Abstract
Glutamatergic signaling through N-methyl-D-aspartate receptors (NMDARs) is important for neuronal development and plasticity and is often dysregulated in psychiatric disorders. Mice mutant for the transcription factor Sp4 have reduced levels of NMDAR subunit 1 (NR1) protein, but not mRNA, and exhibit behavioral and memory deficits (Zhou et al., 2010). In developing cerebellar granule neurons (CGNs), Sp4 controls dendrite patterning (Ramos et al., 2007). Sp4 target genes that regulate dendrite pruning or NR1 levels are not known. Here we report that Sp4 activates transcription of Nervous Wreck 2 (Nwk2; also known as Fchsd1) and, further, that Nwk2, an F-BAR-domain containing protein, mediates Sp4-dependent regulation of dendrite patterning and cell surface expression of NR1. Knockdown of Nwk2 in CGNs increased primary dendrite number, phenocopying Sp4 knockdown, and exogenous expression of Nwk2 in Sp4-depleted neurons rescued dendrite number. We observed that acute Sp4 depletion reduced levels of surface, but not total, NR1, and this was rescued by Nwk2 expression. Furthermore, expression of Nr1 suppressed the increase in dendrite number in Sp4- or Nwk2-depleted neurons. We previously reported that Sp4 protein levels were reduced in cerebellum of subjects with bipolar disorder (BD) (Pinacho et al., 2011). Here we report that Nwk2 mRNA and NR1 protein levels were also reduced in postmortem cerebellum of BD subjects. Our data suggest a role for Sp4-regulated Nwk2 in NMDAR trafficking and identify an Sp4-Nwk2-NMDAR1 pathway that regulates neuronal morphogenesis during development and may be disrupted in bipolar disorder.
INTRODUCTION
Glutamate receptors, including NMDARs, have profound effects on neuronal development. NMDAR signaling regulates dendrite patterning, which determines the way a neuron integrates inputs (Cline, 2001), and contributes to synaptic plasticity including long-term potentiation (LTP) and depression (LTD) (Luscher and Malenka, 2012). In both animal models and human subjects, NMDA receptor inhibition causes psychosis and cognitive impairments resembling symptoms of schizophrenia (Krystal et al., 1994; Olney et al., 1999; Moghaddam and Javitt, 2012). Altered NMDAR function has also been proposed to contribute to other psychiatric disorders including bipolar disorder and autism (Carlson, 2012; Fountoulakis, 2012).
NMDARs are composed of a common NR1 subunit assembled with NR2 or NR3 subunits to form a functional ion channel. Spatially and temporally regulated expression of NMDAR subunits leads to receptors with different channel and trafficking properties (Paoletti et al., 2013). Trafficking of NMDARs is highly regulated during development and in response to experience and the mechanisms regulating spatial distribution of NMDARs have important consequences for neuronal physiology (Perez-Otano and Ehlers, 2005; Lau and Zukin, 2007).
Proteins bearing FER/Cip4 homology-Bin/amphiphysin/Rvs (F-BAR) and Src homology 3 (SH3) domains play important roles in neuronal development and function through regulation of membrane curvature, actin cytoskeleton and endosomal trafficking (Aspenstrom et al., 2006). Nervous wreck (Nwk) is an F-BAR and SH3 domain containing protein first identified in Drosophila where mutations in Nwk resulted in paralysis due to synaptic overgrowth at the neuromuscular junction (Coyle et al., 2004). Nwk and its mammalian homologs, Nwk1 and Nwk2 (also known as Fchsd2 and Fchsd1) have been shown to regulate actin dynamics and induce membrane protrusions in cells (Coyle et al., 2004; Becalska et al., 2013; Cao et al., 2013). Current data suggests that Drosophila Nwk controls synaptic growth through effects on endosomal trafficking of receptors such as the BMP receptor homolog Tkv (Itoh et al., 2005; O’Connor-Giles et al., 2008; Rodal et al., 2008; Rodal et al., 2011). Although functions of F-BAR/SH3-containing proteins such as pacsin1/syndapin1 and WRP/srGAP3, which has been implicated in mental retardation, are well established, the functions of Nwk homologs in mammalian neurons has not been determined.
Transcription factor Sp4 controls dendrite patterning in developing cerebellar granule neurons by limiting branch formation and promoting activity-dependent pruning (Ramos et al., 2007). In humans, variations at the Sp4 locus are associated with schizophrenia and bipolar disorder (Kelsoe et al., 2006; Zhou et al., 2009; Tam et al., 2010). We have reported that Sp4 protein level is reduced in cerebellum and cerebral cortex of subjects with bipolar disorder and associated with severe negative symptoms in patients with schizophrenia (Pinacho et al., 2011; Pinacho et al., 2013). Mice with reduced expression of Sp4 have deficits in contextual and spatial memory, reduced LTP and increased sensitivity to the non-competitive NMDAR antagonist, ketamine (Zhou et al., 2005; Zhou et al., 2010; Ji et al., 2013). Interestingly, Sp4 hypomorphic mice have reduced levels of NR1 protein, but not mRNA, which may contribute to some of these phenotypes (Zhou et al., 2010). Sp4 is a zinc finger transcription factor that binds a GC-rich sequence found in the promoters of many genes (Black et al., 2001), but little is known about the target genes of Sp4 that regulate dendrite development or NMDAR signaling.
Here we report that Nwk2 is an activation target of transcription factor Sp4 that regulates dendrite patterning and NR1 levels in developing cerebellar granule neurons. We found reduced Nwk2 mRNA and protein in CGNs from Sp4 hypomorphs and we determined that depletion of Nwk2 impaired pruning of primary dendrites and reduced surface expression of NR1 in CGNs. We also report that levels of NWK2 mRNA and NR1 protein were reduced in postmortem cerebellum of bipolar disorder subjects. These studies identify an Sp4-Nwk2-NMDAR1 pathway that regulates neuronal morphogenesis in development and is disrupted in bipolar disorder.
MATERIALS AND METHODS
Animals
Hypomorphic Sp4 (Sp4neo-/-) mice in the Black Swiss background were a gift from Dr. Xianjin Zhou at the University of California, San Diego (Zhou et al., 2005). All studies were in this background except RNA for the microarray analysis was from F1 and N1 Black Swiss × CD1 mice. Long Evans Rats were obtained from Charles River. Animals were maintained in American Association for Accreditation of Laboratory Animal Care approved facilities at Tufts University. All protocols for the use of vertebrate animals were approved by the Tufts University Institutional Animal Care and Use Committee.
Cell culture and transfection
CGNs were obtained from postnatal day 5 (mouse) or 6 (rat) pups as described (Bilimoria and Bonni, 2008). Neurons were maintained in Basal Minimum Essential medium supplemented with 10% fetal calf serum, 25 mM KCl, penicillin (50 U/ml), streptomycin (50 μ/ml) and 2 mM glutamine. Cells were transfected at DIV2 with the indicated plasmids and a vector expressing Bcl-xL by calcium phosphate precipitation. Viability of transfected cells in the absence of Bcl-xL was determined at DIV6 by staining with DAPI and visual inspection for fragmented or pyknotic nuclei. RNA and protein were isolated and morphology analysed at DIV6. For biotin labeling studies, CGNs were infected at DIV0, selected and analysed at DIV5. Neuro2A cells were plated at 1.8 × 104 cells/cm2 24 h before transfection with Lipofectamine 2000 (Invitrogen).
Plasmids
Nwk2-Flag was expressed from the CMV promoter in the pcDNA vector. shRNAs were expressed from the U6 promoter in pLKO.1-GFP plasmid (modified from pLKO.1-puro (Stewart et al., 2003)). Sequences targeted by short hairpin RNAs were: Nwk2 #1 gcaatgagtacctgctaaatt for mouse, gcaacgagtacctgctaaatt for rat; Nwk2 #2 gctttgtccctgagcgatatc for mouse, gctttgttcctgaacgatatc for rat; Sp4 gggtgctgcaggtgttcaagt as described in (Ramos et al., 2007), and control gacgacccgcaggcgatacgt. Silent mutations introduced to make RNAi-resistant cDNAs were: Nwk2 gcaatgagtacTtATtGaatt and Sp4 gggCgcGgcGggtgttcaagt.
mRNA analysis
For microarray analysis, total RNA was prepared from postnatal day 21 cerebella of 3 pairs of gender-matched WT and Sp4 hypomorphic mouse littermates using Trizol (Invitrogen) and RNeasy Kit (QiAGEN). Gene expression analysis was performed using the MouseWG-6 v2.0 Expression BeadChip (Illumina). Data were analysed through the use of IPA (Ingenuity®Systems, www.ingenuity.com). The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO Series accession number GSE53061 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE53061). For RT-qPCR, first strand cDNA synthesis from 2 μg total RNA was carried out with Oligo dT primer using Superscript III reverse transcriptase (Invitrogen). Samples were analyzed by quantitative real-time PCR on a CFX96 real-time PCR detection system (BioRad) using the ΔΔCt method. Expression was normalized to 2 internal controls: Gapdh and Hprt1. At least 3 technical repeats were performed using at least 3 independent biological replicates. Primers used were: Nwk1-F: CTCACAGTCCTTTAAACGCC; Nwk1-R: CTTGTGTAACTTTCACCTTCCTC; Nwk2-F: TGAGCTGTCAGAATACTTGAG; Nwk2-R: GTTCCCAGTTTACCTGAGAC; NR1-F: GTACCCATGTCATCCCAAAT; NR1-R: TCTGGTGGACATCTGGTATC NR2A-F:GCAGAGAATAGGACCCACTCCCTAA;NR2A-R:TGGCATGTGGCCCGGCTTGA NR2B-F:CAACGGGACCTGGAACGGCAT; NR2B-R: AAGGGCACAGAGAAGTCAACCACC Gapdh-F: CTGAGGACCAGGTTGTGTCC; Gapdh-R: CATTGTCATACCAGGAAATGAGC. Hprt1-F: CTCTCGAAGTGTTGGATACAG; Hprt1-R: ACAAACGTGATTCAAATCCC
Postmortem human brain tissue samples
Postmortem human brain samples from the cerebellum of subjects with bipolar disorder (n=10) and control subjects with no history of psychiatric episodes (n=10) were obtained from the UPV/EHU brain collection. Demographic data of this cohort is detailed in Table 1 (Pinacho et al., 2011). Samples were obtained at autopsy by forensic pathologists under research policies with postmortem samples. All deaths were subjected to retrospective analysis for previous medical diagnosis. Subjects with antemortem criteria for bipolar disorder according to the Diagnostic and Statistical Manual of Mental Disorders (DSM-IV) were matched to control subjects based on gender, age and postmortem delay (PMD). Toxicological screening for antipsychotics, antidepressants, and other drugs was performed at the National Institute of Toxicology, Madrid, Spain. Specimens of the cerebellum, extending from the pial surface to white matter and only including gray matter were dissected and stored at −80°C.
Table 1. Gene list with differential expression in Sp4 hypomorphic mouse cerebellum.
| Genes with decreased expression | Fold change | P Value |
|---|---|---|
| Gcnt1, glucosaminyl (N-acetyl) transferase 1, core 2 | −5.57 | 2.53E-05 |
| Ccl21b, chemokine (C-C motif) ligand 21B (leucine) | −4.95 | 1.82E-02 |
| Nwk2, FCH and double SH3 domains 1 | −4.15 | 1.84E-04 |
| Ccl21c, chemokine (C-C motif) ligand 21C (leucine) | −3.91 | 1.49E-02 |
| Prkag2, protein kinase, AMP-activated, gamma 2 non-catalytic subunit | −3.38 | 1.46E-02 |
| Znf24, zinc finger protein 24 | −3.33 | 8.16E-03 |
| Comp, cartilage oligomeric matrix protein | −3.28 | 3.78E-05 |
| Spata7, spermatogenesis associated 7 | −3.18 | 2.53E-02 |
| Dhcr24, 24-dehydrocholesterol reductase | −2.73 | 1.84E-02 |
| Tspan17, tetraspanin 17 | −2.55 | 2.95E-04 |
| Zfp191, zinc finger protein 191 | −2.44 | 2.82E-02 |
| Il17re, interleukin 17 receptor E | −2.35 | 3.00E-03 |
| Commd2, COMM domain containing 2 | −2.28 | 7.94E-03 |
| Tmem159, transmembrane protein 159 | −2.28 | 3.56E-05 |
| Ramp3, receptor (G protein-coupled) activity modifying protein 3 | −2.27 | 6.21E-04 |
| Top1mt, topoisomerase (DNA) I, mitochondrial | −2.23 | 8.88E-04 |
| Stxbp2, syntaxin binding protein 2 | −2.13 | 1.22E-03 |
| Cited4, Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4 | −2.13 | 7.75E-05 |
| Psmg2, proteasome (prosome, macropain) assembly chaperone 2 | −2.13 | 4.59E-03 |
| Rom1, retinal outer segment membrane protein 1 | −2.12 | 9.25E-04 |
| Echdc2, enoyl Coenzyme A hydratase domain containing 2 | −2.12 | 1.35E-02 |
| Cd59a, CD59a antigen | −2.09 | 1.64E-02 |
| Irf6, interferon regulatory factor 6 | −2.08 | 5.22E-04 |
| Sp4 | −2.08 | 7.60E-04 |
| Cdh15, cadherin 15 | −2.06 | 6.73E-03 |
| Ly6d, lymphocyte antigen 6 complex, locus D | −2.05 | 7.59E-03 |
| Bmp8a, bone morphogenetic protein 8a | −2.00 | 1.85E-03 |
|
| ||
| Genes with increased expression | Fold change | P Value |
|
| ||
| Zfp35, zinc finger protein 35 | 5.36 | 4.60E-03 |
| Pnlip, pancreatic lipase | 5.19 | 1.86E-02 |
| Mkks, McKusick-Kaufman syndrome | 3.64 | 2.01E-02 |
| Rbbp4, retinoblastoma binding protein 4 | 3.59 | 3.77E-02 |
| Pcdh9, protocadherin 9 | 3.56 | 9.65E-05 |
| Btg4, B-cell translocation gene 4 | 3.24 | 4.59E-05 |
| Serpinb2, serpin peptidase inhibitor, clade B (ovalbumin), member 2 | 3.04 | 1.88E-02 |
| Atp6v1d, ATPase, H+ transporting, lysosomal 34kDa, V1 subunit D | 2.99 | 8.17E-04 |
| Cnksr1, connector enhancer of kinase suppressor of Ras 1 | 2.63 | 8.32E-04 |
| Plekhf2, pleckstrin homology domain containing, family F (with FYVE domain) member 2 | 2.60 | 6.86E-05 |
| Cntfr, ciliary neurotrophic factor receptor | 2.52 | 2.25E-02 |
| Cdk5rap1, CDK5 regulatory subunit associated protein 1 | 2.46 | 1.10E-04 |
| Esd, esterase D | 2.28 | 3.59E-04 |
| Efha1, EF-hand domain family, member A1 | 2.23 | 3.37E-04 |
| Rapgef5, Rap guanine nucleotide exchange factor (GEF) 5 | 2.23 | 5.85E-03 |
| Taf12, TAF12 RNA polymerase II, TATA box binding protein (TBP)-associated factor | 2.18 | 2.23E-02 |
| Got1l1, glutamic-oxaloacetic transaminase 1 -like 1 | 2.07 | 3.37E-03 |
| Slco1a4, solute carrier organic anion transporter family, member 1a4 | 2.01 | 1.28E-03 |
| Xlr4a, X-linked lymphocyte-regulated 4A | 2.00 | 4.17E-03 |
Real-time Quantitative RT-PCR (RT-qPCR) of human samples
Total RNA was extracted using Trizol reagent (Sigma-Aldrich). RNA Integrity number (RIN) was measured with an Agilent Bioanalyzer (Agilent Technologies). First strand cDNA was synthesized using SuperScript III (Invitrogen). Applied Biosystems Taqman® master mix formulation for gene expression, probe and primers were used for RT-qPCR in a randomized and blind manner using the ΔΔCt method as described (Pinacho et al., 2011). Assay identification primers were Hs00607744_m1 for NWK2 and Hs00609557_m1 for NR1 (TaqMan® Gene Expression Assays, Applied Biosystems). Expression levels were referred to a reference sample and to Beta-glucuronidase (GUSB) (Applied Biosystems), as the most stable reference gene for this cohort (Pinacho et al., 2011).
Immunoblotting
Lysates from mouse tissue were prepared using Tissue extraction reagent II (Invitrogen). Lysates from cultured cells were prepared in lysis buffer (50 mM Tris pH 7.4, 150 mM NaCl containing 1% NP-40 and Protease Inhibitor cocktail (Roche)). Surface proteins on intact primary CGNs in culture were biotin labeled on ice for 30 min and isolated using Cell Surface Protein Isolation Kit (Thermo Scientific, Pierce). Human cerebellum samples were homogenized on ice with NP40 lysis buffer as described (Pinacho et al., 2011) and 50 μg of human protein lysates were analyzed in a blinded manner. Lysates were resolved by SDS/PAGE and immunoblotted with indicated antisera including anti-Sp4 (Santa Cruz), anti-Nwk2 (Abcam), anti-NR1 (BD Pharmingen), NR2A (Invitrogen), NR2B (Abcam), GluR2 (NeuroMab) and anti-Gapdh (Chemicon International or Millipore). For human samples, densitometric quantification was performed using Quantity One software (BioRad) in duplicate samples. Values were normalized to GAPDH and to a standard sample (bipolar subject).
Immunofluorescence and Immunohistochemistry
For morphometric analysis, cells were fixed in 4% paraformaldehyde, permeabilized with 0.25% TritonX-100, then labelled with anti-GFP followed by Alexa Fluor® Dye conjugated secondary antibodies and mounted with Prolong Gold antifade reagent with DAPI (Molecular Probes). Images of individual transfected neurons with no overlapping processes from other transfected neurons were captured randomly in a blinded manner at 400× magnification using a Nikon A1R confocal microscope. For every condition, quantitation was performed in at least 3 independent experiments, n = 48-63 per condition. Axons were identified based on morphology including protrusion from the cell body, overall length and characteristic T-shape.
For staining of surface NR1, neurons were fixed in 4% paraformaldehyde for 10 min on ice, but were not permeabilized before incubation with anti-NMDAR1 (BD Pharmingen). Non-permeabilized transfected neurons were identified by GFP autofluorescence. For staining of total NR1, cells were fixed with 4% PFA and permeablized with 0.25% Triton X-100. In these conditions, transfected cells were labelled by anti-GFP. The fluorescence intensity of NR1 was measured using Volocity software and the intensity of NR1 in transfected neurons was normalized to the mean intensity of surrounding untransfected neurons in the field after substraction of background. The relative fluorescence among groups was compared, n = 48-63 per condition.
Chromatin immunoprecipitation
Chromatin immunoprecipitation was performed following Upstate Biotechnology protocol with modifications. 2 × 107 cerebellar granule neurons were crosslinked with 1% formaldehyde for 15 min at 37 °C. Chromatin was sonicated to between 500 and 1000 bp and immunoprecipitated with 3 μg of anti-Sp4 (Santa Cruz) or normal rabbit IgG. Immunoprecipitated DNA was amplified by real-time PCR using specific primers as indicated. Real-time PCR was performed using the SsoFast™ EvaGreen Supermix (Bio-Rad). 10% of total starting material was precipitated and used as input.
Primers for ChIP: Nwk2 promoter-F: ACAGGTTGAGTTCCACTGCTG; Nwk2 promoter-R: CAAAGGCTGCCAGACACTCTA Nwk2 3′UTR-F: CTGGTAACCCATCAGCACAGT; Nwk2 3′UTR-R:TGGAGGTGGGTGAACTGTAAG
Statistical Analysis
For RT-qPCR studies from mice, two-tailed t-test was used to determine the significance between groups (WT vs. Sp4 hypomorph). For morphometric and immunofluorescence assays, statistical differences were determined by ANOVA followed by a post hoc Tukey test.
For postmortem human brain study, Grubbs test and D’Agostino & Pearson omnibus normality test were carried out to detect outliers and to test whether the variables followed a normal distribution, respectively. Non-parametric Mann-Whitney test was used to compare control and bipolar disorder groups and Spearman coefficient was performed for correlations between NWK2 mRNA and NR1 or SP protein levels. Bivariate analyses were carried out to detect association of our variables with potential confounding factors (age, gender, postmortem delay, toxicology, pH and RIN), using Mann-Whitney test for two groups, Kruskal-Wallis for more than two groups and Spearman correlation for quantitative variables. Statistical analysis was performed with GraphPad Prism version 5.00 and SPSS 19. All statistical tests were two-tailed unless otherwise specified and significance level was set to 0.05.
RESULTS
Transcription factor Sp4 activates expression of Nwk2 in neurons
Transcription factor Sp4 is enriched in neurons where it contributes to neuronal development and behavior (Zhou et al., 2005; Zhou et al., 2005; Mao et al., 2007; Ramos et al., 2007), but little is known about genes regulated by Sp4 in brain. In order to identify Sp4 target genes, we compared gene expression between WT and Sp4 hypomorph cerebellum at postnatal day 21. Sp4 hypomorphs express dramatically reduced levels of Sp4 (Figure 1D and (Zhou et al., 2005). By genome-wide expression profiling using microarray, we identified 161 genes decreased and 112 genes increased in expression in Sp4 hypomorph cerebellum compared to WT, using a cutoff of p<0.05 and fold change>1.5 after quantile normalization. A list of genes whose expression was up- or down-regulated two-fold or more is shown in Table 1. Many candidate Sp4 target genes have functions related to nervous system development or disease, with an enrichment of genes related to neurite morphogenesis (Figure 1A). Investigation of one target gene is presented here.
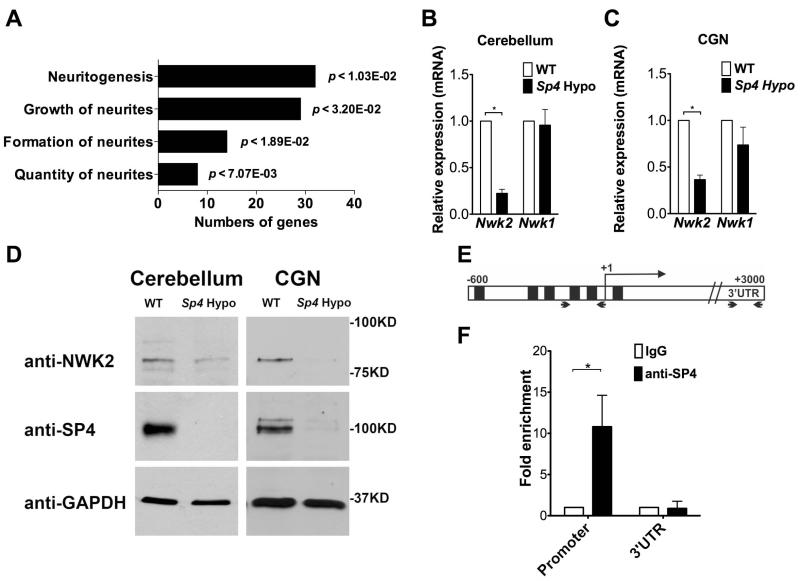
Figure 1. Transcription factor Sp4 regulates expression of Nwk2.
(A) Genes differentially expressed in WT and Sp4 hypomorph cerebellum (p<0.05) were analysed with Ingenuity Systems Interactive Pathway Analysis tool to identify top regulated annotations in nervous system development and function. (B) Total RNA from P21 cerebellum of WT (open bar) and Sp4 hypomorphic (shaded bar) mice was analysed by RT qPCR. Expression of Nwk2 and Nwk1 relative to WT set at 1 is shown. Values represent mean ± SEM from experiments performed in triplicate and normalized to Gapdh and Hprt expression levels. Student’s two-tailed t-test was used to determine the significance between groups. (*) P value <0.05. (C) RNA from primary cultured cerebellar granule neurons (CGNs) from WT and Sp4 hypomorphic mice was analysed by RT-qPCR as described in (B). (D) Protein lysates from cerebellum (left) or CGNs (right) of WT and Sp4 hypomorphic mice were immunoblotted for Nwk2, Sp4 and GAPDH as indicated. (E) A schematic diagram of the Nwk2 gene with shaded boxes representing predicted Sp4 binding sites in the promoter region. Arrows indicate the positions of PCR primers. (F) Chromatin immunoprecipitation (ChIP) from CGNs using antibody against Sp4 or normal rabbit IgG as control. Immunoprecipitated DNA was analyzed by real-time PCR with primers that amplified fragments in the Nwk2 promoter or the 3′UTR. Signal for anti-Sp4 relative to IgG set at 1 is shown.
We identified Nwk2 (also known as Fchsd1) as a candidate Sp4 target gene whose expression was reduced in Sp4 hypomorphic mice (Table 1). Nwk2 is a homolog of Nervous Wreck (Nwk) in Drosophila, an F-BAR and SH3-domain containing protein that acts to limit synaptic growth at the neuromuscular junction (Coyle et al., 2004). We confirmed Nwk2 mRNA and protein were reduced in cerebellum and primary cultured CGNs from Sp4 hypomorphic mice (Figure 1B, C and D). Expression of the other Nwk homolog in mice, Nwk1 (Fchsd2), was not altered in the Sp4 hypomorph (Figure 1B and C). Immunohistochemistry of postnatal day 7 mouse cerebellum also revealed reduced Nwk2 expression in Sp4 mutant mice (Supplemental Figure 1). In order to determine whether reduced Nwk2 mRNA levels were likely to be a direct effect of Sp4 loss, chromatin immunoprecipitation (ChIP) was performed in CGNs. ChIP revealed Sp4 occupancy at the Nwk2 promoter, but not the 3′UTR region (Figure 1E and F). Taken together, these data show that Sp4 binds the Nwk2 promoter and is important for transcription of Nwk2 in CGNs.
Knockdown of Nwk2 in cerebellar granule neurons increased dendrite number
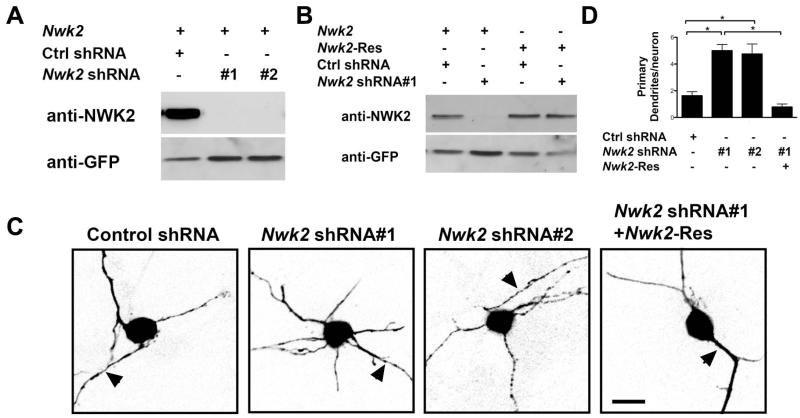
We previously reported that Sp4 promotes pruning of primary dendrites in developing CGNs (Ramos et al., 2007). In order to investigate the effect of reduced Nwk2 on dendrite patterning, we generated two shRNAs each of which robustly knocked down expression of Nwk2 (Figure 2A). We also generated an RNAi resistant form of Nwk2 (Nwk2-Res; Figure 2B). Using these reagents, we examined CGNs 6 days post-transfection to determine the effect of depleting endogenous Nwk2 in these cells. Knockdown of Nwk2 had no effect on viability as judged by the appearance of pyknotic or fragmented nuclei (88.9% +/−7.3% viable vs. 92.7% +/− 4.9% in control, p=0.67), but Nwk2 knockdown led to an increased number of primary dendrites (Figure 2 C and D). To confirm the specificity of the Nwk2 shRNA-induced dendrite phenotype, we performed a rescue experiment. Co-expression of RNAi-resistant Nwk2 suppressed the appearance of excess dendrites upon Nwk2 knockdown (Figure 2C and D). Thus, knockdown of Nwk2 in CGN led to an increased number of primary dendrites, suggesting that, similar to Sp4 (Ramos et al., 2007), Nwk2 acts to limit dendrite number in developing CGNs.
Figure 2. Knockdown of Nwk2 increased primary dendrite numbers.
(A) Lysates from Neuro2A cells cotransfected with a Nwk2-Flag plasmid and a plasmid expressing GFP together with the indicated shRNAs, were immunoblotted with antibodies against Nwk2 and GFP as a loading control. (B) Lysates from Neuro2A cells cotransfected with the indicated shRNA plasmid that also encodes GFP, together with an expression vector encoding Nwk2-Flag WT or an RNAi resistant Nwk2 (Nwk2-Res), were immunoblotted with antibodies against Nwk2 and GFP. (C, D) CGNs were transfected with the indicated Nwk2 shRNA or control shRNA plasmid and, where indicated, an expression vector encoding RNAi-resistant Nwk2. 6 days post-transfection the neurons were immunostained with anti-GFP. (C) Images of representative transfected neurons. Arrowhead indicates axon. Scale bar is 10μm. (D) Quantification of primary dendrite numbers. Values represent mean ± SEM of an experiment repeated 3 times and analysed by ANOVA followed by a post-hoc Tukey test;*P<0.05.
Sp4 regulates dendrite patterning through Nwk2
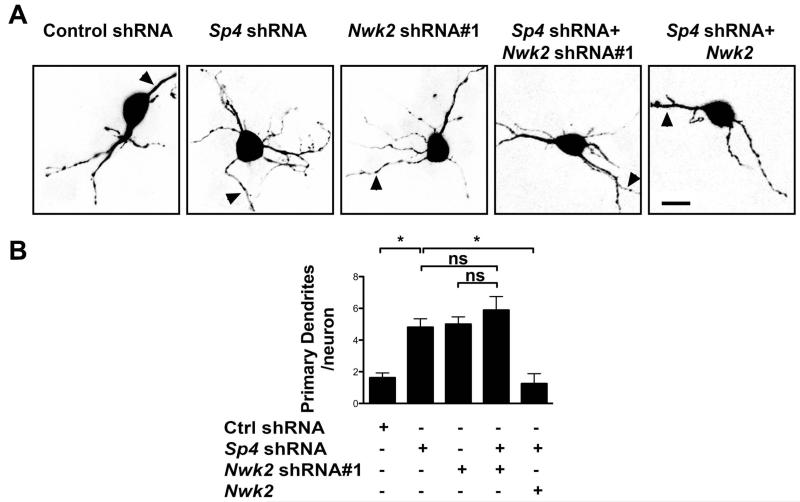
Since knockdown of either Sp4 or Nwk2 alone led to an increased number of primary dendrites (Figure 3 and (Ramos et al., 2007)), we investigated whether Sp4 and Nwk2 act in a common pathway. When Sp4 and Nwk2 were knocked down together, we did not observe any further increase in dendrite number, compared to knockdown of either Sp4 or Nwk2 alone (Figure 3). This finding is consistent with the model that Sp4 and Nwk2 operate in a common pathway to control neuronal dendrite numbers. If, in fact, Sp4-mediated activation of Nwk2 expression were important for Sp4-dependent regulation of dendrite patterning, we predicted that exogenous expression of Nwk2 would suppress the phenotype of Sp4 depletion in CGNs. Indeed, we observed that expression of Nwk2 was sufficient to reduce the appearance of excess dendrites in Sp4 knockdown neurons (Figure 3). These data support the model that Sp4 regulates transcription of Nwk2 to control dendrite number.
Figure 3. Expression of Nwk2 suppresses excess dendrite number in Sp4 knockdown neurons.
CGNs were transfected with plasmids expressing GFP together with shRNAs targeting Sp4, Nwk2 or a control (Ctrl) and, where indicated, an expression vector encoding Nwk2. 6 days post-transfection, the neurons were immunostained with anti-GFP. (A) Images of representative transfected neurons. Arrowhead indicates axon. Scale bar is 10μm. (B) Quantification of primary dendrite numbers. Values represent mean ± SEM of an experiment repeated 3 times and analysed by ANOVA followed by a post-hoc Tukey test;*P<0.05, n.s.= not significant.
Sp4-Nwk2 pathway regulates surface levels of NMDAR1
Interestingly, reduced levels of NR1 protein were observed in the hippocampus and cortex of adult Sp4 hypomorphic mice, and this has been suggested to contribute to impaired spatial learning/memory and defective LTP in these animals (Zhou et al., 2010). We determined that protein levels of NR1, NR2A and NR2B, but not another glutamate receptor, GluR2, were reduced in cultured primary CGNs from Sp4 mutants compared to WT (Supplemental Figure 2A). Although Sp4 has been suggested to regulate transcription of several NMDAR subunits (Okamoto et al., 2002; Liu et al., 2004; Priya et al., 2013), RT-qPCR analysis revealed no change in the mRNA level of Nr1, Nr2A and Nr2B in Sp4 hypomorph CGNs (Supplemental Figure 2B). Thus, consistent with previous reports examining other brain regions (Zhou et al., 2010), our data indicates that, in this animal model, Sp4 regulates NMDAR1 protein levels in neurons by a post-transcriptional mechanism.
In order to investigate a possible role of Sp4 target genes in regulation of NR1 levels, we first determined whether we could recapitulate this phenomenon in cultured neurons. We observed that acute knockdown of Sp4 in CGNs decreased the levels of NR1 protein at the cell surface, as measured by immunofluorescence of non-permeabilized cells (Figure 4A and B). Control Map2 staining confirmed that the cells were not permeabilized in our protocol (Supplemental Figure 3). The observed decrease was a specific result of Sp4 knockdown, as surface NR1 levels were rescued by expression of RNAi-resistant Sp4 (Figure 4A and B). In contrast, the immunofluorescence signal for total NR1 levels, measured in permeabilized cells, was not significantly altered upon Sp4 knockdown. As can be seen in Figure 4 (A and D), we detected a high level of NR1 immunostaining around the cell soma in these immature neurons and this is the signal we measured. Thus, the striking decrease in total NR1 levels observed in CGNs from the Sp4 mutant mouse was not recapitulated under conditions of acute Sp4 knockdown, suggesting that this phenotype depends on a more complex cellular context and/or reduction of Sp4 during an extended period. These data show that depletion of Sp4 acts cell autonomously to reduce neuronal surface levels of NR1.
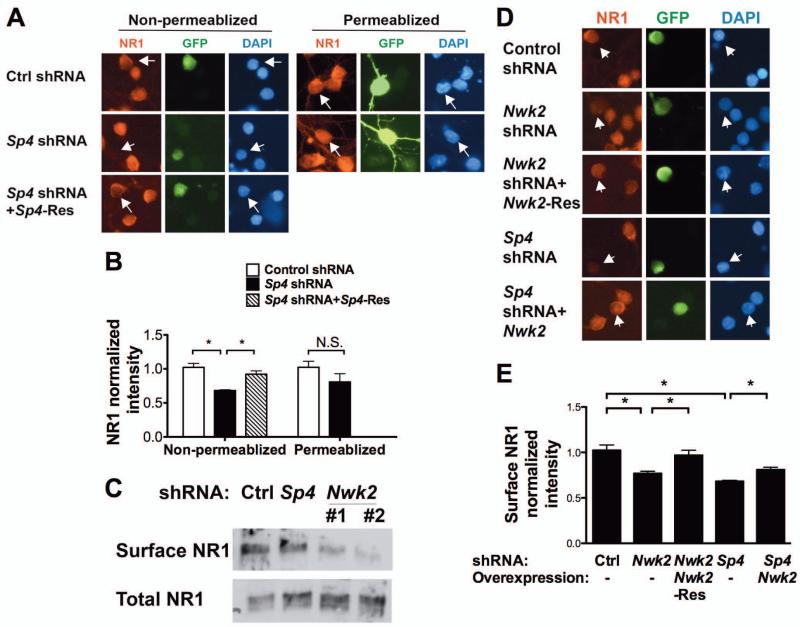
Figure 4. Sp4 regulates NMDAR1 surface levels via Nwk2.
(A,B) CGNs were transfected with plasmids expressing Sp4 shRNA and GFP and, where indicated, RNAi-resistant Sp4 (Sp4-Res). 5 days post-transfection, non-permeabilized or permeablized neurons were immunostained with antibody against NR1. Transfected cells were identified by GFP autofluorescence (non-permeabilized) or staining with anti-GFP (permeabilized). (A) Representative images of neurons are shown. Arrowheads indicate the transfected neurons. (B) Quantification of fluorescence intensity of NR1 immunostaining at the cell body normalized to untransfected neurons in the same field. (C) CGNs infected with lentivirus expressing Sp4 or Nwk2 shRNA and GFP were selected (Supplemental Figure 4) and cell surface proteins labelled with Biotin and purified. Total cell lysates (10%) and surface proteins were analysed by immunoblot with anti-NR1. (D, E) CGNs were transfected with the indicated plasmids and GFP. NR1 immunostaining in non-permeabilized cells was analysed as above. (D) Representative images of neurons are shown. Arrowheads indicate the transfected neurons. (E) Quantification of fluorescence intensity of NR1 immunostaining at the cell body normalized to untransfected neurons in the same field. (B and E) Values represent mean ± SEM of an experiment repeated 3 times and analysed by ANOVA followed by a post-hoc Tukey test; *P<0.05, n.s.= not significant.
Drosophila Nwk has been suggested to regulate synaptic growth through effects on growth factor signaling and endosomal trafficking (O’Connor-Giles et al., 2008; Rodal et al., 2008; Rodal et al., 2011). We therefore investigated the role of the Sp4 target gene Nwk2 in Sp4-dependent regulation of surface NR1 levels. Purification and analysis of biotin labeled cell surface proteins revealed that knockdown of either Sp4 or Nwk reduced the fraction of NR1 on the cell surface to 10 - 40% of control levels (Figure 4C). Immunostaining of NR1 in non-permeabilized cells also revealed that when Nwk2 was knocked down, NR1 surface levels decreased. This effect was specific, as NR1 surface levels were rescued by expression of RNAi-resistant Nwk2 (Figure 4 D and E). Notably, when we expressed Nwk2 in neurons depleted of Sp4, NR1 levels were partially restored (Figure 4 D and E). We conclude that transcription factor Sp4 modulates cell surface expression of NR1 through the regulated transcription of Nwk2 and perhaps additional targets.
NMDAR1 acts as a downstream effector of the Sp4-Nwk2 pathway in dendrite patterning
Our data indicate that Sp4 regulates Nwk2 expression to control primary dendrite numbers and surface expression of NR1. We therefore investigated the hypothesis that altered NR1 protein levels contributes to the dendrite patterning defects in Sp4 and Nwk2 depleted neurons. While transfection of an NR1 expression vector had no effect on primary dendrite number in control cells, NR1 overexpression suppressed the phenotype of excess primary dendrites observed upon Sp4 or Nwk2 depletion (Figure 5). Whether rescue by overexpressed NR1 occured because the knockdowns were partial, because pathways in addition to the Sp4-Nwk2 pathway promote surface expression of NMDAR, or some other mechanism has not been determined. Nonetheless, these data support the view that NR1 plays a major role downstream of Sp4-mediated Nwk2 expression to control dendritic patterning.
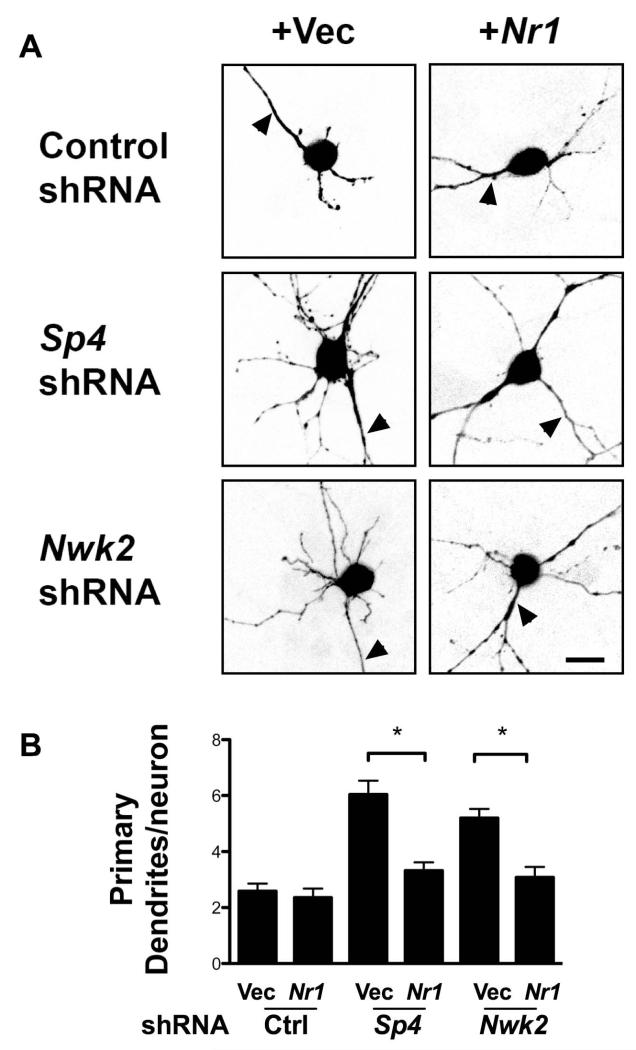
Figure 5. NMDAR1 expression rescues the dendritic phenotype of Sp4 and Nwk2 knockdowns.
CGNs were transfected with plasmids expressing GFP and shRNAs targeting Sp4 or Nwk2 or a control shRNA. Where indicated, a plasmid expressing NR1 was also transfected. 5 days post-transfection, neurons were immunostained with antibody against GFP. (A) Representative images of transfected neurons are shown. Scale bar is 10μm. (B) Quantification of primary dendrite numbers. Values represent mean ± SEM of an experiment repeated 3 times and analysed by ANOVA followed by a post-hoc Tukey test;*P<0.05.
Nwk2 mRNA and NR1 protein expression are reduced in cerebellum of bipolar disorder subjects
We have reported reduced Sp4 levels in cerebellum of BD subjects (Pinacho et al., 2011). We therefore investigated if other components of the Sp4-Nwk2-NR1 pathway are also altered in BD cerebellum. We determined NWK2 mRNA expression levels in the postmortem cerebellum of 10 BD subjects and 10 matched healthy controls. The two groups did not show any significant differences in sociodemographic and tissue-related measures (Table 2). We found that NWK2 mRNA levels were significantly reduced in the postmortem cerebellum in bipolar disorder (Figure 6A). In addition, we found that SP4 protein levels in the same cohort, reported in (Pinacho et al., 2011), correlated with NWK2 mRNA levels in cerebellum (Spearman’s r=0.511, p<0.05, n=20). We used bivariate analyses to evaluate the influence of potential confounders in the significant changes and/or associations detected in the cerebellum. Only RNA Integrity Number (RIN) associated with NWK2 mRNA levels in the cerebellum (Supplemental Table 1). However, our comparison analysis of healthy and bipolar disorder samples with no significant difference in RIN values between groups excludes the influence of this variable on NWK2 mRNA levels (Table 1). These results indicate that Nwk2 gene expression is downregulated in cerebellum of BD subjects and suggest that reduced levels of SP4 may contribute to this downregulation.
Table 2. Demographic characteristics of subjects with bipolar disease (BD) (n=10) and controls (n=10).
| Gender1 |
Age (years)2 |
Postmortem delay (hours)2 |
pH2 |
RNA integrity Cause of death number2 |
Treatment3 |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pairs | BD | C | BD | C | BD | C | BD | C | BD | C | BD | C | BD | C |
| 1 | M | M | 72 | 72 | 22 | 43 | 6.5 | 7.1 | 7.3 | 8.0 | Suicide | Accident | Drug-free | Drug- free |
| 2 | F | F | 44 | 49 | 19 | 40 | 6.5 | 6.8 | 7.4 | 7.3 | Suicide | Accident | APS. BZD. Mirtazapine |
Drug- free |
| 3 | M | M | 58 | 54 | 10 | 23 | 7.1 | 6.5 | 7.6 | 8.5 | Suicide | Accident | APS. Lamotrigine |
BZD |
| 4 | M | M | 40 | 43 | 17 | 11 | 6.8 | 6.5 | 7.2 | 8.3 | Suicide | Accident | BZD | Drug- free |
| 5 | M | M | 27 | 30 | 10 | 11 | 6.8 | 6.5 | 8.1 | 8.3 | Suicide | Accident | Drug-free | Cannabis |
| 6 | M | M | 63 | 60 | 8 | 4 | 6.8 | 7.1 | 8.1 | 8.3 | Suicide | Natural | BZD | Drug- free |
| 7 | M | M | 64 | 61 | 23 | 23 | 7.1 | 6.8 | 7.3 | 6.8 | Suicide | Accident | BZD. Venlafaxine |
Drug- free |
| 8 | M | M | 57 | 55 | 22 | 22 | 6.5 | 6.5 | 7.4 | 8.1 | Natural | Natural | Drug-free | Drug- free |
| 9 | M | M | 63 | 67 | 31 | 19 | 6.5 | 7.1 | 6.7 | 7.6 | Natural | Natural | Drug-free | NSAID |
| 10 | F | F | 73 | 74 | 3 | 19 | 6.8 | 6.8 | 8.1 | 7.8 | Suicide | Accident | Drug-free | NSAID |
|
| ||||||||||||||
| Statistic | 0.00; 14 | 48.504 | 36.004 | 47.004 | 27.004 | N/A | N/A | |||||||
BD, Bipolar Disorder; C, control; M, male; F, female; APS, antipsychotics; BZD,benzodiazepines; NSAID, nonsteroidal anti-inflammatory drug.
Chi-square statistic and degrees of freedom are shown for categorical variables.
Mann Whitney U is shown for non-parametric variables.
Treatment indicates results of toxicology analysis performed by the National Institute of Toxicology, Madrid, Spain.
Not significant pvalue, p>0.05.
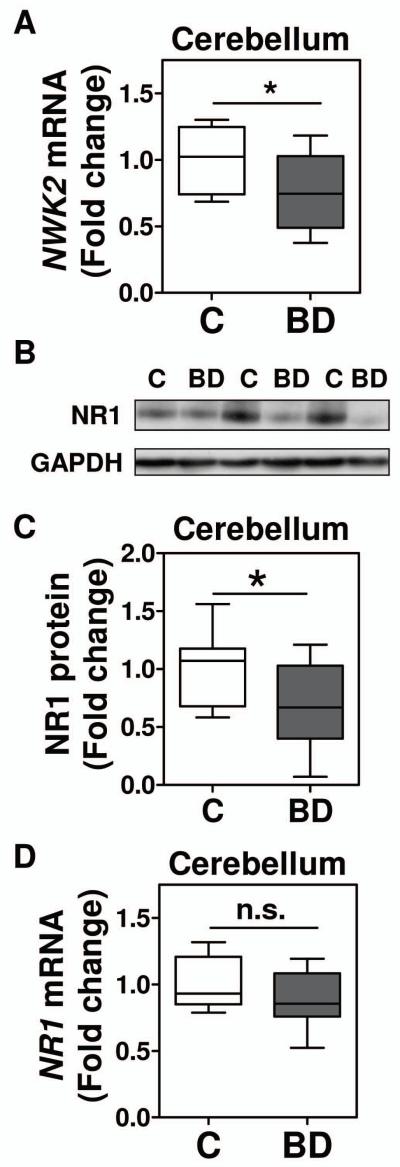
Figure 6. Levels of NWK2 mRNA and NR1 protein are reduced in the postmortem cerebellum from bipolar disorder subjects.
(A) Nwk2 mRNA levels in extracts from cerebellar postmortem tissue of non-psychiatric control subjects (Control, C, n= 10), and subjects with bipolar disease (Bipolar, BD, n=10) were determined by RT-qPCR and normalized to a reference healthy control sample and beta glucuronidase (GUSB). Height of box plots shows interquartile range; horizontal line, median; error bars, range of at least two independent analyses performed in duplicate. Statistical analysis was performed using two-tailed Mann-Whitney test for non parametric values (p<0.05-*). (B, C) NR1 protein levels were determined in protein extracts from cerebellar postmortem tissue of control (Control, C, n= 10), and bipolar disorder subjects (Bipolar, BD, n=10) by immunoblot. The resultant bands were quantified by densitometry. NR1 was normalized to GAPDH values and referred to a standard sample as described in the material and methods. (B) Representative Western blot images for NR1 and GAPDH from three control individuals and three bipolar subjects. (C) The graph shows normalized NR1 protein levels. Each box plot represents median, interquartile range and range of at least two independent analyses. (D) NR1 mRNA levels were determined as in (A). Each box plot represents median, interquartile range and range of an analysis performed in duplicate. An outlier was detected for NR1 mRNA levels in the control group and therefore excluded from the analysis (C, n=9; BP, n=10). Statistical analysis was performed using one-tailed Mann-Whitney test (p<0.05-*; n.s.-not significant).
We also characterized NR1 mRNA and protein levels in the postmortem cerebellum of the same control and BD subjects. We observed that NR1 protein levels were significantly decreased in the cerebellum in BD, whereas NR1 mRNA levels did not change (Figure 6 B, C and D). These findings indicate that, similar to the situation in Sp4 hypomorphic mice (Supplemental Figure 2A and (Zhou et al., 2010), the reduction in NR1 protein level in the cerebellum of bipolar disorder subjects is due to post-transcriptional mechanisms. We also analyzed whether NR1 protein levels were associated with NWK2 mRNA levels in the cerebellum. In this small cohort, we found a trend for NR1 to correlate with NWK2 only in control subjects (r=0.624, p=0.0603), suggesting that analysis of a larger sample size will likely result in a significant correlation of both parameters.
DISCUSSION
In this study, we provide evidence for an Sp4-Nwk2-NMDAR1 pathway that regulates dendrite patterning and NMDAR surface levels in developing cerebellar granule neurons. We identified Nwk2 as an activation target of transcription factor Sp4. Further, we demonstrated that the Sp4-Nwk2 pathway controls dendrite patterning and NR1 levels at the cell surface. Overexpression of NR1 suppressed the phenotype of excess primary dendrites upon Sp4 or Nwk2 knockdown. Notably, in the cerebellum of bipolar disorder subjects, in addition to reduced Sp4 protein (Pinacho et al., 2011), we also observed reduced NWK2 mRNA and NR1 protein. These findings provide new insights into the regulation of neuronal morphogenesis and the pathogenesis of bipolar disorder.
Targets of transcription factor Sp4
Altered levels of transcription factor Sp4 protein have been observed in bipolar disorder, schizophrenia and first episode psychosis (Pinacho et al., 2011; Fuste et al., 2013; Pinacho et al., 2013). Sp4 target genes are likely to vary dependent on cell type and developmental context (St Amand et al., 2006) and little is known about the target genes of Sp4 important for neuronal development or altered in psychiatric disorders. We previously reported that Sp4-dependent repression of neurotrophin-3 is required to limit dendritic branching, but changes in NT3 expression did not appear to mediate the pruning defect in Sp4-depleted CGNs (Ramos et al., 2009). In order to identify additional Sp4 target genes, we carried out a microarray analysis and we identified many genes whose expression was significantly altered between the cerebella of WT and Sp4 mutant mice. Here we describe studies of one of these, Nwk2. We show that Sp4 is required for Nwk2 expression and binds the Nwk2 promoter, suggesting that Nwk2 is a direct activation target gene of Sp4. Importantly, our data indicate that Nwk2 contributes to Sp4-dependent regulation of dendrite patterning and NR1 surface expression in cerebellar granule neurons. We did not observe changes in NMDAR mRNA levels in CGNs of Sp4 mutant mice. Thus, the reported Sp4-mediated transcriptional regulation of NMDAR subunits (Liu et al., 2003; Liu et al., 2004; Priya et al., 2013) is likely context-dependent and/or compensated in this animal model. Other candidate Sp4 target genes identified in our microarray may also contribute to Sp4-dependent regulation of neuron development and may be altered in psychiatric disease with reduced Sp4 function (Figure 1A and Table 1).
Regulation of NMDAR1 levels
Previous work showed that NR1 protein, but not mRNA, was reduced in the brain of Sp4 hypomorphic mice (Zhou et al., 2010). Although no changes in NR1 splicing were observed, it was not determined whether post-transcriptional regulation of NR1 was due to non-cell autonomous effects of Sp4 loss or the consequence of Sp4 loss from a very early stage of development. Here we examined the effects of acute Sp4 and Nwk2 knockdown on NR1 expression in neurons. We found that levels of surface NR1 were reduced upon knockdown of Sp4 or Nwk2 and, further, overexpression of Nwk2 partially restored immunostaining of surface NR1 in Sp4 knockdown neurons (Figure 4). Total levels of NR1 were not significantly affected by Sp4 knockdown, suggesting Sp4 acts in a cell-autonomous manner to regulate NMDAR assembly or trafficking. The finding that total NR1 levels were reduced in CGNs from Sp4 mutant mice but not when Sp4 was knocked down in WT CGNs (Supplemental Figure 2 and Figure 4) suggests that the more complex cellular context in vivo and/or the prolonged loss of Sp4, particularly during early development, contribute to the observed decrease in NR1 levels in the Sp4 hypomorphic animals. These data indicate that the Sp4-Nwk2 pathway is a cell intrinsic mechanism that regulates NMDAR1 levels at the cell surface.
Nwk2 function
Nwk2, like its Drosophila homolog, has an N-terminal FER/Cip4 homology-Bin/amphiphysin/Rvs (F-BAR) domain and 2 Src homology 3 (SH3) domains in the C-terminus. Consistent with roles for proteins with similar domain structures in linking membrane coated vesicles and the actin cytoskeleton (Aspenstrom et al., 2006), Nwk and its mammalian homologs were shown to regulate actin dynamics and the F-BAR domains were shown to induce membrane protrusions in cells (Coyle et al., 2004; Becalska et al., 2013; Cao et al., 2013). Expression of Nwk2 but not Nwk1 was reduced in Sp4 mutant mice (Figure 1), consistent with the idea that these mammalian homologs may have distinct regulation and functions (Cao et al., 2013). We observed that RNAi-mediated knockdown of Nwk2 led to an increased number of primary dendrites in developing CGNs (Figure 2). Several other F-BAR family proteins have also been shown to regulate neurite morphogenesis including CIP4, Pacsin1/syndapin1, Raspotlin and WRP/srGAP3, a protein implicated in mental retardation (Dharmalingam et al., 2009; Guerrier et al., 2009; Carlson et al., 2011; Wakita et al., 2011; Saengsawang et al., 2012). Further, our data suggest that Nwk2 regulates dendrite patterning, at least in part, through promoting surface levels of NR1. Nwk and its homologs interact with components of the endocytic machinery and Drosophila Nwk has been suggested to control synaptic growth through effects on endosomal trafficking, and possibly recycling, of receptors such as the BMP receptor homolog Tkv (Itoh et al., 2005; O’Connor-Giles et al., 2008; Rodal et al., 2008; Rodal et al., 2011). Another F-BAR and SH3 domain-containing protein, Pacsin1/syndapin1, regulates endocytosis and synaptic removal of NR3A-containing NMDARs (Perez-Otano et al., 2006). Thus, although the mechanisms underlying Nwk2’s role in controlling NR1 surface expression remain to be elucidated, we consider it likely that Nwk2 regulates endocytic trafficking of NMDAR via effects on membrane and actin dynamics.
NMDAR1 and dendrites
We observed that overexpression of NR1 rescued the excess dendrite number in developing CGNs depleted of Sp4 or Nwk2. Sp4-dependent effects on dendrites are context specific as, in addition to its roles in promoting pruning and limiting branching in CGNs (Ramos et al., 2007), Sp4 stimulates dendrite growth in hippocampal granule neurons (Zhou et al., 2007). We recently reported that phosphorylation of Sp4 is reduced downstream of NMDAR signaling and may inhibit Sp4 function in dendrite patterning (Saia et al., 2014). The role of NMDA receptor signaling in dendrite patterning is also context dependent and NMDARs have been found to promote both dendrite growth and elimination (Rajan and Cline, 1998; Datwani et al., 2002; Sin et al., 2002). Of particular relevance to our study, knockout of NR2B in dentate gyrus granule neurons was reported to increase numbers of primary dendrites due to a defect in pruning (Espinosa et al., 2009). In CGNs, NMDAR activity has been shown to promote cell survival, migration and, at early times, neurite outgrowth (Pearce et al., 1987; Balazs et al., 1988; Komuro and Rakic, 1993). Our data suggest that NMDARs also play a role in promoting dendrite pruning during CGN maturation.
Sp4-Nwk2-NR1 pathway in disease
We observed decreased Sp4 protein, NWK2 mRNA, and NR1 protein levels in postmortem cerebellum from subjects with bipolar disorder (Figure 6 and (Pinacho et al., 2011)). Sp4 protein levels and NWK2 mRNA correlated, although correlations with NR1 did not reach significance in our small sample. A trend for a significant association between NWK2 mRNA and NR1 protein levels was found only in control subjects, supporting a contribution of NWK2 expression to NR1 protein levels in healthy conditions. Further studies with a larger sample size are needed to confirm this correlation and determine if NWK2 and NR1 expression also correlate in BD subjects or if the NWK2-NR1 pathway is disrupted in this pathological context. While best known for its classical role in motor function, alterations in the cerebellum have been suggested to contribute to affective, psychological and cognitive defficits in psychiatric disorders (Konarski et al., 2005; Schmahmann et al., 2007; Bolbecker et al., 2009; Yeganeh-Doost et al., 2011). Consistent with our observations in the cerebellum, studies of bipolar disorder subjects have reported reduced NMDAR expression in other brain regions including the hippocampus and superior temporal cortex (Nudmamud-Thanoi and Reynolds, 2004; Beneyto et al., 2007).
It is important to note that the use of postmortem brain for the study of molecular mechanisms disrupted in psychiatric disorders is a helpful tool but also has important limitations. For example, RNA quality has to be taken into account. In our study we found a correlation of NWK2 with RIN, however no significant difference in RIN was found between groups, ruling out the influence of RIN on the decrease of NWK2 in the cerebellum of bipolar disorder. Analysis of drugs present in blood at the time of death suggests these treatments did not influence our findings, however, the posible effect of long term treatments cannot be ruled out, since this data was not available. Eight out of ten bipolar disorder subjects examined here commited suicide, suggesting a possible confounder effect of this mechanism of death on our molecular findings. Further studies will be needed in order to explore this possibility, as well as to extend these findings in a larger and independent cohort of patients.
Although more studies are needed, the data presented here suggest that the Sp4-Nwk2-NMDAR1 pathway is conserved and may be altered in bipolar disorder and possibly other psychiatric diseases. NMDA receptor dysfunction, arising through multiple mechanisms, has been implicated in both schizophrenia and bipolar disorder (Fountoulakis, 2012; Ginsberg et al., 2012; Moghaddam and Javitt, 2012). Interestingly, our data suggest NR1 levels are reduced in bipolar disorder cerebellum by a post-transcriptional mechanism (Figure 6). A role for disrupted NMDAR trafficking in psychotic disorders is supported, for example, by findings that the products of the schizophrenia and bipolar disorder susceptibility genes neuregulin1 (NRG1) and dysbindin-1A (DTNBP1) regulate NMDAR trafficking (Lau and Zukin, 2007; Geddes et al., 2011; Jeans et al., 2011). In fact, proteomic and genomic findings implicate endocytosis and endosomal trafficking in the development of bipolar disorder and schizophrenia (Ryder and Faundez, 2009; Schubert et al., 2011). Thus, future studies of Nwk2 function in endosomal trafficking and the contribution of endosomal trafficking disturbance to psychotic disorders, particularly those associated with altered Sp4 activity, are warranted.
Supplementary Material
Acknowledgements
We thank Xianjin Zhou at the University of California, San Diego for providing the Sp4 hypomorphic mouse strain and Lakshmanan Iyer in the Tufts Center for Neuroscience Research (P30 NS047243) for help with microarray analysis. We also thank Jasmin Lalonde, Xiaohu Mei, Alvaro Valin, Jian Ouyang and Michele Jacobs for reagents and discussions. This work was supported by a grant from the National Institutes of Health (R01 HD043364) to G.G., the Marie Curie Program IRG RTD REG/T.2 (2007)D/530573, Plan Nacional de Investigación BFU2008-01103 (MCINN) and the Centro de Investigación Biomédica en Red de Salud Mental (CIBERSAM) to B.R., as well as predoctoral Fellowships from ISCIII and Fondo de Investigación Sanitaria (PFIS) to R.P..
Footnotes
Conflict of Interest: The authors declare no competing financial interests.
REFERENCES
- Aspenstrom P, Fransson A, Richnau N. Pombe Cdc15 homology proteins: regulators of membrane dynamics and the actin cytoskeleton. Trends Biochem Sci. 2006;31:670–679. doi: 10.1016/j.tibs.2006.10.001. [DOI] [PubMed] [Google Scholar]
- Balazs R, Jorgensen OS, Hack N. N-methyl-D-aspartate promotes the survival of cerebellar granule cells in culture. Neuroscience. 1988;27:437–451. doi: 10.1016/0306-4522(88)90279-5. [DOI] [PubMed] [Google Scholar]
- Becalska AN, Kelley CF, Berciu C, Stanishneva-Konovalova TB, Fu X, Wang S, Sokolova OS, Nicastro D, Rodal AA. Formation of membrane ridges and scallops by the F-BAR protein Nervous Wreck. Mol Biol Cell. 2013;24:2406–2418. doi: 10.1091/mbc.E13-05-0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beneyto M, Kristiansen LV, Oni-Orisan A, McCullumsmith RE, Meador-Woodruff JH. Abnormal glutamate receptor expression in the medial temporal lobe in schizophrenia and mood disorders. Neuropsychopharmacology. 2007;32:1888–1902. doi: 10.1038/sj.npp.1301312. [DOI] [PubMed] [Google Scholar]
- Bilimoria PM, Bonni A. Cultures of cerebellar granule neurons. CSH Protoc. 20082008 doi: 10.1101/pdb.prot5107. pdb prot5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black AR, Black JD, Azizkhan-Clifford J. Sp1 and kruppel-like factor family of transcription factors in cell growth regulation and cancer. J Cell Physiol. 2001;188:143–160. doi: 10.1002/jcp.1111. [DOI] [PubMed] [Google Scholar]
- Bolbecker AR, Mehta C, Johannesen JK, Edwards CR, O’Donnell BF, Shekhar A, Nurnberger JI, Steinmetz JE, Hetrick WP. Eyeblink conditioning anomalies in bipolar disorder suggest cerebellar dysfunction. Bipolar Disorders. 2009;11:19–32. doi: 10.1111/j.1399-5618.2008.00642.x. [DOI] [PubMed] [Google Scholar]
- Cao H, Yin X, Cao Y, Jin Y, Wang S, Kong Y, Chen Y, Gao J, Heller S, Xu Z. FCHSD1 and FCHSD2 are expressed in hair cell stereocilia and cuticular plate and regulate actin polymerization in vitro. Plos One. 2013;8:e56516. doi: 10.1371/journal.pone.0056516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson BR, Lloyd KE, Kruszewski A, Kim IH, Rodriguiz RM, Heindel C, Faytell M, Dudek SM, Wetsel WC, Soderling SH. WRP/srGAP3 facilitates the initiation of spine development by an inverse F-BAR domain, and its loss impairs long-term memory. J Neurosci. 2011;31:2447–2460. doi: 10.1523/JNEUROSCI.4433-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson GC. Glutamate receptor dysfunction and drug targets across models of autism spectrum disorders. Pharmacol Biochem Behav. 2012;100:850–854. doi: 10.1016/j.pbb.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline HT. Dendritic arbor development and synaptogenesis. Curr Opin Neurobiol. 2001;11:118–126. doi: 10.1016/s0959-4388(00)00182-3. [DOI] [PubMed] [Google Scholar]
- Coyle IP, Koh YH, Lee WC, Slind J, Fergestad T, Littleton JT, Ganetzky B. Nervous wreck, an SH3 adaptor protein that interacts with Wsp, regulates synaptic growth in Drosophila. Neuron. 2004;41:521–534. doi: 10.1016/s0896-6273(04)00016-9. [DOI] [PubMed] [Google Scholar]
- Datwani A, Iwasato T, Itohara S, Erzurumlu RS. NMDA receptor-dependent pattern transfer from afferents to postsynaptic cells and dendritic differentiation in the barrel cortex. Molecular and Cellular Neuroscience. 2002;21:477–492. doi: 10.1006/mcne.2002.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmalingam E, Haeckel A, Pinyol R, Schwintzer L, Koch D, Kessels MM, Qualmann B. F-BAR proteins of the syndapin family shape the plasma membrane and are crucial for neuromorphogenesis. J Neurosci. 2009;29:13315–13327. doi: 10.1523/JNEUROSCI.3973-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinosa JS, Wheeler DG, Tsien RW, Luo L. Uncoupling dendrite growth and patterning: single-cell knockout analysis of NMDA receptor 2B. Neuron. 2009;62:205–217. doi: 10.1016/j.neuron.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fountoulakis KN. The possible involvement of NMDA glutamate receptor in the etiopathogenesis of bipolar disorder. Curr Pharm Des. 2012;18:1605–1608. doi: 10.2174/138161212799958585. [DOI] [PubMed] [Google Scholar]
- Fuste M, Pinacho R, Melendez-Perez I, Villalmanzo N, Villalta-Gil V, Haro JM, Ramos B. Reduced expression of SP1 and SP4 transcription factors in peripheral blood mononuclear cells in first-episode psychosis. J Psychiatr Res. 2013;47:1608–1614. doi: 10.1016/j.jpsychires.2013.07.019. [DOI] [PubMed] [Google Scholar]
- Geddes AE, Huang XF, Newell KA. Reciprocal signalling between NR2 subunits of the NMDA receptor and neuregulin1 and their role in schizophrenia. Prog Neuropsychopharmacol Biol Psychiatry. 2011;35:896–904. doi: 10.1016/j.pnpbp.2011.02.017. [DOI] [PubMed] [Google Scholar]
- Ginsberg SD, Hemby SE, Smiley JF. Expression profiling in neuropsychiatric disorders: emphasis on glutamate receptors in bipolar disorder. Pharmacol Biochem Behav. 2012;100:705–711. doi: 10.1016/j.pbb.2011.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrier S, Coutinho-Budd J, Sassa T, Gresset A, Jordan NV, Chen K, Jin WL, Frost A, Polleux F. The F-BAR domain of srGAP2 induces membrane protrusions required for neuronal migration and morphogenesis. Cell. 2009;138:990–1004. doi: 10.1016/j.cell.2009.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T, Erdmann KS, Roux A, Habermann B, Werner H, De Camilli P. Dynamin and the actin cytoskeleton cooperatively regulate plasma membrane invagination by BAR and F-BAR proteins. Dev Cell. 2005;9:791–804. doi: 10.1016/j.devcel.2005.11.005. [DOI] [PubMed] [Google Scholar]
- Jeans A, Malins R, Padamsey Z, Reinhart M, Emptage N. Increased expression of dysbindin-1A leads to a selective deficit in NMDA receptor signaling in the hippocampus. Neuropharmacology. 2011;61:1345–1353. doi: 10.1016/j.neuropharm.2011.08.007. [DOI] [PubMed] [Google Scholar]
- Ji B, Wang X, Pinto-Duarte A, Kim M, Caldwell S, Young JW, Behrens MM, Sejnowski TJ, Geyer MA, Zhou X. Prolonged Ketamine Effects in Hypomorphic Mice: Mimicking Phenotypes of Schizophrenia. Plos One. 2013;8:e66327. doi: 10.1371/journal.pone.0066327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelsoe J, Zhou XJ, Guo SZ, Shehktman T, He L, Geyer M. SP4 gene conveys susceptibility to bipolar disorder and schizophrenia. American Journal of Medical Genetics Part B-Neuropsychiatric Genetics. 2006;141B:713–713. [Google Scholar]
- Komuro H, Rakic P. Modulation of neuronal migration by NMDA receptors. Science. 1993;260:95–97. doi: 10.1126/science.8096653. [DOI] [PubMed] [Google Scholar]
- Konarski JZ, McIntyre RS, Grupp LA, Kennedy SH. Is the cerebellum relevant in the circuitry of neuropsychiatric disorders? J Psychiatry Neurosci. 2005;30:178–186. [PMC free article] [PubMed] [Google Scholar]
- Krystal JH, Karper LP, Seibyl JP, Freeman GK, Delaney R, Bremner JD, Heninger GR, Bowers MB, Charney DS. Subanesthetic Effects of the Noncompetitive Nmda Antagonist, Ketamine, in Humans - Psychotomimetic, Perceptual, Cognitive, and Neuroendocrine Responses. Archives of General Psychiatry. 1994;51:199–214. doi: 10.1001/archpsyc.1994.03950030035004. [DOI] [PubMed] [Google Scholar]
- Lau CG, Zukin RS. NMDA receptor trafficking in synaptic plasticity and neuropsychiatric disorders. Nature Reviews Neuroscience. 2007;8:413–426. doi: 10.1038/nrn2153. [DOI] [PubMed] [Google Scholar]
- Liu A, Hoffman PW, Lu W, Bai G. NF-kappaB site interacts with Sp factors and up-regulates the NR1 promoter during neuronal differentiation. J Biol Chem. 2004;279:17449–17458. doi: 10.1074/jbc.M311267200. [DOI] [PubMed] [Google Scholar]
- Liu A, Zhuang Z, Hoffman PW, Bai G. Functional analysis of the rat N-methyl-D-aspartate receptor 2A promoter: multiple transcription starts points, positive regulation by Sp factors, and translational regulation. J Biol Chem. 2003;278:26423–26434. doi: 10.1074/jbc.M211165200. [DOI] [PubMed] [Google Scholar]
- Luscher C, Malenka RC. NMDA receptor-dependent long-term potentiation and long-term depression (LTP/LTD) Cold Spring Harb Perspect Biol. 2012;4 doi: 10.1101/cshperspect.a005710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao X, Yang SH, Simpkins JW, Barger SW. Glutamate receptor activation evokes calpain-mediated degradation of Sp3 and Sp4, the prominent Sp-family transcription factors in neurons. Journal of Neurochemistry. 2007;100:1300–1314. doi: 10.1111/j.1471-4159.2006.04297.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moghaddam B, Javitt D. From revolution to evolution: the glutamate hypothesis of schizophrenia and its implication for treatment. Neuropsychopharmacology. 2012;37:4–15. doi: 10.1038/npp.2011.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nudmamud-Thanoi S, Reynolds GP. The NR1 subunit of the glutamate/NMDA receptor in the superior temporal cortex in schizophrenia and affective disorders. Neuroscience Letters. 2004;372:173–177. doi: 10.1016/j.neulet.2004.09.035. [DOI] [PubMed] [Google Scholar]
- O’Connor-Giles KM, Ho LL, Ganetzky B. Nervous wreck interacts with thickveins and the endocytic machinery to attenuate retrograde BMP signaling during synaptic growth. Neuron. 2008;58:507–518. doi: 10.1016/j.neuron.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto S, Sherman K, Bai G, Lipton SA. Effect of the ubiquitous transcription factors, SP1 and MAZ, on NMDA receptor subunit type 1 (NR1) expression during neuronal differentiation. Brain Res Mol Brain Res. 2002;107:89–96. doi: 10.1016/s0169-328x(02)00440-0. [DOI] [PubMed] [Google Scholar]
- Olney JW, Newcomer JW, Farber NB. NMDA receptor hypofunction model of schizophrenia. Journal of Psychiatric Research. 1999;33:523–533. doi: 10.1016/s0022-3956(99)00029-1. [DOI] [PubMed] [Google Scholar]
- Paoletti P, Bellone C, Zhou Q. NMDA receptor subunit diversity: impact on receptor properties, synaptic plasticity and disease. Nature Reviews Neuroscience. 2013;14:383–400. doi: 10.1038/nrn3504. [DOI] [PubMed] [Google Scholar]
- Pearce IA, Cambray-Deakin MA, Burgoyne RD. Glutamate acting on NMDA receptors stimulates neurite outgrowth from cerebellar granule cells. FEBS Lett. 1987;223:143–147. doi: 10.1016/0014-5793(87)80525-2. [DOI] [PubMed] [Google Scholar]
- Perez-Otano I, Ehlers MD. Homeostatic plasticity and NMDA receptor trafficking. Trends Neurosci. 2005;28:229–238. doi: 10.1016/j.tins.2005.03.004. [DOI] [PubMed] [Google Scholar]
- Perez-Otano I, Lujan R, Tavalin SJ, Plomann M, Modregger J, Liu XB, Jones EG, Heinemann SF, Lo DC, Ehlers MD. Endocytosis and synaptic removal of NR3A-containing NMDA receptors by PACSIN1/syndapin1. Nature Neuroscience. 2006;9:611–621. doi: 10.1038/nn1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinacho R, Villalmanzo N, Lalonde J, Haro JM, Meana JJ, Gill G, Ramos B. The transcription factor SP4 is reduced in postmortem cerebellum of bipolar disorder subjects: control by depolarization and lithium. Bipolar Disorders. 2011;13:474–485. doi: 10.1111/j.1399-5618.2011.00941.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinacho R, Villalmanzo N, Roca M, Iniesta R, Monje A, Haro JM, Meana JJ, Ferrer I, Gill G, Ramos B. Analysis of Sp transcription factors in the postmortem brain of chronic schizophrenia: a pilot study of relationship to negative symptoms. J Psychiatr Res. 2013;47:926–934. doi: 10.1016/j.jpsychires.2013.03.004. [DOI] [PubMed] [Google Scholar]
- Priya A, Johar K, Wong-Riley MT. Specificity protein 4 functionally regulates the transcription of NMDA receptor subunits GluN1, GluN2A, and GluN2B. Biochim Biophys Acta. 2013;1833:2745–2756. doi: 10.1016/j.bbamcr.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajan I, Cline HT. Glutamate receptor activity is required for normal development of tectal cell dendrites in vivo. J Neurosci. 1998;18:7836–7846. doi: 10.1523/JNEUROSCI.18-19-07836.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos B, Gaudilliere B, Bonni A, Gill G. Transcription factor Sp4 regulates dendritic patterning during cerebellar maturation. Proc Natl Acad Sci U S A. 2007;104:9882–9887. doi: 10.1073/pnas.0701946104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos B, Valin A, Sun XX, Gill G. Sp4-dependent repression of neurotrophin-3 limits dendritic branching. Molecular and Cellular Neuroscience. 2009;42:152–159. doi: 10.1016/j.mcn.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodal AA, Blunk AD, Akbergenova Y, Jorquera RA, Buhl LK, Littleton JT. A presynaptic endosomal trafficking pathway controls synaptic growth signaling. J Cell Biol. 2011;193:201–217. doi: 10.1083/jcb.201009052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodal AA, Motola-Barnes RN, Littleton JT. Nervous wreck and Cdc42 cooperate to regulate endocytic actin assembly during synaptic growth. J Neurosci. 2008;28:8316–8325. doi: 10.1523/JNEUROSCI.2304-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder PV, Faundez V. Schizophrenia: the “BLOC” may be in the endosomes. Sci Signal. 2009;2:pe66. doi: 10.1126/scisignal.293pe66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saengsawang W, Mitok K, Viesselmann C, Pietila L, Lumbard DC, Corey SJ, Dent EW. The F-BAR protein CIP4 inhibits neurite formation by producing lamellipodial protrusions. Curr Biol. 2012;22:494–501. doi: 10.1016/j.cub.2012.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saia G, Lalonde J, Sun X, Ramos B, Gill G. Phosphorylation of the transcription factor Sp4 is reduced by NMDA receptor signaling. Journal of Neurochemistry. 2014 doi: 10.1111/jnc.12657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmahmann JD, Weilburg JB, Sherman JC. The neuropsychiatry of the cerebellum - insights from the clinic. Cerebellum. 2007;6:254–267. doi: 10.1080/14734220701490995. [DOI] [PubMed] [Google Scholar]
- Schubert KO, Focking M, Prehn JH, Cotter DR. Hypothesis review: are clathrin-mediated endocytosis and clathrin-dependent membrane and protein trafficking core pathophysiological processes in schizophrenia and bipolar disorder? Mol Psychiatry. 2011 doi: 10.1038/mp.2011.123. [DOI] [PubMed] [Google Scholar]
- Sin WC, Haas K, Ruthazer ES, Cline HT. Dendrite growth increased by visual activity requires NMDA receptor and Rho GTPases. Nature. 2002;419:475–480. doi: 10.1038/nature00987. [DOI] [PubMed] [Google Scholar]
- St Amand TR, Lu JT, Zamora M, Gu Y, Stricker J, Hoshijima M, Epstein JA, Ross JJ, Jr., Ruiz-Lozano P, Chien KR. Distinct roles of HF-1b/Sp4 in ventricular and neural crest cells lineages affect cardiac conduction system development. Dev Biol. 2006;291:208–217. doi: 10.1016/j.ydbio.2005.10.018. [DOI] [PubMed] [Google Scholar]
- Stewart SA, Dykxhoorn DM, Palliser D, Mizuno H, Yu EY, An DS, Sabatini DM, Chen IS, Hahn WC, Sharp PA, Weinberg RA, Novina CD. Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA. 2003;9:493–501. doi: 10.1261/rna.2192803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam GW, van de Lagemaat LN, Redon R, Strathdee KE, Croning MD, Malloy MP, Muir WJ, Pickard BS, Deary IJ, Blackwood DH, Carter NP, Grant SG. Confirmed rare copy number variants implicate novel genes in schizophrenia. Biochem Soc Trans. 2010;38:445–451. doi: 10.1042/BST0380445. [DOI] [PubMed] [Google Scholar]
- Wakita Y, Kakimoto T, Katoh H, Negishi M. The F-BAR protein Rapostlin regulates dendritic spine formation in hippocampal neurons. J Biol Chem. 2011;286:32672–32683. doi: 10.1074/jbc.M111.236265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeganeh-Doost P, Gruber O, Falkai P, Schmitt A. The role of the cerebellum in schizophrenia: from cognition to molecular pathways. Clinics (Sao Paulo) 2011;66(Suppl 1):71–77. doi: 10.1590/S1807-59322011001300009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Kelsoe J, Geyer MA. The Sp4 transcription factor gene is critical for postnatal development of dentate gyrus in mice and associated with bipolar disorder in humans. Neuropsychopharmacology. 2005;30:S158–S158. [Google Scholar]
- Zhou X, Long JM, Geyer MA, Masliah E, Kelsoe JR, Wynshaw-Boris A, Chien KR. Reduced expression of the Sp4 gene in mice causes deficits in sensorimotor gating and memory associated with hippocampal vacuolization. Molecular Psychiatry. 2005;10:393–406. doi: 10.1038/sj.mp.4001621. [DOI] [PubMed] [Google Scholar]
- Zhou X, Nie ZG, Roberts A, Zhang DX, Sebat J, Malhotra D, Kelsoe JR, Geyer MA. Reduced NMDAR1 expression in the Sp4 hypomorphic mouse may contribute to endophenotypes of human psychiatric disorders. Human Molecular Genetics. 2010;19:3797–3805. doi: 10.1093/hmg/ddq298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Qyang Y, Kelsoe JR, Masliah E, Geyer MA. Impaired postnatal development of hippocampal dentate gyrus in Sp4 null mutant mice. Genes Brain and Behavior. 2007;6:269–276. doi: 10.1111/j.1601-183X.2006.00256.x. [DOI] [PubMed] [Google Scholar]
- Zhou X, Tang W, Greenwood TA, Guo SZ, He L, Geyer MA, Kelsoe JR. Transcription Factor SP4 Is a Susceptibility Gene for Bipolar Disorder. Plos One. 2009;4 doi: 10.1371/journal.pone.0005196. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.