Abstract
Analysis of large-scale systems of biomedical data provides a perspective on neuropsychiatric disease that may be otherwise elusive. Described here is an analysis of three large-scale systems of data from using Autism Spectrum Disorder (ASD) and ASD research as exemplar of what might be achieved from study of such data. The first is the biomedical literature that highlights that there are two very successful but quite separate research communities and findings pertaining to genetics and the molecular biology of ASD. That is those studies positing ASD etiologies related to immunological dysregulation and those related to disorders of synaptic function and neuronal connectivity. The second is the emerging use of electronic health record systems and other large clinical databases to allow the data acquired during the course of care to be used to identify distinct subpopulations, clinical trajectories and pathophysiological substructure of ASD. These reveal subsets of patients with distinct clinical trajectories some of which are immunologically related and others which follow pathologies conventionally thought of as neurological. The third is genome-wide genomic and transcriptomic analyses which show molecular pathways that overlap neurological and immunological mechanisms. The convergence of these three large-scale data perspectives illustrates the scientific leverage that large-scale data analyses can provide in guiding researchers in an approach to the diagnosis of neuropsychiatric disease that is inclusive and comprehensive.
Introduction
Perhaps the most successful branch of medicine in achieving a precise diagnosis of disease, one directly linked to etiology, has been that of infectious disease. Only a little over one hundred years passed between the identification of microorganisms as the etiological agents for multiple diseases and the consequent development of dozens of therapies, in immunizations and antibiotics, that have had a greater impact on mortality and morbidity than any other medical intervention (1). It is this understanding on the consequences of etiological and precise diagnostic capabilities that were the main drivers of the recent National Academy of Sciences report on Precision Medicine: to use multiple comprehensive measurement modalities to identify which sub group of patients a given patient most resembles and therefore to be able to both assign a diagnostic label and predict clinical course in response to therapeutic intervention. I review here how a systematic approach to large-scale data can make some preliminary and illuminating strides towards a “precision medicine” of neuropsychiatric disease. I use the autism spectrum disorders (ASD) as a prismatic example of the larger opportunity by illustrating how this approach reveals two richly productive but largely separate avenues of research in ASD defined by apparently distinct mechanistic hypotheses. That is, ASD as a disorder of neural connectivity and specifically synaptic connectivity regulation (2, 3) and ASD as a disorder of immunological signaling (4-6).
First some framing is required regarding the task being addressed: diagnosis of the disorder. Here, diagnosis of ASD will be defined in the probabilistic framework used in decision making: the probability of a disease D, given the findings F summarized by the notation p(D∣F). In ASD, we often attempt to diagnose or rule-out a single disease (i.e. autism) even though it is recognized that there are likely to be multiple diseases (i.e. The set of diseases D comprised of {D1..Dn} that together constitute ASD). A diagnosis will be more useful to the extent that p(D∣F) is high (i.e. close to 1.0) corresponding to the high likelihood of disease or low (i.e. close to 0.0) corresponding to the low likelihood of disease. Further confidence in this likelihood estimate is provided if the error of this estimate is low. The appropriateness of therapy can then be determined by how well it is matched to the disease. This thereby highlights the value of determining which of the diseases that constitute ASD of the set {D1..Dn} have the highest probability as each therapy will have different efficacy for each of them.
The Published Literature for Large Scale Characterization of Research
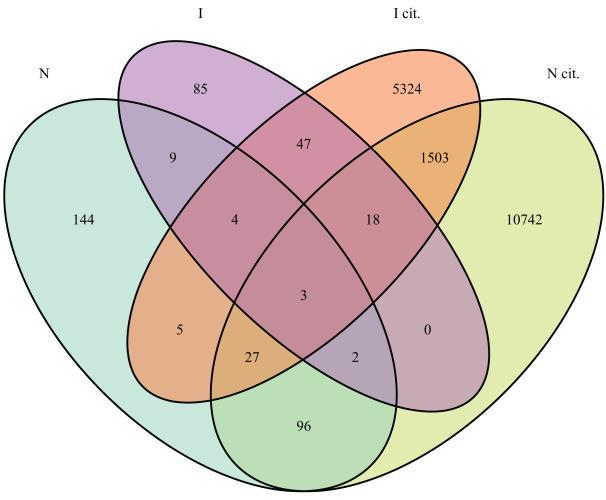
In the recently published Diagnostic Standards Manual 5, Autism Spectrum Disorder is defined as including persistent deficits in social communication and social interaction and restricted, repetitive patterns of behavior, interests, or activities. This new single disorder replaces several previously defined disorders including Autistic Disorder, Asperger’s Disorder, and Pervasive Developmental Disorder Not Otherwise Specified. This redefinition will surely lead to a change in diagnosis for many individuals and possibly a change in funding of support services. The controversy that emerged prior to and after this publication illustrates the challenge posed by the diagnostic and prognostic task when applied to a disease complex that many recognize to be a constellation of heterogeneous pathophysiologies (5, 7-12), some of which have genetic etiologies, some environmental or a combination thereof. A multi-dimensional characterization of the patient population of interest, that measures the multiple genetic, molecular, clinical and environmental-exposure features of each patient to derive the overall landscape of the constellation of heterogeneous diseases that distinguish that population, provides the most comprehensive and systematic viewpoint (13). Of course, such integrative data sets are currently far and few between, with the Simon’s Simplex Collection (14) constituting an notable example of what such integration can yield (and the effort and investments required to bring it together). With the steady accretion of clinical and research data sets, we can anticipate such multidimensional assessment to grow. Therefore it will become essential to determine which of the set of diseases comprising ASD in {D1..Dn} are being diagnostically evaluated. Merely making this determination of which diseases are being considered as part of the ASD set is challenging. This challenge is best illustrated by a large-scale database available to all ASD researchers: that of the published literature. If we focus on those recent publications that were NIH supported and therefore deposited in the Open Access Pubmed Central NIH repository (15), then as illustrated in Figure 1, not only is the primary literature balkanized, but even the citations made by the authors of this literature largely address disparate domains of biology. If we label the autism and genetics literature as pertaining non-exclusively to four sets: neuronal synaptic function (N), immunological function/disorders (I), with N cit. and I cit. denoting the literature cited by these first two sets, then as shown in Figure 1, the overlap is remarkably slight. For example, of 290 publications in N only 18 are also in I and of the 12391 cited by the publications in N, only 1551 are cited by I. At best, this suggests that either the set of findings or the set of diseases considered in developing a precision diagnosis of the ASDs is incomplete, depending on which research community is addressed. This raises the question of what population studies can reveal regarding this apparent dichotomy? By way of example, large-scale population genomics have revealed previously poorly defined or unsuspected subtypes of disease within breast cancer (16), non small cell lung carcinomas (17) and leukemia (18). However preceding the advent of genomics by more than a century, physician-scientists have used observational studies to define disease subtypes. Jean Martin Charcot, for example, systematically and comprehensively studied the patients in a large neurological hospital in Paris and was thereby able to define new and lasting disease entities out of a pool of previously monolithic and broad neurological diagnoses (19). A century and a half after Charcot, can we undertake large scale observational studies of patients enabled by the recent acceleration in electronic health record systems deployment to augment our ability to generate an integrated view of p(D∣F) for ASD?
Figure 1.
Illustration of the incomplete overlap in research of ASD genetics based on investigations of synapses and research in ASD genetics based on investigations of the immune system. Four ellipses are shown corresponding to four corpora all selected from Pubmed Central. N denotes those publications focused on genetics and synapses, I denotes those publications focused on genetics and immune system. I cit. denotes those publications that were cited by those in I. N cit. denotes those publications that were cited by those in N. The intersection between N and I accounts for only 4 percent of the combined publications and the intersection between N cit. and I cit. accounts for only 8 percent of the combined citations.
Electronic Health Records for Large Scale Characterizations
The acceleration of the adoption of electronic health records (EHR’s) in clinical care through the HITECH Act of 2009 (20) may or may not increase the productivity or safety of healthcare delivery but it certainly has provided a large source of detailed clinical documentation of patients. This enables researchers adept in the “secondary use” of EHR data to identify patients with the clinical phenotype of interest and then use the samples acquired in subsequent visits for clinical diagnostics for the purposes of genotyping, resequencing and even epigenetic characterization, as reviewed in (21, 22). In addition to structured or codified data (e.g. laboratory test, medications, diagnostic and procedure billing codes), the development of “natural language processing” (NLP) techniques (23-27) enables the narrative text of clinical notes to be mined to obtain a far more accurate phenotypic assessment of the patients than from the codified data. Given that the codified billing data is well known to be biased for reimbursement and insufficiently fine grained, this is not surprising. However, when the codified data is combined with the NLP-derived data the phenotyping accuracy is higher than with either clinical source alone (22). Furthermore, this automated phenotyping has been shown to be generalizable, portable and reproducible across healthcare systems (28, 29). These very encouraging early studies should not obscure the methodological challenges that these observational data sets entail. The time span covered by most EHRs is of short duration in most systems because of their recent adoption (30). NLP techniques currently require effortful fine-tuning based on iterative comparison of their performance selecting the “right” patients relative to that of experts manually reviewing a subset of the same records. Moreover, whereas the claims data may be biased for reimbursement, they do cover populations through the entirety of their paid health encounters whereas electronic healthcare data may have greater detail but often only pertain to a fraction of these encounters (31). For example, an academic center’s EHR may include documentation of the initial ASD diagnosis and subsequent episodes of acute morbidity. However, they often lack the documentation of the growth and development of these children noted in the community pediatric practices. All these sources of bias and complexity suggest that the use of these data requires at least as much care and multidisciplinary expertise (31) as genomic data analysis early in the adoption of a new sequencing platform.
Importantly, at a time when genomic studies of neuropsychiatric disease require tens of thousands of subjects, EHR-driven phenotyping coupled to the genomic characterization of discarded samples is one to two orders of magnitude faster and less costly in identifying patients of interest than conventional study cohort techniques (21). This EHR-driven phenotyping has been performed successfully for several neuropsychiatric phenotypes including major depressive disorder (32, 33) and bipolar disorder (34) and several groups are currently working on similar approaches to ASD. It remains however, that even for diseases that are as common as 1 in 100, any single healthcare system may not have sufficient numbers of patients to enable a statistically robust characterization of these diseases. This is even more problematic when the disease is not monolithic but rather a constellation of the many rarer diseases {D1..Dn} of which they are composed. This can be addressed by enabling queries that cross multiple healthcare systems. For example, we have developed a system—the Shared Health Information Network (SHRINE) (35, 36)—which has been used, with appropriate governance, oversight and privacy protection measures, to issue real-time queries across multiple healthcare sites comprising records of millions of patients to both identify rare events (14) and enable phenotyping for neuropsychiatric genomic studies to occur at the scale of hundreds of thousands of individuals. SHRINE has been adopted for the Harvard affiliated hospitals (comprising 6 million patients and 10 billion facts), and the University of California for its UC REX system (37) covering over 11 million patients.
In this context, a recent use of SHRINE to study the co morbidity landscape of ASD presages future large EHR system studies of the neuropsychiatric diseases. This study, one of the largest to date, covered over 14,000 patients with ASD over a 15 year period (38) representing at least 0.5% of the hospital populations. Many of the co morbidities fall squarely into categories that are commonly thought as related to neuronal and synaptic function including increased seizure frequency (19.44%) and increased sleep disorders (1.12%), bowel disorders—excluding inflammatory bowel disease (11.74%) and schizophrenia (2.4% increasing to 8.76% after age 18). All these prevalences were highly significantly (often an order of magnitude) higher than either the general population or even utilization-matched hospital populations. Conversely, several diseases that were anecdotally reported to include increased frequency of ASD were confirmed as such, including muscular dystrophy with 5% ASD prevalence. With regard to the aforementioned divide between the immunological and synaptic studies, several diseases with an autoimmune component were identified with much higher prevalence than both the general population and the matched hospital populations: type 1 diabetes (0.67% rising to 2.08% after age 18) and inflammatory bowel disease (0.68% rising to 1.99% after age 18). There have been previously many case reports about these co-morbidities but the absence of a systematic population view has made it understandably easy to treat their biological import with some hesitation. Moreover, because a single developmental medicine specialist seeing 1000’s patients with ASD is unlikely to see more than 10 patients with IBD or type 1 DM, these claims might not be consistent with their impression of their own population. Understandably, this has lead some to question the validity these electronic health record-based diagnoses. Detailed review suggests that they are indeed valid. For example, in a comparison of “gold standard” IBD diagnoses by expert gastroenterologists, the combination of natural language processing and codified data from the electronic health record attained specificities in the 95-97% range (39).
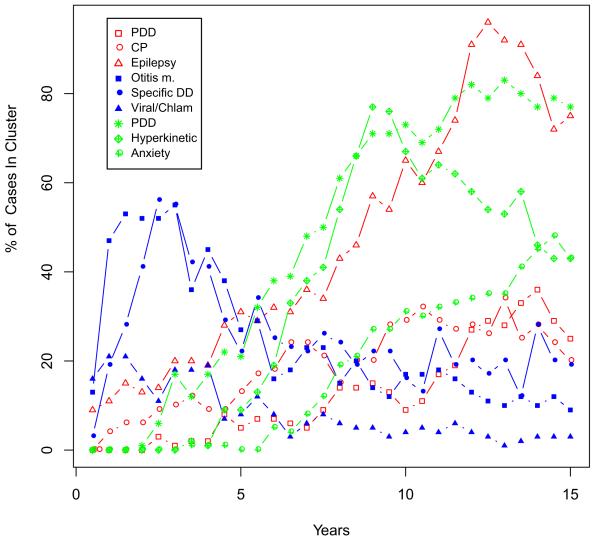
The insight provided by a systematic population perspective is enhanced further, by having longitudinal, if retrospective, follow-up of these patients over 15 or more years (40). Just as in the early expression microarray experiments (41), the patients are hierarchically clustered together based on their similar trajectories but instead of characterized by gene expression, they are characterized by the co-morbidities noted at each six-month interval. As summarized by Figure 2 below there are at least 3 distinct clusters that are currently identifiable. One cluster is highly enriched for seizures with a prevalence as high as 80%. This is in contrast to the alternative hypothesis which would be a homogenous random distribution of epilepsy across the population with autism if the epileptogenesis was due to a common etiology across ASD. Another cluster includes individuals with increased prevalence of ear infections, sinusitis as well as multiple upper respiratory infections and (not shown) inflammatory bowel disease. A third cluster is characterized by multiple neuro-behavioral disorders such as ADHD and anxiety and at a lower frequency (not shown) schizophrenia, the latter becoming much more prevalent in this population after age 18 (42).
Figure 2.
Trajectories of Comorbidities Characterizing Three Distinct Sub-clusters of ASD Defined by Electronic Health Record Data. Shown in red are the top three co-morbidities from cluster 1 where seizures rise to a prevalence of 80%. Shown in blue are the top three co-morbidities from cluster 2 with an early childhood peak of infections. Not shown (because lower ranked) is a rise in inflammatory bowel disease that continues to rise through adolescence. Shown in green are the top three co-morbidities in cluster 3 which are characterized by anxiety and hyperkinetic activity. Not shown (because lower ranked) is a rise in schizophrenia that accelerates with onset of adolescence.
The significance of these clusters here is that they represent two important consequences for diagnosis and prognosis. First of all, they are instances, albeit preliminary, of the distinct pathophysiologies of children who all have the label of autism but in fact appear to have very different diseases. That is the patients who are members of these clusters have clinical manifestations that appear to belong to different underlying diseases in the set {D1..Dn} currently comprising ASD. For example, cluster 3 appears much more as neuropsychiatric clinical manifestation whereas cluster 2 appears more immunological or infectious-related but all of them share in common the manifestations of autism. These immune or infection related etiologies are also supported by large epidemiological studies such as those documenting increased ASD prevalence in children whose parents have rheumatoid arthritis or type 1 DM (43) and increased ASD in pregnancies characterized by high C-reactive protein (44). Of course, these early studies at the population level are encouraging but follow-up studies are required to determine if these distinct clusters correspond to the aforementioned mechanisms previously described in the literature. The trajectories shown are also relevant in that they provide a chronological signature. So for example whereas some of the neuropsychiatric disorders appear to increase with time, some of the immunological disorders such as sinusitis and otitis media peak early in childhood. Others, such as inflammatory bowel disease, type 1 diabetes and schizophrenia increase in prevalence with age. Another contribution to diagnostic precision may be enabled by the identification of these phenotypic subclusters. Genetic studies that are focused on these subgroups, rather than the undifferentiated group of patients that fall under the ASD rubric, may provide greater biological homogeneity and therefore have higher power to find genetic contributions to risk.
EHR data sets are perhaps the fastest growing source of observational clinical data and therefore they will likely overlap and complement the membership of other cohort studies such as the Avon Longitudinal Study of Parents and Children (45). This presents at least two opportunities: the validation and calibration of findings in the EHR-derived populations against the more systematically acquired study cohorts (46) and testing the generality of those cohort studies by comparing them to geographically distant clinical populations from EHR-equipped health systems.
High Throughput Large-Scale Data for Integrative Characterization
Genome-wide assessment of genetic variation (e.g. in exome studies) and function (e.g. transcriptomic or epigenomic measures) promise an unbiased perspective on disease processes. The former captures the heritable component whereas the latter integrates environmental and genetic influences. If these are unbiased then why does the literature derived from them, as described above, appear to have such a dichotomous nature? One argument is that the underlying disorders discussed here, immunological vs. synaptic/neural-connectivity dysregulation are inhomogenously split across the environmental component and inherited component. For example it has been argued that the immunological signature is environmentally mediated rather than inherited (47). However, close analysis of the results of these high-throughput data reveals other potential reasons.
First, is a cognitive bias that results from the history and context within which a gene’s function was discovered. For example, many chemokines and other inflammatory mediators thought to be characteristic of the immune system have been now shown to be powerful and essential morphogens in the normal development of the mammalian brain (48, 49). So much so, that it is likely that if they had been first discovered by neuroscientists, they would be universally called neurokines (50). Similar arguments apply to the regulation of mTOR mediated autophagy processes which might have been labeled as synaptic pruning functions if first discovered in the CNS (51). From this perspective, many of the genes implicated in ASD have both a synaptic or neuronal connectivity function and an immunological function. For example, of the genes implicated in autism by the Simons Foundation (see Table 1), 10% of them overlap with the GO categories covering immunological function. Similarly the genes in T receptor signaling pathway overlap with 21% of the genes in the long term potentiation pathway (one of the mechanisms underlying synaptic plasticity) as do the genes in the GO immune genes. From this perspective, the ASD immunological and synaptic genetics research communities might be much closer in their focus than is apparent from their literature. It also implies that some of the inherited variation could be as easily labeled as immunological as it is labeled synaptic/neuronal connectivity.
Table 1.
Overlap between sets of genes found to be implicated in autism and those in immunological regulation and long-term potentiation.
| Two sets intersected | Genes in the intersection |
|---|---|
| Gene Ontology Immune Genes (http://bit.ly/KIcYOZ) and the Simons Foundation autism gene database (https://gene.sfari.org) |
ADA CD44 HLA-A C4B ITGA4 NOS2A PTGS2 IL1RAPL1 APC ITGB3 ITGB7 ADORA2A ALOX5AP HRAS ADRB2 NRP2 RPS6KA2 LAMB1 (~10% of SFARI genes) |
| Gene Ontology Immune Genes (http://bit.ly/KIcYOZ) and KEGG Long Term Potentiation (hsa:04720) |
RAF1 PRKCA CREBBP MAP2K2 BRAF MAP2K1 RPS6KA2 MAPK3 MAPK1 HRAS EP300 CAMK2A CAMK2G PRKACA ATF4 (~21% of KEGG LTP genes) |
| KEGG T cell receptor signaling (hsa:04660) and KEGG Long Term Potentiation (hsa:04720) |
RAF1 PPP3R2 PPP3CC PPP3R1 MAP2K2 MAP2K1 NRAS CHP2 MAPK3 MAPK1 HRAS KRAS PPP3CB PPP3CA CHP (~21% of KEGG LTP genes) |
It should be acknowledged that in contrast to classical Mendelian disorders, complex diseases such as ASD are fertile ground for the cognitive biases outlined above. With so many genes in common, with the phenotypic pleiomorphy of ASD, and with multiple non-CNS immunological co-morbidities (e.g. type 1 DM, inflammatory bowel disease, rheumatoid arthritis) there are plenty of opportunities for investigators focused on a single system or single organ to observe reflections of the same genetic dysregulation, but in their tissue of interest. Likewise, the overlap is one possible explanation of why peripheral blood RNA or protein expression levels differ in ASD and non-ASD subjects (52, 53) and that the difference can be used to classify these patients characterized by many of the same pathways identified in genomic studies (7, 54).
The consequence of cognitive bias results in another kind of bias: that of narrative bias. For example, in a study summarizing thousands of findings in a whole genome study, there is inevitably a process by which the investigators will choose which mechanisms/genes are highlighted in the limited space available in their publication. In an important study of CNV’s in ASD (55), for example, enrichment was also found in major histocompatibility complex MHC-I related gene-sets as noted in the Supplementary Materials. However, the investigators understandably chose to omit the finding from their main text because it did not relate to the other molecular themes they had chosen to focus on. In the literature that then cites that article, this immunological component rarely appears if at all. The narrative bias thereby leads to another well-known phenomenon: citation bias (56) . Citation bias leads to the insular interpretation of findings that focus on mechanisms that do not fit into that bias. So for example previously early evidence of the familial clustering of autoimmune disorders in families with ASD (57) and HLA-DR4 association with ASD (58, 59) is only cited by the immunologically oriented literature in ASD.
The aforementioned balkanization of neuropsychiatric investigations may be increasingly a phenomenon of the past. Data sources such as the NDAR repository at NIMH (60) and the Psychiatric Genomics Consortium (61) provide investigators with the a set of integrative measurements previously unavailable. These more comprehensive data resources enable analyses across disorders (62-65) which allows the common and distinguishing aspect of the spectrum of these disorders to be studied phenotypically and etiologically. This broader perspective is also reflected in recent reviews (66-68) which bridge the gap illustrated in Figure 1.
Conclusion
As in many other domains of human disease, neuropsychiatric disorders are prone to the natural tendency to focus on specific aspects that do not reflect the entirety of the manifestation or mechanisms of these disorders. Here I have illustrated how three large-scale data sources: the literature, electronic health records and high throughput genome-scale measurements illustrate the extent and balkanization of the study of neuropsychiatric disease, specifically in the case of ASD. At the same time, these large-scale data sources provide the means to attain a comprehensive perspective. That is, by systematically analyzing large-scale data sources, we can identify the molecular and clinical characteristics of the disparate disorders {D1..Dn} of which ASD serves as a unifying, if temporary, label. In doing so, we enable selectivity in our therapeutic trials and ultimately therapeutic decision process.
The three large-scale data sources discussed are only the most currently accessible of those relevant to ASD. There are several others that are highly likely to be informative. Chief among these are unbiased approaches to measuring human environmental exposures (69-71) at the population level as well as the broader instrumentation of behavioral/cognitive performance (72) which is only glimpsed during formal clinical evaluation. Such comprehensive environmental and behavioral assessments are essential if we are to understand the large proportion of the variance in the disorders that lies outside their inherited predispositions, which in the case of ASD is at least 30-40%.
Acknowledgements
I.K. was funded in part by the i2b2 National Center for Biomedical Computing (NIH/NLM U54 LM008748), the Conte Center for Computational System Genomics of Neuropsychiatric Phenotypes (NIH P50MH94267), the MEDSEQ project (NIH U01-HG006500) and the eMERGE network (NIH U01HG006828). Thanks to Dr. Griffin Weber for his advice for bibliometric analysis.
Footnotes
Financial Disclosure I.K. is on the Scientific Advisory Board of SynapDx for which he has not received equity but is paid as a consultant. SynapDx is a company developing a blood-based test for the early detection of Autism Spectrum Disorders (ASDs)
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lederberg J. Infectious history. Science. 2000;288:287–293. doi: 10.1126/science.288.5464.287. [DOI] [PubMed] [Google Scholar]
- 2.Bear MF, Huber KM, Warren ST. The mGluR theory of fragile X mental retardation. Trends Neurosci. 2004;27:370–377. doi: 10.1016/j.tins.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 3.Auerbach BD, Osterweil EK, Bear MF. Mutations causing syndromic autism define an axis of synaptic pathophysiology. Nature. 2011;480:63–68. doi: 10.1038/nature10658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ashwood P, Wills S, Van De Water J. The immune response in autism: a new frontier for autism research. Journal of Leukocyte Biology. 2006;80:1–15. doi: 10.1189/jlb.1205707. [DOI] [PubMed] [Google Scholar]
- 5.Patterson PH. Immune involvement in schizophrenia and autism: etiology, pathology and animal models. Behavioural brain research. 2009;204:313–321. doi: 10.1016/j.bbr.2008.12.016. [DOI] [PubMed] [Google Scholar]
- 6.Malkova NV, Yu CZ, Hsiao EY, Moore MJ, Patterson PH. Maternal immune activation yields offspring displaying mouse versions of the three core symptoms of autism. Brain Behav Immun. 2012;26:607–616. doi: 10.1016/j.bbi.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campbell MG, Kohane IS, Kong SW. Pathway-based outlier method reveals heterogeneous genomic structure of autism in blood transcriptome. BMC Med Genomics. 2013;6:34. doi: 10.1186/1755-8794-6-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu TW, Chahrour MH, Coulter ME, Jiralerspong S, Okamura-Ikeda K, Ataman B, et al. Using whole-exome sequencing to identify inherited causes of autism. Neuron. 2013;77:259–273. doi: 10.1016/j.neuron.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bartlett CW, Goedken R, Vieland VJ. Effects of updating linkage evidence across subsets of data: reanalysis of the autism genetic resource exchange data set. American Journal of Human Genetics. 2005;76:688–695. doi: 10.1086/429345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma D, Cuccaro M, Jaworski J, Haynes C, Stephan D, Parod J, et al. Dissecting the locus heterogeneity of autism: significant linkage to chromosome 12q14. Mol Psychiatry. 2007;12:376–384. doi: 10.1038/sj.mp.4001927. [DOI] [PubMed] [Google Scholar]
- 11.Lai MC, Lombardo MV, Baron-Cohen S. Autism. Lancet. 2013 doi: 10.1016/S0140-6736(13)61539-1. [DOI] [PubMed] [Google Scholar]
- 12.Rosti RO, Sadek AA, Vaux KK, Gleeson JG. The genetic landscape of autism spectrum disorders. Dev Med Child Neurol. 2014;56:12–18. doi: 10.1111/dmcn.12278. [DOI] [PubMed] [Google Scholar]
- 13.Kohane IS, Eran A. Can we measure autism? Sci Transl Med. 2013;5:209ed218. doi: 10.1126/scitranslmed.3007340. [DOI] [PubMed] [Google Scholar]
- 14.Abrahams BS, Arking DE, Campbell DB, Mefford HC, Morrow EM, Weiss LA, et al. SFARI Gene 2.0: a community-driven knowledgebase for the autism spectrum disorders (ASDs) Mol Autism. 2013;4:36. doi: 10.1186/2040-2392-4-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Caelleigh AS. PubMed Central and the new publishing landscape: shifts and tradeoffs [editorial] Acad Med. 2000;75:4–10. doi: 10.1097/00001888-200001000-00004. [DOI] [PubMed] [Google Scholar]
- 16.Liu R, Wang X, Chen GY, Dalerba P, Gurney A, Hoey T, et al. The prognostic role of a gene signature from tumorigenic breast-cancer cells. N Engl J Med. 2007;356:217–226. doi: 10.1056/NEJMoa063994. [DOI] [PubMed] [Google Scholar]
- 17.Raponi M, Dossey L, Jatkoe T, Wu X, Chen G, Fan H, et al. MicroRNA classifiers for predicting prognosis of squamous cell lung cancer. Cancer Res. 2009;69:5776–5783. doi: 10.1158/0008-5472.CAN-09-0587. [DOI] [PubMed] [Google Scholar]
- 18.Kang H, Chen IM, Wilson CS, Bedrick EJ, Harvey RC, Atlas SR, et al. Gene expression classifiers for relapse-free survival and minimal residual disease improve risk classification and outcome prediction in pediatric B-precursor acute lymphoblastic leukemia. Blood. 2010;115:1394–1405. doi: 10.1182/blood-2009-05-218560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Silvester A. Jean Martin Charcot (1825-93) and John Hughlings Jackson (1835-1911): neurology in France and England in the 19th century. J Med Biogr. 2009;17:210–213. doi: 10.1258/jmb.2009.009039. [DOI] [PubMed] [Google Scholar]
- 20.Buntin MB, Jain SH, Blumenthal D. Health information technology: laying the infrastructure for national health reform. Health Aff (Millwood) 2010;29:1214–1219. doi: 10.1377/hlthaff.2010.0503. [DOI] [PubMed] [Google Scholar]
- 21.Murphy S, Churchill S, Bry L, Chueh H, Weiss S, Lazarus R, et al. Instrumenting the health care enterprise for discovery research in the genomic era. Genome Res. 2009;19:1675–1681. doi: 10.1101/gr.094615.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kohane IS. Using electronic health records to drive discovery in disease genomics. Nat Rev Genet. 2011;12:417–428. doi: 10.1038/nrg2999. [DOI] [PubMed] [Google Scholar]
- 23.Perlis RH, Iosifescu DV, Castro VM, Murphy SN, Gainer VS, Minnier J, et al. Using electronic medical records to enable large-scale studies in psychiatry: treatment resistant depression as a model. Psychol Med. 2012;42:41–50. doi: 10.1017/S0033291711000997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoogenboom WS, Perlis RH, Smoller JW, Zeng-Treitler Q, Gainer VS, Murphy SN, et al. Limbic system white matter microstructure and long-term treatment outcome in major depressive disorder: A diffusion tensor imaging study using legacy data. The world journal of biological psychiatry : the official journal of the World Federation of Societies of Biological Psychiatry. 2012 doi: 10.3109/15622975.2012.669499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kullo IJ, Fan J, Pathak J, Savova GK, Ali Z, Chute CG. Leveraging informatics for genetic studies: use of the electronic medical record to enable a genome-wide association study of peripheral arterial disease. Journal of the American Medical Informatics Association : JAMIA. 2010;17:568–574. doi: 10.1136/jamia.2010.004366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chapman WW, Nadkarni PM, Hirschman L, D’Avolio LW, Savova GK, Uzuner O. Overcoming barriers to NLP for clinical text: the role of shared tasks and the need for additional creative solutions. Journal of the American Medical Informatics Association : JAMIA. 2011;18:540–543. doi: 10.1136/amiajnl-2011-000465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Savova GK, Masanz JJ, Ogren PV, Zheng J, Sohn S, Kipper-Schuler KC, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. Journal of the American Medical Informatics Association : JAMIA. 2010;17:507–513. doi: 10.1136/jamia.2009.001560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kho AN, Pacheco JA, Peissig PL, Rasmussen L, Newton KM, Weston N, et al. Electronic medical records for genetic research: results of the eMERGE consortium. Science translational medicine. 2011;3:79re71. doi: 10.1126/scitranslmed.3001807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carroll RJ, Thompson WK, Eyler AE, Mandelin AM, Cai T, Zink RM, et al. Portability of an algorithm to identify rheumatoid arthritis in electronic health records. Journal of the American Medical Informatics Association : JAMIA. 2012 doi: 10.1136/amiajnl-2011-000583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jha AK, DesRoches CM, Campbell EG, Donelan K, Rao SR, Ferris TG, et al. Use of electronic health records in U.S. hospitals. N Engl J Med. 2009;360:1628–1638. doi: 10.1056/NEJMsa0900592. [DOI] [PubMed] [Google Scholar]
- 31.Devoe JE, Gold R, McIntire P, Puro J, Chauvie S, Gallia CA. Electronic health records vs Medicaid claims: completeness of diabetes preventive care data in community health centers. Ann Fam Med. 2011;9:351–358. doi: 10.1370/afm.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perlis RH, Iosifescu DV, Castro VM, Murphy SN, Gainer VS, Minnier J, et al. Using electronic medical records to enable large-scale studies in psychiatry: treatment resistant depression as a model. Psychol Med. 2011:1–10. doi: 10.1017/S0033291711000997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gallagher PJ, Castro V, Fava M, Weilburg JB, Murphy SN, Gainer VS, et al. Antidepressant response in patients with major depression exposed to NSAIDs: a pharmacovigilance study. The American journal of psychiatry. 2012;169:1065–1072. doi: 10.1176/appi.ajp.2012.11091325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pato MT, Sobell JL, Medeiros H, Abbott C, Sklar BM, Buckley PF, et al. The genomic psychiatry cohort: Partners in discovery. American journal of medical genetics Part B, Neuropsychiatric genetics : the official publication of the International Society of Psychiatric Genetics. 2013;162:306–312. doi: 10.1002/ajmg.b.32160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McMurry AJ, Murphy SN, MacFadden D, Weber G, Simons WW, Orechia J, et al. SHRINE: enabling nationally scalable multi-site disease studies. PloS one. 2013;8:e55811. doi: 10.1371/journal.pone.0055811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weber GM, Murphy SN, McMurry AJ, Macfadden D, Nigrin DJ, Churchill S, et al. The Shared Health Research Information Network (SHRINE): A prototype federated query tool for clinical data repositories. J Am Med Inform Assoc. 2009;16:624–630. doi: 10.1197/jamia.M3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.UC ReX Frequently Asked Questions. University of California; San Diego: 2014. http://ctri.ucsd.edu/Informatics/UC-ReX/Pages/ucrex-FAQ.aspx. [Google Scholar]
- 38.Kohane IS, McMurry A, Weber G, Macfadden D, Rappaport L, Kunkel L, et al. The co-morbidity burden of children and young adults with autism spectrum disorders. PloS one. 2012;7:e33224. doi: 10.1371/journal.pone.0033224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ananthakrishnan AN, Cai T, Savova G, Cheng SC, Chen P, Perez RG, et al. Improving Case Definition of Crohn’s Disease and Ulcerative Colitis in Electronic Medical Records Using Natural Language Processing: A Novel Informatics Approach. Inflammatory bowel diseases. 2013;19:1411–1420. doi: 10.1097/MIB.0b013e31828133fd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Doshi-Velez F, Ge Y, Kohane I. Comorbidity clusters in autism spectrum disorders: an electronic health record time-series analysis. Pediatrics. 2014;133:e54–63. doi: 10.1542/peds.2013-0819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Iyer VR, Eisen MB, Ross DT, Schuler G, Moore T, Lee JCF, et al. The transcriptional program in the response of human fibroblasts to serum [see comments] Science. 1999;283:83–87. doi: 10.1126/science.283.5398.83. [DOI] [PubMed] [Google Scholar]
- 42.Kohane I, McMurry A, Weber G, MacFadden D, Rappaport L, Kunkel L, et al. The Co-Morbidity Burden of Children and Young Adults with Autism Spectrum Disorders. PLoS ONE. 2012;7:e33224. doi: 10.1371/journal.pone.0033224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Atladóttir HO, Pedersen MG, Thorsen P, Mortensen PB, Deleuran B, Eaton WW, et al. Association of family history of autoimmune diseases and autism spectrum disorders. PEDIATRICS. 2009;124:687–694. doi: 10.1542/peds.2008-2445. [DOI] [PubMed] [Google Scholar]
- 44.Brown AS, Sourander A, Hinkka-Yli-Salomäki S, McKeague IW, Sundvall J, Surcel H-M. Elevated maternal C-reactive protein and autism in a national birth cohort. Mol Psychiatry. 2013 doi: 10.1038/mp.2012.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sullivan S, Rai D, Golding J, Zammit S, Steer C. The association between autism spectrum disorder and psychotic experiences in the Avon longitudinal study of parents and children (ALSPAC) birth cohort. J Am Acad Child Adolesc Psychiatry. 2013;52:806–814. e802. doi: 10.1016/j.jaac.2013.05.010. [DOI] [PubMed] [Google Scholar]
- 46.Kaelber DC, Foster W, Gilder J, Love TE, Jain AK. Patient characteristics associated with venous thromboembolic events: a cohort study using pooled electronic health record data. J Am Med Inform Assoc. 2012;19:965–972. doi: 10.1136/amiajnl-2011-000782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Voineagu I, Wang X, Johnston P, Lowe JK, Tian Y, Horvath S, et al. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 2011 doi: 10.1038/nature10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cayre M, Canoll P, Goldman JE. Cell migration in the normal and pathological postnatal mammalian brain. Prog Neurobiol. 2009;88:41–63. doi: 10.1016/j.pneurobio.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Streuli CH, Akhtar N. Signal co-operation between integrins and other receptor systems. Biochem J. 2009;418:491–506. doi: 10.1042/BJ20081948. [DOI] [PubMed] [Google Scholar]
- 50.Levi-Montalcini R, Skaper SD, Dal Toso R, Petrelli L, Leon A. Nerve growth factor: from neurotrophin to neurokine. Trends Neurosci. 1996;19:514–520. doi: 10.1016/S0166-2236(96)10058-8. [DOI] [PubMed] [Google Scholar]
- 51.Lee KM, Hwang SK, Lee JA. Neuronal Autophagy and Neurodevelopmental Disorders. Exp Neurobiol. 2013;22:133–142. doi: 10.5607/en.2013.22.3.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Corbett BA, Kantor AB, Schulman H, Walker WL, Lit L, Ashwood P, et al. A proteomic study of serum from children with autism showing differential expression of apolipoproteins and complement proteins. Mol Psychiatry. 2006:15. doi: 10.1038/sj.mp.4001943. [DOI] [PubMed] [Google Scholar]
- 53.Hu V, Sarachana T, Kim K, Nguyen A, Kulkarni S, Steinberg MH, et al. Gene expression profiling differentiates autism case-controls and phenotypic variants of autism spectrum disorders: evidence for circadian rhythm dysfunction in severe autism. Autism research : official journal of the International Society for Autism Research. 2009 doi: 10.1002/aur.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kong SW, Collins CD, Shimizu-Motohashi Y, Holm IA, Campbell MG, Lee IH, et al. Characteristics and predictive value of blood transcriptome signature in males with autism spectrum disorders. PLoS ONE. 2012;7:e49475. doi: 10.1371/journal.pone.0049475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pinto D, Pagnamenta AT, Klei L, Anney R, Merico D, Regan R, et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466:368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Greenberg SA. How citation distortions create unfounded authority: analysis of a citation network. BMJ (Clinical research ed) 2009;339:b2680. doi: 10.1136/bmj.b2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Comi AM, Zimmerman AW, Frye VH, Law PA, Peeden JN. Familial clustering of autoimmune disorders and evaluation of medical risk factors in autism. J Child Neurol. 1999;14:388–394. doi: 10.1177/088307389901400608. [DOI] [PubMed] [Google Scholar]
- 58.Warren RP, Odell JD, Warren WL, Burger RA, Maciulis A, Daniels WW, et al. Strong association of the third hypervariable region of HLA-DR beta 1 with autism. J Neuroimmunol. 1996;67:97–102. doi: 10.1016/0165-5728(96)00052-5. [DOI] [PubMed] [Google Scholar]
- 59.Johnson WG, Buyske S, Mars AE, Sreenath M, Stenroos ES, Williams TA, et al. HLA-DR4 as a risk allele for autism acting in mothers of probands possibly during pregnancy. Arch Pediatr Adolesc Med. 2009;163:542–546. doi: 10.1001/archpediatrics.2009.74. [DOI] [PubMed] [Google Scholar]
- 60.Hall D, Huerta MF, McAuliffe MJ, Farber GK. Sharing heterogeneous data: the national database for autism research. Neuroinformatics. 2012;10:331–339. doi: 10.1007/s12021-012-9151-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sullivan PF. The psychiatric GWAS consortium: big science comes to psychiatry. Neuron. 2010;68:182–186. doi: 10.1016/j.neuron.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cross-Disorder Group of the Psychiatric Genomics C. Smoller JW, Craddock N, Kendler K, Lee PH, Neale BM, et al. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet. 2013;381:1371–1379. doi: 10.1016/S0140-6736(12)62129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cross-Disorder Phenotype Group of the Psychiatric GC. Craddock N, Kendler K, Neale M, Nurnberger J, Purcell S, et al. Dissecting the phenotype in genome-wide association studies of psychiatric illness. Br J Psychiatry. 2009;195:97–99. doi: 10.1192/bjp.bp.108.063156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cross-Disorder Group of the Psychiatric Genomics C. Lee SH, Ripke S, Neale BM, Faraone SV, Purcell SM, et al. Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat Genet. 2013;45:984–994. doi: 10.1038/ng.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Smoller JW. Disorders and borders: psychiatric genetics and nosology. Am J Med Genet B Neuropsychiatr Genet. 2013;162B:559–578. doi: 10.1002/ajmg.b.32174. [DOI] [PubMed] [Google Scholar]
- 66.Dazert E, Hall MN. mTOR signaling in disease. Curr Opin Cell Biol. 2011;23:744–755. doi: 10.1016/j.ceb.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 67.Pardo CA, Eberhart CG. The neurobiology of autism. Brain Pathol. 2007;17:434–447. doi: 10.1111/j.1750-3639.2007.00102.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Voineagu I, Eapen V. Converging Pathways in Autism Spectrum Disorders: Interplay between Synaptic Dysfunction and Immune Responses. Front Hum Neurosci. 2013;7:738. doi: 10.3389/fnhum.2013.00738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Herbert MR, Russo JP, Yang S, Roohi J, Blaxill M, Kahler SG, et al. Autism and environmental genomics. Neurotoxicology. 2006;27:671–684. doi: 10.1016/j.neuro.2006.03.017. [DOI] [PubMed] [Google Scholar]
- 70.Patel CJ, Chen R, Kodama K, Ioannidis JP, Butte AJ. Systematic identification of interaction effects between genome- and environment-wide associations in type 2 diabetes mellitus. Hum Genet. 2013;132:495–508. doi: 10.1007/s00439-012-1258-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Patel CJ, Bhattacharya J, Butte AJ. An Environment-Wide Association Study (EWAS) on type 2 diabetes mellitus. PLoS. ONE. 2010;5:e10746. doi: 10.1371/journal.pone.0010746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wolf G. The New York Times. Sunday Magazine ed New York: 2010. The Data Driven Life. [Google Scholar]