Abstract
Significant left-right (L-R) differences in tumor incidence and disease outcome occur for cancers of paired organs, including the breasts; however, the basis for this laterality is unknown. Here, we show that despite their morphological symmetry, left versus right mammary glands in wild type mice have baseline differences in gene expression that are L-R independently regulated during pubertal development, including genes that regulate luminal progenitor cell renewal, luminal cell differentiation, mammary tumorigenesis, tamoxifen sensitivity, and chemotherapeutic resistance. In MMTV-cNeuTg/Tg mice, which model HER2/Neu amplified breast cancer, baseline L-R differences in mammary gene expression are amplified, sustained, or inverted in a gene-specific manner and the mammary ductal epithelium undergoes L-R asymmetric growth and patterning. Comparative genomic analysis of mouse L-R mammary gene expression profiles with gene expression profiles of human breast tumors revealed significant linkage between right-sided gene expression and decreased breast cancer patient survival. Collectively, these findings are the first to demonstrate that mammary glands are lateralized organs, and moreover, that mammary glands have L-R differential susceptibility to HER2/Neu oncogene-mediated effects on ductal epithelial growth and differentiation. We propose that intrinsic molecular laterality may play a role in L-R asymmetric breast tumor incidence and furthermore, that interplay between the L-R molecular landscape and oncogene activity may contribute to the differential disease progression and patient outcome that are associated with tumor situs.
Keywords: breast cancer, ErbB2, HER2/Neu, laterality, left-right asymmetry, mammary gland
INTRODUCTION
Cancers that initiate in paired organs and other bilaterally symmetric tissues exhibit an unusual feature, which is that tumors occur with non-equivalent incidence on the left versus right sides (1–5). The basis for this laterality has not been addressed at the cellular or molecular level, an oversight that may be significant because patient survival is reported to differ according to primary tumor situs (1–2). The side with elevated tumor incidence is organ-dependent and not necessarily the same side that is associated with poorer disease outcome. For breast cancer, the majority of occurrences are unilateral, with higher tumor incidence on the left (3). Left-side predominance also occurs in bilateral cases, in which more tumors develop first in the left breast or are larger than those on the right (3). Yet, despite the increased incidence and larger average tumor size of left-sided breast cancer, right-sided breast cancer may be associated with worse prognosis. Right-sided breast tumors are prone to earlier onset of bone metastasis and give rise to higher numbers of sites with metastatic involvement (6). This suggests that disease progression is related to the side of tumor formation, which could result in differential patient survival. Although studies directly addressing breast cancer patient survival relative to tumor laterality are limited and have generated contradictory findings (3), there is some indication that lower survival rates occur in patients with right-sided disease (7).
The left-sided excess of breast cancer and potential relationship between tumor laterality and patient prognosis suggests that mammary tissues harbor L-R differences that are relevant to oncogenesis. To address this we have used normal and neoplastic MMTV-cNeuTg/Tg mice to probe for L-R differences at the beginning and end of puberty--a period when the rapidly growing ductal epithelium (8) is vulnerable to genetic, hormonal, and other environmental perturbations that heighten risk for developing breast cancer later in life (9–11). Here we provide evidence that mouse mammary glands have baseline L-R differences in gene expression that are L-R discordantly altered by HER2/Neu and that are accompanied by asymmetric ductal epithelial growth and patterning. Furthermore, we used comparative genomic analysis to show that the L-R differences in gene expression that we identified in mouse mammary glands are predictive of breast cancer patient outcome, with right-side expression profiles associated with significantly poorer long-term patient survival.
RESULTS AND DISCUSSION
Thoracic mammary glands are molecularly L-R asymmetric
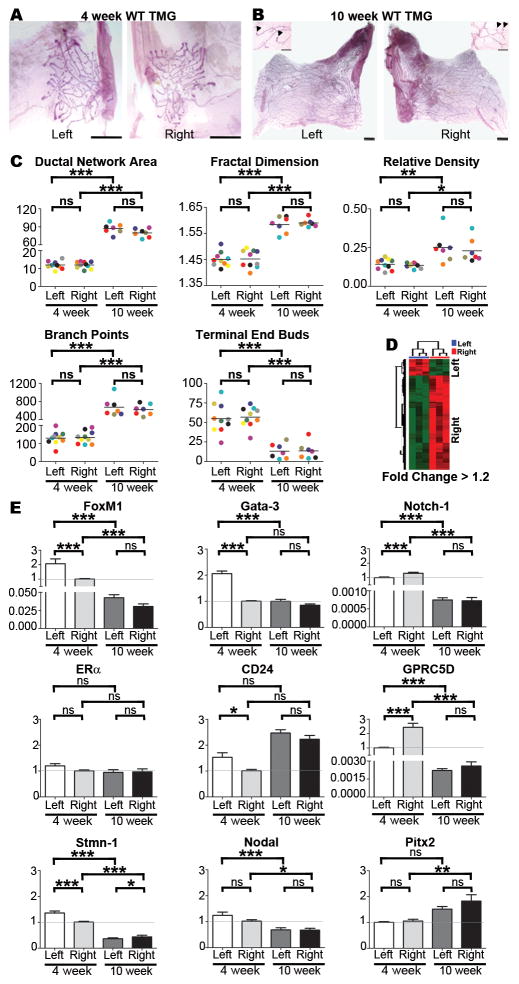
Ductal epithelial networks in thoracic mammary glands (TMGs) of early pubertal (4-week) and post-pubertal (10-week) wild type (WT) mice (Fig. 1A, B) were quantified by image and fractal analysis as described previously (12). Despite increases in network area and number of branch points between weeks 4 and 10, as well as changes in TEBs, which decrease in number and initiate regression by week 10 (13), all of these morphological parameters were statistically equivalent for left and right TMGs at both timepoints, indicative of L-R symmetry (Fig. 1C). By contrast, microarray analysis yielded approximately 161 transcripts that were L-R differentially expressed (i.e., up-regulated or down-regulated) with >1.2 fold change (q-value<0.05, Fig. 1D), including genes and pathways that have established roles in oncogenesis and/or therapeutic sensitivity (Table S1). Several of the transcripts identified in the array were examined by qRT-PCR (Fig. 1E), which confirmed that relative to left-side expression, some genes were increased and others were decreased in expression levels on the right side. For example, Gata-3 and FoxM1, which regulate luminal progenitor cell differentiation and renewal (14–15), and which also have opposing protective and causative roles in tumorigenesis in the breast and other organs (16–17), were more highly expressed on the left side (Fig. 1E). By the end of puberty, both genes were down-regulated; however, the fold decrease was significantly greater for left-side glands which resulted in net symmetric expression (Fig. 1E). Asymmetric expression also was found for notch-1, another regulator of mammary luminal progenitor cell commitment (18) that is involved in breast tumorigenesis (19) (Fig. 1E). Notch-1 was right-side elevated, and by 10-weeks it showed slightly higher fold decrease in right-side glands compared to left (Fig. 1E). To determine if asymmetric expression of genes with dual roles in ductal growth and tumorigenesis is a general property of TMGs, we examined estrogen receptor alpha (ERα). ERα was L-R equivalently expressed at both the start and end of puberty, consistent with it not being identified as a candidate by microarray (Fig. 1E). We also examined CD24, a pan-epithelial marker in mouse mammary glands (20), which showed modest left-side elevation in 4-week TMGs, but not in 10-week TMGs (Fig. 1E), raising the possibility that subtle differences in epithelial cell number could be present during early puberty, despite equivalent ductal network growth and morphology.
Figure 1. Morphological and molecular analysis of TMGs.
Wild type mouse TMGs such as the representative L-R matched pairs shown at 4-weeks (A) and 10-weeks (B) were processed as carmine red stained whole mounts and composite images were manually traced, isolated, and converted to 8-bit gray scale images for analysis with MetaMorph® image analysis software (scale bar = 1mm). Thresholds and the total area of ductal networks were determined by the integrated morphometry analysis subroutine, and the fractal dimension, a measure of complexity of the network’s morphology, was determined using HarFa analytical software by applying the box counting method (12). Branch points and terminal end buds (TEBs), which are shown in higher magnification insets for 10-week glands (arrowheads indicate TEBs; scale bar = 5 μm), were quantified by manual counting. Color-coding can be used to follow matched L-R pairs harvested from the same mouse in all graphs. No significant L-R differences (C) were found in ductal network area, fractal dimension, relative density, branch points or TEBs at 4 or 10 weeks as determined by one-tailed paired student’s t-test. Microarray analysis of left versus right TMGs (D) using left as the baseline reference was performed using RNA pooled from 3–4 intact 4-week TMGs [#3 and #8 glands as diagrammed in Veltmaat et al (41)] for cDNA synthesis and hybridization to Affymetrix GeneChip Mouse Genome 430 2.0 Arrays. The arrays were preprocessed and normalized using RMA (42). Each array experiment was completed in biological and technical triplicate. Differentially expressed probesets were identified based on a fold-change (increase or decrease in right side compared to left) of at least 1.2, and a q-value of less than 0.05. Pathway analysis was carried out for each set of laterality associated genes (left or right) by probing the NCI Pathway interaction database (43). SYBR Green-based qRT-PCR of select array candidates was performed with primers listed in Table S2 (E). Real-time PCR miner was used to calculate Ct values and replication efficiency(44) and fold changes relative to GAPDH mRNA were determined by delta-delta Ct. Fold changes across groups were determined using the lower level of 4-week expression as baseline as indicated by the horizontal grey line. Bars represent mean ± SEM of ≥5 mice; *p<0.05, **p<0.01; *** p<0.001 (two-tailed paired student’s t-tests).
Genes involved in therapeutic sensitivity also were represented in the microarray. Elevated right-side expression was detected for retinoic acid-inducible G-protein coupled receptor 5D (GPRC5D), a gene that enhances sensitivity to an estrogen receptor antagonist, tamoxifen, in MCF7 breast cancer cells (21) and that was decreased by the end of puberty (Fig. 1E). In addition, stathmin-1 (Stmn-1), a microtubule destabilizing protein that confers chemoresistance in breast and other tumor types (22–26), was modestly left-side elevated in 4-week TMGs, followed by inversion to modest right-side elevated expression in 10-week TMGs (Fig. 1E). Given the many L-R differences in gene expression in TMGs, it was surprising that microarray analysis did not uncover connections to any known laterality genes (Table S1), including nodal and Pitx2, regulators of embryonic L-R patterning that also are expressed in breast cancer and other tumor types (4). Thus, we assessed these genes by qRT-PCR, which confirmed symmetric expression (Fig. 1E). Together, these findings demonstrate that despite symmetric nodal and Pitx2 expression, the left and right TMGs of WT mice are molecularly lateralized with asymmetric expression of other genes that may impart differential predisposition to oncogenesis.
HER2/Neu causes L-R asymmetric ductal growth and alters L-R gene expression in TMGs
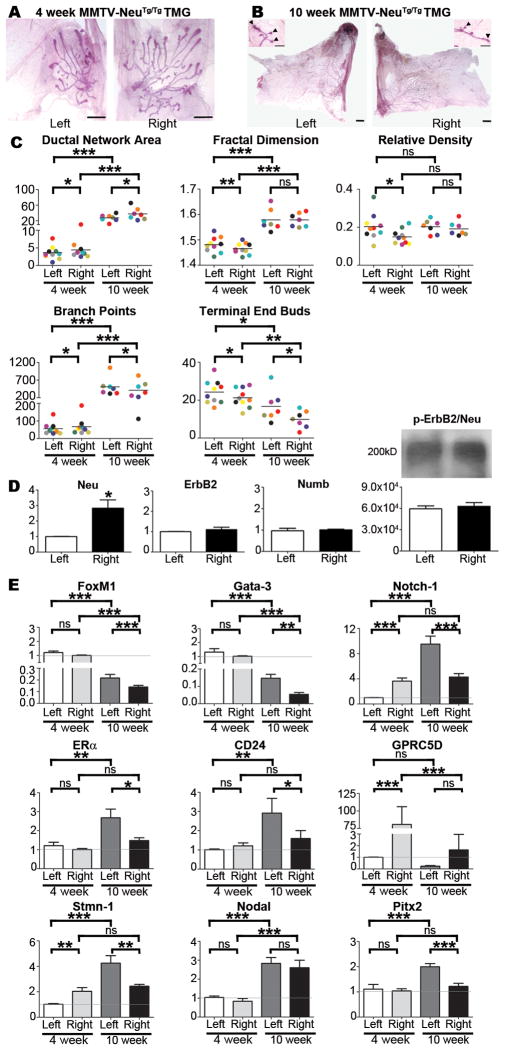
To address the possibility that mammary ductal epithelium might be primed for differential growth during neoplasia, we quantified ductal networks in MMTV-cNeuTg/Tg mice, which are a commonly used model of HER2+ breast cancer (27). Compared to WT, the ductal network area was smaller in 4-week MMTV-cNeuTg/Tg TMGs and in particular, left-sided MMTV-cNeuTg/Tg networks were significantly smaller than their right-sided counterparts (Fig. 2A, C). Left-sided networks also contained fewer branch points, and had higher fractal dimension, relative density, and number of TEBs (Fig. 2A, C). Morphological asymmetry persisted through the end of pubertal development, with left-sided networks maintaining decreased area and higher numbers of branch points and TEBs (Fig. 2B, C). Given the L-R differences in MMTV-cNeuTg/Tg ductal network growth and pattern, we evaluated whether MMTV-cNeuTg/Tg TMGs have asymmetric ErbB2/Neu expression or activity. Although qRT-PCR showed that Neu expression was elevated in right-side TMGs, endogenous ErbB2 expression was L-R equivalent, as was Numb, a notch inhibitor whose expression is regulated by ErbB2 (28) (Fig. 2D). Moreover, phospho-ErbB2/Neu immunoprecipitation showed equivalent levels in left and right side TMGs, suggesting similar activation of ErbB2/Neu signaling on both sides (Fig. 2D).
Figure 2. Morphological and molecular analysis of MMTV-cNeuTg/Tg TMGs.
TMGs from MMTV-cNeuTg/Tg mice such as the representative L-R matched pairs shown at 4-weeks (A) and 10-weeks (B) were processed for morphometric analysis and data for matched L-R pairs in individual mice were color coded as described in Fig. 1 and tested by Grubb’s Outlier test, which indicated an absence of outliers. Ductal network area, fractal dimension, relative density, branch points, and TEBs exhibited significant L-R differences at 4 weeks (C) as determined by one-tailed paired student’s t-test (*p<0.05; **p<0.01). Ductal network area, branch points, and TEBs remained significantly L-R different at 10 weeks (C). SYBR Green-based qRT-PCR showed asymmetric expression of Neu, but symmetric mRNA expression of ErbB2 and Numb (D). Bars represent mean ± SEM. N ≥ 5, *p = 0.003. Results were confirmed with a second primer set listed in Table S1. Total ErbB2/Neu protein was immunoprecipitated from left or right TMGs (Antibody #4290, Cell Signaling), immunoblotted, and probed with anti-phospho-ErbB2/Neu (Antibody #2243, Cell Signaling). Densitometry of triplicate results indicated no significant L-R differences (D). SYBR Green-based qRT-PCR analysis of gene expression in left vs. right TMGs of MMTV-cNeuTg/Tg mice was performed as described in Fig. 1E. Bars represent mean ± SEM of 5 mice, *p<0.05; **p<0.01; *** p<0.001 (two-tailed paired student’s t-tests).
Further analysis of MMTV-cNeuTg/Tg TMGs indicated that molecular laterality was amplified, sustained, or inverted in a gene-specific manner by comparison to WT. Notch-1 expression was approximately 3-fold higher in right-sided 4-week MMTV-cNeuTg/Tg TMGs (Fig. 2E), which is an amplification of the modest Notch-1 asymmetry that was present in WT TMGs (Fig. 1E and Fig. S1). In 10-week MMTV-cNeuTg/Tg TMGs, asymmetric Notch-1 expression was inverted, with approximately 2-fold higher expression in left-side glands (Fig. 2E). Because Notch influences breast cancer cell sensitivity to several therapeutic agents, including trastuzumab, gefitinib, docetaxel, and tamoxifen (29), the L-R uncoupled regulation of Notch-1 expression in MMTV-cNeuTg/Tg TMGs may be important in the context of differential disease progression. In addition, FoxM1 and Gata-3, which were left-side elevated in 4-week WT TMGs (Fig. 1E), were decreased on both sides in MMTV-cNeuTg/Tg TMGs; however, the fold decrease for FoxM1 was greater on the left side (Fig. S1) resulting in net L-R symmetric expression (Fig. 2E). Analysis of 10-week MMTV-cNeuTg/Tg TMGs showed that FoxM1 expression was further decreased, albeit the fold decrease was greater for right-side glands (Fig. 2E and Fig. S1). Given the additional role of FoxM1 in modulating endocrine and chemotherapeutic resistance in breast cancer cells (30–32), the L-R uncoupled regulation of FoxM1 expression in MMTV-cNeuTg/Tg TMGs was notable. We also found similar L-R asymmetric regulation of Gata-3 in MMTV-cNeuTg/Tg TMGs, which resulted in modestly higher left-sided expression by 10-weeks (Fig. 2E and Fig. S1).
Genes with symmetric expression in 4-week MMTV-cNeuTg/Tg TMGs included ERα, CD24, nodal, and Pitx2 (Fig. 2E). However, by 10 weeks their expression was elevated in left-sided glands, with the exception of nodal, which was elevated on both sides (Fig. 2E). GPRC5D, which was right-side elevated in 4-week WT TMGs (Fig. 1E), also was right-side elevated in 4-week MMTV-cNeuTg/Tg TMGs (Fig. 2E), despite an overall marked reduction in expression on both sides (Fig. S1). Stmn-1 was asymmetric in 4-week MMTV-cNeuTg/Tg TMGs (Fig. 2D, Fig. S1) but by week-10 was increased only on the left-side, resulting in inverted asymmetric expression (Fig. 2E).
IMGs are refractory to HER2/Neu-induced asymmetric growth and show delayed L-R asymmetric gene expression
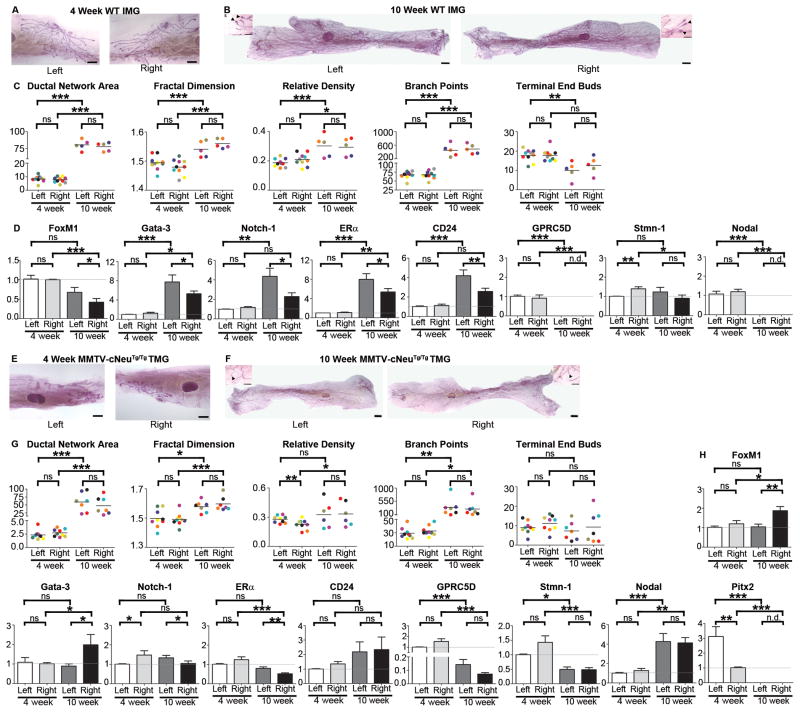
Although mouse TMGs share more similarity with human mammary glands than inguinal mammary glands (IMGs) (3), IMGs are more commonly used in experimentation because of their larger size and easier accessibility (33–34). Therefore, we also examined IMGs. Like TMGs, IMGs showed no significant L-R differences in morphology at either 4-weeks or 10-weeks (Fig. 3A–C). Unlike TMGs, early pubertal IMGs showed an absence of significant molecular asymmetry except for Stmn-1, which was modestly right-side increased (Fig. 3D). However, by the end of puberty, 10-week IMGs had developed molecular L-R asymmetry similar to that observed in 4-week WT TMGs, with left-side elevated expression of FoxM1, Gata-3, Notch-1, ERα, and CD24 (Fig. 3D).
Figure 3. Morphological and molecular analysis of IMGs.
L-R pairs of IMGs from 4-week (A) and 10-week (B) wild type and 4-week (E) and 10-week (F) old MMTV-cNeuTg/Tg mice were processed and analyzed as described in Fig. 1A–C. Ductal network area, fractal dimension, relative density, branch points, and TEBs did not exhibit significant L-R differences (C, G) as determined by one-tailed paired student’s t-test. SYBR Green-based qRT-PCR analysis of gene expression of left vs. right IMGs of wild type (D) and MMTV-cNeuTg/Tg (H) mice was performed as described in Fig. 1E. Pitx2 was not detectable in wild type IMGs at either age nor in MMTV-cNeuTg/Tg IMGs at 4 weeks. Bars represent mean ± SEM of ≥3 mice, *p = 0.01 (two-tailed paired student’s t-tests).
As previously reported (35), ductal networks in MMTV-cNeuTg/Tg IMGs were smaller compared to WT, and we found symmetric morphology at both 4 and 10-weeks (Fig. 3A–C). Although there were overall changes in gene expression relative to WT (Fig. S1), 4-week MMTV-cNeuTg/Tg IMGs did not exhibit molecular L-R asymmetry, with the exception of modest right-side elevation of Notch-1 and a more robust 3.5-fold left-side elevation of Pitx2 (Fig. 3H). Although Pitx2 was not associated with asymmetric ductal growth per se in either TMGs or IMGs, given that altered Pitx2 methylation occurs in breast and other cancer types (4) the overall changes in Pitx2 expression nevertheless suggest a potential role in HER2/Neu-induced neoplasia. By the end of puberty, MMTV-cNeuTg/Tg IMGs showed pronounced molecular asymmetry, as exemplified by right-side elevated FoxM1 and Gata-3 expression and left-side elevated ERα and Notch-1 (Fig. 3H). Thus by comparison to TMGs, both WT and MMTV-cNeuTg/Tg IMGs were temporally delayed in developing molecular asymmetry, which may account for their remaining refractory to HER2/Neu-induced asymmetric epithelial growth and morphogenesis.
TMG molecular laterality is associated with differential breast cancer patient survival
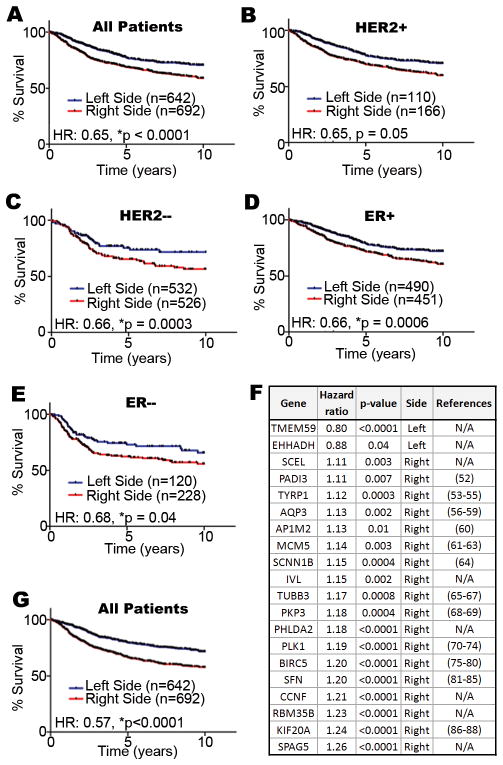
To determine if L-R differences detected in mouse mammary glands are clinically relevant, the genes identified in our microarray experiment (Fig. 1D) were evaluated in a large number of breast tumor gene expression data sets for which corresponding patient outcome is also known (n=1334). Of the 161 transcripts identified in the microarray, we were able to map 96 of them by Unigene ID to their human transcript counterpart for each patient.
Because the sidedness of tumor location was not available in the clinical annotation files, patients were assigned to left (n=642) or right-side (n=692) groups based on whether their tumor gene expression profiles more closely matched with the left or right profiles identified in the mouse microarray. Notably, right-side gene expression was linked to poorer patient survival (Fig. 4A). We next analyzed subsets of patients with HER2+ and HER2− tumors. Whereas the relationship between L-R gene expression and outcome fell just short of significance in the HER2 over-expressing subset (Fig. 4B), the relationship was significant in the HER2− subset (Fig. 4C). It should be noted that because HER2 status was not available in the clinical annotation files, we assigned patients to the HER2+ and HER2− subsets based on mean ERBB2 transcript levels. For this reason, and also because of statistical power limitations due to the HER2+ subset containing far fewer patients (n=276) than the HER2− subset (n=1058), the relationship between HER2+ patient survival and L-R gene expression may be unclear and require additional investigation with a larger HER2+ test cohort.
Figure 4. Comparative genomic analysis of mouse L-R mammary gene expression profiles with human breast tumors and the relationship to breast cancer patient survival.
We compiled a large cohort of breast cancer patients from multiple studies available through the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) to test the association between laterality associated genes and patient survival. Hazard ratios (HR) are indicated for all patients (A), and HER2+ (B), HER2− (C), ER+ (D), ER− (E) subsets. A 20-gene subset of the 96 L-R TMG gene expression set (F) is a robust predictor of outcome among all breast cancer patients (G). References are provided for genes previously implicated in oncogenesis; those with none available (N/A) are indicated. Our combined cohort comprised patients from the GSE2034 (45), GSE7390 (46), GSE4922(47), GSE25055 (48), and GSE3494 (49) cohorts (n=1334). For all patients, clinical outcome data as well as the gene expression profile of their respective tumors was available. Whenever possible we used 10-yr disease free survival as the clinical endpoint in our study; however, when disease free survival was not available we alternatively used either distant metastasis free or overall survival as the clinical endpoint. The arrays for each separate cohort were preprocessed and normalized using RMA(42). ER status was assigned based on the clinical annotation files, and HER2 status was assigned based on the mean ERBB2 transcript levels (probe set ID 216836_s_at) within each study cohort independently. Affymetrix GeneChip Mouse Genome 430 2.0 Arrays probe sets were mapped to their human counterpart genes by Unigene IDs. When multiple probe sets recognized the same gene transcripts, only the probe with the highest mean intensity was used. To assign signature scores to patients, the expression values for each gene were standardized such that the mean and standard deviation were set to 0 and 1 in each individual patient cohort, respectively. Subsequently, we calculated signature scores for each patient as previously described (50–51), where positive scores were considered to indicate that a tumor had ‘right-sided’ gene expression and negative scores were considered to indicate that a tumor had ‘left-sided’ gene expression. Survival curves were graphed using Graphpad Prism® 5 and statistical tests were completed in R.
Since ER status also is tightly linked to breast cancer patient outcome (36), we evaluated L-R gene expression patterns in ER+ and ER− patient subsets. In both subsets, right-side gene expression was associated with decreased survival (Fig. 4D, E). Lastly, we performed univariate Cox-regression survival analyses with each of the L-R transcripts, which identified a 20-gene subset that likely drove the predictive capacity of the complete 96-gene set (*p<0.05, Cox-regression) (Fig. 4F). Indeed, the evaluation of these 20 genes among the 1334 patient cohort outperformed the original 96 L-R gene set (Fig. 4G). Thus, the L-R gene expression profiles identified in mouse mammary glands are significantly linked to breast cancer patient survival rates, and demonstrate that right-sided gene expression is associated with poorer survival.
In summary, our results indicate that despite their morphological symmetry, mammary glands are molecularly lateralized. Although left and right glands express the same genes, the relative levels of gene expression significantly differ and are subject to L-R uncoupled regulation during pubertal development. Our results also demonstrate that many of the genes associated with the left side are down-regulated, yet remain elevated or amplified on the right-side in TMGs of MMTV-cNeuTg/Tg mice, consistent with more aggressive disease progression reported for right-sided breast tumors (6). Moreover, the L-R uncoupled gene expression is accompanied by asymmetric growth and morphogenesis of the ductal epithelium. The molecular laterality of mammary glands at the start of puberty appears to be important in potentiating HER2/Neu oncogene-mediated asymmetric growth since IMGs, which exhibit L-R differences in gene expression at the end of puberty, but not at the start, fail to undergo L-R asymmetric growth in MMTV-cNeuTg/Tg mice. From the perspective of modeling human breast development and cancer, these results confirm there are significant differences between thoracic and inguinal glands and provide the first evidence that each mammary pair is independently L-R regulated regardless of its anterior or posterior location. By analogy to anterior-posterior differences that underlie differential development and neoplastic susceptibility of mouse TMGs versus IMGs (3), as well as the processes that establish molecular L-R differences in other bilaterally symmetric tissues (37–39), we hypothesize that mammary laterality may be rooted in embryonic patterning. Therefore, future investigation to determine the connections between positional differences in gene expression, axial patterning, and the relationship to mammary development and tumorigenesis will be revealing. Furthermore, given the roles of ErbB2/HER2 in normal and neoplastic mammary development (40), as well as the significant link we found between L-R gene expression and breast cancer patient survival, our findings highlight laterality as a parameter that warrants greater consideration in experimental design in mouse mammary models as well as clinical analysis of breast cancer patients.
Supplementary Material
Transcripts identified in the L-R mouse TMG microarray are listed and the side with elevated expression is indicated (A). Results from pathway analysis of microarray data also are indicated (B).
Primer sets used for qRT-PCR are listed. For experiments that were confirmed by alternate primer sets, the primers used to generate the results presented in the Figures are indicated (*).
SYBR Green-based qRT-PCR was used to assess gene expression in L-R paired 4-week TMGs (A), 10-week TMGs (B), 4-week IMGs (C), and 10-week IMGs (D). For 4-week TMGs, data were normalized relative to WT right-sided glands for all genes except Notch-1, ERα, and GPRC5D, which were normalized relative to WT left-sided glands. Comparison of fold differences between WT and MMTV-cNeuTg/Tg glands indicated that FoxM1, Gata-3, Notch-1, CD24, GPRC5D, and Stmn-1 were independently up or down-regulated in left versus right-sided glands. For 10-week TMGs, data were normalized relative to WT left-sided glands for all genes except FoxM1, Gata-3, Nodal, and Pitx2, which were normalized relative to WT right-sided glands. Comparison of fold differences between WT and MMTV-cNeuTg/Tg glands indicated that FoxM1, Gata-3, Notch-1, CD24, GPRC5D, and Pitx2 were independently up or down-regulated in left versus right-sided glands. For 4-week IMGs, data were normalized relative to WT right-sided glands, except for Notch-1, ERα, and Stmn-1, which were normalized to WT left-sided glands. Comparison of fold differences between WT and MMTV-cNeuTg/Tg glands indicated that all genes examined were similarly up or down-regulated in left versus right-sided glands except Notch-1, which was slightly more elevated in MMTV-cNeuTg/Tg right-sided glands compared to WT. SYBR Green-based qRT-PCR showed gene expression in 10-week left vs. right IMGs (D). For 10-week IMGs, data were normalized relative to WT right-sided glands for all genes. Comparison of fold differences between WT and MMTV-cNeuTg/Tg glands indicated that FoxM1, Gata-3, Notch-1, ERα, and CD24 were independently up or down-regulated in left versus right-sided glands. Bars represent mean ± SEM of ≥ 3 mice, *p<0.05; **p<0.01; *** p<0.001, two-tailed paired student’s t-tests.
Acknowledgments
We thank Dr. Scott Argraves, Dr. Jeremey Barth, and Mr. Victor Fresco of the MUSC Proteogenomics Facility for performing the DNA microarray and Dr. Caroline Alexander and Dr. Elizabeth Yeh for helpful discussion.
Funding
Supported by National Institutes of Health K02HL086737 (A.F.R.), R21HD068993 (A.F.R.) and conducted in facilities supported, in part, by Cancer Center Support Grant P30CA138313 to the Hollings Cancer Center, MUSC.
Footnotes
Competing interests statement
The authors declare no competing interests.
References
- 1.Delahunt B, Bethwaite P, Nacey JN. Renal cell carcinoma in New Zealand: a national survival study. Urology. 1994 Mar;43(3):300–9. doi: 10.1016/0090-4295(94)90070-1. [DOI] [PubMed] [Google Scholar]
- 2.Roychoudhuri R, Putcha V, Møller H. Cancer and Laterality: A Study of The Five Major Paired Organs (UK) Cancer Causes and Control. 2006;17(5):655–62. doi: 10.1007/s10552-005-0615-9. [DOI] [PubMed] [Google Scholar]
- 3.Veltmaat JM, Ramsdell AF, Sterneck E. Positional variations in mammary gland development and cancer. J Mammary Gland Biol Neoplasia. 2013;18(2):179–88. doi: 10.1007/s10911-013-9287-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilting J, Hagedorn M. Left-right asymmetry in embryonic development and breast cancer: common molecular determinants? Current medicinal chemistry. 2011;18(36):5519–27. doi: 10.2174/092986711798347252. [DOI] [PubMed] [Google Scholar]
- 5.Yoruk O, Karasen M, Timur H, Erdem T, Dane S, Tan U. Lateralizations of head-neck cancers are not associated with peripheral asymmetry of cell-mediated immunity. The International journal of neuroscience. 2009;119(6):815–20. doi: 10.1080/00207450802329639. [DOI] [PubMed] [Google Scholar]
- 6.Fatima N, Zaman MU, Maqbool A, Khan SH, Riaz N. Lower incidence but more aggressive behavior of right sided breast cancer in pakistani women: does right deserve more respect? Asian Pacific journal of cancer prevention: APJCP. 2013;14(1):43–5. doi: 10.7314/apjcp.2013.14.1.43. [DOI] [PubMed] [Google Scholar]
- 7.Hartveit F, Tangen M, Hartveit E. Side and survival in breast cancer. Oncology. 1984;41(3):149–54. doi: 10.1159/000225812. [DOI] [PubMed] [Google Scholar]
- 8.Watson CJ, Khaled WT. Mammary development in the embryo and adult: a journey of morphogenesis and commitment. Development. 2008 Mar;135(6):995–1003. doi: 10.1242/dev.005439. [DOI] [PubMed] [Google Scholar]
- 9.Biro FM, Deardorff J. Identifying opportunities for cancer prevention during preadolescence and adolescence: puberty as a window of susceptibility. The Journal of adolescent health: official publication of the Society for Adolescent Medicine. 2013 May;52(5 Suppl):S15–20. doi: 10.1016/j.jadohealth.2012.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fenton SE. Endocrine-Disrupting Compounds and Mammary Gland Development: Early Exposure and Later Life Consequences. Endocrinology. 2006 Jun 1;147(6):s18–24. doi: 10.1210/en.2005-1131. [DOI] [PubMed] [Google Scholar]
- 11.Fenton SE, Reed C, Newbold RR. Perinatal environmental exposures affect mammary development, function, and cancer risk in adulthood. Annual review of pharmacology and toxicology. 2012;52:455–79. doi: 10.1146/annurev-pharmtox-010611-134659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fuseler JW, Robichaux JP, Atiyah HI, Ramsdell AF. Morphometric and fractal dimension analysis identifies early neoplastic changes in mammary epithelium of MMTV-cNeu mice. Anticancer Res. 2014;34(3):1171–7. [PubMed] [Google Scholar]
- 13.Richert MM, Schwertfeger KL, Ryder JW, Anderson SM. An atlas of mouse mammary gland development. J Mammary Gland Biol Neoplasia. [Research Support, U.S. Gov’t, Non-P.H.S. Research Support, U.S. Gov’t, P.H.S. Review] 2000 Apr;5(2):227–41. doi: 10.1023/a:1026499523505. [DOI] [PubMed] [Google Scholar]
- 14.Asselin-Labat ML, Sutherland KD, Barker H, Thomas R, Shackleton M, Forrest NC, et al. Gata-3 is an essential regulator of mammary-gland morphogenesis and luminal-cell differentiation. Nature cell biology. [Research Support, Non-U.S. Gov’t] 2007 Feb;9(2):201–9. doi: 10.1038/ncb1530. [DOI] [PubMed] [Google Scholar]
- 15.Carr JR, Kiefer MM, Park HJ, Li J, Wang Z, Fontanarosa J, et al. FoxM1 regulates mammary luminal cell fate. Cell reports. 2012 Jun 28;1(6):715–29. doi: 10.1016/j.celrep.2012.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Teh MT. FOXM1 coming of age: time for translation into clinical benefits? Frontiers in oncology. 2012;2:146. doi: 10.3389/fonc.2012.00146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chou J, Provot S, Werb Z. GATA3 in development and cancer differentiation: cells GATA have it! Journal of cellular physiology. 2010 Jan;222(1):42–9. doi: 10.1002/jcp.21943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bouras T, Pal B, Vaillant F, Harburg G, Asselin-Labat ML, Oakes SR, et al. Notch signaling regulates mammary stem cell function and luminal cell-fate commitment. Cell stem cell. 2008 Oct 9;3(4):429–41. doi: 10.1016/j.stem.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 19.Farnie G, Clarke RB. Mammary stem cells and breast cancer--role of Notch signalling. Stem cell reviews. 2007 Jun;3(2):169–75. doi: 10.1007/s12015-007-0023-5. [DOI] [PubMed] [Google Scholar]
- 20.Visvader JE. Keeping abreast of the mammary epithelial hierarchy and breast tumorigenesis. Genes Dev. 2009 Nov 15;23(22):2563–77. doi: 10.1101/gad.1849509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mendes-Pereira AM, Sims D, Dexter T, Fenwick K, Assiotis I, Kozarewa I, et al. Genome-wide functional screen identifies a compendium of genes affecting sensitivity to tamoxifen. Proc Natl Acad Sci U S A. 2012 Feb 21;109(8):2730–5. doi: 10.1073/pnas.1018872108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baquero MT, Hanna JA, Neumeister V, Cheng H, Molinaro AM, Harris LN, et al. Stathmin expression and its relationship to microtubule-associated protein tau and outcome in breast cancer. Cancer. 2012 Oct 1;118(19):4660–9. doi: 10.1002/cncr.27453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han ZX, Wang HM, Jiang G, Du XP, Gao XY, Pei DS. Overcoming Paclitaxel Resistance in Lung Cancer Cells Via Dual Inhibition of Stathmin and Bcl-2. Cancer biotherapy & radiopharmaceuticals. 2013 Mar 15; doi: 10.1089/cbr.2012.1323. [DOI] [PubMed] [Google Scholar]
- 24.Meng XL, Su D, Wang L, Gao Y, Hu YJ, Yang HJ, et al. Low expression of stathmin in tumor predicts high response to neoadjuvant chemotherapy with docetaxel-containing regimens in locally advanced breast cancer. Genetic testing and molecular biomarkers. 2012 Jul;16(7):689–94. doi: 10.1089/gtmb.2011.0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miceli C, Tejada A, Castaneda A, Mistry SJ. Cell cycle inhibition therapy that targets stathmin in in vitro and in vivo models of breast cancer. Cancer gene therapy. 2013 Apr 26; doi: 10.1038/cgt.2013.21. [DOI] [PubMed] [Google Scholar]
- 26.Su D, Smith SM, Preti M, Schwartz P, Rutherford TJ, Menato G, et al. Stathmin and tubulin expression and survival of ovarian cancer patients receiving platinum treatment with and without paclitaxel. Cancer. 2009 Jun 1;115(11):2453–63. doi: 10.1002/cncr.24282. [DOI] [PubMed] [Google Scholar]
- 27.Hutchinson JN, Muller WJ. Transgenic mouse models of human breast cancer. Oncogene. 2000 Dec 11;19(53):6130–7. doi: 10.1038/sj.onc.1203970. [DOI] [PubMed] [Google Scholar]
- 28.Lindsay J, Jiao X, Sakamaki T, Casimiro MC, Shirley LA, Tran TH, et al. ErbB2 induces Notch1 activity and function in breast cancer cells. Clinical and translational science. 2008 Sep;1(2):107–15. doi: 10.1111/j.1752-8062.2008.00041.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Z, Li Y, Ahmad A, Azmi AS, Banerjee S, Kong D, et al. Targeting Notch signaling pathway to overcome drug resistance for cancer therapy. Biochimica et biophysica acta. 2010 Dec;1806(2):258–67. doi: 10.1016/j.bbcan.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carr JR, Park HJ, Wang Z, Kiefer MM, Raychaudhuri P. FoxM1 mediates resistance to herceptin and paclitaxel. Cancer Res. 2010 Jun 15;70(12):5054–63. doi: 10.1158/0008-5472.CAN-10-0545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kwok JM, Peck B, Monteiro LJ, Schwenen HD, Millour J, Coombes RC, et al. FOXM1 confers acquired cisplatin resistance in breast cancer cells. Molecular cancer research: MCR. 2010 Jan;8(1):24–34. doi: 10.1158/1541-7786.MCR-09-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Millour J, Constantinidou D, Stavropoulou AV, Wilson MS, Myatt SS, Kwok JM, et al. FOXM1 is a transcriptional target of ERalpha and has a critical role in breast cancer endocrine sensitivity and resistance. Oncogene. 2010 May 20;29(20):2983–95. doi: 10.1038/onc.2010.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brill B, Boecher N, Groner B, Shemanko CS. A sparing procedure to clear the mouse mammary fat pad of epithelial components for transplantation analysis. Laboratory animals. 2008 Jan;42(1):104–10. doi: 10.1258/la.2007.06003e. [DOI] [PubMed] [Google Scholar]
- 34.Cardiff RD, Wellings SR. The comparative pathology of human and mouse mammary glands. J Mammary Gland Biol Neoplasia. [Comparative Study Review] 1999 Jan;4(1):105–22. doi: 10.1023/a:1018712905244. [DOI] [PubMed] [Google Scholar]
- 35.Mukherjee S, Louie SG, Campbell M, Esserman L, Shyamala G. Ductal growth is impeded in mammary glands of C-neu transgenic mice. Oncogene. 2000 Dec 7;19(52):5982–7. doi: 10.1038/sj.onc.1203964. [DOI] [PubMed] [Google Scholar]
- 36.Sotiriou C, Pusztai L. Gene-expression signatures in breast cancer. The New England journal of medicine. 2009 Feb 19;360(8):790–800. doi: 10.1056/NEJMra0801289. [DOI] [PubMed] [Google Scholar]
- 37.Golding JP, Partridge TA, Beauchamp JR, King T, Brown NA, Gassmann M, et al. Mouse myotomes pairs exhibit left-right asymmetric expression of MLC3F and alpha-skeletal actin. Developmental dynamics: an official publication of the American Association of Anatomists. 2004 Dec;231(4):795–800. doi: 10.1002/dvdy.20176. [DOI] [PubMed] [Google Scholar]
- 38.Golding JP, Tsoni S, Dixon M, Yee KT, Partridge TA, Beauchamp JR, et al. Heparin-binding EGF-like growth factor shows transient left-right asymmetrical expression in mouse myotome pairs. Gene expression patterns: GEP. 2004 Nov;5(1):3–9. doi: 10.1016/j.modgep.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 39.Chintapalli VR, Terhzaz S, Wang J, Al Bratty M, Watson DG, Herzyk P, et al. Functional correlates of positional and gender-specific renal asymmetry in Drosophila. PloS one. 2012;7(4):e32577. doi: 10.1371/journal.pone.0032577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eccles SA. The epidermal growth factor receptor/Erb-B/HER family in normal and malignant breast biology. The International journal of developmental biology. 2011;55(7–9):685–96. doi: 10.1387/ijdb.113396se. [DOI] [PubMed] [Google Scholar]
- 41.Veltmaat JM, Relaix F, Le LT, Kratochwil K, Sala FG, van Veelen W, et al. Gli3-mediated somitic Fgf10 expression gradients are required for the induction and patterning of mammary epithelium along the embryonic axes. Development. [Research Support, Non-U.S. Gov’t] 2006 Jun;133(12):2325–35. doi: 10.1242/dev.02394. [DOI] [PubMed] [Google Scholar]
- 42.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003 Apr;4(2):249–64. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 43.Schaefer CF, Anthony K, Krupa S, Buchoff J, Day M, Hannay T, et al. PID: the Pathway Interaction Database. Nucleic Acids Res. 2009 Jan;37(Database issue):D674–9. doi: 10.1093/nar/gkn653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao S, Fernald RD. Comprehensive algorithm for quantitative real-time polymerase chain reaction. J Comput Biol. 2005 Oct;12(8):1047–64. doi: 10.1089/cmb.2005.12.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Y, Klijn JG, Zhang Y, Sieuwerts AM, Look MP, Yang F, et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005 Feb 19–25;365(9460):671–9. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- 46.Desmedt C, Piette F, Loi S, Wang Y, Lallemand F, Haibe-Kains B, et al. Strong time dependence of the 76-gene prognostic signature for node-negative breast cancer patients in the TRANSBIG multicenter independent validation series. Clin Cancer Res. 2007 Jun 1;13(11):3207–14. doi: 10.1158/1078-0432.CCR-06-2765. [DOI] [PubMed] [Google Scholar]
- 47.Ivshina AV, George J, Senko O, Mow B, Putti TC, Smeds J, et al. Genetic reclassification of histologic grade delineates new clinical subtypes of breast cancer. Cancer Res. 2006 Nov 1;66(21):10292–301. doi: 10.1158/0008-5472.CAN-05-4414. [DOI] [PubMed] [Google Scholar]
- 48.Hatzis C, Pusztai L, Valero V, Booser DJ, Esserman L, Lluch A, et al. A genomic predictor of response and survival following taxane-anthracycline chemotherapy for invasive breast cancer. JAMA. May 11;305(18):1873–81. doi: 10.1001/jama.2011.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Miller LD, Smeds J, George J, Vega VB, Vergara L, Ploner A, et al. An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. Proc Natl Acad Sci U S A. 2005 Sep 20;102(38):13550–5. doi: 10.1073/pnas.0506230102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hallett RM, Dvorkin-Gheva A, Bane A, Hassell JA. A gene signature for predicting outcome in patients with basal-like breast cancer. Scientific reports. 2012;2:227. doi: 10.1038/srep00227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hallett RM, Pond G, Hassell JA. A target based approach identifies genomic predictors of breast cancer patient response to chemotherapy. BMC medical genomics. 2012;5:16. doi: 10.1186/1755-8794-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mohanan S, Cherrington BD, Horibata S, McElwee JL, Thompson PR, Coonrod SA. Potential role of peptidylarginine deiminase enzymes and protein citrullination in cancer pathogenesis. Biochem Res Int. 2012;2012:895343. doi: 10.1155/2012/895343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.El Hajj P, Journe F, Wiedig M, Laios I, Sales F, Galibert MD, et al. Tyrosinase-related protein 1 mRNA expression in lymph node metastases predicts overall survival in high-risk melanoma patients. Br J Cancer. 2013 Apr 30;108(8):1641–7. doi: 10.1038/bjc.2013.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Falkenius J, Lundeberg J, Johansson H, Tuominen R, Frostvik-Stolt M, Hansson J, et al. High expression of glycolytic and pigment proteins is associated with worse clinical outcome in stage III melanoma. Melanoma Res. 2013 Dec;23(6):452–60. doi: 10.1097/CMR.0000000000000027. [DOI] [PubMed] [Google Scholar]
- 55.Journe F, Id Boufker H, Van Kempen L, Galibert MD, Wiedig M, Sales F, et al. TYRP1 mRNA expression in melanoma metastases correlates with clinical outcome. Br J Cancer. 2011 Nov 22;105(11):1726–32. doi: 10.1038/bjc.2011.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mobasheri A, Barrett-Jolley R. Aquaporin Water Channels in the Mammary Gland: From Physiology to Pathophysiology and Neoplasia. J Mammary Gland Biol Neoplasia. 2013 Dec 13; doi: 10.1007/s10911-013-9312-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cao XC, Zhang WR, Cao WF, Liu BW, Zhang F, Zhao HM, et al. Aquaporin3 is required for FGF-2-induced migration of human breast cancers. PloS one. 2013;8(2):e56735. doi: 10.1371/journal.pone.0056735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li A, Lu D, Zhang Y, Li J, Fang Y, Li F, et al. Critical role of aquaporin-3 in epidermal growth factor-induced migration of colorectal carcinoma cells and its clinical significance. Oncol Rep. 2013 Feb;29(2):535–40. doi: 10.3892/or.2012.2144. [DOI] [PubMed] [Google Scholar]
- 59.Liu W, Wang K, Gong K, Li X, Luo K. Epidermal growth factor enhances MPC-83 pancreatic cancer cell migration through the upregulation of aquaporin 3. Mol Med Rep. 2012 Sep;6(3):607–10. doi: 10.3892/mmr.2012.966. [DOI] [PubMed] [Google Scholar]
- 60.Colas E, Perez C, Cabrera S, Pedrola N, Monge M, Castellvi J, et al. Molecular markers of endometrial carcinoma detected in uterine aspirates. International journal of cancer Journal international du cancer. 2011 Nov 15;129(10):2435–44. doi: 10.1002/ijc.25901. [DOI] [PubMed] [Google Scholar]
- 61.de Wit M, Kant H, Piersma SR, Pham TV, Mongera S, van Berkel MP, et al. Colorectal cancer candidate biomarkers identified by tissue secretome proteome profiling. J Proteomics. 2014 Jan 10;99C:26–39. doi: 10.1016/j.jprot.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 62.Hsu I, Chuang KL, Slavin S, Da J, Lim WX, Pang ST, et al. Suppression of ERbeta signaling via ERbeta knockout or antagonist protects against bladder cancer development. Carcinogenesis. 2013 Nov 30; doi: 10.1093/carcin/bgt348. [DOI] [PubMed] [Google Scholar]
- 63.Gakiopoulou H, Korkolopoulou P, Levidou G, Thymara I, Saetta A, Piperi C, et al. Minichromosome maintenance proteins 2 and 5 in non-benign epithelial ovarian tumours: relationship with cell cycle regulators and prognostic implications. Br J Cancer. 2007 Oct 22;97(8):1124–34. doi: 10.1038/sj.bjc.6603992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dalgin GS, Drever M, Williams T, King T, DeLisi C, Liou LS. Identification of novel epigenetic markers for clear cell renal cell carcinoma. J Urol. 2008 Sep;180(3):1126–30. doi: 10.1016/j.juro.2008.04.137. [DOI] [PubMed] [Google Scholar]
- 65.Li WJ, Zhong SL, Wu YJ, Xu WD, Xu JJ, Tang JH, et al. Systematic expression analysis of genes related to multidrug-resistance in isogenic docetaxel- and adriamycin-resistant breast cancer cell lines. Mol Biol Rep. 2013 Nov;40(11):6143–50. doi: 10.1007/s11033-013-2725-x. [DOI] [PubMed] [Google Scholar]
- 66.Hwang JE, Hong JY, Kim K, Kim SH, Choi WY, Kim MJ, et al. Class III beta-tubulin is a predictive marker for taxane-based chemotherapy in recurrent and metastatic gastric cancer. BMC cancer. 2013;13:431. doi: 10.1186/1471-2407-13-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pasini A, Paganelli G, Tesei A, Zoli W, Giordano E, Calistri D. Specific Biomarkers Are Associated with Docetaxeland Gemcitabine-Resistant NSCLC Cell Lines. Transl Oncol. 2012 Dec;5(6):461–8. doi: 10.1593/tlo.12256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Maynadier M, Chambon M, Basile I, Gleizes M, Nirde P, Gary-Bobo M, et al. Estrogens promote cell-cell adhesion of normal and malignant mammary cells through increased desmosome formation. Mol Cell Endocrinol. 2012 Nov 25;364(1–2):126–33. doi: 10.1016/j.mce.2012.08.016. [DOI] [PubMed] [Google Scholar]
- 69.Valladares-Ayerbes M, Diaz-Prado S, Reboredo M, Medina V, Lorenzo-Patino MJ, Iglesias-Diaz P, et al. Evaluation of plakophilin-3 mRNA as a biomarker for detection of circulating tumor cells in gastrointestinal cancer patients. Cancer Epidemiol Biomarkers Prev. 2010 Jun;19(6):1432–40. doi: 10.1158/1055-9965.EPI-10-0123. [DOI] [PubMed] [Google Scholar]
- 70.Hu K, Law JH, Fotovati A, Dunn SE. Small interfering RNA library screen identified polo-like kinase-1 (PLK1) as a potential therapeutic target for breast cancer that uniquely eliminates tumor-initiating cells. Breast Cancer Res. 2012;14(1):R22. doi: 10.1186/bcr3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wierer M, Verde G, Pisano P, Molina H, Font-Mateu J, Di Croce L, et al. PLK1 signaling in breast cancer cells cooperates with estrogen receptor-dependent gene transcription. Cell reports. 2013 Jun 27;3(6):2021–32. doi: 10.1016/j.celrep.2013.05.024. [DOI] [PubMed] [Google Scholar]
- 72.Maire V, Nemati F, Richardson M, Vincent-Salomon A, Tesson B, Rigaill G, et al. Polo-like kinase 1: a potential therapeutic option in combination with conventional chemotherapy for the management of patients with triple-negative breast cancer. Cancer Res. 2013 Jan 15;73(2):813–23. doi: 10.1158/0008-5472.CAN-12-2633. [DOI] [PubMed] [Google Scholar]
- 73.King SI, Purdie CA, Bray SE, Quinlan PR, Jordan LB, Thompson AM, et al. Immunohistochemical detection of Polo-like kinase-1 (PLK1) in primary breast cancer is associated with TP53 mutation and poor clinical outcom. Breast Cancer Res. 2012;14(2):R40. doi: 10.1186/bcr3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zou J, Rezvani K, Wang H, Lee KS, Zhang D. BRCA1 downregulates the kinase activity of Polo-like kinase 1 in response to replication stress. Cell cycle. 2013 Jun 19;12(14) doi: 10.4161/cc.25349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kaneko N, Yamanaka K, Kita A, Tabata K, Akabane T, Mori M. Synergistic antitumor activities of sepantronium bromide (YM155), a survivin suppressant, in combination with microtubule-targeting agents in triple-negative breast cancer cells. Biol Pharm Bull. 2013;36(12):1921–7. doi: 10.1248/bpb.b13-00515. [DOI] [PubMed] [Google Scholar]
- 76.Cheng Y, Holloway MP, Nguyen K, McCauley D, Landesman Y, Kauffman MG, et al. XPO1 (CRM1) inhibition represses STAT3 activation to drive a survivin-dependent oncogenic switch in triple negative breast cancer. Molecular cancer therapeutics. 2014 Jan 15; doi: 10.1158/1535-7163.MCT-13-0416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhao YC, Wang Y, Ni XJ, Li Y, Wang XM, Zhu YY, et al. Clinical significance of Smac and survivin expression in breast cancer patients treated with anthracyclinebased neoadjuvant chemotherapy. Mol Med Rep. 2014 Feb;9(2):614–20. doi: 10.3892/mmr.2013.1842. [DOI] [PubMed] [Google Scholar]
- 78.Petrarca CR, Brunetto AT, Duval V, Brondani A, Carvalho GP, Garicochea B. Survivin as a predictive biomarker of complete pathologic response to neoadjuvant chemotherapy in patients with stage II and stage III breast cancer. Clin Breast Cancer. 2011 Apr;11(2):129–34. doi: 10.1016/j.clbc.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 79.Zhang M, Zhang X, Zhao S, Wang Y, Di W, Zhao G, et al. Prognostic value of survivin and EGFR protein expression in triple-negative breast cancer (TNBC) patients. Target Oncol. 2013 Nov 15; doi: 10.1007/s11523-013-0300-y. [DOI] [PubMed] [Google Scholar]
- 80.Wang QP, Wang Y, Wang XD, Mo XM, Gu J, Lu ZY, et al. Survivin up-regulates the expression of breast cancer resistance protein (BCRP) through attenuating the suppression of p53 on NF-kappaB expression in MCF-7/5-FU cells. The international journal of biochemistry & cell biology. 2013 Sep;45(9):2036–44. doi: 10.1016/j.biocel.2013.06.026. [DOI] [PubMed] [Google Scholar]
- 81.Flanagan JM, Wilhelm-Benartzi CS, Metcalf M, Kaye SB, Brown R. Association of somatic DNA methylation variability with progression-free survival and toxicity in ovarian cancer patients. Annals of oncology: official journal of the European Society for Medical Oncology / ESMO. 2013 Nov;24(11):2813–8. doi: 10.1093/annonc/mdt370. [DOI] [PubMed] [Google Scholar]
- 82.Boudreau A, Tanner K, Wang D, Geyer FC, Reis-Filho JS, Bissell MJ. 14-3-3 sigma stabilizes a complex of soluble actin and intermediate filament to enable breast tumor invasion. Proc Natl Acad Sci U S A. 2013 Oct 8;110(41):E3937–44. doi: 10.1073/pnas.1315022110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang EY, Cristofanilli M, Robertson F, Reuben JM, Mu Z, Beavis RC, et al. Genome wide proteomics of ERBB2 and EGFR and other oncogenic pathways in inflammatory breast cancer. J Proteome Res. 2013 Jun 7;12(6):2805–17. doi: 10.1021/pr4001527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gheibi A, Kazemi M, Baradaran A, Akbari M, Salehi M. Study of promoter methylation pattern of 14-3-3 sigma gene in normal and cancerous tissue of breast: A potential biomarker for detection of breast cancer in patients. Adv Biomed Res. 2012;1:80. doi: 10.4103/2277-9175.102990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zurita M, Lara PC, del Moral R, Torres B, Linares-Fernandez JL, Arrabal SR, et al. Hypermethylated 14-3-3-sigma and ESR1 gene promoters in serum as candidate biomarkers for the diagnosis and treatment efficacy of breast cancer metastasis. BMC cancer. 2010;10:217. doi: 10.1186/1471-2407-10-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gasnereau I, Boissan M, Margall-Ducos G, Couchy G, Wendum D, Bourgain-Guglielmetti F, et al. KIF20A mRNA and its product MKlp2 are increased during hepatocyte proliferation and hepatocarcinogenesis. The American journal of pathology. 2012 Jan;180(1):131–40. doi: 10.1016/j.ajpath.2011.09.040. [DOI] [PubMed] [Google Scholar]
- 87.Imai K, Hirata S, Irie A, Senju S, Ikuta Y, Yokomine K, et al. Identification of HLA-A2-restricted CTL epitopes of a novel tumour-associated antigen, KIF20A, overexpressed in pancreatic cancer. Br J Cancer. 2011 Jan 18;104(2):300–7. doi: 10.1038/sj.bjc.6606052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yamashita J, Fukushima S, Jinnin M, Honda N, Makino K, Sakai K, et al. Kinesin family member 20A is a novel melanoma-associated antigen. Acta Derm Venereol. 2012 Nov;92(6):593–7. doi: 10.2340/00015555-1416. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Transcripts identified in the L-R mouse TMG microarray are listed and the side with elevated expression is indicated (A). Results from pathway analysis of microarray data also are indicated (B).
Primer sets used for qRT-PCR are listed. For experiments that were confirmed by alternate primer sets, the primers used to generate the results presented in the Figures are indicated (*).
SYBR Green-based qRT-PCR was used to assess gene expression in L-R paired 4-week TMGs (A), 10-week TMGs (B), 4-week IMGs (C), and 10-week IMGs (D). For 4-week TMGs, data were normalized relative to WT right-sided glands for all genes except Notch-1, ERα, and GPRC5D, which were normalized relative to WT left-sided glands. Comparison of fold differences between WT and MMTV-cNeuTg/Tg glands indicated that FoxM1, Gata-3, Notch-1, CD24, GPRC5D, and Stmn-1 were independently up or down-regulated in left versus right-sided glands. For 10-week TMGs, data were normalized relative to WT left-sided glands for all genes except FoxM1, Gata-3, Nodal, and Pitx2, which were normalized relative to WT right-sided glands. Comparison of fold differences between WT and MMTV-cNeuTg/Tg glands indicated that FoxM1, Gata-3, Notch-1, CD24, GPRC5D, and Pitx2 were independently up or down-regulated in left versus right-sided glands. For 4-week IMGs, data were normalized relative to WT right-sided glands, except for Notch-1, ERα, and Stmn-1, which were normalized to WT left-sided glands. Comparison of fold differences between WT and MMTV-cNeuTg/Tg glands indicated that all genes examined were similarly up or down-regulated in left versus right-sided glands except Notch-1, which was slightly more elevated in MMTV-cNeuTg/Tg right-sided glands compared to WT. SYBR Green-based qRT-PCR showed gene expression in 10-week left vs. right IMGs (D). For 10-week IMGs, data were normalized relative to WT right-sided glands for all genes. Comparison of fold differences between WT and MMTV-cNeuTg/Tg glands indicated that FoxM1, Gata-3, Notch-1, ERα, and CD24 were independently up or down-regulated in left versus right-sided glands. Bars represent mean ± SEM of ≥ 3 mice, *p<0.05; **p<0.01; *** p<0.001, two-tailed paired student’s t-tests.