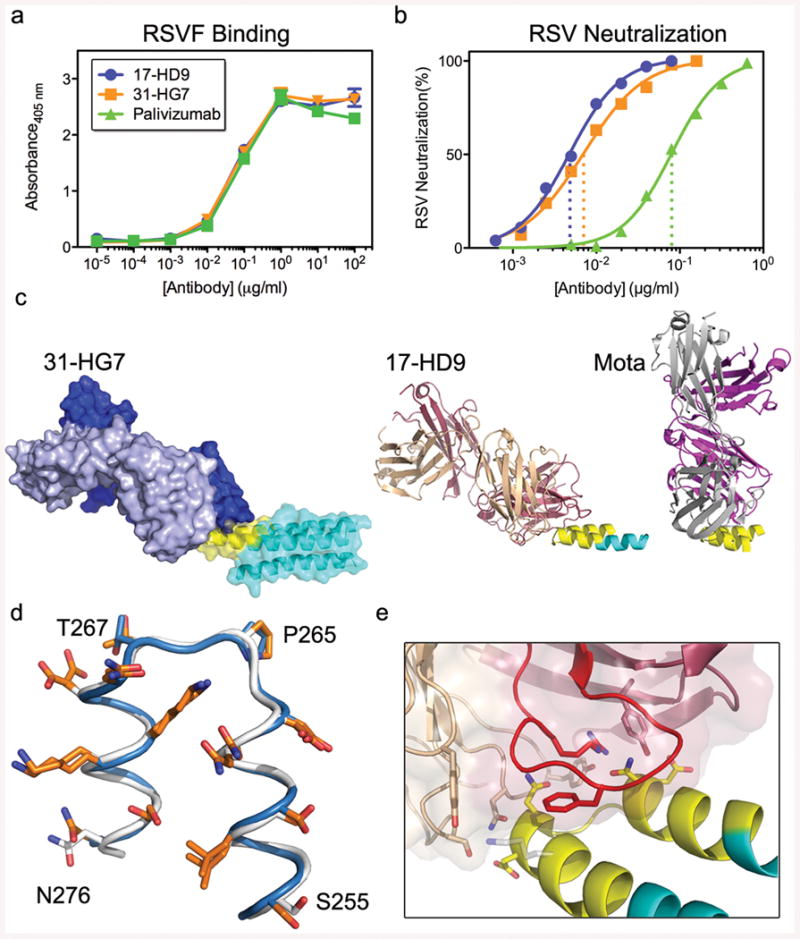
a, ELISA binding of the macaque mAbs (17-HD9 and 31-HG7) and Pali to RSV F. b, Neutralization of RSV by the macaque mAbs and Pali, measured by a microneuralization assay. The IC50s for Pali, 17-HD9 and 31-HG7 were 0.08, 0.005 and 0.007 μg/mL, respectively. c, MR model of 31-HG7 bound to FFL_001 (left), a crystal structure of 17-HD9 bound to a 35-residue helix-turn-helix peptide from FFL_001 (middle), and the crystal structure (PDB: 3ixt) of Mota bound to peptide. The three structures are aligned with respect to the helix-turn-helix epitope. d, Structural alignment of the helix-turn-helix epitopes bound to Mota (blue) and 17-HD9 (white), in which sidechains are colored orange if at least 15% of the total area (backbone+sidechain) of that residue is buried by the respective antibody. 9 positions are buried by both antibodies, two positions in the turn are buried only by 17-HD9 (P265 and T267, RSV residue numbering), and two positions near the peptide termini are buried only by Mota (S255 and N276). e, Close up view of the interface between 17-HD9 and helix-turn-helix epitope. Interaction residues are shown in stick, and the CDRH3 is colored red. K82/272 (scaffold numbering/RSV numbering), at the edge of the interface, is colored gray.
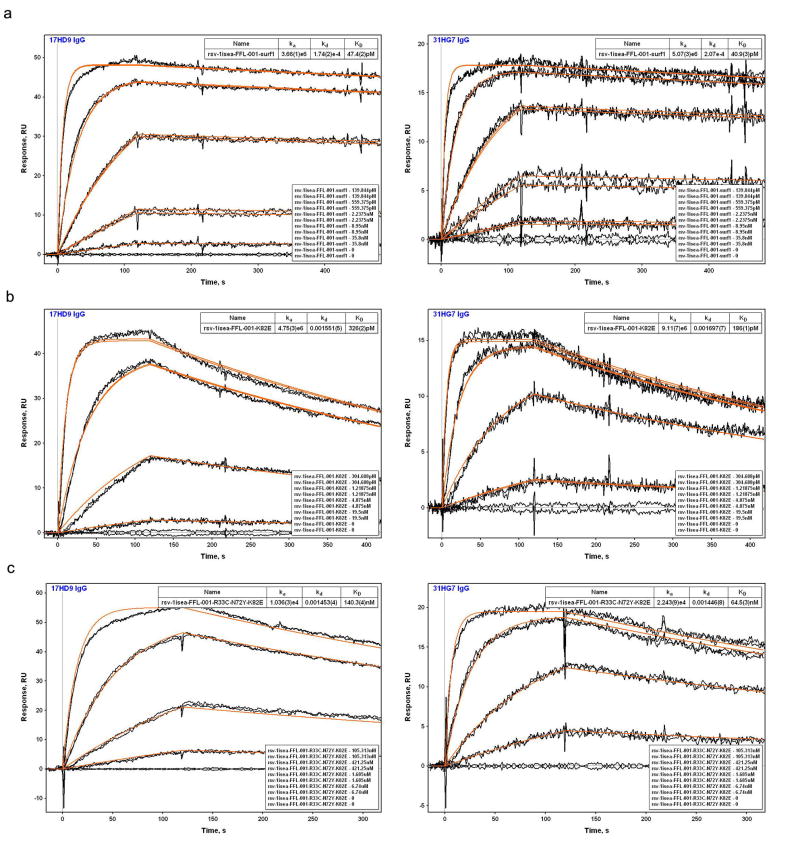
To map the epitopes for 17-HD9 and 31-HG7, we assessed binding to several scaffold variants (Extended Data Fig. 9). Both mAbs: (i) bind with very high affinity (KDs of 40-50 pM) to FFL_001_surf1 that has an antigenically distinct surface from FFL_001 outside the RSV epitope, (ii) retain high affinity (KDs of 180-330 pM) for the FFL_001_K82E Mota escape mutant, (iii) retain modest affinity (KDs of 60-140 nM) for the FFL_001_N72Y_K82E double escape mutant, and (iv) lack detectable affinity for FFL_MPV_001 that swaps RSV residues on FFL_001 to those at the analogous positions on human metapneumovirus that has a similar helix-turn-helix conformation (RMSD 0.9 Å, PDBid: 4DAG29) but very different amino acid sequence. These results indicate that the two macaque mAbs target the same helix-turn-helix epitope as Mota and Pali but have different fine specificities.
To understand the structural basis for the binding and neutralizing potency of these macaque mAbs, we pursued crystallography of 17-HD9 and 31-HG7 complexes with FFL_001. We obtained crystals of the 31-HG7+FFL_001 complex that diffracted to 3.8 Å, which was sufficient to determine a molecular replacement (MR) solution using the FFL_001 crystal structure and a composite Fab model, but insufficient to perform detailed rebuilding and refinement. The MR solution allowed determination of the rigidbody orientation of 31-HG7 relative to FFL_001 and demonstrated that 31-HG7 approaches the helix-turn-helix from a different angle than Mota (angle difference is ∼56°, Fig. 4c). We also obtained crystals and determined the structure of the 17-HD9+FFL_001 complex (dmin = 2.5 Å), which contained four complexes of 17-HD9 bound to a 35 residue helix-turn-helix peptide (scaffold substructure) in the asymmetric unit (Fig. 4c and Extended Data Fig. 10). The 17-HD9 complex structures demonstrated that 17-HD9 recognizes essentially the same helix-turn-helix epitope as Mota and Pali—the conformation of the epitope in the 17-HD9 complexes is very similar to that in the structures of Mota+FFL_001 (RMSD 0.5-0.7 Å), RSV F pre-fusion (RMSD 0.3-0.4 Å) and RSV F post-fusion (RMSD 0.5 Å), and 85% of the epitope residues buried by either Mota or 17-HD9 are also buried by the other (Fig. 4d and Supplementary Table 3). While 17-HD9 and Mota bury a similar amount of area on the epitope (690 Å2 versus 683 Å2), 17-HD9 uses a different paratope to make more hydrogen bonds (15-18 versus 7) that plausibly contribute to its higher scaffold affinity and higher neutralization potency (Fig. 4e and Supplementary Tables 4-5). The 17-HD9 complexes are also consistent with the ability of 17-HD9 to bind to the K82E Mota escape mutant—density for the K82 sidechain is absent in two of four 17-HD9 complexes, and K82 is only 37% buried by 17-HD9 in the other two complexes (Fig. 4e); in contrast K82 is 65% buried by Mota and makes a buried salt bridge to Mota light chain residue D50. Taken together, these results demonstrate that epitope scaffold immunization can “re-elicit” neutralizing antibodies that target with high precision an epitope pre-defined by a protective antibody.
Concluding Remarks
We have demonstrated that small, thermally and conformationally stable protein scaffolds that accurately mimic the structure of a viral neutralization epitope can induce neutralizing activity in a majority of vaccinated macaques. The results establish the feasibility of epitope-focused and scaffold-based vaccine design, and encourage the application of these strategies for a variety of vaccine targets. The biophysical, structural and functional data on the Mota scaffolds validate the computational design method (FFL), and support its continued development and application to other vaccine epitopes and other types of functional sites. Indeed, the data should encourage the general use of methods employing protein backbone flexibility to design novel functional proteins.
The scaffolds themselves represent promising leads for RSV vaccine research and development (particularly the scaffolds presented on virus-like-particles). Non-replicating RSV vaccine candidates are not tested in RSV naive young infants, the highest priority target population, owing to vaccine-mediated disease enhancement in early clinical trials of formalin-inactived RSV10. Scaffold immunogens that focus antibody responses to a known protective epitope but are otherwise unrelated to RSV may have a lower safety barrier to testing in human infants than other non-replicating RSV vaccine approaches. The frequency, magnitude, and durability of neutralizing responses to these scaffolds remain to be optimized, by varying such parameters as adjuvant, dose, schedule, particle display system and mode of delivery. In the context of vaccinating diverse humans (or NHPs) with different, time-dependent B cell repertoires, neutralizing antibody responses to a single epitope may be more variable than responses to whole antigens or pathogens containing multiple neutralization epitopes. (Indeed, our discordant results in mice compared to macaques may reflect differences in species-dependent repertoires.) Thus, epitope-focused or scaffold-based vaccines for RSV or other pathogens may also be improved by inclusion of more than one epitope.
Features of the very potent neutralizing mAbs isolated from one vaccinated macaque offer implications for vaccine design. These mAbs had unusually high affinity for the eliciting antigen, for example when compared to mAbs isolated from macaques that had been immunized with different, more conformationally labile antigens using similar regimens28. This suggests that rigid epitope structures may more efficiently induce extremely high affinity antibodies, a possibility that merits further investigation. In cases of antigenically highly variable pathogens such as HIV, influenza, or Hepatitis C virus, the vaccine challenge is to induce responses to conserved but immunorecessive epitopes instead of the strain-specific epitopes that dominate the response to native antigens. Such conserved epitopes––the sites of vulnerability targeted by broadly neutralizing antibodies––are typically in close physical proximity to variable residues, making precision of immuno-focusing a vaccine requirement. Our crystallographic finding that scaffold-elicited mAbs re-capitulate the Mota neutralization specificity with high precision provides proof of principle that epitope-focused vaccine design can meet this immuno-focusing challenge.
Methods
Fold From Loops
The Fold From Loops (FFL) protocol was implemented in the Rosetta molecular modeling platform30. The four stage protocol is described in the main text, here we provide additional details. In stage i), FFL required two structural inputs in the form of atomic coordinates: a functional motif and a target topology (Extended Data Fig. 1). In stage ii), ab initio folding sampled conformational space using a fragment assembly protocol21,31. Fragments of lengths 3 and 9 were generated based on the sequence and secondary structure of PDBid: 3LHP, chain S using the program NNMAKE31, and were provided as input to FFL. The sequence location of the functional motif in the target topology was defined by a loop file (Supplementary Methods). FFL automatically appended extended polypeptide chains to the termini of the functional motif to match the total number of residues of the template topology. The ab initio simulations were performed in Rosetta centroid mode in which the backbone atoms were explicitly defined along with a pseudo-atom representing the side-chain. The FFL protocol allows optional use of residue distance restraints to bias the folding trajectories towards similar structures to the target topology, using methods described previously31. Here, distance restraints between Cα atoms were extracted from the coordinates of the target topology. Restraints were defined for residue pairs that were at least 6 residues apart, except for the segment composed of the functional motif plus 5 residues up- and down-stream. The FFL restrained simulations were performed allowing standard deviations of 1.5 or 3 Å for each Cα-Cα distance. FFL allows the user to set the termini of the functional motif as moveable in order to favor smooth structural transitions between the structurally rigid functional motif and the rest of the protein. Here, two residues at each terminus of the functional motif were set as moveable, while the rest of the backbone dihedral angles within the functional motif were fixed. In stage iii), low resolution models generated in the folding stage were filtered by RMSD relative to the coordinates of the target topology. Models with RMSD > 5 Å were discarded, and the remaining were subjected to iterative sequence design and conformational relaxation. Side-chain conformations from the functional motif were imposed and kept fixed throughout the design and relaxation simulations. Sequence design was performed with RosettaDesign21 in which each position outside of the functional motif was allowed to mutate to any amino acid except cysteine. After each step of sequence design, a step of all-atom relaxation31 was performed, composed of several rounds of small perturbations in the backbone dihedral angles, side-chain repacking, and energy minimization. The FFL designs were generated by employing 3 cycles of sequence design and structural relaxation, but the number of cycles is user-adjustable. FFL allows the option to perform sequence design at selected positions within the functional motif. In the case of the mota epitope as functional motif, only one face of the helix-turn-helix motif constituted the antibody binding interface, so the side chains of the other face were allowed as designable. In stage iv) designs from four separate FFL simulations of 10000 models each were filtered and subjected to human-guided optimization. Rosetta full-atom energy was the first filter; the 50 lowestenergy designs from each simulation were retained for further examination. Next, we applied a composite filter to select designs with the best structural features according to Ramachandran score 21, counts of buried polar atoms not involved in hydrogen bonds, and core packing as assessed by RosettaHoles32. Designs that scored within the top 25 by each filter were retained; this included 4 to 9 designs for each of the four simulations. Several of the design models were curved helical bundles, unlike the starting topology that was relatively straight, so we employed Helanal33 to identify the straightest designed bundles. FFL_005, FFL_007, and FFL_008 were identified by Helanal. Human-guided computational design and subsequent relaxation was then performed to improve core packing and to modify surfaces, as described in the main text. At this stage, mutations in the core nearly always caused the Rosetta energy to increase, even after relaxation, but mutations were allowed if the post-relaxation energy was within 20% of the pre-mutation energy. The different criteria used to select each design, the number of mutations in the core, and the Rosetta energies at the different stages are summarized in Supplementary Table 1. Example command lines for FFL and other Rosetta modes are provided in Supplementary Information.
Epitope-Scaffolds expression and purification
Epitope-scaffolds were purified as previously described16. For NMR, 15N isotopically labeled samples of FFL_001, FFL_005, FFL_006 and FFL_007 were grown in minimal MOPS medium supplemented with 1g/L of 15N ammonium chloride. The starter cultures were expanded to 1L of MOPS and incubated overnight at 37 °C; 3 ml of 40% 15N glucose was added to continue growth, 250 μM of IPTG was added to the cultures to induce protein expression, and the cells were then incubated overnight at 16 °C.
Circular dichroism
Experiments were performed on Aviv 62A DS and Aviv 420 spectrometers. Far-UV wavelength scans (190-260 nm) of protein samples with concentration ranging from 15 to 25 μM were collected in a 1 mm path length cuvette. Temperature-induced denaturation was monitored by changes in ellipticity at 210 nm, over the temperature range 1 to 99 °C, in 2 °C increments every 3 minutes. The resulting data was converted to mean residue ellipticity, fitted to a two-state model, and melting temperatures (Tm) were obtained34. Chemical denaturations were performed using GuHCl in increments of 0.2 M, and the total GuHCl concentration ranged from 0 to 8 M. The protein concentrations ranged from 1 to 3 μM. The free energy differences for the unfolding transitions (ΔG) were obtained from the denaturation curves by a non-linear least squares fitting using a two-state unfolding and linear extrapolation model35.
Light Scattering
The monodispersivity and molecular weight of purified proteins were assessed by HPLC (Agilent, 1200 series) coupled in-line to a static light scattering device (miniDAWN TREOS, Wyatt). 100 μL of 1-2 mg/mL protein sample was used and the collected data was analyzed with the ASTRA software (Wyatt).
Preparation of FFL_001 conjugated Virus Like Particles
To conjugate FFL_001 to Hepatitis B (HepB) virus like particles we utilized an engineered version of the HepB core antigen (HBcAg) in which a segment containing the lysine for chemical conjugation (GGKGG) was introduced between P79 and A80 26. This places a lysine at the tip of the major immunodominant region of HBcAg. HBcAg constructs were expressed in E. coli using a published protocol16 up to the point of cell lysis. Lysed cells were pelleted and the supernatant was concentrated approximately 4 fold using spin concentrators with 100 kDa membrane cutoff (Vivaspin). Next, 1 mL aliquots of the concentrated supernatant were layered on top of 10%-50% sucrose gradients with the total volume of 10.5 mL. The gradients were spun in an ultracentrifuge at 29,000 rpm for 4 hours, fractionated in twelve 1 mL fractions and analyzed by SDS-PAGE. Fractions containing HBcAg were dialyzed overnight in 4 L of EndoFree 1x PBS in a dialysis membrane of 100 kDa to remove the sucrose and then concentrated using concentrators with 100 kDa cutoff (Vivaspin). Conjugation of FFL_001_R33C to HBcAg particles was achieved with two heterobifunctional chemical crosslinkers (PEG4-PFB and MHPH, Solulink) that react covalently with specific residues in each of the protein counterparts (PEG4-PFB with lysine on HBcAg, MHPH with cysteine on FFL_001_R33C) and also react with each other to form a covalent bond between the formylbenzamide (FB) group of PEG4-PFB and hydrazinonicotinamide (HyNic) group of MHPH. After functionalization of HBcAg with PEG4-PFB and of FFL_001_R33C with MHPH, the HyNic-4FB conjugation was performed to obtain FFL_001_R33C covalently conjugated to HBcAg particles. Dialyzed fractions of HBcAg were buffer-exchanged to modification buffer (100 mM PBS, 150 mM NaCl and pH = 7.4) using Zeba desalting columns (Solulink) according to the manufacturer's protocol. The HBcAg fractions in modification buffer were then incubated with 7-fold molar excess of PEG4-PFB (stock solution of 20 mg/mL) and the conjugation reaction proceeded for 2 hours. Upon the completion of the conjugation, the reaction product was buffer-exchanged to Conjugation buffer (100 mM of PBS, 150 mM NaCl at pH 6.0) using desalting columns. To conjugate MHPH to FFL_001_R33C, the scaffold was incubated under reducing conditions (2 mM DTT) for 15 min to prevent potential dimerization and to maximize the number of cysteines available to react with MHPH. After the reduction step the scaffold sample was buffer-exchanged to conjugation buffer and incubated with 10-fold molar excess of MHPH (stock solution of 20 mg/ml) during 1 hour. The conjugation between the functionalized FFL_001_R33C and HBcAg particles was performed overnight in a scaffold to particle molar ratio of 3:1. The reaction product was split in 1 mL aliquots and scaffold conjugated particles were purified using the sucrose gradient protocol described above. To confirm the conjugation of the FFL_001_R33C scaffold to the HBcAg particles, the fractions resulting from the sucrose purification were analyzed by SDS-PAGE. SPR studies were also used to verify that Mota Fab had high affinity for scaffold-conjugated particles (not shown). Preparation of low-endotoxin protein for immunization studies was carried out as previously described16.
Crystallography
Crystallization conditions and structure determination protocols are provided in supplementary information.
NMR
Samples were prepared in 25mM sodium phosphate, 150mM NaCl, pH 7.0, and 90% H2O/10% D2O at a concentration of 500uM. HSQC spectra for FFL_001, FFL_005, FFL_006 and FFL_007 were recorded on a Bruker Avance 600 MHz NMR spectrometer equipped with an actively shielded z-gradient triple resonance cryo-probe. All spectra were recorded at 25°C. Spectra were processed using NMRPipe36 and NMRView37.
Surface plasmon resonance
All experiments were carried out on a Biacore 2000 (GE Healthcare) at 25 °C with HBSEP (0.01 M HEPES pH 7.4, 0.15 M NaCl, 3 mM EDTA and 0.005% (v/v) Surfactant P20) (GE Healthcare) as running buffer. For binding analysis, 200-500 response units (RUs) of IgG was captured on a CM5 sensor chip containing 8000-9000 RUs of amine-linked mouse anti-human IgG (Human Antibody Capture kit, GE Healthcare). Samples of different protein concentrations were injected in duplicates over this surface at a flow rate of 50-100 μl/min. If necessary, surface regeneration was performed with two 60 seconds injections of 3 M MgCl2 at a flow rate of 10 μl/min. One flow cell contained anti-human IgG only and its interaction with the analyte was used as reference. Alternatively, other formats were employed: i) epitope-scaffolds were amine-coupled to the sensor chip and Mota Fab was flowed as analyte; ii) NHP_ D039 isolated monoclonal (17-HD9 and 31-HG7) IgGs were amine coupled to the chip and the epitope-scaffold was flowed as analyte; iii) Motavizumab Fab was amine-coupled to the chip and the epitope-scaffold was flowed as analyte. The alternative formats were performed with identical flow rates as mentioned above. Data preparation and analysis were performed using Scrubber 2.0 (BioLogic Software).
Mouse Immunization
A total of nine groups of four mice each were immunized three to six times on a monthly schedule, and different doses, adjuvants, and routes of administration were tested in addition to the different immunogens.
Macaque Immunization
Sixteen rhesus macaques (male, 4 years old) were screened for antibody to RSV by enzyme immunosorbent assay prior to initiation of the study. Briefly, 96 well plates were coated with 100 ng of RSV whole virus lysate in carbonate buffer. Serum from NHPs (1:100 and 1:200 dilution) were added to the plates and incubated for 30 minutes at room temperature. Serum was decanted, and plates were washed with PBS-Triton-X100. HRP-conjugated secondary antibody was added to the plates and incubated for 30 minutes at room temperature. The secondary antibody was decanted, plates washed with PMB-Triton-X-100, and TMB substrate was added. Substrate incubated for ten minutes then was stopped with 1N H2SO4. Plates were read immediately on a spectrophotometer at an OD of 450 nm. Positive results were indicated by OD values of greater than 1.000. All animals were seronegative by this assay, none registering an OD value higher than 0.07. Measurements were done in triplicate and compared to positive controls using Synagis antibody. Rhesus macaques were immunized with FFL scaffold monomers and FFL_001-conjugated-HepBcAg-particles. Immunogens were scaffold monomers FFL_001, FFL_005, FFL_007 and FFL_001-conjugated-HepBcAg-particles, in which the latter are referred to as HepBcAg_001. Four animals per immunogen were immunized by the intramuscular route at 0, 4, 8, 12, and 18 weeks. Animals were injected with 1 mL total volume, consisting of antigen and Adjuplex adjuvant diluted into PBS, with 0.5 mL injected into each arm. The first immunization included a total of 200 μg of scaffold equivalent; subsequent immunizations included a total of 100 μg scaffold equivalent. Plasma was taken at weeks 0, 2, 4, 5, 8, 9, 12, 13, 16, 18, 19, 20, 28, 32.
Recombinant protein ELISA assay
384-well plates (Nunc) were coated with antigen (2 μg/mL of FFL scaffolds or RSV F) and incubated overnight at 4°C. The plate was washed 3x, incubated with blocking buffer for 2 h at room temperature, and washed once more. Serum samples were serially diluted in blocking buffer and applied to the plate. Following overnight incubation at 4 °C, the plate was washed 4x. A 1:4000 dilution of alkaline phosphatase-conjugated anti-monkey IgG (Fitzgerald Industries) was applied to the plate and incubated for one hour at room temperature. The plate was washed 4x and then incubated with 4-nitrophenyl phosphate disodium salt hexahydrate substrate solution (Sigma) for thirty minutes. Absorbance was measured at 405 nm using a BioTek Power Wave HT plate reader. All washes were performed using a BioTek EL406 plate washer. For NHP sera, ELISA endpoint titers indicate the serum dilution that resulted in an absorbance two fold over naïve sera at the same dilution. For mouse sera, ELISA end point titers indicate the serum dilution that resulted in an absorbance two fold over background. Background was assigned based on the absorbance of wells that received secondary Ab only, and was ∼0.1 absorbance units. The same assay was used to measure binding of recombinant purified monoclonal antibodies to the different antigens, but binding time of purified monoclonal antibodies to plate bound antigen was reduced to 1 h. The concentrations of the purified monoclonal antibodies ranged from 100 mg/mL to 0.01 mg/mL.
RSV F protein for ELISA
Plasmid pcDNA3.1 encoding a codon-optimized version of RSV A2 F plus a GCN4 trimerization domain and HIS-tag was transfected via polyethleneimine (Polysciences, Inc.) into 293-F cells (Life Technologies). Four days later, recombinant RSV F was purified from 293-F culture supernatant via a HisTrap HP column (GE Healthcare) using an AKTA FPLS (Amersham Biosciences).
Whole virus ELISA assay
Immulon 96-well ultra-high binding plates (Thermo Scientific) were coated with 100 ng/well of purified human RSV viral lysate (ABI) in BupH Carbonate Buffer (pH 9.5) (Pierce Thermo Scientific) in a volume of 100 μL/well for positive wells or negative wells (no antigen) with 100 μL/well of plain BupH Carbonate Buffer overnight at 4°C. The next day, coating buffer was removed and plates were blotted dry before the addition of 275 μL/well of blocking solution (1×PBS, 5% non-fat dry milk, 1% normal goat serum) for 1 hour at room temperature. Sera from immunized animals were diluted 1:100-1:51200 (2-fold dilutions) in sample diluent (1×PBS, 0.1% Triton X-100). Human anti-RSV F (Synagis/Palivizumab) (MedImmune) was used as a positive control and diluted two-fold from 100 mg/mL to 1:40000-1:5120000. Samples and controls were added to the plate (100 μL/well) – into two wells, one coated with the RSV viral lysate and other with no antigen, and incubated for 30 minutes at room temperature. Serum was removed, plates were washed by immersion and blotted dry. Secondary antibody (goat anti-monkey IgG HRP) (Vector) was diluted 1:1000 in 1×PBS, 5% non-fat dry milk, added at 100 μL/well and incubated at room temperature for 30 minutes. Upon removal of the secondary antibody, the plates were washed by immersion and blotted dry. The plates were developed with addition of 1-Step Ultra TMB ELISA substrate (Thermo Scientific) followed by 2N sulfuric acid in equal volumes. The plates were read immediately on an ELISA plate reader at 450 nm (SpectraMax M5, Molecular Devices). ELISA titers were calculated as the last dilution where the absorbance of a sample on the RSV viral lysate well is two times the absorbance of the same sample on the no antigen well. The procedure described above was also used to measure RSV F reactivity of the NHP_ D039 isolated monoclonal antibody samples. These samples were diluted two-fold from their various concentrations to 1:100-1:51200.
Neutralization assays
The plaque reduction assay was performed using RSV subtype A2 as previously described 38. Serum dilutions (1:10, 1:40, 1:160, and 1:640) were mixed 1:1 with virus to yield final dilutions of 20, 80, 320, and 1280. The 50% and 60% plaque reduction neutralizing activity were determined by regression curve analysis. The microneutralization assay method is provided in Supplementary Information. The flow cytometry neutralization assay was performed as previously described39.
Monoclonal isolation from immunized NHP
DNA sequences for antigen-specific monoclonal antibodies were isolated by B cell sorting, RT-PCR, and DNA sequencing as previously described28. The difference in the protocol occurred on the last step of cellsorting where epitope specific B-cells were required to be FFL_001(+) and FFL_001_N72Y_K82E(-). Both FFL scaffolds had an engineered cysteine (R33C) that was conjugated with biotin-maleiemide (Solulink) according to the manufacturer protocol. Complete, high-quality sequences with unambiguous nucleotide identifications were obtained for the variable regions of both heavy and light chains for approximately 24 antibodies, from which 12 were recombinantly expressed and 11 were successfully purified.
IgG and Fab Expression and Purification
IgGs and Fabs were produced in FreeStyleTM 293F (Invitrogen) suspension cultures by co-transfection of pFUSEss (IgG, Invitrogen) or pHLsec (Fab) expression vectors containing the heavy and light chains, using 293Fectin (Invitrogen). Supernatants were harvested 96 h after transfection. IgGs were purified using ProteinA SepharoseTM (GE Healthcare) and dialyzed overnight into PBS (0.01 M sodium phosphate, pH 7.4, 0.137 M sodium chloride). Fab supernatants were concentrated to 100 mL by tangential flow concentration (Milipore), and Fabs were purified by CaptureSelect IgG-CH1 (BAC) affinity chromatography followed by dialysis in HBS overnight.
Extended Data
Extended Data Figure 1.
Overview of the Fold from Loops (FFL) computational procedure. Initially large conformational spaces are sampled by low resolution folding, and subsequently iterative sequence design and small structural optimizations are performed to accommodate the target functional motif.
Extended Data Figure 2.
Properties of designed proteins in this study. a) Sequence alignment of the FFL designs. 3LHP_S is the protein used as the template topology. b) Sequence alignment for the FFL_surf series designed based on the FFL_001 design model. c) Parameters and filtering criteria and results in the design process for FFL designs. a SD - standard deviation allowed on the constraints derived from the target topology; b Design epitope segment- design of residues within the epitope segment that were not part of the epitope-antibody interface; c Filtering criteria based on the helix bend angle; d Rosetta energy after human guided optimization. d) Structural diversity in the FFL design models. Values give the backbone RMSD in Å between two designs or between the template (3LHP_S) and the designs. e) Mutational diversity in the FFL designs. Values give the number of mutations between two designs or between the template (3LHP_S) and the designs.
Extended Data Figure 3.
Structural properties of FFL designs in solution. a) Characterization of the oligomeric state by SECMALS. All molecules that showed a single monodisperse species by SEC had molecular weights computed from MALS that were consistent with expectation for a monomer (approximately 15 kDa). b) Secondary structure and thermal stability of FFL designs assessed by circular dichroism. Wavelength scans at T = 25 ° (left column) show the double minima typical for helical proteins. Thermal denaturation curves (right row) reveal high thermostability. d) HSQC spectra of several 15N labeled FFL designs. The spectra exhibit features typical for properly folded proteins with high alpha-helical content, particularly FFL_006. FFL_005 and FFL_007 exhibited reduced dispersion possibly due to self-association at higher concentrations.
Extended Data Figure 4.
SPR data for FFL designs binding to Mota or Pali. a) Binding of FFL_005 and FFL_007 to Mota, in which the epitope scaffolds were amine-coupled to the sensor chip and Mota Fab was used as analyte. The concentrations of Mota Fab ranged from 950 nM to 436.5 pM and were used in serial dilutions with a dilution factor of three. b) Binding of FFL_001 and FFL_005 to Mota. Mota IgG was captured on the sensor chip by anti-human IgG and epitope scaffolds were used as analytes. The concentrations of scaffold ranged from 6.9 nM to 255.6 pM and were used in serial dilutions with a dilution factor of three. Kinetic fits are shown in red for both panels. c) Binding of FFL_001 and FFL_007 to Pali assessed by SPR. Pali IgG was captured by anti-human IgG on the sensor ship, and scaffolds were analytes. d) Mota binding specificity of FFL_001 assessed by SPR. Mota IgG was the ligand, captured by antihuman IgG on the sensor chip, and FFL_001 (blue) and an epitope point mutant of FFL_001 (FFL_001_K82E, black) were analytes at a concentration of 22 nM. The interaction between FFL_001 and Mota was eliminated by the point mutation.
Extended Data Figure 5.
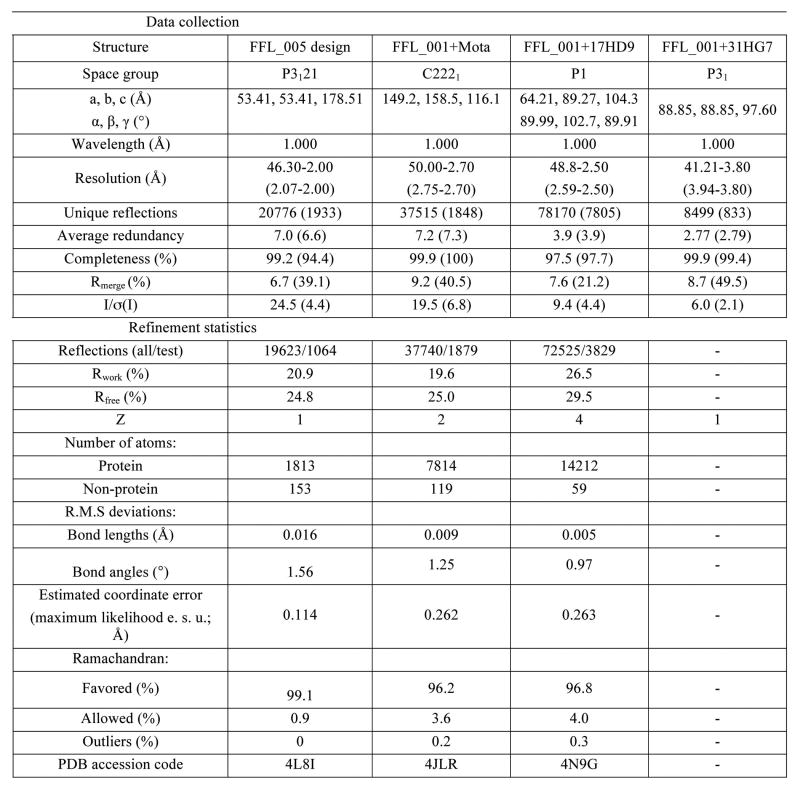
Crystallographic statistics for crystal structures determined. Values in parentheses refer to the highest resolution shell.
Extended Data Figure 6.
Immunological evaluation of FFL scaffolds by different means. a) Evaluation of scaffolds as probes to detect the presence of epitope-specific antibodies in human sera. Sera from six healthy seropositive individuals were tested by ELISA for reactivity to FFL_001, FFL_001 with two different epitope point mutants (FFL_001_K82E and FFL_001_N72Y), and to recombinant RSV F glycoprotein. b) ELISA endpoint titers from mice immunized with immunogens shown on the x-axis. Autologous titers were measured against 001, 005, 007, or HBcAg particles without conjugated scaffold (triangles), and titers were also measured against RSV F protein (red). Titers after 2 immunizations are on the left, titers after 4 immunizations are on the right. c) ELISA endpoint titers for binding to recombinant RSV F protein, from NHPs immunized with 001, 005, 007, and HBcAg-FFL_001. d) RSV microneutralization assay results for NHPs immunized with 001, 005, 007, and HBcAg-FFL_001. In c) and d), values at each timepoint are mean ± standard deviation computed for the four animals per group at that timepoint.
Extended Data Figure 7.
Neutralization of RSV by week 20, post-5 immunization NHP sera assessed by a flow cytometry based assay. a) The neutralization curves for several vaccinated animals are shown. 07C0012 was immunized with FFL_001; 07C004 and 07D039 were immunized with HBcAg-FFL_001; 07C0010 and 07D087 were immunized with FFL_007. b) Table showing 50% neutralization titers measured in two independent assays. c) Flow cytometry assay results for RSV subtypes A and B.
Extended Data Figure 8.
Properties of NHP mAbs isolated by B cell sorting from an animal immunized with HBcAg-FFL_001. a) ELISA binding of recombinant NHP mAbs to FFL_001 (left) and recombinant RSV F glycoprotein (right). b) Sequence alignment of heavy (left) and light (right) chains of the Fv domains of NHP mAbs 17-HD9 and 31-HG7 along with Mota and Pali. c) SPR data for mAbs 17-HD9 and 31HG7 binding to FFL_001. mAb IgGs were captured by anti-human IgG on the sensor chip (mAbs were expressed with human Fc) and FFL_001 was flowed as analyte. d) Head-to-head comparison of the neutralization potency of NHP mAbs, Mota, and Pali in the plaque reduction assay. The data values are shown as mean ± standard deviation from two assays. The data were fit by the equation for one site specific binding with Hill slope, implemented in GraphpadPrism. According to the fits, the IC50s were 0.21 ug/mL (Pali), 0.046 ug/mL (Mota), 0.031 ug/mL (17-HD9), and 0.049 ug/mL (31-HG7). e) EC50 values for neutralization of RSV subtypes A and B by 17-HD9 and 31-HG7 as reported by the flow cytometry assay.
Extended Data Figure 9.
SPR data for the binding of NHP mAbs to FFL_001 variants. a) FFL_001_surf1; b) FFL_001_K82E; c) FFL_001_R33C_N72Y_K82E. mAbs were captured by anti-human IgG on the sensor chip (mAbs were expressed with human Fc) and FFL_001 variants were flowed as analytes.
Extended Data Figure 10.
Four complex structures of 17-HD9+peptide in the asymmetric unit, from PDB: 4N9G. The four complexes in the asymmetric unit consisted of two pairs of nearly identical structures (RMSD within each pair was 0.3 Å), with the pairs differing from each other primarily in the Fv angle of approach to the epitope (angle difference ∼9°) and in the Fab elbow angle (angle difference ∼10°); differences within the peptide between pairs were small (RMSD over peptide between pairs was 0.7 Å). a) Chains A+B+C; b) chains E+F+D; c) chains H+L+Y; d) chains M+N+Z. e) View of crystal packing interaction, in which the “backside” of one peptide interacts with the “backside” of another. Partial scaffolds (peptides) are packed against each other at crystal contacts between complexes through an interface outside of the epitope, with perfect dyad symmetry broken by a translation along the NCS dyad axis to accommodate complementary packing of apolar side-chains. The crystal packing is incompatible with the scaffold being present as a three helix bundle as in the Mota or 31-HG7 complex structures. Clear density was lacking for the scaffold outside the helix-turn-helix peptide. Scaffold missing density is possibly due to partial proteolysis or unfolding of the scaffold that may have occurred while purified Fab+scaffold complexes incubated at high concentration (∼10 mg/mL) in crystallization liquor for three months prior to crystal formation (see Supplementary Methods). The location and size of solvent channels in the crystal could accommodate the disordered region of the scaffold as an extended, flexible peptide unfolded under the conditions of crystallization, but it is also plausible that limited proteolysis has reduced the scaffold to a minimal structure protected by contacts with the antibody.
Supplementary Material
Acknowledgments
We thank Sergey Menis, Dan Kulp, and Dennis Burton for comments on the manuscript, Yih-En Andrew Ban, Darwin Alonso, and Keith E. Laidig for computing assistance, Erin Gribben and Robert Carnahan for assistance with mouse immunizations, and Chris Slaughter for assistance in statistical analysis. Adjuplex adjuvant was a generous gift of Advanced BioAdjuvants, LLC. Coordinates and structure factors for FFL_005, FFL_001+Mota, and FFL_001+17-HD9 structures have been deposited in the Protein Data Bank with accession codes 4L8I, 4JLR, and 4N9G, respectively. The University of Washington has filed patents relating to immunogens in this manuscript. Materials and information will be provided under MTA. Support for this work was provided by Fundação para a Ciência e a Tecnologia fellowship SFRH/BD/32958/2006 (B.E.C.), NIH NRSA Training Grant fellowship T32CA080416 (J.J.), The Children's Hospital of Philadelphia (P.R.J.), a Bill and Melinda Gates Foundation CAVD award (W.R.S., R.K.S. and D.B.), the International AIDS Vaccine Initiative Neutralizing Antibody Consortium (W.R.S. and D.B.), the International AIDS Vaccine Initiative Neutralizing Antibody Center (W.R.S. and R.T.W.), a grant from the March of Dimes (J.E.C.), National Institute of Allergy and Infectious Diseases grants P01AI094419 (W.R.S. and R.K.S.), 5R21AI088554 (W.R.S), 1UM1AI100663 (W.R.S. and R.T.W), 1R01AI102766-01A1 (Y.L. and R.T.W.), P30AI36214 (from the Center for AIDS Research, University of California, San Diego, to Y.L.), and the National Institute of Allergy and Infectious Diseases Intramural Research Program (B.S.G.). This is manuscript XXXXX from The Scripps Research Institute.
Footnotes
Author Contributions: B.E.C., J.T.B, R.J.L., R.E.K., B.S.G., R.T.W., D.B., R.K.S., J.E.C, P.R.J., and W.R.S. designed research. B.E.C. wrote the code for FFL, designed proteins and performed biophysical characterization. J.T.B., R.J.L., M.C. and M.J.C. performed serological analysis. G.B. and E.S. prepared and characterized particle immunogens. C.C., J.G.J., P.R., and M.A.H. performed x-ray crystallography. O.K. performed biophysical characterization. V.V. performed NMR studies. M.J.C. performed NHP immunizations. O.K., A.S., S.M., A.M.S, Y. A., and E.S. performed protein expression and purification. Y.L. performed B-cell sorting and RT-PCR. B.E.C. and W.R.S. wrote, and all co-authors edited, the manuscript.
References
- 1.Plotkin SA, Orenstein WA, Offit PA. Vaccines. 6. Elsevier; 2012. [Google Scholar]
- 2.Rappuoli R, Mandl CW, Black S, De Gregorio E. Vaccines for the twentyfirst century society. Nat Rev Immunol. 2011;11:865–872. doi: 10.1038/nri3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lozano R, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380:2095–2128. doi: 10.1016/S0140-6736(12)61728-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murray CJ, et al. Disability-adjusted life years (DALYs) for 291 diseases and injuries in 21 regions, 1990-2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380:2197–2223. doi: 10.1016/S0140-6736(12)61689-4. [DOI] [PubMed] [Google Scholar]
- 5.Rerks-Ngarm S, et al. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N Engl J Med. 2009;361:2209–2220. doi: 10.1056/NEJMoa0908492. [DOI] [PubMed] [Google Scholar]
- 6.Agnandji ST, et al. A phase 3 trial of RTS, S/AS01 malaria vaccine in African infants. N Engl J Med. 2012;367:2284–2295. doi: 10.1056/NEJMoa1208394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tameris MD, et al. Safety and efficacy of MVA85A, a new tuberculosis vaccine, in infants previously vaccinated with BCG: a randomised, placebocontrolled phase 2b trial. Lancet. 2013 doi: 10.1016/S0140-6736(13)60177-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Osterholm MT, Kelley NS, Sommer A, Belongia EA. Efficacy and effectiveness of influenza vaccines: a systematic review and meta-analysis. Lancet Infect Dis. 2012;12:36–44. doi: 10.1016/S1473-3099(11)70295-X. [DOI] [PubMed] [Google Scholar]
- 9.Bhatt S, et al. The global distribution and burden of dengue. Nature. 2013;496:504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anderson LJ, et al. Strategic priorities for respiratory syncytial virus (RSV) vaccine development. Vaccine. 2013;31 Suppl 2:B209–215. doi: 10.1016/j.vaccine.2012.11.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.http://www.fda.gov/BiologicsBloodVaccines/Vaccines/ApprovedProducts/.
- 12.Burton DR. Antibodies, viruses and vaccines. Nat Rev Immunol. 2002;2:706–713. doi: 10.1038/nri891. [DOI] [PubMed] [Google Scholar]
- 13.Burton DR, Poignard P, Stanfield RL, Wilson IA. Broadly neutralizing antibodies present new prospects to counter highly antigenically diverse viruses. Science. 2012;337:183–186. doi: 10.1126/science.1225416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu H, et al. Development of motavizumab, an ultra-potent antibody for the prevention of respiratory syncytial virus infection in the upper and lower respiratory tract. J Mol Biol. 2007;368:652–665. doi: 10.1016/j.jmb.2007.02.024. [DOI] [PubMed] [Google Scholar]
- 15.McLellan JS, et al. Structural basis of respiratory syncytial virus neutralization by motavizumab. Nat Struct Mol Biol. 2010;17:248–250. doi: 10.1038/nsmb.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Correia BE, et al. Computational design of epitope-scaffolds allows induction of antibodies specific for a poorly immunogenic HIV vaccine epitope. Structure. 2010;18:1116–1126. doi: 10.1016/j.str.2010.06.010. [DOI] [PubMed] [Google Scholar]
- 17.Ofek G, et al. Elicitation of structure-specific antibodies by epitope scaffolds. Proc Natl Acad Sci U S A. 2010;107:17880–17887. doi: 10.1073/pnas.1004728107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McLellan JS, et al. Design and characterization of epitope-scaffold immunogens that present the motavizumab epitope from respiratory syncytial virus. J Mol Biol. 2011;409:853–866. doi: 10.1016/j.jmb.2011.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Azoitei ML, et al. Computation-guided backbone grafting of a discontinuous motif onto a protein scaffold. Science. 2011;334:373–376. doi: 10.1126/science.1209368. [DOI] [PubMed] [Google Scholar]
- 20.Azoitei ML, et al. Computational design of high-affinity epitope scaffolds by backbone grafting of a linear epitope. J Mol Biol. 2011;415:175–192. doi: 10.1016/j.jmb.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuhlman B, et al. Design of a novel globular protein fold with atomic-level accuracy. Science. 2003;302:1364–1368. doi: 10.1126/science.1089427. [DOI] [PubMed] [Google Scholar]
- 22.Koga N, et al. Principles for designing ideal protein structures. Nature. 2012;491:222–227. doi: 10.1038/nature11600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Correia BE, et al. Computational protein design using flexible backbone remodeling and resurfacing: case studies in structure-based antigen design. J Mol Biol. 2011;405:284–297. doi: 10.1016/j.jmb.2010.09.061. [DOI] [PubMed] [Google Scholar]
- 24.Zhu Q, et al. Analysis of respiratory syncytial virus preclinical and clinical variants resistant to neutralization by monoclonal antibodies palivizumab and/or motavizumab. J Infect Dis. 2011;203:674–682. doi: 10.1093/infdis/jiq100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clarke BE, et al. Improved immunogenicity of a peptide epitope after fusion to hepatitis B core protein. Nature. 1987;330:381–384. doi: 10.1038/330381a0. [DOI] [PubMed] [Google Scholar]
- 26.Jegerlehner A, et al. A molecular assembly system that renders antigens of choice highly repetitive for induction of protective B cell responses. Vaccine. 2002;20:3104–3112. doi: 10.1016/s0264-410x(02)00266-9. [DOI] [PubMed] [Google Scholar]
- 27.Jafri HS, Wu X, Makari D, Henrickson KJ. Distribution of respiratory syncytial virus subtypes A and B among infants presenting to the emergency department with lower respiratory tract infection or apnea. Pediatr Infect Dis J. 2013;32:335–340. doi: 10.1097/INF.0b013e318282603a. [DOI] [PubMed] [Google Scholar]
- 28.Sundling C, et al. High-resolution definition of vaccine-elicited B cell responses against the HIV primary receptor binding site. Sci Transl Med. 2012;4:142ra196. doi: 10.1126/scitranslmed.3003752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wen X, et al. Structure of the human metapneumovirus fusion protein with neutralizing antibody identifies a pneumovirus antigenic site. Nat Struct Mol Biol. 2012;19:461–463. doi: 10.1038/nsmb.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leaver-Fay A, et al. ROSETTA3: an object-oriented software suite for the simulation and design of macromolecules. Methods Enzymol. 2011;487:545–574. doi: 10.1016/B978-0-12-381270-4.00019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rohl CA, Strauss CE, Misura KM, Baker D. Protein structure prediction using Rosetta. Methods Enzymol. 2004;383:66–93. doi: 10.1016/S0076-6879(04)83004-0. [DOI] [PubMed] [Google Scholar]
- 32.Sheffler W, Baker D. RosettaHoles2: a volumetric packing measure for protein structure refinement and validation. Protein Sci. 2010;19:1991–1995. doi: 10.1002/pro.458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bansal M, Kumar S, Velavan R. HELANAL: a program to characterize helix geometry in proteins. J Biomol Struct Dyn. 2000;17:811–819. doi: 10.1080/07391102.2000.10506570. [DOI] [PubMed] [Google Scholar]
- 34.Greenfield NJ. Using circular dichroism collected as a function of temperature to determine the thermodynamics of protein unfolding and binding interactions. Nat Protoc. 2006;1:2527–2535. doi: 10.1038/nprot.2006.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Greenfield NJ. Determination of the folding of proteins as a function of denaturants, osmolytes or ligands using circular dichroism. Nat Protoc. 2006;1:2733–2741. doi: 10.1038/nprot.2006.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Delaglio F, et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 37.Johnson BA, Blevins RA. NMR View: A computer program for the visualization and analysis of NMR data. J Biomol NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 38.Murphy BR, Sotnikov AV, Lawrence LA, Banks SM, Prince GA. Enhanced pulmonary histopathology is observed in cotton rats immunized with formalin-inactivated respiratory syncytial virus (RSV) or purified F glycoprotein and challenged with RSV 3-6 months after immunization. Vaccine. 1990;8:497–502. doi: 10.1016/0264-410x(90)90253-i. [DOI] [PubMed] [Google Scholar]
- 39.Chen M, et al. A flow cytometry-based assay to assess RSV-specific neutralizing antibody is reproducible, efficient and accurate. J Immunol Methods. 2010;362:180–184. doi: 10.1016/j.jim.2010.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.