Abstract
Pharmacovigilance involves continually monitoring drug safety after drugs are put to market. To aid this process; algorithms for the identification of strongly correlated drug/adverse drug reaction (ADR) pairs from data sources such as adverse event reporting systems or Electronic Health Records have been developed. These methods are generally statistical in nature, and do not draw upon the large volumes of knowledge embedded in the biomedical literature. In this paper, we investigate the ability of scalable Literature Based Discovery (LBD) methods to identify side effects of pharmaceutical agents. The advantage of LBD methods is that they can provide evidence from the literature to support the plausibility of a drug/ ADR association, thereby assisting human review to validate the signal, which is an essential component of pharmacovigilance. To do so, we draw upon vast repositories of knowledge that has been extracted from the biomedical literature by two Natural Language Processing tools, MetaMap and SemRep. We evaluate two LBD methods that scale comfortably to the volume of knowledge available in these repositories. Specifically, we evaluate Reflective Random Indexing (RRI), a model based on concept-level co-occurrence, and Predication-based Semantic Indexing (PSI), a model that encodes the nature of the relationship between concepts to support reasoning analogically about drug-effect relationships. An evaluation set was constructed from the Side Effect Resource 2 (SIDER2), which contains known drug/ADR relations, and models were evaluated for their ability to “rediscover” these relations. In this paper, we demonstrate that both RRI and PSI can recover known drug-adverse event associations. However, PSI performed better overall, and has the additional advantage of being able to recover the literature underlying the reasoning pathways it used to make its predictions.
Keywords: Pharmacovigilance, Distributional semantics, Literature-based discovery, Predication-based semantic indexing, Reflective random indexing
1. Introduction
An adverse drug reaction (ADR) is an “appreciably harmful or unpleasant reaction, resulting from an intervention related to the use of a medical product” [1]. ADRs were reported to be between the fourth and sixth leading cause of death in the United States in 1994 [2], accounting for 3–7% of medical hospital admissions [3,4] and a substantial number of health care visits [5]. They have a considerable negative impact on health and the healthcare system, despite the fact that extensive pre-marketing clinical trials are designed to test drug safety and efficacy. For example Phase III clinical trials have been estimated to cost 86.3 million U.S. dollars and last 30.5 months on average [6]. Nonetheless, rare ADRs may not be detected due to the limited duration and sample size of such trials, and others may occur on account of idiosyncratic characteristics of individuals excluded from the evaluated sample. The continued monitoring for ADRs after drugs are released into the market, called pharmacovigilance (PV), is therefore an important tool to monitor and improve drug safety.
Over the last decade, drug safety data obtained from spontaneous reporting systems (SRSs) have been analyzed using quantitative data mining procedures to retrieve strongly associated drug/ ADR pairs [7–9]. These highlighted associations are subsequently reviewed and scrutinized by domain experts. Unfortunately, research suggests data collected by SRS are limited by long time latency, incorrect or incomplete clinical information, underreporting and reporting bias [10,11]. Consequently, clinicians and researchers have also utilized existing healthcare data sources such as Electronic Health Records (EHRs) to attempt to identify previously unreported ADRs [12–15]. However, these data are inherently noisy as drugs and potential side effects may co-occur in the EHR for many reasons. In addition, the EHR often contains free-text data, and the accuracy of Natural Language Processing (NLP) tools is not perfect. New methods are required to selectively identify potentially hazardous drug/ADR associations. Consequently, the development of computational approaches to more accurately detect potential side effects is currently an active area of research [16–20]. These approaches have predominantly focused on improving signal detection using statistical methods, machine learning (ML) or some combination thereof.
In this paper, we develop an approach that is conceptually different than, and complementary to, such efforts. Methods of literature- based discovery (LBD) are used to detect potential drug/ ADR associations and to retrieve literature that supports their plausibility. The paper proceeds as follows. First we discuss the significance and challenges of pharmacovigilance, and how LBD methods might address these. We then provide relevant background on recent developments in LBD; and introduce the NLP tools that were used to extract knowledge from the literature for our experiments. We then discuss these experiments, in which we attempt to identify known ADRs using knowledge from the biomedical literature, and discuss their implications for pharmacovigilance practice.
2. Background
2.1. Pharmacovigilance: post-marketing drug surveillance
Vioxx (Rofecoxib) was withdrawn voluntarily from market by Merck in 2004, after it was found that the use of this agent increased the risk of myocardial infarction [21]. Avandia (Rosiglitazone) was suspended from the European market in 2010 [22–24] on account of an increased risk of cardiovascular complications. These high-profile examples illustrate that PV is an important supplement to existing drug safety profiles because clinical drug trials cannot be large or long enough to identify all problems related to a new drug [7]. Additionally, subjects are pre-selected by eligibility criteria and therefore may not fully represent the patient population after the drugs are put to market [25]. Consequently, it is highly unlikely that instances of all possible ADRs will be detected during pre-marketing clinical trials.
The fact that more than 75 drug products were removed from the market due to safety problems between 1964 and 2002 further emphasizes the importance of post-marketing drug monitoring, known as PV – “the science and activities relating to the detection, assessment, understanding and prevention of adverse effects or any other drug-related problem after drugs are on market” [26]. PV is designed to detect any rare or long-term adverse effects over a very large population and a long period of time. To advance this aim, health departments and organizations (such as the World Health Organization (WHO), U.S. Food and Drug Administration (FDA), and European Medicines Agency (EMA)) encourage physicians, other health care professionals, and patients to report voluntarily about any observed ADRs. In addition to voluntarily reporting, pharmaceutical companies are required to report serious adverse events [27]. These bodies have Spontaneous Reporting Systems (SRSs) to enable the efficient submission of reports electronically [28,29].
In general, the PV process proceeds as follows [30,31]:
Reported drug-related problems are collected in SRSs nationally or internationally.
Quantitative data mining procedures are used to analyze these data and retrieve relatively strongly correlated drug/ ADR pairs (drug/ADR associations).
These highlighted associations are then reviewed and evaluated by domain experts making up an expert clinical review panel.
Associations considered to be of clinical interest are then annotated as signals.
Specifically, signal is defined as “reported information on a possible causal relationship between an adverse event and a drug, the relationship being unknown or incompletely documented previously” [32]. Overall, the PV process includes two components – a statistical component (quantitative signal detection, steps (1) and (2)) and a qualitative component (expert clinical review, steps (3) and (4)) [31].
Through PV, international and national health institutions gather large amounts of data from SRS for further analysis. In addition, researchers have leveraged the opportunity provided by broader availability of EHRs by utilizing EHR data for signal detection [12,33]. These authors argue that EHR data can compensate for some of the deficiencies of SRS, such as under-reporting, misclassification, a long lag time between observation and reporting, reporting bias and the provision of incomplete clinical information [7,8]. Regardless of source, statistical algorithms are applied to both SRS [34–39] and EHRs [12] to measure the strength of observed drugevent associations.
It has been argued, though, that causality assessment is lacking in pharmacovigilance practice [25]. While expert clinical review is designed to verify potential ADRs, it is a human-intensive and time-consuming process. The available human resources are inadequate to review the large amount of noisy signal detected in SRS and EHR data, creating a bottleneck in the PV process. More research is needed to develop methods to automate, or assist with, the knowledge-intensive task of expert clinical review.
2.2. Assessment of causality
To address the issue of causality assessment, general principles exist that can be applied to evaluate the causality of potential ADRs [40]. The theoretical basis for these principles was proposed by Sir Austin Bradford-Hill in 1965 [41]. Bradford-Hill, an English epidemiologist and statistician, was the first to demonstrate that cigarette smoking contributes toward lung cancer using what are now referred to as the “Bradford-Hill criteria” [42]. The Bradford- Hill criteria provide viewpoints from which to evaluate evidence indicative of causality. These criteria are named ‘strength’, ‘consistency’, ‘specificity’, ‘temporality’, ‘biological gradient’ (referring to dose–response relationships), ‘plausibility’, ‘coherence’, ‘experimental evidence’, and ‘analogy’ [41,43,44]. Since then, the criteria have been widely used in epidemiology and may be applied to assess the causality of drug/ADR relationships [25,40,45]. Three of these criteria seem particularly pertinent to the development of pharmacovigilance methods:
The strength criterion reflects that strong associations are more likely to be causal than weak associations [40]. Quantitative statistical data mining methods evaluate adverse drug reaction signal from the strength of association point of view.
The plausibility criterion relates to evidence about mechanisms that may be involved to support a causal relationship.
The coherence criterion relates to the consistency of the hypothesis in question with contemporary medical knowledge.
Review by domain experts is required to evaluate a signal from the above points of view using their knowledge and judgment to find a signal with clinical significance. However, on account of the human-intensive nature of this task, automated assistance is desirable. In this study, we attempt to partially automate this aspect of the signal evaluation process. We do so using methods that leverage knowledge extracted from the biomedical literature as a means to assess the plausibility of an observed association. As one of these methods involves automated analogical reasoning, it is interesting to note that Bradford-Hill also permitted reasoning by analogy as an indicator of causality.
2.3. Literature-based discovery
Processing published biomedical literature to uncover implicit relationships among entities is referred to as literature-based discovery (LBD) [46–49]. LBD involves finding new knowledge by analyzing the literature, rather than through scientific experimentation. This is accomplished by identifying hidden connections between entities described in the published literature [46,50]. The origins of LBD may be traced to the serendipitous discovery that fish oils can be therapeutically useful in the treatment of Raynaud’s syndrome (poor circulation in the peripheries) by information scientist Don Swanson [46,50]. Weeber describes two types of LBD [48].
One type, referred to as “open LBD”, starts from a known term or concept (generally called A, although also referred to as C in Swanson’s early work) and tries to find an interesting hypothesis in the form of a previously unrecognized connection to some other term. If an article argues that A is associated with B and a second article mentions that B is associated with C; A may treat C. For example dietary fish oil (A) affects platelet aggregation, blood viscosity and vascular reactivity (B), and these biological factors (B) play a role in Raynaud’s syndrome (C) [50]. Consequently, it is reasonable to hypothesize that A treats C. The open LBD process proceeds from the source term A to an unknown target term C and culminates in the generation of a new hypothesis.
The second type of LBD is referred to as “closed LBD”. In a closed LBD process the goal is to evaluate an existing hypothesis. Closed LBD starts with known terms A and C, with the goal to identify intermediate terms B that provide the bridge between A and C [48]. For example, in 1988 Swanson found intermediate concepts to explain a hypothetical relationship between migraine and magnesium [51]. Smalheiser and Swanson used closed LBD to propose an explanation for epidemiologic evidence that estrogen might protect against Alzheimer’s disease [52].
LBD methodologies generally utilize statistical information derived from the frequency with which terms, or discrete concepts extracted from the literature using automated tools (e.g. MetaMap) or assigned to it by human annotators [53], co-occur [54,55]. This has been referred to as the co-occurrence model [56]. These cooccurrence statistics are interpreted by correlation mining and ranking algorithms [55,57].
A limitation of these methods is that they generally do not consider the nature of the relationship between the terms or concepts concerned. To address this limitation, Hristovski et al. [54] propose using semantic relations to eliminate spurious relationships introduced by frequently co-occurring concepts that are not meaningfully related. In their initial work, the semantic relations concerned were extracted from the literature by two Natural Language Processing (NLP) systems: SemRep [58] and (specifically to extract phenotypic information) BioMedLEE [59]. Their approach involved the specification of “discovery patterns”, patterns of relationships between concepts that may indicate an implicit therapeutic relationship [60]. These conditions can be specified as sets of semantic predicates. For example, Ahlers et al. [61] defined the May_Disrupt pattern as follows:
Substance X 〈inhibits〉 Substance Y
Substance Y 〈causes|predisposes|associated with〉 Pathology Z
Substance X 〈may disrupt〉 Pathology Z
Variants of this approach have been applied to generate or support the hypotheses that fish oil treats Raynaud’s disease [54], insulin treats Huntington disease [54], and antipsychotic agents prevent cancer [61]. Recently, this approach was also adapted to provide evidence to support the plausibility of an observed drug/ ADR association [62,63], providing proof-of-concept that LBD methods can be applied within the problem domain of pharmaco-vigilance. Regardless of the application domain, knowledge used to populate discovery patterns is extracted from the biomedical literature using NLP.
2.4. MetaMap and SemRep
MetaMap is a widely-used NLP tool that identifies concepts from the Unified Medical Language System (UMLS) in biomedical text [64,65]. SemRep [58,66] is a rule-based Natural Language Processing tool [67] that draws on concepts extracted by MetaMap and medical domain knowledge in the UMLS to extract semantic predications [58]. Its input consists of sentences from the literature; its output is a series of semantic predications identified in the respective text. A semantic predication is a subject-predicate-object triple in which the subject and object are UMLS concepts and the predicate is a semantic relationship. For example, metformin (UMLS Concept C0025598) TREATS diabetes mellitus (C0011849) is a semantic predication extracted from the phrase “Treatment of diabetes mellitus with metformin”. Evaluations of SemRep reveal a precision between 0.73 and 0.81, and a recall of 0.55 on the biomedical literature [67–69]. Semantic predications benefit the LBD process in several respects. The additional information provided by semantic predications makes the LBD results easier to interpret. In addition, it has been noted that a large number of uninformative co-occurrences must be manually reviewed when LBD is based on lexical statistics alone [70]. In contrast, semantic predications provide the means to isolate relationships between concepts that are logically connected in a meaningful way.
2.5. Semantic vectors for scalability
Regardless of whether co-occurrence relations or discovery patterns are used, LBD systems must explore large numbers of possible reasoning pathways to identify explanatory hypotheses (for closed discovery) or previously unrecognized relationships (for open discovery). Consequently, the process of LBD can be computationally expensive, and thus faces scalability issues in the context of the rapid growth of the biomedical literature. In contrast, the field of distributional semantics has produced corpus-derived statistical models that can measure the relatedness between two concepts by comparing vector representations of these concepts, called semantic vectors, that are derived from the contexts they have occurred in [71], without the need to explicitly explore cooccurring concepts once the initial model has been generated. Consequently, several authors have explored the use of distributional models for LBD [72–74]. These geometrically motivated models of distributional semantics represent terms or concepts as high-dimensional vectors derived from the contexts in which they have occurred. Relatedness between a pair of terms or concepts is then estimated from the similarity between the vectors [74].
Random indexing (RI), a relatively recent development, further improves the scalability of distributional methods by avoiding computationally intensive approaches to dimensional reduction of the original term-by-context matrix [75,76]. The algorithm’s computational complexity scales linearly with increasing size of the input data. It can be incrementally updated as new documents are added without retraining the whole dataset; thus it is applicable to large corpora such as MEDLINE.
In the experiments that follow, we evaluate the extent to which two variants of RI that have been applied to LBD in our previous work can identify known side effects of pharmaceutical agents. To implement a co-occurrence based approach, we use Reflective Random Indexing (RRI) [74]. To implement a discovery pattern based approach, we use Predication-based Semantic Indexing (PSI) [77]. On account of their scalability, these models permit inference on a scale that would be prohibitively time-consuming if explicit exploration of all possible reasoning pathways were attempted. This is accomplished through a mechanism known as “indirect inference” [78], which enables distributional models to find meaningful connections between terms that do not co-occur with one another directly, without the need to explore intervening terms explicitly.
3. Materials and methods
3.1. Materials
In this study, MetaMapped Medline Baseline (MMB) and Semantic MEDLINE Database (SemMedDB) were used to represent knowledge from the biomedical literature. Side Effect Resource 2 (SIDER2) was used as data set for drug/ADR associations. The Semantic Vectors package was used to build concept-based (RRI) and predication-based (PSI) semantic space models [74,77].
3.1.1. MMB and SemMedDB
We used the 2012 MMB as a repository for concept-based modeling. The MMB contains 20,494,848 articles included in Medline up to November, 2011 and contains 399,701 distinct concepts. We used the SemMedDB V2.2 (semmedVER22) for this experiment, which was processed by SemRep version 1.5. This was the current version when our experiments started. SemMedDB contains 22,252,812 citations included in Medline up to March 31, 2013 and contains 63,795,467 predications. There are 58 distinct predicates and 257,350 distinct concepts in SemMedDB. There are also negated predications in the SemMedDB repository (e.g. anticoagulant_therapy NEG_TREATS (does not TREAT) phlebitis). However, the number of negative predications is relatively small (1.2% of total predications), so we did not include these predications in the PSI model.
3.1.2. SIDER2
SIDER2 is a publicly available database containing information on marketed medicines and their known adverse reactions [79]. SIDER2 was used to construct a dataset for our experiment and as a reference standard to confirm whether a predicted side effect is a true adverse reaction. We normalized SIDER2 terms by mapping drug and side effects terms to UMLS CUI with UMLS Terminology Services (UTS) API 2.0 [80] and then subsequently searching these UMLS CUIs in SemMedDB and MMB to retrieve the mapped UMLS concepts which are represented in SemMedDB and MMB.
SIDER2 contains 996 drugs, 4192 side effects, and 99,423 drug/ ADR pairs. Only those side effects and drugs that were represented in both the RRI and the PSI spaces were retained, so our reference set contains 959 drugs, 3436 side effects, and 90,787 drug/ADR pairs. Each vector model’s search space was composed of vectors representing the SIDER2 side effects. For the PSI model, SIDER2 drug/ADR pairs were also used as training data to infer predicate reasoning pathways.
3.1.3. MEDication–Indication (MEDI)
As we would anticipate connections in the literature between medications and diseases they treat, we evaluated the utility of another knowledge resource, MEDI [81], as a means to eliminate drug indications from consideration as potential side effects. MEDI is a medication indication resource that was extracted from a set of commonly used medication resources, including RxNorm, MedlinePlus, SIDER2, and Wikipedia [81]. MEDI drugs are represented by RxNorm codes, and indications are represented by ICD-9 codes. MEDI contains 3112 medications and 63,343 medication–indication pairs. Additionally, the MEDI high-precision subset (MEDI-HPS) was created by only including indications that are retrieved from RxNorm or at least two of the three other resources. MEDIHPS contains 2136 medications and 13,304 medication–indication pairs. The estimated precision of MEDI-HPS is about 92% [81].
In our experiments, MEDI was used to eliminate drugs’ indications from the side effects search space. To do so, we first needed to normalize all terms representing drugs (RxCUI) and indications (ICD-9 codes) in MEDI to UMLS concepts, and then filtered each drug’s indications from this drug’s search space. In many cases, there exist hierarchical relationships between concepts. For example, C0264702 Acute myocardial infarction of apical–lateral wall is a child node of C0155626 Acute myocardial infarction. So in our experiment, we extended the MEDI list by aggregating the related concepts by different hierarchical relations. We tested these various extensions of the MEDI list as different MEDI interventions.
3.2. Methods
3.2.1. RRI
Reflective Random Indexing (RRI) [74] is a variant of RI adapted to enable the recognition of meaningful indirect associations. The variant of RRI we used for our experiments allows for the estimation of semantic relatedness between UMLS concepts, and proceeds as follows.
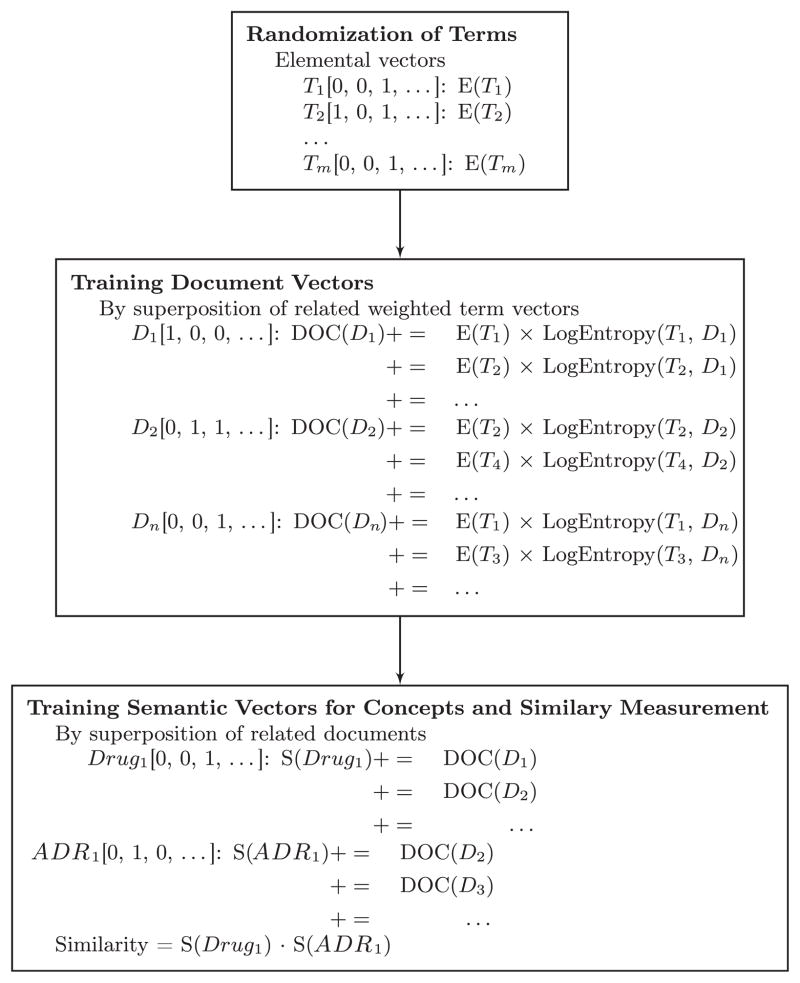
First, all terms in the text corpus are assigned unique vector representations, known as elemental vectors. We will refer to the elemental vector for concept C as E(C) for remainder of this manuscript. In accordance with the RI paradigm [82], elemental vectors are generated stochastically. In this way, RI creates unique fingerprints for all terms in the text corpus. The vector components can be binary, ternary, real, or complex values [82,83]. In our experiments, we use 32,000 dimensional binary vectors constructed in accordance with the Binary Spatter Code (BSC) [84], one of a family of representational approaches known as Vector Symbolic Architectures (VSAs) [84–87]. This dimensionality was selected based on the results of simulation experiments in previous research [88], which suggest that at this dimensionality around 2000 unique elemental vectors can be superposed with low probability of the superposed product being closer to some other elemental vector in the space than its component vectors. However, we did not attempt to optimize this parameter, and would anticipate some improvement in accuracy in exchange for the additional computational work required to perform these experiments at higher dimensionalities. In the BSC, elemental vectors are constructed by distributing an equal number of 1’s and 0’s at random across the dimensions of the vector concerned. Consequently, elemental vectors have a high probability of being orthogonal or close-to-orthogonal to each other, with orthogonality defined as a Hamming Distance (HD) of half the dimensionality of the vectors concerned [75,84,86].
The next step is to generate vector representations of documents, by superposing the elemental vectors of the terms contained in these documents. With binary vectors, superposition is accomplished by keeping track of the number of 1’s and 0’s that have been added in each dimension, and assigning the value in this dimension using the majority rule, with ties split at random. We will refer to this operation by using the “+” symbol, with “+ =“ indicating a superposition that includes the vector on the left of the operator also (so DOC(D)+=E(C) is equivalent to DOC(D) = DOC(D)+E(C)), a common operation during training). In our experiments, this superposition is weighted using the Log-Entropy weighting procedure. The local term weight for term i in document j (lij) is derived from the frequency of a term in a document. The global weight for term i (gi) describes the frequency of the term within the entire text corpus. They are computed with Eq. (1):
| (1) |
This weighting scheme reduces the influence of high frequency terms that may be uninformative, and tempers the influence of terms that recur frequently within a single document [89]. Once document vectors have been generated (Eq. (2)), it is possible to generate vector representations of concepts (in our case, or terms in the general case), known as semantic vectors. We will refer to the semantic vector for concept C as S(C). Semantic vectors are constructed by superposing vector representations of the documents a concept occurs in.
| (2) |
Superposition of binary vectors requires maintaining a “voting record” that keeps track of the number of 1’s and 0’s added in each dimension. When local and global weighting metrics are utilized, the “votes” may not be integer values. So, for example, if the vector 1010 were added with a weight of 0.5, a straightforward implementation of the voting record would add 0.5 to the dimensions of the voting record corresponding to the 1’s, and subtract 0.5 from the dimensions corresponding to the 0’s. Normalization involves tallying these votes. After training is complete, those dimensions of the voting record with positive values would be assigned 1, those with negative values would be assigned 0, and those with a zero value would be assigned either 1 or 0 at random. In practice, however, it is computationally inconvenient to maintain and update 32,000 real values to serve as a voting record for each semantic vector. Consequently, the Semantic Vectors package employs a binary matrix approximation of the voting record, which sacrifices some floating-point precision in exchange for computational efficiency. These implementation details are provided in [83].
These operations are expressed concisely in the pseudo code in Fig. 1, adapted from [74]. A schematic representation for RRI is shown in Fig. 2. We used Semantic Vectors Version 3.7 to build RRI vectors. Once semantic vectors were constructed, the relatedness between drugs and ADRs was estimated as . Therefore, a ranked list of ADRs for each drug was provided.
Fig. 1.
The pseudo code for RRI model training process.
Fig. 2.
Schematic representation of RRI training and inference process.
3.2.2. PSI
3.2.2.1. Operations in PSI
The PSI model provides the means to implement discovery patterns for LBD using distributional semantics [77,90]. This is accomplished by representing concepts and relationships extracted by SemRep as high-dimensional vectors using an adaption of RI. In previous work, PSI has been applied to discover therapeutic relationships [77] using a two-stage process of discovery by analogy: first a geometric operator is used to infer discovery patterns from known treatments, then the identified discovery patterns are used to infer previously unseen therapeutic relationships.
In addition to the superposition operation described previously, the PSI model utilizes a binding operation. Binding (⊗) is a compositional operation that is provided by VSAs, such as the BSC [86,87]. Binding two elemental vectors generates a third vector, which is dissimilar from these two component vectors. The binding operation is reversible (release ø). With binary vectors, pairwise exclusive OR (XOR) is used to accomplish both binding (⊗) and release (ø).
3.2.2.2. PSI training process
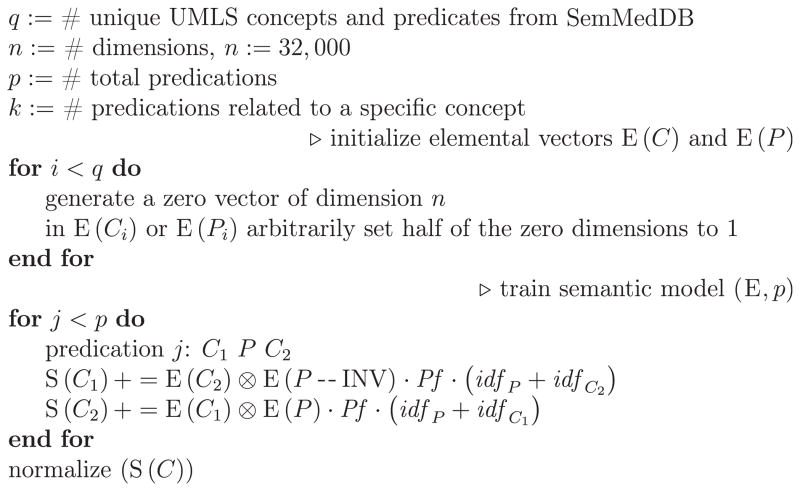
The training process for generating semantic vectors proceeds as follows:
Generate elemental vectors for all concepts and relations occurring in semantic predications.
Generate a semantic vector for each concept, initially empty.
For each predication (concept–predicate–concept), bind the elemental vector of one concept and the elemental vector of the predicate, and add this bound product to the semantic vector for the other concept.
During step (3), a statistical weighting scheme is applied. For the predication C1 P C2, the semantic vector S(C2) is generated as shown in Eq. (3).
| (3) |
The global weight Pf is derived from the number of times that the predication occurs in the SemMedDB. The local weight idf (inverse document frequency of the concept c or the predicate p) reflects the occurrence of the concept across all documents. They are computed as shown in Eq. (4).
| (4) |
The pseudo code for PSI is displayed in Fig. 3.
Fig. 3.
The pseudo code for PSI model training process.
All concepts and relations were assigned a binary elemental vector of 32,000 bits in length. The semantic vector of each concept was generated by superposing bound products related to this concept, where the bound products were produced by binding the elemental vectors for the other concept and predicate elemental vectors in each predication this concept occurs in. The search space of SIDER2 side effects contains 3436 ADRs.
3.2.2.3. Inferring discovery patterns
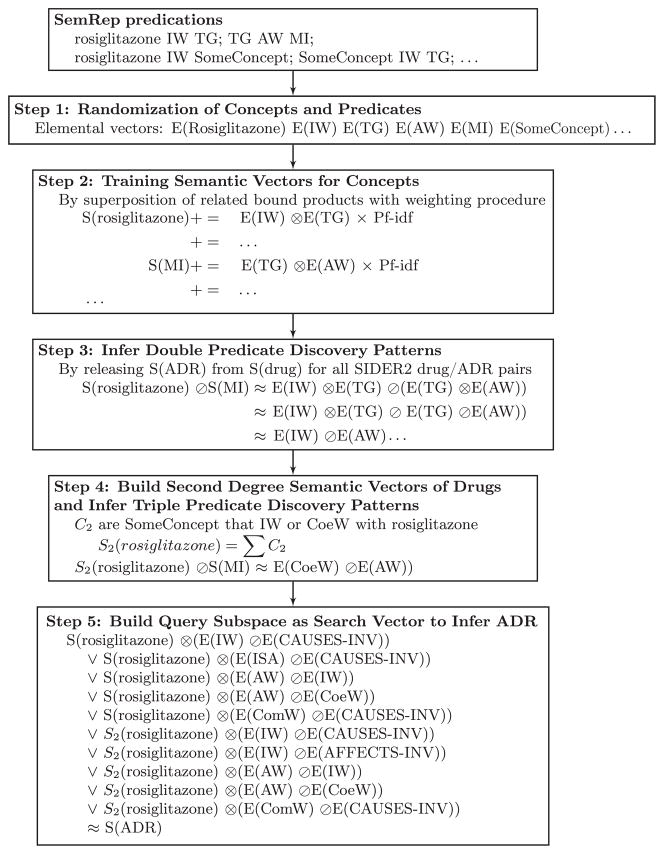
After training the semantic vectors, the PSI model can be used to infer discovery patterns by “releasing” the semantic vector of a drug using the semantic vector of its ADR.
The bound product of the drug’s semantic vector and discovery patterns’ vectors can be subsequently used as a query vector to search the vector space of side effects. In our procedure, discovery patterns were inferred from all known drug/ADR associations. For each drug, the five discovery patterns that were most frequently inferred from all other drugs and their ADRs were retained.
The pathways connecting drugs to side effects may not be restricted to one middle term (and two predicates). In previous experiments predicting therapeutic relationships, performance was improved by including pathways of three predicates and two middle terms [90]. This is accomplished by generating a second-degree semantic vector for a concept, S2(concept), by adding together the (first-degree) semantic vectors of all concepts connected to it by a predicate of interest. In our experiments, the two most popular predicates from inferred double-predicate reasoning pathways – INTERACTS_WITH and COMPARED_WITH – were used to build second-degree semantic vectors S2. This vector is then used as an alternative starting point for the inference procedure. From this point, the five most frequently inferred double-predicate reasoning pathways using the second order semantic vector of all other drugs and the (first order) semantic vectors of their ADRs were retained. As these inferred pathways connect to drugs through either INTERACTS_WITH or COMPARED_WITH, they are referred to as triple-predicate pathways.
3.2.2.4. Applying discovery patterns to find possible ADRs (Step 5 in Fig. 4)
Fig. 4.
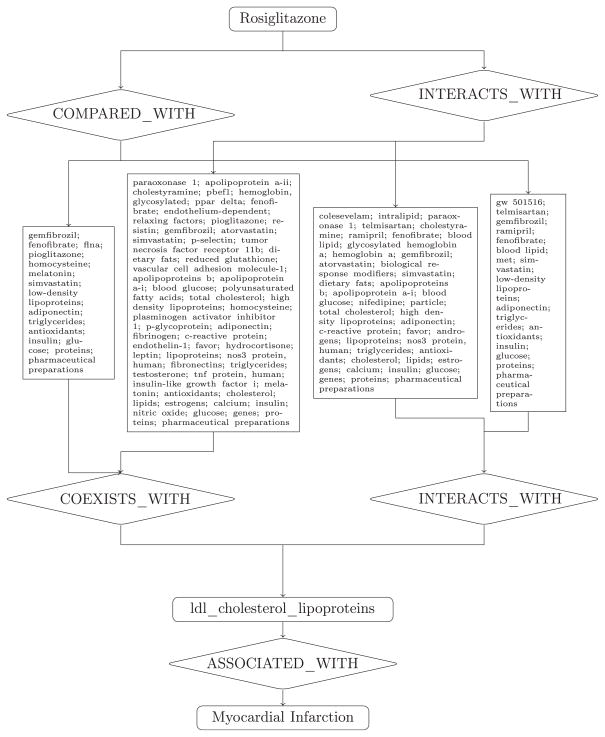
Schematic representation of PSI training and inference process. triglycerides: TG; myocardial infarction: MI; INTERACTS_WITH: IW; COEXISTS_WITH: CoeW; ASSOCIATED_WITH: AW; COMPARED_WITH: ComW.
To combine query vectors for frequently inferred reasoning pathways into one search expression, we use a disjunction operation that originates in the quantum logic of Birkhoff and von Neumann, and was first applied to information retrieval by Widdows and Peters [91,92]. We define the disjunction of these five query vectors as a query subspace derived from them using a binary vector approximation [93] of the Gram–Schmidt orthonormalization procedure [94]. The length of the projection of some other vector in this subspace provides an estimate of vector-subspace similarity.
For the double-predicate discovery patterns model, a drug’s query subspace was constructed from this drug’s first-degree semantic vector bound to the vector representations of the five double predicate reasoning pathways most frequently inferred from other drugs. For the double- and triple-predicate discovery patterns model, a drug’s query subspace also included this drug’s second-degree semantic vector bound to vector representations of the five reasoning pathways most frequently inferred from the second-degree semantic vectors of other drugs.
The length of the projection of the semantic vector for a candidate ADR into a drug’s query subspace was used to estimate the relatedness between these entities, providing a ranked list of potential ADRs for each drug.
Fig. 4 provides an overview of the PSI-based analogical reasoning process in its entirety.
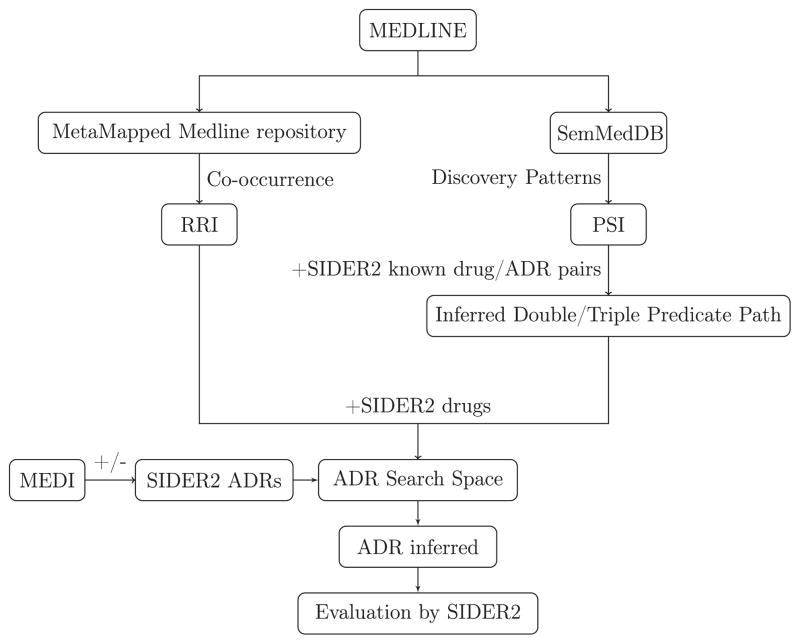
3.3. Experimental design
An overview of the experimental design is shown in Fig. 5. The first experiment was conducted without knowledge of drug indications. The concept-based RRI model and discovery pattern-based PSI model were compared with respect to their ability to identify known drug/ADR associations. In the second experiment, the model with the best performance from the first experiment was used to evaluate the effect of eliminating known indications from the list of predictions.
Fig. 5.
Experimental design in the detection of SIDER2 known ADRs using LBD distributional semantic models.
3.3.1. Experiment 1 design
Distributional semantic vectors were used to model MMB and SemMedDB. RRI vectors and PSI vectors formed the basis for our models of LBD concept-based co-occurrence and LBD discovery patterns, respectively. As MetaMap may retrieve many more concepts from a particular document than SemRep retrieves predications, we varied the RRI model to assess the extent to which observed effects were due to the advantage of a more extensive (albeit less structured) knowledge base. In one case, a RRI space was derived from only those sentences from which predications were extracted. Consequently, there are three distributional semantic models – RRI built from documents (RRI-from-document group), RRI built from predication source sentences (RRI-from-predication group), and PSI built from predications. The PSI model was evaluated with two settings. In the first case, only two-predicate discovery patterns were considered (PSI-double group), while the second case considered both two- and three-predicate patterns (PSI-double + triple group). The elemental vectors for terms, which are not meaningfully related to one another, were used to implement a random baseline (Baseline group).
With the RRI models, for each drug, related problems were sought by comparing each vector in the side effect search space to this drug’s vector representation.
With the PSI model, SIDER2 known drug-side effect pairs were used to infer predicate paths. For the PSI-double group, each drug’s query subspace was built as the disjunction of the bound products between the drug and its five double-predicate reasoning pathways. For the PSI-double + triple group, each drug’s query subspace additionally included the second degree semantic vector of this drug bound to five triple-predicate paths. The five triple-predicate paths were retrieved by the extension of second degree semantic vectors of drugs. Comparing a drug’s query subspace with each vector in the search space allowed us to infer the drug’s possible side effects.
3.3.2. Experiment 2 design (Fig. 5)
From our preliminary results, we found that there were some indications in the inferred ADRs. So we hypothesized that excluding known indications for drugs from the search space would improve performance. We tested this hypothesis in the second experiment utilizing knowledge of drug indications from MEDI. In this experiment, we tested variants of the MEDI indication list using the best performing model from the first experiment (PSI double + triple group). We extended the MEDI-complete and MEDI-HPS lists to include all offspring, or immediate offspring nodes based on the UMLS semantic network utilizing the MRREL.RRF file. This file includes relationships between UMLS concepts found in the UMLS Metathesaurus [95]. By utilizing these ancestor-offspring hierarchical relationships, we define an offspring node as a node that has a MEDI indication as an ancestor (regardless of the number of intervening nodes); and an immediate offspring node as a node that has this MEDI indication as its parent.
In this procedure, we first normalized all MEDI terms. For MEDI drugs, we mapped each drug’s RxCUI to a UMLS CUI with the RxNorm API [96] and then subsequently searched the UMLS CUI in SemMedDB and MMB to retrieve the mapped UMLS concept. For MEDI indications, we mapped each indication’s ICD-9 term to a UMLS CUI using the UTS API 2.0 [80] and then subsequently searched for this UMLS CUI in SemMedDB and MMB to retrieve the mapped UMLS concept. After normalizing MEDI terms, the hierarchical relation of synonym (SY), child (CHD), and sibling (SIB) in MRREL.RRF were used to find drugs’ MEDI indications extended offspring or immediate offspring. Consequently, there were six MEDI lists (Table 1). These MEDI lists were used to exclude indications from the side effect search space and were tested in the second experiment.
Table 1.
Different groups by extending MEDI in experiment 2.
| MEDI intervention | Extension procedure | Experiment group |
|---|---|---|
| No MEDI | None | No-MEDI group |
| MEDI-HPS indications for SIDER2 drugs | All synonym (SY), child (CHD), and sibling (SIB), as well as their offspring | MEDI-HPS-offspring group |
| Immediate synonym (SY), child (CHD), and sibling (SIB) relationships | MEDI-HPS-immediate offspring group | |
| MEDI-complete indications for SIDER2 drugs | None | MEDI-complete group |
| All synonym (SY), child (CHD), and sibling (SIB), as well as their offspring | MEDI-complete-offspring group | |
| Immediate synonym (SY), child (CHD), and sibling (SIB) relationships | MEDI-complete-immediate offspring group |
3.3.3. Performance measurements
To evaluate performance, we used a number of widely used metrics. Precision measures the proportion of accurate ADRs in relation to the total number of ADRs retrieved [97]. To evaluate the precision at different points in a ranked list, we used Average Precision (AP, the average of the precision values measured at the point at which each correct result is retrieved for one example [98]). Mean average precision (MAP) is the average of the AP across all drugs. Precision at k [98] measures the precision at fixed levels of retrieved results and emphasizes the importance of finding relevant results early. We evaluated precision at k = 50 (Pk=50). Recall represents the proportion of ADRs retrieved out of the total number of ADR associations in the reference standard [97].
We define a “rediscovery” (true discovery) as an adverse effect inferred by a vector model and subsequently confirmed by SIDER2 as a true prediction. Consequently, the median rediscovery rank for a particular drug approximates the point in the ranked list produced by a particular model at which half of the known adverse reactions for this drug were recovered.
The AP and median rank of the rediscoveries across drugs were compared by the paired t test and the Wilcoxon matched-pairs signed-rank test, respectively.
To measure the performance with respect to the true positive rate (TPR) and false positive rate (FPR), receiver operating characteristic (ROC) curve was plotted for all drug/ADR pairs for all models. Subsequently, a global area under the ROC curve (AUC, “global” indicates that the scores of all drug-ADR pairs were combined into a single curve) was calculated using AUCCalculator [99]. For the model with the best global AUC, a drug-based AUC was also calculated and compared between drugs.
4. Results
4.1. Experiment 1
4.1.1. Inferring discovery patterns
The most strongly associated double-predicate path was calculated for each known drug/ADR pair. In total, 90,787 predicate paths were inferred. Among them, there were 1485 unique predicate paths. The five most frequently inferred double-predicate paths were selected. Second degree semantic vectors for drugs were constructed by adding together the semantic vector representations of any concept occurring in a semantic predication with the drug in question, where the predicate type was either INTERACTS_ WITH or COMPARED_WITH. The most frequently occurring double predicate paths and inferred triple predicate paths with corresponding examples are shown in Table 2. They are consistent across all drugs. Many of these paths are readily interpretable, and could support a plausible biological mechanism for a predicted effect. For example, INTERACTS_WITH:CAUSES-INV suggests a drug may interfere with some biological factor which may cause a side effect. COMPARED_WITH:CAUSES-INV can be used to identify similar side effects by comparing their drug class information as COMPARED_WITH often indicates a comparative evaluation across different drugs in the same therapeutic category. Triple predicate paths extend the connecting path for drugs and related ADRs.
Table 2.
The most frequent predicate paths in inferred discovery patterns.
| Double/triple-predicate | Example |
|---|---|
| INTERACTS_WITH CAUSES-INV |
Dipyridamole INTERACTS_WITH nitric oxide Bradycardia CAUSES-INV nitric oxide |
| ASSOCIATED_WITH COEXISTS_WITH |
Rosiglitazone COEXISTS_WITH apolipoprotein
a-ii Apolipoprotein a-ii ASSOCIATED_WITH myocardial infarction |
| COMPARED_WITH CAUSES-INV |
Bisoprolol COMPARED_WITH metoprolol Hypotension CAUSES-INV metoprolol |
| ASSOCIATED_WITH INTERACTS_WITH |
Rosiglitazone INTERACTS_WITH
triglycerides Triglycerides ASSOCIATED_WITH myocardial infarction |
| ISA CAUSES-INV |
Naproxen ISA calcineurin inhibitor Toxic nephropathy CAUSES-INV calcineurin inhibitor |
| INTERACTS_WITH INTERACTS_WITH ASSOCIATED_WITH |
Rosiglitazone INTERACTS_WITH lyrm1 Lyrm1 INTERACTS_WITH fatty acids, nonesterified Fatty acides, nonesterified ASSOCIATED_WITH myocardial infarction |
| INTERACTS_WITH ASSOCIATED_WITH COEXISTS_WITH |
Rosiglitazone INTERACTS_WITH
glycerol-3-phosphate dehydrogenase Glycerol-3-phosphate dehydrogenase COEXISTS_WITH succinate dehydrogenase Succinate dehydrogenase ASSOCIATED_WITH myocardial infarction |
| COMPARED_WITH INTERACTS_WITH ASSOCIATED_WITH |
Rosiglitazone COMPARED_WITH
glycerophosphates Glycerophosphates INTERACTS_WITH low-density lipoproteins Low-density lipoproteins ASSOCIATED_WITH myocardial infarction |
| COMPARED_WITH COEXISTS_WITH ASSOCIATED_WITH |
Rosiglitazone COMPARED_WITH gw 501516 Gw 501516 OEXISTS_WITH high-density lipoprotein cholesterol High-density lipoprotein cholesterol ASSOCIATED_WITH myocardial infarction |
4.1.2. Performance
Results for different vector models are shown in Table 3. PSI-based models performed better than RRI-based models and both models perform better than the random baseline. The PSI-double + triple group outperformed all other groups. All differences in median rank and MAP were statistically significant (as estimated by Wilcoxon’s signed rank test and paired t test respectively). Pk=50 for each drug was compared across groups using Pearson’s correlation. For variants of the same model (RRI or PSI), Pk=50 was highly correlated (0.75–0.84). Correlation in Pk=50 between the PSI and RRI models was between 0.52 and 0.57, suggesting the potential to improve performance by combining results.
Table 3.
Results of precision and rank-based measures for different groups.
| Group | MAP | Pk=50 for all drugs (global precision) | Median rank (n
= 3, 436) |
AUC | |
|---|---|---|---|---|---|
| Median | Mean | ||||
| Baseline | 0.0300 | 0.0284 | 1708 | 1711.44 | 0.5021 |
| RRI-from-predication | 0.0365 | 0.0469 | 1629 | 1651.08 | 0.5140 |
| RRI-from-document | 0.0520 | 0.0784 | 1333 | 1454.30 | 0.5508 |
| PSI-double | 0.0591 | 0.0942 | 1233 | 1379.65 | 0.5973 |
| PSI-double + triple | 0.0848 | 0.1410 | 808 | 1108.47 | 0.6841 |
4.1.3. AUC
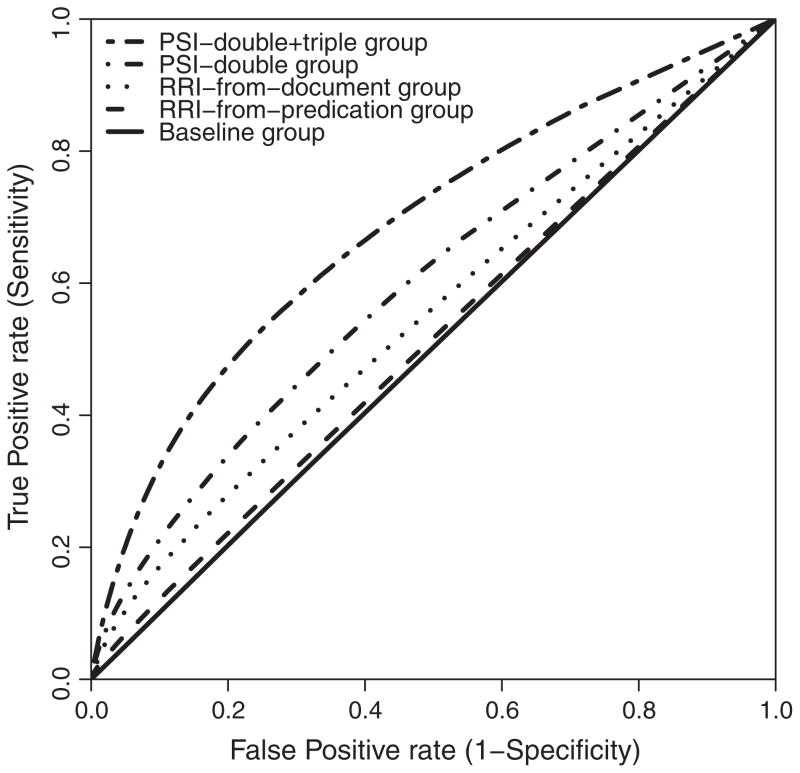
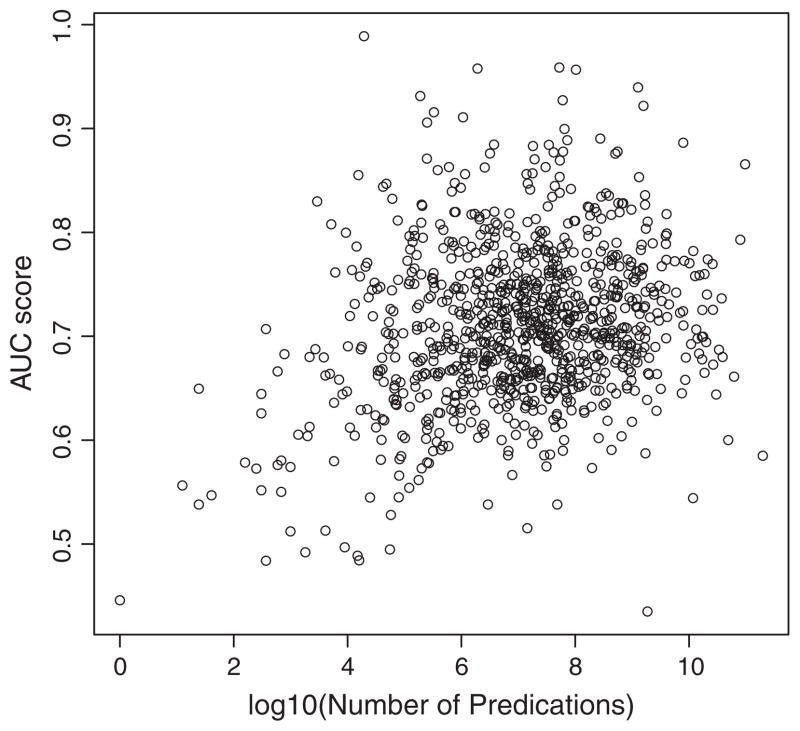
Fig. 6 and Table 3 present the global ROC curves for all models. ROC curve shows the tradeoff between sensitivity and specificity. The global AUC provides a cumulative estimate of accuracy, and is shown for each model in Table 3. PSI-double + triple group has the best global AUC of 0.6841. We measured its AUC at the drug level (Fig. 7). The mean and median AUC are 0.7102 ± 0.0752 and 0.7058 respectively. Fig. 7 shows a plot of the AUC for each drug against the log of the number of predications in SemMedDB with this drug as subject. This suggests a trend in which performance is generally better for those drugs for which more knowledge is available in the database. Those drugs with an AUC of 0.8 or above tend to occur in 10,000 or more predications as subject.
Fig. 6.
ROC plot of true positive rate and false positive rate for all groups.
Fig. 7.
Performance of AUC for all drugs by PSI-double + triple group.
Note that the global AUC as a metric will be inflated by methods that incorporate category bias into their prediction [100,101], a subject we will return to in the discussion section.
4.1.4. Rediscovery results
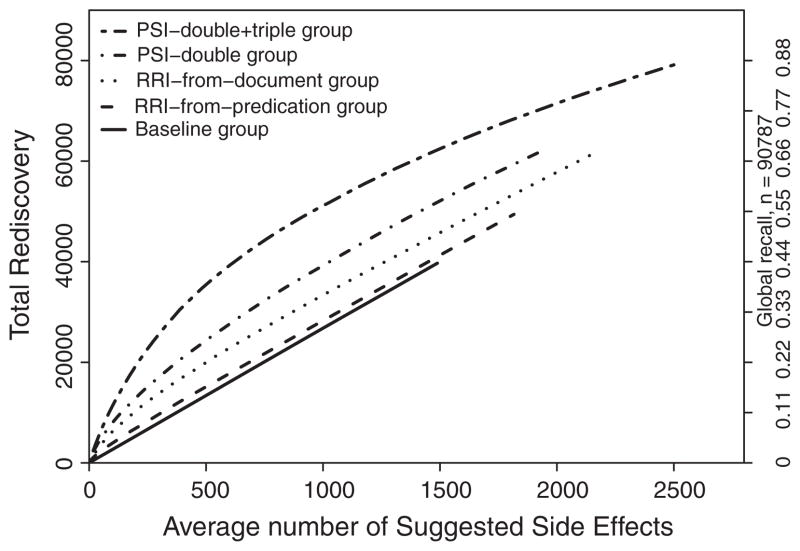
The results of this experiment are illustrated in Fig. 8. This figure plots the number of rediscovered side effects (left Y axis) and the proportion of the valid side effects rediscovered (or global recall, right Y axis) for each model against the mean number of suggested potential ADRs (X axis) at different statistical thresholds. All distributional models outperform the random baseline.
Fig. 8.
Rediscovery plot for experiment groups.
With approximately 100 predictions per drug, baseline, RRI-from- predication, RRI-from-document, PSI-double and PSI-double + triple group have a global recall of 0.029, 0.045, 0.069, 0.088, 0.125, respectively.
4.2. Experiment 2
The PSI-double + triple model was the best performing model in the first experiment, and was selected to test the effects of using variants of the MEDI list as a way to exclude therapeutic relationships to reduce the number of highly ranked false positive predictions.
Table 4 presents the performance of the PSI-double + triple model when different MEDI lists were used. The median rank of true positive predictions was lower when MEDI was used to exclude the indication from the search space for each drug. However, as median rank is based on the rank of true positive results only, it does not consider known side effects that may have been excluded from consideration by the MEDI list. In contrast, MAP also measures whether true side effects have been excluded. Consequently, MAP in the MEDI-complete-immediate offspring was higher than other groups. Overall, AUC was highest for the MEDI-HPS- immediate offspring group. Of the models, only the MEDI-HPS- immediate offspring group outperformed the baseline PSI model by all metrics, and the improvements in performance were small in comparison with the differences in performance between distributional models in experiment 1. All differences between all MEDI intervention groups and No-MEDI group in Pk=50 are statistically significant as measured by the paired t test. However, the improvement in cumulative accuracy is negligible.
Table 4.
PSI-double + triple model performance across all tests with different MEDI lists. Best results are in boldface. Performance exceeding the baseline (results obtained by the best PSI model without MEDI) is marked with an asterisk (*).
| Group | MAP | Pk=50 for all drugs | Median rank (n
= 3, 436) |
AUC | |
|---|---|---|---|---|---|
| Median | Mean | ||||
| No-MEDI | 0.0848 | 0.1410 | 808 | 1108.47 | 0.6841 |
| MEDI-HPS | 0.0849* | 0.1417* | 807* | 1107.60 | 0.6839 |
| MEDI-HPS-offspring | 0.0798 | 0.1283 | 765 | 1050.75* | 0.6813 |
| MEDI-HPS-immediate offspring | 0.0866* | 0.1444* | 795* | 1094.41* | 0.6850* |
| MEDI-complete | 0.0839 | 0.1401 | 809 | 1108.06* | 0.6831 |
| MEDI-complete-offspring | 0.0673 | 0.0917 | 681* | 953.76* | 0.6737 |
| MEDI-complete-immediate offspring | 0.0889* | 0.1467* | 773* | 1068.72* | 0.6821 |
4.3. Plausibility evidence found by PSI discovery patterns approach
In this paper, the association between rosiglitazone and myocardial infarction, a highly publicized ADR discovered after the drug was released to the market, is used to illustrate how evidence from the literature can be retrieved for the evaluation of plausibility by a domain expert. The term “myocardial_infarction” was ranked in the top 1% (rank = 29) and top 1.5% (rank = 50) of potential side effects for rosiglitazone by the PSI-double and PSI-double + triple models respectively.
Rosiglitazone is a thiazolidinedione (TZD) antidiabetic drug, used to treat type 2 diabetes mellitus as an adjunct to lifestyle changes [102–104]. Since its approval by the FDA in 1999, rosiglitazone was prescribed 3.8 million times annually up to June 2009 in the United States [105]. A meta-analysis of clinical trials conducted by Nissen and Wolski [106] in 2007 suggested that the use of rosiglitazone was associated with a significant increase in the risk of myocardial infarction. This led to rosiglitazone’s withdrawal from the European market in 2010 and a rosiglitazone black-box warning in the U.S. [105,107]. In 2013, the FDA lifted some prescription restrictions in the U.S. market based on a reevaluation of the Rosiglitazone Evaluated for Cardiac Outcomes and Regulation of Glycaemia in Diabetes (RECORD) trial (ClinicalTrials.gov Identifier NCT00379769) [108], but the European suspension is still in effect at the time of this writing.
Rosiglitazone is a nuclear peroxisome proliferator-activated receptor (PPAR-gamma) agonist. The mechanism through which rosiglitazone causes cardiovascular events is unclear, but is thought to be related to unfavorable effects on triglycerides, low-density lipoprotein cholesterol (LDL-C) particle size and density, and greater affinity for PPAR-gamma than other TZD drugs [109– 112]. To evaluate the extent to which these hypotheses were consistent with information utilized by the PSI-double + triple model, we reconstructed the pathways of predicates and concepts that were consistent with the inferred discovery patterns used to make this prediction.
For myocardial infarction, each discovery pattern that was used for the inference was used to search the indexed SemMedDB predications and find middle terms that connect rosiglitazone with myocardial infarction through the discovery pattern. The middle terms retrieved were ranked based on their inverse document frequency. Since the indexed SemMedDB predications contain the source literature ID (PMID), we also retrieved related literature evidence that supports the prediction.
Consequently 108,100 unique predication pathways were retrieved through 8 unique predicate paths (Table 5) with distinct middle terms that connect rosiglitazone with myocardial infarction. Table 6 shows some example predication pathways, that were composed of two or three predications. There were around 17 sentences providing evidence to support each predication on average. We analyzed middle terms’ semantic groups [113] and list the sample with distinct predicate paths connecting with different semantic groups (Fig. 9).
Table 5.
Reasoning pathways used to retrieve evidence from the literature for the pair rosiglitazone – myocardial infarction.
| Predicate path |
| COMPARED_WITH : COEXISTS_WITH : ASSOCIATED_WITH |
| COMPARED_WITH : INTERACTS_WITH : ASSOCIATED_WITH |
| INTERACTS_WITH : COEXISTS_WITH : ASSOCIATED_WITH |
| INTERACTS_WITH : INTERACTS_WITH : ASSOCIATED_WITH |
| COEXISTS_WITH : ASSOCIATED_WITH |
| INTERACTS_WITH : ASSOCIATED_WITH |
| COMPARED_WITH : ASSOCIATED_WITH : COEXISTS_WITH |
| INTERACTS_WITH : ASSOCIATED_WITH : COEXISTS_WITH |
Table 6.
Some example predications for possible mechanism of rosiglitazone causing myocardial infarction.
| Middle term “LDL cholesterol lipoprotein”; 123 unique predication pathways; |
Middle term “triglyceride”;
1515 unique predication pathways;
|
Middle term “ppar_gamma”; 992
unique predication pathways; for example:
|
Fig. 9.
The predications retrieved by reasoning pathway for rosiglitazone causing myocardial infarction with specifying semantic groups for concepts.
There were 2618 distinct predication pathways about “triglycerides”, “LDL lipoprotein” and “PPAR-gamma” specifying 247 unique middle terms.
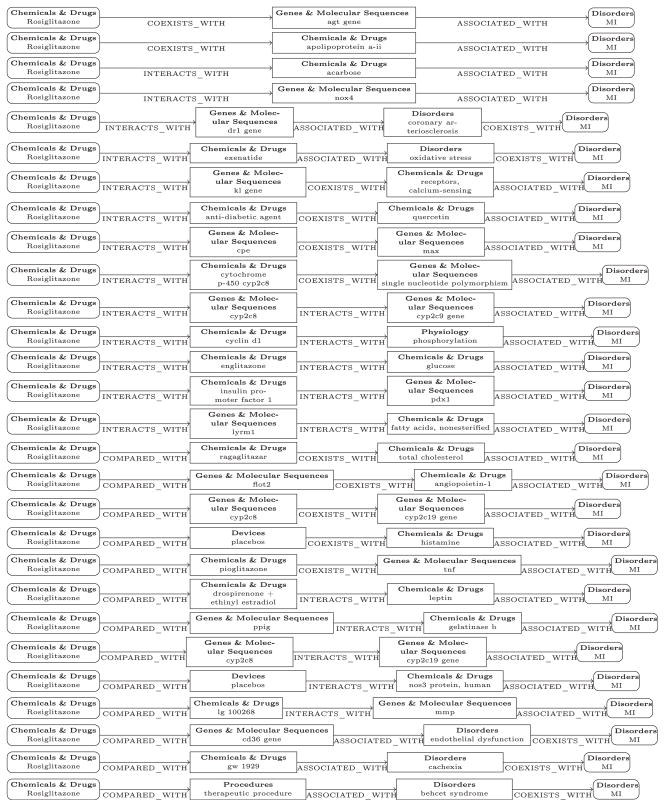
Drilling down, Fig. 10 shows the connecting concepts between LDL-C and myocardial infarction that fall along the reasoning pathways employed by the PSI-double + triple model. In each reasoning pathway, the middle terms were ranked using inverse document frequency, to approximate the weighting used by the predictive model. For each predication in these pathways, the source sentences from the literature were retrieved. For example, the article “A comparison of lipid and glycemic effects of pioglitazone and rosiglitazone in patients with type 2 diabetes and dyslipidemia” [143] explains that rosiglitazone increased triglycerides compared with pioglitazone and has different effect on plasma lipids which may contribute to heart disease. Fig. 10 shows the middle terms retrieved to justify that rosiglitazone may cause myocardial infarction via LDL-C.
Fig. 10.
Middle terms that were retrieved by PSI discovery patterns involving LDL-C.
The capacity to retrieve and organize knowledge in this way suggests a new paradigm for information retrieval in which information supporting a hypothesis of interest is automatically aggregated and organized at the conceptual level. However, as the number of assertions in the literature far exceeds the number of documents, further research is needed to develop methods through which to prioritize these assertions, and present them in a manner conducive to human consumption.
5. Discussion
This study evaluates the ability of scalable LBD methods based on distributional semantics to rank the plausibility of connections between drugs and potential ADRs. We find that both the RRI and PSI models are able to retrieve known side effects of drugs, but PSI performs this task better, as one would anticipate given the additional information beyond co-occurrence that it encodes. The PSI model can further provide the reasoning pathways that were used to link a drug to a predicted side effect. Consequently, relevant literature can be retrieved to support the predictions, and provided to experts for review. However further research is needed to develop approaches through which the assertions underlying the large numbers of reasoning pathways utilized by the model can be prioritized for expert review, as these are too numerous for exhaustive manual review. Ultimately, we aim to provide domain experts with essential evidence while preventing information over-load. Even though it is not the best performing model, the RRI model has the advantages of a simple training process and the availability of more data to draw upon (as MetaMap has higher recall for concepts than SemRep has for predications). Conversely, the PSI model has the advantage of modeling plausibility, a capability with the potential to assist expert clinical review for pharmacovigilance. In addition, the correlation analysis between groups suggests that RRI and PSI complement each other, and can potentially be combined to improve performance on this task.
For predicting ADRs, several statistical models and ML algorithms have been evaluated against an edition of SIDER, or a subset of this repository. In addition to methodological differences, these approaches have leveraged different data sets and a variety of knowledge bases as a basis for making predictions. In the section that follows, we will provide a review of these approaches, and the performance they have documented for the prediction of ADRs in SIDER.
Pauwel’s et al. represented drugs using as features the presence or absence of chemical substructure components described in PubChem [144]. In addition to standard supervised ML approaches, they applied canonical correlation analysis (CCA), including a sparse variant that emphasizes a small number of informative features for each training example. These methods were used to predict SIDER side effects, with a reported global AUC of 0.8932 [145] on a set of 1350 ADR and 888 drugs, using fivefold cross-validation.
Subsequently, Liu et al. applied five supervised ML algorithms to the same SIDER set. In addition to the PubChem-derived chemical substructure features used by Pauwel’s et al., features were drawn from DrugBank [146] (drug targets, transporters, and enzymes), KEGG [147] (pathway information) and SIDER itself (drug indications and side effects). A classifier was built for each SIDER ADR, and the classifiers were then evaluated on 832 SIDER drugs (for which DrugBank IDs could be found) using fivefold cross-validation. The support vector machine (SVM) algorithm performed best with a global AUC of 0.9524 on the full SIDER dataset [101]. The authors attribute much of the improvement in performance by this and other metrics to the effects of incorporating SIDER side effects as features, suggesting that certain side effects have a tendency to co-occur in drug label data.
Other authors have reported performance on subsets of SIDER using similar methods. For cardio-toxicity related ADRs in SIDER, a median AUC of 0.771 using SVM for prediction has been reported [148]. In this case, features were selected from information about intended drug targets in DrugBank, and information about off-target effects from an expanded protein–protein interaction network developed using gene ontology (GO) annotations. A SVM classifier was built for each evaluated ADR and cross-validated on SIDER.
With respect to performance, two of these studies, Pauwels et al. [145] and Liu et al. [101] report a global AUC of close to 0.9 or higher with the best of their methods. Though our results are not directly comparable as we made predictions on a per-drug rather than a per-ADR basis, the difference between the global AUC of these methods and that obtained with our approach seems large. However this difference in global AUC is misleading. As noted by Liu and colleagues in their paper, the imbalance between positive and negative examples across ADRs and the way in which the global AUC was calculated in this work leads to an apparent inconsistency between it and the other evaluation metrics presented. For example, Pauwels and his colleagues display the AUC across different ADRs in a series of box plots, which shows a median AUC for the best-performing method (by this metric) of slightly above 0.6. Acknowledging this issue, Liu et al. also report precision and recall for each evaluated method with, for example, precision of 0.66 and recall of 0.63 for SVMs with their maximal feature set. Notably, the AUC in this case was around 0.95.
This apparent inconsistency can be explained by the effect of the prevalence of positive examples for each ADR on the prediction strength. This is readily apparent for simple algorithms such as Naive Bayes, where the prior probability of a given category is incorporated into the estimate. However, it is also an issue for more sophisticated algorithms such as SVM [100] particularly when the imbalance between categories is severe. This is the case for many of the ADR examples: Liu et al. report a positive to negative ratio of around 1:166 for 554 of the 1135 ADRs. So given the same set of features, instances in these cases are likely to receive a lower prediction score than those in balanced cases. When these scores are aggregated across examples to generate a global AUC, ML methods that incorporate the category bias will obtain an inflated global AUC on account of this tendency to assign lower scores to instances with few positive examples. However, as noted by Liu et al. and demonstrated by the other reported metrics, this AUC is not an accurate reflection of the ability of these models to detect positive examples.
To simulate the effects of category bias on global AUC, we performed a simple experiment in which we multiplied the similarity scores produced by our model by the proportion of positive examples for each drug. This roughly approximates the effects of an accurately estimated prior class probability during cross-validation experiments. This resulted in an increase of our global AUC from 0.68 to 0.88. We do not present this result for the purpose of comparative evaluation, as our experiments are not directly comparable with prior ML work for other reasons we will subsequently discuss. Rather, we present it as an illustration of the disproportionate influence of category bias on global AUC, which underscores the issues with this evaluation metric raised by Liu et al. We trust it will also serve to dispel the misleading impression that the predictive accuracy of our methods is vastly inferior to that reported previously.
As our method does not consider the number of ADRs associated with a particular drug, the global AUC and median AUC approximately agree with one another. Our median AUC (across all drugs) of 0.7058, which falls somewhere in between that reported by Pauwels et al. [145] (across all ADRs) and Huang et al. [148] (across cardio-toxicity related ADRs only). On account of the difference in denominator these results are not directly comparable, but they do further illustrate the discrepancy between global and local AUC in models that are not agnostic to class imbalance. Arguably such agnosticism is desirable from the perspective of an expert review, as it is difficult to justify the assertion that those drugs with fewer known associated side effects should be considered less likely to cause some newly observed side effect (and vice versa).
With respect to methodological differences, all of the above methods are supervised ML methods, and were applied to infer whether or not drugs were associated with each ADR from the features of other drugs known to be associated with this ADR. So the predictive models were generally customized on a per-ADR basis, for example by generating an individual classifier for each ADR in the case of SVM. In contrast, our approach infers a set of abstract reasoning pathways that were consistent across the drugs we evaluated. However, as illustrated by the absence of evidence across certain pathways in the rosiglitazone example, some pathways may be more predictive for particular medications or ADRs. So it seems likely that we could further improve our performance by incorporating supervised ML, a direction we plan to explore in future work.
Our approach differs with respect to the knowledge sources utilized also. For example, KEGG and DrugBank are manually curated databases. Our knowledge base, SemMedDB, contains predications that have been automatically extracted by SemRep from the biomedical literature using NLP. Inaccuracies in language processing, or indeed in the literature itself may introduce sources of error that are not present in manually curated data. However, the scope of the literature is much broader than that of human-curated resources. Furthermore, as there is no agreed-upon gold standard for ADRs, different studies have utilized different datasets as reference sets [149]. Our study employed SIDER2, which includes considerably more drugs and ADRs than SIDER1.
This work has several limitations. The first of these concerns the use of SIDER2 as a reference standard. As SIDER2 consists of recognized side effects only, we cannot reliably distinguish between false positive signals and previously unknown ADRs. Furthermore, SIDER was compiled from package insert information by NLP tools [79], and as such may include side effects that seldom occur in practice or false associations that were caused by text-mining errors [150]. While SIDER2 is sufficient to evaluate the hypotheses of the current work, in future work we plan to incorporate other data sources, such as EHR data and FDA reports. These data sources may provide additional evidence to support the assertion that an unknown drug/ADR pair is worth investigating further. Alternatively, they may provide the means to select a subset of the side effects in SIDER2 that have been observed frequently in practice as an additional evaluation set.
Secondly, the MMB repository contains one year less literature than the SemMedDB dataset. There is a difference of 1,7579,64 citations (7.9% of SemMedDB dataset). These were the newest datasets at the time of the experiment. However the MMB repository has many more data points than SemMedDB. For example, more than 99.99% of citations have concepts extracted by MetaMap and 59.91% of citations have predications extracted by SemRep.
Another concern is the existing knowledge about causal relationships between drugs and related ADRs from the literature. For our dataset (90,787 pairs), 45% of pairs (concerning 953 drugs) co-occur directly in the MMB repository and 5% of pairs (concerning 693 drugs) have direct causal relationship (drug CAUSES ADR) in SemMedDB. So PSI’s accuracy is dependent upon its ability to meaningfully infer connections between concepts that were not previously linked in its database, a capacity that would be particularly useful as a means of assessing novel ADRs that had not previously been documented in the literature. RRI is also able to draw such inferences, but in this case more of its performance may be attributable to direct co-occurrence.
Inspecting the middle terms that our model retrieved for rosiglitazone- MI association (Fig. 10), we found that at times uninformative high-level concepts, such as “genes” and “proteins”, were retrieved. In our study, we addressed the issue of uninformative high level concepts in two ways, both related to their propensity to occur relatively frequently in the corpus. Firstly we used a frequency threshold of 1,000,000 to exclude frequently occurring concepts contained in SemMedDB. The frequency of “genes” and “proteins” is less than the threshold and cannot be filtered. Secondly, we used a weighting procedure to reduce the influence of high-frequency terms on the training process. However, more sophisticated approaches to filtering are possible. Information concerning UMLS semantic types and position in the UMLS hierarchy could be used to develop more sophisticated approaches, to further filter out uninformative high-level concepts, which may improve performance.
The predictions made by PSI depend upon assertions extracted from the biomedical literature. One concern about the extracted predications is that they may be implausible on account of NLP errors. Though SemRep has been optimized for precision, its precision is not perfect. For example, Kilicoglu et al. estimate the precision of SemRep to be around 0.77 [151]. Based on this, and other published evaluations [152,69], it is reasonable to estimate that around three in four predications in the set are perfectly accurate. In many cases, inaccurate predications nonetheless indicate cooccurrence, which is also informative. The PSI-based analogical reasoning approach we have employed is robust to isolated language processing errors, as highly ranked predictions are based on assertions extracted from thousands of unique reasoning pathways. For example, for the rosiglitazone-MI association, 108,100 unique predications were retrieved, spanning eight of the inferred reasoning pathways. On average, individual predications were supported by 17 excerpts from the literature. If we extrapolate from prior published evaluations of SemRep, the predication concerned would have been accurately extracted from around 12 of these excerpts. So it is likely that at least some of the evidence supporting each individual assertion is accurate. Moreover, as this method is distributional in nature, it does not require that these assertions be perfectly accurate. Rather, the frequency with which an assertion is extracted factors into the strength of its contribution to a reasoning pathway. Nonetheless, the biomedical literature may contain controversial assertions, or contradictory conclusions from different experts or different experiments. This is illustrated by the rosiglitazone (brand name: Avandia) case. In 2007, the FDA added a black-box warning for heart-related risks to Avandia based on a meta-analysis [106] and three other studies [153]. In 2013, the FDA lifted certain Avandia prescribing restrictions based on the readjudicated results of the RECORD trial [154,155], claiming the initial concerns were overblown [108]. This decision was condemned by one of the authors of the original meta-analysis [156]. Currently our models weight the contribution of assertions using statistics related to local and global frequency. However, it would also be possible to weight the importance of these assertions based on some assessment of the reliability of the source. For example, in information retrieval experiments, an approach incorporating citation information was better able to identify articles considered as important in a preexisting bibliography [157]. Possibilities include weights derived from the citation count of the source article, the impact factor of the journal, or the nature of the experiment described. It is possible that weighting metrics of this source would improve the predictions of our models, and they also suggest approaches to prioritize the large numbers of assertions supporting our predictions for review by human experts.
6. Conclusion
In this research, an emerging, scalable method of LBD that uses distributional statistics to infer and apply discovery patterns was adapted to evaluate the plausibility of drug/ADR relationships for the purpose of pharmacovigilance. The effective application of large amounts of partially accurate biomedical knowledge to this problem was facilitated by the scalable and robust nature of approximate inference in geometric space. This approach was shown to be more effective than a comparable co-occurrence based baseline, and has the further benefit of permitting the retrieval of evidence underlying the assertions used by the system to make its predictions. Consequently, our approach provides the means to assist with expert clinical review by providing evidence supporting the plausibility of the connection between drugs and ADRs. Furthermore, the models we have developed can be applied to filter drug/ADR signals that are detected in spontaneous reporting systems or EHR data, a direction we plan to explore in future work.
Acknowledgments
The work was supported by the U.S. National Library of Medicine Grant (1R01LM011563), Using Biomedical Knowledge to Identify Plausible Signals for Pharmacovigilance. This work was also supported in part by the Intramural Research Program of the U.S. National Institutes of Health, National Library of Medicine. The authors would like to thank Peter Davies for his comments on the paper, and for discussions regarding the implications of these methods for the evaluation of potential ADRs.
References
- 1.Edwards IR, Aronson JK. Adverse drug reactions: definitions, diagnosis, and management. Lancet. 2000;356(9237):1255–9. doi: 10.1016/S0140-6736(00)02799-9. [DOI] [PubMed] [Google Scholar]
- 2.Lazarou J, Pomeranz B, Corey P. Incidence of adverse drug reactions in hospitalized patients. JAMA: J Amer Med Assoc. 1998;279(15):1200–5. doi: 10.1001/jama.279.15.1200. [DOI] [PubMed] [Google Scholar]
- 3.Hamilton RA, Briceland LL, Andritz MH. Frequency of hospitalization after exposure to known drug–drug interactions in a medicaid population. Pharmacother: J Human Pharmacol Drug Ther. 1998;18(5):1112–20. [PubMed] [Google Scholar]
- 4.Baker GR, Norton PG, Flintoft V, Blais R, Brown A, Cox J, et al. The Canadian adverse events study: the incidence of adverse events among hospital patients in Canada. Can Med Assoc J. 2004;170(11):1678–86. doi: 10.1503/cmaj.1040498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bourgeois FT, Mandl KD, Valim C, Shannon MW. Pediatric adverse drug events in the outpatient setting: an 11-year national analysis. Pediatrics. 2009;124(4):e744–50. doi: 10.1542/peds.2008-3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DiMasi JA, Hansen RW, Grabowski HG. The price of innovation: new estimates of drug development costs. J Health Econ. 2003;22(2):151–85. doi: 10.1016/S0167-6296(02)00126-1. [DOI] [PubMed] [Google Scholar]
- 7.Rawlins MD. Spontaneous reporting of adverse drug reactions. I: The data. Brit J Clin Pharmacol. 1988;26(1):1–5. doi: 10.1111/j.1365-2125.1988.tb03356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rawlins MD. Spontaneous reporting of adverse drug reactions. II:, Uses. Brit J Clin Pharmacol. 1988;26(1):7–11. doi: 10.1111/j.1365-2125.1988.tb03357.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van Puijenbroek EP, Diemont WL, van Grootheest K. Application of quantitative signal detection in the Dutch spontaneous reporting system for adverse drug reactions. Drug Saf. 2003;26(5):293–301. doi: 10.2165/00002018-200326050-00001. [DOI] [PubMed] [Google Scholar]
- 10.Hasford J, Goettler M, Munter K-H, Müller-Oerlinghausen B. Physicians’ knowledge and attitudes regarding the spontaneous reporting system for adverse drug reactions. J Clin Epidemiol. 2002;55(9):945–50. doi: 10.1016/s0895-4356(02)00450-x. [DOI] [PubMed] [Google Scholar]
- 11.Alvarez-Requejo A, Carvajal A, Begaud B, Moride Y, Vega T, Arias LM. Under-reporting of adverse drug reactions estimate based on a spontaneous reporting scheme and a sentinel system. Europ J Clin Pharmacol. 1998;54(6):483–8. doi: 10.1007/s002280050498. [DOI] [PubMed] [Google Scholar]
- 12.Wang X, Hripcsak G, Markatou M, Friedman C. Active computerized pharmacovigilance using natural language processing, statistics, and electronic health records: a feasibility study. J Amer Med Info Assoc. 2009;16(3):328–37. doi: 10.1197/jamia.M3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Trifirò G, Pariente A, Coloma PM, Kors JA, Polimeni G, Miremont-Salamé G, et al. Data mining on electronic health record databases for signal detection in pharmacovigilance: which events to monitor? Pharmacoepidemiol Drug Saf. 2009;18(12):1176–84. doi: 10.1002/pds.1836. [DOI] [PubMed] [Google Scholar]
- 14.Haerian K, Varn D, Vaidya S, Ena L, Chase HS, Friedman C. Detection of pharmacovigilance-related adverse events using electronic health records and automated methods. Clin Pharmacol Ther. 2012;92(2):228–34. doi: 10.1038/clpt.2012.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harpaz R, Vilar S, DuMouchel W, Salmasian H, Haerian K, Shah NH, et al. Combing signals from spontaneous reports and electronic health records for detection of adverse drug reactions. J Amer Med Info Assoc. 2013;20(3):413–9. doi: 10.1136/amiajnl-2012-000930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Friedman C. Discovering novel adverse drug events using natural language processing and mining of the electronic health record. In: Combi C, Shahar Y, Abu-Hanna A, editors. Artificial intelligence in medicine. Lecture notes in computer science. Vol. 5651. Berlin Heidelberg: Springer; 2009. pp. 1–5. [Google Scholar]
- 17.Oliveira JL, Lopes P, Nunes T, Campos D, Boyer S, Ahlberg E, et al. The EU-ADR web platform: delivering advanced pharmacovigilance tools. Pharmacoepidemiol Drug Saf. 2013;22(5):459–67. doi: 10.1002/pds.3375. [DOI] [PubMed] [Google Scholar]
- 18.Deftereos SN, Andronis C, Friedla EJ, Persidis A, Persidis A. Drug repurposing and adverse event prediction using high-throughput literature analysis. Wiley Interdisciplinary Rev: Systems Biol Med. 2011;3(3):323–34. doi: 10.1002/wsbm.147. [DOI] [PubMed] [Google Scholar]
- 19.Bauer-Mehren A, van Mullingen EM, Avillach P, del Carmen Carrascosa M, Garcia-Serna R, Piñero J, et al. Automatic filtering and substantiation of drug safety signals. PLoS Comput Biol. 2012;8(4):e1002457. doi: 10.1371/journal.pcbi.1002457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shetty KD, Dalal SR. Using information mining of the medical literature to improve drug safety. J Amer Med Info Assoc. 2011;18(5):668–74. doi: 10.1136/amiajnl-2011-000096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Merck. Merck announces voluntary worldwide withdrawal of VIOXX. 2004 < http://www.merck.com/newsroom/vioxx/pdf/vioxx_press_release_final.pdf>.
- EMA. European medicines agency recommends suspension of Avandia, Avandamet and Avaglim. 2010. Press release. [Google Scholar]
- 23.Ye J-p. Challenges in drug discovery for thiazolidinedione substitute. Acta Pharmaceut Sinica B. 2011;1(3):137–42. doi: 10.1016/j.apsb.2011.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blind E, Dunder K, De Graeff P, Abadie E. Rosiglitazone: a European regulatory perspective. Diabetologia. 2010;54(2):213–8. doi: 10.1007/s00125-010-1992-5. [DOI] [PubMed] [Google Scholar]
- 25.Anderson N, Borlak J. Correlation versus Causation? Pharmacovigilance of the analgesic flupirtine exemplifies the need for refined spontaneous ADR reporting. PLoS ONE. 2011;6(10):e25221. doi: 10.1371/journal.pone.0025221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.WHO. Pharmacovigilance. 2010 < http://www.who.int/medicines/areas/quality_safety/safety_efficacy/pharmvigi/en/index.html>.
- 27.Ahmad SR. Adverse drug event monitoring at the food and drug administration. J Gener Intern Med. 2003;18(1):57–60. doi: 10.1046/j.1525-1497.2003.20130.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kessler DA, Natanblut S, Kennedy D, Lazar E, Rheinstein P, Anello C, et al. Introducing MEDWatch: a new approach to reporting medication and device adverse effects and product problems. JAMA: J Amer Med Assoc. 1993;269(21):2765–8. doi: 10.1001/jama.269.21.2765. [DOI] [PubMed] [Google Scholar]
- 29.Wysowski DK, Swartz L. Adverse drug event surveillance and drug withdrawals in the United States, 1969–2002: the importance of reporting suspected reactions. Arch Intern Med. 2005;165(12):1363. doi: 10.1001/archinte.165.12.1363. [DOI] [PubMed] [Google Scholar]
- 30.Edwards IR. The WHO Database – I. Drug Info J. 1992;26(4):477–80. [Google Scholar]
- 31.Bate A. Dissertation. Umeå University; 2003. The use of Bayesian confidence propagation neural network in pharmacovigilance. [Google Scholar]
- 32.Edwards IR, Biriell C. Harmonisation in pharmacovigilance. Drug Saf. 1994;10(2):93–102. doi: 10.2165/00002018-199410020-00001. [DOI] [PubMed] [Google Scholar]
- 33.Collins FS. Reengineering translational science: the time is right. Sci Transl Med. 2011;3(90):90cm17. doi: 10.1126/scitranslmed.3002747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bate A, Lindquist M, Edwards I, Olsson S, Orre R, Lansner A, et al. A Bayesian neural network method for adverse drug reaction signal generation. Europ J Clin Pharmacol. 1998;54(4):315–21. doi: 10.1007/s002280050466. [DOI] [PubMed] [Google Scholar]
- 35.Lindquist M, Edwards IR, Bate A, Fucik H, Nunes AM, Ståhl M. From association to alert – a revised approach to international signal analysis. Pharmacoepidemiol Drug Saf. 1999;8(Suppl 1):S15–25. doi: 10.1002/(sici)1099-1557(199904)8:1+<s15::aid-pds402>3.3.co;2-2. [DOI] [PubMed] [Google Scholar]
- 36.Noren GN, Bate A, Orre R, Edwards IR. Extending the methods used to screen the WHO drug safety database towards analysis of complex associations and improved accuracy for rare events. Statist Med. 2006;25(21):3740–57. doi: 10.1002/sim.2473. [DOI] [PubMed] [Google Scholar]
- 37.Lindquist M, Ståhl M, Bate A, Edwards IR, Meyboom RH. A retrospective evaluation of a data mining approach to aid finding new adverse drug reaction signals in the WHO international database. Drug Saf. 2000;23(6):533–42. doi: 10.2165/00002018-200023060-00004. [DOI] [PubMed] [Google Scholar]
- 38.DuMouchel W. Bayesian data mining in large frequency tables, with an application to the FDA spontaneous reporting system. Amer Statist. 1999;53(3):177–90. [Google Scholar]
- 39.DuMouchel W, Pregibon D. Empirical Bayes screening for multi-item associations. Proceedings of the seventh ACM SIGKDD international conference on knowledge discovery and data mining; ACM; 2001. pp. 67–76. [Google Scholar]
- 40.Shakir SA, Layton D. Causal association in pharmacovigilance and pharmacoepidemiology: thoughts on the application of the Austin Bradford-Hill criteria. Drug Saf. 2002;25(6):467–71. doi: 10.2165/00002018-200225060-00012. [DOI] [PubMed] [Google Scholar]
- 41.Hill AB. The environment and disease: association or causation? Proceed Roy Society Med. 1965;58(5):295. [PMC free article] [PubMed] [Google Scholar]
- 42.Doll R. Austin Bradford Hill 8 July 1897–18 April 1991. Biogr Mem Fellows Roy Society. 1994;40:128–40. [Google Scholar]
- 43.Ward AC. The role of causal criteria in causal inferences: Bradford Hill’s “aspects of association”. Epidemiol Perspect Innov. 2009;6(1):2. doi: 10.1186/1742-5573-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kleinberg S, Hripcsak G. A review of causal inference for biomedical informatics. J Biomed Info. 2011;44(6):1102–12. doi: 10.1016/j.jbi.2011.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perrio M, Voss S, Shakir SA. Application of the Bradford Hill criteria to assess the causality of cisapride-induced arrhythmia: a model for assessing causal association in pharmacovigilance. Drug Saf. 2007;30(4):333–46. doi: 10.2165/00002018-200730040-00006. [DOI] [PubMed] [Google Scholar]
- 46.Swanson DR. Undiscovered public knowledge. Libr Quart. 1986:103–18. doi: 10.1353/pbm.1986.0087. [DOI] [PubMed] [Google Scholar]
- 47.Swanson DR. Two medical literatures that are logically but not bibliographically connected. J Amer Society Info Sci. 1987;38(4):228–33. [Google Scholar]
- 48.Weeber M, Klein H, de Jong-van den Berg L, Vos R. Using concepts in literature-based discovery: simulating Swanson’s Raynaud-fish oil and migraine–magnesium discoveries. J Amer Society Info Sci Tech. 2001;52(7):548–57. [Google Scholar]
- 49.Bruza PD, Weeber M. Literature-based discovery. Springer; 2008. [Google Scholar]
- 50.Swanson DR. Fish oil, Raynaud’s syndrome, and undiscovered public knowledge. Perspect Biol Med. 1986;30(1):7. doi: 10.1353/pbm.1986.0087. [DOI] [PubMed] [Google Scholar]
- 51.Swanson DR. Migraine and magnesium: eleven neglected connections. Perspect Biol Med. 1988;31(4):526–57. doi: 10.1353/pbm.1988.0009. [DOI] [PubMed] [Google Scholar]
- 52.Smalheiser NR, Swanson DR. Linking estrogen to Alzheimer’s disease: an informatics approach. Neurology. 1996;47(3):809–10. doi: 10.1212/wnl.47.3.809. [DOI] [PubMed] [Google Scholar]
- 53.Srinivasan P, Rindflesch TC. Exploring text mining from MEDLINE. Proceedings of the AMIA symposium; 2002. pp. 722–6. [PMC free article] [PubMed] [Google Scholar]
- 54.Hristovski D, Friedman C, Rindflesch TC, Peterlin B. Exploiting semantic relations for literature-based discovery. AMIA annual symposium proceeding; 2006. pp. 349–53. [PMC free article] [PubMed] [Google Scholar]
- 55.Yetisgen-Yildiz M, Pratt W. Using statistical and knowledge-based approaches for literature-based discovery. J Biomed Info. 2006;39(6):600–11. doi: 10.1016/j.jbi.2005.11.010. [DOI] [PubMed] [Google Scholar]
- 56.Sehgal AK, Qiu XY, Srinivasan P. Literature-based discovery. Springer; 2008. Analyzing LBD methods using a general framework; pp. 75–100. [Google Scholar]
- 57.Yetisgen-Yildiz M, Pratt W. A new evaluation methodology for literature-based discovery systems. J Biomed Info. 2009;42(4):633–43. doi: 10.1016/j.jbi.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 58.Rindflesch TC, Fiszman M. The interaction of domain knowledge and linguistic structure in natural language processing: interpreting hypernymic propositions in biomedical text. J Biomed Info. 2003;36(6):462–77. doi: 10.1016/j.jbi.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 59.Chen L, Friedman C. Extracting phenotypic information from the literature via natural language processing. Stud Health Tech Info. 2004;107:758–62. [PubMed] [Google Scholar]
- 60.Hristovski D, Friedman C, Rindflesch TC, Peterlin B. Literature-based knowledge discovery using natural language processing. In: Bruza P, Weeber M, editors. Literature-based discovery, no. 15 in information science and knowledge management. Berlin Heidelberg: Springer; 2008. pp. 133–52. [Google Scholar]
- 61.Ahlers CB, Hristovski D, Kilicoglu H, Rindflesch TC. Using the literature-based discovery paradigm to investigate drug mechanisms. AMIA, Annual Symposium Proceedings; 2007. pp. 6–10. [PMC free article] [PubMed] [Google Scholar]
- 62.Hristovski D, Burgun-Parenthoine A, Avillach P, Rindflesch TC. Quality of life through quality of information, MIE2012. 2012a. Towards using literature-based discovery to explain drug adverse effects. [Google Scholar]
- 63.Hristovski D, Burgun-Parenthoine A, Avillach P, Rindflesch TC. Using literature-based discovery to explain drug adverse effects. AMIA, annual symposium proceedings; 2012b. [Google Scholar]
- 64.Aronson AR. Effective mapping of biomedical text to the UMLS metathesaurus: the MetaMap program. Proceedings of the AMIA symposium; 2001. pp. 17–21. [PMC free article] [PubMed] [Google Scholar]
- 65.Aronson AR, Lang F-M. An overview of MetaMap: historical perspective and recent advances. J Amer Med Info Assoc. 2010;17(3):229–36. doi: 10.1136/jamia.2009.002733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rindflesch TC, Fiszman M, Libbus B. Semantic interpretation for the biomedical research literature. In: Chen H, Fuller SS, Friedman C, Hersh W, editors. Medical informatics, no. 8 in integrated series in information systems. US: Springer; 2005. pp. 399–422. [Google Scholar]
- 67.Aronson AR, Rindflesch TC. A report to the Board of Scientific Counselors. 1998. Semantic knowledge representation project. [Google Scholar]
- 68.Rindflesch TC, Aronson AR. Semantic processing for enhanced access to biomedical knowledge. Real World Semantic Web Appl. 2002:157–72. [Google Scholar]
- 69.Kilicoglu H, Fiszman M, Rosemblat G, Marimpietri S, Rindflesch TC. Arguments of nominals in semantic interpretation of biomedical text. Proceedings of the 2010 workshop on biomedical natural language processing; 2010. pp. 46–54. [Google Scholar]
- 70.Lindsay RK, Gordon MD. Literature-based discovery by lexical statistics. J Amer Society Info Sci. 1999;50(7):574–87. [Google Scholar]
- 71.Cohen T, Widdows D. Empirical distributional semantics: methods and biomedical applications. J Biomed Info. 2009;42(2):390–405. doi: 10.1016/j.jbi.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cole RJ, Bruza PD. A bare bones approach to literature-based discovery: an analysis of the Raynaud’s/fish-oil and migraine–magnesium discoveries in semantic space. In: Hoffmann A, Motoda H, Scheffer T, editors. Discovery science. Lecture notes in computer science. Vol. 3735. Berlin/ Heidelberg: Springer; 2005. pp. 84–98. [Google Scholar]
- 73.Gordon MD, Dumais S. Using latent semantic indexing for literature based discovery. J Amer Society Info Sci. 1998;49(8):674–85. [Google Scholar]
- 74.Cohen T, Schvaneveldt R, Widdows D. Reflective random indexing and indirect inference: a scalable method for discovery of implicit connections. J Biomed Info. 2010;43(2):240–56. doi: 10.1016/j.jbi.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 75.Kanerva P, Kristofersson J, Holst A. Random indexing of text samples for latent semantic analysis. Proceedings of the 22nd annual conference of the cognitive science society. 2000;1036:1036–7. [Google Scholar]
- 76.Kanerva P. What we mean when we say “What’s, the dollar of mexico?: prototypes and mapping in concept space”. Proc AAAI, fall symp on quantum informatics for cognitive, social, and semantic processes; 2010. [Google Scholar]
- 77.Cohen T, Widdows D, Schvaneveldt RW, Davies P, Rindflesch TC. Discovering discovery patterns with predication-based semantic indexing. J Biomed Info. 2012;45(6):1049–65. doi: 10.1016/j.jbi.2012.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Landauer TK, Dumais ST. A solution to Plato’s problem: the latent semantic analysis theory of acquisition, induction, and representation of knowledge. Psych Rev. 1997;104(2):211. [Google Scholar]
- 79.Kuhn M, Campillos M, Letunic I, Jensen LJ, Bork P. A side effect resource to capture phenotypic effects of drugs. Mol Systems Biol. 2010;6:343. doi: 10.1038/msb.2009.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.U.S. National Library of Medicine, National Institutes of Health. UMLS Terminology Services (UTS) API 2.0. 2012 < http://uts.nlm.nih.gov/metathesaurus.html>.
- 81.Wei W-Q, Cronin RM, Xu H, Lasko TA, Bastarache L, Denny JC. Development and evaluation of an ensemble resource linking medications to their indications. J Amer Med Info Assoc. 2013;20(5):954–61. doi: 10.1136/amiajnl-2012-001431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kanerva P. Hyperdimensional computing: an introduction to computing in distributed representation with high-dimensional random vectors. Cognitive Computat. 2009;1(2):139–59. [Google Scholar]
- 83.Widdows D, Cohen T. Real, complex, and binary semantic vectors. Proc sixth intl symp on quantum interactions; Paris, France: Springer; 2012. pp. 24–35. [Google Scholar]
- 84.Kanerva P. ICANN’94. Springer; 1994. The spatter code for encoding concepts at many levels; pp. 226–9. [Google Scholar]
- 85.Plate TA. Holographic reduced representations. IEEE Trans Neural Networks. 1995;6(3):623–41. doi: 10.1109/72.377968. [DOI] [PubMed] [Google Scholar]
- 86.Kanerva P. Artificial neural networks – ICANN 96. Springer; 1996. Binary spatter-coding of ordered K-tuples; pp. 869–73. [Google Scholar]
- 87.Gayler RW. Vector symbolic architectures answer Jackendoff’s challenges for cognitive neuroscience. arXiv preprint cs/0412059. [Google Scholar]
- 88.Wahle M, Widdows D, Herskovic JR, Bernstam EV, Cohen T. Deterministic binary vectors for efficient automated indexing of MEDLINE/PubMed abstracts. AMIA annual symposium proceedings; American Medical Informatics Association; 2012. p. 940. [PMC free article] [PubMed] [Google Scholar]
- 89.Dumais ST. Improving the retrieval of information from external sources. Behav Res Methods Instruments Computer. 1991;23(2):229–36. [Google Scholar]
- 90.Cohen T, Widdows D, Schvaneveldt RW, Rindflesch TC. Discovery at a distance: farther journeys in predication space. Bioinformatics and biomedicine workshops (BIBMW), 2012 IEEE international conference on; 2012b. pp. 218–25. [Google Scholar]
- 91.Birkhoff G, Von Neumann J. The logic of quantum mechanics. Ann Math. 1936:823–43. [Google Scholar]
- 92.Widdows D, Peters S. Word vectors and quantum logic: experiments with negation and disjunction. Math Lang. 2003;8(141) [Google Scholar]
- 93.Cohen T, Widdows D, De Vine L, Schvaneveldt R, Rindflesch TC. Quantum interaction. Springer; 2012. Many paths lead to discovery: analogical retrieval of cancer therapies; pp. 90–101. [Google Scholar]
- 94.Golub GH, Van Loan CF. Matrix computations. Vol. 3. JHU Press; 2012. [Google Scholar]
- 95.National Center for Biotechnology Information, U.S. National Library of Medicine 8600 Rockville Pike, Bethesda MD, 20894 Usa. Metathesaurus – Rich Release Format (RRF) 2009 < http://www.ncbi.nlm.nih.gov/books/NBK9685/>.
- 96.U.S. National Library of Medicine, National Institutes of Health. RxNorm API. 2013 < http://rxnav.nlm.nih.gov/RxNormAPI.html>.
- 97.Salton G, Fox EA, Wu H. Extended Boolean information retrieval. Communications ACM. 1983;26(11):1022–36. [Google Scholar]
- 98.Manning CD, Raghavan P, Schütze H. Introduction to information retrieval. Vol. 1. Cambridge: Cambridge University Press; 2008. [Google Scholar]
- 99.Davis J, Goadrich M. The relationship between precision–recall and ROC curves. Proceedings of the 23rd international conference on Machine learning; 2006. pp. 233–40. [Google Scholar]
- 100.Akbani R, Kwek S, Japkowicz N. Machine learning: ECML 2004. Springer; 2004. Applying support vector machines to imbalanced datasets; pp. 39–50. [Google Scholar]
- 101.Liu M, Wu Y, Chen Y, Sun J, Zhao Z, Chen X-w, et al. Large-scale prediction of adverse drug reactions using chemical, biological, and phenotypic properties of drugs. J Amer Med Info Assoc. 2012;19(e1):e28–35. doi: 10.1136/amiajnl-2011-000699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hamblin M, Chang L, Fan Y, Zhang J, Chen YE. PPARs and the cardiovascular system. Antioxidants Redox Signaling. 2009;11(6):1415–52. doi: 10.1089/ars.2008.2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cheung BM. Behind the rosiglitazone controversy. Exper Rev Clin Pharmacol. 2010;3(6):723–5. doi: 10.1586/ecp.10.126. [DOI] [PubMed] [Google Scholar]
- 104.Lygate CA, Hulbert K, Monfared M, Cole MA, Clarke K, Neubauer S. The PPARγ-activator rosiglitazone does not alter remodeling but increases mortality in rats post-myocardial infarction. Cardiov Res. 2003;58(3):632–7. doi: 10.1016/s0008-6363(03)00289-x. [DOI] [PubMed] [Google Scholar]
- 105.Shah ND, Montori VM, Krumholz HM, Tu K, Alexander GC, Jackevicius CA, et al. Responding to an FDA, warning – geographic variation in the use of rosiglitazone. New England J Med. 2010;363(22):2081–4. doi: 10.1056/NEJMp1011042. [DOI] [PubMed] [Google Scholar]
- 106.Nissen SE, Wolski K. Effect of rosiglitazone on the risk of myocardial infarction and death from cardiovascular causes. New England J Med. 2007;356:2457–71. doi: 10.1056/NEJMoa072761. [DOI] [PubMed] [Google Scholar]
- 107.Berthet S, Olivier P, Montastruc J-L, Lapeyre-Mestre M. Drug safety of rosiglitazone and pioglitazone in France: a study using the French PharmacoVigilance database. BMC Pharmacol Toxicol. 2011;11(1):5. doi: 10.1186/1472-6904-11-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.U.S. Food and Drug Administration. Press announcements – FDA, requires removal of certain restrictions on the diabetes drug Avandia. 2013 < http://www.fda.gov/NewsEvents/Newsroom/PressAnnouncements/ucm376516.htm>.
- 109.Loke YK, Kwok CS, Singh S. Comparative cardiovascular effects of thiazolidinediones: systematic review and meta-analysis of observational studies. BMJ: Brit Med J. 2011;342(7):d1309. doi: 10.1136/bmj.d1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Goldberg RB, Kendall DM, Deeg MA, Buse JB, Zagar AJ, Pinaire JA, et al. A comparison of lipid and glycemic effects of pioglitazone and rosiglitazone in patients with type 2 diabetes and dyslipidemia. Diabetes Care. 2005;28(7):1547–54. doi: 10.2337/diacare.28.7.1547. [DOI] [PubMed] [Google Scholar]
- 111.Khanderia U, Pop-Busui R, Eagle KA. Thiazolidinediones in type 2 diabetes: a cardiology perspective. Ann Pharmacother. 2008;42(10):1466–74. doi: 10.1345/aph.1K666. [DOI] [PubMed] [Google Scholar]
- 112.Bourg CA, Phillips BB. Rosiglitazone, myocardial ischemic risk, and recent regulatory actions. Ann Pharmacother. 2012;46(2):282–9. doi: 10.1345/aph.1Q400. [DOI] [PubMed] [Google Scholar]
- 113.Bodenreider O, McCray AT. Exploring semantic groups through visual approaches. J Biomed Info. 2003;36(6):414–32. doi: 10.1016/j.jbi.2003.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Brackenridge AL, Jackson N, Jefferson W, Stolinski M, Shojaee-Moradie F, Hovorka R, et al. Effects of rosiglitazone and pioglitazone on lipoprotein metabolism in patients with Type 2 diabetes and normal lipids. Diabetic Med: J Brit Diabetic Assoc. 2009;26(5):532–9. doi: 10.1111/j.1464-5491.2009.02729.x. [DOI] [PubMed] [Google Scholar]
- 115.Sarafidis PA, Lasaridis AN, Nilsson PM, Mouslech TF, Hitoglou-Makedou AD, Stafylas PC, et al. The effect of rosiglitazone on novel atherosclerotic risk factors in patients with type 2 diabetes mellitus and hypertension. An open-label observational study. Meta: Clin Exp. 2005;54(9):1236–42. doi: 10.1016/j.metabol.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 116.Vessby B, Kostner G, Lithell H, Thomis J. Diverging effects of cholestyramine on apolipoprotein B and lipoprotein Lp(a). A dose-response study of the effects of cholestyramine in hypercholesterolaemia. Atherosclerosis. 1982;44(1):61–71. doi: 10.1016/0021-9150(82)90053-3. [DOI] [PubMed] [Google Scholar]
- 117.van Wijk J, Coll B, Cabezas MC, Koning E, Camps J, Mackness B, et al. Rosiglitazone modulates fasting and post-prandial paraoxonase 1 activity in type 2 diabetic patients. Clin Exper Pharmacol Physiol. 2006;33(12):1134–7. doi: 10.1111/j.1440-1681.2006.04505.x. [DOI] [PubMed] [Google Scholar]
- 118.Gupta N, Singh S, Maturu VN, Sharma YP, Gill KD. Paraoxonase 1 (PON1), polymorphisms, haplotypes and activity in predicting CAD, risk in North-West Indian Punjabis. PLoS One. 2011;6(5):e17805. doi: 10.1371/journal.pone.0017805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yoshida T, Kato K, Yokoi K, Oguri M, Watanabe S, Metoki N, et al. Association of genetic variants with myocardial infarction in Japanese individuals with different lipid profiles. Int J Mol Med. 2010;25(4):607–16. doi: 10.3892/ijmm_00000383. [DOI] [PubMed] [Google Scholar]
- 120.Phillips GB. Relationship between serum sex hormones and glucose, insulin and lipid abnormalities in men with myocardial infarction. Proceed Natl Acad Sci. 1977;74(4):1729–33. doi: 10.1073/pnas.74.4.1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kuller L, Arnold A, Tracy R, Otvos J, Burke G, Psaty B, et al. Nuclear magnetic resonance spectroscopy of lipoproteins and risk of coronary heart disease in the cardiovascular health study. Arterioscler Throm Vas Biol. 2002;22(7):1175–80. doi: 10.1161/01.atv.0000022015.97341.3a. [DOI] [PubMed] [Google Scholar]
- 122.Friis-Moller N, Sabin CA, Weber R, d’Arminio Monforte A, El-Sadr WM, Reiss P, et al. Combination antiretroviral therapy and the risk of myocardial infarction. New England J Med. 2003;349(21):1993–2003. doi: 10.1056/NEJMoa030218. [DOI] [PubMed] [Google Scholar]
- 123.Lekhal S, Børvik T, Brodin E, Nordøy A, Hansen J-B. Tissue factor-induced thrombin generation in the fasting and postprandial state among elderly survivors of myocardial infarction. Thromb Res. 2010;126(4):353–9. doi: 10.1016/j.thromres.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 124.De Caterina R, Talmud PJ, Merlini PA, Foco L, Pastorino R, Altshuler D, et al. Strong association of the APOA5-1131T>C gene variant and early-onset acute myocardial infarction. Atherosclerosis. 2011;214(2):397–403. doi: 10.1016/j.atherosclerosis.2010.11.011. [DOI] [PubMed] [Google Scholar]
- 125.Chao L, Marcus-Samuels B, Mason MM, Moitra J, Vinson C, Arioglu E, et al. Adipose tissue is required for the antidiabetic, but not for the hypolipidemic, effect of thiazolidinediones. J Clin Invest. 2000;106(10):1221–8. doi: 10.1172/JCI11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Nadeau KJ, Ehlers LB, Aguirre LE, Reusch JEB, Draznin B. Discordance between intramuscular triglyceride and insulin sensitivity in skeletal muscle of Zucker diabetic rats after treatment with fenofibrate and rosiglitazone. Diabetes Obe Metab. 2007;9(5):714–23. doi: 10.1111/j.1463-1326.2006.00696.x. [DOI] [PubMed] [Google Scholar]
- 127.Suzuki S, Suzuki M, Sembon S, Fuchimoto D, Onishi A. Characterization of actions of octanoate on porcine preadipocytes and adipocytes differentiated in vitro. Biochem Biophys Res Communications. 2013;432(1):92–8. doi: 10.1016/j.bbrc.2013.01.080. [DOI] [PubMed] [Google Scholar]
- 128.Im MJ, Hoopes JE. Increases in dihydronicotinamide adenine dinucleotide (NADH) content and alpha-glycerophosphate dehydrogenase activity in epidermal wound healing. Proceedings of the society for experimental biology and medicine. Society for Experimental Biology and Medicine (New York, NY) 1983;173(1):17–20. doi: 10.3181/00379727-173-41602. [DOI] [PubMed] [Google Scholar]
- 129.Johnson BA, Wilson EM, Li Y, Moller DE, Smith RG, Zhou G. Ligand-induced stabilization of PPARγ monitored by NMR spectroscopy: implications for nuclear receptor activation. J Mol Biol. 2000;298(2):187–94. doi: 10.1006/jmbi.2000.3636. [DOI] [PubMed] [Google Scholar]
- 130.Risérus U, Tan GD, Fielding BA, Neville MJ, Currie J, Savage DB, et al. Rosiglitazone increases indexes of stearoyl-CoA desaturase activity in humans link to insulin sensitization and the role of dominant-negative mutation in peroxisome proliferator-activated receptor-γ. Diabetes. 2005;54(5):1379–84. doi: 10.2337/diabetes.54.5.1379. [DOI] [PubMed] [Google Scholar]
- 131.Otake K, Azukizawa S, Fukui M, Shibabayashi M, Kamemoto H, Miike T, et al. 4-tetrahydroisoquinoline-3-carboxylic acids: peroxisome proliferator-activated receptor α/γ dual agonists with protein-tyrosine phosphatase 1B inhibitory activity. Chem Pharmaceut Bull. 2010;59(10):1233–42. doi: 10.1248/cpb.59.1233. [DOI] [PubMed] [Google Scholar]
- 132.Egerod FL, Svendsen JE, Hinley J, Southgate J, Bartels A, Brünner N, et al. PPARα and PPARγ coactivation rapidly induces Egr-1 in the nuclei of the dorsal and ventral urinary bladder and kidney pelvis urothelium of rats. Toxicol Pathol. 2009;37(7):947–58. doi: 10.1177/0192623309351723. [DOI] [PubMed] [Google Scholar]
- 133.Fliegner D, Westermann D, Riad A, Schubert C, Becher E, Fielitz J, et al. Up-regulation of PPARγ in myocardial infarction. Europ J Heart Fail. 2008;10(1):30–8. doi: 10.1016/j.ejheart.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 134.Lee C-H, Olson P, Evans RM. Minireview: lipid metabolism, metabolic diseases, and peroxisome proliferator-activated receptors. Endocrinology. 2003;144(6):2201–7. doi: 10.1210/en.2003-0288. [DOI] [PubMed] [Google Scholar]
- 135.Gao D-F, Niu X-L, Hao G-H, Peng N, Wei J, Ning N, et al. Rosiglitazone inhibits angiotensin II-induced CTGF expression in vascular smooth muscle cells – role of PPAR-γ in vascular fibrosis. Biochem Pharmacol. 2007;73(2):185–97. doi: 10.1016/j.bcp.2006.09.019. [DOI] [PubMed] [Google Scholar]
- 136.Tzameli I, Fang H, Ollero M, Shi H, Hamm JK, Kievit P, et al. Regulated production of a peroxisome proliferator-activated receptor-γ ligand during an early phase of adipocyte differentiation in 3T3-L1 adipocytes. J Biol Chem. 2004;279(34):36093–102. doi: 10.1074/jbc.M405346200. [DOI] [PubMed] [Google Scholar]
- 137.Tsukahara T, Tsukahara R, Yasuda S, Makarova N, Valentine WJ, Allison P, et al. Different residues mediate recognition of 1-O-oleyllysophosphatidic acid and rosiglitazone in the ligand binding domain of peroxisome proliferator-activated receptor. J Biol Chem. 2005;281(6):3398–407. doi: 10.1074/jbc.M510843200. [DOI] [PubMed] [Google Scholar]
- 138.Kim KY, Cheon HG. Antiangiogenic effect of rosiglitazone is mediated via peroxisome proliferator-activated receptor-activated Maxi-K channel opening in human umbilical vein endothelial cells. J Biol Chem. 2006;281(19):13503–12. doi: 10.1074/jbc.M510357200. [DOI] [PubMed] [Google Scholar]
- 139.Muhlhausler BS, Duffield JA, McMillen IC. Increased maternal nutrition increases leptin expression in perirenal and subcutaneous adipose tissue in the postnatal lamb. Endocrinology. 2007;148(12):6157–63. doi: 10.1210/en.2007-0770. [DOI] [PubMed] [Google Scholar]
- 140.Ding J, Nagai K, Woo J-T. Insulin-dependent adipogenesis in stromal ST2 cells derived from murine bone marrow. Biosci Biotech Biochem. 2003;67(2):314–21. doi: 10.1271/bbb.67.314. [DOI] [PubMed] [Google Scholar]
- 141.Jung HS, Youn B-S, Cho YM, Yu K-Y, Park HJ, Shin CS, et al. The effects of rosiglitazone and metformin on the plasma concentrations of resistin in patients with type 2 diabetes mellitus. Metab: Clin Exper. 2005;54(3):314–20. doi: 10.1016/j.metabol.2004.05.019. [DOI] [PubMed] [Google Scholar]
- 142.Patel L, Buckels AC, Kinghorn IJ, Murdock PR, Holbrook JD, Plumpton C, et al. Resistin is expressed in human macrophages and directly regulated by PPAR gamma activators. Biochem Biophys Res Communications. 2003;300(2):472–6. doi: 10.1016/s0006-291x(02)02841-3. [DOI] [PubMed] [Google Scholar]
- 143.Boyle PJ, King AB, Olansky L, Marchetti A, Lau H, Magar R, et al. Effects of pioglitazone and rosiglitazone on blood lipid levels and glycemic control in patients with type 2 diabetes mellitus: a retrospective review of randomly selected medical records. Clin Ther. 2002;24(3):378–96. doi: 10.1016/s0149-2918(02)85040-8. [DOI] [PubMed] [Google Scholar]
- 144.Chen B, Wild D, Guha R. PubChem as a source of polypharmacology. J Chem Info Model. 2009;49(9):2044–55. doi: 10.1021/ci9001876. [DOI] [PubMed] [Google Scholar]
- 145.Pauwels E, Stoven V, Yamanishi Y. Predicting drug side-effect profiles: a chemical fragment-based approach. BMC Bioinfo. 2011;12(1):169. doi: 10.1186/1471-2105-12-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Knox C, Law V, Jewison T, Liu P, Ly S, Frolkis A, et al. DrugBank 3. 0: a comprehensive resource for ‘omics’ research on drugs. Nucl Acids Res. 2011;39(Suppl 1):D1035–41. doi: 10.1093/nar/gkq1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucl Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Huang L-C, Wu X, Chen JY. Predicting adverse side effects of drugs. BMC Genom. 2011;12(Suppl 5):S11. doi: 10.1186/1471-2164-12-S5-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Tatonetti NP, Patrick PY, Daneshjou R, Altman RB. Data-driven prediction of drug effects and interactions. Sci Transl Med. 2012;4(125):125ra31. doi: 10.1126/scitranslmed.3003377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kuhn M, Al Banchaabouchi M, Campillos M, Jensen LJ, Gross C, Gavin A-C, et al. Systematic identification of proteins that elicit drug side effects. Mol Syst Biol. 2013;9(1) doi: 10.1038/msb.2013.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kilicoglu H, Fiszman M, Rodriguez A, Shin D, Ripple A, Rindflesch TC. Semantic MEDLINE: a web application for managing the results of PubMed Searches. Proceedings of the third international symposium for semantic mining in biomedicine; 2008. pp. 69–76. [Google Scholar]
- 152.Ahlers CB, Fiszman M, Demner-Fushman D, Lang F-M, Rindflesch TC. Extracting semantic predications from Medline citations for pharmacogenomics. Pac symp biocomput. 2006;12:209–20. [PubMed] [Google Scholar]
- 153.U.S. Food and Drug Administration. 2007 – FDA Adds Boxed Warning for Heart-related Risks to Anti-Diabetes Drug Avandia. 2007 Nov 14; < http://www.fda.gov/NewsEvents/Newsroom/PressAnnouncements/2007/ucm109026.htm>.
- 154.Mahaffey KW, Hafley G, Dickerson S, Burns S, Tourt-Uhlig S, White J, et al. Results of a reevaluation of cardiovascular outcomes in the RECORD trial. Amer Heart J. 2013;166(2):240–9. doi: 10.1016/j.ahj.2013.05.004. [DOI] [PubMed] [Google Scholar]
- 155.Lopes RD, Dickerson S, Hafley G, Burns S, Tourt-Uhlig S, White J, et al. Methodology of a reevaluation of cardiovascular outcomes in the RECORD, trial: study design and conduct. Amer Heart J. 2013;166(2):208–216.e28. doi: 10.1016/j.ahj.2013.05.005. [DOI] [PubMed] [Google Scholar]
- 156.Thompson D. FDA to lift restrictions on diabetes drug Avandia. 2013 < http://health.usnews.com/health-news/news/articles/2013/11/25/fda-to-lift-restrictions-on-diabetes-drug-avandia>.
- 157.Herskovic JR, Bernstam EV. Using incomplete citation data for MEDLINE, results ranking. AMIA, annual symposium proceedings; 2005. pp. 316–20. [PMC free article] [PubMed] [Google Scholar]