Highlights
-
•
Identifies components required for CoV 2′O-MTase activity including structural motifs and interaction partners.
-
•
Demonstrates attenuation of NSP16 mutants in multiple CoV strains.
-
•
Defines innate immune components including MDA5 and IFIT proteins that mediate the attenuation of 2′O-MTase CoV mutants.
-
•
Provides approaches to exploit 2′O-MTase pathways for antiviral treatment of CoVs and other viruses.
Keywords: CoV, SARS-CoV, 2′O-Methyl-transferase, 2′O-MTase, NSP16, MDA5
Abstract
The recent emergence of Middle East Respiratory Syndrome Coronavirus (MERS-CoV), nearly a decade after the Severe Acute Respiratory Syndrome (SARS) CoV, highlights the importance of understanding and developing therapeutic treatment for current and emergent CoVs. This manuscript explores the role of NSP16, a 2′O-methyl-transferase (2′O-MTase), in CoV infection and the host immune response. The review highlights conserved motifs, required interaction partners, as well as the attenuation of NSP16 mutants, and restoration of these mutants in specific immune knockouts. Importantly, the work also identifies a number of approaches to exploit this understanding for therapeutic treatment and the data clearly illustrate the importance of NSP16 2′O-MTase activity for CoV infection and pathogenesis.
1. Introduction
Coronaviruses represent important human pathogens that have emerged multiple times from zoonotic reservoirs over the past few centuries (Becker et al., 2008, Graham et al., 2013, Li et al., 2005, Nicholls et al., 2003, Perlman and Netland, 2009). In the aftermath of severe acute respiratory syndrome coronavirus (SARS-CoV) emergence in 2003, substantial efforts were made to improve our understanding of CoV infection and pathogenesis in order to develop novel therapeutics for current and future CoV mediated outbreaks (Perlman and Netland, 2009). Despite significant advances over the past decade, broadly effective treatments for SARS-CoV remain elusive. Coupled with the recent emergence of both Middle East Respiratory Syndrome (MERS) CoV in 2012 (Corman et al., 2012, Josset et al., 2013) and a virulent strain of porcine epidemic diarrhea virus (PEDV) (Huang et al., 2013), the lack of progress underscores the continued need to study and develop treatment for this family of viruses.
As the SARS-CoV outbreak progressed, computational models predicted a number of enzymatic functions conserved in the viral poly-protein including a conserved 2′O-methyl-transferase activity (2′O-MTase) for CoV non-structural protein 16 (NSP16) (Snijder et al., 2003). While unknown at the time, subsequent immunity research revealed a role for 2′O-methylation of viral mRNA in distinguishing between self/non-self (Daffis et al., 2010, Zust et al., 2011). The presence of these host and virus effectors suggested the importance of 2′O-methylation as a virulence determinant during CoV infection. In this review, we examine NSP16 interaction partners and activity as well as the factors that mediate host responses. In addition, the manuscript explores approaches to exploit these pathways for therapeutic treatment of both current and emergent disease. Overall, the review seeks to update the current state of the CoV NSP16 field and possibilities for future therapeutics based on targeting 2′O-MTase activity and corresponding immune responses.
2. mRNA capping and host recognition
Overcoming the host immune response is paramount to the success of any viral infection. Since the immune response is predicated on recognition, viruses have evolved means to disrupt host sensing through either direct antagonism of pathway components or molecular mimicry of host processes (Decroly et al., 2012). An important example of the latter is the duplication of capping elements for viral mRNAs (Decroly et al., 2012). Eukaryotic hosts utilize a 5′-terminal capping system to promote efficient nuclear export, robust translation, and enhanced stability of host mRNA. In addition, mRNA capping structure also helps to distinguish between self/non-self RNA and can lead to initiation of the host immune response. In recent years, unprotected 5′-triphosphates on nascent RNA (Fig. 1A) has been identified as part of the recognition trigger for host sensor molecule retinoic acid inducible gene I (RIG-I) leading to down stream type I IFN induction (Hornung et al., 2006, Myong et al., 2009). Similarly, both IFN-induced protein with tetratricopeptide repeats (IFIT) 1 and IFIT5, highly induced interferon-stimulated genes (ISGs), also bind exposed 5′-triphosphate on viral RNA, interfering with their activity through competition with EIF4E (Diamond, 2014, Pichlmair et al., 2011, Habjan et al., 2013). Triphosphate cleavage and addition of the 7-methylguanosine cap effectively protect host mRNA from this particular targeting (Fig. 1B–D). However, unmethylated 2′O on the ribose of cap-0 RNA has also been identified as a pathogen associated molecular pattern (PAMP); recognized by host sensor molecule Melanoma Differentiation-Associated protein 5 (MDA5) as well the effector IFIT family, the absence of 2′O-methylation on viral RNA induces a more robust type I IFN response that attenuates viral replication and infection in vitro and in vivo (Fig. 1E) (Daffis et al., 2010, Zust et al., 2011). In contrast, viral mRNA maintaining both the 7-methylguanosine cap and 2′O-methylation (caps 1 and 2, Fig. 1F) remains viable to levels similar to host mRNA. Together, recognition of these moieties and the subsequent host response indicate the importance of mRNA capping restrictions as a barrier to viral infection. These findings also raise the possibility that capping activities may preferentially function as a defense mechanism against cell intrinsic antiviral effectors, suggesting the possibility that viral mRNA are preferentially translated by cap-independent mechanisms.
Fig. 1.
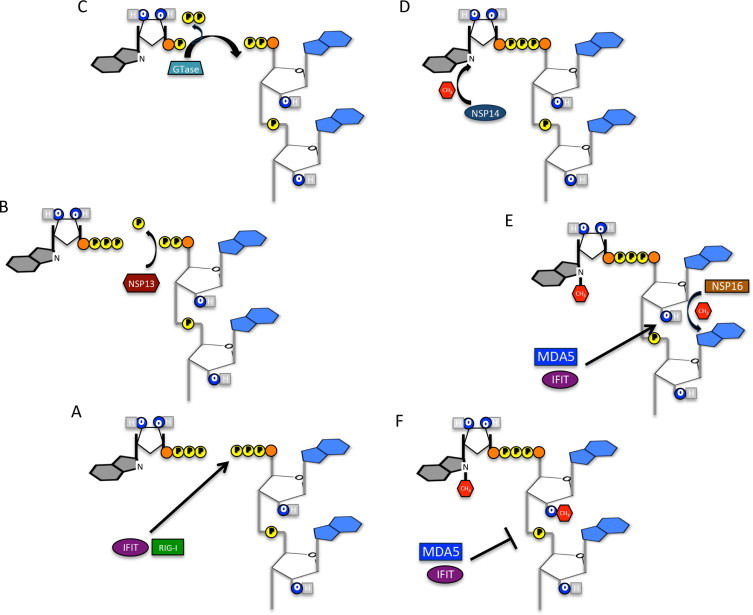
Viral mRNA capping. (A) Nascent mRNA incorporates an unprotected 5′-triphosphate that can be recognized by both immune sensor RIG-I and ISG IFIT molecules if left unmodified. (B) Triphosphate cleavage initiates the first step in mRNA capping and is predicted to be mediated by CoV NSP13, a 5′–3′ helicase/RTPase. (C) Guanyly transferase (Gtase) cleaves GTP to GMP and mediates attachment to diphosphate linked nascent RNA. Gtase has yet to be identified in CoV capping process. (D) CoV NSP14, an SAM dependent methyl-transferase (MTase), methylates the N7 guanosine, resulting in cap-0 structure. (E) The cap-0 structure, while protected from RIG-I recognition, still stimulates MDA5 and IFIT proteins via the unprotected 2′OH. NSP16, a 2′O-MTase, recognizes and binds to cap-0 CoV RNA, resulting in methylation. (F) Cap-1 viral mRNA fails to induced either MDA or IFIT proteins, permitting robust viral replication in the absence of the innate immune response.
3. Coronavirus capping
Naturally, viruses have evolved a variety of mechanisms to overcome these capping restrictions. For viruses that replicate within the nucleus including most DNA viruses and retroviruses, using the host capping machinery provides the primary means of protecting mRNA from restriction (Decroly et al., 2012). Similarly, other viral families including orthomyxoviruses, arenaviruses, and bunyaviruses, employ “cap snatching” to excise cap structures from early host RNAs and incorporated them into nascent viral RNAs. Finally, many viral families encode their own capping machinery that either mimics the sequential eukaryotic approach or an alternative approach to effectively maintain translation and block aspects of host immune recognition. Examples of the later approach include VpG-like viral proteins that mimic cap structures like picornaviruses and caliciviruses that bind 5′ viral RNA acting as a cap (Decroly et al., 2012); similarly, RNA secondary structure including internal ribosome entry sites (IRES) elements permit translation of viral RNA and may offer minor protection from host recognition (Hyde et al., 2014). Importantly, a significant portion of viruses encode their own viral capping proteins that directly copy the host machinery, making it indistinguishable from host mRNA and neutralizing an important barrier to viral infection.
For coronaviruses, a series of highly conserved non-structural proteins (NSPs) have been identified in capping viral RNA. Detailed reports have identified or predicted roles for NSP13 (Ivanov et al., 2004, Ivanov and Ziebuhr, 2004), NSP14 (Chen et al., 2009), and most recently, the NSP10/NSP16 complex (Bouvet et al., 2010, Chen et al., 2011, Debarnot et al., 2011, Decroly et al., 2008, Decroly et al., 2011, Lugari et al., 2010) in capping of CoV RNA. Similar to the host process, the capping is believed to be initiated by a RNA phosphatase that removes a phosphate group from the nascent mRNA (Fig. 1A); for CoVs, this process has been attributed to the 5′–3′ helicase/NTPase NSP13, although not yet confirmed by experimental examination (Fig. 1B) (Snijder et al., 2003). Thereafter, an undetermined host or viral guanyly transferase (GTase) mediates cleavage of GTP to GMP and covalent attachment to the diphosphate linked mRNA (Fig. 1C). Next, NSP14, an SAM-dependent MTase, facilitates the addition of a methyl group to the guanosine at N7, producing a viral mRNA cap-0 structure (Fig. 1D). Following this step, the viral RNA is effectively protected from RIG-I recognition of the free 5′ triphosphate; however, absent 2′O-methylation, the viral mRNA will still trigger the sensor MDA5 and IFIT effectors (Fig. 1E). The mRNA cap for CoVs is completed by NSP16, an SAM-dependent nucleoside-2′O-methyl-transferase, that ensures formation of a protective cap-1 structure that prevent recognition by either MDA5 or IFIT proteins (Fig. 1F). Finally, the NSP16/NSP10 complex finishes CoV capping process permitting viral infection with reduced host recognition.
4. Structural conservation and catalytic tetrad
CoV 2′O-MTase belongs to the RrmJ/fibrillarin superfamily of ribose 2′O-methyl-transferases conserved in a number of host cellular homologues as well as viral orthologs in Flaviviruses, Alphaviruses, and Nidoviruses (Feder et al., 2003). This family of proteins catalyzes the transfer of S-adenosylmethionine (SAM) methyl group to methyl acceptors and relies on a conserved K-D-K-E tetrad within the substrate binding pocket for activity. Substitution at any of three residues (K-D-K) resulted in ablation of catalytic activity (Feder et al., 2003). In addition to the tetrad, the canonical members of this enzyme family including catechol O-MTase, maintain a conserved structural motif of seven-stranded B-sheets flanked by three alpha helices on each side that constitute both the acceptor substrate and SAM-binding domains (Martin and McMillan, 2002). These structural and catalytic elements are required for SAM-dependent MTase activity.
For CoV NSP16, the requirement for a viral co-factor, NSP10, distinguishes them from the more canonical 2′O-MTase family members. Early computational studies had predicted NSP16 as a 2′O-MTase (Snijder et al., 2003, von Grotthuss et al., 2003) and subsequent work confirmed a low, but present 2′O-MTase activity with feline CoV (FCoV) NSP16 (Decroly et al., 2008). However, recombinant SARS-CoV NSP16 failed to replicate a similar activity despite noteworthy homology with FCoV NSP16. Examination of yeast-two hybrid screens later revealed a strong interaction between NSP16 and NSP10 suggesting a possible role in capping (Imbert et al., 2008, Pan et al., 2008). Further work demonstrated co-expression of NSP10 with NSP16 permitted SARS-CoV 2′O-MTase activity in the presence of already capped N7-methylated RNA (Bouvet et al., 2010). Similarly, co-expression permitted capture of a purified NSP16-NSP10 crystal structure unstable with NSP16 alone (Decroly et al., 2011). Based on this structure, the authors postulated the absence of two the flanking alpha helices resulted in a less stable CoV 2′O-MTase than canonical family members; the addition of NSP10 potentially alleviates this instability permitting 2′O-MTase activity to occurs. Additional structural and biochemical analysis suggested that NSP10 binding within this region provided support for SAM-binding of NSP16 as well as extending the RNA binding groove of the complex (Chen et al., 2011). Overall, the data demonstrates the complex interaction between NSP16 and NSP10 that is required for stability and 2′O-MTase activity.
5. NSP16 conservation, CoV mutants, and attenuation
The amino acid sequence of NSP16 is highly conserved across the entire CoV family, suggesting similar structural domains and functional activities. This conservation, illustrated in the structural similarities between SARS-CoV and MERS-CoV NSP16/NSP10 complexes (Fig. 2 ), indicate mutations that ablate or alter activity would be conserved across the entire viral family, resulting in similar phenotypic mutants. Therefore, therapeutics and treatments that target the activity and function of NSP16/NSP10 may prove effective against previous strain like SARS-CoV and HCoV 229E, as well as emergent viruses like MERS-CoV and PEDV.
Fig. 2.
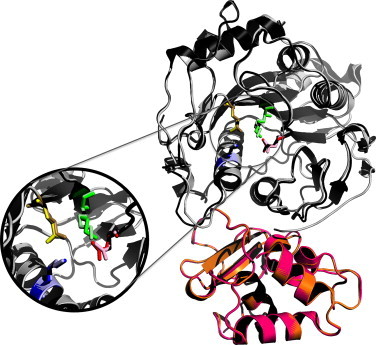
Conservation of CoV NSP16. Overlay of NSP16 structures from SARS-CoV (black) and emergent MERS-CoV (gray). The conserved KDKE motif is represented in the inset for SARS-CoV (primary colors) and MERS-CoV (pastel colors). Also shown, required NSP10 scaffold for SARS-CoV (pink) and MERS-CoV (orange). Homology models were created using Modeler in the Max-Planck Institute's Bioinformatics Toolkit. The known crystal structure for the NSP10/16 complex (3R24 in the RCSB protein data bank) was used as the template structure (Chen et al., 2011). Homology models were then manipulated using MacPyMol.
The initial attempts to alter NSP16 function focused on deletion and the conserved KDKE tetrad (Chen et al., 2011). Deletion of the NSP16 coding sequence blocked RNA synthesis; similarly, deletion of up stream components (Exonuclease-NSP14 and N-terminal zinc binding domain endoribonuclease-NSP15) also ablated viral replication and RNA synthesis (Almazan et al., 2006). However, mutation at D130A (D6905A) within the conserved KDKE tetrad of NSP16 resulted in a severely attenuated, but partially viable mutant virus. In addition, temperature sensitive mutants in mouse hepatitis virus (MHV) established a role for NSP16 in synthesis or stability of positive strand synthesis (Sawicki et al., 2005). Together, these early studies implied a critical role for NSP16 in the CoV life cycle.
However, these results may have been influenced by the underlying immune response captured by the cell cultures system. Subsequent attempts with HCoV 229E, MHV, and SARS-CoV were able to generate mutants that reduced or ablated NSP16 activity, but retain robust viral replication in vitro (Menachery et al., 2014b, Zust et al., 2011). Substitution of aspartic acid for alanine (D129A) within the KDKE motif of HCoV 229E resulted in ablation of 2′O-MTase activity; importantly, the 229E mutant was attenuated in fibroblast multistep growth curves, induced more type I IFN in macrophages, and was more sensitive to IFNβ pretreatment that WT 229E (Zust et al., 2011). However, the same mutation (D130A) in MHV resulted in resulted in minimal attenuation 17Cl1 murine fibroblasts, but robust replication deficits and increased type I IFN induction in macrophages, suggesting a cell type specific contribution to NSP16 attenuation. Notably, NSP16 mutations that targeted the putative cap-0 binding site (MHV-Y14A) maintained partial 2′O-MTase activity, had reduced immune induction, and minimal replication attenuation in macrophages compared to the D130A mutant. Overall, the data suggested that NSP16 activity is required for efficient replication in certain, immune competent cells.
For SARS-CoV, mutations in NSP16 also target the conserved KDKE motif; focusing on the trio required for catalytic activity, SARS-CoV NSP16 mutants were generated with single amino acid changes at K46A, D130A, and K170A (Menachery et al., 2014b). All three mutant viruses were viable and had no replication defects in Vero cells, contrasting previous deletion studies in BHK cells (Almazan et al., 2006). Underlying innate immunity may account for these differences, as all three SARS-CoV mutants were attenuated in immune competent Calu3 cells during multistep replication cycles (MOI 0.01) (Menachery et al., 2014b). The SARS-CoV D130A mutant, the least attenuated in Calu3 cells, was characterized further in vivo demonstrating robust attenuation at late times (D4 and later) in regards to weight loss, viral lung titers, lung histology, and breathing function. Notably, SARS-CoV ΔNSP16 attenuation was delayed both in vitro and in vivo, suggesting that initial infection failed to stimulate a more robust immune response than WT. While the ΔNSP16 displayed increased type I IFN sensitivity similar to mutants in MHV and 229E (Zust et al., 2011), the SARS-CoV mutant virus failed to induce more type I IFN either in vitro or in vivo (Menachery et al., 2014b). Similarly, analysis of known interferon-stimulated genes (Menachery et al., 2014a) including IFIT1 and MDA5 found no significant change induction magnitude or kinetics following ΔNSP16 challenge as compared to WT (Fig. 3A–C). Globably, the data indicates that while SARS-CoV mutants are attenuated in vitro and in vivo, the immune response induced by the host plays a critical role in this attenuation. Differences between the NSP16 mutants across the viral families may be explained by the baseline immune response in the target cell, which may be in higher in some (macrophages) and less robust in others (fibroblasts, epithelial cells).
Fig. 3.
SARS-CoV 2′O-MTase mutant maintains similar ISG profile to WT. (A, B) Consensus interferon-stimulated gene expression (as defined in (Menachery et al., 2014a)) following (A) WT SARS-CoV and (B) ΔNSP16 mutant infection of Calu3 cells (MOI 5). Values represent log2 fold change compared to time-matched mocks. (C) Subtraction of WT SARS-CoV expression from ΔNSP16 mutant virus expression provides relative change between WT and 2′O-MTase mutant. RNA expression and proteomics data were derived from previous in vitro with SARS-CoV (Sims et al., 2013, Menachery et al., 2014b). The raw microarray data are accessible through the NCBI Gene Expression Omnibus (Edgar et al., 2002) through GEO Series accession number GSE33267 and GSE33266.
While targeting the KDKE catalytic tetrad has been the main approach to ablate 2′O-MTase activity in a variety of CoVs, several other approaches may also be effective in knocking out NSP16 activity. As mentioned previously, the NSP16/NSP10 complex only binds to 0′ capped RNA, thus permitting activity; therefore, mutation to the N7-methyl-transferase activity of NSP14 is also be expected to ablate NSP16 and may capture similar phenotypic outcomes. In addition, recent efforts to characterize the residues required for NSP16/NSP10 interaction also provide another means to target 2′O-MTase function. Prior work has demonstrated that alanine replacement of central core amino acids in MHV NSP10 resulted in lethal phenotypes (Donaldson et al., 2006); these mutations track within the interface of the NSP16/NSP10 complex and thus may alter or ablate 2′O-MTase function (Decroly et al., 2011, Lugari et al., 2010). Other approaches, including altering the substrate binding site or taking advantage of temperature sensitive mutants, represent novel means to target and diminish 2′O-MTase activity in the CoV family of viruses.
6. Restoration of NSP16 mutants
Sensitivity to type I IFN has been a hallmark of NSP16 mutants across the CoV family. For HCoV 229E and MHV mutants encoding the D to A substitution (D129A and D130A, respectively), infection also results in augmented type I IFN production and attenuation in macrophages (Zust et al., 2011). In contrast, while the same mutation in SARS-CoV (D130A) resulted in ablated 2′O-MTase activity and IFN sensitivity, surprisingly, infection did not augment type I IFN induction relative to WT virus (Menachery et al., 2014b). This fact suggests that despite producing high levels of recognizable non-self RNA, SARS-CoV is able to control type I IFN induction pathways. Illustrating this point, IFNAR−/− mice have augmented viral replication, but fail to restore virulence to NSP16 deficient SARS-CoV. Similarly, while not restored to WT levels, the D130A mutation in MHV results in robust replication within the liver and the spleen of IFNAR−/− mice (Zust et al., 2011). Together, the data indicate that while type I IFN plays a critical role, NSP16 attenuation is primarily mediated by additional host immune responses.
Contrasting type I IFN deficiency, the absence of MDA5 results in a more robust restoration of CoV 2′O-MTase mutants. Studies in murine macrophages demonstrated that absent MDA5, MHV D130A mutant had restored viral replication and reduced IRF3 nuclear localization (Zust et al., 2011). While sensitivity to type I IFN was maintained, the results suggested that the absence of the sensor molecule results in reduced IFN induction (Zust et al., 2011). However, MDA5−/− mice failed to have any restoration in replication of the MHV D130A, but had partial restoration of the cap-0 binding mutant, MHV Y15A. When complemented with TLR7 deficiency, replication of MHV D130A was restored to IFNAR−/− levels in vivo, suggesting that both pathways contribute to attenuation of MHV 2′O-MTase mutants. In contrast, SARS-CoV D130A mutants were found to have restored virulence in MDA5−/− mice in terms of weight loss, histology, and breathing function (Menachery et al., 2014b). This discrepancy is likely explained by differences in cell tropism, as SARS-CoV is limited to ACE2 expressing cells within the lung; in contrast, MHV infects a variety of immune competent cells and likely results in more robust, systemic responses. Importantly, while the absence of MDA5 restores SARS-CoV 2′O-MTase mutants in vivo, no evidence exists to suggest it plays an effector role. Knockdown studies utilizing shRNA indicated no augmented replication for the SARS-CoV ΔNSP16 mutant in the absence of MDA5, contrasting IFIT family results (Fig. 4 ) (Menachery et al., 2014b). The results suggest that MDA5 functions in primary recognition of CoV 2′O-MTase mutants and initiates an interferon-stimulated gene cascade that leads to attenuation of replication in vitro and in vivo.
Fig. 4.
IFIT proteins, not MDA5, provide effector function against ΔNSP16 mutant viruses. Vero cells expressing shRNA (as described in (Menachery et al., 2014b)) targeting IFIT1 (blue), IFIT2 (red), MDA5 (green, two constructs) or no shRNA control (black) were pretreated with IFNβ (PBL Laboratories) and infected with (A) SARS-CoV or (B) ΔNSP16. P-values based on Student's T-test and are marked as indicated: *<0.05, **<0.01, ***<0.001.
While MDA5 and type I IFN deficiency led to restoration of CoV 2′O-MTase mutants, neither immune component provides direct effector function. Instead, the host response requires the activity of the IFIT family of proteins to mediate attenuation of CoV NSP16 mutants. For the NSP16 mutants of MHV and HCoV 229E, the absence of IFIT1 resulted in restored replication relative to WT control in vitro (Habjan et al., 2013, Zust et al., 2011); similar results were seen for SARS-CoV ΔNSP16 with both IFIT1 and IFIT2 knockdown (Menachery et al., 2014b). However, while replication is augmented in IFIT1−/− mice, MHV D130A remains significantly attenuated relative to WT MHV (Habjan et al., 2013). These results indicate that both IFIT restriction and type I IFN induction via MDA5/TLR7 contribute to attenuation to MHV 2′O-MTase mutants. In contrast, the SARS-CoV ΔNSP16 mutant was restored to near WT levels in IFIT1−/− mice (Menachery et al., 2014b); in terms of viral replication, weight loss, and breathing function, IFIT1 deficiency resulted in robust infection with the 2′O-MTase mutant and only modest augmentation of WT SARS-CoV. These results are consistent with restored virulence observed with WNV 2′O-MTase mutants in IFIT1−/− mice (Daffis et al., 2010) and indicate that IFIT1 mediates primary attenuation of SARS-CoV 2′O-MTase mutant. To date, neither CoV mutant has been tested in IFIT2−/− mice which display increased sensitivity to neurotropic viruses including wild-type MHV and VSV (Butchi et al., 2014, Fensterl et al., 2014). Similar to the MDA5 deficiency, differences in cell type and tissue tropism between MHV and SARS-CoV likely contribute to differential 2′O-MTase mutant restoration in IFIT1−/− deficient hosts.
The robust attenuation of CoV 2′O-MTase mutants highlights the importance of specific aspect of the host innate immune response in limiting virus infection. The absence of both type I IFN induction through MDA5/TLR7 and subsequent ISG effector responses through the IFIT family are necessary components in restoration of CoV NSP16 mutants. Notably, the presence of the functional NSP16 and other viral antagonists rendered these innate immune pathways ineffective against WT viruses, as noted by minimal augmentation of disease in IFNAR−/−, MDA5−/− or IFIT1−/− KO mice (Frieman et al., 2010, Habjan et al., 2013, Menachery et al., 2014b). The results also suggest that the rapid and robust innate mediated attenuation observed with CoV NSP16 mutants may preclude the necessity for an adaptive immune response for clearance. While prior work had demonstrated a requirement for both B and T-cells in clearance of both WT and SARS-CoV mutants (Graham et al., 2012, Sheahan et al., 2008), the presence of functional MDA5 and IFIT1 might be sufficient to clear SARS-CoV ΔNSP16 without an additional adaptive response. To test this theory, RAG−/− deficient mice were infected with either WT SARS-CoV or SARS-CoV ΔNSP16 (Fig. 5 ). The results demonstrated that while attenuation was similar in regards to weight loss, replication of SARS-CoV ΔNSP16 was maintained through day 10 with no significant difference relative to WT SARS-CoV. Together, the results indicate that despite innate mediated attenuation, SARS-CoV 2′O-MTase mutants still require an adaptive immune response for sterilizing immunity and clearance.
Fig. 5.
Adaptive immune response still required for ΔNSP16 clearance. Ten-week-old mice deficient in adaptive immunity (RAG−/−) were infected with either SARS-CoV MA15 (black) or ΔNSP16 (red). (A) Weight loss and (B) viral titer results demonstrate continued attenuation of ΔNSP16, but failed clearance of the either virus ten day post-infection. This study was carried out in accordance with the recommendations for care and use of animals by the Office of Laboratory Animal Welfare (OLAW), NIH. The UNC Institutional Animal Care and Use Committee (IACUC) (UNC, Permit Number A-3410-01) approved the animal study protocol (IACUC #13-033) followed in this experiment.
7. 2′O-MTase based therapeutic development
Having identified the host processes that mediate 2′O-MTase mutant attenuation, the possibility exists to leverage this knowledge for the development of therapeutic treatment options for human disease. While more standard treatments target essential viral processes like viral proteases or replication complexes formation, this approach impairs aspects of viral infection that improve the efficacy of the host immune response. By targeting and disrupting 2′O-MTase activity, viral mRNAs become susceptible to the activities of MDA5 and IFIT family members, attenuating viral infection through natural innate immune responses. Importantly, the broad conservation of 2′O-MTase in CoVs, as well as in a number of other viral families, provides a broadly applicable approach ideal for targeting both current and emerging viral infections. With this in mind, both vaccine and drug treatment approaches have been conceived to target 2′O-MTase activity for CoVs and other emergent viral infections.
7.1. 2′O-MTase based vaccines
Employing 2′O-MTase mutants as a live attenuated vaccine platform has proved effective in developing vaccine strategies for a variety of viral families. Several flaviviruses including West Nile, Dengue, and Japanese Encephalitis viruses have utilized mutation in the 2′O-MTase active domain of NS5 to attenuate and protect animals from lethal challenge (Dong et al., 2008, Zust et al., 2013). A similar vaccination approach has recently been described for paramyxoviruses including human metapneumovirus and targets the L protein which mediates 2′O-MTase activity (Zhang et al., 2014). In addition, recent work with Venezuelan Equine Encephalitis virus (VEE) revealed a single nucleotide mutation in the 5′UTR of the vaccine strain that ablated the RNA structure that mimicked 2′O-Methylations and allowed recognition of viral RNA by IFIT1 (Hyde et al., 2014). The broad efficacy of the approach across a variety viral families lends credence to application to CoVs and other emerging viruses that employ 2′O-MTase to overcome the host immune response (Ma et al., 2014).
Paralleling the success found for other RNA virus vaccines, the 2′O-MTase mutant of SARS-CoV proved effective as a live attenuated vaccine platform (Menachery et al., 2014b). Young, susceptible Balb/C mice (10 week old) were inoculated with 100pfu of SARS-CoV ΔNSP16 or PBS mock control; following challenge with a lethal dose of mouse-adapted SARS-CoV (MA15 1 × 105), mock vaccinated animals endured rapid weight loss and death. In contrast, ΔNSP16 vaccinated animals presented no clinical signs of disease and survived the lethal challenge. Importantly, plaque reduction neutralization titers (PRNT50) indicated a very high level of protective antibody (>1:1600) produced following ΔNSP16 vaccination. This PRNT50 value represents an improvement over other live attenuated platforms (Fett et al., 2013, Graham et al., 2012) and supports the idea that the more robust replication induced early during ΔNSP16 infection may improve the overall immune performance (Fett et al., 2013, Menachery et al., 2014b). Alternatively, the augmented IFN induction via MDA5 may result in a greater TH1 skew thus improving the quality of the protective adaptive response. Supporting this idea, MDA5 deficiency has been reported to alter the early inflammatory response in the lung following Sendai virus infection (Kim et al., 2014); decreased neutrophil and increased alternatively activated macrophage populations are indicative of altered T-cell composition and may impact immune memory response (Page et al., 2012). While further testing is required, the results suggest that targeting 2′O-MTase activity via vaccine may serve dual roles of both attenuating the virus and improving the overall immune response.
While initial studies are promising, several significant questions remain to be addressed in regards to the efficacy and utility of a ΔNSP16-based vaccine platform. First among them is the ability of this mutant to protect aged animals. To date, multiple vaccine platforms have failed or caused immune induced pathology in aged populations. While live attenuated SARS-CoV platforms have had greater efficacy (Bolles et al., 2011, Sheahan et al., 2011), these protection parameters must be explored in the context of the senescent host for any ΔNSP16-based vaccine to move forward. In addition, while the D130A mutation has been maintained in tissue culture, more studies are required to assess the possibility of reversion of the two nucleotide change in vivo as previously described (Graham et al., 2012); the results may indicate the need to generate and evaluate more reversion proof mutants employing either a three nucleotide change or multiple mutations within the KDKE motif. Finally, like other vaccine platforms, ΔNSP16 vaccination must evaluate efficacy against heterologous challenge with potential emergent SARS-like CoV strains including newly discovered bat strains (Ge et al., 2013, He et al., 2014). Despite these questions, utilizing 2′O-MTase mutation as the basis for a live attenuated CoV platform has great promise for not only SARS-CoV, but other emergent viruses like MERS-CoV and PEDV.
7.2. Drug targeting CoV 2′O-MTase activity
In addition to the vaccine platform, efforts to impair 2′O-MTase activity in the context of a normal wild-type infection may also provide an effective treatment option. Over the years, a number of small molecules have been identified for their ability to inhibit the activity of CoV NSP16 including S-adenosyl-l-homocysteine, sinefungin, and aurintricarboxylic acid (ATA) (Bouvet et al., 2010, Decroly et al., 2011, He et al., 2004). These molecules, all believed to interfere with substrate binding, effectively ablated 2′O-MTase activity of both NSP16/NSP10 and NSP14 (Bouvet et al., 2010). Given the robust attenuation of 2′O-MTase mutants, this finding provided a mechanistic explanation for ATA inhibition of SARS-CoV replication that was previously described (He et al., 2004). In contrast, sinefungin pretreatment alone proved ineffective in significantly reducing SARS-CoV replication (Fig. 6 ); however, in the context of type I IFN induction the addition of sinefungin improved the overall inhibition. Collectively, the data suggest that 2′O-MTase inhibition can be used as a CoV treatment, but requires the activation of the ISG response for efficacy. Notably, targeting the SAM-binding site would also be expected to work against other virally encoded 2′O-MTase providing a broad based antiviral therapeutic. However, the impact on host 2′O-MTase activity may induce cytotoxic effects that limit its utility. Overall, the data argues that targeting 2′O-MTase via the SAM-binding site remains a viable treatment approach but requires further refinement and testing.
Fig. 6.
Targeting 2′O-MTase activity effective in the context of IFNβ response. WT SARS-CoV replication following pretreatment of Vero cells with either of IFNβ (100 units/mL), sinefungin (2.56 mM), or combination of both 16 h prior to infection. Viral replication evaluated 24 h post-infection. P-values based on Student's T-test and are marked as indicated: *<0.05, ***<0.001.
In addition to targeting the 2′O-MTase active site, other approaches exploit indirect factors necessary for NSP16 activity. As highlighted earlier, NSP16 requires binding with NSP10 for stability and 2′O-MTase activity (Bouvet et al., 2010). Therefore, drugs that interfere with this interaction may prove to be an effective treatment strategy for SARS and other CoVs. Illustrating this point, two short peptides derived from the interaction domains of SARS-CoV NSP10 have been shown to ablate NSP16 2′O-MTase activity (Ke et al., 2012). The inverse approach, designing peptides based on the NSP16 binding domain, may also provide an additional avenue for therapy. In both cases, disruption of NSP10/NSP16 interaction should attenuate the wild-type virus if a robust type I IFN/ISG response has been induced. Similarly, the sequential nature of CoV capping (Fig. 1) requires recognition of upstream modifications for activity. Therefore, disrupting elements of N7-methyl capping via NSP14 or other CoV mRNA process should also ablate NSP16 activity, thus rendering enhanced susceptibility. These CoV specific approaches have the potential to significantly impact viral infection and have minimal toxicity concerns compared to approaches that target the 2′O-MTase active site. Importantly, all of the drug approaches outlined require induction of the host ISG response for efficacy and may vary greatly depending on the IFN antagonists encoded by the specific CoV in question.
8. Conclusion and future directions
Together, the studies of NSP16 have defined the critical elements and illustrated the importance of 2′O-MTase activity for successful coronavirus infection. In the absence of functional NSP16, SARS-CoV, HCoV 229E, and MHV infections are significantly attenuated both in vitro and in vivo. This deficit is mediated by both increased recognition of viral RNA by host sensor molecules as well as the effector responses of the IFIT family of ISGs. Understanding these precise viral/host interactions permits the development of novel therapeutics that impairs the virus while also increasing the efficacy of the existing host immune response. Contrasting methods that target viral specific factors like replication, fidelity, or structural protein function, the approach has broad application potential to not only CoVs, but other viral families that employ 2′O-MTase approaches to capping viral RNA. Importantly, with the continued threat of viral emergence highlighted by MERS-CoV and PEDV in the past few years, targeting the highly conserved NSP16 activity provides a therapeutic approach applicable to any novel CoV. While significant research questions remain, the understanding and targeting of 2′O-MTase activity represents an important avenue for treating current and future CoV mediated diseases.
Acknowledgements
Research reported in this publication was supported by the National Institute Of Allergy & Infectious Diseases of the NIH under contract no. HHSN272200800060C and award numbers U19AI100625, U19AI109761, and F32AI102561. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
- Almazan F., Dediego M.L., Galan C., Escors D., Alvarez E., Ortego J., Sola I., Zuniga S., Alonso S., Moreno J.L., Nogales A., Capiscol C., Enjuanes L. Construction of a severe acute respiratory syndrome coronavirus infectious cDNA clone and a replicon to study coronavirus RNA synthesis. J. Virol. 2006;80(21):10900–10906. doi: 10.1128/JVI.00385-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker M.M., Graham R.L., Donaldson E.F., Rockx B., Sims A.C., Sheahan T., Pickles R.J., Corti D., Johnston R.E., Baric R.S., Denison M.R. Synthetic recombinant bat SARS-like coronavirus is infectious in cultured cells and in mice. Proc. Natl. Acad. Sci. U. S. A. 2008;105(50):19944–19949. doi: 10.1073/pnas.0808116105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolles M., Deming D., Long K., Agnihothram S., Whitmore A., Ferris M., Funkhouser W., Gralinski L., Totura A., Heise M., Baric R.S. A double-inactivated severe acute respiratory syndrome coronavirus vaccine provides incomplete protection in mice and induces increased eosinophilic proinflammatory pulmonary response upon challenge. J. Virol. 2011;85(23):12201–12215. doi: 10.1128/JVI.06048-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvet M., Debarnot C., Imbert I., Selisko B., Snijder E.J., Canard B., Decroly E. In vitro reconstitution of SARS-coronavirus mRNA cap methylation. PLoS Pathog. 2010;6(4):e1000863. doi: 10.1371/journal.ppat.1000863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butchi N.B., Hinton D.R., Stohlman S.A., Kapil P., Fensterl V., Sen G.C., Bergmann C.C. Ifit2 deficiency results in uncontrolled neurotropic coronavirus replication and enhanced encephalitis via impaired alpha/beta interferon induction in macrophages. J. Virol. 2014;88(2):1051–1064. doi: 10.1128/JVI.02272-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Cai H., Pan J., Xiang N., Tien P., Ahola T., Guo D. Functional screen reveals SARS coronavirus nonstructural protein nsp14 as a novel cap N7 methyltransferase. Proc. Natl. Acad. Sci. U. S. A. 2009;106(9):3484–3489. doi: 10.1073/pnas.0808790106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Su C., Ke M., Jin X., Xu L., Zhang Z., Wu A., Sun Y., Yang Z., Tien P., Ahola T., Liang Y., Liu X., Guo D. Biochemical and structural insights into the mechanisms of SARS coronavirus RNA ribose 2′-O-methylation by nsp16/nsp10 protein complex. PLoS Pathog. 2011;7(10):e1002294. doi: 10.1371/journal.ppat.1002294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Eckerle I., Bleicker T., Zaki A., Landt O., Eschbach-Bludau M., van Boheemen S., Gopal R., Ballhause M., Bestebroer T.M., Muth D., Muller M.A., Drexler J.F., Zambon M., Osterhaus A.D., Fouchier R.M., Drosten C. Detection of a novel human coronavirus by real-time reverse-transcription polymerase chain reaction. Euro Surveill. 2012;17(39) doi: 10.2807/ese.17.39.20285-en. [DOI] [PubMed] [Google Scholar]
- Daffis S., Szretter K.J., Schriewer J., Li J., Youn S., Errett J., Lin T.Y., Schneller S., Zust R., Dong H., Thiel V., Sen G.C., Fensterl V., Klimstra W.B., Pierson T.C., Buller R.M., Gale M., Jr., Shi P.Y., Diamond M.S. 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature. 2010;468(7322):452–456. doi: 10.1038/nature09489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debarnot C., Imbert I., Ferron F., Gluais L., Varlet I., Papageorgiou N., Bouvet M., Lescar J., Decroly E., Canard B. Crystallization and diffraction analysis of the SARS coronavirus nsp10-nsp16 complex. Acta Crystallogr. Sect. F: Struct. Biol. Cryst. Commun. 2011;67(Pt 3):404–408. doi: 10.1107/S1744309111002867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decroly E., Debarnot C., Ferron F., Bouvet M., Coutard B., Imbert I., Gluais L., Papageorgiou N., Sharff A., Bricogne G., Ortiz-Lombardia M., Lescar J., Canard B. Crystal structure and functional analysis of the SARS-coronavirus RNA cap 2′-O-methyltransferase nsp10/nsp16 complex. PLoS Pathog. 2011;7(5):e1002059. doi: 10.1371/journal.ppat.1002059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decroly E., Ferron F., Lescar J., Canard B. Conventional and unconventional mechanisms for capping viral mRNA. Nature reviews. Microbiology. 2012;10(1):51–65. doi: 10.1038/nrmicro2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decroly E., Imbert I., Coutard B., Bouvet M., Selisko B., Alvarez K., Gorbalenya A.E., Snijder E.J., Canard B. Coronavirus nonstructural protein 16 is a cap-0 binding enzyme possessing (nucleoside-2′O)-methyltransferase activity. J. Virol. 2008;82(16):8071–8084. doi: 10.1128/JVI.00407-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond M.S. IFIT1: a dual sensor and effector molecule that detects non-2′-O methylated viral RNA and inhibits its translation. Cytokine Growth Factor Rev. 2014 doi: 10.1016/j.cytogfr.2014.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson E.F., Sims A.C., Deming D.J., Baric R.S. Mutational analysis of MHV-A59 replicase protein-nsp10. Adv. Exp. Med. Biol. 2006;581:61–66. doi: 10.1007/978-0-387-33012-9_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H., Zhang B., Shi P.Y. Flavivirus methyltransferase: a novel antiviral target. Antiviral Res. 2008;80(1):1–10. doi: 10.1016/j.antiviral.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R., Domrachev M., Lash A.E. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30(1):207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feder M., Pas J., Wyrwicz L.S., Bujnicki J.M. Molecular phylogenetics of the RrmJ/fibrillarin superfamily of ribose 2′-O-methyltransferases. Gene. 2003;302(1–2):129–138. doi: 10.1016/s0378-1119(02)01097-1. [DOI] [PubMed] [Google Scholar]
- Fensterl V., Wetzel J.L., Sen G.C. The interferon-induced protein, Ifit2, protects mice from infection of the peripheral nervous system by Vesicular stomatitis virus. J. Virol. 2014;88(18):10303–10311. doi: 10.1128/JVI.01341-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fett C., DeDiego M.L., Regla-Nava J.A., Enjuanes L., Perlman S. Complete protection against severe acute respiratory syndrome coronavirus-mediated lethal respiratory disease in aged mice by immunization with a mouse-adapted virus lacking E protein. J. Virol. 2013;87(12):6551–6559. doi: 10.1128/JVI.00087-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frieman M.B., Chen J., Morrison T.E., Whitmore A., Funkhouser W., Ward J.M., Lamirande E.W., Roberts A., Heise M., Subbarao K., Baric R.S. SARS-CoV pathogenesis is regulated by a STAT1 dependent but a type I, II and III interferon receptor independent mechanism. PLoS Pathog. 2010;6(4):e1000849. doi: 10.1371/journal.ppat.1000849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X.Y., Li J.L., Yang X.L., Chmura A.A., Zhu G., Epstein J.H., Mazet J.K., Hu B., Zhang W., Peng C., Zhang Y.J., Luo C.M., Tan B., Wang N., Zhu Y., Crameri G., Zhang S.Y., Wang L.F., Daszak P., Shi Z.L. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 2013;503(7477):535–538. doi: 10.1038/nature12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham R.L., Becker M.M., Eckerle L.D., Bolles M., Denison M.R., Baric R.S. A live, impaired-fidelity coronavirus vaccine protects in an aged, immunocompromised mouse model of lethal disease. Nat. Med. 2012;18(12):1820–1826. doi: 10.1038/nm.2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham R.L., Donaldson E.F., Baric R.S. A decade after SARS: strategies for controlling emerging coronaviruses. Nat. Rev. Microbiol. 2013;11(12):836–848. doi: 10.1038/nrmicro3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habjan M., Hubel P., Lacerda L., Benda C., Holze C., Eberl C.H., Mann A., Kindler E., Gil-Cruz C., Ziebuhr J., Thiel V., Pichlmair A. Sequestration by IFIT1 impairs translation of 2′O-unmethylated capped RNA. PLoS Pathog. 2013;9(10):e1003663. doi: 10.1371/journal.ppat.1003663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B., Zhang Y., Xu L., Yang W., Yang F., Feng Y., Xia L., Zhou J., Zhen W., Feng Y., Guo H., Zhang H., Tu C. Identification of diverse alphacoronaviruses and genomic characterization of a novel severe acute respiratory syndrome-like coronavirus from bats in china. J. Virol. 2014;88(12):7070–7082. doi: 10.1128/JVI.00631-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He R., Adonov A., Traykova-Adonova M., Cao J., Cutts T., Grudesky E., Deschambaul Y., Berry J., Drebot M., Li X. Potent and selective inhibition of SARS coronavirus replication by aurintricarboxylic acid. Biochem. Biophys. Res. Commun. 2004;320(4):1199–1203. doi: 10.1016/j.bbrc.2004.06.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung V., Ellegast J., Kim S., Brzozka K., Jung A., Kato H., Poeck H., Akira S., Conzelmann K.K., Schlee M., Endres S., Hartmann G. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314(5801):994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- Huang Y.W., Dickerman A.W., Pineyro P., Li L., Fang L., Kiehne R., Opriessnig T., Meng X.J. Origin, evolution, and genotyping of emergent porcine epidemic diarrhea virus strains in the United States. MBio. 2013;4(5) doi: 10.1128/mBio.00737-13. e00737-00713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde J.L., Gardner C.L., Kimura T., White J.P., Liu G., Trobaugh D.W., Huang C., Tonelli M., Paessler S., Takeda K., Klimstra W.B., Amarasinghe G.K., Diamond M.S. A viral RNA structural element alters host recognition of nonself RNA. Science. 2014;343(6172):783–787. doi: 10.1126/science.1248465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imbert I., Snijder E.J., Dimitrova M., Guillemot J.C., Lecine P., Canard B. The SARS-Coronavirus PLnc domain of nsp3 as a replication/transcription scaffolding protein. Virus Res. 2008;133(2):136–148. doi: 10.1016/j.virusres.2007.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov K.A., Thiel V., Dobbe J.C., van der Meer Y., Snijder E.J., Ziebuhr J. Multiple enzymatic activities associated with severe acute respiratory syndrome coronavirus helicase. J. Virol. 2004;78(11):5619–5632. doi: 10.1128/JVI.78.11.5619-5632.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov K.A., Ziebuhr J. Human coronavirus 229E nonstructural protein 13: characterization of duplex-unwinding, nucleoside triphosphatase, and RNA 5′-triphosphatase activities. J. Virol. 2004;78(14):7833–7838. doi: 10.1128/JVI.78.14.7833-7838.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josset L., Menachery V.D., Gralinski L.E., Agnihothram S., Sova P., Carter V.S., Yount B.L., Graham R.L., Baric R.S., Katze M.G. Cell host response to infection with novel human coronavirus EMC predicts potential antivirals and important differences with SARS coronavirus. MBio. 2013;4(3) doi: 10.1128/mBio.00165-13. e00165–00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke M., Chen Y., Wu A., Sun Y., Su C., Wu H., Jin X., Tao J., Wang Y., Ma X., Pan J.A., Guo D. Short peptides derived from the interaction domain of SARS coronavirus nonstructural protein nsp10 can suppress the 2′-O-methyltransferase activity of nsp10/nsp16 complex. Virus Res. 2012;167(2):322–328. doi: 10.1016/j.virusres.2012.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W.K., Jain D., Sanchez M.D., Koziol-White C.J., Matthews K., Ge M.Q., Haczku A., Panettieri R.A., Jr., Frieman M.B., Lopez C.B. Deficiency of melanoma differentiation-associated protein 5 results in exacerbated chronic postviral lung inflammation. Am. J. Respir. Crit. Care Med. 2014;189(4):437–448. doi: 10.1164/rccm.201307-1338OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Shi Z., Yu M., Ren W., Smith C., Epstein J.H., Wang H., Crameri G., Hu Z., Zhang H., Zhang J., McEachern J., Field H., Daszak P., Eaton B.T., Zhang S., Wang L.-F. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310(5748):676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- Lugari A., Betzi S., Decroly E., Bonnaud E., Hermant A., Guillemot J.C., Debarnot C., Borg J.P., Bouvet M., Canard B., Morelli X., Lecine P. Molecular mapping of the RNA Cap 2′-O-methyltransferase activation interface between severe acute respiratory syndrome coronavirus nsp10 and nsp16. J. Biol. Chem. 2010;285(43):33230–33241. doi: 10.1074/jbc.M110.120014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Wei Y., Zhang X., Zhang Y., Cai H., Zhu Y., Shilo K., Oglesbee M., Krakowka S., Whelan S.P., Li J. mRNA cap methylation influences pathogenesis of vesicular stomatitis virus in vivo. J. Virol. 2014;88(5):2913–2926. doi: 10.1128/JVI.03420-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin J.L., McMillan F.M. SAM (dependent) I AM: the S-adenosylmethionine-dependent methyltransferase fold. Curr. Opin. Struct. Biol. 2002;12(6):783–793. doi: 10.1016/s0959-440x(02)00391-3. [DOI] [PubMed] [Google Scholar]
- Menachery V.D., Eisfeld A.J., Schafer A., Josset L., Sims A.C., Proll S., Fan S., Li C., Neumann G., Tilton S.C., Chang J., Gralinski L.E., Long C., Green R., Williams C.M., Weiss J., Matzke M.M., Webb-Robertson B.J., Schepmoes A.A., Shukla A.K., Metz T.O., Smith R.D., Waters K.M., Katze M.G., Kawaoka Y., Baric R.S. Pathogenic influenza viruses and coronaviruses utilize similar and contrasting approaches to control interferon-stimulated gene responses. MBio. 2014;5(3) doi: 10.1128/mBio.01174-14. e01174–01114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menachery V.D., Yount B.L., Jr., Josset L., Gralinski L.E., Scobey T., Agnihothram S., Katze M.G., Baric R.S. Attenuation and restoration of severe acute respiratory syndrome coronavirus mutant lacking 2′-o-methyltransferase activity. J. Virol. 2014;88(8):4251–4264. doi: 10.1128/JVI.03571-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myong S., Cui S., Cornish P.V., Kirchhofer A., Gack M.U., Jung J.U., Hopfner K.P., Ha T. Cytosolic viral sensor RIG-I is a 5′-triphosphate-dependent translocase on double-stranded RNA. Science. 2009;323(5917):1070–1074. doi: 10.1126/science.1168352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls J., Dong X.-P., Jiang G., Peiris M. SARS: clinical virology and pathogenesis. Respirology. 2003;8(Suppl.):6–8. doi: 10.1046/j.1440-1843.2003.00517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page C., Goicochea L., Matthews K., Zhang Y., Klover P., Holtzman M.J., Hennighausen L., Frieman M. Induction of alternatively activated macrophages enhances pathogenesis during severe acute respiratory syndrome coronavirus infection. J. Virol. 2012;86(24):13334–13349. doi: 10.1128/JVI.01689-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J., Peng X., Gao Y., Li Z., Lu X., Chen Y., Ishaq M., Liu D., Dediego M.L., Enjuanes L., Guo D. Genome-wide analysis of protein–protein interactions and involvement of viral proteins in SARS-CoV replication. PLoS ONE. 2008;3(10):e3299. doi: 10.1371/journal.pone.0003299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman S., Netland J. Coronaviruses post-SARS: update on replication and pathogenesis. Nat. Rev. Microbiol. 2009;7(6):439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichlmair A., Lassnig C., Eberle C.A., Gorna M.W., Baumann C.L., Burkard T.R., Burckstummer T., Stefanovic A., Krieger S., Bennett K.L., Rulicke T., Weber F., Colinge J., Muller M., Superti-Furga G. IFIT1 is an antiviral protein that recognizes 5′-triphosphate RNA. Nat. Immunol. 2011;12(7):624–630. doi: 10.1038/ni.2048. [DOI] [PubMed] [Google Scholar]
- Sawicki S.G., Sawicki D.L., Younker D., Meyer Y., Thiel V., Stokes H., Siddell S.G. Functional and genetic analysis of coronavirus replicase–transcriptase proteins. PLoS Pathog. 2005;1(4):e39. doi: 10.1371/journal.ppat.0010039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan T., Morrison T.E., Funkhouser W., Uematsu S., Akira S., Baric R.S., Heise M.T. MyD88 is required for protection from lethal infection with a mouse-adapted SARS-CoV. PLoS Pathog. 2008;4(12):e1000240. doi: 10.1371/journal.ppat.1000240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan T., Whitmore A., Long K., Ferris M., Rockx B., Funkhouser W., Donaldson E., Gralinski L., Collier M., Heise M., Davis N., Johnston R., Baric R.S. Successful vaccination strategies that protect aged mice from lethal challenge from influenza virus and heterologous severe acute respiratory syndrome coronavirus. J. Virol. 2011;85(1):217–230. doi: 10.1128/JVI.01805-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims A.C., Tilton S.C., Menachery V.D., Gralinski L.E., Schafer A., Matzke M.M., Webb-Robertson B.J., Chang J., Luna M.L., Long C.E., Shukla A.K., Bankhead A.R., 3rd, Burkett S.E., Zornetzer G., Tseng C.T., Metz T.O., Pickles R., McWeeney S., Smith R.D., Katze M.G., Waters K.M., Baric R.S. Release of severe acute respiratory syndrome coronavirus nuclear import block enhances host transcription in human lung cells. J. Virol. 2013;87(7):3885–3902. doi: 10.1128/JVI.02520-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J., Bredenbeek P.J., Dobbe J.C., Thiel V., Ziebuhr J., Poon L.L., Guan Y., Rozanov M., Spaan W.J., Gorbalenya A.E. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 2003;331(5):991–1004. doi: 10.1016/S0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Grotthuss M., Wyrwicz L.S., Rychlewski L. mRNA cap-1 methyltransferase in the SARS genome. Cell. 2003;113(6):701–702. doi: 10.1016/S0092-8674(03)00424-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Wei Y., Zhang X., Cai H., Niewiesk S., Li J. Rational design of human metapneumovirus live attenuated vaccine candidates by inhibiting viral messenger RNA cap methyltransferase. J. Virol. 2014;88(19):11411–11429. doi: 10.1128/JVI.00876-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zust R., Cervantes-Barragan L., Habjan M., Maier R., Neuman B.W., Ziebuhr J., Szretter K.J., Baker S.C., Barchet W., Diamond M.S., Siddell S.G., Ludewig B., Thiel V. Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat. Immunol. 2011;12(2):137–143. doi: 10.1038/ni.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zust R., Dong H., Li X.F., Chang D.C., Zhang B., Balakrishnan T., Toh Y.X., Jiang T., Li S.H., Deng Y.Q., Ellis B.R., Ellis E.M., Poidinger M., Zolezzi F., Qin C.F., Shi P.Y., Fink K. Rational design of a live attenuated dengue vaccine: 2′O-methyl-transferase mutants are highly attenuated and immunogenic in mice and macaques. PLoS Pathog. 2013;9(8):e1003521. doi: 10.1371/journal.ppat.1003521. [DOI] [PMC free article] [PubMed] [Google Scholar]


